Introduction
Colorectal cancer (cancer of the colon and/or
rectum) is the second leading cause of cancer-related mortality in
the US according to the National Comprehensive Cancer Network
(NCCN) in 2015 (1). It has been
reported that in 15–25% of patients with colorectal cancer, hepatic
metastasis has already occurred prior to diagnosis and metastasis
occurs in more than half of the patients with this disease.
Although the 5-year survival rate of patients with colon cancer
following radical surgery is often >50%, this decreases to
<12% with the occurrence of metastasis (2,3).
Metastasis has thus become the key obstacle to the effective
treatment of colon cancer and remains a big challenge for
clinicians due to the wide disparities in survival rates.
The cancer stem cell (CSC) theory is a new theory
which has appeared in recent years as regards the occurrence,
development, metastasis and recurrence of tumors. CSCs are defined
as a subpopulation of cancer cells which have the characteristics
of self-renewal, differentiation abilities, metastatic potential
and the ability to resist conventional chemoradiotherapeutics
(4–6). The discovery of markers of CSCs has
facilitated the screening and studying of a number of types of
cancer, including leukemia, brain cancer, breast cancer, abdominal
cancer and cancers of the reproductive system (7–15).
CD133 and CD44 are considered markers of the surface membrane of
cells and have been used to identify colon CSCs (CCSCs) (11–13) and have been recently reported to
be co-expressed in colon cancer with hepatic metastases (16).
Epithelial-mesenchymal transition (EMT) is
considered to occur during cancer invasion and migration, or tumor
progression. In this process, epithelial cells lose their
epithelial characteristics and adopt a mesenchymal-like appearance
or characteristics (17,18). The downregulation or loss of
E-cadherin, a transmenbrane protein important for cell-cell
junctions, is treated as the hallmark of EMT (19). Since CSCs and EMT both play
significant roles in cancer development, understanding the link
between them may enhance our knowledge of the pathogenesis and
mechanisms responsible for metastasis in cancer.
The polycomb group (PcG) of proteins are a family of
transcriptional repressors that orchestrate alterations in
chromatin structure to regulate gene activity (20,21). A number of PcG proteins have been
confirmed to be altered in human cancers, such as Bmi-1 (22–24). Bmi-1 is known as transcriptional
repressor targeting the Ink4a/Arf gene locus, and has also been
described as an oncogene in many solid tumors, playing a critical
role in the maintenance of CSCs (25). However, the role of Bmi-1 in the
functions CCSCs has rarely been reported, at least to the best of
our knowledge.
In our previous study (26), we found that the expression of
Bmi-1 in colon cancer tissues closely correlated with the clinical
stage, invasion depth and metastatic ability of the tumors. In the
present study, we screened and identified CCSCs using the HCT116
colon cancer cell line. We also wished to determine the role that
Bmi-1 plays in CCSCs and to elucidate the underlying mechanisms. It
would be of clinical and therapeutic significance to provide a
novel target for colon cancer therapy.
Materials and methods
Cell line and cell culture
The HCT116 colon cancer cell line was obtained from
the Cell Bank of the Committee on Type Culture Collection of the
Chinese Academic of Science (CCTCC; Shanghai, China). The cells
were cultured in RPMI-1640 (Corning Inc., Corning, New York, NY,
USA) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL,
Grand Island, NY, USA)at 37°C with 5% CO2. The culture
conditions for the HCT116 cells to form tumor spheres in suspension
were as previously described (27–29). The sphere formation medium (SFM)
used was RPMI-1640 supplemented with 20 ng/ml basic fibroblast
growth factor (bFGF) and 20 ng/ml epidermal growth factor (EGF)
(both from PeproTech, Inc., Rocky Hill, NJ, USA), 2% B27 (1:50
dilution; Gibco-BRL), and 0.4% bovine serum albumin (BSA;
Invitrogen Life Technologies, Carlsbad, CA, USA). Enzymatically
dissociated single cells were diluted to a density of
2×104/ml and gradually replaced with SFM after plating
into 24-well plates. The cells were cultured in an incubator at
37°C with 5% CO2.
Flow cytometry
The HCT116 cells cultured in SFM were washed twice
with phosphate-buffered saline (PBS; Corning, Inc.) and resuspended
in PBS at a density of 1×107 cells/100 µl. The
dissolved cells were stained using anti-human CD133-PE antibody
(1:10 dilution; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany)
[no need for cells after magnetic-activated cell sorting (MACS)],
and anti-human/mouse CD44 antibody (1:50 dilution; eBioscience,
Inc., San Diego, CA, USA), followed by incubation for 20 min on ice
and subsequent washing with PBS again twice. The respective isotype
controls were used at the same concentrations according to the
manufacturer's instructions (eBioscience, Inc.). The cells were
analyzed on a flow cytometer (FACSVerse; BD Biosciences, Franklin
Lakes, NJ, USA) at the Sun Yat-Sen Memorial Hospital of the Sun
Yat-Sen University (Guangzhou, China).
MACS
The HCT116 cells cultured in SFM were resuspended in
PBS with 2% FBS to a total volume of 1 ml at a density of
1×108 cells/ml in a 12×75 mm polystyrene tube to
properly fit into the magnet (EasySep; STEMCELL Technologies,
Inc.). This was followed by the addition of 100 µl
anti-human CD32 (FcγRII) blocker, 50 µl of anti-human
CD133-PE, 100 µl of PE selection cocktail and 50 µl
of magnetic nanoparticles in turn according to the manufacturer's
instructions (EasySep). The cell suspension was then brought to a
total volume of 2.5 ml by the addition of PBS with 2% FBS (Step A).
The tube was then placed into the magnet for 5 min and the
supernatant fraction was poured off (Step B). Steps A and B were
repeated twice. The magnetically labeled cells remained inside the
tube and held by the magnetic field of the magnet. The cells in the
tube were then used flow cytometric analysis or other assays.
Colony formation assay
In total, numbers of 50/100/200
CD133+CD44+ and
CD133−CD44− HCT116 cells were seeded in each
well in 6-well plates. The medium was discarded when colonies were
observed by the naked eye. The colonies were fixed in methanol and
stained with 10% Giemsa (Teaching and Research Section of Pathology
of Shantou Medical College, Shantou, China). A microscope (Leica
DMI, Leica Microsystems Inc., Buffalo Grove, IL, USA) was used to
confirm that the colonies were made up of >50 cells and to
measure the diameters.
Cell Counting Kit-8 (CCK-8) assay for the
analysis of cell proliferation and cell viability
One hundred microliters of suspension (RPMI-1640
only, CD133+CD44+ and
CD133−CD44− HCT116 cells) was respectively
plated in 96-well plates and cultured for approximately 4 h until
the cells had completely attached to the bottom of the wells. The
cells were cultured at a population of 1,000, 2,000, 4,000 and
8,000 cells per well. Subsequently, 10 µl of
2-(2-methoxyl-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,
monosodium salt (WST-8; (Dojindo Molecular Technologies Inc.,
Kumamoto, Japan) were added to each well followed by incubation at
37°C for approximately 4 h. The absorption (OD value) was read at
450 nm using a spectrophotometer (Multiskan GO; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The OD values were corrected
after the value of the control (cells cultured in RPMI-1640 only)
was subtracted. To examine cell viability, 8,000 cells were used
from each group, and the experiment was repeated 30 times. Cell
proliferation and viability assays were performed by CCK-8 assay as
previously described (30–34).
Tumor transplantation assay using
BALB/c-nu/nu mice
CD133+CD44+ and
CD133−CD44− HCT116 cells were trypsinized,
pelleted and resuspended in RPMI-1640 with Matrigel (1:10 dilution;
BD Biosciences). The mice (license no. SCXK 2011–0029; Sun Yat-Sen
University; n=42; age, 28–35 days; weight, 12–13 g) were randomly
divided into 7 groups by the breeder. The mice were first divided
into 3 main groups (CD133+CD44+,
CD133−CD44− and NaCl solution). The
CD133+CD44+ and
CD133−CD44− groups were further respectively
divided into 3 subgroups (2,000, 20,000 and 200,000 cells). Thus,
there were 7 subgroups, with 6 mice in each group. Only the mice in
the CD133+CD44+ main group formed tumors. Two
hundred microliters of cell suspension (2,000/20,000/200,000
CD133+CD44+ or
CD133−CD44− cells) or NaCl solution were then
injected subcutaneously into the BALB/C-nu/nu mice. Tumor volumes
and the body weight of the mice were measured at regular time
intervals using an electronic balance. After 4 weeks, the mice were
administered an intraperitoneal anesthesia with 4% chloral hydrate
(400 mg/kg), and sacrificed by cervical dislocation and the tumors
were removed. Following removal, the tumors were stored in formalin
and were sent to Google Biological Technology Co., Ltd. (Wuhan,
China) for processing (the tumors were fixed in 10% neutral
buffered formalin, and then subjected to conventional methods of
dehydration, paraffin-embedding and H&E staining). All animal
experiments were conducted in accordance with the protocol of the
Institutional Animal Care and Use Committee of Sun Yat-Sen
University (IACUC-DB-15-1210; Guangzhou, China). and following the
approval of the Research Ethics Committee of Guangdong General
Hospital, Guangdong Academy of Medical Sciences (no. GDREC
2015268A).
Transfection with small interfering RNA
(siRNA) targeting Bmi-1 (Bmi-1-siRNA)
The sequences of the siRNAs used to suppress Bmi-1
were as follows: forward, 5′-GCGGUAACCACCAAUCUUCdTdT-3′ and
reverse, 3′-dTd TCGCCAUUGGUGGUUAGAAG-5′, which targeted the
sequence of GCGGTAACCACCAATCTTC. The control siRNA sequence was
custom ordered and provided by Shanghai SBO Medical Biotechnology
Co., Ltd. (Shanghai, China). The CD133+CD44+
HCT116 cells were transfected with 100 nM of siRNA using SunBio
Trans-EZ (SBO) according instructions provided by the manufacturer.
Cells only transfected with reagent (normal cells) were used as
negative controls.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from thye cultured cells
using TRIzol reagent [Takara Biotechnology (Dalian) Co., Ltd.,
Dalian, China] and 1.0 µg of total RNA was used for cDNA
synthesis using the PrimeScript RT reagent Master mix (Takara
Biotechnology (Dalian) Co., Ltd.). Quantitative (real-time) PCR
(qPCR) was performed using SYBR Premix Ex Taq II (Tli RNaseH Plus)
(Takara Biotechnology (Dalian) Co., Ltd.) with an ABI PRISM 7500
Fast Real-Time PCR system (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) with the following program: 95°C for 30 sec, 95°C for 5
sec, 60°C for 1 min, and 95°C for 30 sec, for 40 cycles. The
results were analyzed using the 2−∆∆CT method. β-actin
gene expression was measured as an endogenous control. Experiments
were carried out in technical triplicates and were repeated at
least twice independently. Primers were custom ordered (Boshang
Biotechnology Co., Ltd, Shanghai, China) with the following
sequences: Bmi-1 forward, 5′-TCTGGGAGTGACAAGG-3′ and
reverse, 5′-AAACAAGAAGAGGTGGA-3′; E-cadherin forward,
5′-TGCCCAGAAAATGAAAAAGG-3′ and reverse, 5′-GTGTATGTGGCAATGCGTTC-3′;
β-actin forward, 5′-GCCAACACAGTGCTGTCTG-3′ and reverse,
5′-TACTCCTGCTTGCTGATCCA-3′.
Western blot analysis
The cells were lysed in lysis buffer (50 mM Tris pH
7.4, 150 mM NaCl, 0.1% NP-40, 0.5% sodium deoxycholate). The
protein concentration of the lysate was quantitated using the BSA
method. Equal amounts of lysate were loaded and separated by
SDS-polyacrylamide gels, and transferred onto nitrocellulose
membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked
with 5% non-fat milk powder in TBS for 1 h and probed with primary
antibodies against Bmi-1 (D20B7) rabbit monoclonal antibody (mAb)
(#6964, 1:1,000 dilution) and E-cadherin (4A2) mouse mAb (#14472,
1:1,000 dilution) (both from Cell Signalling Technology, Inc.,
Danvers, MA, USA), and GAPDH (KC-5G4, 1:8,000 dilution; Kangchen
Biotech, Inc., Shanghai, China). After washing with TBS-T, the
membranes were incubated with secondary antibodies (1:6,000
dilution, A21020, HRP goat anti-rabbit; A21010, HRP goat
anti-mouse; Abbkine, Redlands, CA, USA) and visualized using
chemiluminescence with ImageQuant LAS 500 software (GE Healthcare
Life Sciences, Buckinghamshire, UK).
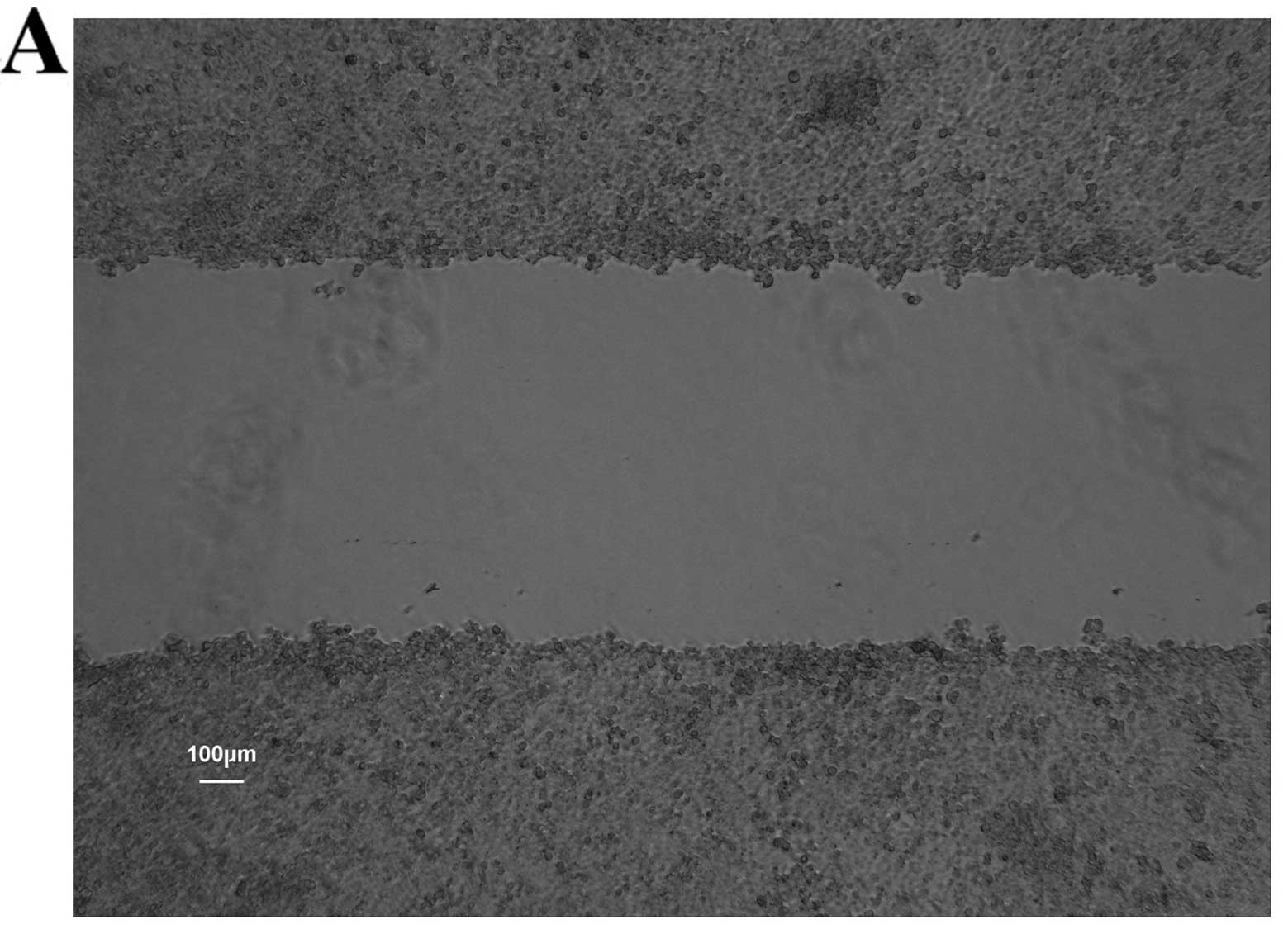
Wound healing assay
The cells (5×105/well) were plated in
6-well plates and cultured until they reached confluence. A
diametric scratch was created using a pipette tip and washed with
PBS 3 times. The cells were photographed under a microscope (Leica
DMI1, Leica Microsystems Inc.) in several pre-marked spots as 0 h.
Images were then acquired at 24 h in the same spots for comparison.
The scratch width was measured and the migration rates of each
group cells were compared on average using Image-Pro Plus 6.0
software (Media Cybernetics, Inc., Rockville, MD, USA).
Transwell migration assay
A Matrigel matrix (BD Biosciences) was used at a
working concentration of 300 µg/ml; the cells were plated in
24-well plates at 100 µl/well and cultured in an incubator
for 1 h before the Falcon cell culture inserts (Corning, Inc.) were
added. The cells were resuspended in RPMI-1640 at a concentration
of 1×105/ml. The upper chamber was loaded with 100
µl of cell suspension and the lower chamber was loaded with
500 µl of RPMI-1640 with 20% FBS. Following incubation for
24 h at 37°C with 5% CO2, the filter was fixed with
methanol and stained with 10% Giemsa. The cells on the upper side
of the filter were wiped off using a cotton swab. The cells that
had migrated to the undersurface of the membrane were counted under
a microscope (Leica DMI1, Leica Microsystems Inc.). Nine
microscopic fields (×100 magnification) were randomly selected to
count the cells. Each assay was carried out in triplicate.
Statistical analyses
All data are presented as the means ± standard error
of the mean (SEM). Statistical significance of differences between
mean values was assessed by the Student's t-test for unpaired data.
Comparisons of data between multiple groups were performed using
analysis of variance (ANOVA). A value of P<0.05 was considered
to indicate a statistically significant differene.
Results
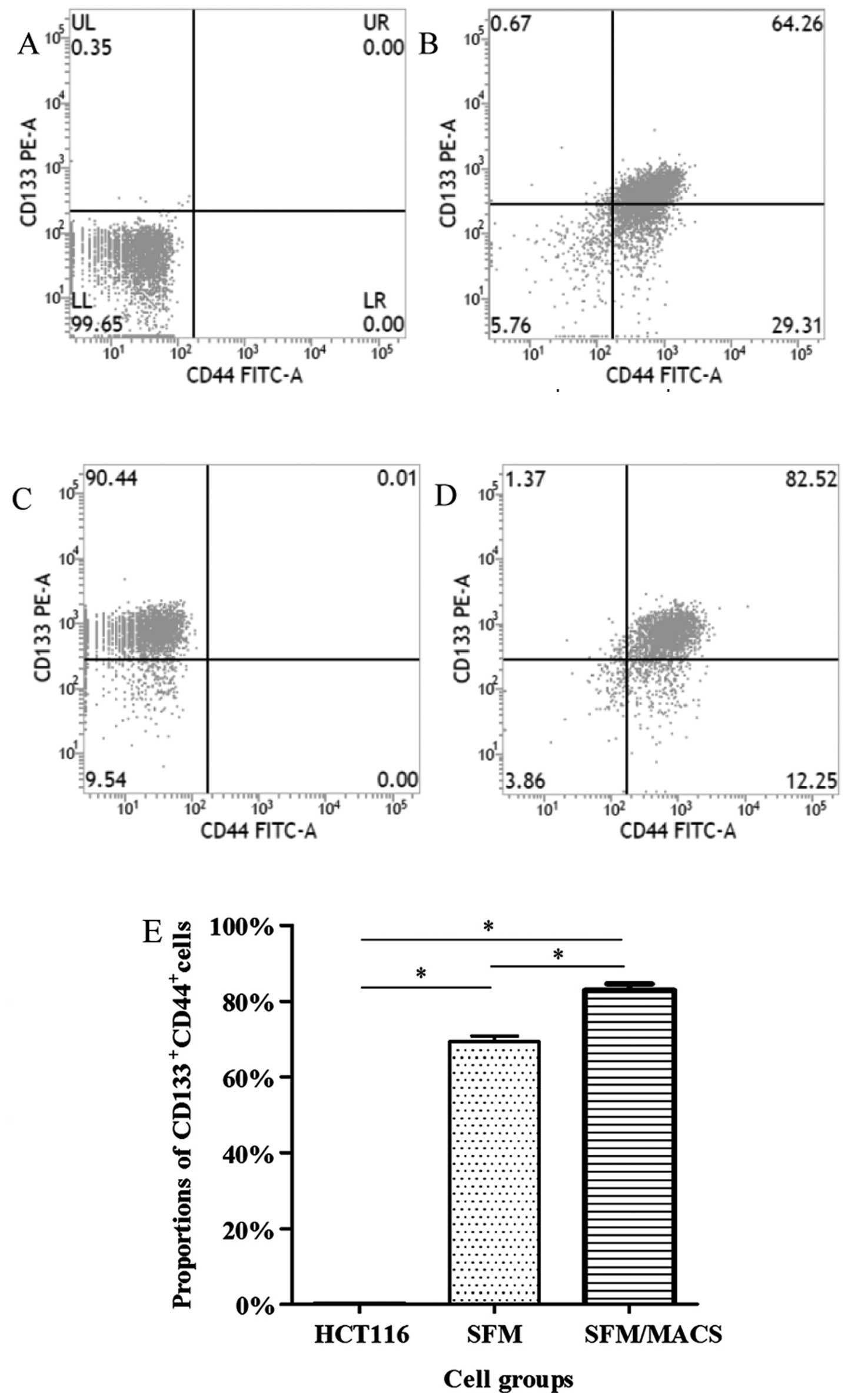
Enrichment and screening of CCSCs by the
use of SFM and MACS
The surface markers, CD133 and CD44, have been
widely used for the selection and isolation of CCSCs from colon
cancer cells (11–13). In our study, the
CD133+CD44+ subpopulation of HCT116 cells
before the experiment only accounted for <1.00%; thus, these
cells were defined as CD133−CD44− cells.
Following culture in SFM and subsequent MACS, we found that the
proportion of CD133+CD44+ cells greatly
increased and these cells were defined as CCSCs (Fig. 1). Furthermore, it seemed that the
proportion of CD133+CD44+ cells depended on
the amount of CD133+ cells. In addition, there was no
significant difference in the frequency of the CD133+
and CD44+ subpopulation between the CCSCs transfected
with the control siRNA or these transfected with Bmi-1-siRNA (data
not shown).
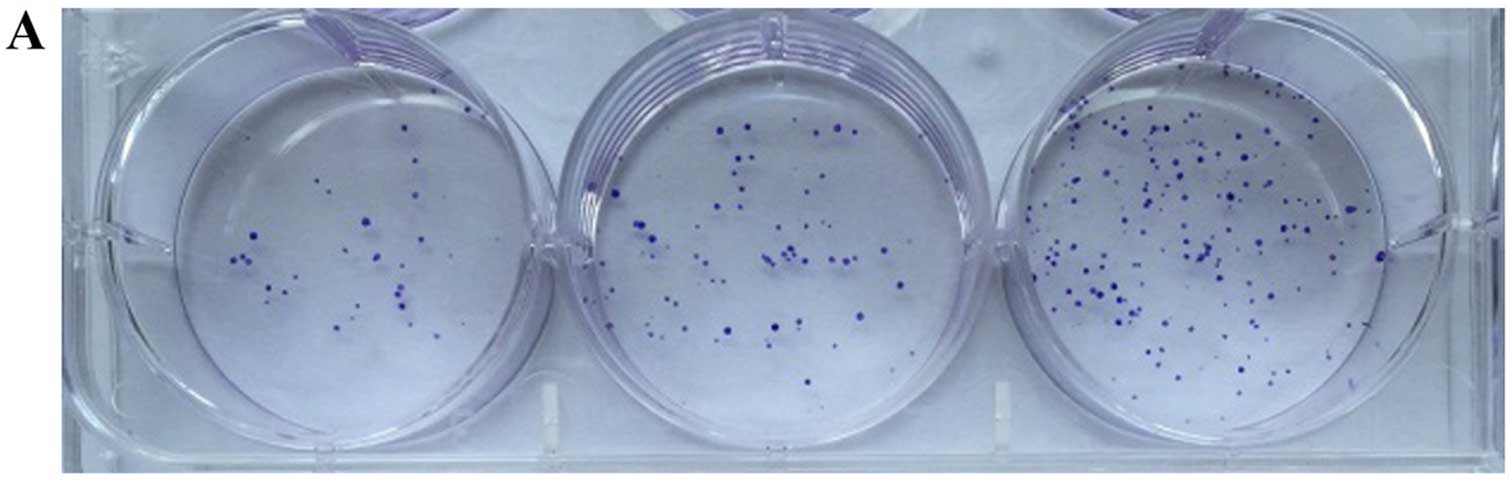
Colony-forming ability of CCSCs in
vitro
Colony formation assay in vitro was used to
identify the CSCs, which reflected the self-renewal and
differentiation abilities of the CSCs. Six-well plates seeded with
cells were photographed following culture for 1 week and the
cloning efficiency of the CD133+CD44+ cells
was markeldy higher than that of the
CD133−CD44− cells (Fig. 2). The biggest and smallest
colonies were almost 10.0 and 5.00 µm in diameter as
observed under a microscope (×100 magnification) after being
stained with 10% Giemsa.
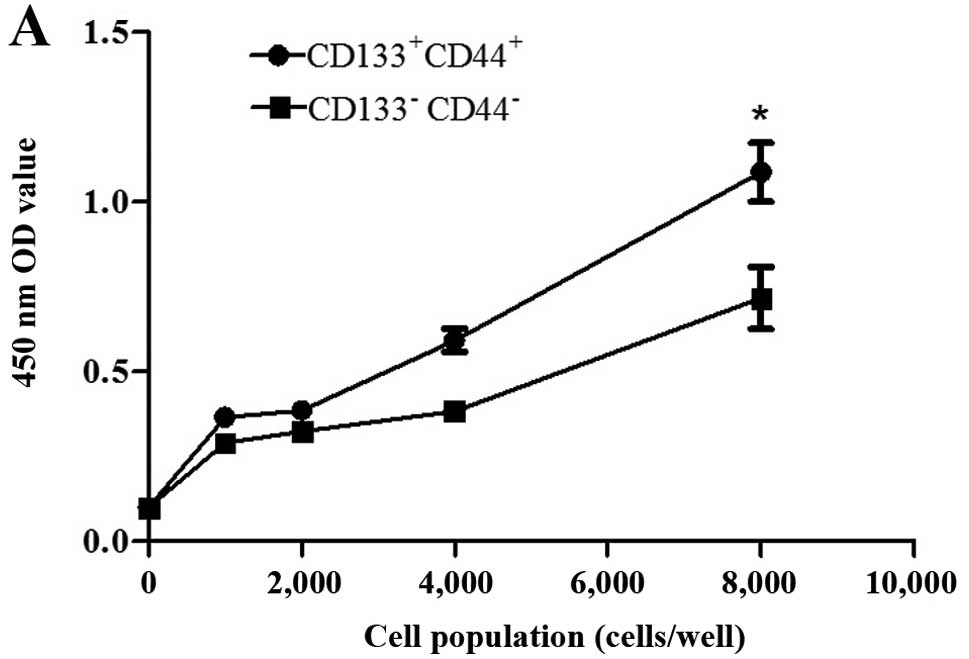
Proliferative ability and viability of
CCSCs in vitro
Cell proliferative ability and cell viability assays
were performed using CCK-8 assay as previously described (30–34). The distinction between the 2
groups of cells was most conspicuously reflected with 8,000 cells
after different gradients of cell populations were designed
according to the manufacturer's instructions (Fig. 3A). Subsequently, another 8,000
cells from each group were cultured as above to compare cell
viability, and the assay was repeated 30 times. The
CD133+CD44+ cells exhibited a greater
viability compared with the CD133−CD44− cells
(Fig. 3B).
Tumor transplantation assay using
BALB/c-nu/nu mice in vivo
We used a tumor transplantation assay to confirm
that the CCSCs had a stronger tumorigenicity in vivo, which
is also considered to be an important characteristic of CSCs. At 4
weeks after the injection, the BALB/c-nu/nu mice were sacrificed
and the tumors were removed. The body weights of the mice injected
with the CD133+CD44+ cells were markedly
lower than those of the mice injected with the
CD133−CD44−cells or with NaCl (Fig. 4A). Only the mice injected with the
CD133+CD44+ cells developed tumors (100%),
which were concentration-dependent (Fig. 4B). The shortest tumor formation
time was approximately 10 days with the injection of 200,000 cells
and the tumor growth curves in the mice in the
CD133+CD44+-injected group are shown in
Fig. 4C. All tumors underwent
routine pathological section examinations to confirm that the assay
was successful (Fig. 4D).
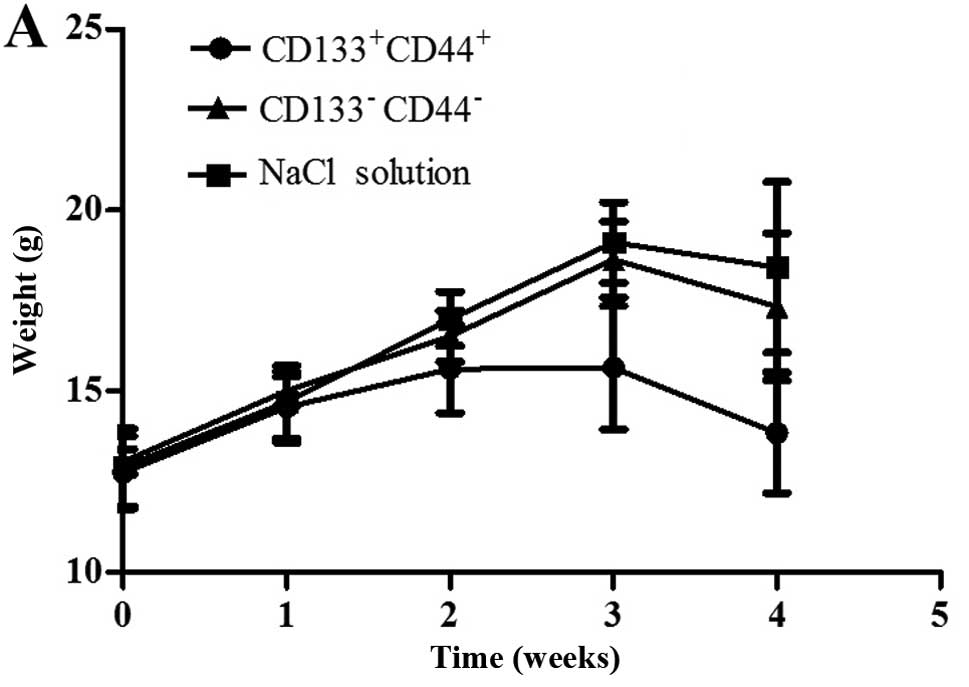 | Figure 4Tumor transplantation assay using
BALB/c-nu/nu mice. (A) The body weights of the mice injected with
NaCl, or the CD133−CD44− cells or
CD133+CD44+ cells were measured over a period
of 4 weeks. (B) Tumors of the mice in the
CD133+CD44+ group injected with various
concentrations of the cells (B1, 200,000 cells; B2, 20,000 cells;
B3, 2,000 cells) were measured using a ruler (cm). (C) Tumor growth
curves of the mice injected with various concentrations of
CD133+CD44+ cells (200,000, 20,000 or 2,000
cells); *P<0.05. (D) Pathological section examination
stained with hematoxylin and eosin (H&E) under a microscope
(×400 magnification). |
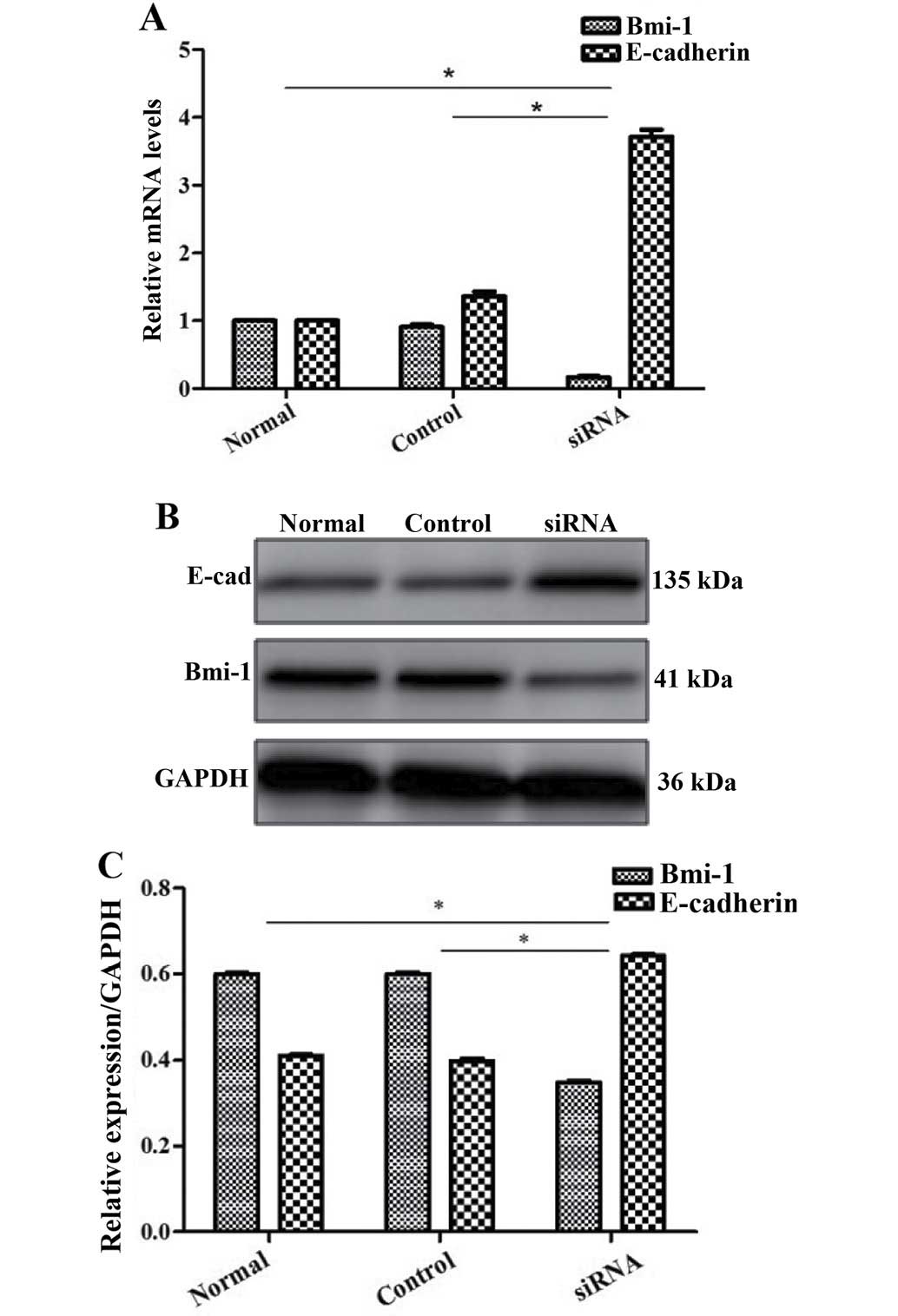
CCSCs exhibit decreased invasive and
migratory abilities after the silencing of Bmi-1
The oncogene Bmi-1 plays a critical role in the
maintenance of CSCs (25) and we
found that it was expressed in the HCT116 colon cancer cell line.
Therefore, we investigated whether Bmi-1 affects the metastatic
potential of CCSCs by gene silencing, using siRNA transfection and
then confirmed our findings by RT-qPCR and western blot analysis
(Fig. 5). By performing wound
healing and Transwell migration assays, we found that the CCSCs
exhibited decreased invasive and migratory abilities, which are
often representative of metastatic potential, after the silencing
of Bmi-1 (Figs. 6 and 7).
We further explored the possible mechanisms
responsible for the effects Bmi-1 on CCSCs. Since EMT occurs during
cancer metastasis and interacts with CSCs (17,18), we focused on E-cadherin (a
hallmark of EMT; the loss of E-cadherin is indicative of EMT) as a
target protein. We found that Bmi-1 had a negative impact on
E-cadherin. In the cells not transfected with Bmi-1-siRNA, the
expression of E-cadherin was low. However, after the silencing of
Bmi-1, E-cadherin expression markedly increased, as shown by
RT-qPCR and western blot analysis (Fig. 5), which may indicate that Bmi-1
promotes the invasion and migration of CCSCs through the
downregulation of E-cadherin, possibly by inducing EMT. The
silencing of Bmi-1 increases E-cadhein expression, thus inhibiting
EMT.
Discussion
The morbidity associated with colorectal cancer is
ranked 3rd among malignant tumors in recent years on the basis of
the treatment guidelines of early colorectal cancer in China
published in 2015 (3). The
prognosis of patients with colorectal cancer is closely related to
early diagnosis and the 5-year survival rate decreases to <12%
at the advanced stage of the disease (2,3).
The incidence and metastasis associated with colorectal cancer are
major concerns and obstacles to effective treatment in both Western
and Eastern countries. It is thus crucial that further research be
carried out to identify methods with which to prevent the
development and metastasis of colon cancer.
The isolation and acquisition of CSCs is a major
achievement in basic and clinical medicine. SFM and MACS can be
used for the enrichment and selection of CSCs. Functional
experiments in vitro and in vivo are often performed
for the identification of CSCs (35–37). In this study, we found that the
CD133+CD44+ HCT116 cells had a greater
cloning efficiency, an enhanced proliferative ability and increased
viability, as well as a stronger tumorigenicity; therefore, they
were used as CCSCs for subsequent experiments. The successful
separation and identification of CCSCs in ours and other studies
strongly supports the CSC theory in colon cancer.
CD133 and CD44 were discovered as important surface
markers of CCSCs (11–13). It is recommended that the
screening and identification of CSCs be performed with more than
one marker. Different markers of cells may represent different
functions and may prove helpful to the understanding of the overall
features. For instance, CD133 may be associated with cloning
efficiency and proliferative ability, while CD44 may be related to
metastasis and survival prediction (16). It has been reported that other
markers of CCSCs include membrane proteins, such as EpCAM (39), Lgr5 (40–42), CD24 (43), CD26 (44,45), CD29 (46) and CD166 (38,47); cytosolic enzymes, such as ALDH1
(48,49); transcription factors, such as as
Oct4 (50), Sox2 (51), Ascl2 (52–54) and Hes1 (55,56); and even the Wnt (57) and Notch (55) signaling pathways. Different
markers may reflect different functions of CCSCs from diverse
perspectives and provide more targets for study and therapy.
However, the critical one and interconnections among them have not
yet been clearly elaborated.
The discovery and eradication of CSCs hold promise
in cancer therapy, as well as genes acting on CSCs and the
regulatory mechanisms. The epigenetic regulator, Bmi-1, is
considered to play essential roles in the self-renewal and
propagation of normal cells and CSCs (25). We found that CCSCs exhibited a
decreased invasive and migratory abilities after Bmi-1 was silenced
by siRNA. This suggests that Bmi-1 is a positive regulator of cell
invasion and migration in colon cancer and that the downregulation
of Bmi-1 may help to prevent metastasis or the progression of colon
cancer. Since the gene targeted therapy of cancer has important
theoretical significance and clinical application prospects, the
knockdown of Bmi-1 is expected to become supplementary treatment
for colon cancer.
In addition, we found E-cadherin was upregulated
when Bmi-1 was silenced, while vimentin and N-cadherin were
downregulated (although we could not make a statistical conclusion
as they were weakly expressed in CCSCs originally; data not shown).
This may indicate that Bmi-1 functions through the downregulation
of E-cadhein, possibly by inducing EMT. This has been previously
demonstrated in nasopharyngeal (58), breast (59,60), melanoma (61), endometrial (62), prostate (63), and bladder (64) cancers. E-cadherin is a
transmenbrane protein important for cell-cell junctions, it
suppresses tumorigenesis and the metastasis of cancers, and its
downregulation or loss is regarded as a hallmark of EMT. The
downregulation or loss of E-cadherin on the surface leads to the
shedding of many types of cancer cells from the tumor mass, which
is the precondition of cancer invasion and metastasis (65,66). The molecular mechanisms
responsible for cancer metastasis, regarded as a promising target
for cancer chemotherapy, are currently receiving increased
attention. Bmi-1 was considered to play an important role in the
pathogenesis of nasopharyngeal cancer by inducing EMT, partly by
targeting the tumor suppressor, PTEN, thus activating the PI3K/Akt
pathway (58). It was also
demonstrated that Bmi-1 induced cell invasion with the activation
of the Akt pathway in breast cancer cells (60). Alternatively, it was shown that
Bmi-1 acted negatively on PTEN and E-cadherin in colorectal cancer
from a histological perspective by Liao et al (67). Therefore, we speculate there is a
strong possibility that Bmi-1 promotes the invasion and migration
of CCSCs through the downregulation of E-cadherin and the induction
of EMT, and through the PI3K/Akt pathway. However, the crosstalk
between different pathways for expanding the cellular communication
signaling network should be also given several considerations. In
this study, we put forward the hypothesis of the role of Bmi-1 and
EMT in CCSCs for the first time, to the best of our knowledge.
Namely, the eradication of CCSCs, and the blocking of EMT and the
downregulation of Bmi-1 may together prevent the metastasis of
colon cancer at an early stage and may thus improve the 5-year
survival rate when applied clinically.
In conclusion, our study demonstrates that
CD133+CD44+ HCT116 cells can be used as CCSCs
for subsequent medical studies. Bmi-1 promotes the invasion and
migration of CCSCs through the downregulation of E-cadherin,
possibly by inducing EMT. This finding may provide a new target for
colon cancer therapy. To this end, it would be of great interest
for us to further investigate the signaling pathways and regulatory
mechanisms of the interaction between Bmi-1 and EMT in colon
cancer.
Acknowledgments
We sincerely thank other colleagues in our
laboratory for their active help in this study. This study was
supported by a grant from the Natural Science Foundation of
Guangdong Province (2014A030313543).
References
|
1
|
Provenzale D, Jasperson K, Ahnen DJ,
Aslanian H, Bray T, Cannon JA, David DS, Early DS, Erwin D, Ford
JM, et al: Colorectal cancer screening, version 1.2015. J Natl
Compr Canc Netw. 13:959–968. 2015.PubMed/NCBI
|
|
2
|
Gastrointestinal surgery group; Colorectal
and anal surgery group; Colorectal cancer Specialized Committee:
Guidelines for diagnosis and comprehensive treatment of colorectal
cancer liver metastases (V 2013). Zhonghua wei chang wai ke za zhi.
16:780–788. 2013.In Chinese.
|
|
3
|
Association of Digestive Endoscopy, Cancer
endoscopy Specialized Committee: Guidelines for early colorectal
cancer screening and endoscopic diagnosis and treatment in China
(2014). Nat Med J China. 95:2235–2252. 2015.In Chinese.
|
|
4
|
Polyak K and Hahn WC: Roots and stems:
stem cells in cancer. Nat Med. 12:296–300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li F, Tiede B, Massagué J and Kang Y:
Beyond tumorigenesis: cancer stem cells in metastasis. Cell Res.
17:3–14. 2007. View Article : Google Scholar
|
|
6
|
Kakarala M and Wicha MS: Implications of
the cancer stem-cell hypothesis for breast cancer prevention and
therapy. J Clin Oncol. 26:2813–2820. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bonnet D and Dick JE: Human acute myeloid
leukemia is organized as a hierarchy that originates from a
primitive hematopoietic cell. Nat Med. 3:730–737. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
9
|
Ponti D, Costa A, Zaffaroni N, Pratesi G,
Petrangolini G, Coradini D, Pilotti S, Pierotti MA and Daidone MG:
Isolation and in vitro propagation of tumorigenic breast cancer
cells with stem/progenitor cell properties. Cancer Res.
65:5506–5511. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar
|
|
12
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar
|
|
13
|
Chu P, Clanton DJ, Snipas TS, Lee J,
Mitchell E, Nguyen ML, Hare E and Peach RJ: Characterization of a
subpopulation of colon cancer cells with stem cell-like properties.
Int J Cancer. 124:1312–1321. 2009. View Article : Google Scholar
|
|
14
|
Laganà AS, Colonese F, Colonese E, Sofo V,
Salmeri FM, Granese R, Chiofalo B, Ciancimino L and Triolo O:
Cytogenetic analysis of epithelial ovarian cancer's stem cells: an
overview on new diagnostic and therapeutic perspectives. Eur J
Gynaecol Oncol. 36:495–505. 2015.PubMed/NCBI
|
|
15
|
López J, Valdez-Morales FJ,
Benítez-Bribiesca L, Cerbón M and Carrancá AG: Normal and cancer
stem cells of the human female reproductive system. Reprod Biol
Endocrinol. 11:532013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jing F, Kim HJ, Kim CH, Kim YJ, Lee JH and
Kim HR: Colon cancer stem cell markers CD44 and CD133 in patients
with colorectal cancer and synchronous hepatic metastases. Int J
Oncol. 46:1582–1588. 2015.PubMed/NCBI
|
|
17
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Birchmeier W and Birchmeier C:
Epithelial-mesenchymal transitions in development and tumor
progression. EXS. 74:1–15. 1995.PubMed/NCBI
|
|
19
|
Onder TT, Gupta PB, Mani SA, Yang J,
Lander ES and Weinberg RA: Loss of E-cadherin promotes metastasis
via multiple downstream transcriptional pathways. Cancer Res.
68:3645–3654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jacobs JJ and van Lohuizen M: Polycomb
repression: from cellular memory to cellular proliferation and
cancer. Biochim Biophys Acta. 1602:151–161. 2002.PubMed/NCBI
|
|
21
|
Piunti A and Pasini D: Epigenetic factors
in cancer development: polycomb group proteins. Future Oncol.
7:57–75. 2011. View Article : Google Scholar
|
|
22
|
Honig A, Weidler C, Häusler S,
Krockenberger M, Buchholz S, Köster F, Segerer SE, Dietl J and
Engel JB: Overexpression of polycomb protein BMI-1 in human
specimens of breast, ovarian, endometrial and cervical cancer.
Anticancer Res. 30:1559–1664. 2010.PubMed/NCBI
|
|
23
|
Vékony H, Raaphorst FM, Otte AP, van
Lohuizen M and Leemans CR: High expression of Polycomb group
protein EZH2 predicts poor survival in salivary gland adenoid
cystic carcinoma. J Clin Pathol. 61:744–749. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Silva J, García JM, Peña C, García V,
Domínguez G, Suárez D, Camacho FI, Espinosa R, Provencio M, España
P and Bonilla F: Implication of polycomb members Bmi-1, Mel-18, and
Hpc-2 in the regulation of p16INK4a, p14ARF, h-TERT, and c-Myc
expression in primary breast carcinomas. Clin Cancer Res.
12:6929–6936. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Molofsky AV, Pardal R, Iwashita T, Park
IK, Clarke MF and Morrison SJ: Bmi-1 dependence distinguishes
neural stem cell self-renewal from progenitor proliferation.
Nature. 425:962–967. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu XH, Sha WH, Lin F, Cen RY and Liao SY:
The correlation between genetic expressions of Bmi-1 and
clinicopathological parameters of colorectal cancer. Modern
Digestion & Intervention. 16:1–5. 2011.In Chinese.
|
|
27
|
Yang J, Liu J, Lyu X and Fei S:
Resveratrol inhibits cell proliferation and up-regulates MICA/B
expression in human colon cancer stem cells. Xi Bao Yu Fen Zi Mian
Yi Xue Za Zhi. 31:889–893. 2015.In Chinese. PubMed/NCBI
|
|
28
|
Xiong B, Ma L, Hu X, Zhang C and Cheng Y:
Characterization of side population cells isolated from the colon
cancer cell line SW480. Int J Oncol. 45:1175–1183. 2014.PubMed/NCBI
|
|
29
|
Feng Y, Dai X, Li X, Wang H, Liu J, Zhang
J, Du Y and Xia L: EGF signalling pathway regulates colon cancer
stem cell proliferation and apoptosis. Cell Prolif. 45:413–419.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou M, Lu Y, Yuan L, Zheng L, Liu Y, Hong
M, Zhang C and Li X: Preliminary screening of downstream proteins
of Sox2 and role of Sox2 in colonic cancer cell migration and
invasion. Nan Nan Fang Yi Ke Da Xue Xue Bao. 34:1594–1600. 2014.In
Chinese.
|
|
31
|
Zhang M, Cui F, Lu S, Lu H, Xue Y, Wang J,
Chen J, Zhao S, Ma S, Zhang Y, et al: Developmental
pluripotency-associated 4: a novel predictor for prognosis and a
potential therapeutic target for colon cancer. J Exp Clin Cancer
Res. 34:602015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie X, Zhao Y, Ma CY, Xu XM, Zhang YQ,
Wang CG, Jin J, Shen X, Gao JL, Li N, et al: Dimethyl fumarate
induces necroptosis in colon cancer cells through GSH depletion/ROS
increase/MAPKs activation pathway. Br J Pharmacol. 172:3929–3943.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin YU, Wu T, Yao Q, Zi S, Cui L, Yang M
and Li J: LGR5 promotes the proliferation of colorectal cancer
cells via the Wnt/β-catenin signaling pathway. Oncol Lett.
9:2859–2863. 2015.PubMed/NCBI
|
|
34
|
Peng HX, Wu WQ, Yang DM, Jing R, Li J,
Zhou FL, Jin YF, Wang SY and Chu YM: Role of B7-H4 siRNA in
proliferation, migration, and invasion of LOVO colorectal carcinoma
cell line. BioMed Res Int. 2015:3269812015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zeng L, Qiu L, Yang XT, Zhou YH, Du J,
Wang HY, Sun JH, Yang C and Jiang JX: Isolation of lung multipotent
stem cells using a novel microfluidic magnetic activated cell
sorting system. Cell Biol Int. 39:1348–1353. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xue ZX, Zheng JH, Zheng ZQ, Cai JL, Ye XH,
Wang C, Sun WJ, Zhou X, Lu MD, Li PH and Cai ZZ: Latexin inhibits
the proliferation of CD133+ miapaca-2 pancreatic cancer
stem-like cells. World J Surg Oncol. 12:1–11. 2014. View Article : Google Scholar
|
|
37
|
Zhang DG, Jiang AG, Lu HY, Zhang LX and
Gao XY: Isolation, cultivation and identification of human lung
adenocarcinoma stem cells. Oncol Lett. 9:47–54. 2015.
|
|
38
|
Levin TG, Powell AE, Davies PS, Silk AD,
Dismuke AD, Anderson EC, Swain JR and Wong MH: Characterization of
the intestinal cancer stem cell marker CD166 in the human and mouse
gastrointestinal tract. Gastroenterology. 139:2072–2082. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Femia AP, Dolara P, Salvadori M and
Caderni G: Expression of LGR-5, MSI-1 and DCAMKL-1, putative stem
cell markers, in the early phases of 1,2-dimethylhydrazine-induced
rat colon carcinogenesis: correlation with nuclear β-catenin. BMC
Cancer. 13:482013. View Article : Google Scholar
|
|
41
|
Yui S, Nakamura T, Sato T, Nemoto Y,
Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K,
et al: Functional engraftment of colon epithelium expanded in vitro
from a single adult Lgr5+ stem cell. Nat Med.
18:618–623. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Barker N, van Es JH, Kuipers J, Kujala P,
van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H,
Peters PJ and Clevers H: Identification of stem cells in small
intestine and colon by marker gene Lgr5. Nature. 449:1003–1007.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vermeulen L, Todaro M, de Sousa Mello F,
Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G and Medema
JP: Single-cell cloning of colon cancer stem cells reveals a
multi-lineage differentiation capacity. Proc Natl Acad Sci USA.
105:13427–13432. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fric P, Sovová V, Sloncová E, Lojda Z,
Jirásek A and Cermák J: Different expression of some molecular
markers in sporadic cancer of the left and right colon. Eur J
Cancer Prev. 9:265–268. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pang R, Law WL, Chu AC, Poon JT, Lam CS,
Chow AK, Ng L, Cheung LW, Lan XR, Lan HY, et al: A subpopulation of
CD26+ cancer stem cells with metastatic capacity in
human colorectal cancer. Cell Stem Cell. 6:603–615. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fujimoto K, Beauchamp RD and Whitehead RH:
Identification and isolation of candidate human colonic clonogenic
cells based on cell surface integrin expression. Gastroenterology.
123:1941–1948. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lugli A, Iezzi G, Hostettler I, Muraro MG,
Mele V, Tornillo L, Carafa V, Spagnoli G, Terracciano L and Zlobec
I: Prognostic impact of the expression of putative cancer stem cell
markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer.
Br J Cancer. 103:382–390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang EH, Hynes MJ, Zhang T, Ginestier C,
Dontu G, Appelman H, Fields JZ, Wicha MS and Boman BM: Aldehyde
dehydrogenase 1 is a marker for normal and malignant human colonic
stem cells (SC) and tracks SC overpopulation during colon
tumorigenesis. Cancer Res. 69:3382–3389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dylla SJ, Beviglia L, Park IK, Chartier C,
Raval J, Ngan L, Pickell K, Aguilar J, Lazetic S, Smith-Berdan S,
et al: Colorectal cancer stem cells are enriched in xenogeneic
tumors following chemotherapy. PLoS One. 3:e24282008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Park IH, Zhao R, West JA, Yabuuchi A, Huo
H, Ince TA, Lerou PH, Lensch MW and Daley GQ: Reprogramming of
human somatic cells to pluripotency with defined factors. Nature.
451:141–146. 2008. View Article : Google Scholar
|
|
51
|
Saigusa S, Tanaka K, Toiyama Y, Yokoe T,
Okugawa Y, Ioue Y, Miki C and Kusunoki M: Correlation of CD133,
OCT4, and SOX2 in rectal cancer and their association with distant
recurrence after chemoradiotherapy. Ann Surg Oncol. 16:3488–3498.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ziskin JL, Dunlap D, Yaylaoglu M, Fodor
IK, Forrest WF, Patel R, Ge N, Hutchins GG, Pine JK, Quirke P, et
al: In situ validation of an intestinal stem cell signature in
colorectal cancer. Gut. 62:1012–1023. 2013. View Article : Google Scholar
|
|
53
|
Zhu R, Yang Y, Tian Y, Bai J, Zhang X, Li
X, Peng Z, He Y, Chen L, Pan Q, et al: Ascl2 knockdown results in
tumor growth arrest by miRNA-302b-related inhibition of colon
cancer progenitor cells. PLoS One. 7:e321702012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jubb AM, Chalasani S, Frantz GD, Smits R,
Grabsch HI, Kavi V, Maughan NJ, Hillan KJ, Quirke P and Koeppen H:
Achaete-scute like 2 (ascl2) is a target of Wnt signalling and is
upregulated in intestinal neoplasia. Oncogene. 25:3445–3457. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Reedijk M, Odorcic S, Zhang H, Chetty R,
Tennert C, Dickson BC, Lockwood G, Gallinger S and Egan SE:
Activation of Notch signaling in human colon adenocarcinoma. Int J
Oncol. 33:1223–1229. 2008.PubMed/NCBI
|
|
56
|
Gao F, Zhang Y, Wang S, Liu Y, Zheng L,
Yang J, Huang W, Ye Y, Luo W and Xiao D: Hes1 is involved in the
self-renewal and tumourigenicity of stem-like cancer cells in colon
cancer. Sci Rep. 4:39632014.PubMed/NCBI
|
|
57
|
Vermeulen L, De Sousa E, Melo F, van der
Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M,
Merz C, Rodermond H, et al: Wnt activity defines colon cancer stem
cells and is regulated by the microenvironment. Nat Cell Biol.
12:468–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu
LJ, Kong QL, Xu LH, Zhang X, Liu WL, et al: The polycomb group
protein Bmi-1 represses the tumor suppressor PTEN and induces
epithelial-mesenchymal transition in human nasopharyngeal
epithelial cells. J Clin Invest. 119:3626–3636. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li H, Song F, Chen X, Li Y, Fan J and Wu
X: Bmi-1 regulates epithelial-to-mesenchymal transition to promote
migration and invasion of breast cancer cells. Int J Clin Exp
Pathol. 7:3057–3064. 2014.PubMed/NCBI
|
|
60
|
Guo BH, Feng Y, Zhang R, Xu LH, Li MZ,
Kung HF, Song LB and Zeng MS: Bmi-1 promotes invasion and
metastasis, and its elevated expression is correlated with an
advanced stage of breast cancer. Mol Cancer. 10:102011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu S, Tetzlaff MT, Cui R and Xu X:
miR-200c inhibits melanoma progression and drug resistance through
down-regulation of BMI-1. Am J Pathol. 181:1823–1835. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dong P, Kaneuchi M, Watari H, Hamada J,
Sudo S, Ju J and Sakuragi N: MicroRNA-194 inhibits epithelial to
mesenchymal transition of endometrial cancer cells by targeting
oncogene BMI-1. Mol Cancer. 10:992011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Nanta R, Kumar D, Meeker D, Rodova M, Van
Veldhuizen PJ, Shankar S and Srivastava RK: NVP-LDE-225
(Erismodegib) inhibits epithelial-mesenchymal transition and human
prostate cancer stem cell growth in NOD/SCID IL2Rγ null mice by
regulating Bmi-1 and microRNA-128. Oncogenesis. 2:e422013.
View Article : Google Scholar
|
|
64
|
Liu L, Qiu M, Tan G, Liang Z, Qin Y, Chen
L, Chen H and Liu J: miR-200c inhibits invasion, migration and
proliferation of bladder cancer cells through down-regulation of
BMI-1 and E2F3. J Transl Med. 12:3052014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dass SD, Cheah PL, Ong DB, Teoh KH and
Looi LM: E-cadherin downregulation at the infiltrating tumour front
is associated with histological grade and stage in colorectal
carcinoma of Malaysians. Malays J Pathol. 37:19–24. 2015.PubMed/NCBI
|
|
66
|
Le Bras GF, Taubenslag KJ and Andl CD: The
regulation of cell-cell adhesion during epithelial-mesenchymal
transition, motility and tumor progression. Cell Adh Migr.
6:365–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liao WT, Cui YM and Ding YQ: Expression
and Significance of Bmi-1 PTEN and E-Cadherin in Colorectal Cancer.
Chin J Clin Oncol. 39:559–563. 2012.In Chinese.
|