Introduction
The area of chronic wound healing has drawn much
attention from many healthcare professionals due to the high cost
of healthcare resources for patients seeking appropriate treatment,
as well as the complexity of its underlying mechanism. It has been
suggested that a reduction in tissue growth factors, an imbalance
between proteinases and their inhibitors, and the presence of
senescent cells is of importance in chronic wounds (1). A variety of treatments, such as
dressings, application of topical growth factors, autologous skin
grafting and bioengineered skin equivalents have been applied to
deal with certain types of chronic wounds in addition to the basic
treatments (1). However, the
specific mechanisms of each treament remain unclear and are under
investigation. Therefore, more insight into the mechanisms
responsible are required to gain a better understanding of the
wound-healing process. Further clarification of this complex system
may contribute to the emergence of a prognositc marker to predict
the healing potential of chronic wounds, enhancing patient
management.
Wound healing is a dynamic and interactive process
that is immediately activated by the damage of skin upon injury.
Primary targets for the treatment of wounds are reducing the
healing time and generating an aesthetical scar or scar-free tissue
without comprimising any normal function. The process of wound
healing consists of three overlapping orchestrated stages, known as
inflammation, proliferation, and re-epithelialization and tissue
remodelling (2). These complex
processes are regulated by numerous signalling pathways involving
several cell types including keratinocytes, macrophages,
fibroblasts and endothelial cells (3). A vast array of cytokines and growth
factors play key roles in the process of wound healing through the
initiation of a variety of signalling cascades. Disruption of these
tightly regulated processes can lead to a variety of pathological
conditions. The Janus kinase/signal transducer and activator of the
transcription (JAK/STAT) pathway is a key pathway, responding to a
wide variety of cytokines and growth factors. Any mutation that
holds the potential to compromise the regular function of JAK/STAT
pathway may affect cytokine stimulated signalling (4). By contrast, loss of regulation to
this signalling pathway may cause inflammatory disease,
erythrocytosis, gigantism and leukaemia (5). Therefore, the mechanism regulating
appropriate JAK/STAT signalling is of great importance. This target
can be achieved by so-called negative regulators, such as
Src-homology 2 (SH2) containing phosphatase (SHP), protein
inhibitors against STATs (PIAS) and suppressor of cytokine
signalling (SOCS), which all comprise specific SH2 domain in their
structure (6). However, compared
to the other two cytokine signalling inhibitors which are
constitutively expressed, SOCS is the only cytokine inducible
inhibitor that may hold the potential to be utilised as a regulator
for the dysregulation of cell metabolism.
SOCS protein was origninally identified in 1997
independently by three different groups as SOCS1, the JAK-binding
protein (JAB) and the STAT-induced STAT inhibitor (SSI) (7–9).
It was subsequently confirmed that all three belong to the SOCS
family, which consists of eight members [SOCS1 to 7 and cytokine
inducible SH2-containing protein (CIS)] in mammals (10). Generally, there are three major
domains contributing to the function of SOCS, identified as a
central conserved SH2 domain, a N-terminal domain of variable
length and divergent sequence, and a carboxy-terminal,
40-amino-acid module known as SOCS box (11). Some of the structurally related
eight SOCS family members are capable of attenuating cytokine
signalling by blocking JAK tyrosine kinase activity, competing for
a docking site with STAT protein and/or binding to the respective
target proteins for subsequent proteosomal degradation (11). Of the eight SOCS family members,
CIS/SOCS1 to SOCS3 are the most well characterised, whereas, the
precise function of the remaining four SOCS proteins, SOCS4 to 7,
have yet to be elucidated due to their extensive expression and
inconsistent protein targets (12). However, evidence from existing
studies show that SOCS family members can be classified according
to their target proteins. CIS/SOCS1 to SOCS3 regulate cytokine
receptor signalling through the JAK-STAT pathway, whereas SOCS4 to
7 are associated with the regulation of growth factor receptor
signalling (13–15).
The SOCS family members have been found to regulate
a number of cytokines and growth factors that are essential in the
wound-healing process (16). With
the absence of SOCS3, interleukin-6 (IL-6), a proinflammaroty
cytokine, acts as an immunosuppressive cytokine that reduces tumour
necrosis factor (TNF) induced by lipopolysaccharides (LPSs)
(17). Additionally, SOCS3
selectively inhibits the activation of STAT3 signalling by IL-6,
but does not influence IL-10-activated STAT3 signalling due to
inefficient binding to the IL-10 receptor. This potentially
explains the opposing function of the proinflammatory cytokine IL-6
and the anti-inflammatory cytokine IL-10, and indicates the
regulatory role of SOCS3 to IL-6-induced STAT3 signalling,
highlighting a potential role for SOCS3 in the treatment of
IL-6-mediated inflammatory diseases (17). The expression of SOCS4 and 5 in
HeLa cells is induced at the mRNA level following treatment with
epidermal growth factor (EGF), which is frequently induced during
the wound-healing process. In addition, either SOCS4 or 5 markedly
reduces EGFR expression (13).
This result suggests that SOCS4 and 5 may have an effect on EGF
signalling through the regulation of EGFR, the respective receptor
of EGF. SOCS5 is also capable of reducing the expression level of
EGFR in a ligand- and c-Cbl-independent manner to enhance its
proteasome degradation. In addition, SH2 and SB domains of SOCS5
are required for the regulation of EGFR, which enables SOCS5 to
attenuate EGF-induced signalling by trans-locating EGFR to
intracellular vesicles (13).
The present study examined the transcript expression
profile of SOCS1 to 7 in a clinical cohort of healing and
non-healing chronic wounds. This expression was further examined in
clinical tissue sections to demonstrate their expression pattern
and localisation in these contrasting subtypes of chronic
wounds.
Materials and methods
Tissue collection and processing
A clinical chronic wound tissue cohort was used in
the present study. Tissues were collected from patients attending
the University Hospital of Wales Wound Healing Clinic. Wound edge
tissues were collected from chronic venous leg ulcer patients
following ethics approval from a Local Committee (South East Wales
Research Ethics Committee). Wound size was recorded at the time of
initial biopsy and was re-measured after 3 months, during which
wounds were treated as per best medical treatment. Wounds that had
become reduced in size over this period were described as
'healing/healed' (n=20) and those which remained static or grew in
size were described as 'non-healing' (n=51) chronic wounds. This
cohort has been previously described (18).
Sample biopsies were snap frozen and stored at −80°C
before being placed in liquid nitrogen until required for
processing and analysis. The samples were sectioned (7 µm)
on a cryostat (Leica Microsystems, Ltd., Milton Keynes, UK), for
immunohistochemical (IHC) staining and multiple sections (20
µm), were processed for the extraction of RNA, reverse
transcription and transcript expression analysis using quantitative
polymerase chain reaction (qPCR).
RNA extraction and reverse
transcription
Multiple sections from the same patient sample
biopsy were combined and homogenised in ice-cold TRIzol reagent
(Sigma-Aldrich, Poole, UK) using a handheld homogeniser
(Cole-Parmer, London, UK). RNA was subsequently extracted following
the manufacturer's instructions. Following extraction, RNA was
resuspended in diethyl pyrocarbonate (DEPC) H2O and
quantified using a spectrophotometer (WPA UV 1101; Biotech
Photometer, Cambridge, UK). The samples were then standardised
prior to undertaking the reverse transcription reaction using an RT
kit (Bio-Rad, Hemel Hemstead, UK) to generate cDNA.
qPCR
qPCR was undertaken to analyse transcript expression
of the SOCS family members in the chronic healing and non-healing
wound samples. This process has been previously described by our
laboratory (18,19). In brief, primers were designed for
each of the SOCS family members using the Beacon Designer software
(Bio-Rad, Hercules, CA, USA). A Z sequence was added to one of each
of the primer pairs (ACTGAACCTGACCGTACA), to allow incorporation of
the FAM tagged UniPrimer probe (Intergen, Inc., New York, NY, USA)
and thus fluorescent detection. Expression of the target sequence
was detected in conjunction with an internal standard used to
generate a standard curve. The samples were amplified and detected
on a StepOnePlus qPCR machine (Life Technologies, Paisley, UK),
using qPCR mastermix (iQ supermix; Bio-Rad), with the specific
forward primer (10 pM), reverse primer containing the Z sequence (1
pM) and the FAM-tagged UniPrimer probe (10 pM). SOCS family members
were detected in conjunction with the cytokeratin 19 (CK19)
housekeeping gene and values were subsequently normalised against
this gene. Primer details are provided in Table I.
 | Table IPrimers used in the present study. |
Table I
Primers used in the present study.
| Primers | Forward | Reverse |
|---|
| SOCS1 |
5′-GATGGTAGCACACAACCAG |
5′-ACTGAACCTGACCGTACAGAGGAGGAGGAGGAAGGTT |
| SOCS2 |
5′-GGATGGTACTGGGGAAGTAT |
5′-ACTGAACCTGACCGTACATGGGAGCTATCTCTAATCAA |
| SOCS3 |
5′-TCAAGACCTTCAGCTCCA |
5′-ACTGAACCTGACCGTACAGTCACTGCGCTCCAGTAG |
| SOCS4 |
5′-GGCAGTGTTTTCCAATAAAG |
5′-ACTGAACCTGACCGTACAAGGTGGGAAAGGACACTTAT |
| SOCS5 |
5′-AGTCAAAGCCTCTCTTTTCC |
5′-ACTGAACCTGACCGTACACATTTTGGGCTAAATCTGA |
| SOCS6 |
5′-CCTTACAGAGGAGCTGAAAA |
5′-ACTGAACCTGACCGTACACGAAAAGAAAAGAACCATC |
| SOCS7 |
5′-CAGGCCCTGAATTACCTC |
5′-ACTGAACCTGACCGTACAGAGGTTGCTGCTGCTGCT |
| CK19 |
5′-AGCCACTACTACACGACCAT |
5′-ACTGAACCTGACCGTACATCGATCTGCAGGACAATC |
IHC staining
Primary antibodies: anti-SOCS1 (sc-9021), anti-SOCS2
(sc-9022), anti-SOCS3 (sc-9023), anti-SOCS4 (sc-68827), anti-SOCS5
(sc-100858), anti-SOCS6 (sc-5608) and anti-SOCS7 (sc-8291 and
sc-137241) were purchased from Insight Biotechnology (Wembley, UK).
All the antibodies were used at a final concentration of 2
µg/ml.
IHC analysis was performed on a subset of chronic
wound tissues that included healing/healed (n=10) and non-healed
(n=10) biopsies. Staining was performed using a standard
avidin-biotin peroxidase technique. In brief, the frozen sections
were fixed for 15 min in dried acetone (Thermo Fisher Scientific,
Ltd., Loughborough, UK) and then air dried for 15 min. The sections
were washed three times in Tris-buffered saline (TBS) for 5 min
each, followed by a 30-min permeabilisation wash with 0.1%
saponin/TBS (Sigma-Aldrich). Since this reaction is reversible,
subsequent washes were performed with 0.1% saponin/TBS. The
sections were then incubated with a blocking solution in a
humidified box at room temperature (20–22°C) for 1 h. The blocking
solution, which contained 0.1% BSA/0.1% saponin/10% horse serum in
TBS, was also used to dilute all subsequent reagents. Excess
blocking solution was removed and the sections were incubated with
the relevant, previously optimised, primary antibody for 1 h.
Following washing with 0.1% saponin/TBS, the sections were then
incubated for 30 min with the relevant biotinylated secondary
antibody (Vector Laboratories, Peterborough, UK), followed by a
30-min incubation with the avidin-biotin complex (ABC) reagent. The
3,3′-diamino-benzidine (DAB) substrate (5 mg/ml) was used to
develop the final reaction product, the sections were rinsed in tap
water, counterstained with Gill's hematoxylin (Vector
Laboratories), and rehydrated through a series of graded alcohols,
cleared in xylene (Thermo Fisher Scientific, Ltd.) and mounted in
DPX (Merck Millipore, UK). Negative controls were performed where
primary antibody was replaced with wash buffer.
Staining was visualised using a microscope, and the
localisation and intensity of staining for each antibody was
assessed by two independent investigators, at both the wound edge
and an area distal to the wound edge. Intensity of staining was
qualitatively graded as strong, moderate or negative.
Statistical analysis
Statistical analysis was undertaken using the
SigmaPlot 11 statistical software package (Systat Software Inc.,
London, UK). Data were analysed using a two-sample, two-tailed,
t-test or a Mann-Whitney test depending on data normality. Values
of P<0.05 were regarded as statistically significant.
Results
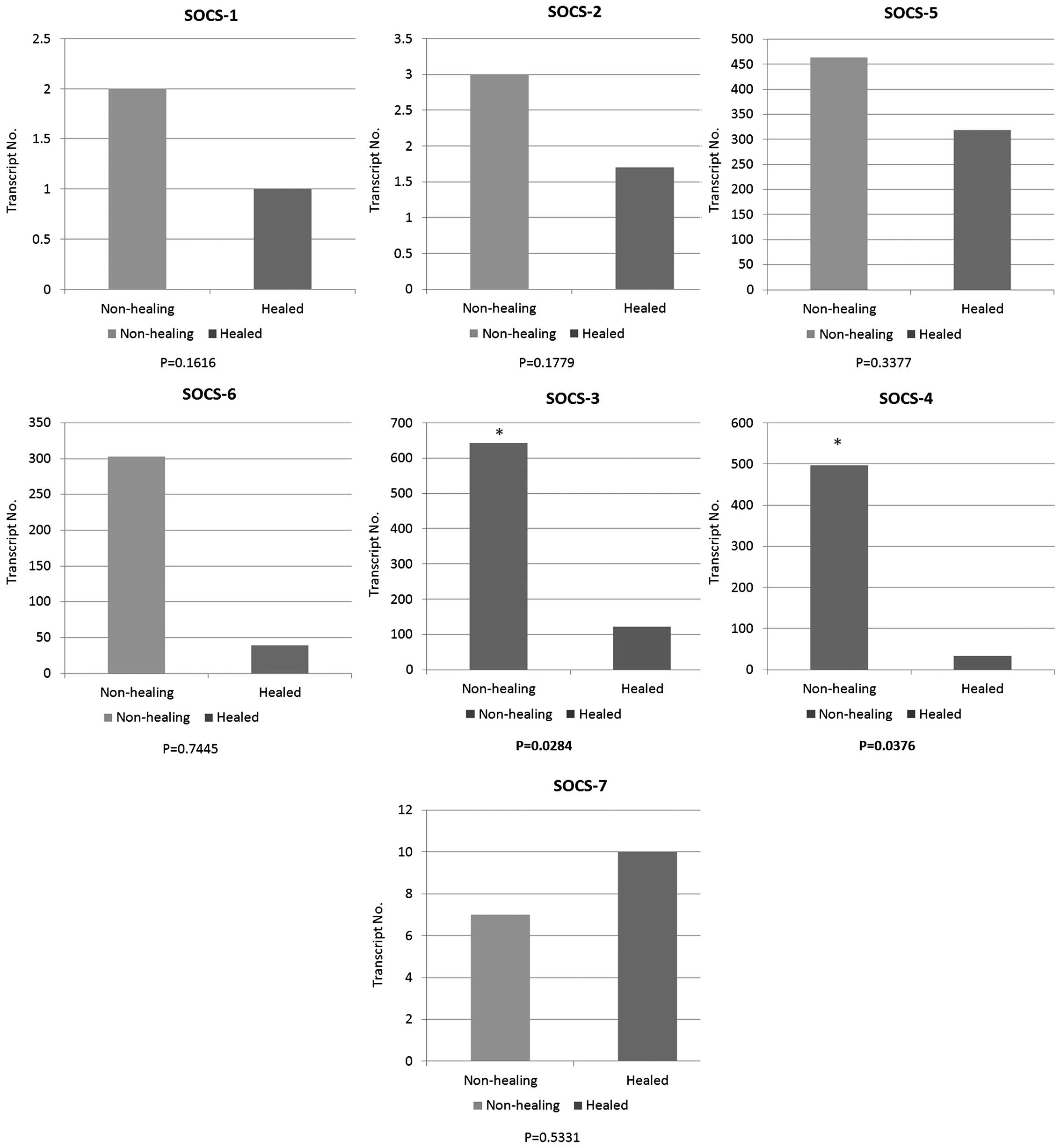
Expression levels of seven SOCS family
members in healing/healed and non-healing chronic wounds
The expression levels of seven SOCS family members
were quantified in a cohort of venous leg ulcer patients with
chronic wounds using qPCR (Fig.
1). The expression of each member of the SOCS family was
obtained between healing/healed and non-healing chronic wounds.
Compared to healing/healed chronic wounds, the non-healing chronic
wound tissues had significantly higher levels of SOCS3 (P=0.0284)
and SOCS4 (P=0.0376) transcripts. Similar to the expression levels
of SOCS3 and 4, the enhanced expression levels of SOCS1, 2, 5 and 6
were observed in the non-healing chronic wounds compared to the
healing/healed chronic wounds. However, the results did not show
any significant differences (P>0.05). In contrast to the trend
of expression levels in SOCS1 to 6, the expression level of SOCS7
was elevated in the healing/healed chronic wounds when compared to
that of the non-healing chronic wounds, albeit the result was not
statistically significant (P>0.05).
Protein expression levels of seven SOCS
family members in chronic tissues SOCS1 staining in chronic
tissues
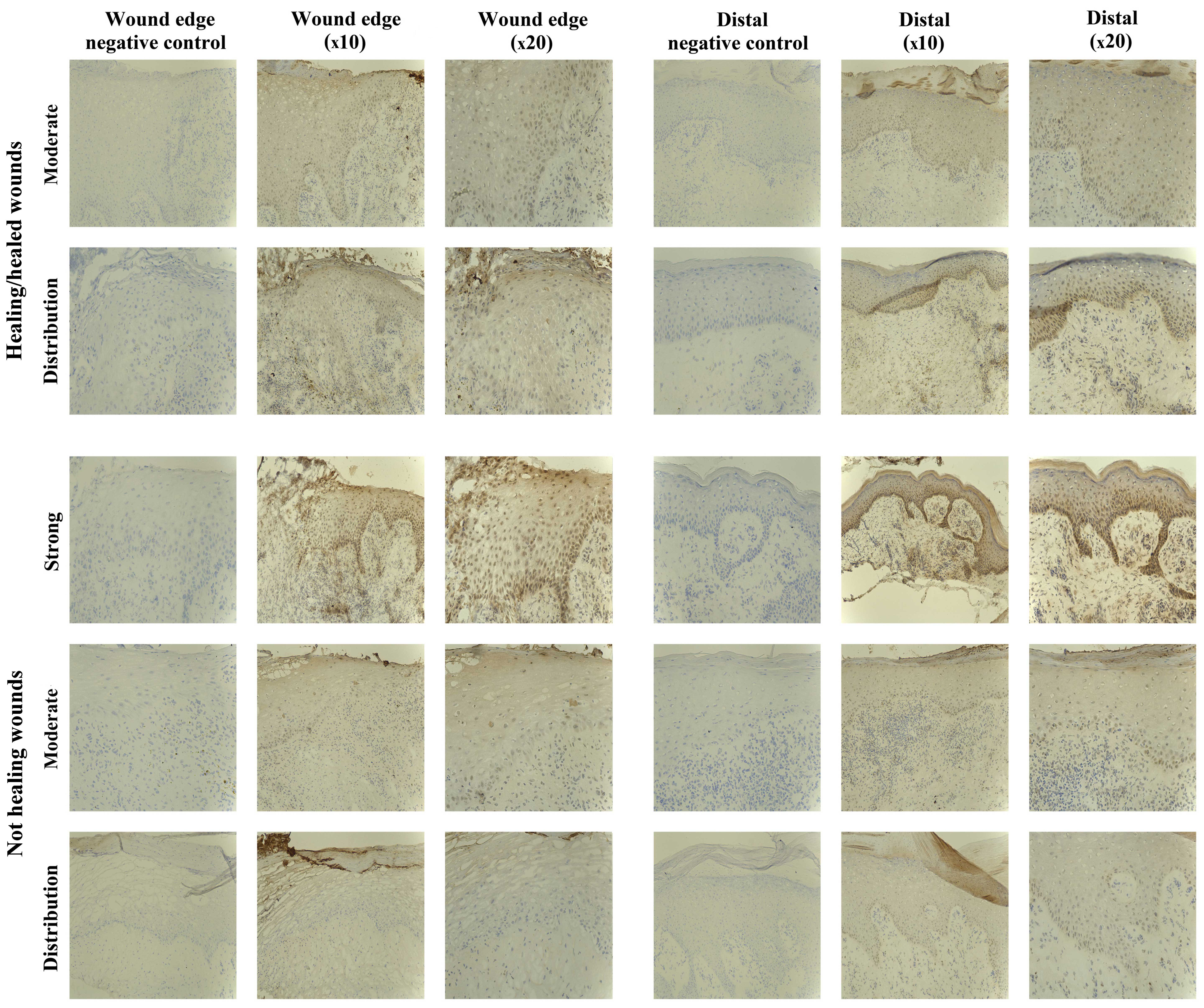
The IHC analysis revealed the majority of chronic
wounds stained positively for SOCS1 protein (15/20 chronic wounds).
Expression of SOCS1 was observed in 7/10 healing/healed chronic
wounds and 8/10 non-healing chronic wounds, and therefore there
were no distinct differences between the two groups. The majority
of the positive biopsies expressed moderate levels of SOCS1, with
the exception of one non-healing chronic wound where the expression
was defined as strong (Fig.
2).
A comparison of SOCS1 expression at the wound edge
to that observed distal to the wound (towards normal tissue)
revealed that 6/20 biopsies had an increased expression of SOCS1
distal to the wound edge, although no difference was evident
between the healing/healed and non-healing groups (3/10
healing/healed and 3/10 non-healed). Generally, in the chronic
wounds, diffuse cytoplasmic staining was observed in the mature
keratinocytes of the epidermis, although increasing nuclear and
cytoplasmic intensity in the basal layer distal to the wound edge
was identified. Most blood vessels were weakly positive for SOCS1
in all the biopsies analysed. Positive staining was also observed
in the dermal inflammatory infiltrate of some biopsies, but dual
fluorescent staining was required to determine whether these were
T-lymphocytes, macrophages, fibroblasts or other cells.
SOCS2 staining in chronic tissues
SOCS2-positive staining was found in 17/20 chronic
wound biopsies studied, 8/10 healing/healed and 9/10 non-healing
biopsies, respectively. The majority of these positively stained
biopsies exhibited SOCS2 expression of moderate intensity, albeit
3/20 revealed stronger intensity (2/10 healing/healed, 1/10
non-healing) (Fig. 3). Three of
the positively stained biopsies showed SOCS2 expression in the
dermal cell infiltrate, while no expression was observed in the
epidermis.
SOCS2 protein expression was uniform throughout the
whole biopsy, with only four chronic wounds showing differences
between the wound edge and distal to the wound (3/10
healing/healed, 1/10 non-healing). Within the healing/healed
subset, two of these biopsies showed an increased expression distal
to the wound edge, whereas the other subset demonstrated an
increased SOCS2 in the dermal infiltrate directly below the leading
migratory epidermis (which was negatively stained).
Expression of SOCS2 protein was generally observed
as diffuse cytoplasmic in mature keratinocytes, or nuclear in the
lower keratinocytes of the basal layer. Membranous staining was
observed in the very mature keratinocytes of some biopsies.
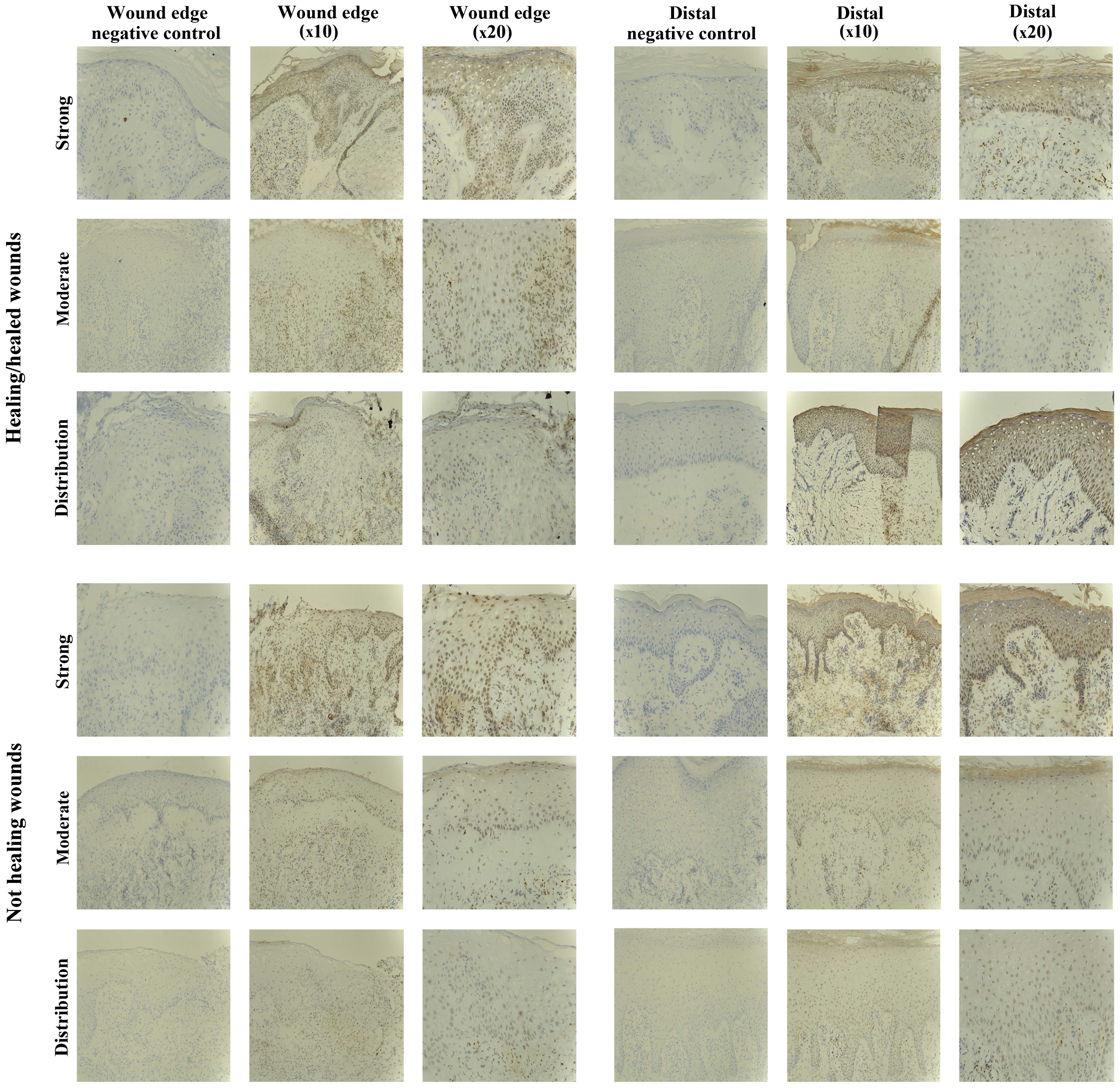
SOCS3 staining in chronic tissues
Positive SOCS3 staining was observed in the
non-healing chronic wounds (10/10), and in the majority of
healing/healed chronic wounds (8/10). Most of the positively
stained biopsies exhibited SOCS3 staining of moderate intensity
(5/10 healing/healed, 8/10 non-healing), but strong staining was
observed in 3/10 healing/healed and 2/10 non-healing wounds
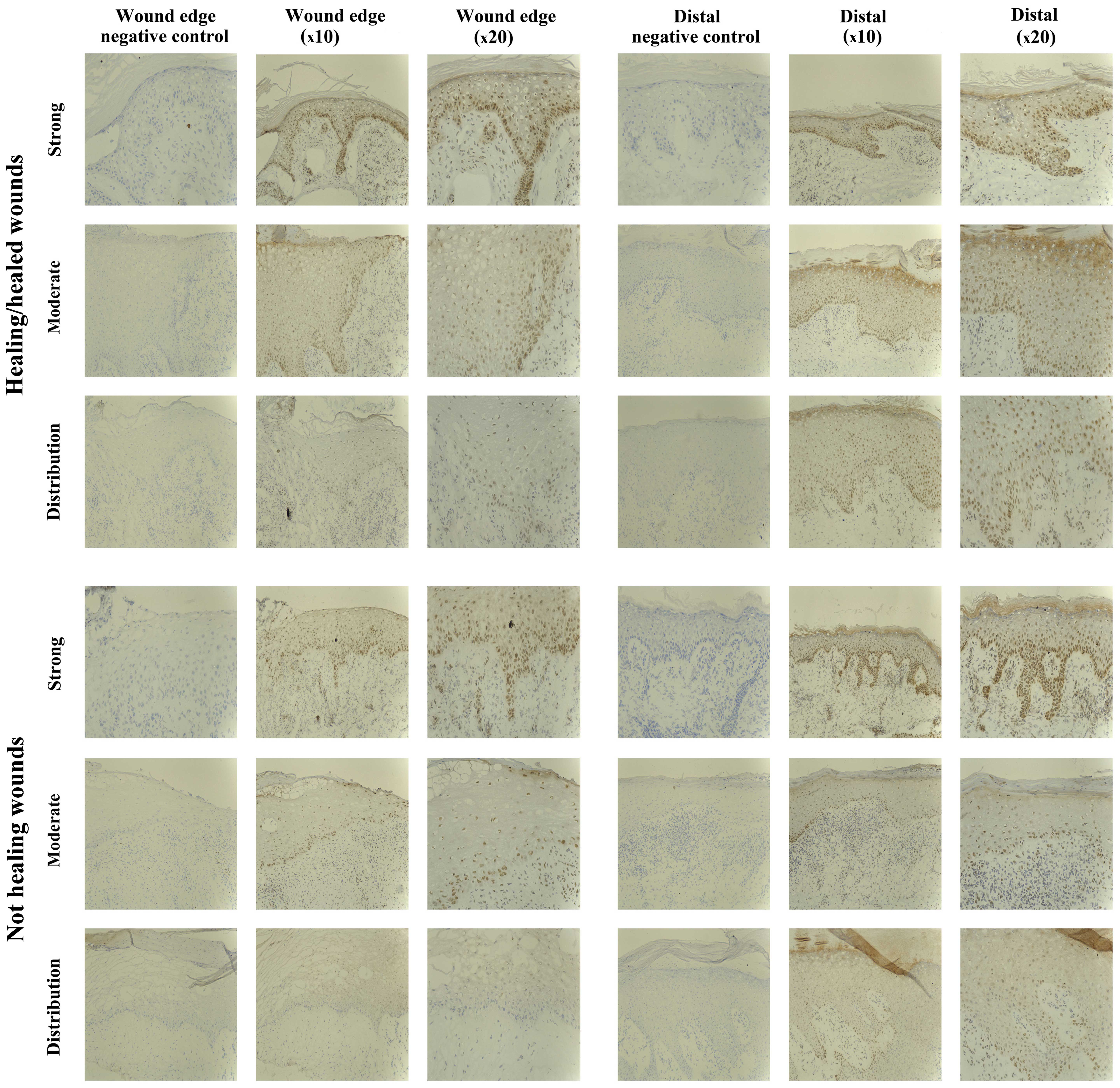
(Fig. 4).
Five of the chronic wounds studied showed increased
expression of SOCS3 protein in the epidermis distal to the wound
edge, and interestingly the majority of these were non-healing
chronic wounds (1/10 healing/healed, 4/10 non-healing), suggesting
a trend of increased expression and relocalisation of SOCS3 in the
non-healing wound environment. The remaining 15/20 chronic wounds
studied had uniform epidermal staining throughout the biopsies.
SOCS3 expression was seen as diffuse cytoplasmic
staining in the mature keratinocytes or nuclear in the lower basal
and suprabasal layers, particularly distal to the wound edge. Many
inflammatory cells within the dermis also expressed SOCS3,
particularly below the leading wound edge (migratory epidermis) and
around the blood vessels.
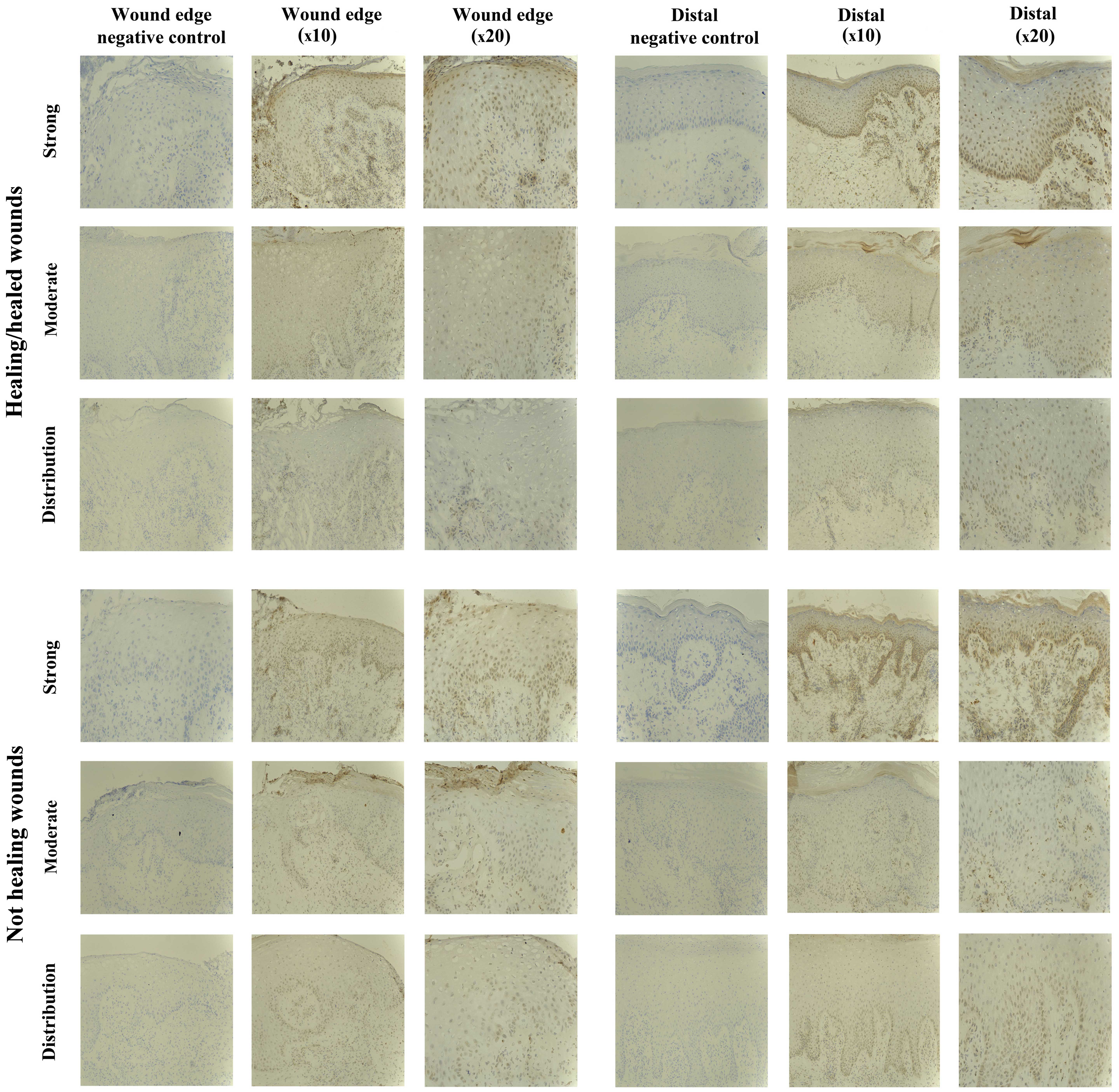
SOCS4 staining in chronic tissues
The SOCS4 protein expression was observed in almost
all the chronic wound biopsies studied (18/20), and no distinct
differences were evident between the two subsets (9/10
healing/healed, 9/10 non-healing). The majority of chronic wounds
had uniform staining of moderate intensity throughout the biopsies
(16/20), and only two samples exhibited increased (strong) staining
for SOCS4 (1/10 healing/healed, 1/10 non-healing) (Fig. 5).
Increased expression of SOCS4 distal to the wound
edge was evident in 5/10 chronic wounds examined, and this was
observed in the healing and non-healing wounds (3/10
healing/healed, 2/10 non-healing).
SOCS4 expression was mostly localised to the nucleus
of the lower basal and suprabasal cells of the epidermis, but
diffuse cytoplasmic staining was also observed in the very mature
upper keratinocyte layers.
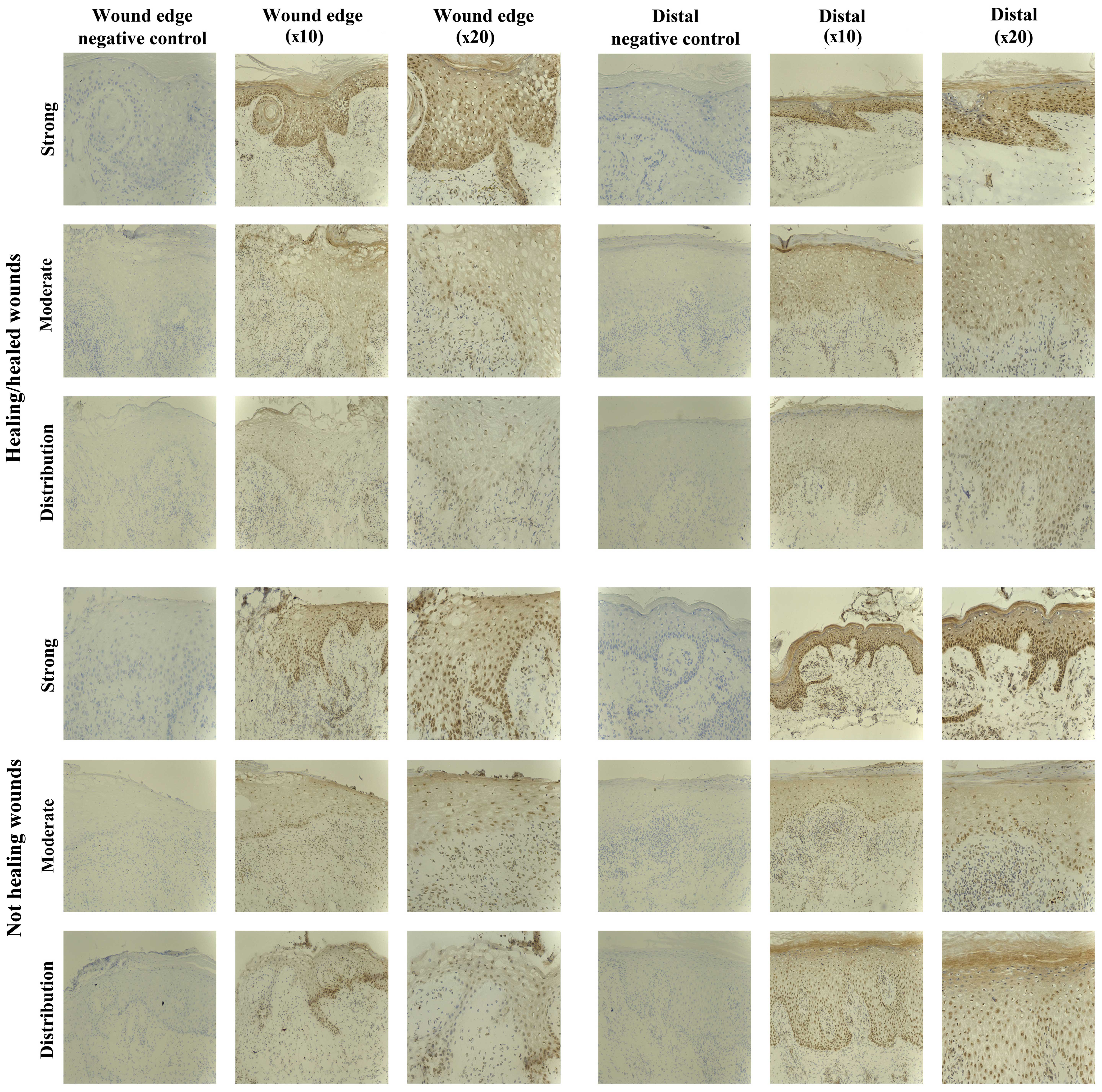
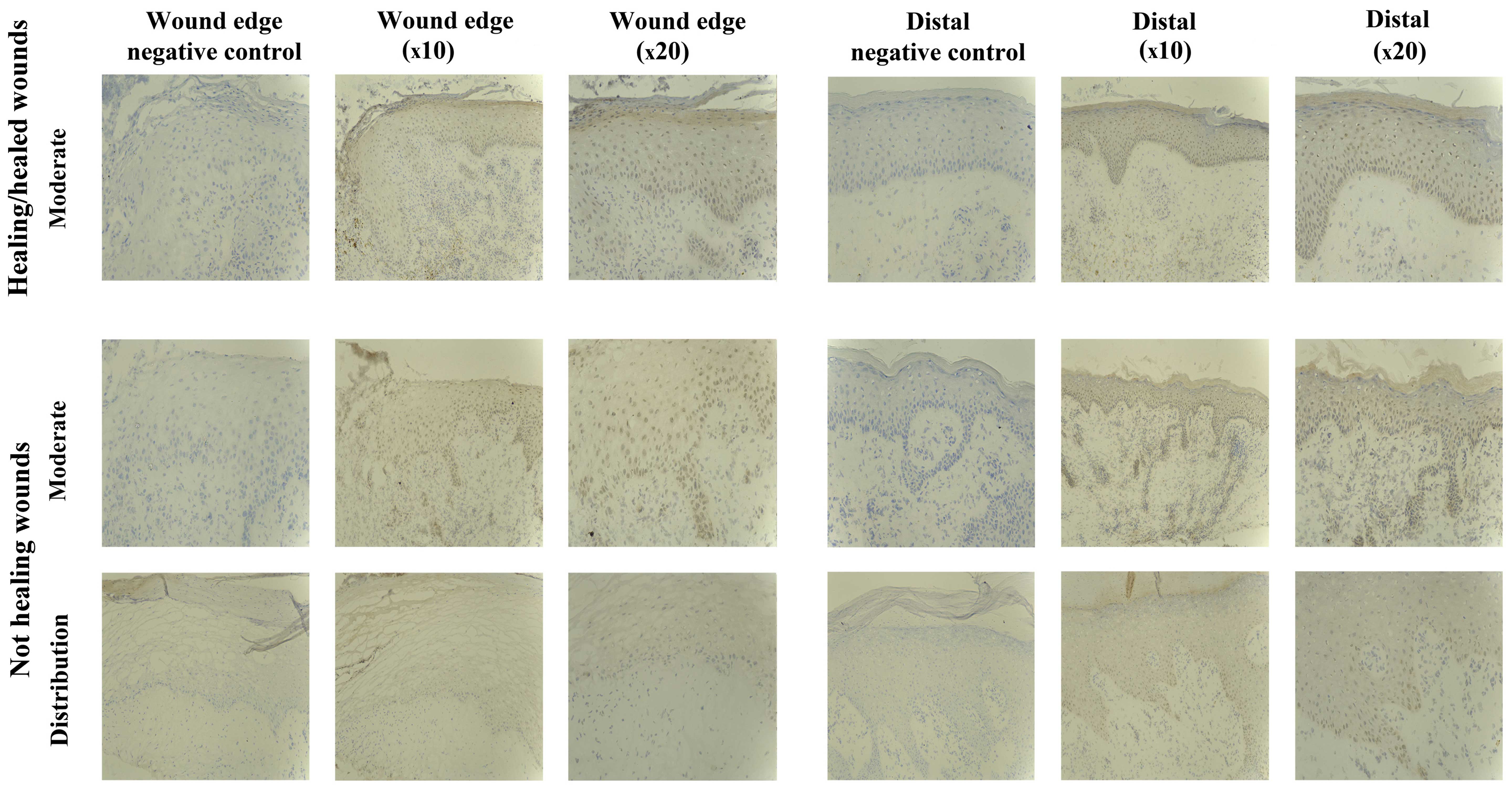
SOCS5 staining in chronic tissues
Staining of SOCS5 protein was generally less
compared to that observed for the other SOCS proteins, but was
obseved in almost all the chronic wounds studied (8/10
healing/healed, 8/10 non-healing) (Fig. 6). Uniform expression was observed
in the two subsets, albeit 2/10 non-healing biopsies showed an
increased SOCS5 expression distal to the wound edge. SOCS5 staining
was mainly cytoplasmic, and observed in all the layers of the
epidemis.
SOCS6 staining in chronic tissues
SOCS6 protein expression was observed in 12/20
chronic wounds, equally in the healing/healed (6/10) and
non-healing chronic wounds (6/10). The IHC analysis revealed the
staining to be of moderate intensity in most biopsies (5/10
healing/healed, 5/10 non-healing), with only two biopsies showing
particularly stronger expression of SOCS6 (1/10 healing/healed,
1/10 non-healing (Fig. 7).
Increased SOCS6 expression was identified in the
epidermal layers distal to the wound edge in 4/20 biopsies examined
(2/10 healing/healed, 2/10 non-healing), although the remaining
biopsies showed uniform staining throughout the tissue samples.
The nucleus of basal and suprabasal keratinocyte
cells were particularly positive for SOCS6. However, cytoplasmic
staining in other layers was also observed.
SOCS7 staining in chronic tissues
SOCS7 expression was examined throughout the
healing/healed and the non-healed chronic wound tissue sections.
The sections demonstrated no significant staining profiles when
tested with either of the two antibodies available within our
laboratories (data not shown). Thus, this molecule appears to
demonstrate weak to no expression in the tissues tested in the
present study.
Discussion
Cytokines play important roles in the wound-healing
process by attracting inflammatory cells, inducing cell
proliferation, differentiation and maturation through various
signalling pathways (2,3). Due to these essential roles, some
cytokine intervention therapies, such as the usage of
platelet-derived growth factor (PDGF) for non-infected foot ulcers,
have been licensed and clinically used on diabetic patients
(1). However, the mixed presence
of cytokines and growth factors at the wound site is concentration-
and time-dependent. Therefore, more focused therapies using
multiple cytokines and growth factors and/or in a time-specific
manner may provide an effective strategy. Such strategies are
beginning to be realised and one therapeutic method, termed
cytokine gene therapy, has shown the ability to topically deliver
strategic cytokines and growth factors at precise time points
during the wound-healing process and demonstrates effectiveness in
improving the healing of the wounds that have specific genetic
defects (20,21). Given the significance of cytokine
signalling in wound healing, dysregulation of such signalling may
enhance the risk of pathologic and abnormal conditions, potentially
leading to delayed healing or non-healing chronic wounds. The
JAK-STAT pathway is utilised by most cytokines for signal
transduction and is subject to regulation by a number of other
molecules including SOCS proteins (5). Therefore, the aim of the present
study was to assess the expression pattern of the seven SOCS family
members within a clinical cohort of healing and non-healing chronic
wounds to examine their potential to act as biomarkers for
predicting healing progression.
Our present data, involving the screening of a
clinical cohort for the transcript expression of seven SOCS
members, show that differences in expression of SOCS family members
were observed between the healing/healed and non-healing chronic
wounds. Notably, significant differences were obtained for the
expression of SOCS3 and 4 between the two groups of chronic wounds.
In addition, gene level screening performed in the present study
suggested that higher, albeit not significant, expression of SOCS1,
2, 5 and 6 in chronic venous leg ulcer patients may indicate poor
healing prognosis, whereas a decreased expression of SOCS7
transcript may suggest higher healing potential. However, the low
levels of SOCS7 protein expression detected in our study may limit
this significance and requires further clarification. Additionally,
screening seven members of the SOCS family in chronic wound biopsy
sections using IHC analysis did not reveal any obviously distinct
differences between the non-healing and healing/healed chronic
wounds and in some ways contrasts our transcript analysis. These
differences may be due to the smaller subset of samples used in the
IHC analysis than in the qPCR analysis. The discrepancy may also
have arisen due to the qualitative nature of the IHC study or may
be some aspect of post-transcriptional regulation.
SOCS family members have been previously linked to
inflammatory disorders and diseases. For example, studies on
SOCS1-deficient mouse models have suggested that SOCS1 is an
important modulator of interferon-γ (IFN-γ) and allows it to exert
protective effects without the risk of hyper-response to viral
infection (22). Some of the
biopsies in the current study had positive SOCS1 staining in the
dermal inflammatory infiltrate. Such results suggested that SOCS1
helps inflammatory cell infiltrate, leading to a prolonged
inflammation phase, a major cause of chronic wounds (2). However, dual fluorescent staining
may be required to identify whether these positively stained cells
were T-lymphocytes, macrophages, fibroblasts or other cells.
SOCS3, acting as an anti-inflammatory regulator,
mediates IL-6-gp130 signalling by preventing the activation of
STAT3 in macrophages (23,24).
Given that chronic inflammatory diseases are promoted by IL-6 and
IL-6-related cytokine-mediated STAT3 pathways, and that the
expression of SOCS3 is able to inhibit STAT3 activation, it appears
that SOCS3 holds the potential to regulate inflammatory diseases
related to IL-6 activation (25).
Additionally, SOCS3 was found to be a negative regulator of
IFN-γ-induced signalling through the suppression of activated STAT1
(26). In the present study,
SOCS3 expression was observed in the cytoplasmic region of mature
keratinocytes or the nuclear region in the lower basal and
suprabasal layers, particularly distal to the wound edge. SOCS3
expression was also found in inflammatory cells in the dermis below
the migratory epidermis and around blood vessels in chronic wound
biopsies. This differential SOCS3 expression pattern over different
areas of the wound biopsies may indicate a differential regulatory
role for SOCS3 in different stages of the wound healing process,
although further investigation is required to identify the specific
molecule(s) or downstream signalling pathway(s) that may be
regulated by SOCS3.
Concerning SOCS5, findings of in vitro
studies on the development of helper T cells suggest that this
protein is involved in impairing IL-4-induced STAT6 activation by
preferentially interacting with the IL-4 receptor, irrespective of
receptor tyrosine phosphorylation. The development of helper T2
(Th2) cells is known to be regulated by cytokine signalling through
the IL-4 receptor. Therefore, SOCS5 may act as a negative regulator
of Th2 development by blocking IL-4 signalling (27). There was only moderate staining
for SOCS5 protein in the healing/healed and non-healing chronic
wound biopsies compared to that of the other SOCS family members
examined in the present study. This result suggests a lower
expression pattern of SOCS5 in chronic wound healing relative to
the other members. Additionally, SOCS5 staining was mainly observed
in the cytoplasmic region in all the layers of the epidemis, which
may indicate its regulatory role in keratinocyte behaviour during
wound healing.
Furthermore, the proliferation and
re-epithelialization phases of wound healing consist of
neoangiogenesis, granulation tissue formation, extracellular matrix
deposition and re-epithelialization (2). Keratinocytes are important in this
stage through the function of migration, proliferation and
differentiation. They are defined as major cell components of the
epidermis, not only for the function of barrier maintenance, but
for its restoration following injury (28). Dysregulation of keratinocyte
migratory function is associated with the clinical phenotype of
chronic non-healing wounds (29).
Direct evidence indicates that SOCS proteins may hold the potential
to influence the wound-healing process by regulating the wound site
resident cell function. Studies have shown that keratinocyte
proliferation and migration are strongly disturbed by the presence
of SOCS3, which contributes to impaired wound healing (30), whereas the exacerbated
inflammation that characterises chronic wounds is shown to be
associated with the overexpression of SOCS3 (31). This evidence may explain the
significantly increased expression of SOCS3 transcript levels in
the non-healing chronic wound biopsies compared to that in the
healing/healed chronic wound biopsies, indicating the upregulation
of SOCS3 in non-healing chronic wounds. The IHC staining results on
the profile of SOCS3 protein expression, and the distinct
proportion (1:4) of the increased SOCS3 protein expression in the
epidermis distal to the wound edge throughout 20 chronic wound
biospies (1/10 healing/healed, 4/10 non-healing) are noteworthy.
They suggest loss of SOCS3 expression on the leading migratory
epidermis in the healing/healed chronic wound and/or the relocation
of SOCS3 protein between healing/healed and non-healing chronic
wounds.
Taken together, SOCS proteins are attractive
molecules that may act as novel biomarkers and/or therapeutics to
manage chronic wounds, although additional study is required to
fully elucidate this role. Thus, establishment of a larger cohort
of these tissues, as well as exploring SOCS function and expression
across acute and chronic wound tissue and normal skin is required.
Additionally, future investigations are required, using in
vitro and in vivo models, to determine the potential
functional traits of SOCS3 and 4 in representative cell types which
play essential roles in the wound-healing process. Such studies may
provide additional detail with regard to the precise nature of the
SOCS family in the wound-healing process and their true potential
to act as novel strategies to combat and manage wound
chronicity.
Acknowledgments
The present study was supported by GlaxoSmithKline
and Cancer Research Wales.
References
|
1
|
Harding KG, Morris HL and Patel GK:
Science, medicine and the future: healing chronic wounds. BMJ.
324:160–163. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Behm B, Babilas P, Landthaler M and
Schreml S: Cytokines, chemokines and growth factors in wound
healing. J Eur Acad Dermatol Venereol. 26:812–820. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barrientos S, Stojadinovic O, Golinko MS,
Brem H and Tomic-Canic M: Growth factors and cytokines in wound
healing. Wound Repair Regen. 16:585–601. 2008. View Article : Google Scholar
|
|
4
|
Igaz P, Tóth S and Falus A: Biological and
clinical significance of the JAK-STAT pathway; lessons from
knockout mice. Inflamm Res. 50:435–441. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rawlings JS, Rosler KM and Harrison DA:
The JAK/STAT signaling pathway. J Cell Sci. 117:1281–1283. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wormald S and Hilton DJ: Inhibitors of
cytokine signal transduction. J Biol Chem. 279:821–824. 2004.
View Article : Google Scholar
|
|
7
|
Starr R, Willson TA, Viney EM, Murray LJ,
Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola
NA, et al: A family of cytokine-inducible inhibitors of signalling.
Nature. 387:917–921. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Endo TA, Masuhara M, Yokouchi M, Suzuki R,
Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H,
et al: A new protein containing an SH2 domain that inhibits JAK
kinases. Nature. 387:921–924. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Naka T, Narazaki M, Hirata M, Matsumoto T,
Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et
al: Structure and function of a new STAT-induced STAT inhibitor.
Nature. 387:924–929. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croker BA, Kiu H and Nicholson SE: SOCS
regulation of the JAK/STAT signalling pathway. Semin Cell Dev Biol.
19:414–422. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alexander WS: Suppressors of cytokine
signalling (SOCS) in the immune system. Nat Rev Immunol. 2:410–416.
2002.PubMed/NCBI
|
|
12
|
Linossi EM, Babon JJ, Hilton DJ and
Nicholson SE: Suppression of cytokine signaling: the SOCS
perspective. Cytokine Growth Factor Rev. 24:241–248. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kario E, Marmor MD, Adamsky K, Citri A,
Amit I, Amariglio N, Rechavi G and Yarden Y: Suppressors of
cytokine signaling 4 and 5 regulate epidermal growth factor
receptor signaling. J Biol Chem. 280:7038–7048. 2005. View Article : Google Scholar
|
|
14
|
Krebs DL, Uren RT, Metcalf D, Rakar S,
Zhang JG, Starr R, De Souza DP, Hanzinikolas K, Eyles J, Connolly
LM, et al: SOCS-6 binds to insulin receptor substrate 4, and mice
lacking the SOCS-6 gene exhibit mild growth retardation. Mol Cell
Biol. 22:4567–4578. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Banks AS, Li J, McKeag L, Hribal ML,
Kashiwada M, Accili D and Rothman PB: Deletion of SOCS7 leads to
enhanced insulin action and enlarged islets of Langerhans. J Clin
Invest. 115:2462–2471. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feng Y, Sanders AJ, Morgan LD, Harding KG
and Jiang WG: Potential roles of suppressor of cytokine signaling
in wound healing. Regen Med. 11:193–209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yasukawa H, Ohishi M, Mori H, Murakami M,
Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, et al:
IL-6 induces an anti-inflammatory response in the absence of SOCS3
in macrophages. Nat Immunol. 4:551–556. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bosanquet DC, Harding KG, Ruge F, Sanders
AJ and Jiang WG: Expression of IL-24 and IL-24 receptors in human
wound tissues and the biological implications of IL-24 on
keratinocytes. Wound Repair Regen. 20:896–903. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Parr C, Watkins G, Mansel RE and Jiang WG:
The hepatocyte growth factor regulatory factors in human breast
cancer. Clin Cancer Res. 10:202–211. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eriksson E: Gene transfer in wound
healing. Adv Skin Wound Care. 13(Suppl 2): 20–22. 2000.PubMed/NCBI
|
|
21
|
Branski LK, Pereira CT, Herndon DN and
Jeschke MG: Gene therapy in wound healing: present status and
future directions. Gene Ther. 14:1–10. 2007. View Article : Google Scholar
|
|
22
|
Alexander WS, Starr R, Fenner JE, Scott
CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R,
Owczarek CM, et al: SOCS1 is a critical inhibitor of interferon
gamma signaling and prevents the potentially fatal neonatal actions
of this cytokine. Cell. 98:597–608. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Croker BA, Krebs DL, Zhang JG, Wormald S,
Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Förster I, Clausen
BE, et al: SOCS3 negatively regulates IL-6 signaling in vivo. Nat
Immunol. 4:540–545. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lang R, Pauleau AL, Parganas E, Takahashi
Y, Mages J, Ihle JN, Rutschman R and Murray PJ: SOCS3 regulates the
plasticity of gp130 signaling. Nat Immunol. 4:546–550. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kubo M, Hanada T and Yoshimura A:
Suppressors of cytokine signaling and immunity. Nat Immunol.
4:1169–1176. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song MM and Shuai K: The suppressor of
cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins
inhibit interferon-mediated antiviral and antiproliferative
activities. J Biol Chem. 273:35056–35062. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seki Y, Hayashi K, Matsumoto A, Seki N,
Tsukada J, Ransom J, Naka T, Kishimoto T, Yoshimura A and Kubo M:
Expression of the suppressor of cytokine signaling-5 (SOCS5)
negatively regulates IL-4-dependent STAT6 activation and Th2
differentiation. Proc Natl Acad Sci USA. 99:13003–13008. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pastar I, Stojadinovic O, Yin NC, Ramirez
H, Nusbaum AG, Sawaya A, Patel SB, Khalid L, Isseroff RR and
Tomic-Canic M: Epithelialization in wound healing: a comprehensive
review. Adv Wound Care (New Rochelle). 3:445–464. 2014. View Article : Google Scholar
|
|
29
|
Raja, Sivamani K, Garcia MS and Isseroff
RR: Wound re-epithelialization: modulating keratinocyte migration
in wound healing. Front Biosci. 12:2849–2868. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Linke A, Goren I, Bösl MR, Pfeilschifter J
and Frank S: The suppressor of cytokine signaling (SOCS)-3
determines keratinocyte proliferative and migratory potential
during skin repair. J Invest Dermatol. 130:876–885. 2010.
View Article : Google Scholar
|
|
31
|
Linke A, Goren I, Bösl MR, Pfeilschifter J
and Frank S: Epithelial overexpression of SOCS-3 in transgenic mice
exacerbates wound inflammation in the presence of elevated
TGF-beta1. J Invest Dermatol. 130:866–875. 2010. View Article : Google Scholar
|