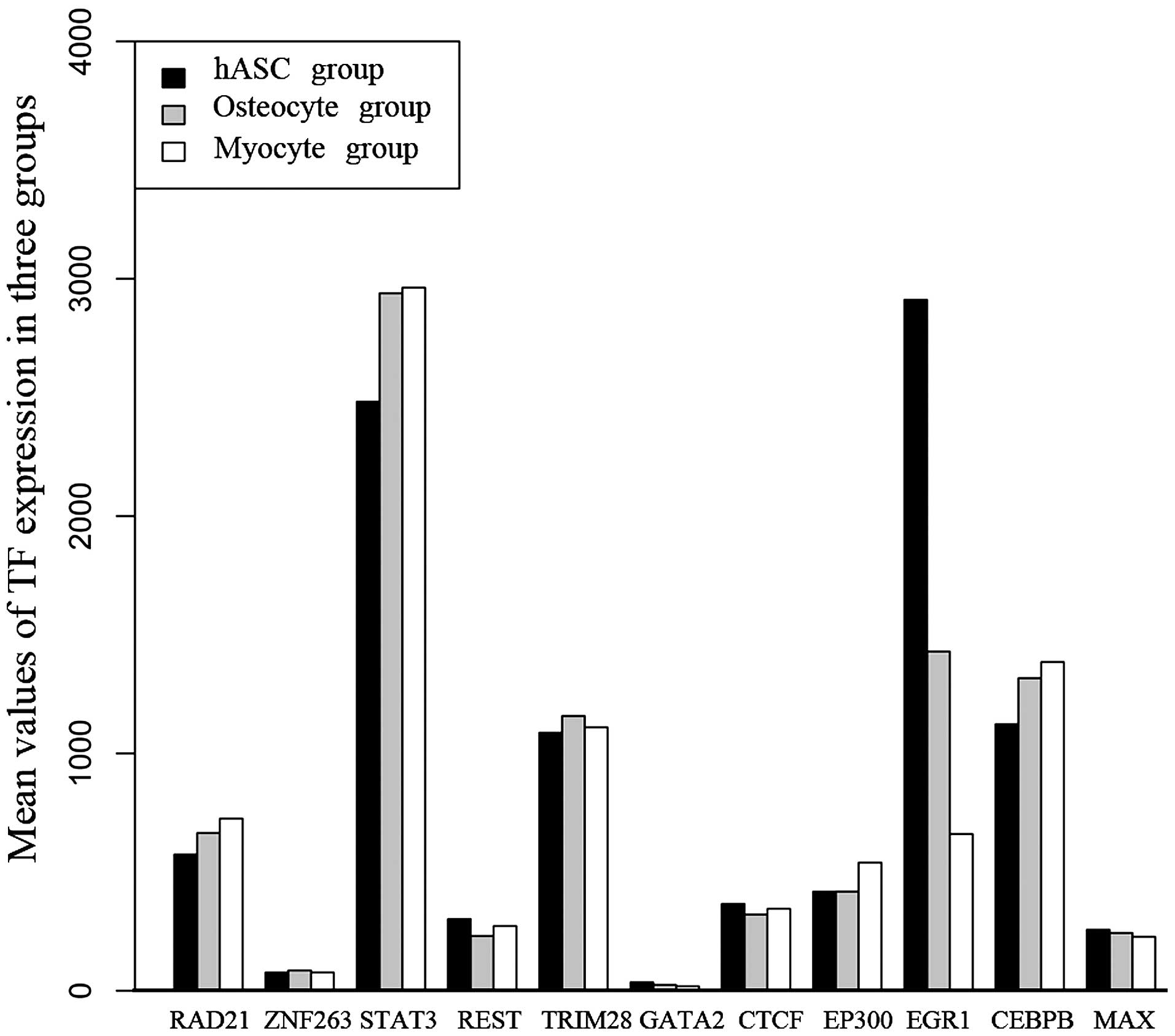
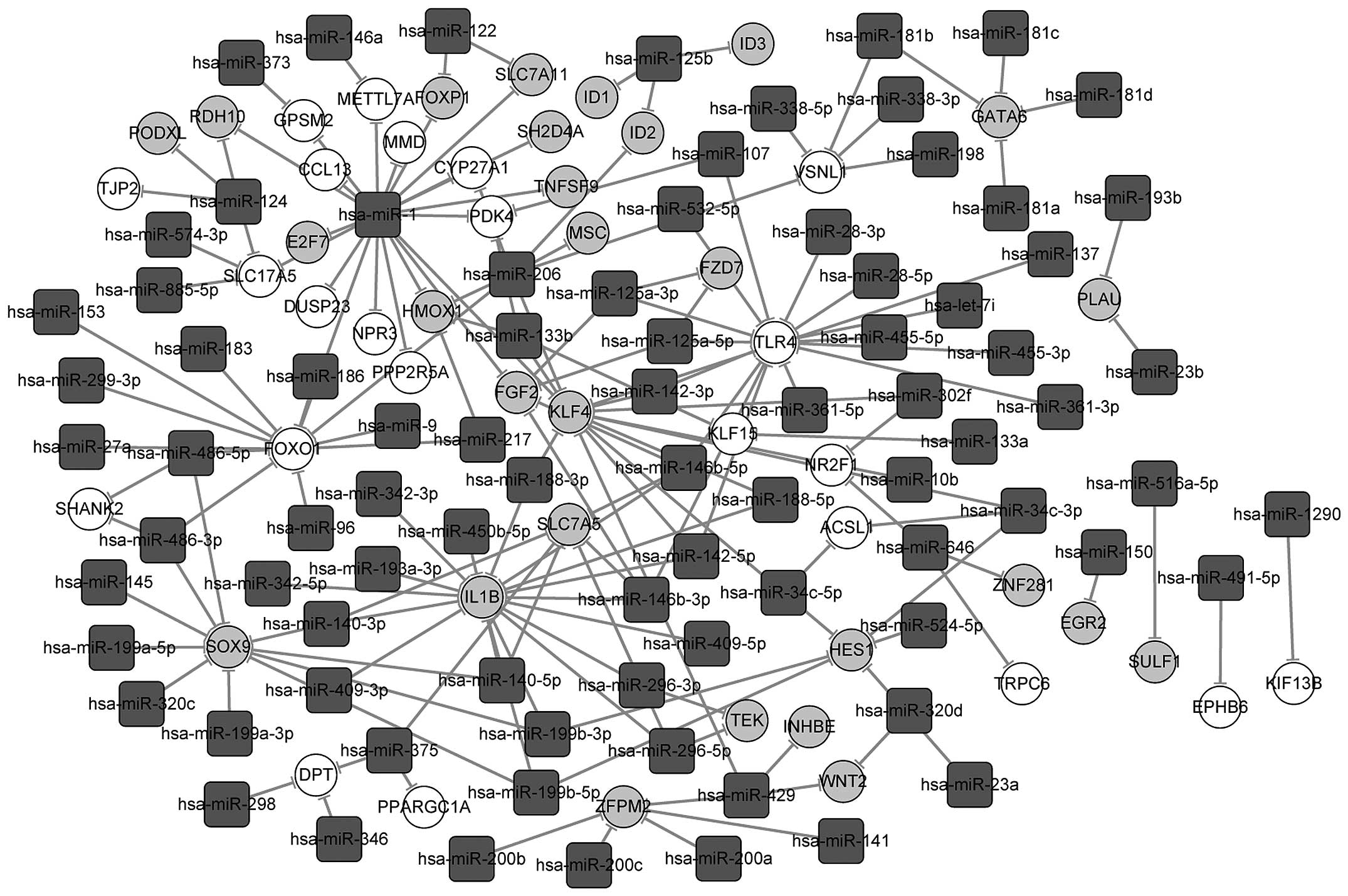
|
1
|
Zuk PA, Zhu M, Mizuno H, Huang J, Futrell
JW, Katz AJ, Benhaim P, Lorenz HP and Hedrick MH: Multilineage
cells from human adipose tissue: implications for cell-based
therapies. Tissue Eng. 7:211–228. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Halvorsen YD, Bond A, Sen A, Franklin DM,
Lea-Currie YR, Sujkowski D, Ellis PN, Wilkison WO and Gimble JM:
Thiazolidinediones and glucocorticoids synergistically induce
differentiation of human adipose tissue stromal cells: biochemical,
cellular, and molecular analysis. Metabolism. 50:407–413. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zuk PA: The adipose-derived stem cell:
looking back and looking ahead. Mol Biol Cell. 21:1783–1787. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luzi E, Marini F, Sala SC, Tognarini I,
Galli G and Brandi ML: Osteogenic differentiation of human adipose
tissue-derived stem cells is modulated by the miR-26a targeting of
the SMAD1 transcription factor. J Bone Miner Res. 23:287–295. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai L, Johnstone BH, Cook TG, Tan J,
Fishbein MC, Chen PS and March KL: IFATS collection: human adipose
tissue-derived stem cells induce angiogenesis and nerve sprouting
following myocardial infarction, in conjunction with potent
preservation of cardiac function. Stem Cells. 27:230–237. 2009.
View Article : Google Scholar
|
|
6
|
Nambu M, Ishihara M, Nakamura S, Mizuno H,
Yanagibayashi S, Kanatani Y, Hattori H, Takase B, Ishizuka T,
Kishimoto S, et al: Enhanced healing of mitomycin C-treated wounds
in rats using inbred adipose tissue-derived stromal cells within an
atelocollagen matrix. Wound Repair Regen. 15:505–510. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilson A and Trumpp A: Bone-marrow
haematopoietic-stem-cell niches. Nat Rev Immunol. 6:93–106. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luu YK, Capilla E, Rosen CJ, Gilsanz V,
Pessin JE, Judex S and Rubin CT: Mechanical stimulation of
mesenchymal stem cell proliferation and differentiation promotes
osteogenesis while preventing dietary-induced obesity. J Bone Miner
Res. 24:50–61. 2009. View Article : Google Scholar :
|
|
9
|
Lodish H, Flygare J and Chou S: From stem
cell to erythroblast: regulation of red cell production at multiple
levels by multiple hormones. IUBMB Life. 62:492–496. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kakudo N, Shimotsuma A and Kusumoto K:
Fibroblast growth factor-2 stimulates adipogenic differentiation of
human adipose-derived stem cells. Biochem Biophys Res Commun.
359:239–244. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stewart AA, Byron CR, Pondenis H and
Stewart MC: Effect of fibroblast growth factor-2 on equine
mesenchymal stem cell monolayer expansion and chondrogenesis. Am J
Vet Res. 68:941–945. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang S, Wang S, Bian C, Yang Z, Zhou H,
Zeng Y, Li H, Han Q and Zhao RC: Upregulation of miR-22 promotes
osteogenic differentiation and inhibits adipogenic differentiation
of human adipose tissue-derived mesenchymal stem cells by
repressing HDAC6 protein expression. Stem Cells Dev. 21:2531–2540.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mizuno H, Zuk PA, Zhu M, Lorenz HP,
Benhaim P and Hedrick MH: Myogenic differentiation by human
processed lipoaspirate cells. Plast Reconstr Surg. 109:199–111.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Planat-Bénard V, Menard C, André M, Puceat
M, Perez A, Garcia-Verdugo JM, Pénicaud L and Casteilla L:
Spontaneous cardiomyocyte differentiation from adipose tissue
stroma cells. Circ Res. 94:223–229. 2004. View Article : Google Scholar
|
|
15
|
Jeon ES, Moon HJ, Lee MJ, Song HY, Kim YM,
Bae YC, Jung JS and Kim JH: Sphingosylphosphorylcholine induces
differentiation of human mesenchymal stem cells into
smooth-muscle-like cells through a TGF-beta-dependent mechanism. J
Cell Sci. 119:4994–5005. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee WC, Rubin JP and Marra KG: Regulation
of alpha-smooth muscle actin protein expression in adipose-derived
stem cells. Cells Tissues Organs. 183:80–86. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim YJ, Bae SW, Yu SS, Bae YC and Jung JS:
miR-196a regulates proliferation and osteogenic differentiation in
mesenchymal stem cells derived from human adipose tissue. J Bone
Miner Res. 24:816–825. 2009. View Article : Google Scholar
|
|
18
|
Maroni P, Brini AT, Arrigoni E, de
Girolamo L, Niada S, Matteucci E, Bendinelli P and Desiderio MA:
Chemical and genetic blockade of HDACs enhances osteogenic
differentiation of human adipose tissue-derived stem cells by
oppositely affecting osteogenic and adipogenic transcription
factors. Biochem Biophys Res Commun. 428:271–277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berdasco M, Melguizo C, Prados J, Gómez A,
Alaminos M, Pujana MA, Lopez M, Setien F, Ortiz R, Zafra I, et al:
DNA methylation plasticity of human adipose-derived stem cells in
lineage commitment. Am J Pathol. 181:2079–2093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smyth GK: Limma: linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. Springer; New York: pp. 397–420. 2005,
View Article : Google Scholar
|
|
22
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium Nat Genet. 25:25–29. 2000.
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
24
|
Wang J, Zhou X, Zhu J, Gu Y, Zhao W, Zou J
and Guo Z: GO-function: deriving biologically relevant functions
from statistically significant functions. Brief Bioinform.
13:216–227. 2012. View Article : Google Scholar
|
|
25
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar :
|
|
26
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: software for visualization and analysis of biological
networks. Methods Mol Biol. 696:296–303. 2011.
|
|
27
|
Meyer LR, Zweig AS, Hinrichs AS, Karolchik
D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al:
The UCSC Genome Browser database: extensions and updates 2013.
Nucleic Acids Res. 41:D64–D69. 2013. View Article : Google Scholar :
|
|
28
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: an integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009. View Article : Google Scholar
|
|
29
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: prediction of possible miRNA binding sites by
'walking' the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rehman J, Traktuev D, Li J, Merfeld-Clauss
S, Temm-Grove CJ, Bovenkerk JE, Pell CL, Johnstone BH, Considine RV
and March KL: Secretion of angiogenic and antiapoptotic factors by
human adipose stromal cells. Circulation. 109:1292–1298. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Olsson AK, Dimberg A, Kreuger J and
Claesson-Welsh L: VEGF receptor signalling - in control of vascular
function. Nat Rev Mol Cell Biol. 7:359–371. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Street J, Bao M, deGuzman L, Bunting S,
Peale FV Jr, Ferrara N, Steinmetz H, Hoeffel J, Cleland JL,
Daugherty A, et al: Vascular endothelial growth factor stimulates
bone repair by promoting angiogenesis and bone turnover. Proc Natl
Acad Sci USA. 99:9656–9661. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Clarkin CE, Emery RJ, Pitsillides AA and
Wheeler-Jones CP: Evaluation of VEGF-mediated signaling in primary
human cells reveals a paracrine action for VEGF in
osteoblast-mediated crosstalk to endothelial cells. J Cell Physiol.
214:537–544. 2008. View Article : Google Scholar
|
|
34
|
Street J and Lenehan B: Vascular
endothelial growth factor regulates osteoblast survival - evidence
for an autocrine feedback mechanism. J Orthop Surg. 4:192009.
View Article : Google Scholar
|
|
35
|
Song YH, Gehmert S, Sadat S, Pinkernell K,
Bai X, Matthias N and Alt E: VEGF is critical for spontaneous
differentiation of stem cells into cardiomyocytes. Biochem Biophys
Res Commun. 354:999–1003. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu C, Rosler E, Jiang J, Lebkowski JS,
Gold JD, O'Sullivan C, Delavan-Boorsma K, Mok M, Bronstein A and
Carpenter MK: Basic fibroblast growth factor supports
undifferentiated human embryonic stem cell growth without
conditioned medium. Stem Cells. 23:315–323. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dvorak P, Dvorakova D, Koskova S, Vodinska
M, Najvirtova M, Krekac D and Hampl A: Expression and potential
role of fibroblast growth factor 2 and its receptors in human
embryonic stem cells. Stem Cells. 23:1200–1211. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Martin I, Muraglia A, Campanile G,
Cancedda R and Quarto R: Fibroblast growth factor-2 supports ex
vivo expansion and maintenance of osteogenic precursors from human
bone marrow. Endocrinology. 138:4456–4462. 1997.PubMed/NCBI
|
|
39
|
Solchaga LA, Penick K, Porter JD, Goldberg
VM, Caplan AI and Welter JF: FGF-2 enhances the mitotic and
chondrogenic potentials of human adult bone marrow-derived
mesenchymal stem cells. J Cell Physiol. 203:398–409. 2005.
View Article : Google Scholar
|
|
40
|
Quarto N and Longaker MT: FGF-2 inhibits
osteogenesis in mouse adipose tissue-derived stromal cells and
sustains their proliferative and osteogenic potential state. Tissue
Eng. 12:1405–1418. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chiou M, Xu Y and Longaker MT: Mitogenic
and chondrogenic effects of fibroblast growth factor-2 in
adipose-derived mesenchymal cells. Biochem Biophys Res Commun.
343:644–652. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo J, Lin GS, Bao CY, Hu ZM and Hu MY:
Anti-inflammation role for mesenchymal stem cells transplantation
in myocardial infarction. Inflammation. 30:97–104. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sariola H: The neurotrophic factors in
non-neuronal tissues. Cell Mol Life Sci. 58:1061–1066. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nitzsche A, Paszkowski-Rogacz M, Mata rese
F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L,
Huebner N, Mann M, et al: RAD21 cooperates with pluripotency
transcription factors in the maintenance of embryonic stem cell
identity. PLoS One. 6:e194702011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang M, Chen B, Lin T, Li Z, Pardo C,
Pampo C, Chen J, Lien CL, Wu L, Ai L, et al: Restraint of
angiogenesis by zinc finger transcription factor CTCF-dependent
chromatin insulation. Proc Natl Acad Sci USA. 108:15231–15236.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Roelen BA and Dijke P: Controlling
mesenchymal stem cell differentiation by TGFBeta family members. J
Orthop Sci. 8:740–748. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen JF, Mandel EM, Thomson JM, Wu Q,
Callis TE, Hammond SM, Conlon FL and Wang DZ: The role of
microRNA-1 and microRNA-133 in skeletal muscle proliferation and
differentiation. Nat Genet. 38:228–233. 2006. View Article : Google Scholar
|
|
48
|
Xie C, Huang H, Sun X, Guo Y, Hamblin M,
Ritchie RP, Garcia-Barrio MT, Zhang J and Chen YE: MicroRNA-1
regulates smooth muscle cell differentiation by repressing
Kruppel-like factor 4. Stem Cells Dev. 20:205–210. 2011. View Article : Google Scholar :
|
|
49
|
Datta J, Kutay H, Nasser MW, Nuovo GJ,
Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et
al: Methylation mediated silencing of MicroRNA-1 gene and its role
in hepatocellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kubo H, Shimizu M, Taya Y, Kawamoto T,
Michida M, Kaneko E, Igarashi A, Nishimura M, Segoshi K, Shimazu Y,
et al: Identification of mesenchymal stem cell (MSC)-transcription
factors by microarray and knockdown analyses, and signature
molecule-marked MSC in bone marrow by immunohistochemistry. Genes
Cells. 14:407–424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Crop MJ, Baan CC, Korevaar SS, Ijzermans
JN, Pescatori M, Stubbs AP, van Ijcken WF, Dahlke MH, Eggenhofer E,
Weimar W and Hoogduijn MJ: Inflammatory conditions affect gene
expression and function of human adipose tissue-derived mesenchymal
stem cells. Clin Exp Immunol. 162:474–486. 2010. View Article : Google Scholar : PubMed/NCBI
|