Introduction
According to the US Food and Drug Administration,
nonunion is established when a minimum of 9 months have elapsed
since injury, and no signs of progression of fracture healing are
demonstrable for 3 consecutive months. Non-union is also defined as
the failure of a broken bone to heal at 6 months following
fracture. The diagnosis of non-union is made upon the radiographic
finding of a persistent radiolucent gap at the fracture site.
Approximately 5–10% of all fractures are complicated by delayed
union or non-union (1). Despite
the substantial progress made in the understanding of the
mechanisms involved in fracture healing (2), non-union continues to be a challenge
for orthopedic surgeons. In particular, atrophic non-union can be
recalcitrant, resulting not only in chronic pain, but also in
severe functional impairment (3).
The cause of atrophic non-union appears to be
multifactorial and is the subject of intensive ongoing research.
The factors associated with the development of non-union, include
local factors, such as excessive mobility at the fracture site,
fracture displacement, an impaired blood supply, open fractures,
soft tissue interposition and infection, we well as systemic
factors, such as osteoporosis, malnourishment, genetic
predisposition, the administration of pharmacological agents (e.g.,
non-steroidal anti-inflammatory drugs), smoking and alcohol
consumption (4).
Fracture healing is a dynamic physiological process
involving a complex interplay of cells, mediators and growth
factors. The healing process involves cellular recruitment,
proliferation and differentiation under the guidance of signaling
molecules, and the deposition of extracellular matrix components,
that serve as a foundation for a successful bone healing response.
The underling cellular and molecular mechanisms of this process are
complex and are not yet completely understood. The disruption of
any of the involved processes may lead to an impaired healing
response, manifesting as delayed healing or non-union. The
unraveling of the molecular mechanisms responsible for non-union
fractures will pave the way for the development of novel
therapeutic modalities.
MicroRNAs (miRNAs or miRs) are small non-coding RNA
molecules that consists are 21–25 nucleotides in length and are
typically excised from 60–110 nucleotide foldback RNA precursors.
These molecules negatively regulate gene expression at the
post-transcriptional level in a sequence-specific manner either by
mRNA decay (predominant mechanism in plants) or by translational
repression (predominant mechanism throughout the animal kingdom
(5). Diverse expression patterns
of miRNAs and altered miRNA expression under certain physiological
conditions appear to indicate a range of hitherto unknown
functions. Over the past two decades, research has indicated that
miRNAs play a critical role in various physiological processes,
such as development, cell proliferation, differentiation and
apoptosis, as well as in many pathophysiological processes, such as
inflammation and oncogenesis (6).
Approximately 2,000 human miRNAs have been identified which may
target approximately 60% of total human mRNAs; however, only a
fraction of the identified miRNAs have been studied thus far
(7–9).
Previous studies have documented that numerous
miRNAs are associated with orthopedic disorders. Many miRNA, such
as the miR-34 family, miR-143, miR-145 and miR-200b/c are
associated with osteosarcoma, whereas the ones associated with
osteosarcoma progression and metastases include miR-20a, miR-181c,
miR-27a, miR-183, miR-93, miRNA-33b and miRNA-26a (10–17). Some miRNAs have been identified to
have prognostic significance. For example, miR-451 is an
unfavorable prognostic factor for both overall and disease-free
survival in osteosarcoma (18).
Low serum levels of miRNA-133b and miRNA-206 are predictors of a
poor prognosis, whereas the combined elevation of miRNA-196a and
miRNA-196b is predictor of unfavorable prognosis in patients with
osteosarcoma (19,20). Apart from osteosarcoma, miRNAs
have also been associated with other orthopedic disorders.
miRNA-210 has been shown to be involved in the regulation of
post-menopausal osteoporosis (21). The expression of mature miR-17-5p
has been shown to be significantly lower in non-traumatic
osteonecrosis samples (22).
Studies have also indicated that several miRNAs are differentially
expressed in patients with intervertebral disc degeneration
(23), and are thought to
participate in the pathological process. For instance, miRNA-10b
has been shown to promote nucleus pulposus cell proliferation by
targeting HOXD10 in intervertebral disc degeneration (24). In addition, the knockdown of
miR-140 has been shown to be associated with osteoarthritis-like
changes, while miR-140 transgenic mice seem to develop resistance
to joint damage due to osteoarthritis (25). Furthermore, miR-124 and miR-146a
have been shown to be associated with rheumatoid arthritis; the
administration of miR-146a has been shown to protect the joints
from inflammatory damage in arthritic mice (26). Although the association between
miRNAs and various other orthopedic disorders has also been
investigated, the relevance of miRNAs in the pathogenesis of
atrophic non-union remains to be elucidated.
In this study, specimens from patients with atrophic
nonunion or bone healing following fracture were obtained for
microarray analysis to characterize the differential miRNA
expression profiles. The differentially expressed miRNAs were
further verified by quantitative-polymerase chain reaction (qPCR)
and these miRNA expression patterns during osteogenesis were also
explored. miR-628-3p expression was high at the fracture site in
patients with atrophic non-union, while it was decreased during
osteoblast differentiation. Gain- of-function analysis revealed
that miR-628-3p inhibited the differentiation of MG63 osteoblasts.
In addition, in silico analysis and molecular analyses
revealed that runt-related transcription factor 2 (RUNX2), the
master gene of osteoblast differentiation, was the target gene of
miR-628-3p, which strongly suggested the role of miR-628-3p/RUNX2
in atrophic non-union.
Materials and methods
Screening for differentially expressed
miRNAs in patients with atrophic non-union
After obtaining approval from the Medical Ethics
Committee of the General Hospital of People's Liberation Army (no.
301 Hospital of People's Liberation Army), 3 samples from patients
with atrophic non-union and 3 samples from patients with normal
fracture healing were collected and were stored at −80°C until
further testing. The samples were taken from the scar tissues of
atrophic non-union and from the bony callus formed around the steel
plate from fracture patients after healing. Written informed
consent was obtained from each of the patients enrolled in this
study.
RNA was extracted using TRIzol reagent (Life
Technologies, Carlsbad, CA, USA) as per the manufacturers'
instructions. The samples were grinded to a fine powder under
liquid nitrogen. The RNA quality was assessed using a Bioanalyzer
2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). miRNA
expression profiling was carried out by KangChen Bio-tech
(Shanghai, China) utilizing the Sanger miRBase 11.0 which contains
1,700 probes.
Cell culture, induction of
differentiation and transfection
Human osteoblastic MG63 cells (American Type Culture
Collection, Rockville, MD, USA) were routinely maintained in
Dulbecco's modified Eagle's medium (DMEM) high glucose (Life
Technologies) supplemented with 10% heat-inactivated fetal bovine
serum (FBS; HyClone, Logan, UT, USA) at 37°C in a humidified
atmosphere of 5% CO2. The culture medium was changed
every other day. For differentiation, the culture medium was
replaced by differentiation medium containing bisphosphonates
(alendronate 50 µM, pamidronate 50 µM and zoledronate
50 mΜ) for 7 days, as described elsewhere (27). Cells cultured in a normal medium
were used as a control. Cell transfection was performed using
Lipofectamine 2000 (Life Technologies) as per the manufacturer's
instructions. The cells were seeded in 6-well plates and cultured
to 70–80% confluence in antibiotic-free fresh medium. The cells
were washed twice with DMEM prior to transfection. Subsequently, a
mixture of Lipofectamine 2000 (Life Technologies) and miR-628-3p
miRNA mimics or miR-654-5p mimics or miRNA mimic negative controls
(Genepharma, Shanghai, China) were added followed by incubation for
6 h. miRNA mimics negative control were used as a negative control
(NC).
Detection of alkaline phosphatase (ALP)
activity
ALP activity was measured using the alkaline
phosphatase assay kit (Beyotime, Hangzhou, China) as per the
manufacturer's instructions. Total protein was extracted using
radioimmuno-precipitation assay (RIPA) lysis buffer. Following
centrifugation of the cell lysate at 14,000 × g for 5 min at 4°C,
the total protein concentration in the lysate was determined by
bicinchoninic acid assay. Equal volumes of substrate and cell
lysate were added to each well of the 96-well plate and incubated
for 10 min. Following the addition of stop solution, ALP activity
was evaluated by measuring the absorbance of the mixture with a
test wavelength of 405 nm using an automicroplate reader (Infinite
M200; Tecan, Grödig, Austria).
Alizarin Red S staining
The cells in the 6-well plate were washed with
phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde.
After washing with PBS 3 times, Alizarin Red staining solution
(Sigma-Aldrich, St. Louis, MO, USA) was added followed by
incubation for 30 min. The cells was washed for another 3 times
with PBS, and images were recorded using a Nikon CoolPix 950
digital camera (Nikon Instruments Inc., Shanghai, China).
3′ Untranslated region (3′UTR) luciferase
reporter assay
The 3′UTR luciferase reporter constructs of RUNX2
were made by cloning the 3′UTR region of the RUNX2 mRNA into the
pGL3-promoter vector (Promega, Madison, WI, USA). The primers for
RUNX2 3′UTR-1 were 5′-acgtctagaagcatgaaatgctggagtga-3′ (sense) and
5′-gatcatatgctgggtggcctacaaaggt-3′ (antisense); for RUNX2 3′UTR-2
were 5′-acgtctagatggtccttctcaaacccacctt-3′ (sense) and
5′-gatcatatgttcgttttctaaaaacaaca-3′ (antisense); for RUNX2
3′UTR-1-mut were
5′-caaggggctgtggagtttggtgtcctagcttgtgtatgaatttgagctaga-3′ (sense)
and 5′-tctagctcaaattcatacacaagctaggacaccaaactccacagcccc-3′
(antisense); and for RUNX2 3′UTR-2-mut were
5′-gtcttactactactgtggaaccatgctagcattcctgggaattaaaat-3′ and
5′-accacgctattttaattcccaggaatgctagcatggttccacagtagtagtaagac-3′. All
the constructs were confirmed by sequencing analysis. 293 cells
(ATCC, Manassas, VA, USA) were co-transfected with the luciferase
reporter plasmid and the miR-628-3p mimics. After 24 h, the
luciferase activities were measured using the Dual-Luciferase
reporter assay system (Promega) according to the manufacturer's
instructions.
RNA quantification
Total RNA was extracted from the tissues or cultured
cells using TRIzol reagent (Life Technologies) as per the
manufacturer's instructions. A total of 1.5 µg of RNA was
used for reverse transcription. qPCR was performed using an ABI
7500 real-time PCR system and was executed using the SYBR-Green PCR
master mix (both from Applied Biosystems, Foster City, CA, USA)
with 2 µl cDNA template in a 40 µl final reaction
mixture (95°C for 15 min; 95°C for 15 sec, 56°C for 30 sec, 68°C
for 30 sec, 40 cycles). The average threshold cycle (Ct) for each
gene was determined from triplicate reactions; the relative
expression level of mRNA or miRNAs was normalized to that of the
internal controls, glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
or U6, using the 2−ΔΔCt method. The primers for ALP were
5′-ctaactccttagtgccagag-3′ (sense) and 5′-catgatgacattcttagccac-3′
(antisense); for collagen, type I, α1 (COL1A1) were
5′-gtgctaaaggtgccaatggt-3′ (sense) and 5′-ctcctcgctttccttcctct-3′
(antisense); for osteocalcin (OC) were 5′-ctcacactcctcgccctattg-3′
(sense) and 5′-cttggacacaaaggctgcac-3′ (antisense); for GAPDH were
5′-tcagtggtggacctgacctg-3′ (sense) and 5′-tgctgtagccaaattcgttg-3′
(antisense); for miR-149* were
5′-agggagggacgggggctgtgc-3′ (sense) and
5′-gcgagcacagaattaatacgac-3′ (antisense); for miR-221 were
5′-agctacattgtctgctgggtttc-3′ (sense) and
5′-gcgagcacagaattaatacgac-3′ (antisense); for miR-628-3p were
5′-tctagtaagagtggcagtcga-3′ (sense) and
5′-gcgagcacagaattaatacgac-3′ (antisense); for miR-654-5p were
5′-tggtgggccgcagaacatgtgc-3′ (sense) and
5′-gcgagcacagaattaatacgac-3′ (antisense); and for U6 were
5′-cgcttcggcagcacatatacta-3′ (sense) and
5′-cgcttcacgaatttgcgtgtca-3′ (antisense).
Western blot analysis
The cells were harvested and lysed with RIPA lysis
buffer. Equal amounts of protein were loaded and separated via 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE). Separated proteins were transferred onto 0.4-µm
polyvinylidene difluoride (PVDF) membranes. After blocking with 5%
skim milk for 3 h, the PVDF membranes were incubated overnight with
primary antibody against RUNX (H00000860-M06) or GAPDH
(H00002597-M01) (1:1,000; Abnova, Taipei, China) diluted in
blocking solution. Subsequently, the membranes were washed 3 times
with Tris-buffered saline (TBS) with Tween-20 (TBST) [50 mM
Tris-HCl (pH 7.5), 150 mM NaCl, and 0.05% Tween-20] and horseradish
peroxidase-conjugated secondary antibodies (ZB-2305; ZSGB-BIO,
Beijing, China) were incubated with the membranes for 1 h at room
temperature. The membranes were washed twice with TBST and once
with TBS, and soaked in enhanced chemiluminescence (ECL) reagent.
Protein bands were visualized using the Western Blotting Systems
ECL kit (Amersham, Piscataway, NJ, USA). Data obtained from western
blot analysis were analyzed using Bio-Rad Quantity One 1-D Analysis
software (Bio-Rad, Hercules, CA, USA).
Screening for target genes
we used the online software program, microRNA.org, to identify the candidate target genes
of miR-628-3p. This was carried out by typing the name of microRNAs
in the search bar.
Statistical analysis
All data are expressed as the means ± standard
deviation (SD). Statistical analyses were performed with SPSS
(version 15.0; SPSS Inc., Chicago, IL, USA). The normality of
distribution of all variables was verified before conducting the
data analyses. Upon performing the Kolmogorov-Smirnov test, no
significant differences were detected in the variance between the
groups (F test). A one-way ANOVA was performed to detect
statistically significant differences between more than 2 groups.
Otherwise, a two-tailed Student's t-test was used to evaluate the
differences between 2 groups. A P-value of <0.05 was considered
to indicate a statistically significant difference.
Results
Screening for differentially expressed
miRNAs in patients with atrophic non-union
Scar tissue from the 3 patients with atrophic
non-union, and tissues from the bony callus from 3 patients with
healed fractures were collected. The differentially expressed
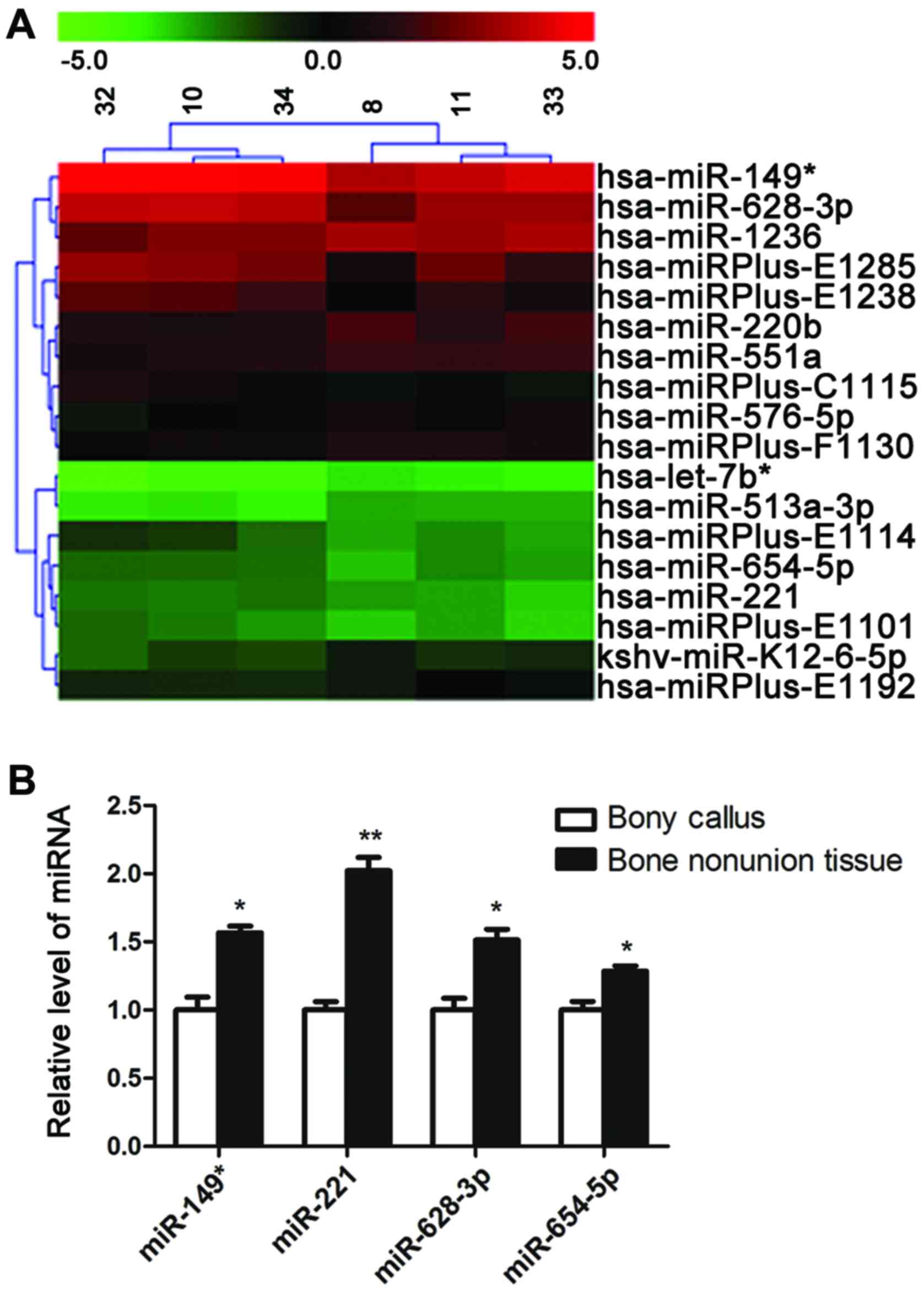
miRNAs were screened with miRNA microarray. A total of 4 miRNAs
(miR-149*, miR-221, miR-628-3p and miR-654-5p) were was
found to be signifi-cantly upregulated, whereas 7 miRNAs
(let-7b*, miR-220b, miR-513a-3p, miR-551a, miR-576-5p,
miR-1236 and kshv-miR-K12-6-5p) were found to be downregulated in
the patients with atrophic non-union (Fig. 1A).
The upregulation of miRNAs in the patients with
atrophic non-union was verified by qPCR, which reconfirmed that the
expression of the 4 miRNAs (miR-149*, miR-221,
miR-628-3p and miR-654-5p) was increased in the scar tissues of
patients with atrophic non-union (Fig. 1B).
Expression patterns of miRNAs upregulated
in patients with atrophic non-union during osteoblast
differentiation
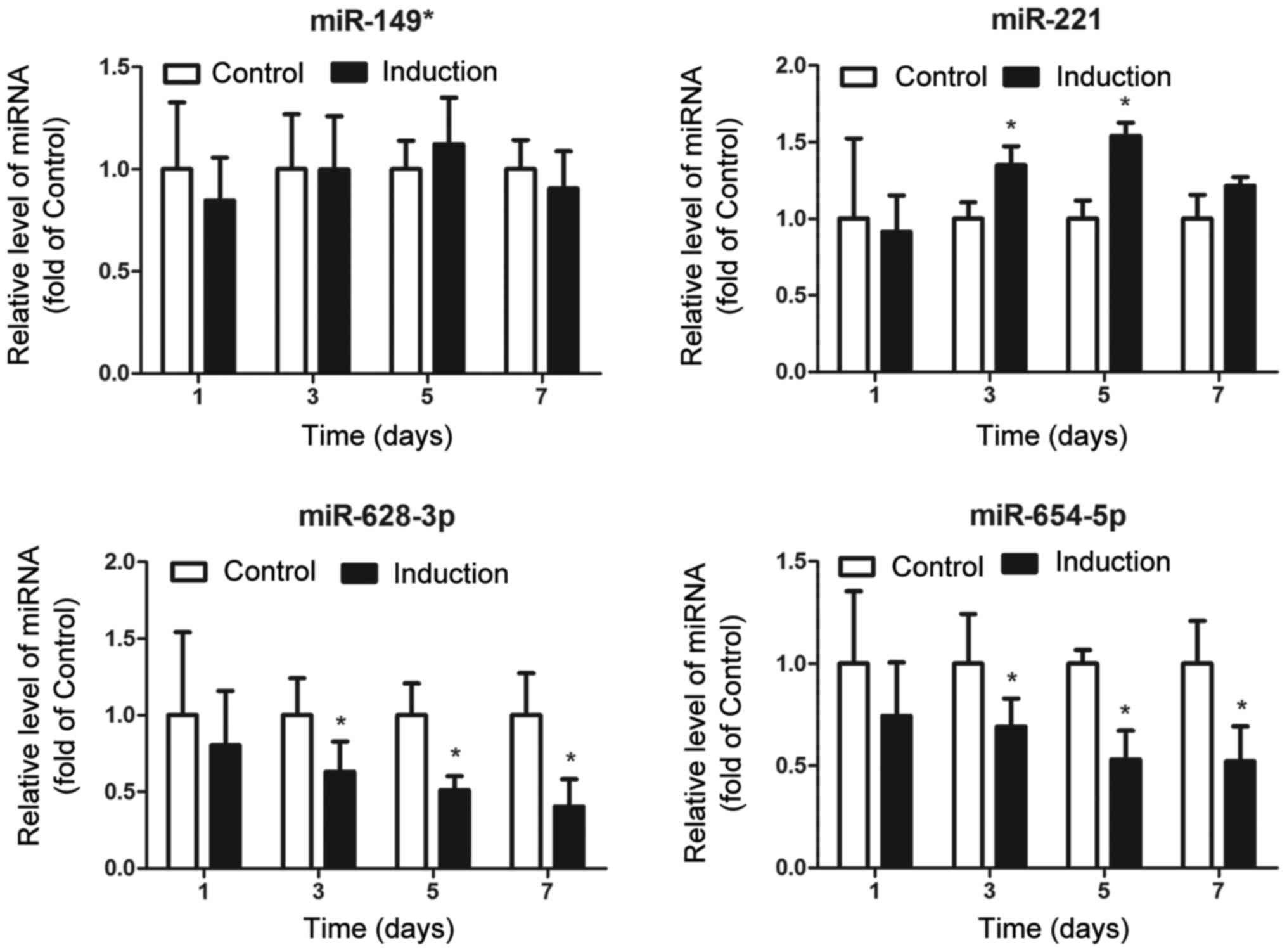
To assess the association between the 4 upregulated
miRNAs and osteoblast differentiation, we detected alterations in
the expression levels of these miRNAs during the differentiation of
MG63 osteoblasts. As shown in Fig.
2, miR-149* expression was consistent during the
differentiation of MG63 osteoblasts; miR-221 expression was
increased at the beginning of differentiation (on day 3), but was
decreased at the end of differentiation (on day 7); the expression
of both miR-628-3p and miR-654-5p was found to be persistently
downregulated during the differentiation process. These results
suggest that miR-628-3p and miR-654-5p may have an inhibitory
regulatory function in osteoblast differentiation.
miR-628-3p, but not miR-654-5p,
attenuates osteoblast differentiation
To clarify the exact roles of miR-628-3p and
miR-654-5p in the differentiation of osteoblasts, both miR-628-3p
and miR-654-5p mimics were introduced into the MG63 cells and
osteoblast differentiation was assessed. The expression levels of
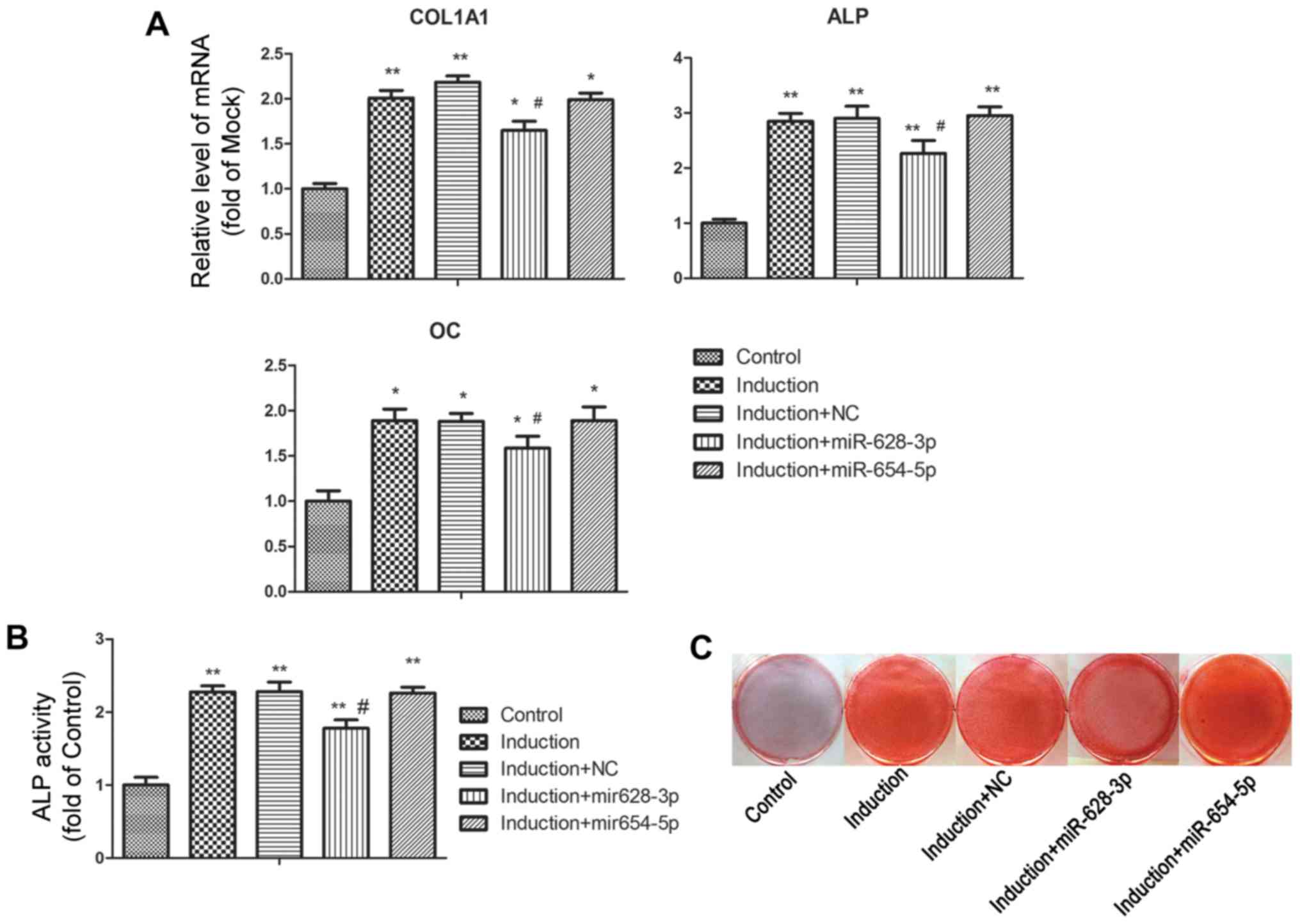
osteoblastic-related genes, including ALP, COL1A1 and OC were
significantly upregulated following culture in osteoblastic
differentiation medium (Fig. 3A).
Under the differentiation-inducing conditions, transfection with
miR-628-3p mimics markedly attenuated the increased expression of
ossification-related genes, whereas the miR-654-5p mimics had no
effect on the expression of these marker genes, compared with the
group transfected with miRNA mimics negative control (induction +
NC). Further results revealed that ALP activity in the
differentiated MG63 cells was inhibited by transfection with
miR-628-3p mimics, but not by miR-654-5p 654-5p mimics (Fig. 3B). The results of in vitro
cell mineral characteristics, as assessed by Alizarin Red S
staining, were in line with the results mentioned above; namely
transfection with miR-628-3p, but not miR-654-5p, mimics diminished
MG63 mineralization induced by osteoblastic differentiation
(Fig. 3C).
RUNX2 is a target gene of miR-628-3p
Osteogenesis is a complex biological process
involving multiple transcription factors. To identify the target
gene by which miR-628-3p abates osteoblast differentiation
potential, we searched the candidate target genes of miR-628-3p
using the miRNA gene prediction software, microRNA.org. The master transcription factor of
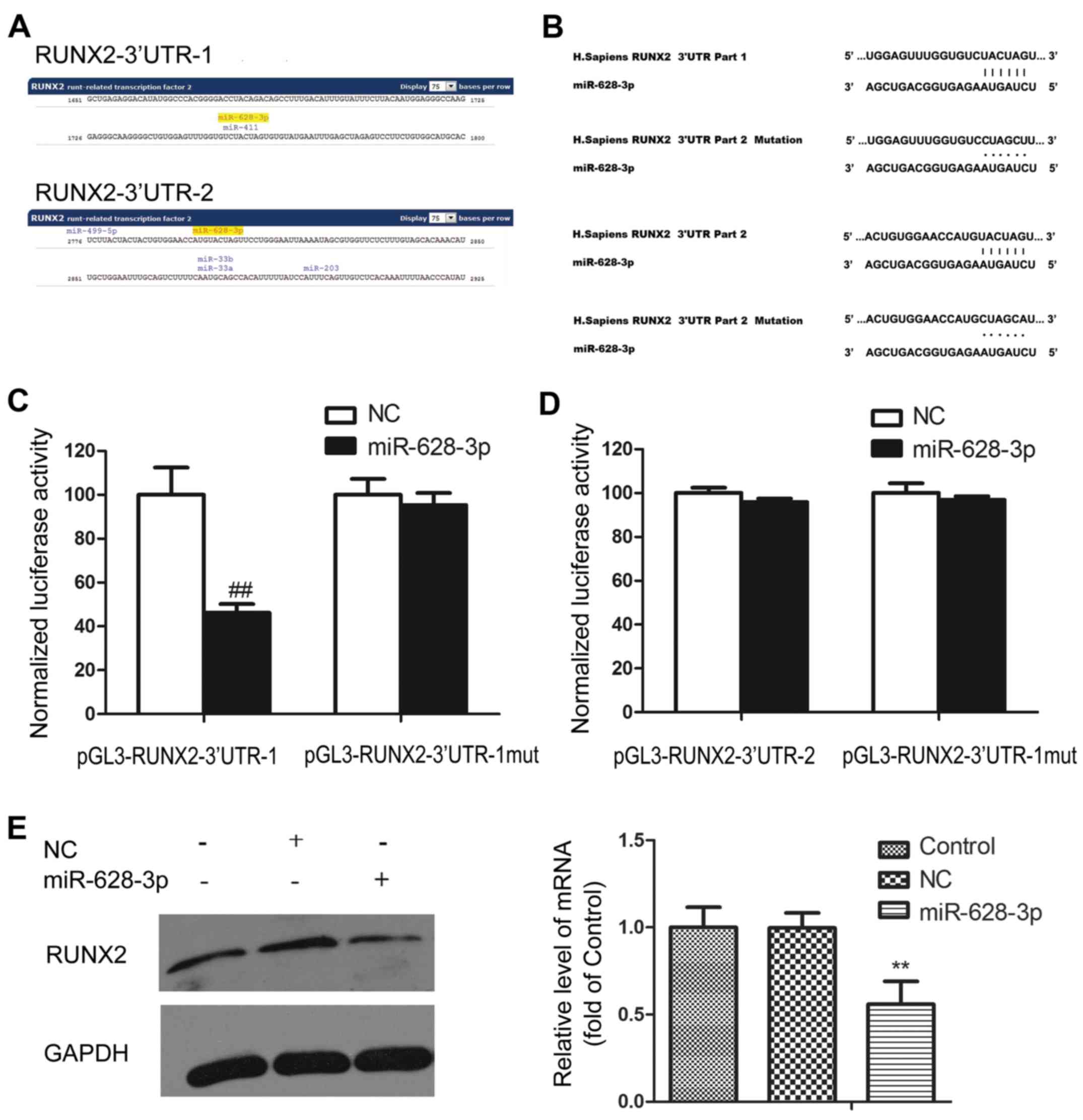
osteoblast differentiation, RUNX2, was predicted as a target gene
of miR-628-3p. Two pairing regions were identified in the RUNX2 3′
non-coding region, named RUNX2-3′UTR-1 and RUNX2-3′UTR-2 (Fig. 4A). To verify whether miR-628-3p
can bind to these regions, luciferase reporter vectors
(pGL3-RUNX2-3′UTR-1 and pGL3-RUNX2-3′UTR-2) were constructed. In
addition, the control vector were also generated with the mutated
binding region (pGL3-RUNX2-3′UTR-1-mut and pGL3-RUNX2-3′UTR-2-mut)
(Fig. 4B). miR-628-3p mimics were
transfected into 293 cells with either the reporter vector or
control vector. The results revealed that luciferase activity
specifically weakened in the cells transfected with miR-628-3p
mimics and pGL3-RUNX2-3′UTR-1, but not in other group, indicating
that miR-628-3p can specifically bind to RUNX2- 3′UTR-1 and may
play a role in the suppression of RUNX2 expression (Fig. 4C and D).
To verify this hypothesis, we examined the RUNX2
protein level following transfection with miR-628-3p mimics. The
results exhibited a markedly decreased RUNX2 expression as compared
with the control group. We further detected the effects of
miR-628-3p on the RUNX2 mRNA levels, which demonstrated that
miR-628-3p decreased the RUNX2 mRNA levels (Fig. 4E).
Discussion
In this study, we demonstrated the differential
expression of miRNAs between patients with atrophic non-union and
patients with bone healing following fracture through screening
with miRNA microarray. A total of 4 upregulated and 7 downregulated
miRNAs were identified. Amongst the upregulated miRNAs, miR-628-3p
and miR-654-5p expression was decreased persistently during
osteoblast differentiation. miR-628-3p mimics, but not miR-654-5p
mimics, attenuated the osteogenic differentiation of MG63 cells.
Furthermore, RUNX2 was predicted as a candidate target gene by
in silico analysis. Further results verified that miR-628-3p
decreased the RUNX2 level at both the protein and mRNA level.
Atrophic non-union is triggered by multiple causes.
Impaired blood circulation is considered to be the most importance
factor (28). However, with the
progress in research on macrovascular techniques, this view has
been challenged in recent years. Multiple studies have indicated
that blood supply is not reduced in atrophic non-union and
angiogenesis also seems unimpaired. In a study by Santavirta et
al (29), pathology specimens
from 10 patients with non-union did not reveal any evidence of
decreased vascular density. Besides, in another study, the blood
supply in atrophic non-union was reported as being more than
adequate, when compared with hypertrophic non-union (30). Increasingly, researchers are
accepting the opinion that insufficient osteogenesis plays a
crucial role in non-union. Bone fracture healing involves hematoma
formation and its filling in by a fibrin network; subsequently,
cells grow along a fibrin meshwork to from procallus and
fibrocartilaginous callus, followed by the deposition of calcium
salt that converts the fibrocartilaginous callus into the spongy
bone that is characteristic of the bony callus. In the final step,
the bony callus remodels into a normal shape. During this process,
impaired osteogenic capability results in atrophic non-union. Many
factors and/or signaling pathways involved in osteogenesis seem to
improve bone healing (30). The
transplantation of pluripotent stem cells, such as mesenchymal stem
cells, which can undergo osteogenic differentiation, has an obvious
therapeutic benefit in patients with non-union (31). As shown in this study, miR-628-3p
is highly expressed in atrophic non-union and can inhibit
osteogenesis, indicating its negative role in atrophic non-union
modulated via the suppression of osteogenesis.
miRNAs control osteoblastogenesis. A general null
mutation of Dicer has been shown to prevent the formation of a
normal vertebrate body plan at gastrulation, suggesting a critical
role of miRNAs in the formation of bone. Furthermore, the knockdown
of Dicer and Drosha using lentiviruses expressing shRNAs, has been
shown to inhibit the osteogenic differentiation of human
multipotent stromal cells (32).
Multiple miRNAs regulate osteoblast differentiation, with some of
them exerting an inhibitory effect. For instance, miR-34s inhibit
osteoblast proliferation and differentiation in mice by targeting
SATB homeobox 2 (SATB2) (33).
Furthermore, miRNA-100 regulates the osteogenic differentiation of
human adipose-derived mesenchymal stem cells by targeting bone
morphogenetic protein receptor type II (BMPR2) (34). miR-214 targets activating
transcription factor 4 (ATF4) to inhibit bone formation (35). The miR-93/Sp7 function loop
mediates osteoblast mineralization in osteogenesis (36). Other microRNAs have been shown to
have a strengthening function. For instance, the upregulation of
miR-22 promotes osteogenic differentiation by repressing histone
deacetylase (HDAC)6 protein expression in human adipose
tissue-derived mesenchymal stem cells (37), and the overexpression of miR-2861
promotes osteoblast differentiation by suppressing HDAC5 in primary
osteoblasts (38). In this study,
miR-628-3p was a found to be involved in osteoblast
differentiation.
Until now, studies investigating the function of
miR-628-3p, have been exceedingly rare, with only a few studies
investigating a possible association between this miRNA and human
diseases. Cui et al reported a marked alteration of serum
miR-628-3p levels in patients with hand-foot-and-mouth disease,
suggesting its potential value as a candidate molecular marker to
differentiate patients with enterovirus infections from healthy
individuals (39). Another recent
study highlighted the potential of this miRNA for helping
distinguish sera of patients with pancreatic cancer from those of
healthy persons (40).
Furthermore, another study indicated that miR-628-3p in platelets
may influence mRNA processing in patients with diabetes mellitus
(41). In the present study, we
demonstrated that miR-628-3p expression was increased in the scar
tissue of patients with atrophic non-union, and that its expression
was downregulated during osteoblast differentiation. Further
experiments proved that this miRNA suppresses osteoblast
differentiation. To the best of our knowledge, this is the first
study to profile the functional aspects of miR-628-3p.
The master transcription factors, such as RUNX2 and
SP7-Osterix, have autonomous effects on osteoblast differentiation.
The protein coding sequence of RUNX2 is approximtely 1,500
nucleotides, whereas, its 3′UTR is much longer (of the order of
3,000 nucleotides), which make this gene the common targeting gene
for miRNAs regulating osteogenesis. With the established 3′UTR
sequence of RUNX2, Zhang et al identified >11 miRNAs
binding to the 3′UTR of RUNX2, i.e., miR-23a, miR-30c, miR-34c,
miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-217, miR-218 and
miR-338. All these miRNA differ in their expression level in
osteogenic and non-osteogenic cells, and exhibit a regulatory
function in osteoblast differentiation (42). Some of these miRNAs were also
identified in parallel studies (43–46). Apart from these 11 miRNAs, it has
also been demonstrated that RUNX2 is also controlled by other
miRNAs. For example, miR-204 has been shown to regulate RUNX2
protein levels in mesenchymal progenitor cells, while miRNA-103a
acts as a mechano-sensitive miRNA suppressing Runx2 to inhibit bone
formation (47). This study
demonstrated that the binding of miR-628-3p to the RUNX2 3′UTR
region suppressed RUNX2 expression, thus adding a new member to the
growing list of miRNAs that regulate RUNX2 expression.
A major limitation of this study was the fact that
we did not verify the function of miR-628-3p in vivo, and,
whether this miRNA has a potential therapeutic role in atrophic
non-union. Whether the overexpression of RUNX2 can reverse the
inhibitory effects of miR-628-3p on osteoblast differentiation also
needs to be verified. In addition, the functions of the miRNAs
found to be downregulated in atrophic non-union also need to be
assessed further.
In conclusion, to the best of our knowledge, the
present study is the first to elucidate the role of miRNAs in the
pathological process of atrophic non-union. We identified an miRNA
i.e., miR-628-3p, which was upregulated in patients with atrophic
non-union and exerted an inhibitory effect on osteogenesis through
the suppression of its target gene, RUNX2. The findings of our
study may provide insight into the development of novel therapeutic
targets for the management of atrophic non-union.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (no. 81000803 to H.C.).
Glossary
Abbreviations
Abbreviations:
|
GADPH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
RUNX2
|
runt-related transcription factor
2
|
References
|
1
|
Tzioupis C and Giannoudis PV: Prevalence
of long-bone non-unions. Injury. 38(Suppl 2): S3–S9. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Einhorn TA and Gerstenfeld LC: Fracture
healing: mechanisms and interventions. Nat Rev Rheumatol. 11:45–54.
2015. View Article : Google Scholar :
|
|
3
|
Dimitriou R, Kanakaris N, Soucacos PN and
Giannoudis PV: Genetic predisposition to non-union: evidence today.
Injury. 44(Suppl 1): S50–S53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Copuroglu C, Calori GM and Giannoudis PV:
Fracture non-union: Who is at risk? Injury. 44:1379–1382. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lim LP, Glasner ME, Yekta S, Burge CB and
Bartel DP: Vertebrate microRNA genes. Science. 299:15402003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar :
|
|
9
|
Friedländer MR, Lizano E, Houben AJ,
Bezdan D, Báñez-Coronel M, Kudla G, Mateu-Huertas E, Kagerbauer B,
González J, Chen KC, et al: Evidence for the biogenesis of more
than 1,000 novel human microRNAs. Genome Biol. 15:R572014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu N, Li Z, Yu Z, Yan F, Liu Y, Lu X and
Yang W: MicroRNA-33b suppresses migration and invasion by targeting
c-Myc in osteosarcoma cells. PLoS One. 9:e1153002014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He C, Xiong J, Xu X, Lu W, Liu L, Xiao D
and Wang D: Functional elucidation of MiR-34 in osteosarcoma cells
and primary tumor samples. Biochem Biophys Res Commun. 388:35–40.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhuo W, Ge W, Meng G, Jia S, Zhou X and
Liu J: MicroRNA-20a promotes the proliferation and cell cycle of
human osteosarcoma cells by suppressing early growth response 2
expression. Mol Med Rep. 12:4989–4994. 2015.PubMed/NCBI
|
|
13
|
Mori F, Sacconi A, Canu V, Ganci F,
Novello M, Anelli V, Covello R, Ferraresi V, Muti P, Biagini R, et
al: miR-181c associates with tumor relapse of high grade
osteosarcoma. Oncotarget. 6:13946–13961. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salah Z, Arafeh R, Maximov V, Galasso M,
Khawaled S, Abou-Sharieha S, Volinia S, Jones KB, Croce CM and
Aqeilan RI: miR-27a and miR-27a* contribute to metastatic
properties of osteosarcoma cells. Oncotarget. 6:4920–4935. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mu Y, Zhang H, Che L and Li K: Clinical
significance of microRNA-183/Ezrin axis in judging the prognosis of
patients with osteosarcoma. Med Oncol. 31:8212014. View Article : Google Scholar
|
|
16
|
Kawano M, Tanaka K, Itonaga I, Ikeda S,
Iwasaki T and Tsumura H: microRNA-93 promotes cell proliferation
via targeting of PTEN in Osteosarcoma cells. J Exp Clin Cancer Res.
34:762015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qu F, Li CB, Yuan BT, Qi W, Li HL, Shen
XZ, Zhao G, Wang JT and Liu YJ: MicroRNA-26a induces osteosarcoma
cell growth and metastasis via the Wnt/β-catenin pathway. Oncol
Lett. 11:1592–1596. 2016.PubMed/NCBI
|
|
18
|
Yuan J, Lang J, Liu C, Zhou K, Chen L and
Liu Y: The expression and function of miRNA-451 in osteosarcoma.
Med Oncol. 32:3242015. View Article : Google Scholar
|
|
19
|
Zhang C, Yao C, Li H, Wang G and He X:
Serum levels of microRNA-133b and microRNA-206 expression predict
prognosis in patients with osteosarcoma. Int J Clin Exp Pathol.
7:4194–4203. 2014.PubMed/NCBI
|
|
20
|
Zhang C, Yao C, Li H, Wang G and He X:
Combined elevation of microRNA-196a and microRNA-196b in sera
predicts unfavorable prognosis in patients with osteosarcomas. Int
J Mol Sci. 15:6544–6555. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liua XD, Caia F, Liu L, Zhang Y and Yang
AL: microRNA-210 is involved in the regulation of postmenopausal
osteoporosis through promotion of VEGF expression and osteoblast
differentiation. Biol Chem. 396:339–347. 2015.
|
|
22
|
Jia J, Feng X, Xu W, Yang S, Zhang Q, Liu
X, Feng Y and Dai Z: miR-17-5p modulates osteoblastic
differentiation and cell proliferation by targeting SMAD7 in
non-traumatic osteonecrosis. Exp Mol Med. 46:e1072014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao B, Yu Q, Li H, Guo X and He X:
Characterization of microRNA expression profiles in patients with
intervertebral disc degeneration. Int J Mol Med. 33:43–50.
2014.
|
|
24
|
Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X
and Qiu G: MicroRNA-10b promotes nucleus pulposus cell
proliferation through RhoC-Akt pathway by targeting HOXD10 in
intervetebral disc degeneration. PLoS One. 8:e830802013. View Article : Google Scholar :
|
|
25
|
Miyaki S, Sato T, Inoue A, Otsuki S, Ito
Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al:
MicroRNA-140 plays dual roles in both cartilage development and
homeostasis. Genes Dev. 24:1173–1185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakasa T, Shibuya H, Nagata Y, Niimoto T
and Ochi M: The inhibitory effect of microRNA-146a expression on
bone destruction in collagen-induced arthritis. Arthritis Rheum.
63:1582–1590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee YJ, Jeong JK, Seol JW, Xue M, Jackson
C and Park SY: Activated protein C differentially regulates both
viability and differentiation of osteoblasts mediated by
bisphosphonates. Exp Mol Med. 45:e92013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brinker MR and Bailey DE Jr: Fracture
healing in tibia fractures with an associated vascular injury. J
Trauma. 42:11–19. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Santavirta S, Konttinen YT, Nordström D,
Mäkelä A, Sorsa T, Hukkanen M and Rokkanen P: Immunologic studies
of nonunited fractures. Acta Orthop Scand. 63:579–586. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moghaddam A, Elleser C, Biglari B,
Wentzensen A and Zimmermann G: Clinical application of BMP 7 in
long bone non-unions. Arch Orthop Trauma Surg. 130:71–76. 2010.
View Article : Google Scholar
|
|
31
|
Fayaz HC, Giannoudis PV, Vrahas MS, Smith
RM, Moran C, Pape HC, Krettek C and Jupiter JB: The role of stem
cells in fracture healing and nonunion. Int Orthop. 35:1587–1597.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Oskowitz AZ, Lu J, Penfornis P, Ylostalo
J, McBride J, Flemington EK, Prockop DJ and Pochampally R: Human
multipotent stromal cells from bone marrow and microRNA: Regulation
of differentiation and leukemia inhibitory factor expression. Proc
Natl Acad Sci USA. 105:18372–18377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wei J, Shi Y, Zheng L, Zhou B, Inose H,
Wang J, Guo XE, Grosschedl R and Karsenty G: miR-34s inhibit
osteoblast proliferation and differentiation in the mouse by
targeting SATB2. J Cell Biol. 197:509–521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zeng Y, Qu X, Li H, Huang S, Wang S, Xu Q,
Lin R, Han Q, Li J and Zhao RC: MicroRNA-100 regulates osteogenic
differentiation of human adipose-derived mesenchymal stem cells by
targeting BMPR2. FEBS Lett. 586:2375–2381. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Guo B, Li Q, Peng J, Yang Z, Wang
A, Li D, Hou Z, Lv K, Kan G, et al: miR-214 targets ATF4 to inhibit
bone formation. Nat Med. 19:93–100. 2013. View Article : Google Scholar
|
|
36
|
Yang L, Cheng P, Chen C, He HB, Xie GQ,
Zhou HD, Xie H, Wu XP and Luo XH: miR-93/Sp7 function loop mediates
osteoblast mineralization. J Bone Miner Res. 27:1598–1606. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang S, Wang S, Bian C, Yang Z, Zhou H,
Zeng Y, Li H, Han Q and Zhao RC: Upregulation of miR-22 promotes
osteogenic differentiation and inhibits adipogenic differentiation
of human adipose tissue-derived mesenchymal stem cells by
repressing HDAC6 protein expression. Stem Cells Dev. 21:2531–2540.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li H, Xie H, Liu W, Hu R, Huang B, Tan YF,
Xu K, Sheng ZF, Zhou HD, Wu XP and Luo XH: A novel microRNA
targeting HDAC5 regulates osteoblast differentiation in mice and
contributes to primary osteoporosis in humans. J Clin Invest.
119:3666–3677. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cui L, Qi Y, Li H, Ge Y, Zhao K, Qi X, Guo
X, Shi Z, Zhou M, Zhu B, et al: Serum microRNA expression profile
distinguishes enterovirus 71 and coxsackievirus 16 infections in
patients with hand-foot-and-mouth disease. PLoS One. 6:e270712011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li A, Yu J, Kim H, Wolfgang CL, Canto MI,
Hruban RH and Goggins M: MicroRNA array analysis finds elevated
serum miR-1290 accurately distinguishes patients with low-stage
pancreatic cancer from healthy and disease controls. Clin Cancer
Res. 19:3600–3610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Stratz C, Nührenberg T, Fiebich BL, Amann
M, Kumar A, Binder H, Hoffmann I, Valina C, Hochholzer W, Trenk D,
et al: Controlled type II diabetes mellitus has no major influence
on platelet micro-RNA expression. Results from micro-array
profiling in a cohort of 60 patients. Thromb Haemost. 111:902–911.
2014. View Article : Google Scholar
|
|
42
|
Zhang Y, Xie RL, Croce CM, Stein JL, Lian
JB, van Wijnen AJ and Stein GS: A program of microRNAs controls
osteogenic lineage progression by targeting transcription factor
Runx2. Proc Natl Acad Sci USA. 108:9863–9868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li Z, Hassan MQ, Volinia S, van Wijnen AJ,
Stein JL, Croce CM, Lian JB and Stein GS: A microRNA signature for
a BMP2-induced osteoblast lineage commitment program. Proc Natl
Acad Sci USA. 105:13906–13911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li Z, Hassan MQ, Jafferji M, Aqeilan RI,
Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS and Lian JB:
Biological functions of miR-29b contribute to positive regulation
of osteoblast differentiation. J Biol Chem. 284:15676–15684. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hassan MQ, Gordon JA, Beloti MM, Croce CM,
van Wijnen AJ, Stein JL, Stein GS and Lian JB: A network connecting
Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the
osteoblast differentiation program. Proc Natl Acad Sci USA.
107:19879–19884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang
W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL,
et al: miR-218 directs a Wnt signaling circuit to promote
differentiation of osteoblasts and osteomimicry of metastatic
cancer cells. J Biol Chem. 287:42084–42092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zuo B, Zhu J, Li J, Wang C, Zhao X, Cai G,
Li Z, Peng J, Wang P, Shen C, et al: microRNA-103a functions as a
mechno-sensitive microRNA to inhibit bone formation through
targeting Runx2. J Bone Miner Res. 30:330–245. 2015. View Article : Google Scholar
|