Introduction
Mesenchymal stem cells (MSCs) are capable of
self-renewal and differentiation, and are therefore a valuable
source of cells for the replacement of diseased or damaged organs.
Compared with embryonic stem cells or induced pluripotent stem
cells, MSCs have an ability to regulate the immune system and do
not form teratomas. They can be isolated from various sources,
including the tonsils (1,2), bone marrow (3), adipose tissue (4), amniotic fluid (5), umbilical cord blood (6), the placenta (7) and dental pulp (8). Previously, we reported that
tonsil-derived MSCs (T-MSCs) can be differentiated into skeletal
myocytes, and are therefore a promising candidate in cell therapies
for skeletal muscle (SKM)-related disease (2). However, the mechanisms of
differentiation from T-MSCs to myocytes remain unclear.
Myogenic differentiation is regulated by a variety
of biochemical pathways, including those involving growth factors
fibroblast growth factor (FGF), transforming growth factor-β
(TGF-β), insulin-like growth factor 1 (IGF1) (9), Janus kinase 2/signal transducer and
activator of transcription (STAT)2/STAT3 (10), Notch (11), ER stress, and autophagy (12). FGF-2 and HGF promote proliferation
of myogenic progenitors and delay their differentiation, in part by
inhibiting the expression of myogenic regulatory factors, such as
MyoD (13,14). IGF1 promotes myogenic
differentiation and enhances protein synthesis in differentiated
myofibers by activating the translation factor 4E-BP and the
ribosomal protein S6 kinase (p70S5K), and by inhibiting
muscle-specific E3 ligases that promote protein degradation
(15). ER stress has a positive
effect on myofiber formation in vitro, possibly mimicking
the action of signals that drive apoptosis and differentiation
in vivo (16). Recently,
it was shown that autophagy is induced during muscle
differentiation despite the concomitant activation of mammalian
target of rapamycin (mTOR) using the mouse myoblast cell line,
C2C12 (12). Autophagy plays an
essential role in cellular development and differentiation
(17), and human bone marrow MSCs
may differentiate to become early osteocytes and adipocytes by
consumption of autophagosomes (18). In addition, mitophagy, the
selective degradation of mitochondria by autophagy, has been
reported to be required for mitochondrial biogenesis and myogenic
differentiation of C2C12 myoblasts (19).
In the present study, we analyzed transcriptomes
using microarrays and then performed large-scale expression
profiling of T-MSCs during differentiation into myocytes compared
with hSKMCs and the candidate genes identified as associated with
myogenic differentiation. Immunocytochemistry, reverse
transcriptase-polymerase chain reaction (RT-qPCR), and western
blotting confirmed the pathway of interest that was obtained from
the candidate gene. The results suggest that autophagy has an
important role in the myogenic differentiation of the T-MSCs.
Materials and methods
Ethics statement
The Institutional Review Board (ECT-11-53-02) of
Ewha Womans University Mokdong Hospital (Seoul, Korea) approved all
of the experimental procedures used in this study. Informed written
consent was secured from each patient and/or their legal
representatives.
Isolation of T-MSCs
Isolation of T-MSCs from tonsil tissue was performed
as described previously (1,2).
Tonsillar tissues were extracted during tonsillectomy, and minced
and digested in Dulbecco's modified Eagle's medium (DMEM)
containing 210 U/ml collagenase type I (both from Invitrogen,
Carlsbad, CA, USA) and DNase (10 mg/ml; Sigma-Aldrich, St. Louis,
MO, USA). After cells were passed through a cell strainer (BD
Biosciences, San Jose, CA, USA), mononuclear cells were obtained by
Ficoll-Paque (GE Healthcare, Chalfont St. Giles, UK) density
gradient centrifugation. Cells were cultured for 48 h at 37°C in
low-glucose DMEM containing 10% fetal bovine serum (FBS;
Invitrogen) and 1% penicillin/streptomycin (Sigma-Aldrich) in a
humidified chamber with 5% CO2, followed by removal of
nonadherent cells and a supply of fresh medium for the T-MSCs.
These freshly cultured cells were expanded over 3-5 passages and
subsequently used in the present study.
Differentiation
To induce the myogenic differentiation of the
T-MSCs, 3–4×106 cells were plated in a 10-cm Petri dish
in low-glucose DMEM supplemented with 10% FBS. At 1–3 days, the
cells spontaneously aggregated to form spheres 50–100 mm in
diameter. Once the spheres had formed, the medium was replaced with
DMEM/nutrient mixture F-12 (DMEM/F-12; Invitrogen) supplemented
with 1 ng/ml TGF-β (R&D Systems, Minneapolis, MN, USA),
nonessential amino acids (NEAA; Invitrogen) and
insulin-transferrin-selenium (ITS; Gibco Life Technologies, Grand
Island, NY, USA) for a further 4 days to allow their
differentiation into myoblasts. The T-MSCs expanded beyond the
spheres when transferred to a collagen-coated dish in the
abovementioned myoblast differentiation medium, and formed a
rosette-like spread. To induce terminal differentiation into
myocytes, the myoblasts were cultured for 2 weeks in myogenic
induction medium, which consisted of low-glucose DMEM containing 10
ng/ml IGF1 (R&D Systems) and 2% FBS (Fig. 1A–E). The hSKMCs (Fig. 1F) were differentiated in
serum-free medium (DMEM F-12; Invitrogen) containing 10 ng/ml IGF1
(R&D Systems).
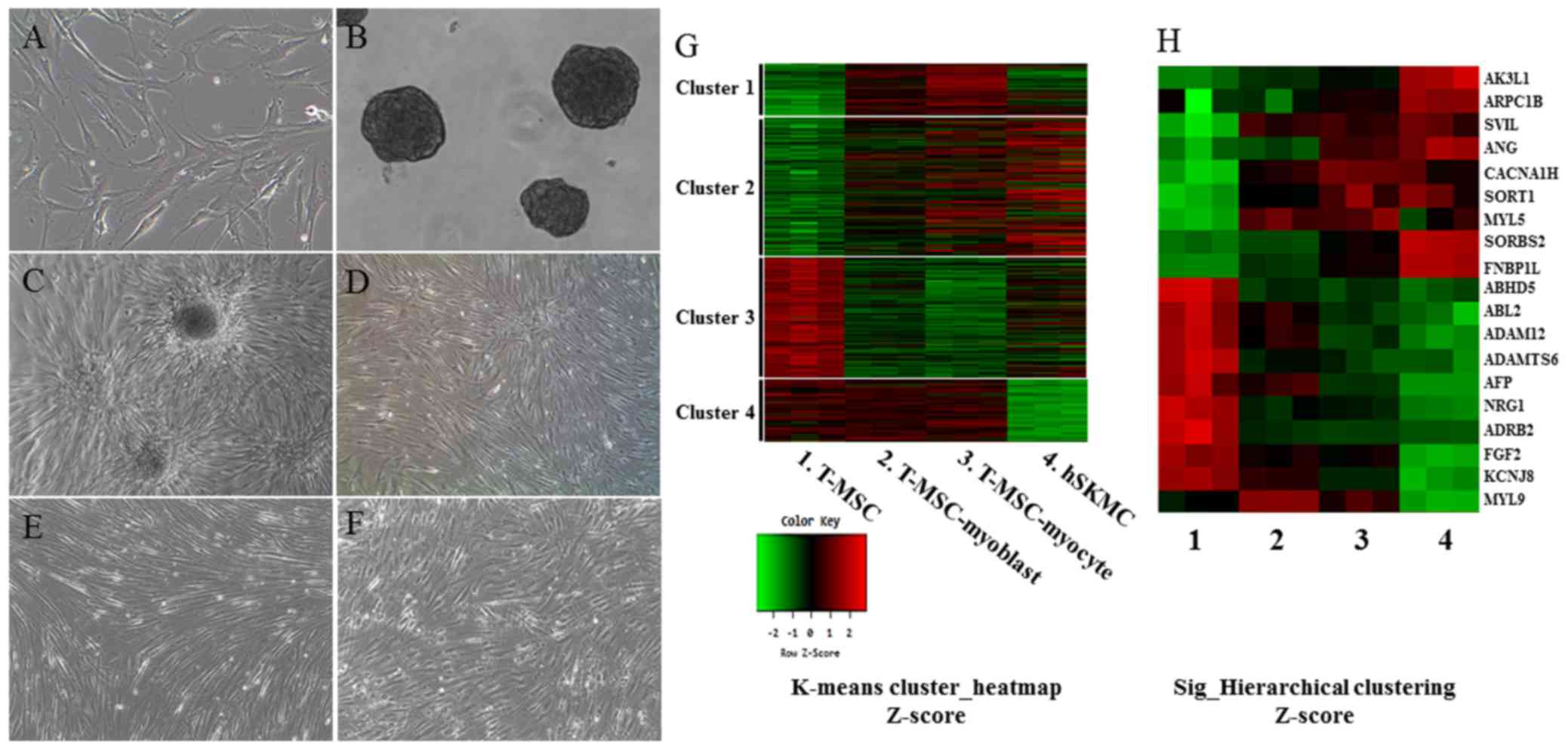 | Figure 1Myogenic differentiation of human
T-MSCs and gene expression analysis by microarray. Undifferentiated
T-MSCs (A) were induced to form spheres 50–100 mm in diameter (B)
in a Petri dish. (C) The T-MSCs expanded beyond the spheres when
transferred to a collagen-coated dish in replating medium and
formed a rosette-like spread. (D) The plated cells cultured for 4
days in myoblast induction medium. (E) The myoblasts after 14 days
in myocyte induction medium showing their altered morphology; they
underwent fusion to generate nascent myotubes. (F) hSKMCs cultured
for 4 days in serum-free medium containing IGF1. Original
magnification, ×100. (G) Heatmap of global mRNA expression
comparing undifferentiated T-MSCs (sample 1), T-MSC-derived
myoblasts (sample 2), T-MSC-derived myocytes (sample 3) and hSKMCs
(sample 4). (H) Myogenic gene profile and unsupervised clustering
based on markers associated with muscle for undifferentiated
T-MSCs, differentiated myogenic cells, and hSKMCs. Lanes 1–4
indicate undifferentiated T-MSCs (sample 1), differentiated
myogenic cells (sample 2, T-MSC-myoblast; sample 3, T-MSC-myocyte)
and hSKMCs (sample 4). Red indicates upregulated genes and green
indicates downregulated genes in G and H. T-MSCs, tonsil-derived
mesenchymal stem cells; hSKMCs, human skeletal muscle cells; IGF1,
insulin-like growth factor 1. |
Chemical reagents
Autophagy was confirmed with 5-azacytidine as an
inducer and bafilomycin A1 (Baf1) as an inhibitor to identify the
mechanism of differentiation to myogenic cells. To produce a stock
solution (1,000X), 1 mM 5-azacytidine and 1 mM Baf1 (Sigma-Aldrich)
were dissolved in 1 ml dimethylsulfoxide in water and acetic acid,
respectively. After the inducer or inhibitor was added to the
differentiation medium for 1 h, the medium was replaced with
chemical reagent-free medium.
Total RNA preparation
The cells were seeded in 100-mm culture dishes under
individual culture medium. Total RNA was extracted from three
replicates per group using a Qiagen (Germantown, MD, USA) RNeasy
Mini kit. RNA purity and integrity were evaluated using a ND-1000
spectrophotometer (NanoDrop, Wilmington, NC, USA), and Agilent 2100
Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Microarray analysis
Total RNA was amplified and purified using a
TargetAmp-Nano Labeling kit for Illumina Expression BeadChip
(Epicentre, Madison, WI, USA) to yield biotinylated cRNA according
to the manufacturer's instructions. Briefly, 200 ng of total RNA
was reverse-transcribed to cDNA using a T7-oligo(dT) primer.
Second-strand cDNA was synthesized, transcribed in vitro,
and labeled with biotin-NTP. After purification, the cRNA was
quantified using an ND-1000 spectrophotometer. An amount 750 ng of
the labeled cRNA samples was hybridized to each Human HT-12 version
4.0 Expression BeadChip for 18 h at 58°C, according to the
manufacturer's instructions (Illumina Inc., San Diego, CA, USA).
Detection of an array signal was achieved using Amersham fluorolink
streptavidin-Cy3 (GE Healthcare Bio-Sciences) following the bead
array manual. Arrays were scanned with an Illumina bead array
reader confocal scanner according to the manufacturer's
instructions. The quality of hybridization and overall chip
performance were monitored by visual inspection of both internal
quality control checks and the raw scanned data. Raw data were
extracted using the software provided by the manufacturer [Illumina
GenomeStudio, version 2011.1 (Gene Expression Module, version
1.9.0)]. Array probes were transformed using a logarithm and
normalized using a quantile method. Statistical significance of the
expression data was determined using an LPE test and fold-change in
which the null hypothesis was that no difference exists between
groups. False discovery rate (FDR) was controlled by adjusting the
P-value using a Benjamini-Hochberg algorithm. For a DEG set,
hierarchical cluster analysis was performed using complete linkage
and Euclidean distance as a measure of similarity. Gene-Enrichment
and Functional Annotation analysis for the significant probe list
was performed using DAVID (http://david.abcc.ncifcrf.gov/home.jsp). All data
analysis and visualization of differentially expressed genes were
conducted using R version 3.0.2 (www.r-project.org).
Real-time quantitative PCR
Total RNA was extracted from cells using a Qiagen
RNeasy Mini kit. Complementary DNA (cDNA) was synthesized using
Superscript II (Invitrogen) and oligo(dT)20 primers at
42°C for 1 h followed by incubation at 72°C for 15 min. From the
results of the microarray analysis, we determined genes that were
either upregulated or downregulated, or related to autophagy.
RT-qPCR was performed using SYBR Premix Ex Taq (Takara Bio Inc.,
Shiga, Japan) on an ABI 7500 Fast Real-Time PCR system (PE Applied
Biosystems, Foster City, CA, USA) to confirm the relative levels of
expression of genes in the T-MSCs, T-MSC-derived myoblasts and
myocytes. The total volume of the PCR reaction was 20 ml,
containing 0.8 ml of each primer (5 mM), 1 ml cDNA, 10 ml 2X SYBR
Premix Ex Taq II, 0.4 ml Rox dye, and 7.8 ml sterile
ddH2O. PCR cycling conditions were as follows: initial
30 sec denaturation at 95°C, followed by 40 cycles of amplification
at 95°C for 3 sec and 60°C for 30 sec, and a subsequent melting
curve analysis, where the temperature was increased from 60 to
95°C. To quantify the expression of each candidate gene, the mRNA
expression levels were normalized to the level of glyceraldehyde
3-phosphate dehydrogenase (GAPDH) mRNA. Relative gene expression
was analyzed using a comparative cycle threshold (Ct) method
(2−ΔΔCt) (20).
RT-qPCR was performed in triplicate for each sample and was
repeated three times for each assay. Primers of the forward and
reverse primers used were as follows: supervillin (SVIL) forward,
5′-GAAGTGCTCCCTTCTGCAAC-3′ and reverse, 5′-AGTGCTTTGCCAGCTGAAAT-3′;
sortilin 1 (SORT1) forward, 5′-TAACAG CTGCGTGGAAAGTG-3′ and
reverse, 5′-GCACTCCAGCCCTAACCATA-3′; angiogenin (ANG) forward,
5′-CTCACCCTGCAAAGACATCA-3′ and reverse, 5′-TCCATGTAGCTTGCAAGTGG-3′;
formin-binding protein (FNBP1L) forward, 5′-AAAGGTGACGGATGGACAAG-3′
and reverse, 5′-TTGCCCATTTCCTCAGAAAC-3′; beclin-1 forward,
5′-AGGTTGAGAAAGGCGAGACA-3′ and reverse, 5′-GCTTTTGTCCACTGCTCCTC-3′;
autophagy-related 5 (Atg5) forward, 5′-AAAGATGTGCTTCGAGATGTGT-3′
and reverse, 5′-CACTTTGTCAGTTACCAACGTCA-3′; autophagy-related 3
(Atg3) forward, 5′-GACCCCGGTCCTCAAGGAA-3′ and reverse,
5′-TGTAGCCCATTGCCATGTTGG-3′; autophagy-related 14 (Atg14) forward,
5′-GCGCCAAATGCGTTCAGAG-3′ and reverse,
5′-AGTCGGCTTAACCTTTCCTTCT-3′; GAPDH forward,
5′-ACACCCACTCCTCCACCTTT-3′ and reverse,
5′-TGCTGTAGCCAAATTCGTTG-3′.
Immunocytochemistry
The cells grown on coverslips were fixed in 4% (v/v)
paraformaldehyde (Sigma-Aldrich) for 15 min at room temperature or
overnight at 4°C. After rinsing in PBS, the fixed cells were
permeabilized and nonspecific epitopes were blocked using 2% bovine
serum albumin (Bovogen Biologicals, Keilor East, Vic, Australia) in
0.1% Tween-20/PBS, followed by incubation in the diluted primary
antibody for 1 h at room temperature or overnight at 4°C. After
three washes in PBS, samples were incubated for 1 h at room
temperature with secondary antibodies diluted in PBS. Prepared
samples were then mounted using Vectashield medium containing
4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories,
Burlingame, CA, USA) and photographed using a fluorescence
microscope (Nikon Corporation, Tokyo, Japan). The manufacturers and
catalog numbers of the antibodies employed were as follows: mouse
anti-myosin heavy chain (MHC; cat. no. MAB4470; R&D Systems)
mouse anti-myogenin (cat. no. ab-1835; Abcam, Cambridge, MA, USA),
mouse anti-desmin (cat. no. D1033; Sigma-Aldrich, St. Louis, MO,
USA), Alexa-568 goat anti-mouse IgG (cat. no. A-11004), Alexa-488
goat anti-rabbit IgG (cat. no. A-11008) (both from Life
Technologies). Quantification of immunofluorescence staining
confirmed that four slides were used for each condition. Graphs
represent the average of multiple tests from three independent
experiments.
Western blotting
Protein concentration was determined using the
Bradford assay reagent (Bio-Rad, Hercules, CA, USA) after lysing
cells in Pro-Prep buffer (Intron Biotechnology, Seongnam, Korea)
supplemented with phosphatase inhibitor cocktail solution
(Dawinbio, Hanam, Korea). The cells were washed with ice-cold PBS
and exposed to Pro-Prep buffer supplemented with phosphatase
inhibitor cocktail solution for 30 min on ice. Insoluble material
was removed by centrifugation at 12,000 × g for 10 min at 4°C.
Proteins (30–80 mg) were separated by 7.5–13.5% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and transferred to a
polyvinylidene fluoride or nitrocellulose membrane (Millipore,
Billerica, MA, USA). The membranes were blocked with 5% skim milk
in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 2 h at
room temperature. The blots were then incubated with primary
antibody overnight at 4°C. Antibodies used for western blot
analysis were rabbit anti-LC3B (cat. no. 14600-1-AP; Proteintech,
Chicago, IL, USA), rabbit anti-Beclin-1 (cat. no. 3738), and rabbit
anti-Atg5 (cat. no. 2630) (both from Cell Signaling Technology,
Beverly, MA, USA), rabbit anti-Bcl-2 (cat. no. SC-492; Santa Cruz
Biotechnology, Inc.). The blots were washed three times for 5 min
with TBST and then incubated with horseradish peroxidase-labeled
secondary antibody for 1 h at room temperature. Goat anti-mouse IgG
(1:2,500; cat. no. SC-2005; Santa Cruz Biotechnology, Inc.) and
goat anti-rabbit IgG (1:2500; cat. no. 7074; Cell Signaling
Technology) were used as the secondary antibodies. After additional
washes, the signal was detected using a West Save Gold western blot
detection kit (Youngin Frontier, Seoul, Korea). The protein signals
were visualized by exposing the membranes in a luminescent image
analyzer (LAS-3000; Fujifilm, Tokyo, Japan). The level of
expression of each protein was normalized to that of GAPDH
(Sigma-Aldrich). The results were quantified using Multi Gauge,
version 3.0 software (Science Laboratory, Tokyo, Japan).
Statistical analysis
The results are presented as mean ± standard error
of the mean (SEM). Statistical comparisons were made with a one-way
ANOVA and the Tukey multiple comparison test using GraphPad Prism
software, version 5.01 (GraphPad Software Inc., San Diego, CA, USA)
to identify significant differences. P-values <0.05 were
considered statistically significant and P-values <0.001 were
considered very significant. All experiments were performed at
least three times.
Results
Expression profiling of differentiated
myogenic cells derived from T-MSCs
T-MSCs have already been reported to have the
characteristics of MSCs (1,2,21).
To monitor the molecular events in myogenically differentiated
T-MSCs, we performed gene expression profiling using microarrays.
To obtain RNA samples, the undifferentiated T-MSCs (Fig. 1A) were induced to form spheres in
Petri dishes (Fig. 1B) and then
the cells expanded beyond the spheres when transferred to a
collagen-coated dish in a myoblast induction medium (Fig. 1C), and were cultured for 4 days in
myoblast induction medium (Fig.
1D). The T-MSC-derived myoblasts were differentiated for 14
days in myocyte induction medium and their morphology was altered;
they fused with one another to generate nascent myotubes (Fig. 1E). As a positive control for
myogenic differentiation, hSKMCs (Fig. 1F) that were cultured for 4 days in
serum-free medium containing IGF1 were sampled. Global mRNA
microarray analysis of T-MSCs was performed at three stages of
myogenic differentiation (T-MSC, T-MSC-derived myoblasts and
myocytes) and hSKMCs. The raw data (47,323 probes) were tested and
19,765 probes were filtered, normalized, and replicated clones
merged and were further taken into account. Subsequently, we found
the expression of 3,755 unique genes to be significantly different
between the four clusters on the whole (P<0.05; LPE test). The
clusters featured four primary expression patterns: an increase in
mRNA level during differentiation only (cluster 1); an increase in
mRNA level during the differentiation of T-MSCs and hSKMCs (cluster
2); a decrease in mRNA level during the differentiation of T-MSCs
and hSKMCs (cluster 3); and a decrease in mRNA level only at the
hSKMCs (cluster 4). A heatmap (Fig.
1G) showed that the gene expression was increased (cluster 1)
and maintained (cluster 4) compared with T-MSCs during myogenic
differentiation, regardless of the gene expression by hSKMCs.
Therefore, the genes expressed in cluster 1 and 4 were not
associated with the genes expressed by hSKMCs. However, the overall
trends of microarray profiles of T-MSC-derived myoblasts and
myocytes were similar to those of hSKMCs and quite different from
those of undifferentiated T-MSCs (clusters 2 and 3) (Fig. 1G). Furthermore, myogenically
differentiated T-MSCs showed a trend of increase or decrease of
selected muscle-associated gene expression, similar to hSKMCs
(Fig. 1H).
Enrichment of functional categories
within muscle metabolism-related genes
The present study classified 3,755 genes into
functional categories according to muscle metabolism. Ultimately,
19 known genes related to muscle differentiation were selected,
which were upregulated and downregulated in T-MSC-derived myogenic
cells and hSKMCs compared with T-MSCs (Tables I and II). These genes were: adenylate kinase
3-like 1 (AK3L1); actin-related protein 2/3 complex, subunit
1B (ARPC1B); supervillin (SVIL); formin-binding
protein 1-like (FNBP1L); angiogenin (ANG); calcium
channel, voltage-dependent, T-type, a1H subunit (CACNA1H);
sortilin 1 (SORT1); myosin, light chain 5 (MYL5);
sorbin and SH3 domain containing 2 (SORBS2); abhydrolase
domain containing 5 (ABHD5); v-abl Abelson murine leukemia
viral oncogene homolog 2 (ABL2); ADAM metallopeptidase
domain 12 (ADAM12); ADAM metallopeptidase with
thrombospondin type 1 motif 6 (ADAMTS6); α-fetoprotein
(AFP); neuroregulin 1 (NRG1); adrenergic, β2,
receptor, surface (ADRB2); (fibroblast growth factor 2
(FGF2); potassium inwardly-rectifying channel, subfamily J,
member 8 (KCNJ8); myosin, light chain 9, regulatory
(MYL9). These are related to muscle cell differentiation,
muscle tissue and organ development, autophagy, regulation of
muscle organ development, myofibrils, actin cytoskeleton, and
negative regulation of smooth muscle cell proliferation. Moreover,
the expression level (fold-change) in the T-MSCs of those genes was
found to be different in the three cell types (P<0.05; LPE
test). Thus, we were able to verify that the properties of the
T-MSC-derived myocytes were more similar to those of hSKMCs than
T-MSC-derived myoblasts. In particular, formin binding protein
1-like (FNBP1L), which has an essential role in antibacterial
autophagy, was selected among the upregulated genes, since
autophagy is related to skeletal muscle metabolism and myogenesis.
Furthermore, FNBP1L has not been reported in autophagy signaling in
stem cells. As a result, we demonstrated the effect of autophagy in
the sequential differentiating of T-MSCs toward skeletal myocytes
in vitro.
 | Table IList of upregulated myogenic genes in
the T-MSC-derived myocytes. |
Table I
List of upregulated myogenic genes in
the T-MSC-derived myocytes.
| Fold-change (per
T-MSC)
|
|---|
| Functional
category | Gene | Accession no.
symbol | Gene name | T-MSC-derived
myoblasts | T-MSC-derived
myocytes | SKMCs |
|---|
| Muscle tissue | AK3L1 | NM_203464.1 | Adenylate kinase
3-like 1 | 1.39 | 1.63 | 3.89 |
| development | ARPC1B | NM_005720.2 | Actin related
protein 2/3 complex, subunit 1B | 1.18 | 1.67 | 2.71 |
| SVIL | NM_003174.3 | Supervillin | 2.43 | 2.43 | 2.77 |
| Autophagy | FNBP1L | NM_001024948.1 | Formin binding
protein 1-like | 1.95 | 3.49 | 12.97 |
| Negative regulation
of smooth muscle cell proliferation | ANG | NM_003174.3 | Angiogenin,
ribonuclease, RNase A family, 5 | 1.21 | 1.88 | 2.45 |
| Muscle cell
differentiation | CACNA1H | NM_021098.2 | Calcium channel,
voltage-dependent, T type, α 1H subunit | 1.93 | 2.45 | 2.02 |
| SORT1 | NM_002959.4 | Sortilin 1 | 1.76 | 2.35 | 2.33 |
| Actin
cytoskeleton | MYL5 | NM_002477.1 | Myosin, light chain
5 | 1.95 | 2.02 | 1.56 |
| SORBS2 | NM_003603.4 | Sorbin and SH3
domain containing 2 | 1.57 | 4.04 | 30.82 |
 | Table IIList of downregulated myogenic genes
in the T-MSC-derived myocytes. |
Table II
List of downregulated myogenic genes
in the T-MSC-derived myocytes.
| Functional
category | Gene | Accession no.
symbol | Gene name | Fold-change (per
T-MSC)
|
|---|
| T-MSC-derived
myoblasts | T-MSC-derived
myocytes | SKMCs |
|---|
| Muscle tissue
development | ABHD5 | NM_016006.3 | Abhydrolase domain
containing 5 | −2.09 | −2.14 | −2.37 |
| ABHD5 | NM_007314.2 | v-abl Abelson
murine leukemia viral oncogene homolog 2 | −1.38 | −1.69 | −2.06 |
| ADAM12 | NM_021641.2 | ADAM
metallopeptidase 12 domain | −1.40 | −1.76 | −2.16 |
| ADAMTS6 | NM_197941.2 | ADAM
metallopeptidase with thrombospondin type 1 motif 6 | −1.84 | −2.07 | −2.38 |
| AFP | NM_001134.1 | α-fetoprotein | −1.41 | −2.03 | −2.87 |
| NRG1 | NM_013962.2 | Neuregulin 1 | −2.42 | −2.41 | −3.70 |
| Regulation of
muscle organ | ADRB2 | NM_000024.3 | Adrenergic, β-2-,
receptor, surface | −4.02 | −4.50 | −5.21 |
| development | FGF2 | NM_002006.3 | Fibroblast growth
factor 2 (basic) | −1.95 | −2.04 | −5.30 |
| Myofibril | KCNJ8 | NM_004982.2 | Potassium
inwardly-rectifying channel, subfamily J, member 8 | −1.67 | −2.42 | −4.31 |
| Actin
cytoskeleton | MYL9 | NM_006097.3 | Myosin, light chain
9, regulatory | 2.00 | 1.34 | −2.15 |
Confirmation of gene expression profiles
of T-MSC-derived myogenic cells
To confirm the microarray findings and their
relationship with autophagy, qPCR was analyzed. Consistent with the
results of the microarray analysis, candidate genes associated with
myogenic differentiation such as SVIL (22), SORT1 (23), ANG (24) and FNBP1L (25), which are muscle metabolism-related
genes, were upregulated in the T-MSC-derived myoblasts and
myocytes, but not in the undifferentiated T-MSCs with the exception
of SORT1. The expression of SORT1 was only upregulated in
T-MSC-derived myocytes (Fig. 2A).
In particular, FNBP1L was expected to be the instrument for
tracking the relationship between autophagy and differentiation
into myocytes. We therefore confirmed the higher expression of
autophagy-related genes such as beclin-1, Atg5, Atg3
and Atg14 in the T-MSC-derived myocytes compared with
expression in the T-MSCs or T-MSC-derived myoblasts (Fig. 2B). Taken together, T-MSC-derived
myogenic cells appeared to have characteristics similar to
differentiated hSKMCs and affect the mechanism of autophagy.
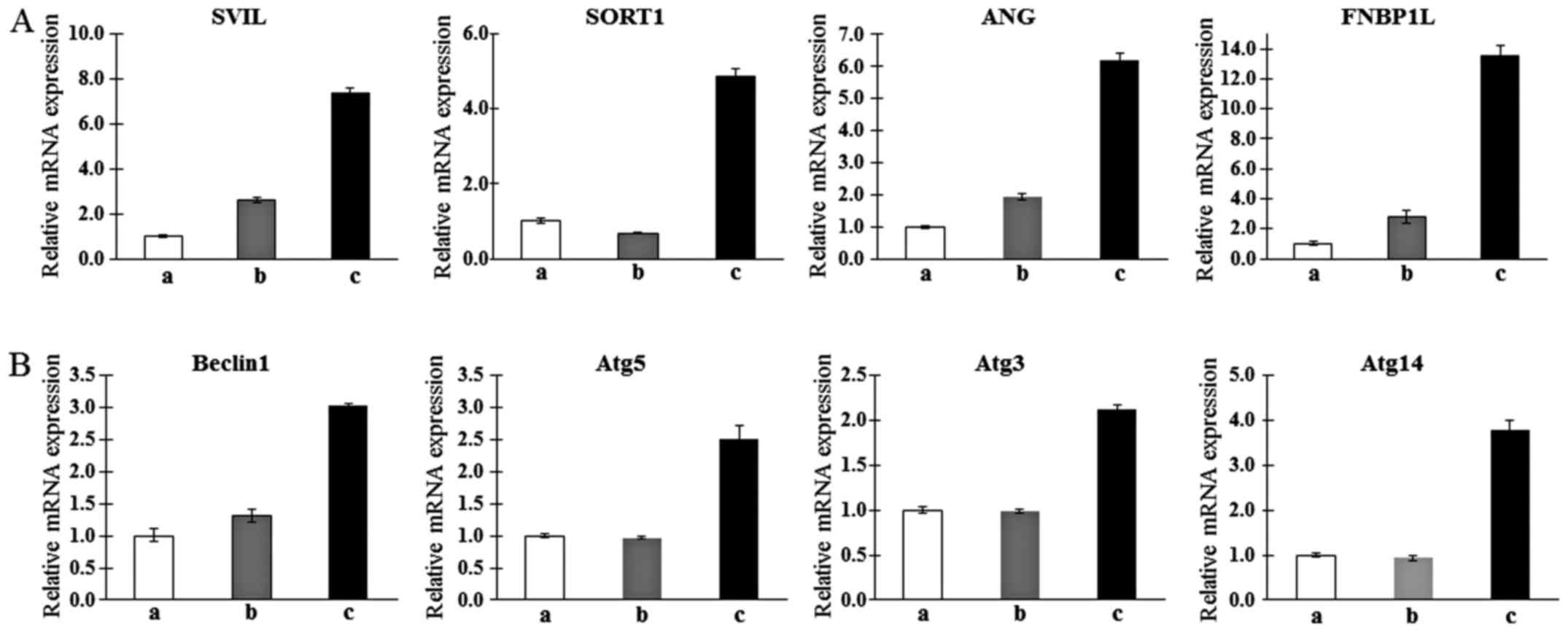 | Figure 2Determination of the mRNA expression
levels of upregulated muscle metabolism (A) and autophagy-related
markers (B) as myogenic markers in human T-MSCs during the process
of myogenic induction (a, T-MSCs; b, T-MSC-derived myoblasts; c,
T-MSC-derived myocytes). GAPDH served as a loading control.
Data are expressed as mean ± standard error of the mean (SEM).
Triplicate independent mRNA samples were used in an RT-PCR
experiment. T-MSCs, tonsil-derived mesenchymal stem cells;
SVIL, supervillin; SORT1, sortilin 1; ANG,
angiogenin; FNBP1L, formin binding protein 1-like;
Atg5, autophagy-related gene 5; Atg3,
autophagy-related gene 3; Atg14, autophagy-related gene 14;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase. |
Expression of autophagy-related genes
under states that induce myogenic differentiation
As determine by western blotting, expression of
Beclin-1 and Atg5 was increased gradually from T-MSC-derived
myoblasts to myocytes (Fig. 3A, C, E
and G). However, the expression of Bcl-2, which binds with
beclin-1 resulting in the inhibition of autophagic initiation
(26), was decreased at all
stages of differentiation (Fig. 3B
and F). The expression of LC3II, which is a well-characterized
protein specifically localized to autophagic structures throughout
the process from phagophores to lysosomal degradation (27), was increased gradually from
T-MSC-derived myoblasts to myocytes, but was not significantly
different between the T-MSC-derived myoblast and myocyte stages
(Fig. 3D and H). These results
indicate that myogenic differentiation of T-MSCs is associated with
the autophagic pathway.
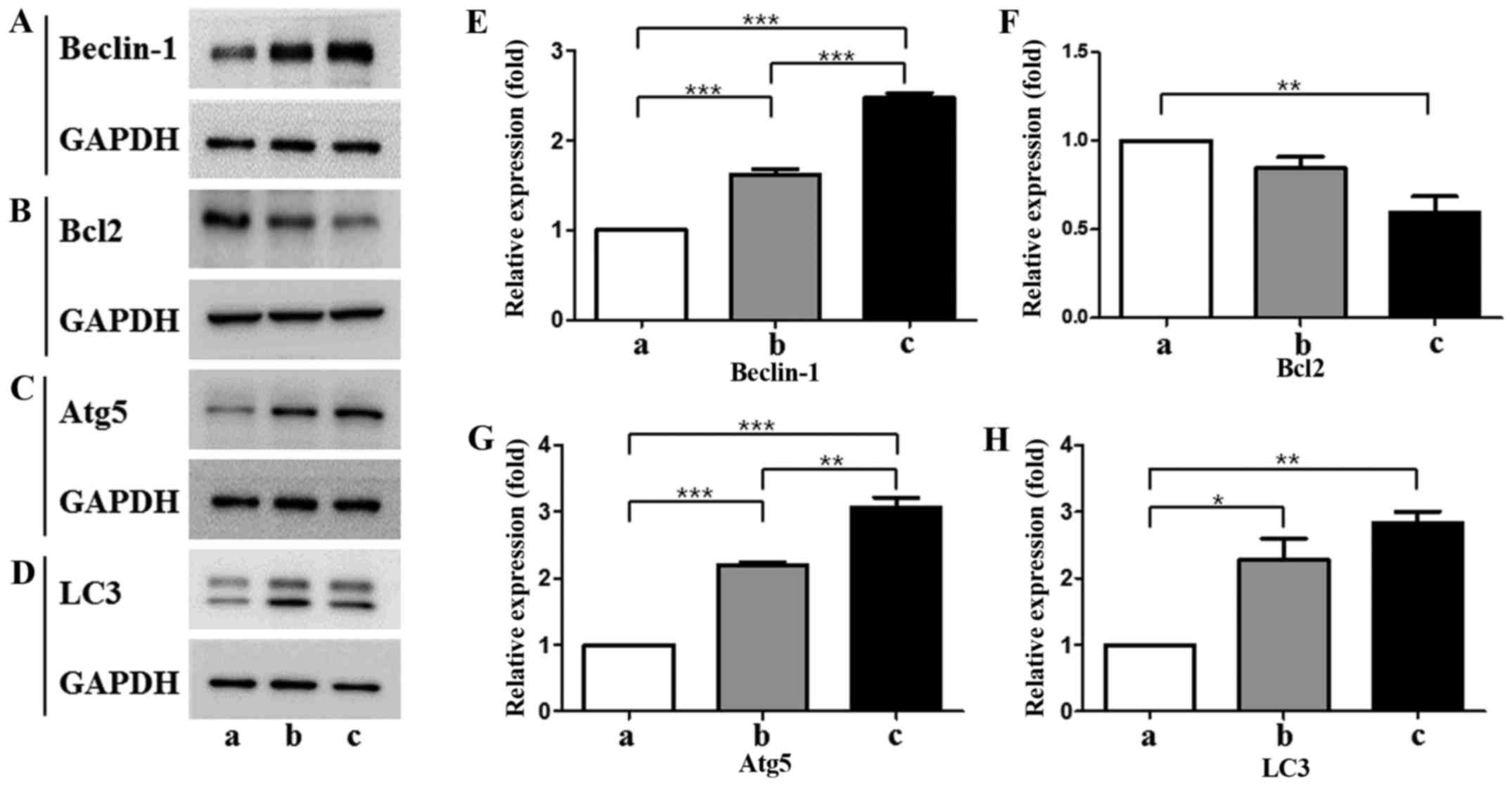 | Figure 3(A–D) Protein expression levels and
(E–H) quantification of autophagic markers in T-MSCs during the
process of myogenic induction (a, T-MSCs; b, T-MSC-derived
myoblasts; c, T-MSC-derived myocytes). The levels of GAPDH were
measured as a loading control (A–D). All signals were analyzed by
densitometric scanning [(E–H) LAS-3000; Fujifilm, Tokyo, Japan)].
Data are expressed as means ± SEM of experiments performed in
triplicate. *P<0.05, **P<0.01 and
***P<0.001. T-MSCs, human tonsil-derived mesenchymal
stem cells; Bcl-2, B cell lymphoma 2; Atg5,
autophagy-related gene 5; LC3, microtubule-associated protein
1A/1B-light chain 3; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase. |
Detection of the role of autophagy in
T-MSC-derived myocytes by immunostaining
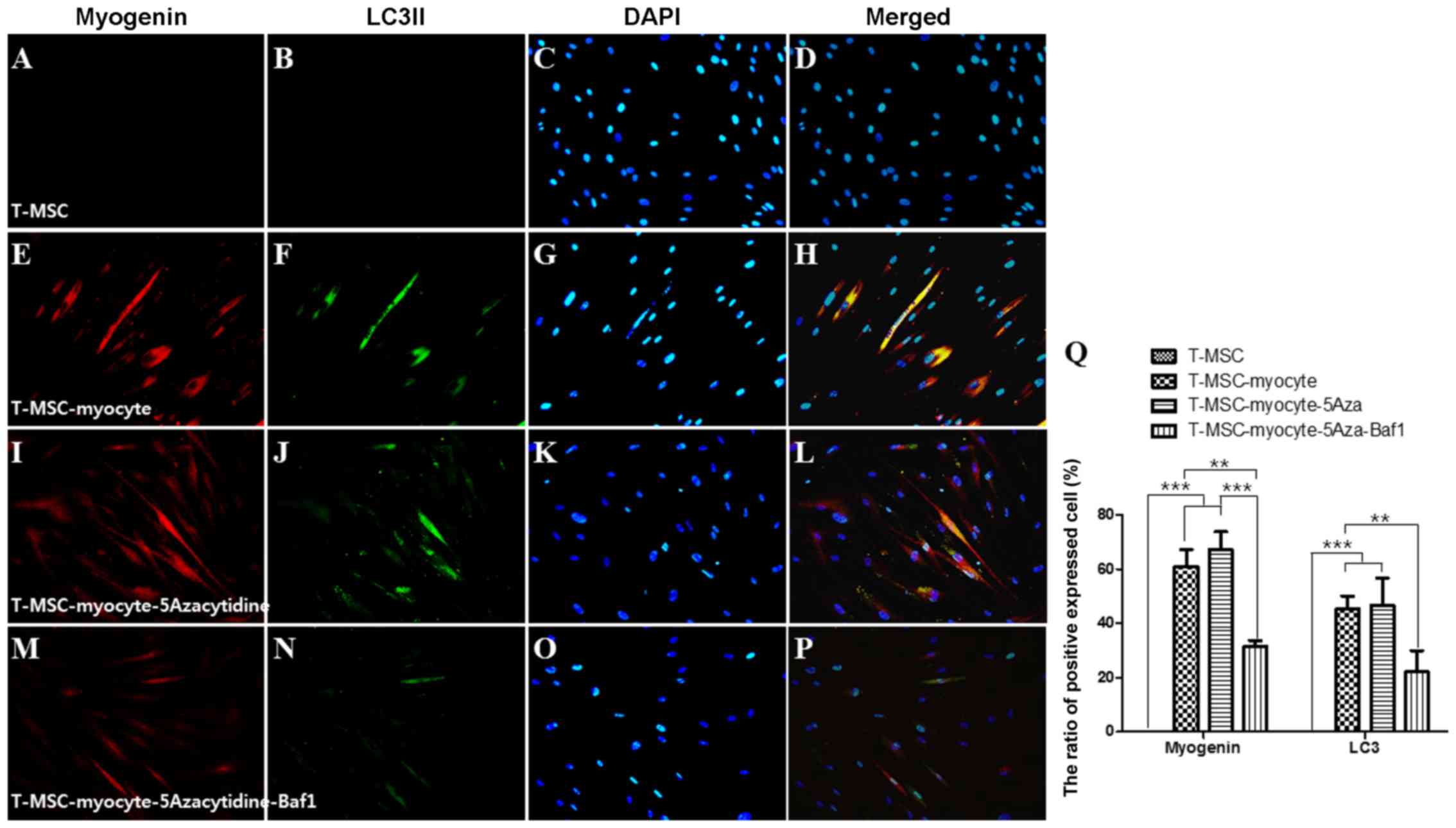
We used immunocytochemistry to investigate whether
autophagy participates in myogenic differentiation of T-MSCs. To
measure their innate ability to differentiate into myocytes,
T-MSC-derived myoblasts were plated for myogenic differentiation to
the skeletal myocytes onto coverslips and cultured sequentially in
myogenic differentiation medium, and treated with an autophagy
inducer (5-azacytidine) and inhibitor (Baf1). These cells were then
fixed and co-labeled with antibodies against the markers of
autophagy (LC3) and skeletal myogenic cells (MHC, myogenin,
desmin), respectively. Following differentiation into myocytes,
60-70% of the cells expressed MHC (Fig. 4E), myogenin (Fig. 5E), and desmin (Fig. 6E), and 45–55% of the cells
co-expressed autophagic marker LC3II (Figs. 4F, 5F and 6F). 5-Azacytidine was added to
T-MSC-derived myoblasts to activate autophagy for powerful myogenic
differentiation. When treated with 5-azacytidine, the expression of
myogenic markers was increased (67–77%) (Figs. 4I, 5I and 6I), while LC3 was not significantly
increased (40–55%)(Figs. 4J,
5J and 6J). By contrast, the expression of
myogenic markers was decreased after treatment with Baf1 during the
differentiation of T-MSC-derived myocytes (13–32%) (Figs. 4M, 5M and 6M) and LC3II was significantly more
decreased than in T-MSC-derived myocytes and T-MSC-derived myocytes
treated with 5-azacytidine (16–23%) (Figs. 4N, 5N and 6N). These results showed that autophagy
has an effect on the mechanism of skeletal myogenic differentiation
in T-MSCs.
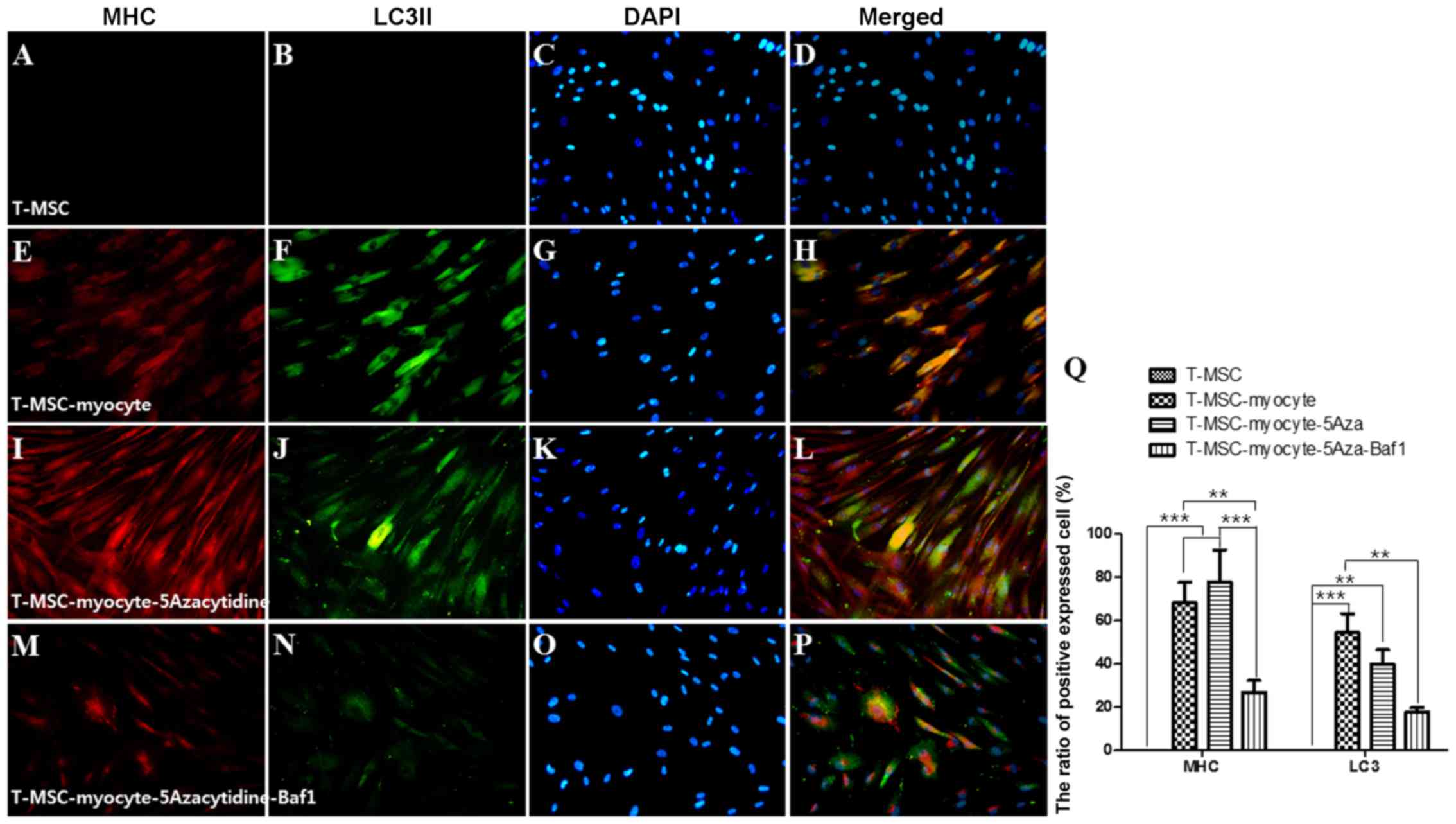 | Figure 4Effect of 5Aza and Baf1 on the
skeletal myogenic differentiation of T-MSCs. The expression of MHC
(red) and LC3II (green) in the T-MSCs (A–D) during the process of
myogenic differentiation [T-MSC-derived myocytes (E–H)]. The role
of autophagy was evaluated with an inducer, 5Aza [T-MSC-derived
myocytes-5-azacytidine (I–L)], and an inhibitor, Baf1
[T-MSC-derived myocytes-5-azacytidine-Baf1 (M–P)], during skeletal
myogenic differentiation. The cells were counterstained with DAPI
(blue). The samples were analyzed under a fluorescence microscope
using appropriate filters. Original magnification, ×200. (Q) The
ratio of cells immunoreactive for anti-MHC and anti-LC3. Data are
expressed as means ± SEM. For each condition, four slides were used
for quantification. Graphs represent the average of multiple tests
from three independent experiments. 5Aza, 5-azacytidine; Baf1,
bafilomycin A1; MHC, myosin heavy chain; LC3II,
microtubule-associated protein 1A/1B-light chain 3; T-MSCs,
tonsil-derived mesenchymal stem cells. |
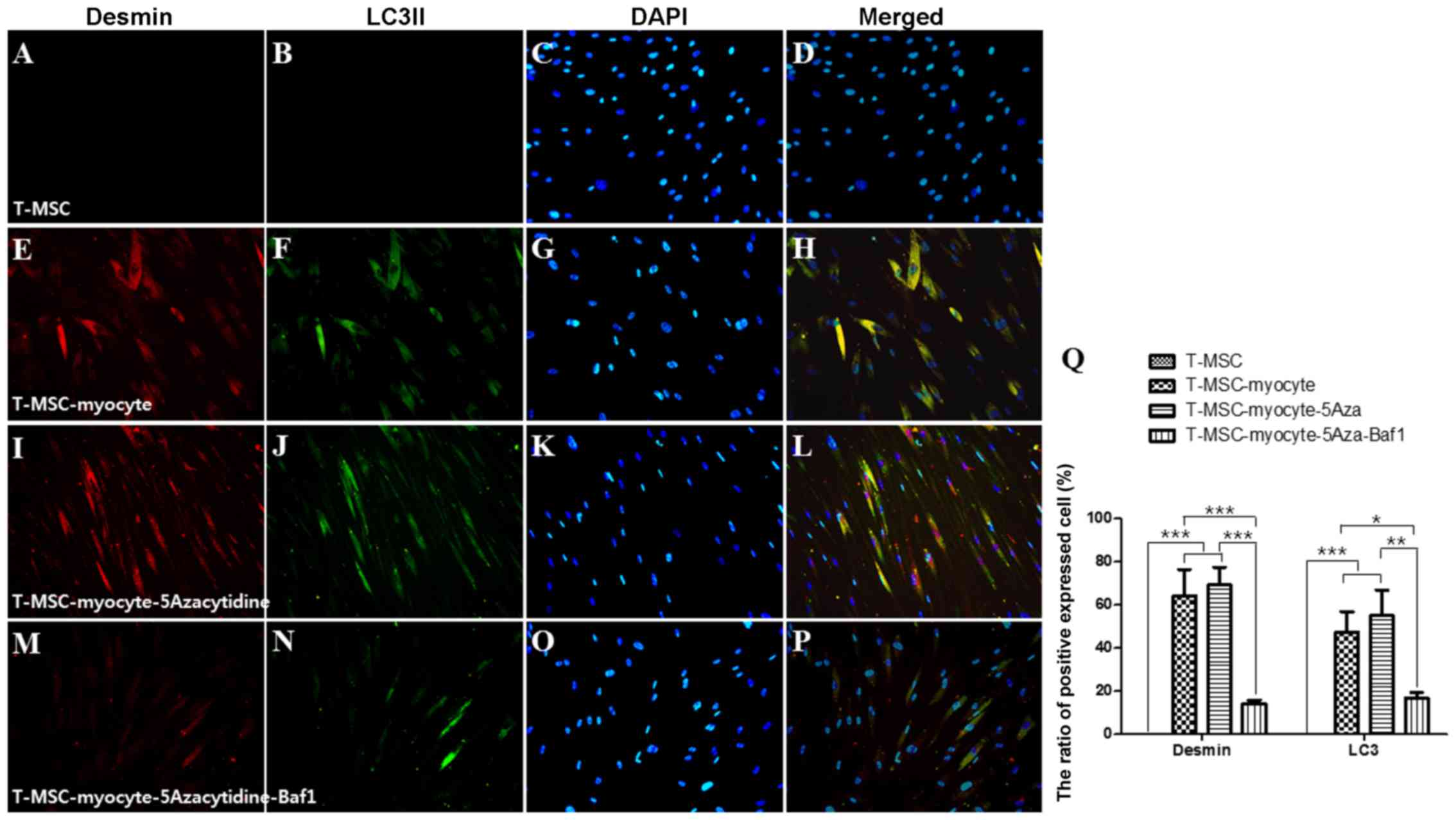 | Figure 6Effect of 5Aza and Baf1 on the
skeletal myogenic differentiation of human T-MSCs. The expression
of desmin and LC3II (green) in the T-MSCs (A–D) during the process
of myogenic differentiation [T-MSC-derived myocytes; (E–H)]. The
role of autophagy was evaluated with an inducer, 5Aza
[T-MSC-derived myocytes-5-azacytidine (I–L)], and an inhibitor,
Baf1 [T-MSC-derived myocytes-5-azacytidine-Baf1 (M–P)], during
skeletal myogenic differentiation. The cells were counterstained
with DAPI (blue). The samples were analyzed under a fluorescence
microscope using appropriate filters. Original magnification, ×200.
(Q) The ratio of cells immunoreactive for anti-desmin and anti-LC3.
Data are expressed as means ± SEM. For each condition, four slides
were used for quantification. Graphs represent the average of
multiple tests from three independent experiments. 5Aza,
5-azacytidine; Baf1, bafilomycin A1; LC3II, microtubule-associated
protein 1A/1B-light chain 3; Baf1, bafilomycin A1; T-MSCs,
tonsil-derived mesenchymal stem cells. |
Discussion
Autophagy plays an essential role in the
differentiation of human bone marrow MSCs into osteocytes and
adipocytes (18). Mitophagy is
required for mitochondrial biogenesis and myogenic differentiation
of C2C12 myoblasts (19). The
microarray analysis followed by immunocytochemistry used in the
present study demonstrated that autophagy is related to the
differentiation of T-MSCs into myocytes.
Microarray analysis revealed the potential for the
differentiation of T-MSCs into skeletal myocytes. All clusters
expressed cell proliferation and motion-related genes in common
(Fig. 1G). In cluster 2, the
muscle cell differentiation and tissue development-related genes
were highly expressed, and the gene expression was gradually
increased from the differentiated T-MSCs to SKMCs. The genes of
cluster 3 showed a tendency for decreased expression and were
associated with the cell cycle or components within the cell and
smooth or cardiac muscle markers. Related to cell cycle progression
and metabolism, these genes are downregulated upon the initiation
of myogenic differentiation (28). The genes related to mesenchymal or
MSC development or gastrulation were particularly expressed in
cluster 4, which showed a pattern of high gene expression except
for SKMCs. It can be assumed that the genes of cluster 4 are
associated with the characteristics of stem cells. In addition, all
the clusters, except cluster 4, expressed mesoderm or
angiogenesis-related genes. In cluster 1, gene expression was
increased in the differentiating T-MSCs (T-MSC-derived myoblasts
and myocytes) and only the TGF-β or collagen gene was expressed.
TGF-β and collagen were used to differentiate myoblasts in the
present study. TGF-β and collagen have a role in the
differentiation of the stem cells to myoblasts (1,29,30).
We selected 9 upregulated genes and 10 downregulated
genes in the T-MSC-derived myogenic cells and hSKMCs compared with
T-MSCs as related to muscle differentiation (Tables I and II). Among the 19 genes selected,
several genes (SVIL, SORT1, ANG and
FNBP1L) appeared as important factors in the mechanism of
skeletal myogenic differentiation, as the RT-PCR analysis data were
consistent with the microarray data. The expression of all of these
genes was significantly increased in the differential stage in the
T-MSC-derived myocytes. Supervillin (SVIL) is implicated in the
direct or indirect control of cell adhesion (22) and localizes within nuclei and with
dystrophin at costameres, regions of F-actin membrane attachment in
skeletal muscle (31).
Archvillin, a muscle-specific isoform of SVIL, is among the first
costameric proteins to assemble during myogenesis and it
contributes to myogenic membrane structure and differentiation.
Sortilin 1 (SORT1) was originally purified from human brain
extracts and has been identified as a major component of
GLUT4-containing vesicles from rat adipocytes (23). SORT1 also has functional roles in
the development of the insulin-responsive glucose uptake system in
muscle cells (32). Angiogenin
(ANG) is involved in blood vessel formation. However, ANG was
originally shown to bind to a 42-kDa binding protein and a smooth
muscle-type α-actin before the identification of endothelial
receptors. ANG has been demonstrated in human arterial smooth
muscle cell proliferation, through the binding of α-actinin-2,
which is traditionally designated as an SKM isoform due to its
presence in both skeletal and cardiac muscles (24,33). FNBP1L is essential for
antibacterial autophagy and bridges the autophagic membrane
extension machinery and its bacterial cargo. An autophagosome is
built around bacterial cargo by a coordinated series of autophagy
protein complexes; therefore, the HR1 domain of FNBP1L is
biochemically essential for its interaction with human Atg3 protein
(25). The pathway of these genes
(FNBP1L and Atg3) and Atg5 causes the
Atg12-Atg5 conjugate to interact with Atg3 (34), which is consistent with our
results in which the gene expression was increased in T-MSC-derived
myocytes (Fig. 2).
Nutrients are the main physiological regulators and
initiators of autophagy; therefore, the lack of nutrients is an
important inducer of SKM autophagy (35). In the present study, we used a
method that minimizes serum in the culture medium to differentiate
stem cells into skeletal myocytes. Autophagy is induced by such a
fasting method and in turn promotes myogenic differentiation.
5-Azacytidine is already used as a factor for the differentiation
of muscle stem cells into muscle cells, and autophagy has relevance
to myogenic differentiation (36,37).
Three main pathways (macrophagy, microphagy and
chaperone-mediated autophagy) are involved in autophagy and these
are mediated by autophagy-related genes and their enzymes (38). Levels of proteins including
Beclin-1, Bcl-2, LC3 and Atg5 were increased and decreased in the
present study, and they are the key factors in the autophagy signal
pathway related to macrophagy. Depending on the myogenic
differentiation of T-MSCs, the expression of Beclin-1, LC3 and Atg5
was increased, while Bcl-2 was decreased. LC3 was coexpressed with
myogenic markers (MHC, myogenin and desmin) in the T-MSC-derived
myocytes, which was confirmed by the treatment of T-MSC-derived
myocytes with an inducer and an inhibitor. The role of autophagy as
a metabolic regulator in muscle is well known. In particular,
macrophagy represents the physiological process that skeletal
muscle uses to transport cytoplasm, organelles, and proteins to
lysosomes for degradation (39).
Various SKM diseases that cause atrophy and dystrophy have shed
light on a significant common feature, namely the build-up of
autophagosomes within myofibers (40). Comparing our study with these
previous reports, autophagy appears to influence skeletal myogenic
differentiation of T-MSCs.
In the present study, we classified genes into
functional categories according to muscle metabolism, and specific
genes related to muscle differentiation were selected in the
T-MSC-derived myogenic cells and hSKMCs. Functional studies
demonstrated that autophagy plays a role during the differentiation
of T-MSCs into hSKMCs. Furthermore, as a result of treatment of
T-MSC-derived myocytes with an autophagy inducer and inhibitor, we
were able to reconfirm that autophagy has an effect on the
mechanism of myogenic differentiation of T-MSCs. Our study suggests
that autophagy plays an important role during the differentiation
of T-MSCs into skeletal myogenic cells and knowledge of this
mechanism is valuable for treating SKM injuries and the damage
caused by other degenerative disorders, including congenital
defects, trauma or tumor removal.
Acknowledgments
The present study was supported by a grant
(HI12C0135) from the Korean Health Technology R&D Project,
Ministry of Health and Welfare, Republic of Korea and intramural
research promotion grants from Ewha Womans University School of
Medicine.
References
|
1
|
Ryu KH, Cho KA, Park HS, Kim JY, Woo SY,
Jo I, Choi YH, Park YM, Jung SC, Chung SM, et al: Tonsil-derived
mesenchymal stromal cells: Evaluation of biologic, immunologic and
genetic factors for successful banking. Cytotherapy. 14:1193–1202.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park S, Choi Y, Jung N, Yu Y, Ryu KH, Kim
HS, Jo I, Choi BO and Jung SC: Myogenic differentiation potential
of human tonsil-derived mesenchymal stem cells and their potential
for use to promote skeletal muscle regeneration. Int J Mol Med.
37:1209–1220. 2016.PubMed/NCBI
|
|
3
|
Dezawa M, Ishikawa H, Itokazu Y, Yoshihara
T, Hoshino M, Takeda S, Ide C and Nabeshima Y: Bone marrow stromal
cells generate muscle cells and repair muscle degeneration.
Science. 309:314–317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Di Rocco G, Iachininoto MG, Tritarelli A,
Straino S, Zacheo A, Germani A, Crea F and Capogrossi MC: Myogenic
potential of adipose-tissue-derived cells. J Cell Sci.
119:2945–2952. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim JA, Shon YH, Lim JO, Yoo JJ, Shin HI
and Park EK: MYOD mediates skeletal myogenic differentiation of
human amniotic fluid stem cells and regeneration of muscle injury.
Stem Cell Res Ther. 4:1472013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nunes VA, Cavaçana N, Canovas M, Strauss
BE and Zatz M: Stem cells from umbilical cord blood differentiate
into myotubes and express dystrophin in vitro only after exposure
to in vivo muscle environment. Biol Cell. 99:185–196. 2007.
View Article : Google Scholar
|
|
7
|
Park S, Kim E, Koh SE, Maeng S, Lee WD,
Lim J, Shim I and Lee YJ: Dopaminergic differentiation of neural
progenitors derived from placental mesenchymal stem cells in the
brains of Parkinson's disease model rats and alleviation of
asymmetric rotational behavior. Brain Res. 1466:158–166. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kerkis I, Kerkis A, Dozortsev D,
Stukart-Parsons GC, Gomes Massironi SM, Pereira LV, Caplan AI and
Cerruti HF: Isolation and characterization of a population of
immature dental pulp stem cells expressing OCT-4 and other
embryonic stem cell markers. Cells Tissues Organs. 184:105–116.
2006. View Article : Google Scholar
|
|
9
|
Husmann I, Soulet L, Gautron J, Martelly I
and Barritault D: Growth factors in skeletal muscle regeneration.
Cytokine Growth Factor Rev. 7:249–258. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang K, Wang C, Xiao F, Wang H and Wu Z:
JAK2/STAT2/STAT3 are required for myogenic differentiation. J Biol
Chem. 283:34029–34036. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Conboy IM and Rando TA: The regulation of
Notch signaling controls satellite cell activation and cell fate
determination in postnatal myogenesis. Dev Cell. 3:397–409. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fortini P, Ferretti C, Iorio E, Cagnin M,
Garribba L, Pietraforte D, Falchi M, Pascucci B, Baccarini S,
Morani F, et al: The fine tuning of metabolism, autophagy and
differentiation during in vitro myogenesis. Cell Death Dis.
7:e21682016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maley MA, Fan Y, Beilharz MW and Grounds
MD: Intrinsic differences in MyoD and myogenin expression between
primary cultures of SJL/J and BALB/C skeletal muscle. Exp Cell Res.
211:99–107. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miller KJ, Thaloor D, Matteson S and
Pavlath GK: Hepatocyte growth factor affects satellite cell
activation and differentiation in regenerating skeletal muscle. Am
J Physiol Cell Physiol. 278:C174–C181. 2000.PubMed/NCBI
|
|
15
|
Heszele MF and Price SR: Insulin-like
growth factor I: The yin and yang of muscle atrophy. Endocrinology.
145:4803–4805. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakanishi K, Dohmae N and Morishima N:
Endoplasmic reticulum stress increases myofiber formation in vitro.
FASEB J. 21:2994–3003. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Levine B and Klionsky DJ: Development by
self-digestion: Molecular mechanisms and biological functions of
autophagy. Dev Cell. 6:463–477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nuschke A, Rodrigues M, Stolz DB, Chu CT,
Griffith L and Wells A: Human mesenchymal stem cells/multipotent
stromal cells consume accumulated autophagosomes early in
differentiation. Stem Cell Res Ther. 5:1402014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sin J, Andres AM, Taylor DJ, Weston T,
Hiraumi Y, Stotland A, Kim BJ, Huang C, Doran KS and Gottlieb RA:
Mitophagy is required for mitochondrial biogenesis and myogenic
differentiation of C2C12 myoblasts. Autophagy. 12:369–380. 2016.
View Article : Google Scholar :
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Yu Y, Park YS, Kim HS, Kim HY, Jin YM,
Jung SC, Ryu KH and Jo I: Characterization of long-term in vitro
culture-related alterations of human tonsil-derived mesenchymal
stem cells: Role for CCN1 in replicative senescence-associated
increase in osteogenic differentiation. J Anat. 225:510–518. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pestonjamasp KN, Pope RK, Wulfkuhle JD and
Luna EJ: Supervillin (p205): A novel membrane-associated,
F-actin-binding protein in the villin/gelsolin superfamily. J Cell
Biol. 139:1255–1269. 1997. View Article : Google Scholar
|
|
23
|
Morris NJ, Ross SA, Lane WS, Moestrup SK,
Petersen CM, Keller SR and Lienhard GE: Sortilin is the major
110-kDa protein in GLUT4 vesicles from adipocytes. J Biol Chem.
273:3582–3587. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu H, Gao X, Sun Y, Zhou J, Yang M and Xu
Z: Alpha-actinin-2, a cytoskeletal protein, binds to angiogenin.
Biochem Biophys Res Commun. 329:661–667. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huett A, Ng A, Cao Z, Kuballa P, Komatsu
M, Daly MJ, Podolsky DK and Xavier RJ: A novel hybrid yeast-human
network analysis reveals an essential role for FNBP1L in
antibacterial autophagy. J Immunol. 182:4917–4930. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marquez RT and Xu L: Bcl-2:Beclin 1
complex: Multiple, mechanisms regulating autophagy/apoptosis toggle
switch. Am J Cancer Res. 2:214–221. 2012.PubMed/NCBI
|
|
27
|
Nakatogawa H, Suzuki K, Kamada Y and
Ohsumi Y: Dynamics and diversity in autophagy mechanisms: Lessons
from yeast. Nat Rev Mol Cell Biol. 10:458–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moran JL, Li Y, Hill AA, Mounts WM and
Miller CP: Gene expression changes during mouse skeletal myoblast
differentiation revealed by transcriptional profiling. Physiol
Genomics. 10:103–111. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Osses N and Brandan E: ECM is required for
skeletal muscle differentiation independently of muscle regulatory
factor expression. Am J Physiol Cell Physiol. 282:C383–C394. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hinz B, Phan SH, Thannickal VJ, Galli A,
Bochaton-Piallat ML and Gabbiani G: The myofibroblast: One
function, multiple origins. Am J Pathol. 170:1807–1816. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oh SW, Pope RK, Smith KP, Crowley JL, Nebl
T, Lawrence JB and Luna EJ: Archvillin, a muscle-specific isoform
of supervillin, is an early expressed component of the costameric
membrane skeleton. J Cell Sci. 116:2261–2275. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ariga M, Nedachi T, Katagiri H and Kanzaki
M: Functional role of sortilin in myogenesis and development of
insulin-responsive glucose transport system in C2C12 myocytes. J
Biol Chem. 283:10208–10220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kostin S, Hein S, Arnon E, Scholz D and
Schaper J: The cytoskeleton and related proteins in the human
failing heart. Heart Fail Rev. 5:271–280. 2000. View Article : Google Scholar
|
|
34
|
Sakoh-Nakatogawa M, Matoba K, Asai E,
Kirisako H, Ishii J, Noda NN, Inagaki F, Nakatogawa H and Ohsumi Y:
Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging
its catalytic site. Nat Struct Mol Biol. 20:433–439. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Neel BA, Lin Y and Pessin JE: Skeletal
muscle autophagy: A new metabolic regulator. Trends Endocrinol
Metab. 24:635–643. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wakitani S, Saito T and Caplan AI:
Myogenic cells derived from rat bone marrow mesenchymal stem cells
exposed to 5-azacytidine. Muscle Nerve. 18:1417–1426. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng JK, Wang Y, Karandikar A, Wang Q,
Gai H, Liu AL, Peng C and Sheng HZ: Skeletal myogenesis by human
embryonic stem cells. Cell Res. 16:713–722. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mizushima N, Ohsumi Y and Yoshimori T:
Autophagosome formation in mammalian cells. Cell Struct Funct.
27:421–429. 2002. View Article : Google Scholar
|
|
39
|
Jackson MJ: Reactive oxygen species and
redox-regulation of skeletal muscle adaptations to exercise. Philos
Trans R Soc Lond B Biol Sci. 360:2285–2291. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Malicdan MC, Noguchi S, Nonaka I, Saftig P
and Nishino I: Lysosomal myopathies: An excessive build-up in
autophagosomes is too much to handle. Neuromuscul Disord.
18:521–529. 2008. View Article : Google Scholar : PubMed/NCBI
|