Introduction
Breast cancer is a hormone-dependent disease of
complex multifactorial etiology and is the main cause of death in
females due to cancer. Approximately 1.15 million new cases of
breast cancer are diagnosed every year and it accounts for 14% of
deaths due to cancer in women worldwide (1). The recent trend of a decreased rate
of death due to breast cancer is the result of improvements in the
early diagnosis of the disease, although the therapeutic options
for more advanced stages are still quite limited. Docetaxel, a
taxane with antimicrotubule activity, is a very effective drug for
the treatment of metastatic breast cancer. The use of adjuvant
taxane therapy also offers significant benefits for patients with
early stage breast cancer (2).
Despite the effectiveness of docetaxel in the treatment of breast
cancer, the exact mechanisms and/or markers associated with
chemosensitivity or resistance to docetaxel remain unclear
(3).
The PKC apoptosis WT1 regulator (PAWR) gene, also
called prostate apoptosis response 4 (PAR4), encodes a 342-amino
acid pro-apoptotic protein that plays a central role in cancer cell
survival and can be considered a candidate for tumor cell-selective
therapy (4). PAR4 protein
comprises two nuclear localization signals (NLS): a leucine zipper
domain at its carboxy terminal region, and a selective for
apoptosis induction in cancer cells (SAC) domain in the central
region of the molecule (5,6).
PAR4 plays a role in both the intrinsic and
extrinsic apoptotic pathways. Intracellular PAR4 induces apoptosis
due to its ability to activate the Fas/FasL/FADD/caspase 8 pathway
by inhibiting the NF-κB survival pathway, which requires
PKA-mediated PAR4 phosphorylation at the T155 residue, and by
downregulation of Bcl-2 expression (7–9).
On the other hand, PAR4 protein is spontaneously secreted by normal
and tumor cells in culture via the classical endoplasmic reticulum
(ER)/Golgi pathway, stimulated by stress-inducing factors of the
ER. The PAR4/GRP78 interaction induces apoptosis via the ER stress
pathway and activation of the FADD/caspase-8/caspase-3 pathway
(10).
PAR4 is expressed in cells from different tissues,
including mammary gland epithelial cells (11,12). Alterations in PAR4 expression have
been observed in prostate tumor (13), melanoma (14), renal tumor (15) and hematopoietic (16) cell lines. Decreased PAR4
expression has been demonstrated in different types of cancers,
including neuroblastoma (17),
endometrial cancer (18), renal
cell carcinoma (15), pancreatic
tumors (19), and breast cancer
(20,21), and PAR4-knockout mice were found
to develop spontaneous tumors in various tissues (22).
PAR4 expression is not enough to induce cell death,
but it increases the sensitivity of most tumor cells to secondary
apoptotic stimulus (8,23,24). Research indicates that PAR4
presents attractive anti-tumoral therapeutic potential, especially
due to its tumor cell selectivity. In mouse tumor models, PAR4
overexpression was found to result in tumor regression due to
activation of apoptosis via the Fas/FADD receptor (12). Zhao et al (25) also observed that transgenic mice
expressing a SAC domain are normal regarding their development and
life expectancy in addition to being resistant to tumor growth. In
a study on K562 myeloid (Bcr-Abl-positive) cells, PAR4
overexpression enhanced sensitivity to imatinib and inhibitors of
histone deacetylases (HDACs), increasing the apoptosis rate of
cells resistant to conventional treatment with doxorubicin and
agonist Trail and Fas (16).
Increased PAR4 expression also increased apoptosis induction in
response to ER stress agents in CAKi renal tumor cells through
decreased XIAP anti-apoptotic protein levels and p-Akt kinase
(26). An in vivo study by
Kline et al (27)
demonstrated the possible role of PAR4 in colon cancer treatment.
Mice receiving nanoliposomes containing the PAR4 expression vector
were more susceptible to 5-fluorouracil (5-FU) chemotherapy. Wang
et al (28) also showed
that PAR4 sensitized colon cancer cells to 5-FU inhibiting NF-κB
and deregulating the miRNA pathway. Increased expression of PAR3 in
SW480 and HT29 tumor cells led to interaction between PAR4 and
NF-κB in the cytoplasm, avoided nuclear translocation of p65 and
p50 subunits, and reduced cell survival. Furthermore, cells with
increased PAR4 expressed reduced levels of DROSHA, a regulator
protein in the miRNA pathway, leading to increased pro-apoptotic
(Bim) target translation and decreased translation of
anti-apoptotic targets, such as Bcl-2 (28).
In rats, Alvarez et al (29) demonstrated that treatment of
mammary tumors with adriamycin and cyclophosphamide, followed by
paclitaxel, led to significant tumor regression and that the tumors
that relapsed after chemotherapy exhibited markedly reduced PAR4
expression. The same was found for patients with breast cancer;
PAR4 levels were associated with increased recurrence over 5 years
compared to women with breast tumors expressing higher levels of
PAR4 (29). Previous results from
our group indicated that the expression of PAR4 modulates
proliferation and cell death and affects the response of MCF7
breast cancer cells to treatment with docetaxel (23). However, the possible mechanisms
involved in the PAR4-mediated chemosensitivity to docetaxel remain
largely unknown. In the present study, we provide evidence that
PAR4 modulates several genes involved in the wingless-type MMTV
integration 1 (WNT) signaling pathway that could take part in
docetaxel chemosensitivity in breast cancer cells.
Materials and methods
Cell line, plasmid transfection, and
docetaxel treatment
The MCF7 cell line derived from breast
adenocarcinoma is representative of luminal A breast tumors and was
obtained from the American Type Culture Collection
(ATCC®, HTB-22; Manassas, VA, USA). MCF7 cells were
stably transfected with pcCMV6-XL6-PAR4 plasmid expressing
the full-length PAR4 cDNA, named MCF7pcPAR4 or with empty
vector pcCMV6-XL6-NEO, named MCF7pcNEO (OriGene
Technologies, Inc., Rockville, MD, USA). Transfection was performed
using TurboFectin 8.0 (OriGene Technologies, Inc.) according to the
manufacturer's recommendations. Twenty-four hours after
transfection, the cells were selected with geneticin (Gibco,
Gaithersburg, MD, USA). The selected clones were screened for PAR4
mRNA and protein expression by real-time PCR and western blotting,
respectively. MCF7pcNEO and MCF7pcPAR4 cells were
treated with 100 nM docetaxel (BD Pharmingen, San Jose, CA, USA)
for 24 h for the cDNA microarray analysis or 50 nM docetaxel for 24
h for the PCR array analysis. The MCF7pcNEO and
MCF7pcPAR4 control cells were maintained with the vehicle
solution (absolute ethanol) for 24 h.
RNA extraction
Before and after docetaxel treatment for 24 h, total
RNA was extracted from the MCF7pcNEO and MCF7pcPAR4
cells using the acid guanidinium thiocyanate-phenol-chloroform
method (30). The RNA
concentration was determined using a NanoDrop ND-1000
spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The
sample integrity was also determined by RNA Nano Chips using the
Agilent 2100 Bioanalyzer platform (Agilent Technologies, Inc.,
Santa Clara, CA, USA). High quality RNA was purified using the
RNeasy Mini kit (Qiagen, Germantown, MD, USA).
cDNA microarray analysis
The cDNA microarray analysis was performed using the
Low Input RNA Linear Amplification kit 2-color (GE Healthcare
Bio-Sciences, Piscataway, NJ, USA) for the amplification of 200 ng
of purified RNA. T7 RNA polymerase was added to generate
complementary RNA (cRNA) and the test samples were labeled with
cyanine 5 (Cy-5) fluorochrome. The reference sample was labeled
with cyanine 3 (Cy-3) fluorochrome. The cRNAs were purified using
the RNeasy Mini kit (Qiagen, Germantown, MD, USA), and 825 ng of
labeled cRNAs were hybridized with the Human GE 4×44K V2 slides
(Agilent Technologies, Inc.) for 17 h at 65°C. The slides were
washed with GE Wash Buffer 1 and GE Wash Buffer 2 (Agilent
Technologies, Inc.) following the manufacturer's instructions and
digitized by the Agilent Microarrays Scanner (Agilent Technologies,
Inc.). The data were quantified and the quality control was
performed using Agilent FE Software (Agilent Technologies, Inc.).
The extracted data were imported to Agilent Gene Spring 12.5-GX
Analyze Program (Agilent Technologies, Inc.). The IPA
Ingenuity® system (Qiagen) was used to identify and
construct gene networks.
The data discussed in this publication have been
deposited in NCBI's gene Expression Omnibus (31) and are accessible through GEO
Series accession number GSE81064 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81064).
Human WNT signaling pathway
RT2 profiler™ PCR array
The Human WNT signaling pathway RT2
profiler™ PCR array plate is a panel containing 84 key genes
involved in the canonical and non-canonical WNT pathway and 5
endogenous genes for reaction control. The reaction was performed
according to the manufacturer's instructions except for the cDNA
synthesis, which was carried out using the High capacity archive
kit (Applied Biosystems Life Technologies, Foster City, CA, USA).
Real-time PCR was performed using SYBR-Green Master Mix (Qiagen)
and processed in the GeneAmp 5700 Sequence Detection system
(Applied Biosystems Life Technologies, Singapore). The data were
exported to and analyzed by RT2 profiler PCR array data
analysis template v 3.3 (SABiosciences, Germantown, MD, USA).
Statistical analysis
Gene expression data derived by Agilent was compared
for MCF7pcPAR4 vs. MCF7pcNEO cells before and after
treatment with docetaxel. According to our experimental design, the
statistical test selected was two-way ANOVA. The P-value
computation algorithm and the P-value correction test applied were
asymptotic and Benjamini Hochberg FDR, respectively. A Venn diagram
appears when two-way ANOVA tests are performed and reflects the
union and intersection of entities passing the chosen cut-off. For
Gene Ontology (GO) analysis the P-value cut-off was set at 1.0. All
tests were two-sided and an adjusted P<0.05 was considered to
indicate a statistically significant difference.
Results
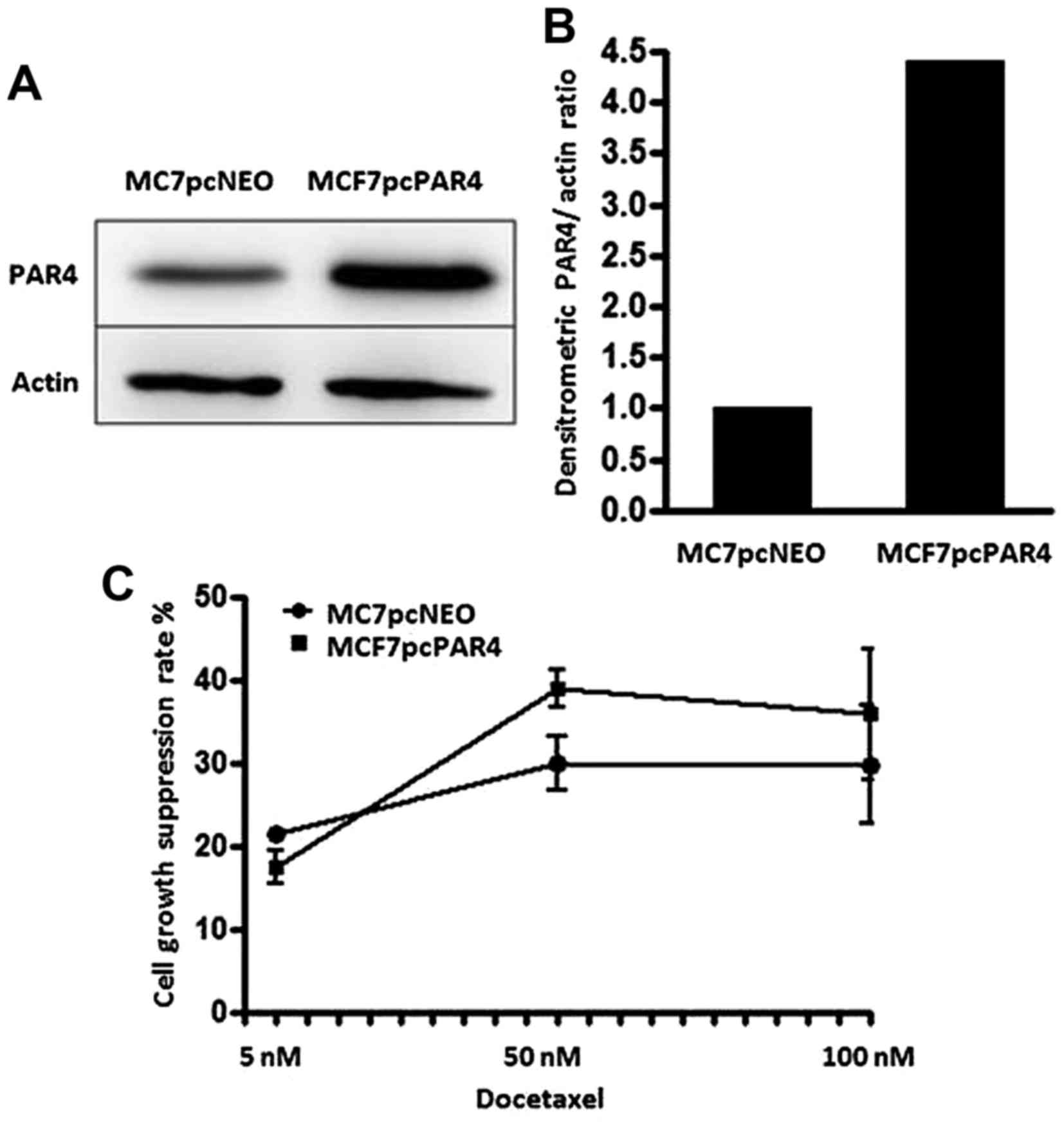
In a previous study, we showed that PAR4
upregulation increases the sensitivity of MCF7 breast cancer cells
to docetaxel (23). MCF7 cells
transfected with the expression vector for PAR4 (MCF7pcPAR4)
exhibited an ~4.5-fold increase in PAR4 protein expression in
comparison with the level noted in the MCF7pcNEO cells,
which were transfected with the empty vector (Figs. 1A and B). After treatment with
docetaxel, the MCF7 cells with increased PAR4 expression exhibited
increased proliferation restraint when compared with the
MCF7pcNEO cells. Similar effects on cell proliferation were
observed following treatment with docetaxel at 50 and 100 nM for 24
h (Fig. 1C). To better understand
the mechanisms involved in PAR4-mediated chemosensitivity, we
evaluated the effect of PAR4 on the expression profile of MCF7
breast cancer cells before and after treatment with docetaxel. A
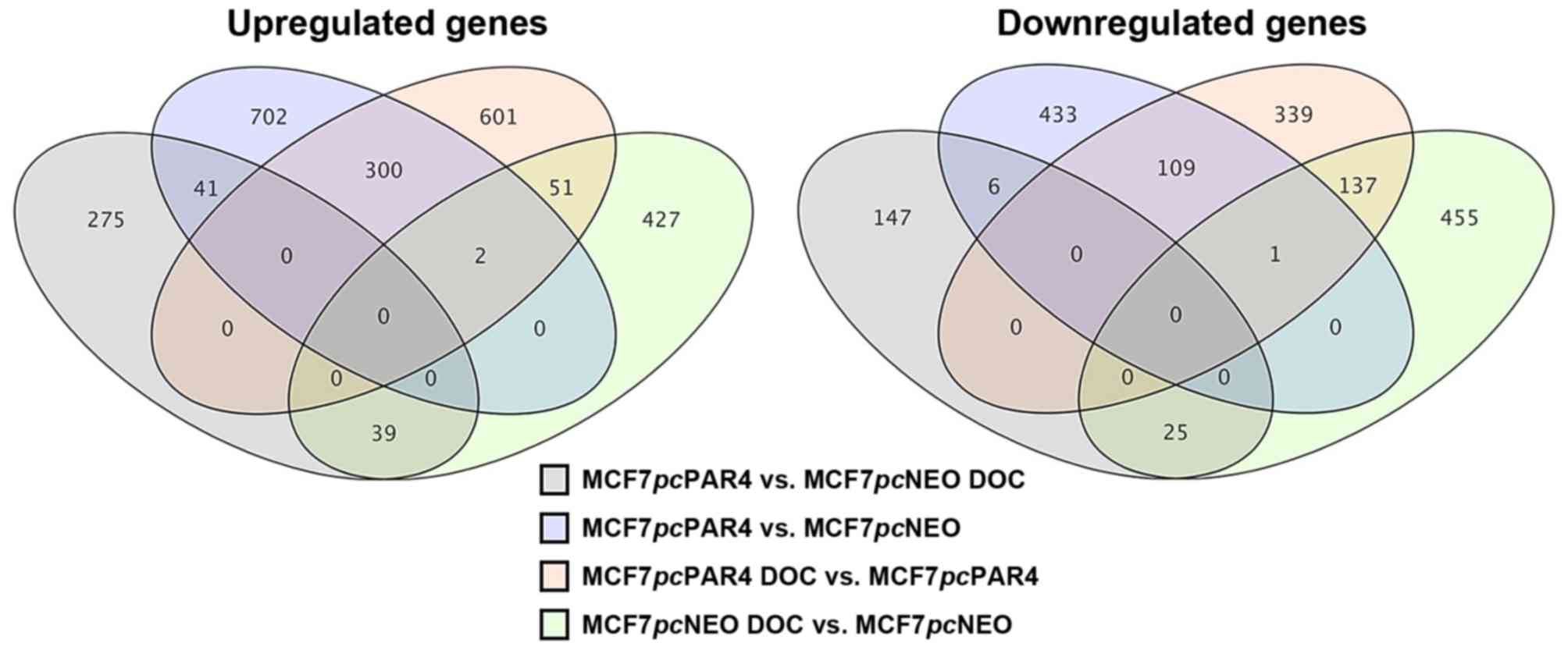
Venn diagram was used to represent the number of differentially
expressed genes that were common or exclusive in the different
comparisons (Fig. 2).
Before docetaxel treatment, 1,045 genes were
upregulated and 549 were downregulated in the MCF7pcPAR4
cells vs. MCF7pcNEO cells, with 702 of the upregulated genes
and 433 of the downregulated genes found exclusively in this
comparison. After docetaxel treatment, a total of 355 genes were
upregulated and 178 were downregulated in the MCF7pcPAR4
cells vs. MCF7pcNEO cells, with 275 of the upregulated genes
and 147 of the downregulated genes found exclusively in this
comparison. Forty-one of the upregulated genes were identified both
before and after docetaxel treatment, but only 6 downregulated
genes were identified both before and after docetaxel
treatment.
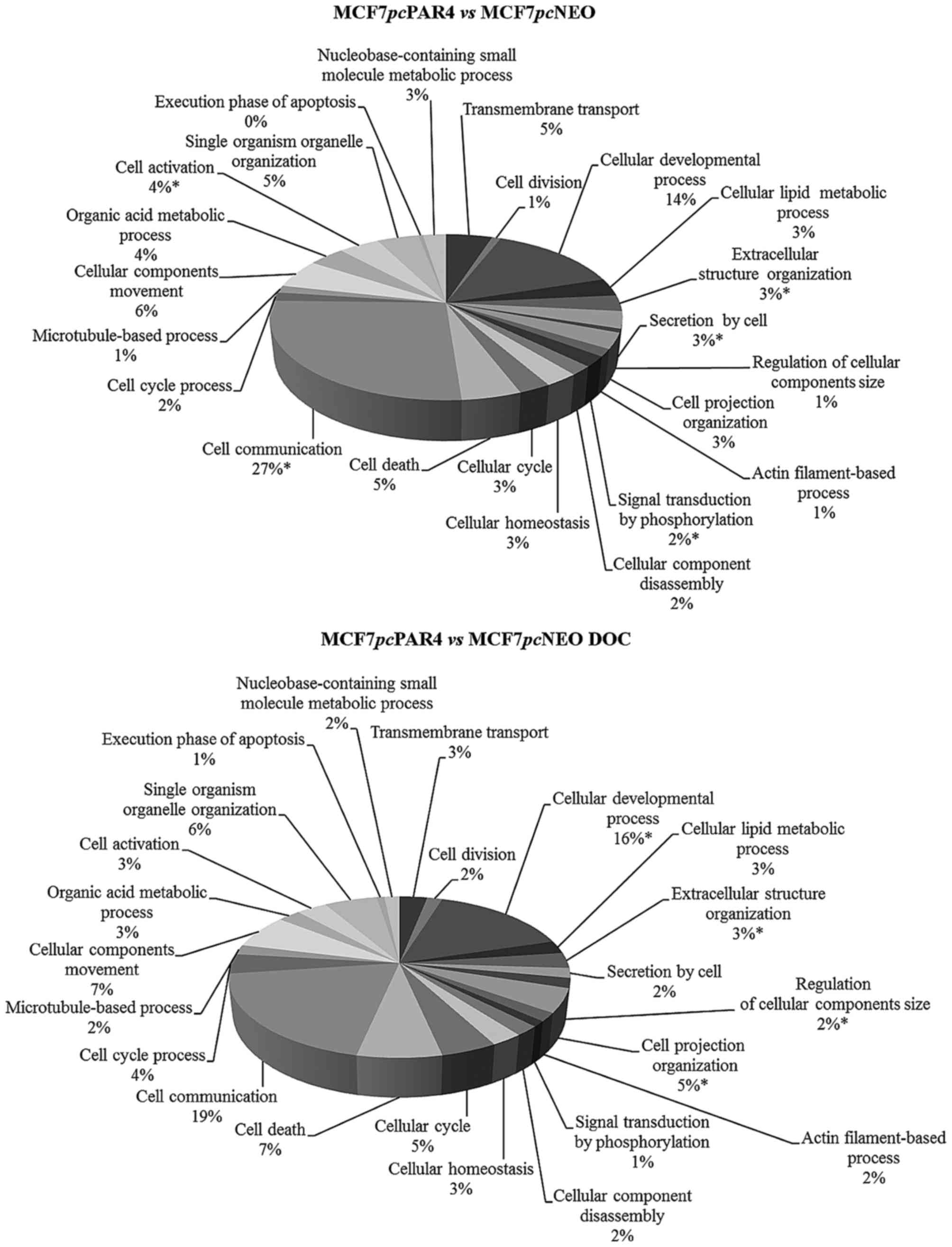
Gene Ontology (GO) analysis was performed to
categorize the differentially expressed genes in terms of the
cellular processes in which they participate. Considering the
upregulated genes, cell communication, extracellular structure
organization and cellular developmental processes were the top
categories in the comparisons of MCF7pcPAR4 and
MCF7pcNEO cells before and after docetaxel treatment.
Regardless of the observed enrichment, the number of genes
represented varied by ≥50% in some categories after treatment with
docetaxel. Among the categories we highlight secretion by cell,
signal transduction by phosphorylation, cell cycle, cell death,
cell cycle processes, microtubule-based processes, and execution
phase of apoptosis, which are related to the pro-apoptotic function
of PAR4 and anti-microtubule and anti-mitotic actions of docetaxel
(Fig. 3).
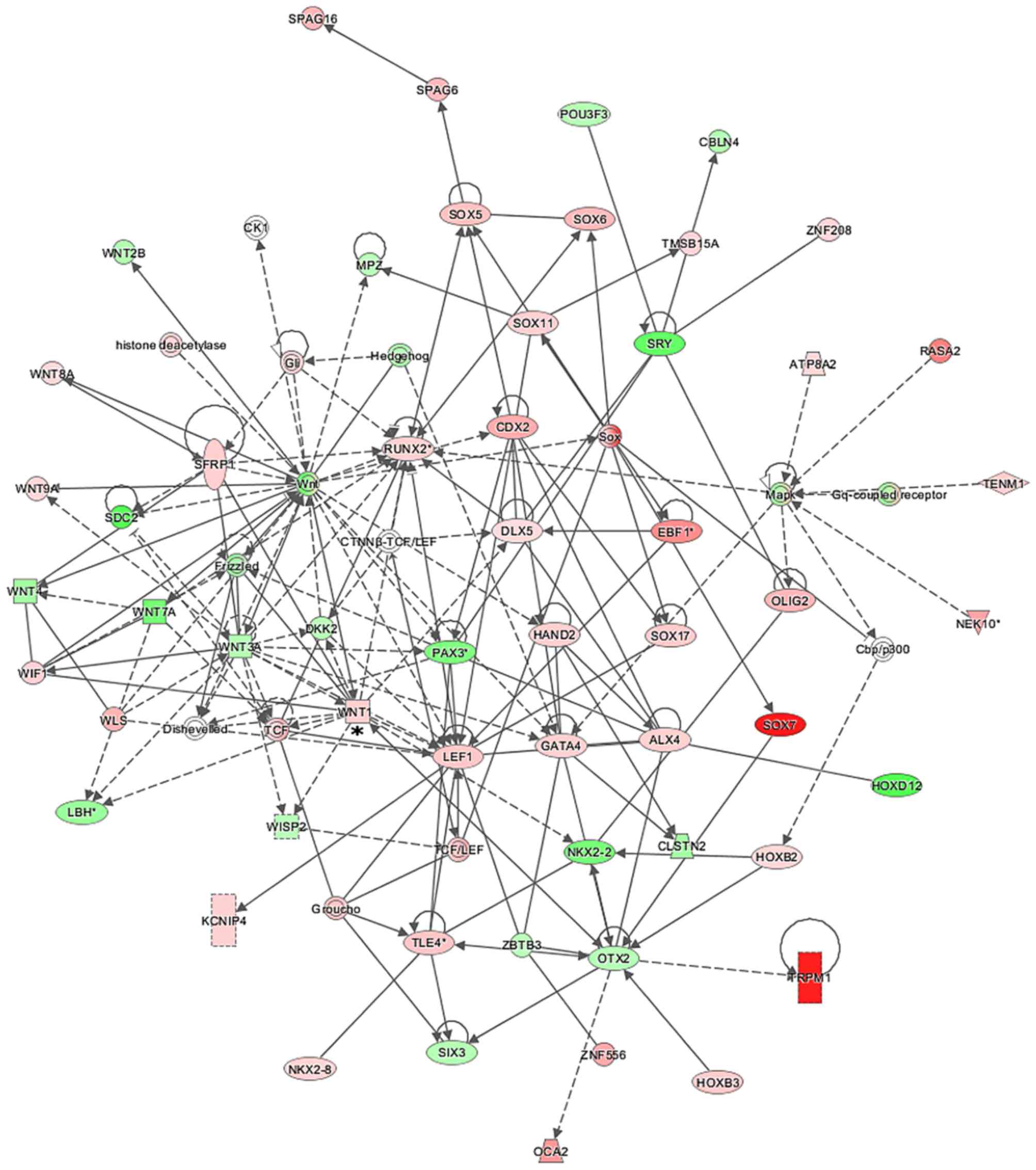
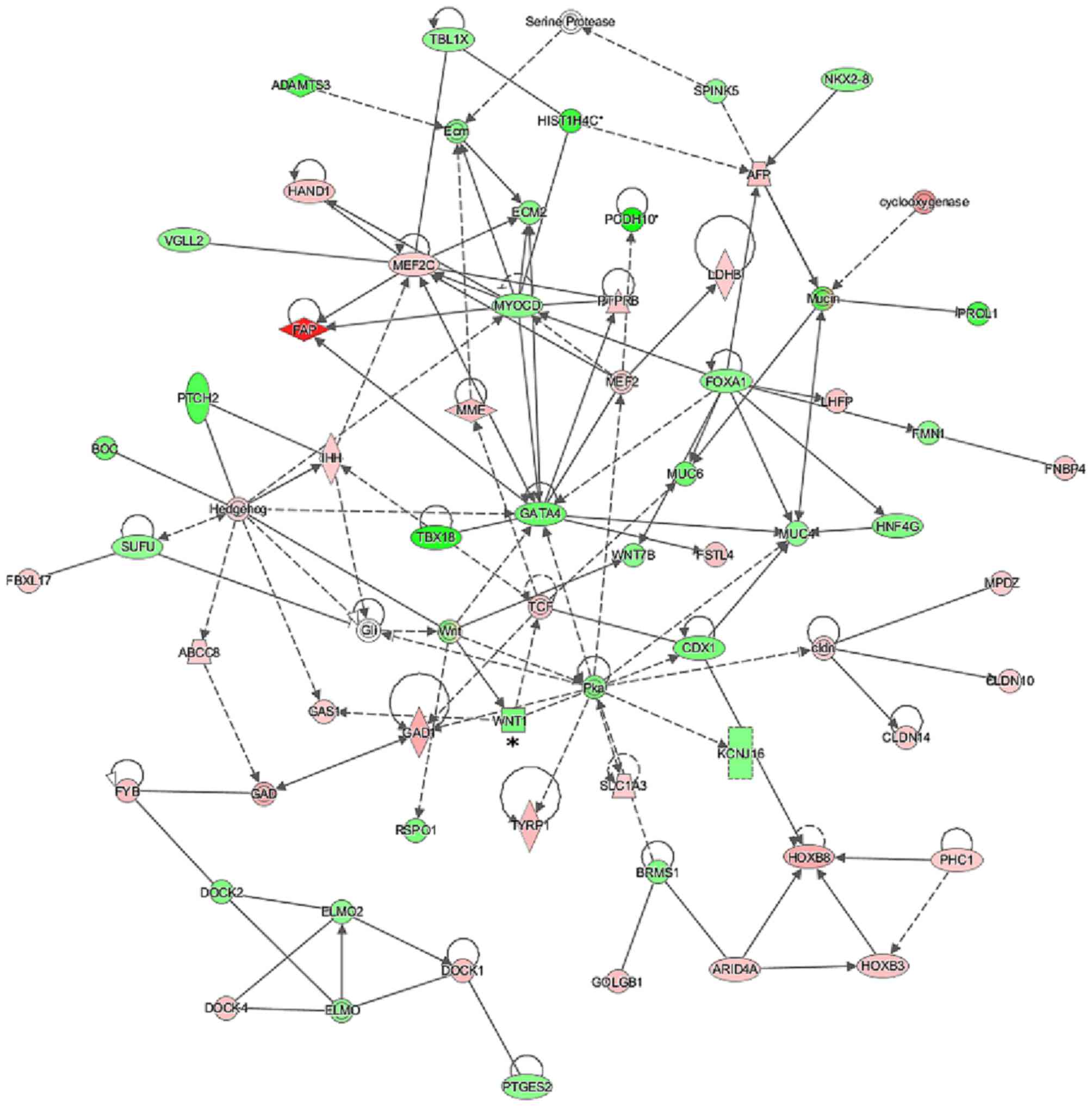
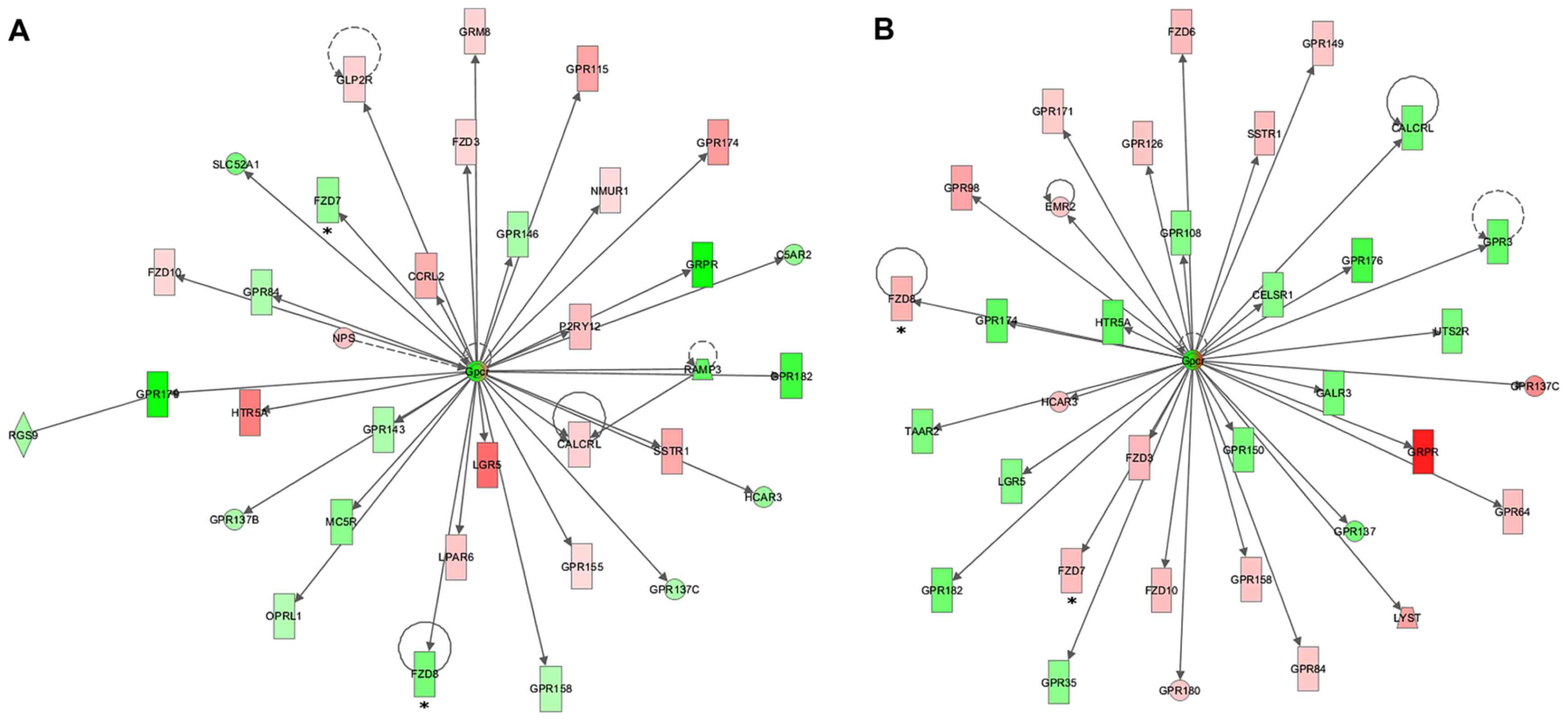
To assess the possible interactions between the
differentially expressed genes, numerous gene networks were
generated using the IPA Ingenuity® program for the
comparisons between MCF7pcPAR4 and MCF7pcNEO cells
before and after treatment with docetaxel. Many of the gene
networks generated revealed the modulation of several genes
directly or indirectly related to the WNT signaling pathway, such
as WNT1, WNT7A, WNT8A, WNT9A,
FZD8, TCF, LEF1, SOX5, SOX7,
SOX11, SOX17, WISP2 and RUNX2 (Figs. 4Figure 5–6). We observed an inversion in the
expression patterns of some genes related to the WNT pathway after
docetaxel treatment, including WNT1, which was upregulated
in the MCF7pcPAR4 cells before treatment (Fig. 4) but downregulated after treatment
with docetaxel (Fig. 5), and
members of the Frizzled gene family, which were found to be
downregulated in the MCF7pcPAR4 cells before treatment and
upregulated after docetaxel treatment (Fig. 6).
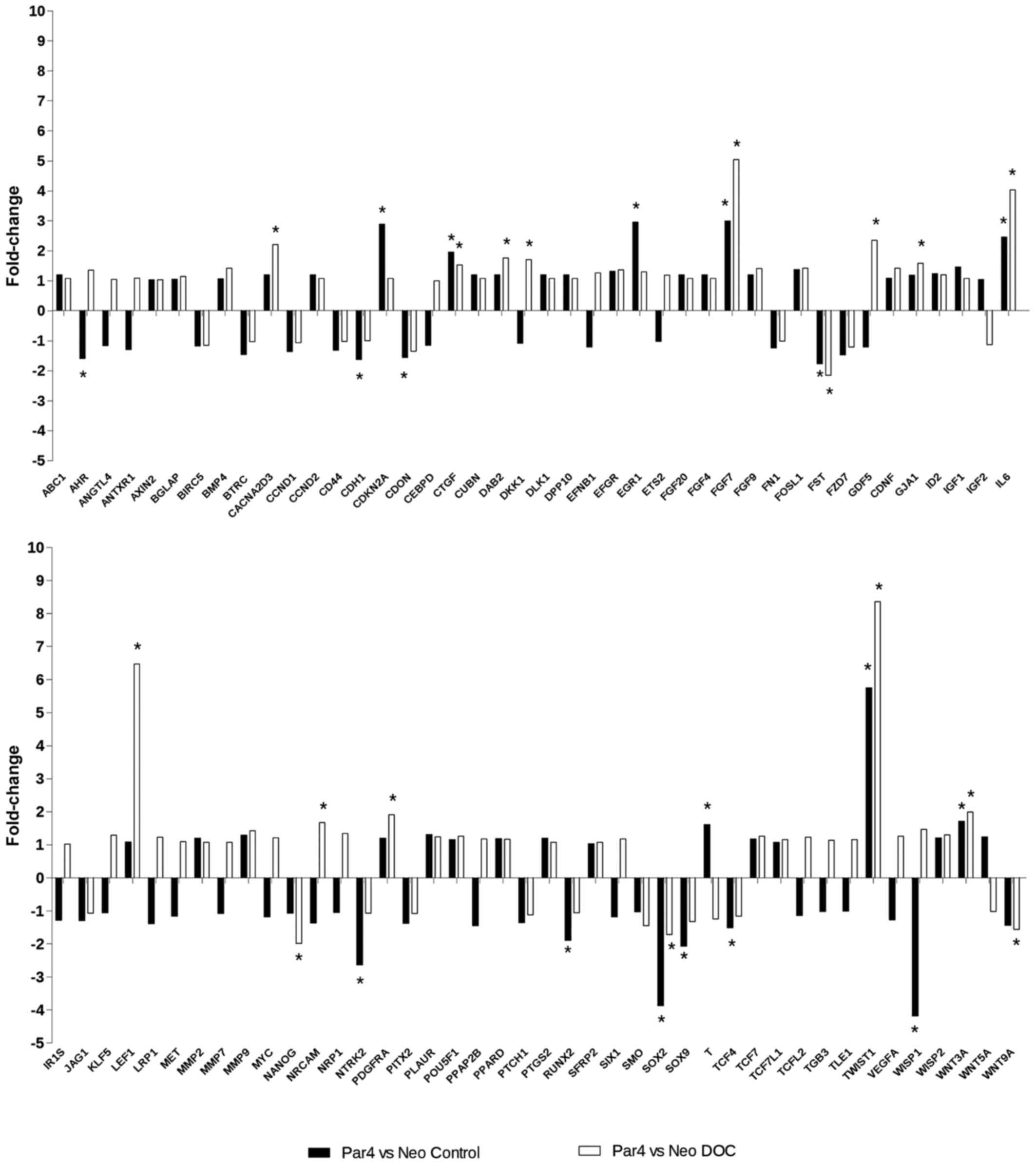
In order to validate the effects of PAR4 expression
on the regulation of genes involved in the WNT signaling pathway in
our experimental model, we used the human WNT signaling pathway
RT2 profiler™ PCR array. Fig. 7 shows the expression profile of 84
genes involved in the WNT signaling pathway in MCF7pcPAR4
vs. MCF7pcNEO cells before and after treatment with
docetaxel. Four transcripts (CDON, CTGF, TCF4
and WNT3A) were regulated by increased PAR4 expression and
exhibited no significant modulation after docetaxel treatment.
Seven transcripts (CACNA2D3, DAB2, GJA1,
LEF1, NANOG, PDGFRA and WNT9A) were
modulated in MCF7 cells expressing increased levels of PAR4 only in
the presence of docetaxel. Seventeen transcripts (AHR,
CDH1, CDKN2A, DKK1, EGR1, FGF7,
FST, GDF5, IL6, NRCAM, NTRK2,
RUNX2, SOX2, SOX9, T, TWIST1 and
WISP1) were regulated by increased PAR4 expression and
further modulated in opposite or incremental directions by
docetaxel treatment.
Discussion
Previous data from our group showed that PAR4
inhibits the growth of breast cancer cells and increases their
sensitivity to docetaxel (23).
In the present study, to identify differentially expressed genes
potentially involved in PAR4-mediated chemosensitivity to
docetaxel, we evaluated the expression profiles of MCF7 breast
cancer cells expressing different levels of PAR4, before and after
docetaxel exposure.
Notably, the GO analysis of the differentially
expressed genes revealed that some of the cellular processes
modulated by PAR4 overexpression in the presence and absence of
docetaxel are related to the pro-apoptotic functions of PAR4 and
anti-microtubule and anti-proliferative actions of docetaxel (e.g.,
cell division, cell cycle processes, microtubule-based processes
and execution phase of apoptosis), confirming the functionality of
our experimental model.
Using the IPA Ingenuity® program, several
gene networks were generated from the differentially expressed
genes. Many of the top gene networks generated were related to the
canonical and non-canonical WNT signaling pathway. The WNT
signaling pathway is highly conserved and can be divided into two
signaling pathways: the WNT/β-catenin pathway referred to as the
canonical pathway and the β-catenin-independent WNT signaling
referred to as the non-canonical pathway. The WNT signaling
pathways control important biological processes, such as embryonic
development, proliferation, cell migration, cell polarity,
morphogenesis and tissue regeneration, and alterations in this
signaling pathway have been shown to be involved in the tumorigenic
process of a number of malignancies, including breast cancer
(32,33).
WNT1 and members of the Frizzled family of
genes, such as FZD8, were upregulated and downregulated,
respectively, in cells expressing increased levels of PAR4 before
docetaxel treatment and were inversely modulated after docetaxel
treatment. The FZDs are transmembrane cell surface receptors
activated by the WNT family of lipoglycoproteins to activate
canonical and/or non-canonical WNT signaling (34). In invasive breast tumors, WNT1
expression seems to promote differentiation and apoptosis but
inhibits proliferation, and is associated with a better prognosis
in a subgroup of patients with stage II disease (35). FZD8 was shown to be overexpressed
in human lung cancer tissue and cell lines. Furthermore, FZD8
knockdown by siRNA has been shown to inhibit cell proliferation,
decrease the activity of the WNT pathway in in vitro/in
vivo models, and sensitize lung cancer cells to docetaxel
chemotherapy (36). In breast
cancer, inhibition of FZD8 expression in CRL2335 cells in the
presence of cisplatin plus TRAIL was found to reduce β-catenin and
survivin levels, leading to increased apoptosis. FZD8 expression
was also observed in residual triple-negative breast tumors treated
with cisplatin and TRAIL, suggesting that FZD8 expression may play
a role in chemoresistance in triple-negative breast tumors
(37). Taken together, the data
suggest that the modulation of WNT1 and FZD8 by PAR4
may be one of the mechanisms involved in PAR4-mediated
chemosensitivity. Although some attempts have been made to classify
the canonical and non-canonical WNT pathways according to the
activated WNT receptors or co-receptors, to what extent these
pathways may overlap is still controversial. In addition, as WNT
signaling is implicated in various diseases, efforts are being made
to identify molecules that interfere with the pathway in a cell
type-specific manner (38).
We used the human WNT signaling pathway
RT2 profiler™ PCR array to evaluate the effects of PAR4
on the expression of a number of genes involved in the canonical
and non-canonical WNT signaling pathways in our experimental model.
The expression profiles of EGR1, IL6, SOX2 and
WISP1 in MCF7 cells with increased PAR4 expression before
treatment with docetaxel likely indicate inactivation of the
WNT/β-catenin signaling pathway (39–42). However, the expression profiles of
CDKN2A, FGF7, TWIST, NTRK2 and
SOX9 in the same cell conditions, likely suggest activation
of the WNT/β-catenin signaling pathway (43–47). Furthermore, the observed
expression profiles of CACNAD2A3, GDF5, IL6
and NANOG in MCF7 cells with increased PAR4 expression after
treatment with docetaxel suggest inactivation of the WNT/β-catenin
pathway (41,48–50), whereas the expression of
FGF7, TWIST and LEF1 transcripts, under the
same cell conditions, suggests that WNT/β-catenin signaling is
active. These are target genes of the canonical pathway and have
been associated with epithelial-mesenchymal transition (EMT) and
chemoresistance (44,51,52).
Alterations in the WNT/β-catenin signaling pathway
have been shown to be a frequent event in the tumorigenic process
and chemoresistance in various types of cancer (53,54). In breast cancer, alterations in
the WNT/β-catenin signaling pathway are critical to tumor
development and progression (55). In addition, increased expression
of the Frizzled receptors or aberrant activation of β-catenin have
been associated with resistance to radiotherapy and chemotherapy in
breast cancer (56,57). Furthermore, downregulation of the
WNT/β-catenin signaling pathway inhibits EMT and reduces invasion
in breast cancer cell lines (58,59). However, to the best of our
knowledge no studies have addressed the involvement of PAR4 in the
WNT signaling pathway and treatment with docetaxel in breast
cancer.
Here we provide evidence that increased PAR4
expression is able to modulate the expression of genes involved in
the canonical and non-canonical WNT signaling pathways. Our
findings are still preliminary and do not allow us to state that
the WNT pathway would be active or inactive in MCF7 cells in the
presence of PAR4 with or without treatment with docetaxel, and they
do not allow us to predict the exact biological role of these
pathways in chemosensitivity to docetaxel in breast cancer
cells.
The role of Wnt and its downstream effectors have
been demonstrated in primary human breast tumors as well as breast
cancer cell lines and are suggested to play an important role in
breast tumor development and progression (60–62). Collectively, our study also opens
up the possibility to speculate that PAR4 increases breast cancer
cell chemosensitivity by the modulation of the WNT signaling
pathway. However, investigations using different breast cancer cell
lines with different expression of PAR4 treated with different
chemotherapeutic agents in the presence and absence of WNT pathway
inhibitors will need to be performed to better understand the
relationship between the expression of PAR4 and the WNT pathway in
chemosensitivity to docetaxel and other chemotherapeutic agents;
issues that we are currently addressing.
Acknowledgments
The present study was supported by Fundação de
Amparo a Pesquisa do Estado de São Paulo (FAPESP; 2013/07035-4) and
Conselho Nacional de Desenvolvimento Científico e Tecnológico
(CNPq; 303134/2013-5).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Laurentiis M, Cancello G, D'Agostino D,
Giuliano M, Giordano A, Montagna E, Lauria R, Forestieri V,
Esposito A, Silvestro L, et al: Taxane-based combinations as
adjuvant chemotherapy of early breast cancer: A meta-analysis of
randomized trials. J Clin Oncol. 26:44–53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Murray CJ, Vos T, Lozano R, Naghavi M,
Flaxman AD, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S,
et al: Disability-adjusted life years (DALYs) for 291 diseases and
injuries in 21 regions, 1990–2010 a systematic analysis for the
Global Burden of Disease Study 2010. Lancet. 380:2197–2223. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y and Rangnekar VM: Apoptosis and
tumor resistance conferred by Par-4. Cancer Biol Ther. 7:1867–1874.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sells SF, Han SS, Muthukkumar S, Maddiwar
N, Johnstone R, Boghaert E, Gillis D, Liu G, Nair P, Monnig S, et
al: Expression and function of the leucine zipper protein Par-4 in
apoptosis. Mol Cell Biol. 17:3823–3832. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hebbar N, Wang C and Rangnekar VM:
Mechanisms of apoptosis by the tumor suppressor Par-4. J Cell
Physiol. 227:3715–3721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boehrer S, Chow KU, Beske F,
Kukoc-Zivojnov N, Puccetti E, Ruthardt M, Baum C, Rangnekar VM,
Hoelzer D, Mitrou PS, et al: In lymphatic cells par-4 sensitizes to
apoptosis by down-regulating bcl-2 and promoting disruption of
mitochondrial membrane potential and caspase activation. Cancer
Res. 62:1768–1775. 2002.PubMed/NCBI
|
|
8
|
Chakraborty M, Qiu SG, Vasudevan KM and
Rangnekar VM: Par-4 drives trafficking and activation of Fas and
Fasl to induce prostate cancer cell apoptosis and tumor regression.
Cancer Res. 61:7255–7263. 2001.PubMed/NCBI
|
|
9
|
Gurumurthy S, Goswami A, Vasudevan KM and
Rangnekar VM: Phosphorylation of Par-4 by protein kinase A is
critical for apoptosis. Mol Cell Biol. 25:1146–1161. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Burikhanov R, Zhao Y, Goswami A, Qiu S,
Schwarze SR and Rangnekar VM: The tumor suppressor Par-4 activates
an extrinsic pathway for apoptosis. Cell. 138:377–388. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Boghaert ER, Sells SF, Walid AJ, Malone P,
Williams NM, Weinstein MH, Strange R and Rangnekar VM:
Immunohistochemical analysis of the proapoptotic protein Par-4 in
normal rat tissues. Cell Growth Differ. 8:881–890. 1997.PubMed/NCBI
|
|
12
|
Gurumurthy S and Rangnekar VM: Par-4
inducible apoptosis in prostate cancer cells. J Cell Biochem.
91:504–512. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sells SF, Wood DP Jr, Joshi-Barve SS,
Muthukumar S, Jacob RJ, Crist SA, Humphreys S and Rangnekar VM:
Commonality of the gene programs induced by effectors of apoptosis
in androgen-dependent and -independent prostate cells. Cell Growth
Differ. 5:457–466. 1994.PubMed/NCBI
|
|
14
|
Lucas T, Pratscher B, Krishnan S, Fink D,
Günsberg P, Wolschek M, Wacheck V, Muster T, Romirer I, Wolff K, et
al: Differential expression levels of Par-4 in melanoma. Melanoma
Res. 11:379–383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cook J, Krishnan S, Ananth S, Sells SF,
Shi Y, Walther MM, Linehan WM, Sukhatme VP, Weinstein MH and
Rangnekar VM: Decreased expression of the pro-apoptotic protein
Par-4 in renal cell carcinoma. Oncogene. 18:1205–1208. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brieger A, Boehrer S, Schaaf S, Nowak D,
Ruthardt M, Kim SZ, Atadja P, Hoelzer D, Mitrou PS, Weidmann E, et
al: In bcr-abl-positive myeloid cells resistant to conventional
chemotherapeutic agents, expression of Par-4 increases sensitivity
to imatinib (STI571) and histone deacetylase-inhibitors. Biochem
Pharmacol. 68:85–93. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kögel D, Reimertz C, Mech P, Poppe M,
Frühwald MC, Engemann H, Scheidtmann KH and Prehn JH: Dlk/ZIP
kinase-induced apoptosis in human medulloblastoma cells:
Requirement of the mitochondrial apoptosis pathway. Br J Cancer.
85:1801–1808. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moreno-Bueno G, Fernandez-Marcos PJ,
Collado M, Tendero MJ, Rodriguez-Pinilla SM, Garcia-Cao I,
Hardisson D, Diaz-Meco MT, Moscat J, Serrano M, et al: Inactivation
of the candidate tumor suppressor par-4 in endometrial cancer.
Cancer Res. 67:1927–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ahmed MM, Sheldon D, Fruitwala MA,
Venkatasubbarao K, Lee EY, Gupta S, Wood C, Mohiuddin M and Strodel
WE: Downregulation of PAR-4, a pro-apoptotic gene, in pancreatic
tumors harboring K-ras mutation. Int J Cancer. 122:63–70. 2008.
View Article : Google Scholar
|
|
20
|
Nagai MA, Gerhard R, Salaorni S, Fregnani
JH, Nonogaki S, Netto MM and Soares FA: Downregulation of the
candidate tumor suppressor gene PAR-4 is associated with poor
prognosis in breast cancer. Int J Oncol. 37:41–49. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Méndez-López LF, Zapata-Benavides P,
Zavala-Pompa A, Aguado-Barrera ME, Pacheco-Calleros J,
Rodríguez-Padilla C, Cerda-Flores RM, Cortés-Gutiérrez EI and
Dávila-Rodríguez MI: Immunohistochemical analysis of prostate
apoptosis response-4 (Par-4) in Mexican women with breast cancer: A
preliminary study. Arch Med Res. 41:261–268. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
García-Cao I, Duran A, Collado M,
Carrascosa MJ, Martín-Caballero J, Flores JM, Diaz-Meco MT, Moscat
J and Serrano M: Tumour-suppression activity of the proapoptotic
regulator Par4. EMBO Rep. 6:577–583. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pereira MC, de Bessa-Garcia SA, Burikhanov
R, Pavanelli AC, Antunes L, Rangnekar VM and Nagai MA: Prostate
apoptosis response-4 is involved in the apoptosis response to
docetaxel in MCF-7 breast cancer cells. Int J Oncol. 43:531–538.
2013.PubMed/NCBI
|
|
24
|
Jagtap JC, Parveen D, Shah RD, Desai A,
Bhosale D, Chugh A, Ranade D, Karnik S, Khedkar B, Mathur A, et al:
Secretory prostate apoptosis response (Par)-4 sensitizes
multicellular spheroids (MCS) of glioblastoma multiforme cells to
tamoxifen-induced cell death. FEBS Open Bio. 5:8–19. 2014.
View Article : Google Scholar
|
|
25
|
Zhao Y, Burikhanov R, Qiu S, Lele SM,
Jennings CD, Bondada S, Spear B and Rangnekar VM: Cancer resistance
in transgenic mice expressing the SAC module of Par-4. Cancer Res.
67:9276–9285. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee TJ, Lee JT, Kim SH, Choi YH, Song KS,
Park JW and Kwon TK: Overexpression of Par-4 enhances
thapsigargin-induced apoptosis via down-regulation of XIAP and
inactivation of Akt in human renal cancer cells. J Cell Biochem.
103:358–368. 2008. View Article : Google Scholar
|
|
27
|
Kline CL, Shanmugavelandy SS, Kester M and
Irby RB: Delivery of PAR-4 plasmid in vivo via nanoliposomes
sensitizes colon tumor cells subcutaneously implanted into nude
mice to 5-FU. Cancer Biol Ther. 8:1831–1837. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang BD, Kline CL, Pastor DM, Olson TL,
Frank B, Luu T, Sharma AK, Robertson G, Weirauch MT, Patierno SR,
et al: Prostate apoptosis response protein 4 sensitizes human colon
cancer cells to chemotherapeutic 5-FU through mediation of an NF
kappaB and microRNA network. Mol Cancer. 9:982010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alvarez JV, Pan TC, Ruth J, Feng Y, Zhou
A, Pant D, Grimley JS, Wandless TJ, Demichele A and Chodosh LA;
I-SPY 1 TRIAL Investigators: Par-4 downregulation promotes breast
cancer recurrence by preventing multinucleation following targeted
therapy. Cancer Cell. 24:30–44. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar :
|
|
32
|
Michaelson JS and Leder P: beta-catenin is
a downstream effector of Wnt-mediated tumorigenesis in the mammary
gland. Oncogene. 20:5093–5099. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Amin N and Vincan E: The Wnt signaling
pathways and cell adhesion. Front Biosci (Landmark Ed). 17:784–804.
2012. View Article : Google Scholar
|
|
34
|
Dijksterhuis JP, Petersen J and Schulte G:
WNT/Frizzled signalling: receptor-ligand selectivity with focus on
FZD-G protein signalling and its physiological relevance: IUPHAR
Review 3. Br J Pharmacol. 171:1195–1209. 2014. View Article : Google Scholar :
|
|
35
|
Mylona E, Vamvakaris I, Giannopoulou I,
Theohari I, Papadimitriou C, Keramopoulos A and Nakopoulou L: An
immunohistochemical evaluation of the proteins Wnt1 and glycogen
synthase kinase (GSK)-3β in invasive breast carcinomas.
Histopathology. 62:899–907. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang HQ, Xu ML, Ma J, Zhang Y and Xie CH:
Frizzled-8 as a putative therapeutic target in human lung cancer.
Biochem Biophys Res Commun. 417:62–66. 2012. View Article : Google Scholar
|
|
37
|
Yin S, Xu L, Bonfil RD, Banerjee S, Sarkar
FH, Sethi S and Reddy KB: Tumor-initiating cells and FZD8 play a
major role in drug resistance in triple-negative breast cancer. Mol
Cancer Ther. 12:491–498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Niehrs C: The complex world of WNT
receptor signalling. Nat Rev Mol Cell Biol. 13:767–779. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tice DA, Soloviev I and Polakis P:
Activation of the Wnt pathway interferes with serum response
element-driven transcription of immediate early genes. J Biol Chem.
277:6118–6123. 2002. View Article : Google Scholar
|
|
40
|
Ji J, Wei X and Wang Y: Embryonic stem
cell markers Sox-2 and OCT4 expression and their correlation with
WNT signal pathway in cervical squamous cell carcinoma. Int J Clin
Exp Pathol. 7:2470–2476. 2014.PubMed/NCBI
|
|
41
|
Lam SP, Luk JM, Man K, Ng KT, Cheung CK,
Rose-John S and Lo CM: Activation of interleukin-6-induced
glycoprotein 130/signal transducer and activator of transcription 3
pathway in mesenchymal stem cells enhances hepatic differentiation,
proliferation, and liver regeneration. Liver Transpl. 16:1195–1206.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Venkatesan B, Prabhu SD, Venkatachalam K,
Mummidi S, Valente AJ, Clark RA, Delafontaine P and Chandrasekar B:
WNT1-inducible signaling pathway protein-1 activates diverse cell
survival pathways and blocks doxorubicin-induced cardiomyocyte
death. Cell Signal. 22:809–820. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ayyanan A, Civenni G, Ciarloni L, Morel C,
Mueller N, Lefort K, Mandinova A, Raffoul W, Fiche M, Dotto GP, et
al: Increased Wnt signaling triggers oncogenic conversion of human
breast epithelial cells by a Notch-dependent mechanism. Proc Natl
Acad Sci USA. 103:3799–3804. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Katoh M and Katoh M: FGF signaling network
in the gastrointestinal tract (Review). Int J Oncol. 29:163–168.
2006.PubMed/NCBI
|
|
45
|
Vadnais C, Shooshtarizadeh P, Rajadurai
CV, Lesurf R, Hulea L, Davoudi S, Cadieux C, Hallett M, Park M and
Nepveu A: Autocrine activation of the Wnt/β-catenin pathway by CUX1
and GLIS1 in breast cancers. Biol Open. 3:937–946. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Barreto RA, Walker FR, Dunkley PR, Day TA
and Smith DW: Fluoxetine prevents development of an early
stress-related molecular signature in the rat infralimbic medial
prefrontal cortex. Implications for depression? BMC Neurosci.
13:1252012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Akiyama H, Lyons JP, Mori-Akiyama Y, Yang
X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR,
et al: Interactions between Sox9 and beta-catenin control
chondrocyte differentiation. Genes Dev. 18:1072–1087. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wong AM, Kong KL, Chen L, Liu M, Wong AM,
Zhu C, Tsang JW and Guan XY: Characterization of CACNA2D3 as a
putative tumor suppressor gene in the development and progression
of nasopharyngeal carcinoma. Int J Cancer. 133:2284–2295. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Enochson L, Stenberg J, Brittberg M and
Lindahl A: GDF5 reduces MMP13 expression in human chondrocytes via
DKK1 mediated canonical Wnt signaling inhibition. Osteoarthritis
Cartilage. 22:566–577. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yin D, Tian L, Ye Y, Li K, Wang J, Cheng
P, Chen A, Guo F and Huang H: Nanog and β-catenin: A new
convergence point in EpSC proliferation and differentiation. Int J
Mol Med. 29:587–592. 2012.PubMed/NCBI
|
|
51
|
Nguyen DX, Chiang AC, Zhang XH, Kim JY,
Kris MG, Ladanyi M, Gerald WL and Massagué J: WNT/TCF signaling
through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis.
Cell. 138:51–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Je EC, Lca BS and Ga GA: The role of
transcription factor TWIST in cancer cells. J Genet Syndr Gene
Ther. 4:1242013.
|
|
53
|
Loh YN, Hedditch EL, Baker LA, Jary E,
Ward RL and Ford CE: The Wnt signalling pathway is upregulated in
an in vitro model of acquired tamoxifen resistant breast cancer.
BMC Cancer. 13:1742013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ren J, Wang R, Song H, Huang G and Chen L:
Secreted frizzled related protein 1 modulates taxane resistance of
human lung adenocarcinoma. Mol Med. 20:164–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lamb R, Ablett MP, Spence K, Landberg G,
Sims AH and Clarke RB: Wnt pathway activity in breast cancer
sub-types and stem-like cells. PLoS One. 8:e678112013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang H, Zhang X, Wu X, Li W, Su P, Cheng
H, Xiang L, Gao P and Zhou G: Interference of Frizzled 1 (FZD1)
reverses multidrug resistance in breast cancer cells through the
Wnt/β-catenin pathway. Cancer Lett. 323:106–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Woodward WA, Chen MS, Behbod F, Alfaro MP,
Buchholz TA and Rosen JM: WNT/beta-catenin mediates radiation
resistance of mouse mammary progenitor cells. Proc Natl Acad Sci
USA. 104:618–623. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu Y, Ginther C, Kim J, Mosher N, Chung S,
Slamon D and Vadgama JV: Expression of Wnt3 activates Wnt/β-catenin
pathway and promotes EMT-like phenotype in trastu-zumab-resistant
HER2-overexpressing breast cancer cells. Mol Cancer Res.
10:1597–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ahmad A, Sarkar SH, Bitar B, Ali S,
Aboukameel A, Sethi S, Li Y, Bao B, Kong D, Banerjee S, et al:
Garcinol regulates EMT and Wnt signaling pathways in vitro and in
vivo, leading to anticancer activity against breast cancer cells.
Mol Cancer Ther. 11:2193–2201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhao Z, Lu P, Zhang H, Xu H, Gao N, Li M
and Liu C: Nestin positively regulates the Wnt/β-catenin pathway
and the proliferation, survival and invasiveness of breast cancer
stem cells. Breast Cancer Res. 16:4082014. View Article : Google Scholar
|
|
61
|
Mukherjee N, Bhattacharya N, Alam N, Roy
A, Roychoudhury S and Panda CK: Subtype-specific alterations of the
Wnt signaling pathway in breast cancer: clinical and prognostic
significance. Cancer Sci. 103:210–220. 2012. View Article : Google Scholar
|
|
62
|
Xiang T, Li L, Yin X, Zhong L, Peng W, Qiu
Z, Ren G and Tao Q: Epigenetic silencing of the WNT antagonist
Dickkopf 3 disrupts normal Wnt/β-catenin signalling and apoptosis
regulation in breast cancer cells. J Cell Mol Med. 17:1236–1246.
2013. View Article : Google Scholar : PubMed/NCBI
|