Introduction
Active neurogenesis occurs throughout life in the
subventricular zone (SVZ) of the lateral ventricle, as well as in
the subgranular zone (SGZ) of the dentate gyrus (DG) (1). Currently, scientific interest in
endogenous repair has mostly focused on these two regions that
harbor multipotent neural stem cells (NSCs), which can give rise to
interneurons and contribute to neural plasticity (1,2).
However, it is unclear how distinct pools of NSCs may react to
neurodegeneration. In addition, the molecular mechanisms underlying
the process are not yet well understood.
Parkinson's disease (PD) is a common
neurodegenerative disorder, characterized by the progressive loss
of dopaminergic neurons and consequent dopamine (DA) deficit. In
the adult brain, DA contributes to the regulation of endogenous
NSCs, and DA depletion may affect neurogenesis (2–4).
However, neurogenesis profiling that is activated or inhibited has
been a subject of heated debate. Several findings have provided
evidence that DA denervation reduces cell proliferation,
differentiation and migration in the SVZ and SGZ in individuals
with PD, as well as in mice administered
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) (2,5),
while others have emphasized that the proliferative capacity of the
SVZ and SGZ is not diminished (4,5) or
even increased (2,6,7) in
the parkinsonian brain. Notably, there are several essential
signaling pathways regulating proliferation, differentiation and
migration, as well as the survival of neural precursor cells
(NPCs), and their integration into the neuronal circuitry network
in the adult brain, e.g., Notch, Sonic hedgehog (Shh),
mitogen-activated protein kinase (MAPK) and Eph:ephrin signaling
pathways (5,8–10).
In addition to signaling transduction pathways, metabolic factors
serve as important modulators of neurogenesis (9). The alterations in these modulators,
such as growth factors during glycolysis, oxidative
phosphorylation, lipogenesis and protein synthesis may be
associated with the development of neurodegenerative diseases
(10–13).
In our study, adult mice were exposed to MPTP. We
then carried out transcriptomic analysis on the tissues of the SVZ
and DG using RNA sequencing (RNA-Seq) technology, and observed
significantly deregulated gene expression following the MPTP
administration. Bioinformatics analysis using the Gene Ontology
(GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG)
databases was applied to interpret the related function of the
differentially expressed genes. Future studies using single-cell
sequencing are required to further investigate the molecular
targets for neuroplasticity in the SVZ and SGZ.
Materials and methods
MPTP administration and behavioral
tests
All animal experiments were performed in accordance
with the guidelines on the Care of Laboratory Animals, and have
been approved by the Ethics Committee of Peking Union Medical
College Hospital, Beijing, China. A total of 52 adult C57BL/6J
female mice (10 weeks old) weighing approximately 25 g were
randomly assigned to 2 groups: i) MPTP-exposed group (n=29) and ii)
saline-treated group (n=23). As previously described, MPTP
hydrochloride (20 mg/kg; Sigma-Aldrich, St. Louis, MO, USA) in
saline was injected intraperitoneally and handled in accordance
with the published guidelines (14,15). The control mice received
intraperitoneal injection of an equal volume of saline. The rotarod
test was performed on day 21 after the MPTP injection. All mice
were pre-trained approximately 1 week before the tests. One day
prior to MPTP administration, the baseline was obtained, and the
test series then began at a speed of 26 rpm. Each animal received 2
trials with a rest interval of 5–10 min, and the latencies to fall
off the rod within 180 sec were recorded and averaged. For the pole
test, mice were placed head upwards on the top of one upright pole
approximate 30 cm long and 8 mm in diameter. Once laid upon the
top, mice oriented themselves downwards and descended to the
ground. Animals also received training that consisted of 3
consecutive trials of each session. After establishing the
baseline, each animal experienced 2 trials and the time to descend
was recorded.
Immunostaining and immunoelectron
microscopy
As previously described (14,16), 21 days following exposure to MPTP,
under deep anesthesia, the animals were sacrificed.
Immunoperoxidase staining for tyrosine hydroxylase (TH) (n=12) and
immunoelectron for ultrastructure (n=10) were performed. Briefly,
the mice were perfused intracardially with cold 0.1 M
phosphate-buffered saline (PBS) followed by 4% paraformaldehyde
(PFA). The brains were carefully removed and post-fixed at 4°C
overnight, cryoprotected in 30% sucrose for 3 days. A complete set
of coronal sections containing the substantia nigra (SN) were then
created at a thickness of 30 µm. The brain slices were then
pre-treated for peroxidase activity using 3% hydrogen peroxide
(H2O2) for 20 min, and blocked with 5% normal
horse serum (NHS) (Vector Laboratories, Burlingame, CA, USA) for 1
h. Thereafter, the selected slices were incubated with primary
antibodies to TH (1:800; #AB152; Millipore, Billerica, MA, USA) at
4°C overnight, and treated with biotinylated horse anti-rabbit IgG
secondary antibody (1:200; #PK-6200) for 1 h, followed by
biotinylated protein A and avidinbiotinylated horseradish
peroxidase complex (ABC kit; #PK-6200; all from Vector
Laboratories) for a further 1 h at room temperature. Immunoreactive
cells were visualized with 3,3′-diaminobenzidine (DAB). Finally,
the sections were dehydrated through a graded series of ethanol,
cleared in xylene, coverslipped, and captured on a Nikon Eclipse
90i microscope (Nikon, Tokyo, Japan). Analysis was performed using
the Stereo Investigator (MBF Bioscience Inc., Williston, VT, USA)
to quantify TH-positive cells in the SN (14).
For direct electron microscopy, the mice were deeply
anesthetized and sacrificed by intracardiac perfusion with 2%
glutaraldehyde. Anatomically matched coronal sections of the SN and
striatum were removed from each mouse and further fixed overnight
in 2% glutaraldehyde at 4°C before embedding for electron
microscopy. The tissues were then washed in 0.1 M sodium cacodylate
buffer (pH 7.4), post-fixed with 2% osmium tetroxide for 30 min,
quickly dehydrated through graded ethanol solutions. The brain
regions of interest (SN and striatum) were then cut out from
tissues identically by the same individual, and covered with fresh
Epon. The resulting blocks containing the regions of interest were
then trimmed, and viewed on a JEOL JEM-1400Plus transmission
electron microscope (JEOL, Ltd., Tokyo, Japan).
Messenger RNA (mRNA) sequencing
Total RNA was extracted from the tissues of the SVZ
and DG, and RNA integrity was examined using a NanoDrop 2000/2000c
(Thermo Fisher Scientific Inc., Waltham, MA, USA). The RNA library
was constructed using TruSeq® RNA LT Sample Prep kit v2
(Illumina Inc., San Diego, CA, USA) according to the manufacturer's
instructions. Briefly, after poly(A)-based mRNA enrichment and RNA
fragmentation, first-strand complementary DNA (cDNA) was
synthesized using the First Strand Master Mix and SuperScript II,
followed by second-strand cDNA synthesis using the Second Strand
Master Mix. Double-stranded cDNA was end repaired, and then 3′ ends
were adenylated and the illumina adaptors were ligated. The
ligation product was amplified with 15 cycles of polymerase chain
reaction. After measures of yield and fragment length distribution,
libraries were sequenced using TrusSeq SBS kit v3-HS, and were
generated on the HiSeq 2500 sequencing system (both from Illumina
Inc.). Each sequencing library generated minimally 20 million
uniquely mapping reads. Images from the instrument were processed
using the manufacturer's software to generate FASTQ sequence
files.
Statistical analysis
The data from the behavioral tests and
immunostaining are presented as the means ± SEM and analyzed using
GraphPad Prism 6.0 software (GraphPad Software, San Diego, CA, USA)
for Mac OS X. The difference between 2 groups was analyzed using a
two-tailed Student's t-test, while two-way ANOVA followed by
Tukey's post-hoc analysis were conducted for multiple comparisons
between 2 or more groups. A p-value <0.05 was defined as a
threshold for statistical significance.
To characterize the transcriptomic profiles, RNA-Seq
was performed. Raw reads were trimmed the adaptor sequences with
trim Galore. Sequencing reads were mapped to the mouse genome using
TopHat 2.1.0. Cufflinks was used to calculate the gene expression
values represented as fragments per kilo-base per million mapped
reads (FPKM). The differential gene expression analysis of RNA-Seq
data was performed using Cuffdiff (17). The differential expression genes
were filtered with FPKM >1, q-value <0.05 and log2 fold
change >1 or <−1, and clustered using Gene Cluster 3.0. In
addition, GO and KEGG analysis were conducted using the DAVID
bioinformatics database. WEGO was used to plot the GO annotation
results. Expression distribution and scatter plots were generated
with CummeRbund package in R (18)
Results
MPTP-induced parkinsonism and
pathological changes in mice
The rotarod and pole tests were compared between
mice administered MPTP and animals receiving saline. No significant
difference in behavioral performance was observed prior to MPTP.
However, 21 days following exposure to MPTP, a statistically
significant deterioration in locomotor activity was observed in the
mice given the neurotoxin compared to the saline-treated controls.
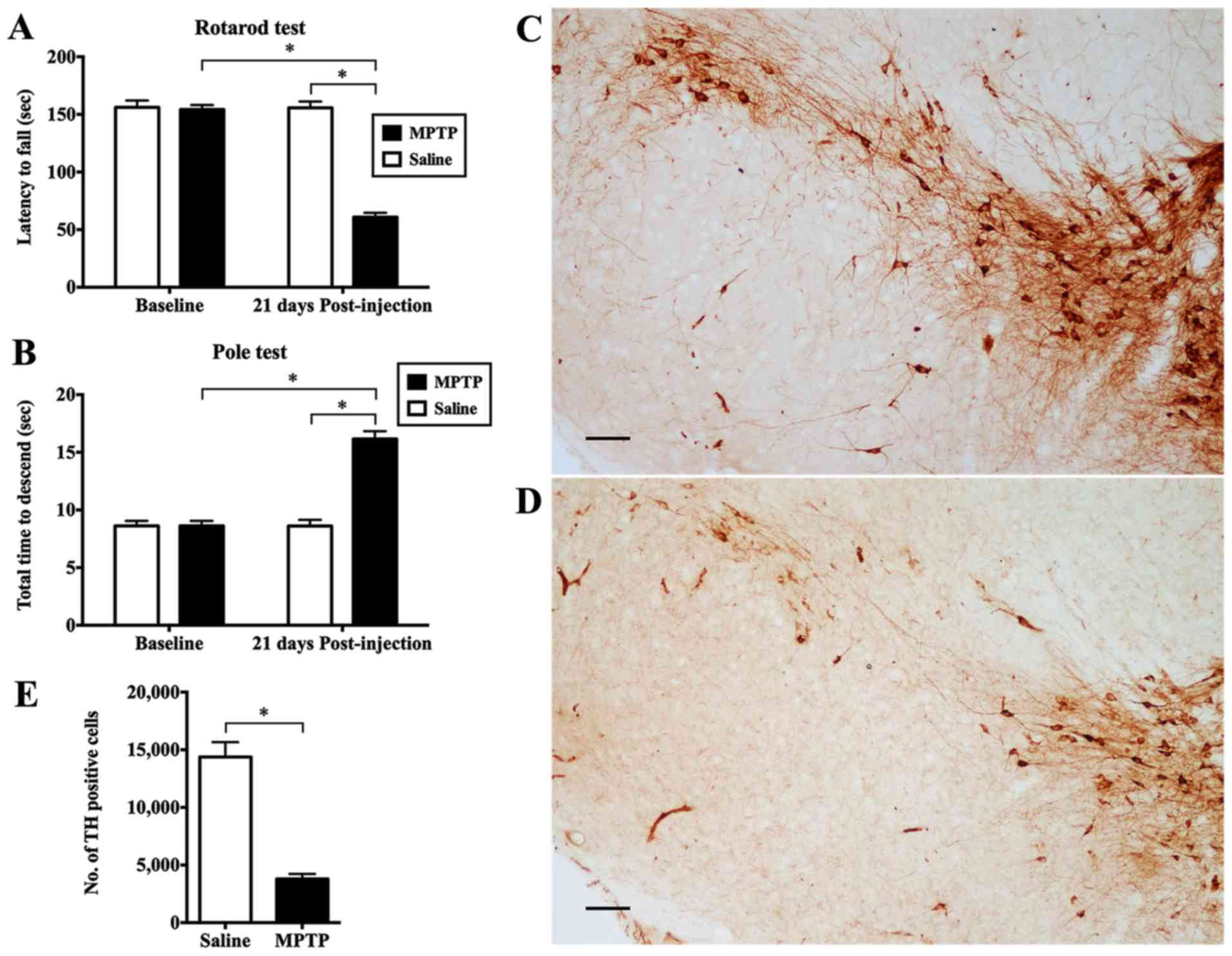
The mice with PD remained on the rotarod for significantly shorter
period of time than the control animals (p<0.05; Fig. 1A). For the pole test, the mice
intoxicated with MPTP took a significantly longer time to complete
the test (p<0.05; Fig.
1B).
We stereologically counted the number of
dopaminergic neurons in the SN stained for TH. When compared with
the saline-injected control (Fig.
1C), there was a marked reduction in TH immunoreactivity in the
mice adminstered MPTP (Fig. 1D).
The MPTP-exposed mice had an average of only 3,803.29±423.96
dopaminergic neurons in the SN compared with the saline-infused
animals which had 14,373.53±1289.03 cells expressing TH (p<0.05;
Fig. 1E). Furthermore, we
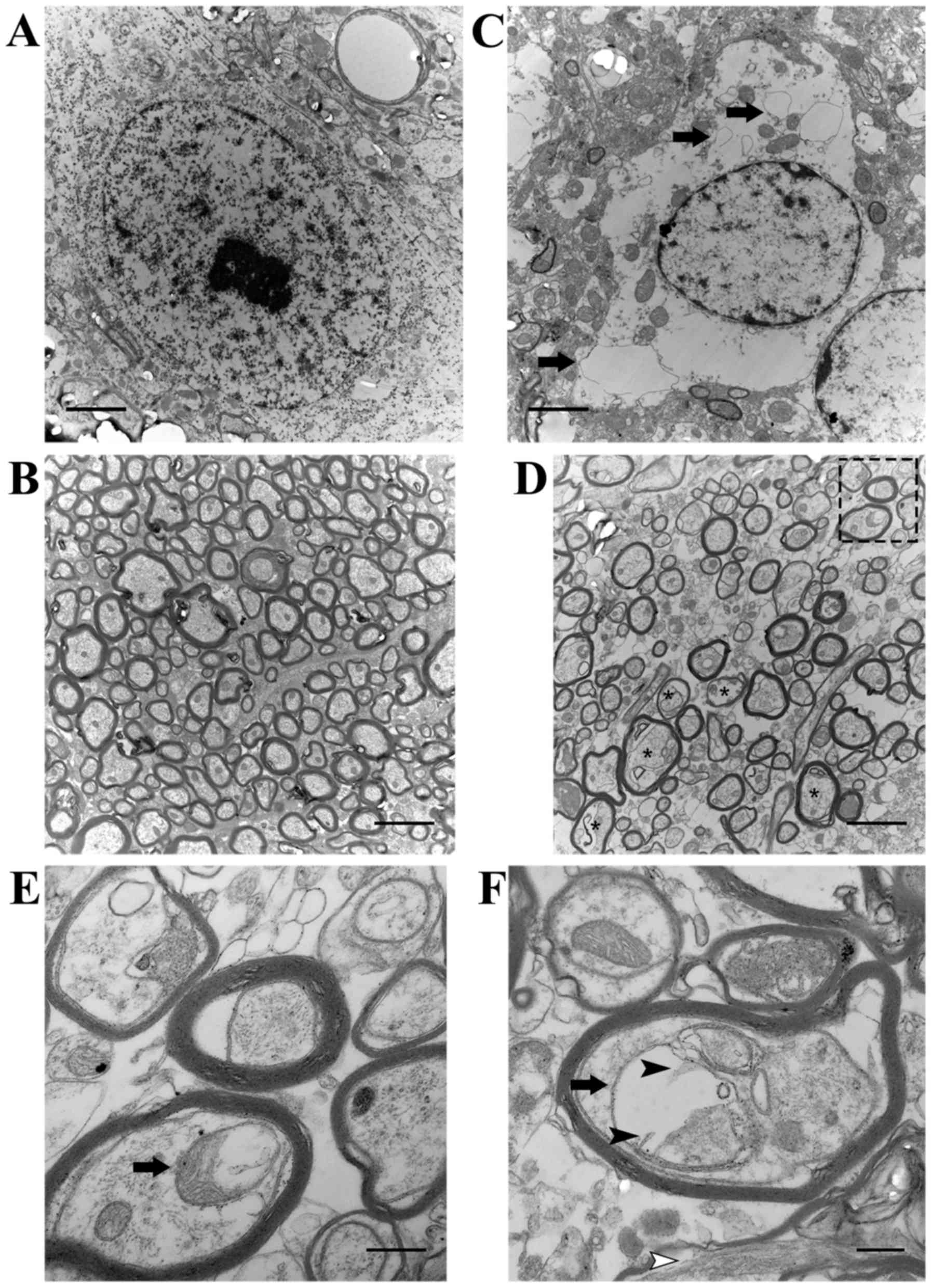
observed the ultrastructure of neurons in the SN and neurites in
the striatum. Striking changes did not occur in the saline group
(Fig. 2A and B). In the
MPTP-exposed mice, the plasma membrane of dying neurons appeared
ruptured, and the perinuclear cytoplasm was severely dilated,
including the endoplasmic reticulum (ER) (arrows in Fig. 2C). In addition, there were
numerous enlarged dystrophic myelinated axons (asterisks in
Fig. 2D) swelled, containing
mitochondria with varying degrees of degeneration in the striatum
of the mice administered MPTP. Notably, one greatly enlarged
mitochondrion grossly deformed into a ring-shaped structure (arrow
in Fig. 2E), while another one
had prominently residual cristae (black arrowheads in Fig. 2F).
Differential expression of genes in the
SVZ and DG of mice following exposure to MPTP
To characterize the transcriptomic profiles in mice
at 4 days and 3 weeks after the MPTP administration, RNA-Seq was
performed. In the SVZ, we found 106 genes significantly deregulated
at different time points: 65 were upregulated and 41 were
downregulated (q-value <0.05 compared with the normal groups;
Fig. 3A). In the DG, the
expression levels of a total of 98 genes were significantly
modulated: 43 were upregulated, and 55 were downregulated (q-value
<0.05 compared with the normal groups; Fig. 3B). We identified two different
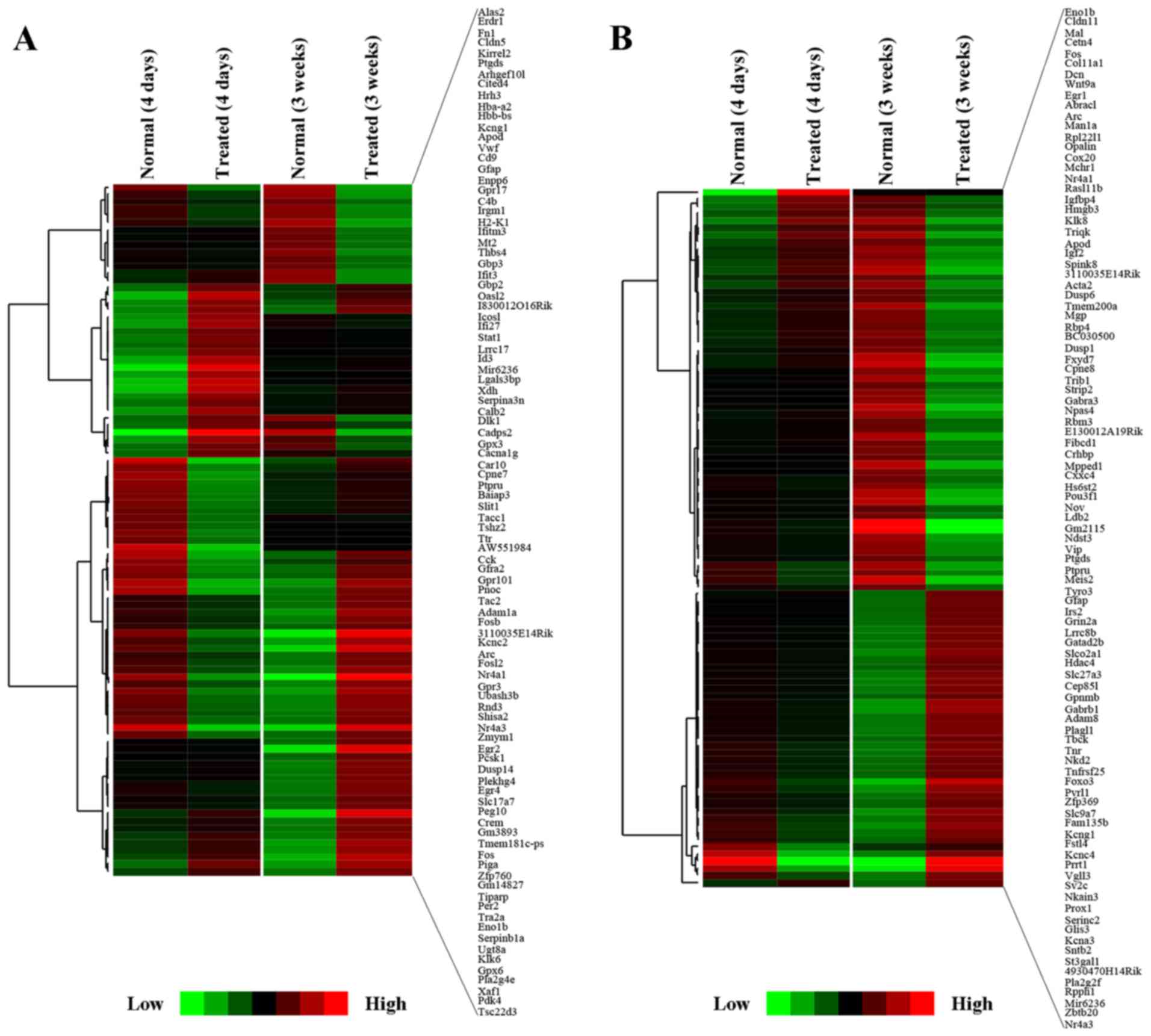
temporal profiles of gene expression in the SVZ (Fig. 4A): i) cluster A, genes with
long-lasting upregulation [from day 4 to week 3, e.g., pyruvate
dehydrogenase kinase, isozyme 4 (Pdk4) and G protein-coupled
receptor 17 (Gpr17)]; and ii) cluster B, genes with a marked
but transient decrease and further oppositely regulated [e.g.,
nuclear receptor subfamily 4, group A, member 3 (Nr4a3),
Fos-like antigen 2 (Fosl2), early growth response 2
(Egr2), Egr4, and solute carrier family 17 (vesicular
glutamate transporter), member 7 (Slc17a7)].
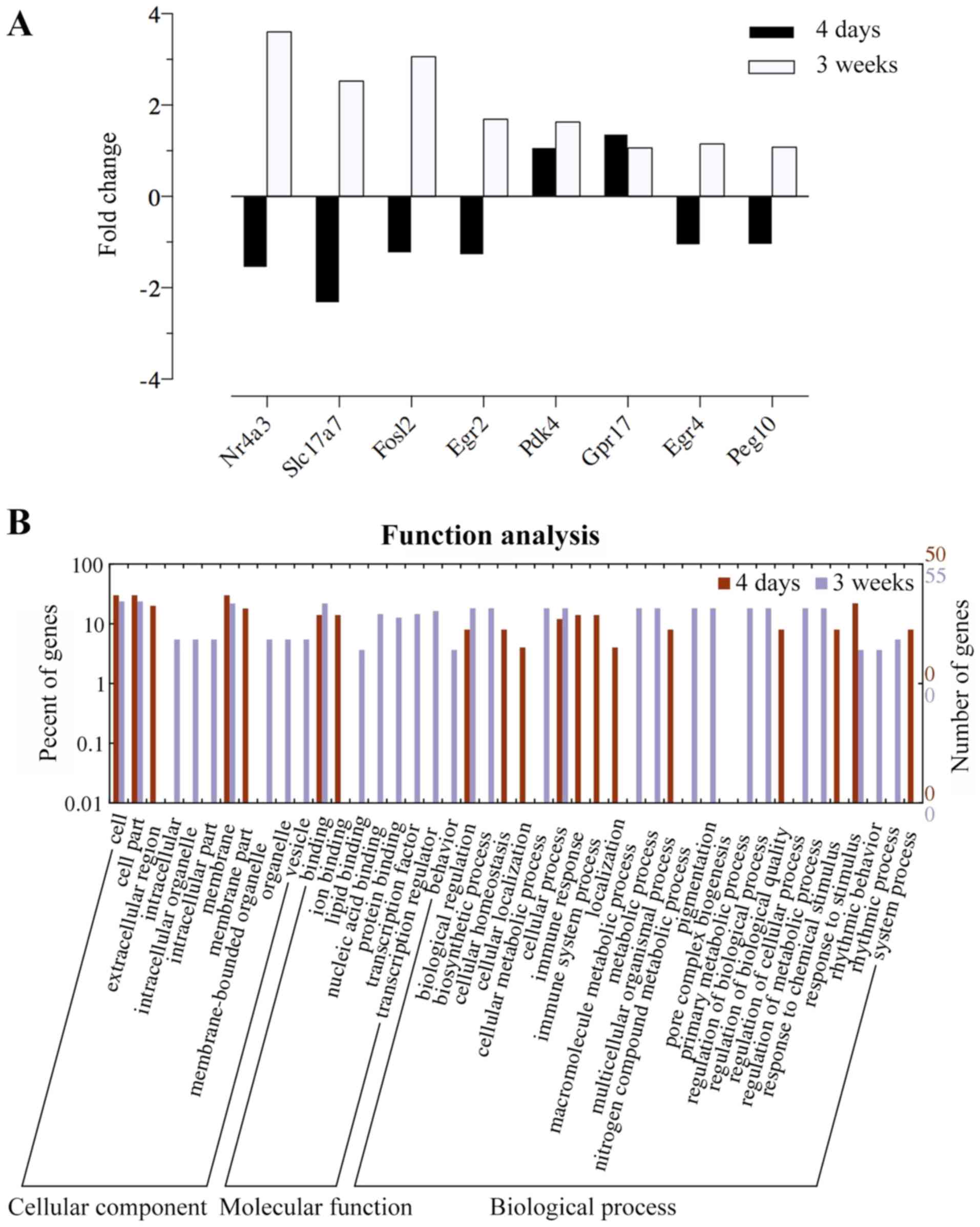 | Figure 4Gene expression patterns and Web Gene
Ontology Annotation Plot (WEGO) in the
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-exposed
subventricular zone (SVZ) at different time points. (A)
Identification of MPTP-induced changes in gene expression in the
SVZ corresponding to the different temporal expression profiles.
There were two different gene expression patterns: i) cluster A,
genes with long-lasting upregulation throughout the experimental
periods [pyruvate dehydrogenase kinase, isozyme 4 (Pdk4) and
G protein-coupled receptor 17 (Gpr17)]; followed by ii)
cluster B, genes with a significant decrease at the early stage but
further overrepresented 3 weeks after MPTP [nuclear receptor
subfamily 4, group A, member 3 (Nr4a3), Fos-like antigen 2
(Fosl2), early growth response 2 (Egr2), Egr4
and solute carrier family 17 (vesicular glutamate transporter),
member 7 (Slc17a7)]. (B) The annotation analysis of
transcriptomic changes in the SVZ was plotted and compared between
two datasets of day 4 and week 3 by WEGO. Gene Ontology (GO)
pathways with >3 genes were considered significant and included
in the figure. In this histogram, significantly differential genes
(FPKM >1, fold change >1 or <−1, q-value <0.05) were
compared between the day 4 and week 3 group. |
Biological functions and pathway analysis
of gene expression changes in the SVZ and DG
To interpret biological functions that may be
altered by differential gene expression after MPTP administration,
we performed GO analysis using the DAVID bioinformatics database.
In the SVZ of the day 4 group, some of the biological functions
were downregulated, including transcription factor activity and the
transmission of nerve impulse, while the significantly enriched GO
term associated with MPTP-induced changes was immune response
(Fig. 4B). The increased
expression of genes included in immune response indicated that this
function was upregulated in the SVZ at the early stage. However, we
found a significant enrichment in transcription factor activity,
the transmission of nerve impulse and myelination, which were
declining at day 4 but presented an opposing direction of changes 3
weeks following exposure (Fig.
5A; Table I). Additionally,
other functions were observed with significant downregulation, such
as cell adhesion (Fig. 5A). In
the DG, a number of biological functions with considerable
activation were found (Fig. 5B;
Table II), including cell
junction, synapse and fatty acid transport. Notably, the
downregulation of the functions, such as transcription activator
activity, MAPK phosphatase activity, extracellular region and
positive regulation of transcription, suggested a potential
negative effect following exposure to MPTP (Fig. 5B).
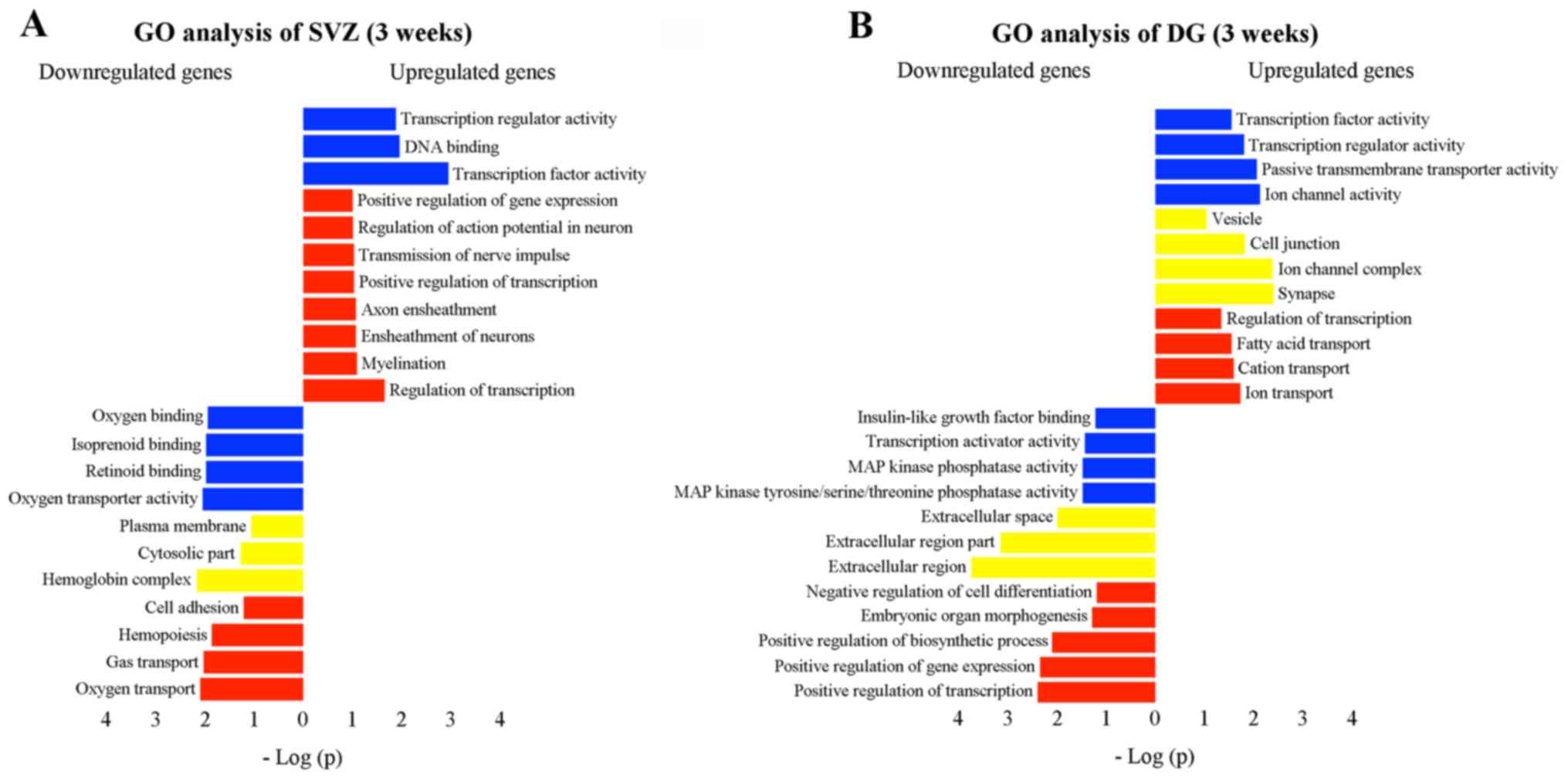 | Figure 5Gene Ontology (GO) analysis,
including cellular component (yellow), molecular function (blue)
and biological process (red) of transcriptomic changes in the (A)
subventricular zone (SVZ) and (B) dentate gyrus (DG) 3 weeks after
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) administration.
The analysis was performed using the functional annotation tool of
the David bioinformatics database. GO pathways with >3 genes
were considered significant if fold enrichment was >2-fold and
the p-value <0.05. The vertical and horizontal axes were the GO
terms and the logarithm of p-values, respectively. Improtantly,
some functions observed in the SVZ after MPTP intoxication were
enhanced (e.g., positive regulation of transcription, transmission
of nerve impulse, and myelination), while others were inversely
affected (e.g., cell adhesion). In the DG, those functions with
considerable activation were identified, including cell junction,
synapse and fatty acid transport. Nevertheless, transcription
activator activity, mitogen-activated protein (MAP) kinase
phosphatase activity, extracellular region, and positive regulation
of transcription were significantly downregulated. |
 | Table ISelected genes differentially
expressed in the SVZ of mice 3 weeks after MPTP exposure. |
Table I
Selected genes differentially
expressed in the SVZ of mice 3 weeks after MPTP exposure.
| Accession no. | Gene symbol | Gene name | FC | q-value |
|---|
| Genes related with
molecular function |
| GOTERM_MF_FAT
transcription factor activity, 8 genes, p-value 7.76E-3 |
| NM_010234 | Fos | FBJ osteosarcoma
oncogene | 1.30 | 0.011 |
| NM_001077364 | Tsc22d3 | TSC22 domain
family, member 3 | 1.19 | 0.011 |
| NM_010118 | Egr2 | Early growth
response 2 | 1.69 | 0.011 |
| NM_008037 | Fosl2 | Fos-like antigen
2 | 3.06 | 0.011 |
| NM_001110850 | Crem | cAMP responsive
element modulator | 1.06 | 0.040 |
| NM_010444 | Nr4a1 | Nuclear receptor
subfamily 4, group A, member 1 | 1.68 | 0.011 |
| NM_001307989 | Nr4a3 | Nuclear receptor
subfamily 4, group A, member 3 | 3.60 | 0.011 |
| NM_008036 | Fosb | FBJ osteosarcoma
oncogene B | 1.14 | 0.011 |
| Genes related with
biological process |
| GOTERM_BP_FAT
transmission of nerve impulse, 5 genes, p-value 0.033 |
| NM_182993 | Slc17a7 | Solute carrier
family 17, member 7 | 2.52 | 0.011 |
| NM_007657 | Cd9 | CD9 antigen | 2.06 | 0.011 |
| NM_010118 | Egr2 | Early growth
response 2 | 1.69 | 0.011 |
| NM_001205075 | Pnoc |
Prepronociceptin | 1.44 | 0.019 |
| NM_011674 | Ugt8a | UDP
galactosyltransferase 8A | 1.17 | 0.011 |
| GOTERM_BP_FAT
regulation of membrane potential, 4 genes, p-value 0.026 |
| NM_007657 | Cd9 | CD9 antigen | 2.06 | 0.011 |
| NM_010118 | Egr2 | Early growth
response 2 | 1.69 | 0.011 |
| NM_011674 | Ugt8a | UDP
galactosyltransferase 8A | 1.17 | 0.011 |
| NM_001112813 | Cacna1g | Calcium channel,
voltage-dependent, T type, α 1G subunit | −1.05 | 0.019 |
| GOTERM_BP_FAT axon
ensheathment, 3 genes, p-value 0.019 |
| NM_007657 | Cd9 | CD9 antigen | 2.06 | 0.011 |
| NM_010118 | Egr2 | Early growth
response 2 | 1.69 | 0.011 |
| NM_011674 | Ugt8a | UDP
galactosyltransferase 8A | 1.17 | 0.011 |
| GOTERM_BP_FAT
myelination, 3 genes, p-value 0.017 |
| NM_007657 | Cd9 | CD9 antigen | 2.06 | 0.011 |
| NM_010118 | Egr2 | Early growth
response 2 | 1.69 | 0.011 |
| NM_011674 | Ugt8a | UDP
galactosyltransferase 8A | 1.17 | 0.011 |
 | Table IISelected genes differentially
expressed in the DG of mice 3 weeks after MPTP exposure. |
Table II
Selected genes differentially
expressed in the DG of mice 3 weeks after MPTP exposure.
| Accession no. | Gene symbol | Gene name | FC | q-value |
|---|
| Genes related with
molecular function |
| GOTERM_MF_FAT
transcription activator activity, 7 genes, p-value 2.22E-3 |
| NM_007913 | Egr1 | Early growth
response 1 | −1.55 | 0.006 |
| NM_207225 | Hdac4 | Histone deacetylase
4 | 1.16 | 0.010 |
| NM_010444 | Nr4a1 | Nuclear receptor
subfamily 4, group A, member 1 | −1.77 | 0.006 |
| NM_019740 | FoxO3 | Forkhead box
O3 | 1.20 | 0.006 |
| NM_001307989 | Nr4a3 | Nuclear receptor
subfamily 4, group A, member 3 | 1.01 | 0.010 |
| NM_011141 | Pou3f1 | POU domain, class
3, transcription factor 1 | −3.65 | 0.006 |
| NM_153553 | Npas4 | Neuronal PAS domain
protein 4 | −2.04 | 0.006 |
| Genes related with
cellular component |
| GOTERM_CC_FAT
synapse, 7 genes, p-value 4.14E-3 |
| NM_207225 | Hdac4 | Histone deacetylase
4 | 1.16 | 0.010 |
| NM_001276684 | Arc | Activity regulated
cytoskeletal-associated protein | −1.67 | 0.006 |
| NM_008067 | Gabra3 | Gamma-aminobutyric
acid (GABA) A receptor, subunit alpha 3 | −1.12 | 0.006 |
| NM_008069 | Gabrb1 | Gamma-aminobutyric
acid (GABA) A receptor, subunit beta 1 | 1.02 | 0.006 |
| NM_008170 | Grin2a | Glutamate receptor,
ionotropic, NMDA2A (ε1) | 1.02 | 0.006 |
| NM_009229 | Sntb2 | Syntrophin, basic
2 | 1.18 | 0.006 |
| NM_029210 | Sv2c | Synaptic vesicle
glycoprotein 2c | 1.46 | 0.006 |
| GOTERM_CC_FAT
extracellular region, 19 genes, p-value 6.37E-4 |
| NM_001313969 | Vip | Vasoactive
intestinal polypeptide | −1.06 | 0.033 |
| NM_001159487 | Rbp4 | Retinol binding
protein 4, plasma | −2.16 | 0.027 |
| NM_008940 | Klk8 | Kallikrein
related-peptidase 8 | −1.33 | 0.020 |
| NM_198408 | Crhbp | Corticotropin
releasing hormone binding protein | −1.99 | 0.006 |
| NM_177059 | Fstl4 | Follistatin-like
4 | 1.06 | 0.043 |
| NM_008597 | Mgp | Matrix Gla
protein | −1.43 | 0.022 |
| NM_001122736 | Igf2 | Insulin-like growth
factor 2 | −2.00 | 0.006 |
| NM_001190451 | Dcn | Decorin | −1.77 | 0.006 |
| NM_183136 | Spink8 | Serine peptidase
inhibitor, Kazal type 8 | −2.29 | 0.006 |
| NM_009177 | St3gal1 | ST3
beta-galactoside α-2,3-sialyltransferase 1 | 1.02 | 0.006 |
| NM_010930 | Nov | Nephroblastoma
overexpressed gene | −4.57 | 0.006 |
| NM_008963 | Ptgds | Prostaglandin
D2 synthase (brain) | −1.73 | 0.006 |
| NM_001301353 | Apod | Apolipoprotein
D | −1.31 | 0.006 |
| NM_022312 | Tnr | Tenascin-R | 1.02 | 0.006 |
| NM_030890 | Prrt1 | Proline-rich
transmembrane protein 1 | 1.01 | 0.006 |
| NM_139298 | Wnt9a | Wingless-type MMTV
integration site family, member 9A | −1.06 | 0.006 |
| NM_007729 | Col11a1 | Collagen, type XI,
α1 | −1.08 | 0.020 |
| NM_010517 | Igfbp4 | Insulin-like growth
factor binding protein 4 | −1.22 | 0.006 |
| NM_012045 | Pla2g2f | Phospholipase A2,
group IIF | 1.28 | 0.006 |
| GOTERM_CC_FAT cell
junction, 8 genes, p-value 7.00E-3 |
| NM_001276684 | Arc | Activity regulated
cytoskeletal-associated protein | −1.67 | 0.006 |
| NM_021424 | Pvrl1 | Poliovirus
receptor-related 1 | 1.18 | 0.006 |
| NM_008067 | Gabra3 | Gamma-aminobutyric
acid (GABA) A receptor, subunit alpha 3 | −1.12 | 0.006 |
| NM_008069 | Gabrb1 | Gamma-aminobutyric
acid (GABA) A receptor, subunit beta 1 | 1.02 | 0.006 |
| NM_008170 | Grin2a | Glutamate receptor,
ionotropic, NMDA2A (ε1) | 1.02 | 0.006 |
| NM_009229 | Sntb2 | Syntrophin, basic
2 | 1.18 | 0.006 |
| NM_001083119 | Ptpru | Protein tyrosine
phosphatase, receptor type, U | −1.31 | 0.006 |
| NM_029210 | Sv2c | Synaptic vesicle
glycoprotein 2c | 1.46 | 0.006 |
| Genes related with
biological process |
| GOTERM_BP_FAT
embryonic organ development, 5 genes, p-value 0.024 |
| NM_001159487 | Rbp4 | Retinol binding
protein 4, plasma | −2.16 | 0.027 |
| NM_001307989 | Nr4a3 | Nuclear receptor
subfamily 4, group A, member 3 | 1.01 | 0.010 |
| NM_139298 | Wnt9a | Wingless-type MMTV
integration site family, member 9A | −1.06 | 0.006 |
| NM_008937 | Prox1 | Prospero homeobox
1 | 1.01 | 0.006 |
| NM_007729 | Col11a1 | Collagen, type XI,
α1 | −1.08 | 0.020 |
| GOTERM_BP_FAT
positive regulation of transcription, 11 genes, p-value
5.21E-5 |
| NM_009538 | Plagl1 | Pleiomorphic
adenoma gene-like 1 | 1.28 | 0.006 |
| NM_007913 | Egr1 | Early growth
response 1 | −1.55 | 0.006 |
| NM_010234 | Fos | FBJ osteosarcoma
oncogene | −1.77 | 0.006 |
| NM_207225 | Hdac4 | Histone deacetylase
4 | 1.16 | 0.010 |
| NM_001305671 | Glis3 | GLIS family zinc
finger 3 | 1.25 | 0.006 |
| NM_001136072 | Meis2 | Meis homeobox
2 | −2.37 | 0.006 |
| NM_010444 | Nr4a1 | Nuclear receptor
subfamily 4, group A, member 1 | −1.77 | 0.006 |
| NM_019740 | Fox03 | Forkhead box
O3 | 1.20 | 0.006 |
| NM_001307989 | Nr4a3 | Nuclear receptor
subfamily 4, group A, member 3 | 1.01 | 0.010 |
| NM_011141 | Pou3f1 | POU domain, class
3, transcription factor 1 | −3.65 | 0.006 |
| NM_153553 | Npas4 | Neuronal PAS domain
protein 4 | −2.04 | 0.006 |
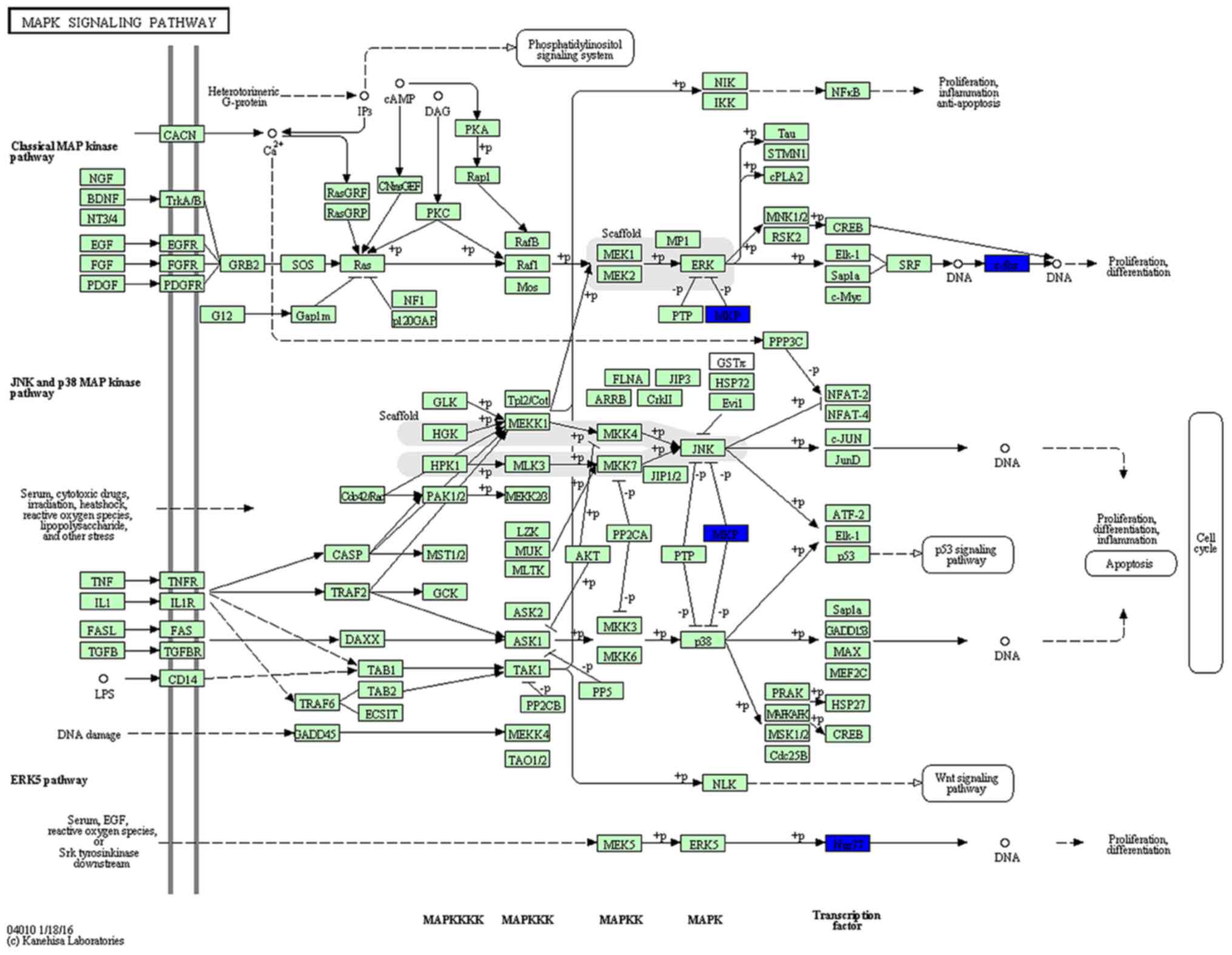
KEGG pathway analysis was then used to reveal
diverse functional entities involved in inter- as well as
intra-cellular signaling cascades. Genes involved in the MAPK
pathway [e.g., FBJ murine osteosarcoma viral oncogene homolog
(Fos), and nuclear receptor subfamily 4, group A, member 1
(Nr4a1)], were significantly upregulated in the SVZ 3 weeks
after the MPTP administration, suggesting active signaling in this
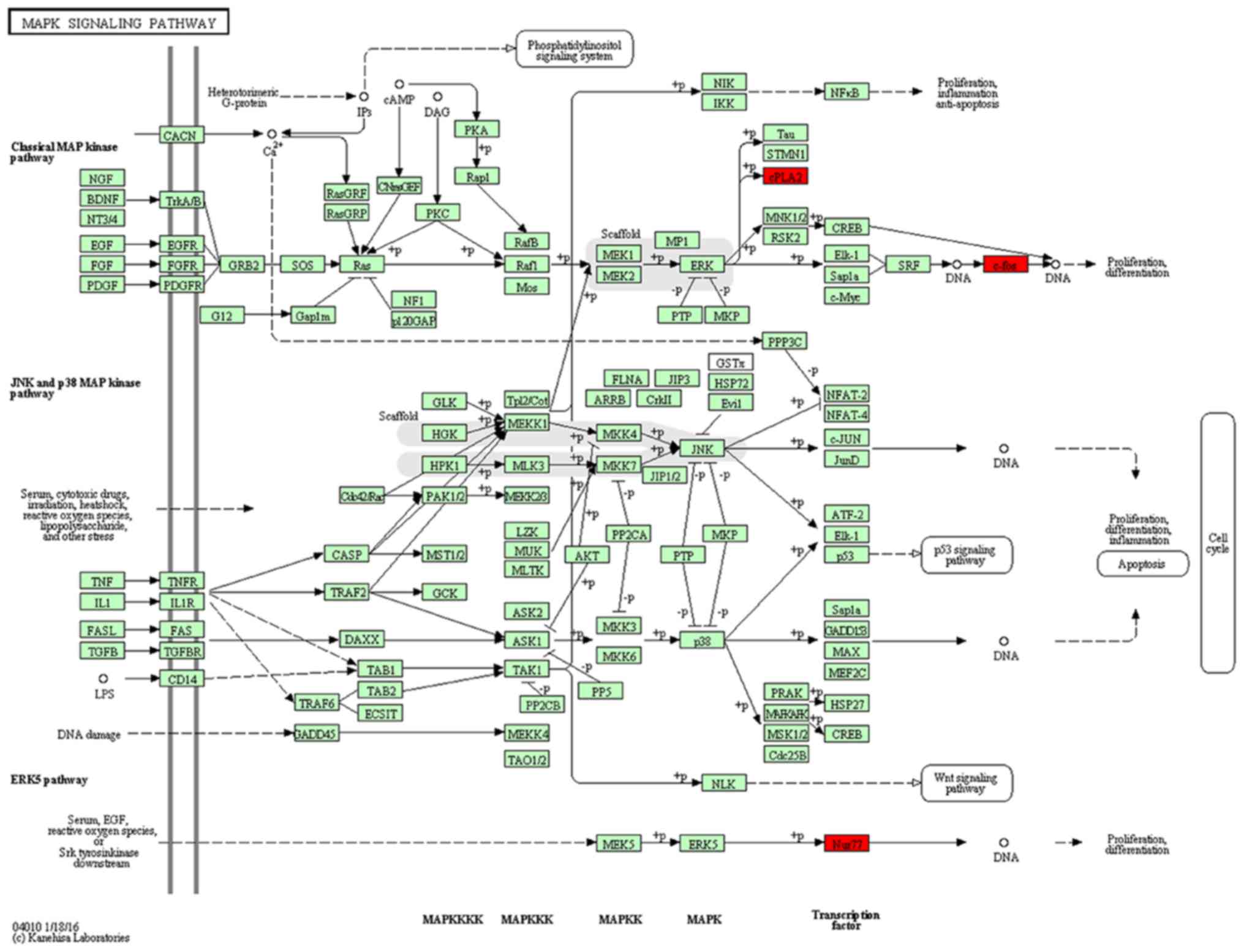
subregion (Fig. 6). In the DG, we
represented significantly downregulated genes in a model of the
MAPK pathway. Surprisingly, the molecular signature in response to
MPTP resulted in a negative effect upon the cascade (Fig. 7). In addition to the
downregulation of transcription factors (Fos and
Nr4a1), a marked decrease of dual-phosphatases mRNA [dual
specificity phosphatase 1 (Dusp1) and Dusp6] was
observed.
Discussion
In this study, using a transcriptomic approach and
taking into account the time course of the effects, we identified
gene expression signature in the SVZ, as well as in the DG induced
by an intraperitoneal injection of MPTP. Our results demonstrated
that the expression of 106 genes in the SVZ and 98 genes in the DG
were modulated in the mice with PD. Immune response, transcription
factor activity, transmission of nerve impulse, myelination and
cell adhesion in the SVZ, and cell junction, fatty acid transport,
extracellular region, and positive regulation of transcription in
the DG were biological functions specifically enriched in our
deregulated gene list. Overall, our data indicated tha expossure to
MPTP regulated MAPK signaling cascades through changes in
transcription factors, as well as MAPK phosphatases.
Genes related to neurogenesis in the
SVZ
Based on the temporal effects, the expression
profiles of more significantly altered genes in the SVZ were
divided into two clusters. Cluster A was composed of genes with
long-lasting upregualtion (e.g., Pdk4 and Gpr17).
Pdk4 encodes pyruvate dehydrogenase kinase that inactivates
pyruvate dehydrogenase complex in the mitochondria (19), shunting the oxidative
phophorylation of glucose to a glycolytic pathway, particularly
after MPTP-induced neurodegeneration (20). Our results revealed that the
MPTP-mediated increase in Pdk4 mRNA expression was sustained
in the SVZ, indicating a metabolic switch to maintain energy
demands. A recent study suggested that the selective activation of
Gpr17 promoted neuronal differentiation and neurite
elongation (21). In addition,
Gpr17 can activate the intracellular phosphorylation of both
extracellular signal-regulated kinase (ERK) and p38 cascades
(21), which have been identified
as members of the MAPK signaling pathway involved in cell
proliferation, differentiation and survival (8,9).
Likewise, we also identified that Gpr17 exhibits persistent
upregulation, which may indicate the active signaling in the
SVZ.
Cluster B consisted of genes with a marked decrease
at the early stage, but further overrepresented 3 weeks after MPTP
exposure. Of note, this cluster included several immediate-early
genes (IEGs) (e.g., Nr4a3, Fosl2, Egr2 and
Egr4) induced by multiple stressors (22–24). It is well known that DA serves as
a regulatory molecule in the SVZ based on the localization of
dopaminergic projections along and within this region (25,26). More recent studies on dopaminergic
neurons have indicated that haloperidol rapidly induces
Nr4a3 in the ventral tegmental area (VTA), and this is
accompanied by the induction of TH and the DA transporter-mRNA
(27). When genes were ranked by
fold change in our study, Nr4a3 was at the top of the list
of genes, in agreement with previous studies showing that the
induction of Nr4a3 was associated with repeated L-DOPA
administration that led to motor improvement (22). The sum of the therapeutic effects
indicated that IEGs, such as Nr4a3 cab be regarded as a
hallmark of recovery in the DA-depleted brain. Although the most
enriched GO term was immune response at the early stage, in our
study, the expression profiles of IEGs (Nr4a3, Egr2
and Egr4) in the SVZ demonstrated that dysfunctional
dopaminergic innervation may be partially rescued. In terms of
temporal dynamics, some functions referring to the genes we
discussed before were also negatively impacted by MPTP at the early
stage and further returned to the normal state, including
transcription factor activity [Fos, Fosl2, FBJ murine
osteosarcoma viral oncogene homolog B (Fosb), Nr4a1,
Nr4a3 and Egr2], transmission of nerve impulse
(Slc17a7 and Egr2) and myelination (Egr2).
Firstly, the upregulation of transcription factors (Fos,
Fosl2, and Fosb) may influence the expression of
various genes involved in cell proliferation and differentiation
after extracellular stimuli. Particularly, Fos is currently
used as a marker of neuronal activity and has been associated with
a number of neural responses to acute stimuli expression. A
decrease in the level of Fos was observed in age-related
degeneration, as well as impaired brain development (28). Llorens-Bobadilla et al
found that the active state of NSCs in the SVZ was associated with
the expression of IEGs including Fos (10). Moreover, other IEGs such as
Nr4a1 and Nr4a3 encode orphan nuclear receptors that
function as transcription factors (23). Nr4a1 has been linked to
synaptic remodeling, behavioral changes, and dopaminergic loss
after administrating MPTP in mice (27). The possible therapeutic role for
Nr4a1 has been manifested since neural damage was rescued by
Nr4a1 overexpression that decreased cell apoptotic rates
(29) and was essential for
neuronal differentiation (30).
In our study, genes involved in the MAPK pathway (Fos and
Nr4a1) were significantly upregulated in the SVZ 3 weeks
after MPTP administration. There are three major groups of MAPKs:
i) ERK which is mainly activated by mitogenic stimuli, such as
growth factors and hormones; ii) c-Jun N-terminal kinase (JNK) and
p38 which are predominantly activated by stress stimuli, such as
oxidative stress and inflammatory cytokines; and iii) ERK5 which is
activated by both stress stimuli as well as growth factors
(9,31). Notably, our results displayed that
since the enhanced transcription of Fos was accompanied by
Nr4a1, MPTP may selectively activate the ERK and ERK5
cascades in the SVZ at later stage, which may lead to the
biological outcomes of cell proliferation and differentiation.
Secondly, Slc17a7 and Egr2 genes may be involved in
the function of glutamatergic synapse and myelination,
respectively. Slc17a7 encodes the protein of vesicular
glutamate transporter 1 (Vglut1) and functions in glutamate
transport (32). Sconce et
al suggested that the alleviation of motor deficits in mice
exposed to MPTP was attributed to the recovery of extracellular
Vglut1 and glutamatergic synapse (33). Others have identified a
contribution of Vglut1 to glutamate release which could promote
neuroblast migration and neurogenesis (34). In addition to neurotransmitter
transporter, myelination that occurs primarily post-natally is
essential for the efficient transmission of nerve impulses.
Egr2 activates several genes that are involved in the
formation and maintenance of myelin at early stage of myelination
(35). Recent studies have
provided evidence that oligodendrocyte progenitor cells in the SVZ
migrate following exposure to stimuli and form glutamatergic
synapse during the early stage of myelination process (35,36). Hence, the fact that the expression
of Slc17a7 and Egr2 was time-dependent has led us to
speculate that neurogenesis, including synaptic formation may be
initially inhibited and further improved to partially rescue the
dysfunctional dopaminergic system in our parkinsonian mice. Future
studies shall attempt to reveal the mechanisms underlying the
temporal profiles of gene expression.
Genes related to neurogenesis in the
DG
Contrary to the expression profiles in the SVZ, the
biological function of positive regulation of transcription in the
DG was negatively affected by MPTP. In support of this hypothesis,
the upregulation of histone deacetylase 4 (Hdac4) and the
downregulation of Fos and Nr4a1 were observed and
annotated to this function. In fact, Hdac4 encodes a protein
to repress transcription, and the hight expression of Hdac4
in granule cells of the DG is associated with neurodegeneration
(37). Furthermore, these changes
can impact several major targets of the MAPK pathway. Specifically,
the ERK and ERK5 cascades may be inactivated by MPTP at a later
stage as the downstream transcription factors, Fos and
Nr4a1, were significantly downregulated after DA depletion.
Indeed, the loss of the ERK cascade results in the depletion of
neural progenitors, impairing granule cell neurogenesis in the DG
(9,38). Jiang et al demonstrated
that the inhibition of MAPK/ERK signaling aggravated hippocampal
neuronal apoptosis, decreased neurogenesis and impaired cognitive
performances (39). Thus, our
results support the previous findings that neurogenesis in the SGZ
of the DG decreased accompanying dopaminergic denervation after
MPTP epxosure (2,38). However, we identified significant
decreases in the levels of Dusp1, as well as Dusp6,
which were MAPK phospha-tases and negative feedback regulators of
ERK cascade (9,40). Consequently, by the MPTP-induced
downregulation of Dusp1 and Dusp6, the inactivation
of ERK could be reduced, leading to the compensatory reactions to
the DA depletion.
In addition to the regulation of transcription,
exposure to MPTP also affected genes mediating extracellular region
remodeling, and cell junction, which may impact neural plasticity
via cell migration or neurite outgrowth. We identified significant
decreases in the expression of insulin-like growth factor 2
(Igf2), insulin-like growth factor binding protein 4
(Igfbp4), prostaglandin D2 synthase
(Ptgds) and nephroblastoma overexpressed (Nov) which
could be annotated to the extracellular region. Currently, it is
well accepted that Igf2 encodes a member of the insulin-like
growth factor family of proteins that can facilitate efficient
signal transduction from extracellular region into the cell and
promote growth and development (11,31). It also acts as a physiological
amplifier of glucose-mediated insulin secretion (11,13), indicating a metabolic function.
Moreover, the gene of Igfbp4 encodes the IGF-binding
proteins that have been shown to stimulate the growth promoting
effects of the IGFs on cell culture (11,12). On the one hand, the MPTP-mediated
downregulation of Igf2 and Igfbp4 may inhibit signal
transduction, causing cell growth dysfunction. On the other hand,
due to the insufficient secretion of insulin, an increase in fatty
acid metabolism may begin, which was confirmed by our GO analysis.
Shin et al found that quiescent NSCs in the DG maintained an
active fatty acid metabolism which began to wane when these cells
were activated (13). Our
findings of active biological process of fatty acid transport may
partially lead to the speculation that NSCs may be inactivated in
the DG. Other downregulated genes related to extracellular region
included collagen, type XI, α1 (Col11a1) (41) and decorin (Dcn) (42) that may have a negative impact on
collagen metabolism and assembly after dopaminergic denervation.
Apart from a negative effect on extracellular region remodeling,
the considerable activation of cell junction and synapse were
observed following exposure to MPTP, which suggested that there
were some compensatory mechanisms in the DG to enable communication
between neighboring cells and reduce stress. Overall, we
demonstrated that epxosure to MPTP may have the potential to
inhibit the expression of genes encoding proteins that could
regulate extracellular matrix structure to prevent cell migration
within the DG.
In conclusion, our findings demonstrated that
exposure to MPTP affects gene expression in the SVZ and DG. The
time-dependent expression profiles in the SVZ have led us to
speculate that neurogenesis may be initially inhibited and further
improved in our mice with PD. Nevertheless, expression analysis has
shown the dysfunction of cell proliferation and migration in the
DG. Several deregulated genes that participate in the glucose and
lipid metabolisms may result in metabolic switch in these two
regions. It is worth emphasizing that the MAPK signaling pathway
has been activated in the SVZ, while negatively regulated in the DG
after MPTP administration. Future studies shall attempt to reveal
the mechanisms underlying neuroplasticity in the SVZ and SGZ using
single-cell sequencing to avoid cellular heterogeneity within mixed
cell populations.
Acknowledgments
This study was supported by the National High
Technology Research and Development Program of China (2013AA020106
and 2014AA020513), National Basic Research Program of China
(2014CB541603), and National Natural Science Foundation of China
(81200916).
References
|
1
|
Ming GL and Song H: Adult neurogenesis in
the mammalian brain: significant answers and significant questions.
Neuron. 70:687–702. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Höglinger GU, Rizk P, Muriel MP,
Duyckaerts C, Oertel WH, Caille I and Hirsch EC: Dopamine depletion
impairs precursor cell proliferation in Parkinson disease. Nat
Neurosci. 7:726–735. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohtani N, Goto T, Waeber C and Bhide PG:
Dopamine modulates cell cycle in the lateral ganglionic eminence. J
Neurosci. 23:2840–2850. 2003.PubMed/NCBI
|
|
4
|
van den Berge SA, van Strien ME, Korecka
JA, Dijkstra AA, Sluijs JA, Kooijman L, Eggers R, De Filippis L,
Vescovi AL, Verhaagen J, et al: The proliferative capacity of the
subventricular zone is maintained in the parkinsonian brain. Brain.
134:3249–3263. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marxreiter F, Regensburger M and Winkler
J: Adult neurogenesis in Parkinson's disease. Cell Mol Life Sci.
70:459–473. 2013. View Article : Google Scholar
|
|
6
|
Aponso PM, Faull RL and Connor B:
Increased progenitor cell proliferation and astrogenesis in the
partial progressive 6-hydroxydopamine model of Parkinson's disease.
Neuroscience. 151:1142–1153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winner B, Geyer M, Couillard-Despres S,
Aigner R, Bogdahn U, Aigner L, Kuhn G and Winkler J: Striatal
deafferentation increases dopaminergic neurogenesis in the adult
olfactory bulb. Exp Neurol. 197:113–121. 2006. View Article : Google Scholar
|
|
8
|
Horgusluoglu E, Nudelman K, Nho K and
Saykin AJ: Adult neurogenesis and neurodegenerative diseases: a
systems biology perspective. Am J Med Genet B Neuropsychiatr Genet.
174:93–112. 2017. View Article : Google Scholar
|
|
9
|
Faigle R and Song H: Signaling mechanisms
regulating adult neural stem cells and neurogenesis. Biochim
Biophys Acta. 1830:2435–2448. 2013. View Article : Google Scholar :
|
|
10
|
Llorens-Bobadilla E, Zhao S, Baser A,
Saiz-Castro G, Zwadlo K and Martin-Villalba A: Single-cell
transcriptomics reveals a population of dormant neural stem cells
that become activated upon brain injury. Cell Stem Cell.
17:329–340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Agis-Balboa RC, Arcos-Diaz D, Wittnam J,
Govindarajan N, Blom K, Burkhardt S, Haladyniak U, Agbemenyah HY,
Zovoilis A, Salinas-Riester G, et al: A hippocampal insulin-growth
factor 2 pathway regulates the extinction of fear memories. EMBO J.
30:4071–4083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou R, Diehl D, Hoeflich A, Lahm H and
Wolf E: IGF-binding protein-4: biochemical characteristics and
functional consequences. J Endocrinol. 178:177–193. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shin J, Berg DA, Zhu Y, Shin JY, Song J,
Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al:
Single-cell RNA-seq with waterfall reveals molecular cascades
underlying adult neurogenesis. Cell Stem Cell. 17:360–372. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zuo FX, Bao XJ, Sun XC, Wu J, Bai QR, Chen
G, Li XY, Zhou QY, Yang YF, Shen Q, et al: Transplantation of human
neural stem cells in a parkinsonian model exerts neuroprotection
via regulation of the host microenvironment. Int J Mol Sci.
16:26473–26492. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jackson-Lewis V and Przedborski S:
Protocol for the MPTP mouse model of Parkinson's disease. Nat
Protoc. 2:141–151. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Giasson BI, Duda JE, Quinn SM, Zhang B,
Trojanowski JQ and Lee VM: Neuronal alpha-synucleinopathy with
severe movement disorder in mice expressing A53T human
alpha-synuclein. Neuron. 34:521–533. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ye J, Fang L, Zheng H, Zhang Y, Chen J,
Zhang Z, Wang J, Li S, Li R, Bolund L, et al: WEGO: a web tool for
plotting GO annotations. Nucleic Acids Res. 34(Web server):
W293–W297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kolobova E, Tuganova A, Boulatnikov I and
Popov KM: Regulation of pyruvate dehydrogenase activity through
phosphorylation at multiple sites. Biochem J. 358:69–77. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee DW, Rajagopalan S, Siddiq A, Gwiazda
R, Yang L, Beal MF, Ratan RR and Andersen JK: Inhibition of prolyl
hydroxylase protects against
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced neurotoxicity:
model for the potential involvement of the hypoxia-inducible factor
pathway in Parkinson disease. J Biol Chem. 284:29065–29076. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Daniele S, Lecca D, Trincavelli ML, Ciampi
O, Abbracchio MP and Martini C: Regulation of PC12 cell survival
and differentiation by the new P2Y-like receptor GPR17. Cell
Signal. 22:697–706. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Charbonnier-Beaupel F, Malerbi M, Alcacer
C, Tahiri K, Carpentier W, Wang C, During M, Xu D, Worley PF,
Girault JA, et al: Gene expression analyses identify Narp
contribution in the development of L-DOPA-induced dyskinesia. J
Neurosci. 35:96–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Södersten E, Feyder M, Lerdrup M, Gomes
AL, Kryh H, Spigolon G, Caboche J, Fisone G and Hansen K: Dopamine
signaling leads to loss of Polycomb repression and aberrant gene
activation in experimental parkinsonism. PLoS Genet.
10:e10045742014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Safe S, Jin UH, Morpurgo B, Abudayyeh A,
Singh M and Tjalkens RB: Nuclear receptor 4A (NR4A) family -
orphans no more. J Steroid Biochem Mol Biol. 157:48–60. 2016.
View Article : Google Scholar
|
|
25
|
Björklund A and Dunnett SB: Dopamine
neuron systems in the brain: an update. Trends Neurosci.
30:194–202. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Baker SA, Baker KA and Hagg T:
Dopaminergic nigrostriatal projections regulate neural precursor
proliferation in the adult mouse subventricular zone. Eur J
Neurosci. 20:575–579. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eells JB, Wilcots J, Sisk S and Guo-Ross
SX: NR4A gene expression is dynamically regulated in the ventral
tegmental area dopamine neurons and is related to expression of
dopamine neurotransmission genes. J Mol Neurosci. 46:545–553. 2012.
View Article : Google Scholar :
|
|
28
|
Velazquez FN, Prucca CG, Etienne O,
D'Astolfo DS, Silvestre DC, Boussin FD and Caputto BL: Brain
development is impaired in c-fos −/− mice. Oncotarget.
6:16883–16901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiao G, Sun T, Songming C and Cao Y: NR4A1
enhances neural survival following oxygen and glucose deprivation:
an in vitro study. J Neurol Sci. 330:78–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tomioka T, Maruoka H, Kawa H, Yamazoe R,
Fujiki D, Shimoke K and Ikeuchi T: The histone deacetylase
inhibitor trichostatin A induces neurite outgrowth in PC12 cells
via the epigenetically regulated expression of the nur77 gene.
Neurosci Res. 88:39–48. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang SH, Sharrocks AD and Whitmarsh AJ:
Transcriptional regulation by the MAP kinase signaling cascades.
Gene. 320:3–21. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guo Z, Zhang L, Wu Z, Chen Y, Wang F and
Chen G: In vivo direct reprogramming of reactive glial cells into
functional neurons after brain injury and in an Alzheimer's disease
model. Cell Stem Cell. 14:188–202. 2014. View Article : Google Scholar
|
|
33
|
Sconce MD, Churchill MJ, Greene RE and
Meshul CK: Intervention with exercise restores motor deficits but
not nigrostriatal loss in a progressive MPTP mouse model of
Parkinson's disease. Neuroscience. 299:156–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sánchez-Mendoza E, Burguete MC,
Castelló-Ruiz M, González MP, Roncero C, Salom JB, Arce C, Cañadas
S, Torregrosa G, Alborch E, et al: Transient focal cerebral
ischemia significantly alters not only EAATs but also VGLUTs
expression in rats: relevance of changes in reactive astroglia. J
Neurochem. 113:1343–1355. 2010.PubMed/NCBI
|
|
35
|
Etxeberria A, Mangin JM, Aguirre A and
Gallo V: Adult-born SVZ progenitors receive transient synapses
during remyelination in corpus callosum. Nat Neurosci. 13:287–289.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Aguirre A, Dupree JL, Mangin JM and Gallo
V: A functional role for EGFR signaling in myelination and
remyelination. Nat Neurosci. 10:990–1002. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Whitehouse A, Doherty K, Yeh HH, Robinson
AC, Rollinson S, Pickering-Brown S, Snowden J, Thompson JC,
Davidson YS and Mann DM: Histone deacetylases (HDACs) in
frontotemporal lobar degeneration. Neuropathol Appl Neurobiol.
41:245–257. 2015. View Article : Google Scholar
|
|
38
|
Vithayathil J, Pucilowska J, Goodnough LH,
Atit RP and Landreth GE: Dentate gyrus development requires ERK
activity to maintain progenitor population and MAPK pathway
feedback regulation. J Neurosci. 35:6836–6848. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang P, Zhu T, Xia Z, Gao F, Gu W, Chen
X, Yuan T and Yu H: Inhibition of MAPK/ERK signaling blocks
hippocampal neurogenesis and impairs cognitive performance in
prenatally infected neonatal rats. Eur Arch Psychiatry Clin
Neurosci. 265:497–509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Banzhaf-Strathmann J, Benito E, May S,
Arzberger T, Tahirovic S, Kretzschmar H, Fischer A and Edbauer D:
MicroRNA-125b induces tau hyperphosphorylation and cognitive
deficits in Alzheimer's disease. EMBO J. 33:1667–1680. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Majava M, Hoornaert KP, Bartholdi D, Bouma
MC, Bouman K, Carrera M, Devriendt K, Hurst J, Kitsos G, Niedrist
D, et al: A report on 10 new patients with heterozygous mutations
in the COL11A1 gene and a review of genotype-phenotype correlations
in type XI collagenopathies. Am J Med Genet A. 143A:258–264. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Buraschi S, Neill T, Goyal A, Poluzzi C,
Smythies J, Owens RT, Schaefer L, Torres A and Iozzo RV: Decorin
causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci
USA. 110:E2582–E2591. 2013. View Article : Google Scholar : PubMed/NCBI
|