Introduction
Rheumatic heart disease (RHD) is an autoimmune
disease that arises following infection of the throat by S.
pyogenes in young individuals (3–19 years old) and children,
who present genetic components that confer susceptibility to this
disease (1). Both in developing
countries and China, RHD remains to be a major public health
concern that contributes to significant cardiac-related mortality
and morbidity (2). RHD accounts
for up to 250,000 premature deaths every year in the world and is
considered as a physical manifestation of poverty and social
inequality (3). A previous study
indicated that molecular mimicry mediates the cross-reactions
between streptococcal antigens and human proteins. Several
autoantigens including vimentin, cardiac myosin epitopes, and other
intracellular proteins have been identified (4).
It is well-accepted that both T and B lymphocytes
can recognize pathogens and self-antigens through different types
of molecular mimicry (4).
Particularly, the role of T lymphocytes in the development of RHD
was previously described in the 1980s (5). The epitope spreading, molecular
mimicry and T cell receptor degeneracy were demonstrated as
mechanisms that mediate rheumatic heart lesions through the
identification of streptococcal and heart-tissue proteins
cross-reactive intralesional T cell clones (4). Cytokines are important secondary
signals following an infection since they cause deleterious
responses in patients with autoimmune disease and trigger effective
immune responses in most individuals (1). Antigen-activated CD4+ T
cells polarize to the T helper type 1 (Th1), Th2 or Th17 subsets,
three subsets of T helper cytokines, depending on the cytokine
secreted. Th1 can produce interleukin-2 (IL-2), interferon-γ
(IFN-γ), and tumor necrosis factor-α (TNF-α), and is involved with
the cellular immune response. Th2 cells can produce IL-4, IL-5 and
IL-13 and mediate humoral and allergic immune responses. Th17 is a
type of proinflammatory response mediated by IL-17. In addition,
the cytokines, transforming growth factor-β (TGF-β), IL-6 and
IL-23, are the factors that induce the Th17 lineage. Among them, a
significant role for IL-4 in Th2 development was recognized early
on in the analysis of the CD4+ T cell subset and was
also further confirmed by in vitro experimentation (6,7).
Moreover, a previous study suggested that mimicry between
streptococcal antigens and heart-tissue proteins, combined with
high inflammatory cytokines and low IL-4 production, lead to the
development of autoimmune reactions and cardiac tissue damage in
RHD patients (8).
Interferon regulatory factor 4 (IRF4) is an immune
system-restricted interferon regulatory factor that is required for
lymphocyte activation. Researchers have demonstrated that IRF4 may
be a component of such a Th2-specific protein complex serving to
enhance the function of nuclear factor of activated T cell-2
(NFATc2) via its physical interaction with NFATc2 at the IL-4
locus, indicating the important role of IRF4 in Th2 development
(9). It seems that IRF4 may play
an important role in RHD progression by regulating IL-4 production
and Th2 development. Deubiquitinating enzymes (DUBs) are proteases
that process ubiquitin or ubiquitin-like gene products, reverse the
modification of proteins by a single ubiquitin(-like) protein, and
remodel polyubiquitin(-like) chains on target proteins (10). The ubiquitin-specific proteases
(USP) have more than 50 members and represent the largest subclass
of DUBs (11). Like other USPs,
USP4 also mediates the removal and processing of ubiquitin. USP4
binds TGF-activated kinase 1 (TAK1), TNF receptor-associated factor
2 (TRAF2) and TRAF6 for deubiquitination and negatively regulates
TNF-α-induced nuclear factor NF-κB (NF-κB) activation (12,13). USP4 directly interacts with and
deubiquitinates TGF-β type I receptor (TβRI), regulating TGF-β
signaling by controlling TβRI levels at the plasma membrane
(14). USP4 also interacts
directly with and deubiquitinates ARF-BP1, leading to the
stabilization of ARF-BP1 and subsequent reduction of p53 levels
(15).
Here, we speculated that USP4 may deubiquitinate
IRF4, and affect Th2 cell function and may be associated with the
pathogenesis of RHD. This study aimed to validate this
hypothesis.
Materials and methods
Human samples
Twenty RHD patients (average age, 50 years) were
enrolled from July, 2013 to January, 2014 at the First Affiliated
Hospital of Anhui Medical University (Anhui, China). Preoperative
echocardiography and postoperative pathological research were used
to confirm RHD. Patients with dysfunction of liver and kidney,
coronary heart disease, infectious diseases, other autoimmune
diseases and malignant tumors were excluded. Ten healthy volunteers
that were randomly selected from a population-based sample living
in the same region with the same age distribution as the patient
group, were considered as controls. Five milliliters of peripheral
blood was drawn from every patient and control. This study was
approved by the Ethics Committee of Anhui Medical University and
the Institute Pasteur of Shanghai, the Chinese Academy of
Sciences.
Plasmids and antibodies
The plasmids, Myc-IRF4 and Ha-USP4 and His-ubiquitin
plasmids used in this study were provided by Dr Li Bin from the
Institute Pasteur of Shanghai. Anti-flag and anti-USP4 (cat. no.
U0635MSDS) antibodies were purchased from Sigma-Aldrich (St. Louis,
MO, USA). The anti-Ha (cat. no. sc-7392 AC) and anti-ubiquitin
(P4D1; cat. no. sc-8017) antibodies were purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). The anti β-actin
antibody (cat. no. LK9001T) was purchased from Tianjin Sungene
Biotech Co., Ltd. (Tianjin, China). PerCP/Cy5.5 anti-human CD45RA
(cat. no. 304121), FITC anti-human CD4 (cat. no. 300506), PE
anti-human CD25 (cat. no. 302606), Pb anti-human/mouse IRF4 (cat.
no. 48-9858-82) was purchased from eBioscience (San Diego, CA,
USA). PerCP/Cy5.5 anti-human IL-4 (cat. no. 500822) was purchased
from BioLegend, Inc. (San Diego, CA, USA).
Flow cytometry-based cell sorting
Peripheral blood mono-nuclear cells (PBMCs) were
prepared by density gradient centrifugation according to standard
Ficoll-Hypaque procedures (Pharmacia Biotech, Uppsala, Sweden).
PBMCs were diluted with phosphate-buffered saline (PBS) and were
stained with FITC anti-human CD4, PE-anti-human CD25, PerCP/Cy5.5
anti-human CD45RA (BD Biosciences, San Jose, CA, USA) or
isotype-matched controls. The concentration of antibodies in each
flow tube was ~20 µl/1–1.5×108 million cells, and
the single-dyed and the non-dyed groups were set at the same time.
The antibody concentration was ~1 µl/l×106 cells
(dissolved in 50 µl PBS). After incubation on ice for 40
min, the cells were washed, resuspended in fluorescence-activated
cell sorter (FACS) staining buffer (BD Biosciences). Naïve
CD4+ T cells were gated on CD4, CD25 and CD45RA. After
that, cells were centrifuged for 10 min, and resuspended in
X-VIV015 medium (Lonza Co., Basel, Switzerland), and were cultured
in an incubator (Thermo Fisher Scientific, Wilmington, DE, USA),
with an atmosphere of 37°C and 5% CO2.
CD4+ T cell
differentiation
Naïve CD4+ cells were seeded at a density
of 2×105/well in 96-well plates, and were cultured in
DMEM, containing 10% fetal bovine serum (FBS). For Th2 cell
differentiation, cells were simulated with CD3/CD28 beads
(Invitrogen, Carlsbad, USA) and the skewing conditions were as
follows: IL-2 (50 U/ml), 20 ng/ml rIL-4 and anti-IFN-γ (10
µg/ml; BioLegend, Inc.). The cells were incubated for 3 days
and further experimental analysis was performed after 7 days.
Cell culture
293T cells were purchased from the American Type
Culture Collection (ATCC, Manassas, VA, USA), and were cultured in
Dulbecco's modified Eagle's medium (DMEM) (HyClone, Logan, UT, USA)
containing 10% FBS. The naïve CD4+ T cells and Th2 cells
were obtained from peripheral blood of healthy samples, and were
cultured in X-VIVO15 medium (Lonza Co.) containing 10% human AB
serum (Weihong Biotechnology Co., Ltd., Shanghai, China), 1%
non-essential amino-acid, 1% glutamine, 1% sodium pyruvate, 1%
penicillin and streptomycin (all from Gibco, Grand Island, NY,
USA). The cells were subsequently cultured in an incubator (Thermo
Scientific), with an atmosphere of 37°C and 5% CO2.
Cell transfection
293T cells were transfected with vectors using
linear polyethyleneimine (PEI; Polysciences, Inc., Warrington, PA,
USA). Cells (1×106) and plated in 6-well plates for 24 h
before transfection. The plasmid was mixed with 100 µl
OPTI-MEM (Gibco) low serum medium, and 12 µl of PEI was
added and incubated for 10 min. The cell supernatant was then
replaced with 900 µl fresh DMEM containing 10% FBS, and the
transfection mixture was added, and then incubated in an incubator,
with an atmosphere of 37°C and 5% CO2. After 4 h, the
cell supernatant was replaced with 10% fresh FBS/fresh medium, and
was cultured for 36–48 h for cell collection.
Transduction of shRNA lentiviruses
The lentiviral vector pLKO.1-puro was purchased from
Addgene (Cambridge, MA, USA), and sh-USP41, sh-USP42 and sh-CK were
obtained as described in a previous study (14). Helper plasmids were cotransfected
with pLKO.1-based plasmids carrying shRNAs in 293T cells to package
recombinant lentiviruses. HIV-1 virus titers were quantified by
determining the TCID50 in 293T cells. Lentiviral vector
supernatant was concentrated by centrifugation at 50,000 × g for
2.5 h directly after harvesting. Cell supernatants were harvested
48 h after transfection and were used to infect Th2 cells.
Polybrene (8 µg/ml) was added and fresh culture solution was
replaced on the next day. Cells were retrieved on day 7 for the
detection of the transcriptional levels of USP4, IRF4, IL-4, IL-5,
IL-10 and IL-13.
Co-immunoprecipitation
After 36–48 h of transfection, the cells was
collected for co-immunoprecipitation analysis as previously
described (16). Briefly, cells
were washed, harvested, and centrifuged. The cell pellet was
homogenized and cell lysis buffer was added. The homogenate was
centrifuged, and the supernatant was incubated with
anti-Ha/anti-Myc antibody and Protein A/G-Plus beads overnight. The
resins were recovered and washed followed by centrifugation. The
pellet was then washed twice with Tris-HCL (pH 7.5) containing 0.1%
(w/v) SDS and 150 mM NaCl. After that, it was resuspended in 1X SDS
loading buffer, heated at 100°C for 3 min and subjected to sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
followed by western blot analysis.
Ni-NTA pull down assay
Before being harvested for Ni-NTA pull down assay,
the cells were treated with MG132 (10 nM) for 4 h to inhibit
proteasomal degradation and prevent the ubiquitination of proteins.
The cells were lysed with urea lysis buffer (8 M urea, 100 mM
Na2HPO4, 10 mM Tris-HCl (pH 8.0), 10 mM
imidazole 0.2% and Triton X-100) and fully disrupted by
ultrasonication. After that, the cell lysate was incubated with
Ni-NTA agarose beads for 3 h to obtain His-tag combined with
Ni-NTA, followed by washing with wash buffer. Then, 2X SDS loading
buffer was added to obtain samples for western blotting. The
ubiquitination modification was analyzed using the indicated
antibodies.
Construction of luciferase reporter gene
constructs
The sequences of the promoter of IL-4 were cloned
into pGL3 basic expression plasmid. pGL3-plasmid with promoter
fragments were sequenced using automated DNA sequencing methods.
293T cells were transfected with pGL3 constructs along with 3 mg of
pSV-β-galactosidase control vector. Lipofectamine 2000 (Invitrogen)
was used according to the manufacturer's instructions. Luciferase
and β-gal assays were conducted 24 h post-transfection using the
Luciferase assay system and the β-galactosidase enzyme assay system
(both from Promega, Madison, WI, USA), respectively.
Flow cytometric analysis
PBMCs were prepared by density gradient
centrifugation from RHD patients and healthy controls according to
standard Ficoll-Hypaque procedures (Pharmacia Biotech, Uppsala,
Sweden). PBMCs were then treated with phorbol myristate acetate
(PMA, 25 ng/ml) and ionomycin (1 µg/ml) for 4 h and PBMCs
were stained and processed by flow cytometry.
Naïve CD4+ T cells were sorted using the
cytometry-based analysis and were cultured for Th2 cell
differentiation, and were treated with the inhibitor of USP4,
vialinin A, for 12 h. Th2 cells were then treated with phorbol
myristate acetate (PMA, 25 ng/ml) and ionomycin (1 µg/ml)
for 4 h and processed by flow cytometry.
Western blot analysis
Following 36–48 h of cell transfection, cell lysates
(30 mg/well) were subjected to sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a
polyvinylidine difluoride membrane (Millipore, Darmstadt, Germany).
The blots were incubated with the primary antibodies overnight at
4°C. The membranes were washed and then incubated with horseradish
peroxidase-conjugated secondary antibodies (Sigma-Aldrich). The
immunoreactive proteins were visualized by the SuperSignal ECL
(Pierce, Rockford, IL, USA) and the densities of the bands were
analyzed by ImageJ software (National Institutes of Health,
Bethesda, MD, USA).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells with TRIzol
reagent (Invitrogen). The amount of each RNA sample with an
A260/A280 ratio between 1.8 and 2.2 were considered for suitable
use. cDNA was reverse-transcribed using PrimeScript® RT
reagent kit (Takara, Tokyo, Japan). The primers used in this study
were as follows: USP4 upstream, 5′-TCAGCCGCTATGTGAAACAG-3′ and
downstream primer, 5′-GTGGTCTCACTGGGGTCATT-3′; actin upstream,
5′-CTCTTCCAGCCTTCCTTCCT-3′ and downstream primer,
5′-CAGGGCAGTGATCTCCTTCT-3′; IL-4 upstream,
5′-GCCACCATGAGAAGGACACT-3′ and downstream primer,
5′-ACTCTGGTTGGCTTCCTTCA-3′; IL-5 upstream,
5′-GGCACTGCTTTCTACTCATCGA-3′ and downstream primer,
5′-AGTTGGTGATTTTTATGTACAGGAACA-3′; IL-10 upstream,
5′-TGCAAAACCAAACCACAAGA-3′ and downstream primer, 5′-TCTC
GGAGATCTCGAAGCAT-3′; IL-13 upstream, 5′-CTATGCATCCGCTCCTCAAT-3′ and
downstream primer, 5′-GGTGATG TTGACCAGCTCCT-3′; IRF4 upstream,
5′-AGAACGAGGAGAAGAGCATC-3′ and downstream primer,
5′-CCTTTAAACAGTGCCCAAG-3′. Quantitative PCR was carried out with
the SYBR® Premix Ex Taq™ kit (Takara). The 20-µl
reaction mix consisted of 2 µl 30-fold diluted first-strand
cDNA, 10 µl 2X SYBR® Premix Ex Taq™, 0.4
µl 10 µM forward and reverse primer, 0.4 µl
50X ROX Reference Dye and 6.8 µl DEPC-treated water.
Reactions were performed in an ABI 7300 real-time quantitative
instrument (Applied Biosystems, Foster City, CA, USA). Specificity
of the PCR products was confirmed by melting curve analysis
followed by the verification of the amplicon length on 1.5% agarose
gels stained with ethidium bromide.
Statistical analysis
Data are expressed as the means ± standard error of
the mean. Paired t-test was used for calculating statistically
significant difference (p<0.05) by GraphPad Prism software
(GraphPad Software Inc., San Diego, CA, USA).
Results
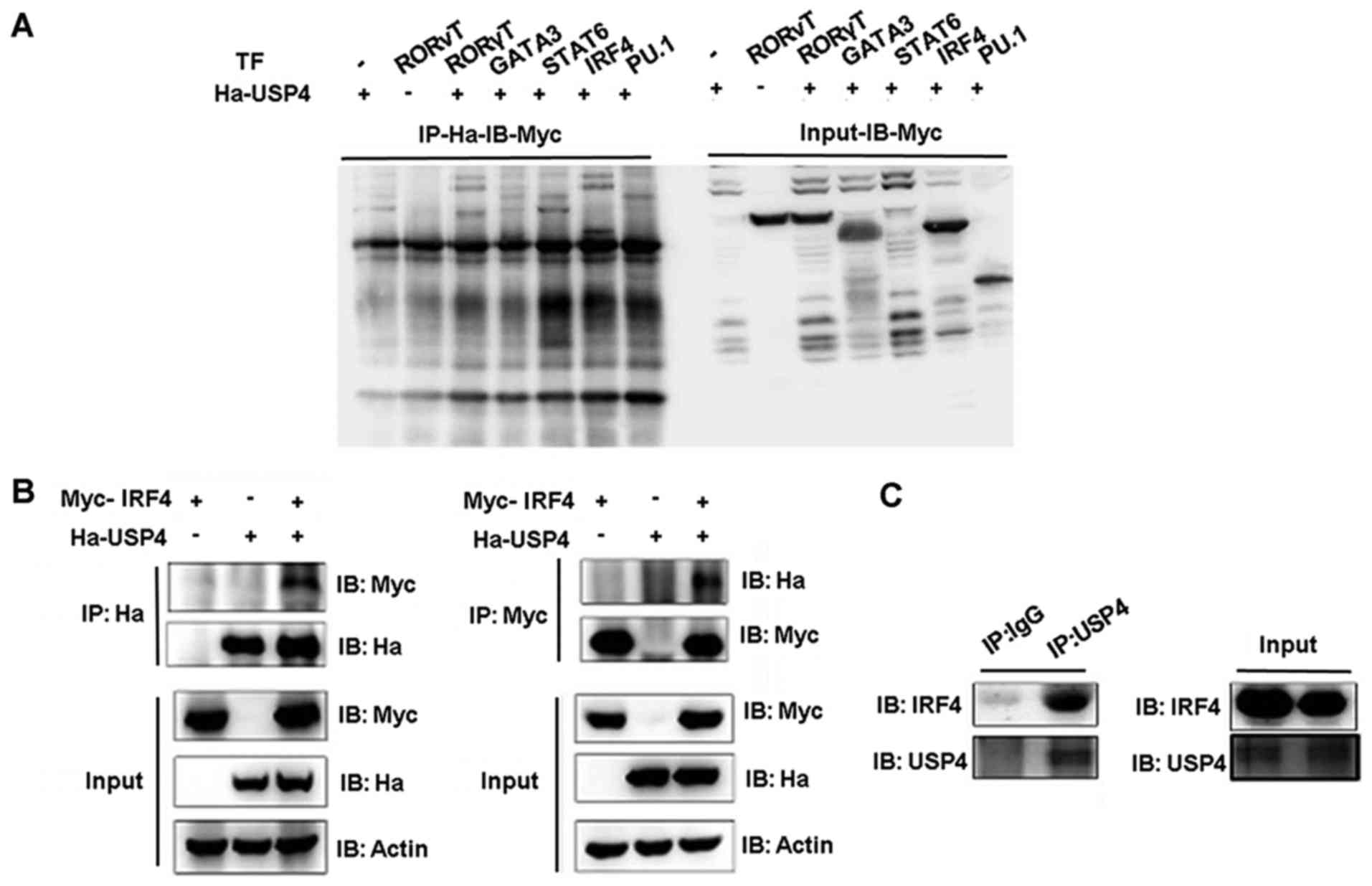
Interaction between USP4 and IRF4
Co-immunoprecipitation assays were performed to
assess the functional interaction between USP4 and transcription
factors GATA3, STAT6, IRF4 and PU.1. USP4 fused to the
hemagglutinin (Ha) epitope (Ha-USP4) and Myc-Flag tagged -GATA3,
-STAT6, -IRF4 or -PU.1 were ectopically coexpressed in 293T cells,
and total protein lysates were prepared after stimulation with PMA
and ionomycin. Immunoprecipitation was conducted with anti-Ha and
immunoblotting was used to detect the expression level.
Immunoprecipitation of Ha-USP4 using the anti-Ha antibody resulted
in the co-immunoprecipitation of Myc-IRF4 as detected by
immunoblotting with anti-Myc antibodies (Fig. 1A). These results demonstrated that
USP4 and IRF4 interact directly with each other. Then, we conducted
an immunoprecipitation assay using anti-Myc or anti-Ha separately.
Compared with the cells that were transfected with Ha-labeled-USP4
or Myc-labeled-IRF4 alone, when Ha-USP4 and Myc-IRF4 were
coexpressed, immunoprecipitation of Ha-USP4 using anti-Ha antibody
resulted in the co-immunoprecipitation of Myc-IRF4 as detected by
immunoblotting with anti-Myc antibodies. The association of USP4
and IRF4 was corroborated by reverse co-immunoprecipitation
experiment using the anti-Myc antibody to immunoprecipitate IRF4
and anti-Ha to detect the associated Ha-IRF4 by western blot
analysis (Fig. 1B). Th2 cell
lysate was then immunoprecipitated separately by using normal IgG
and anti-USP4. We found that immunoprecipitation of USP4 using the
anti-USP4 antibody resulted in the co-immunoprecipitation of
Myc-IRF4 as detected by immunoblotting with anti-Myc antibodies,
indicating that USP4 interacts with IRF4 in Th2 cells (Fig. 1C).
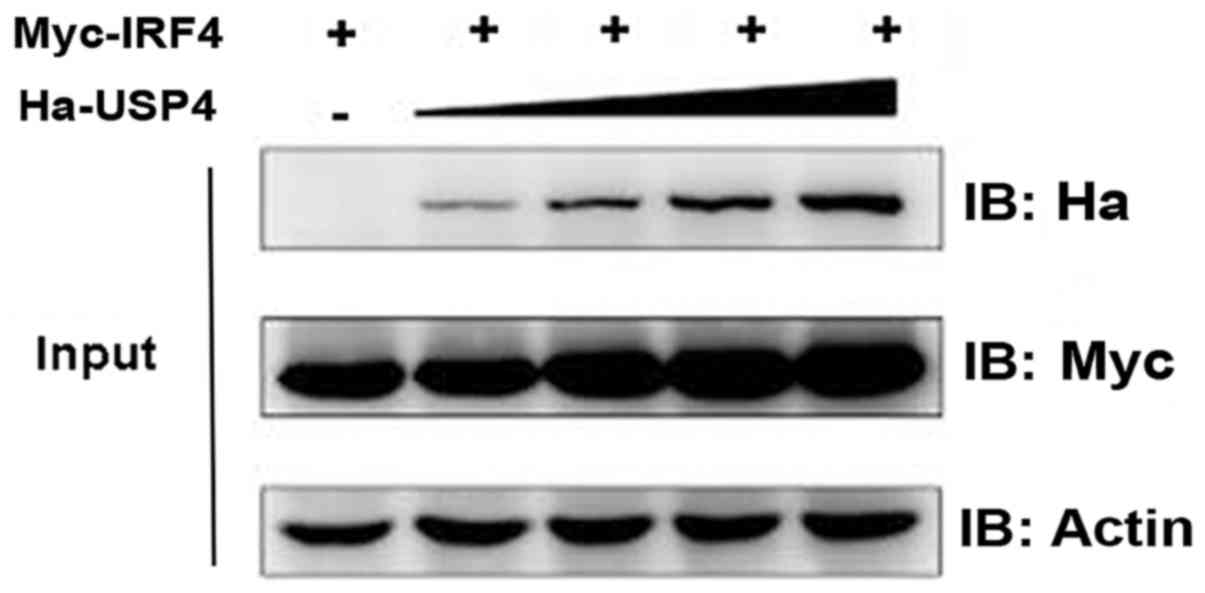
Effect of USP4 on stabilization of
IRF4
We then aimed to test and verify the influence of
USP4 on the stabilization of IRF4. Myc-IRF4 and different doses of
Ha-USP4 were transfected into 293T cells, respectively, and
immunoblotting was used. The results showed that the IRF4 protein
expression level was influenced by USP4 in a dose-dependent manner
(Fig. 2).
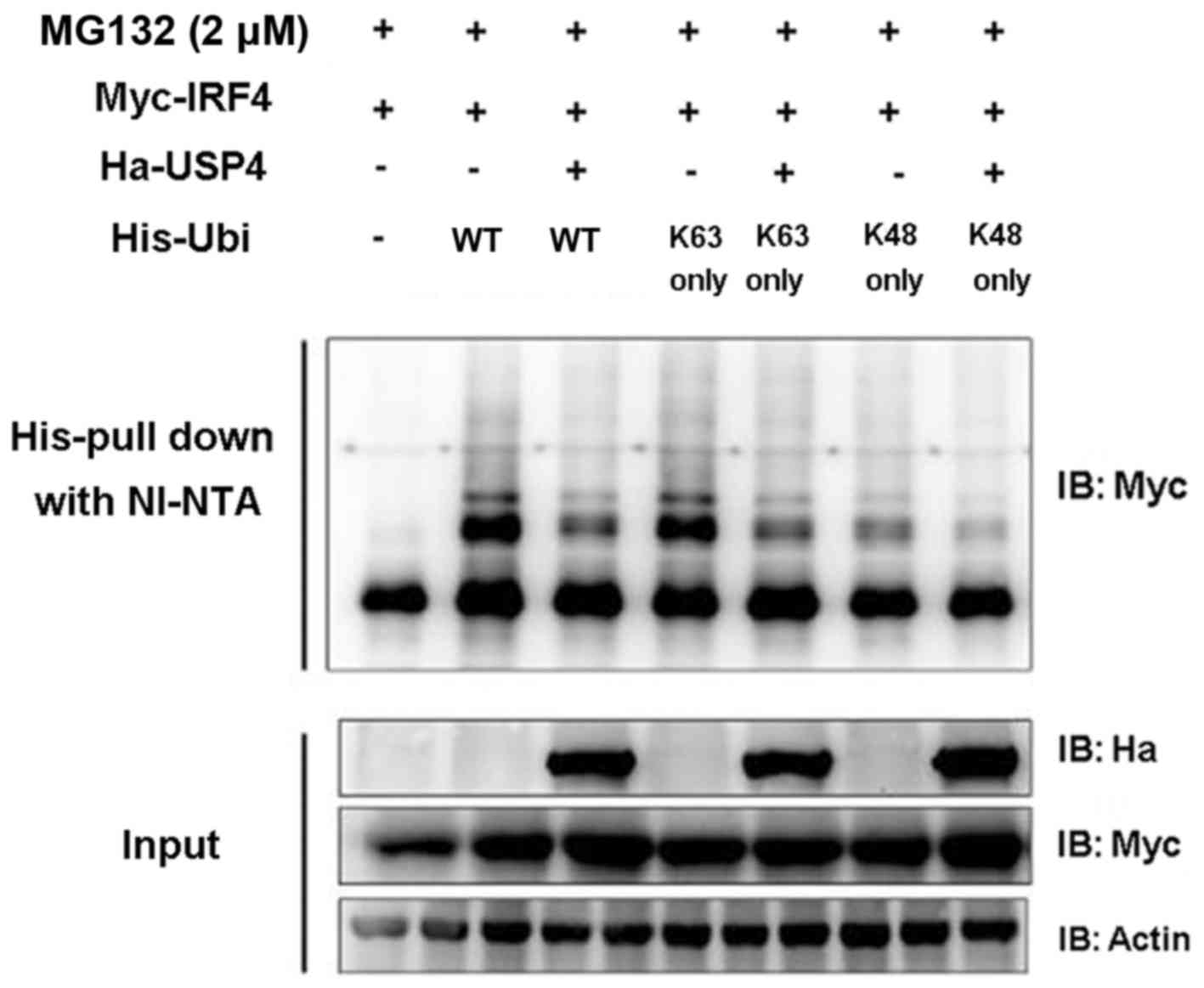
USP4 deubiquitinates IRF4
In order to validate whether USP4 exerts its effects
on IRF4 through deubiquitination, the coding sequence of ubiquitin
and its mutants pIPHis-K48 only and K63 only were cloned into the
pIP-His vector. Myc-IRF4, Ha-USP4, Ha-USP4C311A, His-ubiquitin and
its mutants were transiently expressed in 293T cells, respectively,
and Ni-NTA was used for purified precipitation and then was
detected by immunoblotting analysis. The results showed that USP4
simultaneously mediated the deubiquitination of IRF4 on K48 and
K63. The experimental results (Fig.
3) proved that USP4 can mediate the deubiquitination of IRF4 on
K48 and K63 sequences (amino acid residue sequence), revealing that
USP4 exerts its effects on IRF4 through deubiquitination.
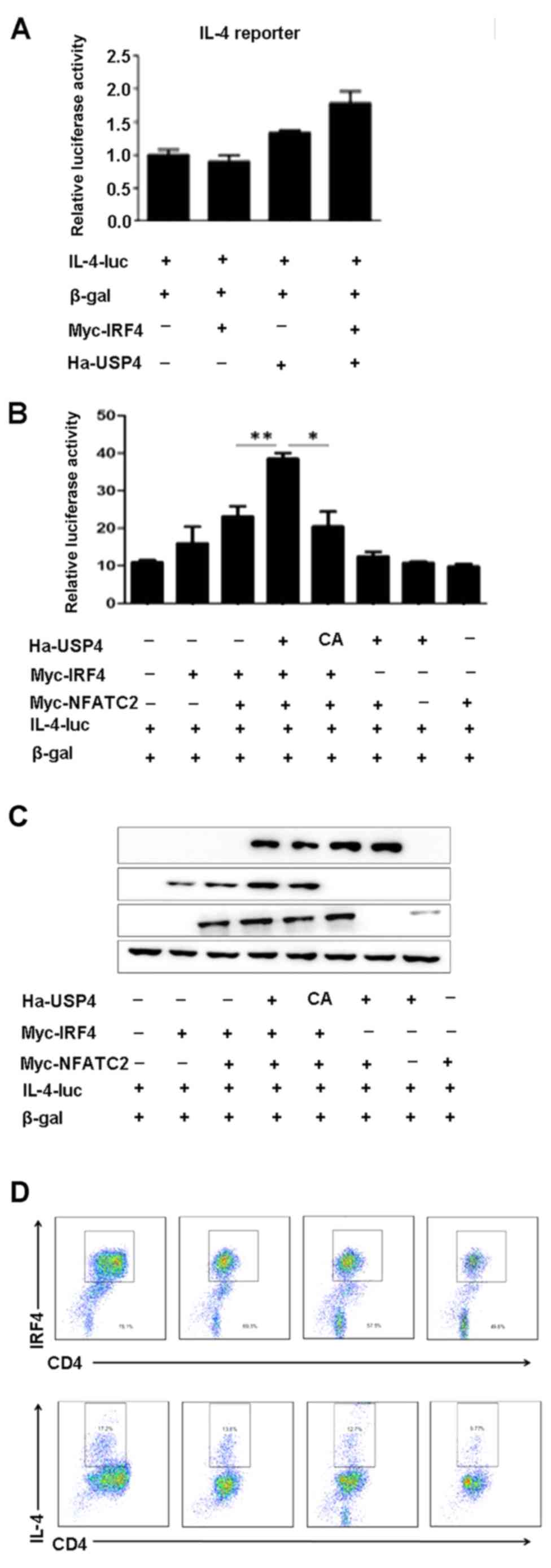
USP4 and IRF4 synergize with NFATc2 to
specifically enhance NFAT-mediated activation of the IL-4
promoter
To assess the ability of USP4 and IRF4 to
transactivate the IL-4 promoter, either on their own or in
conjunction with NFATc2, 293T cells were transfected with an IL-4
luciferase reporter construct. We found that cotransfection with
USP4 and IRF4 had no significant effects on the luciferase activity
of IL-4-luc (Fig. 4A), whereas
cotransfection with USP4, IRF4 and NFATc2 markedly increased the
luciferase activity of IL-4-luc compared with that of the controls
(Fig. 4B and C). Naïve
CD4+ T cells were sorted using flow cytometry-based
analysis and were induced for Th2 cell differentiation, and were
treated with the inhibitor of USP4, vialinin A, and then processed
by flow cytometry. The results showed that the expression levels of
IL-4 and IRF4 were decreased by the treatment of vialinin A
(Fig. 4D).
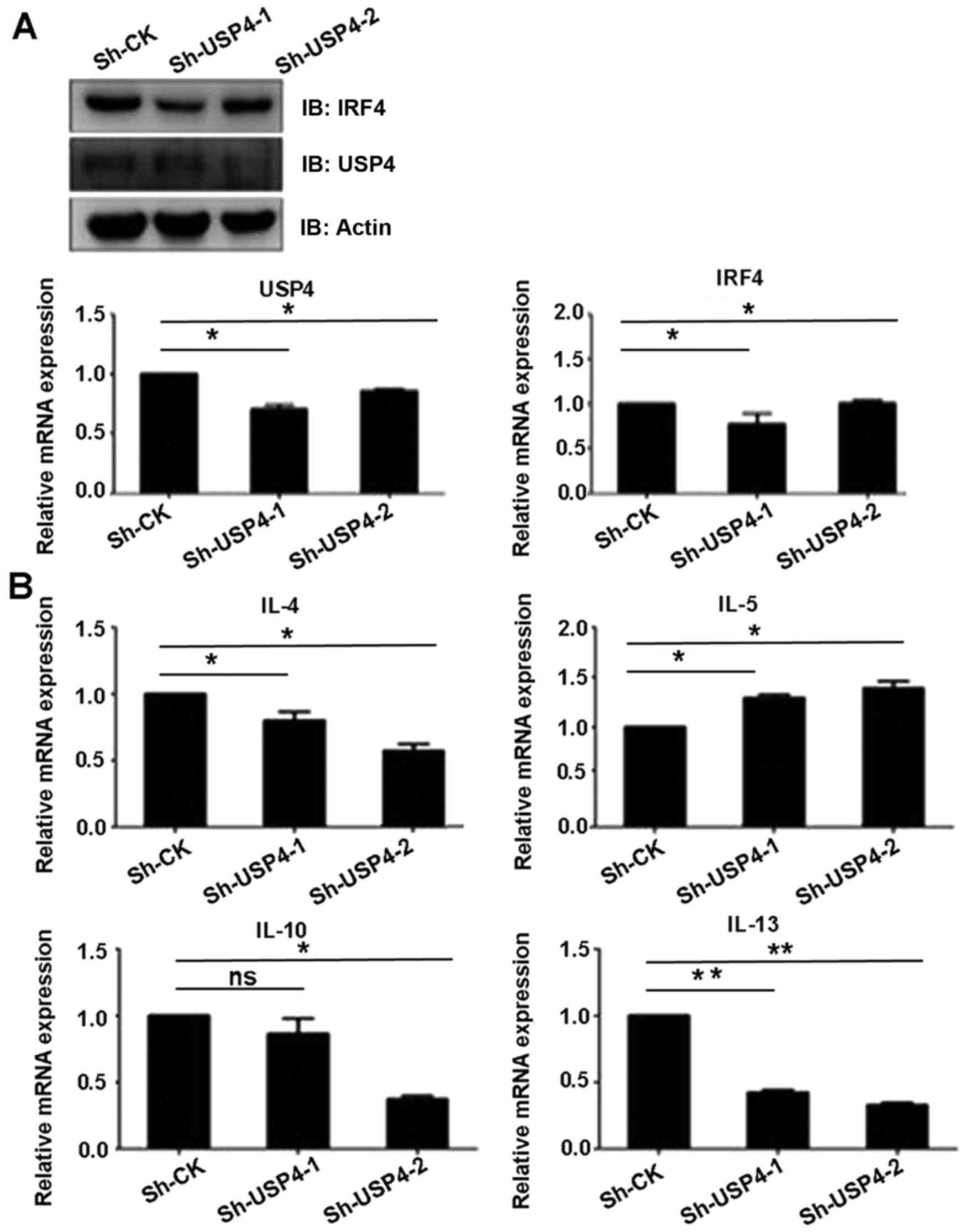
USP4 knockdown affects the expression
level of Th2-related cytokines
Since IRF4 is an important transcription factor of
Th2 cells, we ascertained whether deubiquitinating enzyme USP4 can
affect the function of Th2 cells. Sh-USP4-1, Sh-USP4-2 and the
corresponding control were cloned into the lentivirus vector
pLKO.1, and transfected into 293T cells, and then Th2 cells were
infected in vitro. Firstly, we found that the mRNA and
protein expression levels of USP4 were significantly decreased
after knockdown of USP4. The results also showed that Sh-USP4
significantly decreased the mRNA and protein expression levels of
IRF4 in Th2 cells as compared to the control (Fig. 5A). In addition, the transcription
levels of Th2-related cytokines IL-4, IL-10 and IL-13 were
significantly decreased, whereas the transcription level of IL-5
was significantly increased as compared to the control (Fig. 5B).
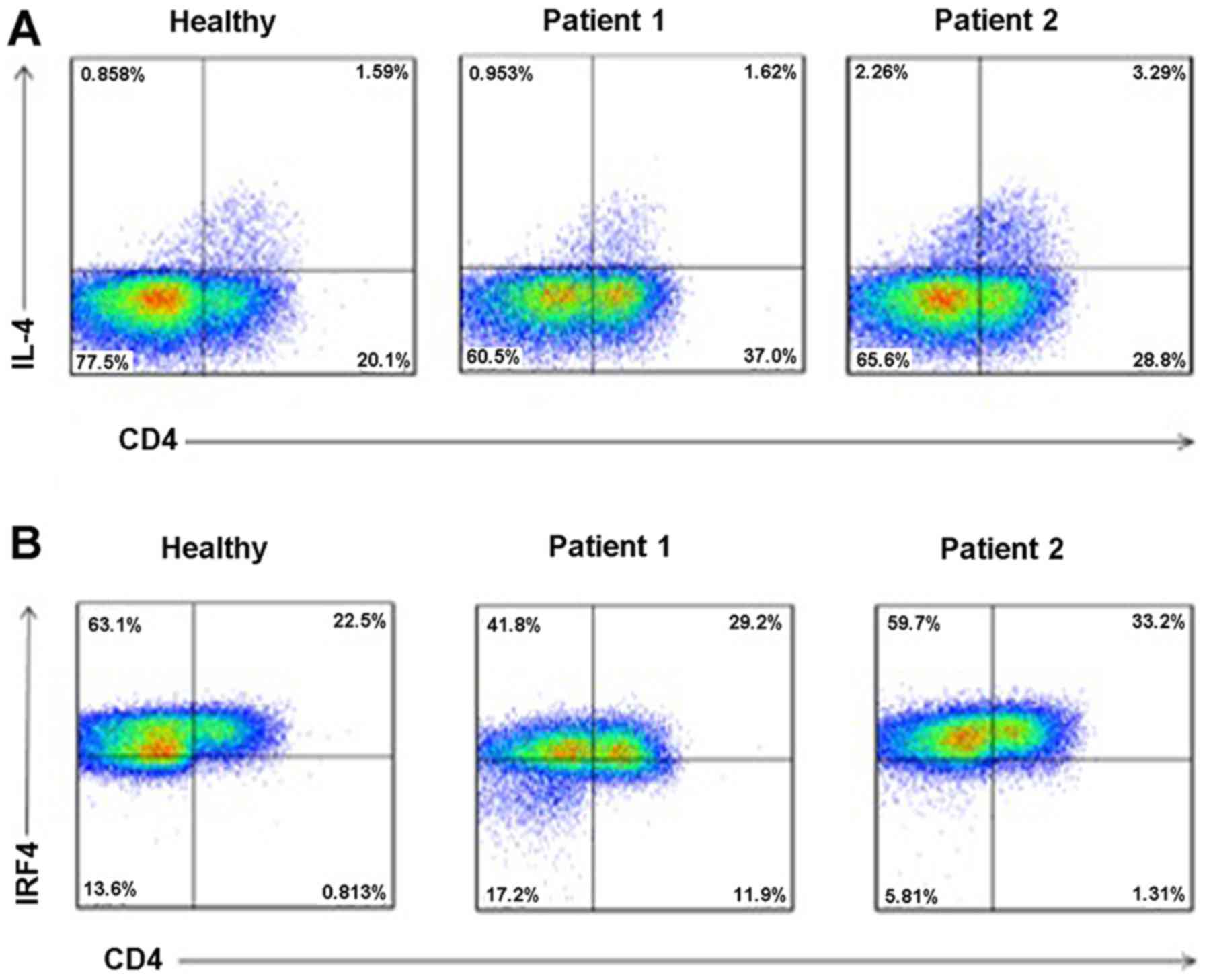
Increased levels of IL-4 and IRF4 in
serum of RHD patients
To determine the levels of IL-4 and IRF4 in serum of
RHD patients, we collected peripheral blood of healthy controls and
RHD patients. PBMCs were separated using density gradient
centrifugation, and flow cytometric analysis was performed. As
shown in Fig. 6, when compared to
the healthy controls, the levels of IL-4 (Fig. 6A) and IRF4 (Fig. 6B) in the serum were all increased
in the RHD patient group.
Discussion
RHD is a complex autoimmune inflammatory disease.
IL-4 is a Th2-type cytokine and plays a regulatory role in the
inflammatory response mediated by Th1 cytokines. Infiltrating
mononuclear cells secreting inflammatory cytokines IFN-γ and TNF-α
are found in valvular and myocardium tissues. In mononuclear cells,
IL-10 and IL-4 (regulatory cytokines) are also secreted in
myocardium tissue. However, in valvular tissue, only a few cells
secrete IL-4, suggesting that these low numbers of IL-4-producing
cells may contribute to the progression of valvular RHD lesions
(17). In this study, when
compared to the healthy control group, the level of IL-4 in the
serum was significantly increased in the RHD group.
We also found that the level of IRF4 in the serum
was significantly increased in the RHD group when compared to the
healthy control group. IRF4 is clearly critical for lymphocyte
function since IRF4-deficient mice exhibit cell intrinsic defects
in both B and T cells (18). A
previous study indicated that naïve Th cells lacking IRF4 are
unable to differentiate into IL-4-producing Th2 effector cells
in vitro (9). They also
identified IRF4 and NFATc2 as transcriptional partners that
together regulate inducible IL-4 gene expression upon T cell
activation (9). The IRF4 protein
(full-length 1-450) possesses an NH2-terminal (1-134) DNA binding
domain, a activation domain region that includes a proline-rich
segment, and the COOH-terminal contains the regulatory domain
(19,20). The previous study has proven that
the amino acids 150–340 and the COOH-terminal region of the IRF4
protein are important for transcriptional synergy as well its
interaction with NFATc2.
Gao et al found that USP22 interacts with and
deubiquitinates NFATc2, and also stabilizes NFATc2 protein and
promotes NFATc2 function to facilitate IL-2 expression in T cells
(21). Chen et al found
that the ubiquitin ligase Stub1 negatively modulates regulatory T
cell suppressive activity by promoting degradation of the
transcription factor Foxp3 (22).
Hou et al found that the E3 deubiquitinase USP21 is a
positive regulator of GATA3 in asthma (23). Other studies indicate that the
deubiquitinase USP17 regulates the stability and nuclear function
of IL-33, and is also a positive regulator of retinoic acid-related
orphan nuclear receptor γt (RORγt) in Th17 cells (24,25). In this study, we not only found an
interaction between USP4 and IRF4, but also proved the effect of
USP4 on the stabilization and deubiquitination of IRF4. Our results
also showed that sh-USP4 significantly decreased the mRNA and
protein expression levels of IRF4 in Th2 cells as compared to the
control. In addition, the transcription levels of Th2-related
cytokines IL-4, IL-10 and IL-13 were significantly decreased as
compared to the control.
There are seven lysines in ubiquitin and
poly-ubiquitin chains can be assembled through linkage to any of
the seven. The distinct chains are known as K6, K11, K27, K29, K33,
K48 and K63 chains, depending upon the lysine through which the
monomers are linked (26). The
experimental results in our study proved that USP4 can mediate
deubiquitination of IRF4 on K48 and K63 sequences (amino acid
residue sequences), revealing that USP4 exerts its effects on IRF4
through deubiquitination.
To conclude, this study indicated that USP4
interacts with and deubiquitinates IRF4, and also stabilizes IRF4
protein and promotes IRF4 function to facilitate IL-4 expression in
Th2 cells, which may be associated with the pathological process of
RHD. Small-molecule inhibitors of USP4 may have a potential role in
providing a new therapeutic strategy for the treatment of RHD.
Acknowledgments
This study was in part supported by grants from the
Key Scientific Research Project of Anhui Provincial Science and
Technology Department (no. 1301043025).
References
|
1
|
Guilherme L and Kalil J: Rheumatic heart
disease: Molecules involved in valve tissue inflammation leading to
the autoimmune process and anti-S. pyogenes vaccine. Front Immunol.
4:352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chopra P and Gulwani H: Pathology and
pathogenesis of rheumatic heart disease. Indian J Pathol Microbiol.
50:685–697. 2007.
|
|
3
|
Rothenbühler M, O'Sullivan CJ, Stortecky
S, Stefanini GG, Spitzer E, Estill J, Shrestha NR, Keiser O, Jüni P
and Pilgrim T: Active surveillance for rheumatic heart disease in
endemic regions: A systematic review and meta-analysis of
prevalence among children and adolescents. Lancet Glob Health.
2:e717–e726. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guilherme L, Köhler KF and Kalil J:
Rheumatic heart disease: Mediation by complex immune events. Adv
Clin Chem. 53:31–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Köhler G and Milstein C: Continuous
cultures of fused cells secreting antibody of predefined
specificity. 1975. Biotechnology. 24:524–526. 1992.PubMed/NCBI
|
|
6
|
Le Gros G, Ben-Sasson SZ, Seder R,
Finkelman FD and Paul WE: Generation of interleukin-4
(IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are
required for in vitro generation of IL-4-producing cells. J Exp
Med. 172:921–929. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hsieh CS, Heimberger AB, Gold JS, O'Garra
A and Murphy KM: Differential regulation of T helper phenotype
development by interleukins 4 and 10 in an alpha beta
T-cell-receptor transgenic system. Proc Natl Acad Sci USA.
89:6065–6069. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Faé KC, Oshiro SE, Toubert A, Charron D,
Kalil J and Guilherme L: How an autoimmune reaction triggered by
molecular mimicry between streptococcal M protein and cardiac
tissue proteins leads to heart lesions in rheumatic heart disease.
J Autoimmun. 24:101–109. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rengarajan J, Mowen KA, McBride KD, Smith
ED, Singh H and Glimcher LH: Interferon regulatory factor 4 (IRF4)
interacts with NFATc2 to modulate interleukin 4 gene expression. J
Exp Med. 195:1003–1012. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Reyes-Turcu FE, Ventii KH and Wilkinson
KD: Regulation and cellular roles of ubiquitin-specific
deubiquitinating enzymes. Annu Rev Biochem. 78:363–397. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nijman SM, Luna-Vargas MP, Velds A,
Brummelkamp TR, Dirac AM, Sixma TK and Bernards R: A genomic and
functional inventory of deubiquitinating enzymes. Cell.
123:773–786. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiao N, Li H, Luo J, Wang R, Chen H, Chen
J and Wang P: Ubiquitin-specific protease 4 (USP4) targets TRAF2
and TRAF6 for deubiquitination and inhibits TNFα-induced cancer
cell migration. Biochem J. 441:979–986. 2012. View Article : Google Scholar
|
|
13
|
Fan YH, Yu Y, Mao RF, Tan XJ, Xu GF, Zhang
H, Lu XB, Fu SB and Yang J: USP4 targets TAK1 to downregulate
TNFα-induced NF-κB activation. Cell Death Differ. 18:1547–1560.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang L, Zhou F, Drabsch Y, Gao R,
Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu
CX, et al: USP4 is regulated by AKT phosphorylation and directly
deubiquitylates TGF-β type I receptor. Nat Cell Biol. 14:717–726.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Berger FG, Yang J and Lu X: USP4
inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO
J. 30:2177–2189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu C, Zheng F, Sun T, Duan Y, Cao J, Feng
H, Shang L, Zhu Y and Liu H: Interaction of avian influenza virus
NS1 protein and nucleolar and coiled-body phosphoprotein 1. Virus
Genes. 46:287–292. 2013. View Article : Google Scholar :
|
|
17
|
Guilherme L, Cury P, Demarchi LM, Coelho
V, Abel L, Lopez AP, Oshiro SE, Aliotti S, Cunha-Neto E,
Pomerantzeff PM, et al: Rheumatic heart disease: Proinflammatory
cytokines play a role in the progression and maintenance of
valvular lesions. Am J Pathol. 165:1583–1591. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mittrücker HW, Matsuyama T, Grossman A,
Kündig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS
and Mak TW: Requirement for the transcription factor LSIRF/IRF4 for
mature B and T lymphocyte function. Science. 275:540–543. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eisenbeis CF, Singh H and Storb U: Pip, a
novel IRF family member, is a lymphoid-specific, PU.1-dependent
transcriptional activator. Genes Dev. 9:1377–1387. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brass AL, Kehrli E, Eisenbeis CF, Storb U
and Singh H: Pip, a lymphoid-restricted IRF, contains a regulatory
domain that is important for autoinhibition and ternary complex
formation with the Ets factor PU.1. Genes Dev. 10:2335–2347. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao Y, Lin F, Xu P, Nie J, Chen Z, Su J,
Tang J, Wu Q, Li Y, Guo Z, et al: USP22 is a positive regulator of
NFATc2 on promoting IL-2 expression. FEBS Lett. 588:878–883. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen Z, Barbi J, Bu S, Yang HY, Li Z, Gao
Y, Jinasena D, Fu J, Lin F, Chen C, et al: The ubiquitin ligase
Stub1 negatively modulates regulatory T cell suppressive activity
by promoting degradation of the transcription factor Foxp3.
Immunity. 39:272–285. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hou X, Zhang J, Chen C, Li B and Shi G:
Identification of the E3 deubiquitinase USP21 as a positive
regulator Of GATA3 in Asthma. Am J Respir Crit Care Med.
185:A64782012.
|
|
24
|
Ni Y, Tao L, Chen C, Song H, Li Z, Gao Y,
Nie J, Piccioni M, Shi G and Li B: The Deubiquitinase USP17
Regulates the Stability and Nuclear Function of IL-33. Int J Mol
Sci. 16:27956–27966. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han L, Yang J, Wang X, Wu Q, Yin S, Li Z,
Zhang J, Xing Y, Chen Z, Tsun A, et al: The E3 deubiquitinase USP17
is a positive regulator of retinoic acid-related orphan nuclear
receptor γt (RORγt) in Th17 cells. J Biol Chem. 289:25546–25555.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dikic I and Robertson M: Ubiquitin ligases
and beyond. BMC Biol. 10:222012. View Article : Google Scholar : PubMed/NCBI
|