Introduction
The loss of β-cell mass in pancreatic islets is
closely associated with type 1 and 2 diabetes mellitus. The
regeneration of β-cells for curative treatment of the disease is an
important topic of research. New β-cells are derived from either
exogenous sources, such as stem cells or endogenous sources, such
as facultative pancreatic progenitors (1–3).
Pancreatic injury models (4–7)
have suggested several endogenous sources for β-cell regeneration.
Surviving β-cells may generate new β-cells by self-duplication
(8), and new β-cells may arise
through transdifferentiation from other pancreatic cells, such as
α- and δ-cells (9,10). Moreover, β-cell neogenesis may
develop from multipotent pancreatic progenitor cells (11,12). These studies indicate the
plasticity of the pancreas. However, the molecular mechanisms of
β-cell neogenesis are not well known.
A number of studies have identified genes
differentially expressed in pancreatic neogenesis by using 'omics'
methods, such as cDNA microarray analysis and two-dimensional
electrophoresis (2-DE) (13–18). De León et al suggested
endoderm transcription factor Foxa2 (also known as HNF3β) as a
potential candidate for the improvement of pancreatic growth and
function (15). Moreover,
lymphocyte cytosolic protein 1 (LCP1), which is upregulated by
partial pancreatectomy, has been suggested as an important protein
for pancreatic regeneration owing to an increase in pancreatic cell
proliferation and regulation of islet markers (18). In addition, various transcription
factors, such as neurogenin 3 (19), paired related homeobox 1 (Prrx1)
(20) and pancreatic and duodenal
homeobox 1 (Pdx1) (21) regulate
pancreatic regeneration. However, protein activity or function is
regulated by the expression level, as well as post-translational
modifications (PTMs). Mutations of PTM sites cause various diseases
(22); in particular, differences
in PTMs between chronic pancreatitis and non-pancreatitis have been
reported (23). Pdx1 is known to
be an important regulator of β-cell maturation, and its activity is
a prerequisite for the regulation of blood glucose homeostasis.
Various studies have suggested that Pdx1 activity is regulated by
phosphorylation as one of the types of PTMs (21,24–28). Frogne et al recently
identified a single phosphorylation site in Pdx1 by isoelectric
focusing (IEF) (29). In a
partial hepatectomy model, Yes-associated protein (YAP) activity
was found to regulate liver regeneration by phosphorylation
(30). Moreover, the
phosphorylation of RelA (p65), a subunit of nuclear factor-κB
(NF-κB), has been reported as an important mechanism in liver
regeneration and cancer (31).
Therefore, the identification of PTM alterations during injury and
regeneration may provide novel mechanistic insight into organ
regeneration. In this study, to analyze the potential association
between PTMs and β-cell replication, we investigated PTMs
associated with pancreatic regeneration using 2-DE analysis and
further examined the tyrosine phosphorylation level of CPB1.
Materials and methods
Partial pancreatectomy
To investigate proteins associated with pancreatic
regeneration following pancreatectomy, 8-week-old male
Sprague-Dawley rats (Daehan Experimental Animals, Seoul, Korea)
were anesthetized with isoflurane (Santa Cruz Animal
Health®, Paso Robles, CA, USA) and divided into 3
treatment groups as follows: partial pancreatectomy (n=2;
approximately 90% pancreatectomy), sham surgery (n=2) and no
surgery (n=2). The animals were allowed free access to standard
diet and water before and after the operation. Partial
pancreatectomy was executed according to Bonner-Weir et al
(32). In brief, 8-week-old male
Sprague-Dawley rats (Daehan Experimental Animals) were anesthetized
with 3% isoflurane in oxygen (O2)/nitrous oxide
(N2O) mixtures, and 90% of the pancreatic tissue was
then removed by gentle abrasion with cotton swabs, leaving the
tissue between the common bile duct and the loop of the duodenum
intact. For sham operation, the same surgical procedure was carried
out without the removal of pancreatic tissue. On the 3rd day after
surgery, the remnant pancreatic tissues were isolated under inhaled
anesthesia (3% isoflurane in O2/N2O mixture)
and the rats were then euthanized. The tissues were fixed with
neutral buffered formalin (NBF) for histological examination, and
immediately stored at −80°C. The study protocol was approved by the
Institutional Animal Care and Use Committee (IACUC) at Konyang
University in accordance with the National Institutes of Health
(NIH) Guidelines for the Care and Use of Laboratory Animals.
2-DE
Isolated pancreatic tissues were solubilized using a
previously described method (33). Solubilized proteins were
quantified with a Bradford protein assay kit (Bio-Rad, Hercules,
CA, USA) and then stored at −80°C until use. For resolution across
the pH 4–7 range, 1 mg of protein extracted from the sham-operated
and pancreatectomized pancreases was mixed with modified
rehydration buffer [6 M urea, 2 M thiourea, 4% CHAPS, 2.5% DTT, 10%
isopropanol, 5% glycerol and 2% immobilized pH gradient (IPG)
buffer pH 4–7], loaded into a cup on the anodic side of a pH 4–7
IPG strip (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA), and
incubated for 12 h. Following rehydration, first-dimension IEF was
performed on a Multiphor II IEF system (GE Healthcare
Bio-Sciences). To separate the second dimension, an equilibrated
IPG gel strip was placed on an 8–16% linear gradient SDS
polyacrylamide gel and the gels were subjected to electrophoresis
using the Ettan-DALT system (GE Healthcare Bio-Sciences). After
2-DE, the gels were stained using colloidal Coomassie brilliant
blue (CBB) G-250 (Sigma-Aldrich, St. Louis, MO, USA). The stained
gel was scanned with a GS-710 calibrated imaging densitometer
(Bio-Rad), and proteins differentially expressed between
pancreatectomized and sham operation samples were identified using
the Melanie III image analysis software (Swiss Institute for
Bioinformatics, Geneva, Switzerland). Two-DE analysis was performed
three times independently. Differences among protein spots were
analyzed using a two-way Student's t-test.
Matrix-assisted laser
desorption/ionization time-of-flight mass spectrometry
(MALDI-TOF/MS) and protein identification
For the identification of differentially expressed
proteins, MALDI-TOF/MS analysis of selected spots was performed as
previously described (33).
Selected spots sliced from the gel were stained with CBB and then
digested with trypsin. Extracted peptides were subjected to
MALDI-TOF/MS analysis on a Voyager DE-STR MALDI-TOF mass
spectrometer (Applied Biosystems, Foster City, CA, USA). All
acquired spectra of samples were processed using Voyager™ 5.1
software (Applied Biosystems) in the default mode. Averages of 500
spectra were obtained for each sample, and scans were performed
twice. Spectra were calibrated automatically upon acquisition using
an external 3-point calibration. Peaks were manually assigned using
the DATA Explorer™ software package (Applied Biosystems), and
spectra were used to search against non-redundant protein sequence
databases available online (SWISS-PROT and/or NCBInr Data Bank).
The peptide mass fingerprinting data were applied to ProFound and
MASCOT search engines (http://prowl.rockefeller.edu/; http://www.matrixscience.com/search_form_select.html)
for protein identification based on ProFound and MASCOT scores. The
Z score of ProFound is the distance to the population mean in units
of the standard deviation.
Western blot analysis
Pancreatic tissues and cell lines were lysed using
RIPA buffer containing 50 mM Tris-HCl (pH 7.4), 1% NP-40, 0.25%
sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, and protease inhibitor
cocktail (Roche, Basel, Switzerland). To investigate differences in
CPB1 phosphorylation, we performed immunoprecipitation. Briefly,
800 µg of lysate was precleared with protein A-agarose for 2
h at 4°C. The supernatant was incubated with anti-CPB1 (GenWay
Biotech, San Diego, CA, USA) by shaking overnight at 4°C, followed
by incubation with protein A-agarose for 2 h. The beads were
resuspended in 2× sample buffer and boiled for 10 min. The proteins
were resolved on 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) gels and transferred onto polyvinylidene
difluoride membranes (Millipore, Billerica, MA, USA). Immunoblots
were incubated in 5% skim milk (Difco, Franklin Lakes, NJ, USA) for
1 h and probed with anti-tyrosine (1:1000; sc-7020; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), anti-CPB1 (1:1000;
GWB-31DED0; GenWay Biotech, San Diego, CA, USA) and anti-β-actin
(1:3000; sc-47778; Santa Cruz Biotechnology, Inc.) primary
antibodies overnight, followed by incubation with a horseradish
peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody
for 2 h. Immunoreactive bands were detected using an enhanced
chemiluminescence kit (ECL; Thermo Scientific, Waltham, MA,
USA).
Immunohistochemical analysis
To identify alterations in CPB1 protein
phosphorylation levels in the pancreas following pancreatectomy,
the sham-operated and partially pancreatectomized pancreases were
removed and fixed with NBF overnight at room temperature. Fixed
tissues were embedded in paraffin and the paraffin blocks were
sliced at 5 µm thicknesses using a tissue microtome (Leica,
Nussloch, Germany). The slides were blocked with 10% goat serum
(Vector Laboratories, Inc., Burlingame, CA, USA) and incubated with
phospho-Tyr antibody (sc-7020; Santa Cruz Biotechnology, Inc.), and
CPB1 antibody (GWB-31DED0; GenWay Biotech) as primary antibodies at
4°C overnight and then with Cy2-conjugated goat anti-rabbit IgG
(111-225-144) and Cy3-conjugated goat anti-mouse IgG (115-165-146)
secondary antibodies (both from Jackson Immuno Research, West
Grove, PA, USA) as secondary antibodies at room temperature for 1
h. Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole
dihydrochloride (Molecular Probes, Waltham, MA, USA). The stained
cells were examined under a confocal microscope (Carl Zeiss,
Oberkochen, Germany).
DNA constructs and mutagenesis
Total RNA was isolated from the rat pancreases using
TRI Reagent® (Ambion, Waltham, MA, USA) according to the
manufacturer's instructions. Approximately 1.2 kb cDNA of CPB1
(NM_012533.1) was obtained from rat pancreatic RNA by RT-PCR using
the following primers: 5′-GCC GCC ACC ATG TTG CTG CTA CTG GCC CT-3′
(forward) and 5′-GGT CCA ATT GGT CAA CAC ACC CA-3′ (reverse), and
the cDNA was cloned (CPB1OE) into the EcoRI site of
pcDNA3-EGFP (Addgene, Cambridge, MA, USA). Mutations of 6 tyrosine
sites of the CPB1 protein (Y298F, Y304F, Y310F, Y312F, Y314F and
Y316F) were introduced (CPB1OE-mutY) using the
QuikChange® Site-Directed Mutagenesis kit (Stratagene,
La Jolla, CA, USA) according to the manufacturer's instructions.
Mutated sites were confirmed by sequencing.
Cell culture and transfection
RIN-m insulinoma cells obtained from the American
Type Culture Collection (ATCC, Manassas, VA, USA) were cultured in
RPMI-1640 (ATCC) supplemented with 10% fetal bovine serum (Gibco,
Carlsbad, CA, USA) and 1% penicillin/streptomycin (Lonza, Basel,
Switzerland). The cells were maintained at 37°C in a humidified
atmosphere containing 5% CO2. The RIN-m cells were
transfected with control pcDNA3-EGFP (Addgene), a rat CPB1
full-length cDNA vector (CPB1OE) and mutant rat CPB1
full-length cDNA vector (CPB1OE-mutY) using Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer's
instructions. To establish stable cell lines, transfected clones
were cultured in selective media containing 150 µg/ml G418
(Sigma-Aldrich).
Cell viability
To identify the association between the
phosphorylation of CPB1 protein and cell viability, stable cell
lines (control, CPB1OE and CPB1OE-mutY) were seeded in 12-well
plates at a density of 5×105 cells/well. Cell viability
was measured at 6, 24 and 48 h. Cells were collected and mixed with
0.4% trypan blue stain solution (Gibco), after which unstained
cells were counted under a microscope (Olympus, Tokyo, Japan).
Statistical analyses
All graphed data are presented as the means ±
standard deviation. The results were analyzed using analysis of
variance (ANOVA) or a Student's t-test. P-values of <0.05 and
<0.01 were considered to indicate statistically significant and
highly statistically significant differences, respectively.
Results
Identification of proteins up- or
downregulated by partial pancreatectomy
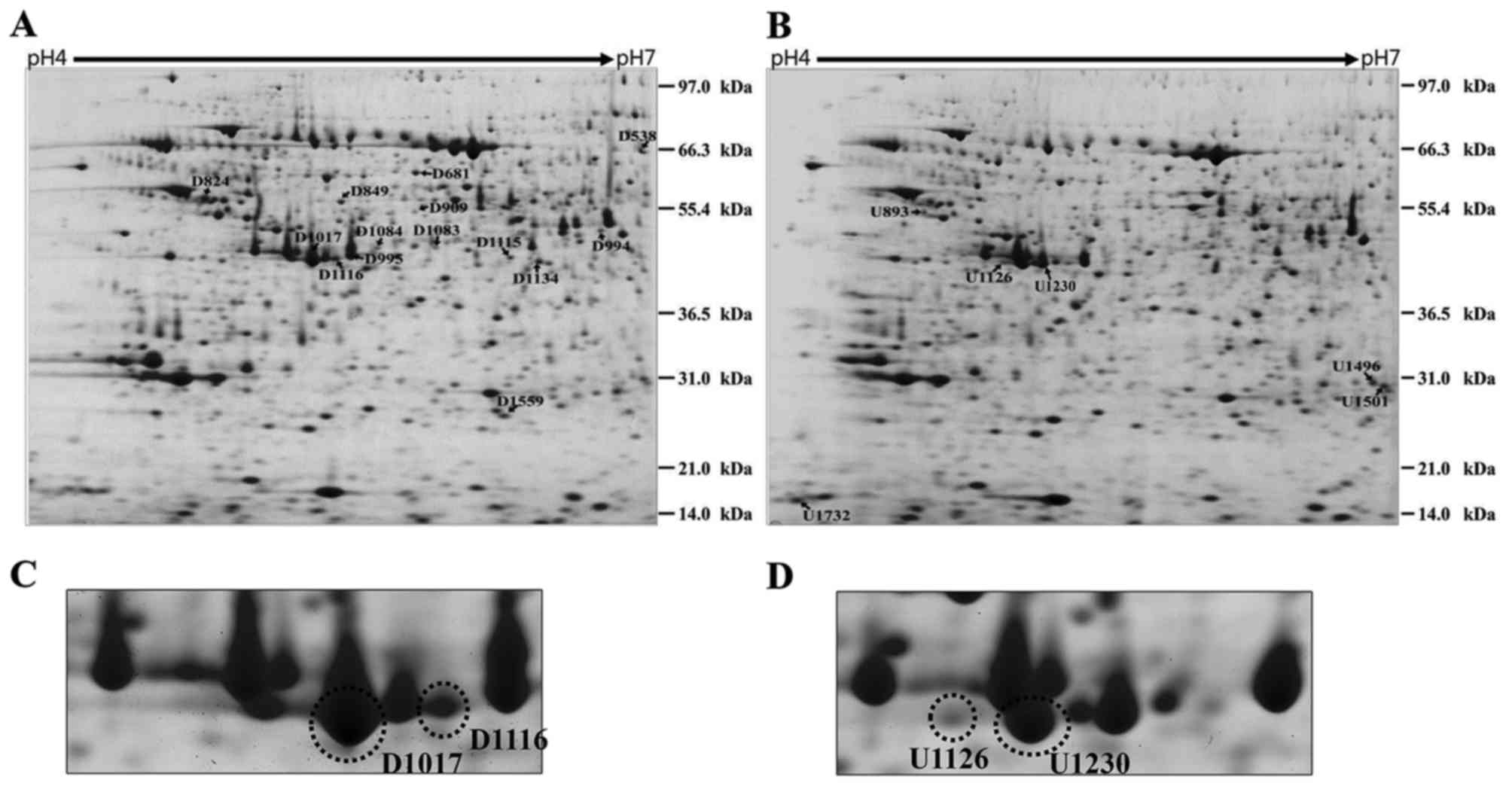
To identify proteins associated with pancreatic
regeneration, we compared proteins in pancreatic tissues obtained
from rats at 3 days after partial pancreatectomy with those of
sham-operated rats by 2-DE analysis with IEF gel electrophoresis on
pH 4–7 linear IPG strips. Independent experiments were each carried
out in duplicate. Melanie III detected differential intensity in 20
spots; 14 proteins were downregulated and 6 were upregulated in the
rats subjected to compared with the sham-operated rats (Fig. 1A and B). The differentially
expressed proteins were identified by MALDI-TOF/MS (Table I). Among the identified proteins,
phosphoenolpyruvate carboxykinase 2 (PCK2) has been reported to be
differentially expressed in pancreatectomy (18). Amylase is also specifically
expressed in the pancreas (34).
Succinate-CoA ligase ADP-forming β subunit (Sucla2) and
isovaleryl-CoA dehydrogenase (Ivd), which are involved in the
mitochondrial Krebs cycle, were downregulated upon pancreatectomy.
Mitochondrial function and plasticity are associated with glucose
circulation (35). Interestingly,
carboxypeptidase B (CPB1) precursor protein was identified from 4
spots at different PIs among the 20 spots (Fig. 1C and D). Therefore, we selected
CPB1 for further study.
 | Table IIdentification of proteins
differentially expressed in partial pancreatectomy in the pH 4–7
range. |
Table I
Identification of proteins
differentially expressed in partial pancreatectomy in the pH 4–7
range.
| Spot ID | Est'd Za | Accession no. | Protein
information | Coverage (%) | pI | kDa |
|---|
| Downregulated
proteins |
| D538 | 2.39 | NP_001101847.2 | Phosphoenolpyruvate
carboxykinase 2 (mitochondrial) | 33 | 8.4 | 71.79 |
| D681 | 2.4 | NP_599153.2 | Serum albumin
precursor | 43 | 6.1 | 71.23 |
| D824 | 1.55 | AAA40788.1 | α-1-antitrypsin
precursor | 50 | 5.7 | 46.03 |
| D849 | 2.39 | AAA41082.1 | Vitamin D-binding
protein | 46 | 5.8 | 55.49 |
| D909 | 1.85 | EDL88549.1 | Rattus
norvegicus (Norway rat) albumin, isoform CRA_a | 27 | 6.8 | 53.46 |
| D994 | 2.22 | NP_001004206.1 |
Proliferation-associated protein 2G4 | 47 | 6.4 | 44.07 |
| D995 | 2.4 | P00731.2 | Carboxypeptidase
A1 | 40 | 5.5 | 47.19 |
| D1017 | 2.18 | NP_036665.1 | Carboxypeptidase B
precursor | 26 | 5.4 | 48 |
| D1083 | 1.47 | NP_001101857.2 | Succinyl-CoA ligase
[ADP-forming] subunit β, mitochondrial | 33 | 6.1 | 47.8 |
| D1084 | 1.82 | NP_599153.2 | Serum albumin
precursor | 24 | 6.1 | 71.23 |
| D1115 | 2.3 | EDL77312.1 | Rattus
norvegicus (Norway rat) rCG25777, isoform CRA_a | 41 | 5.9 | 42.36 |
| D1116 | 2.3 | NP_036665.1 | Carboxypeptidase B
precursor | 49 | 5.4 | 48 |
| D1134 | 2.4 | NP_036724.1 | Isovaleryl-CoA
dehydrogenase, mitochondrial precursor | 34 | 8.5 | 46.44 |
| D1559 | 2.2 | NP_476484.1 | Protein DJ-1 | 71 | 6.3 | 20.24 |
| Upregulated
proteins |
| U893 | 2.3 | NP_112402.1 | Vimentin | 46 | 5.1 | 53.75 |
| U1126 | 2.04 | NP_036665.1 | Carboxypeptidase B
precursor | 39 | 5.4 | 48 |
| U1230 | 55b | NP_036665.1 | Carboxypeptidase B
precursor | 39 | 5.4 | 48 |
| U1496 | 2.36 | XP_345279.4 | Amylase 2a5,
pancreatic | 27 | 9.3 | 50.58 |
| U1501 | 2.36 | NP_445742.1 | Phosphoglycerate
mutase 1 | 56 | 7.1 | 28.97 |
| U1732 | 2.16 | XP_573831.1 | Ribosomal protein
P2-like | 84 | 4.4 | 11.69 |
Post-translational modification of
CPB1
PTMs of proteins such as phosphorylation or
dephosphorylation induce shifts in the PI and regulate the
functional activity of enzymes (36). Therefore, we investigated
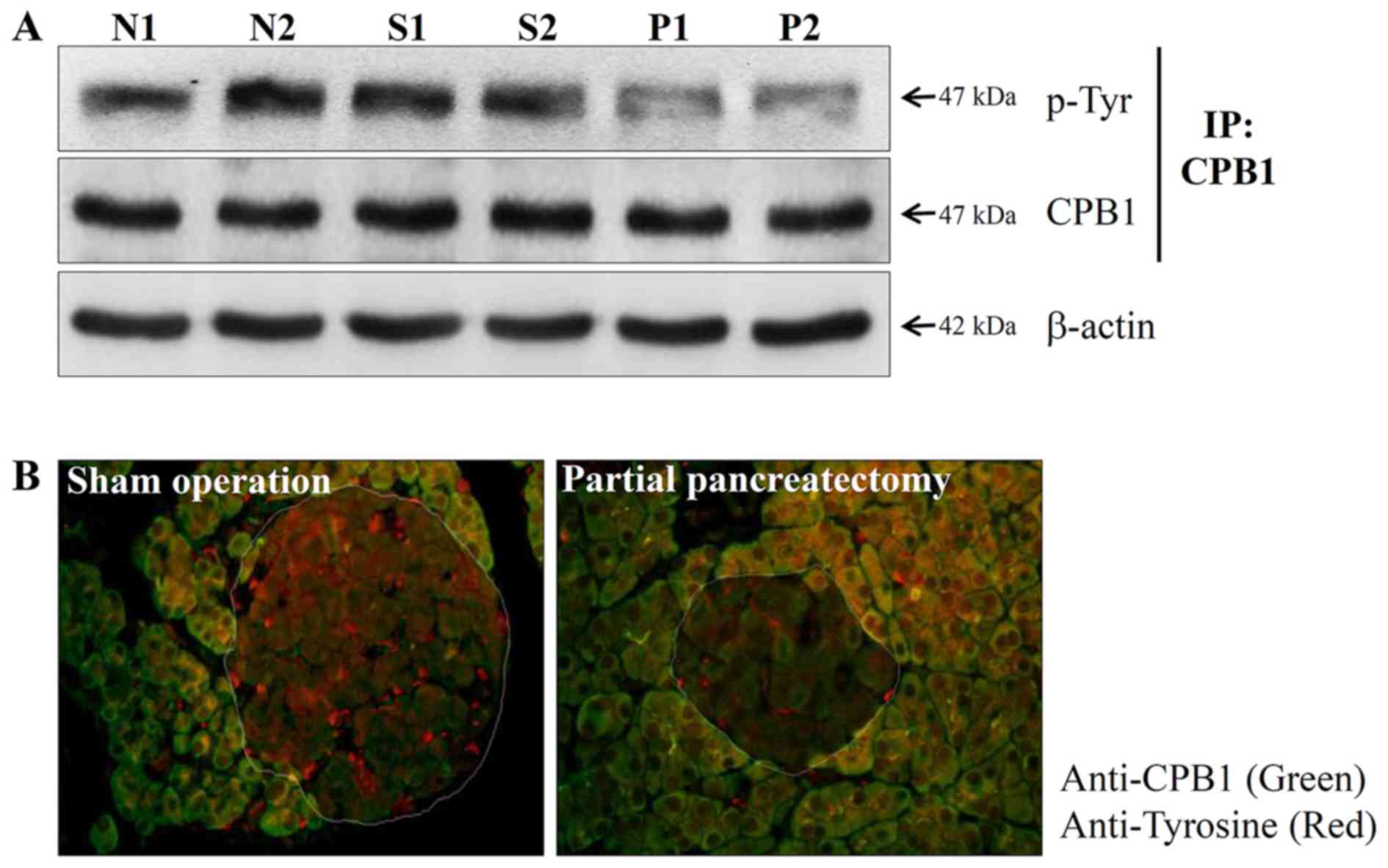
alterations in the phosphorylation level of CPB1 in normal,
sham-operated and partially pancreatectomized pancreases using the
immunoprecipitation method. Phosphorylation of tyrosine residues of
the CPB1 protein was significantly reduced by partial
pancreatectomy (Fig. 2A).
Immunohistochemistry confirmed that the phosphorylation of the
tyrosine residue of CPB1 in partial pancreatectomy decreased
(Fig. 2B). These results
suggested that PTM of the CPB1 protein may be associated with
pancreatic regeneration.
Reduced phosphorylation of CPB1 induces
pancreatic cell proliferation
An increase in β-cell mass is derived from β-cell
neogenesis, proliferation and hypertrophy, and β-cell proliferation
gradually declines with age (37). However, partial pancreatectomy
models in mice and rats have been reported to enhance β-cell
replication (38). Therefore, we
investigated whether the PTM of CPB1 was involved in β-cell
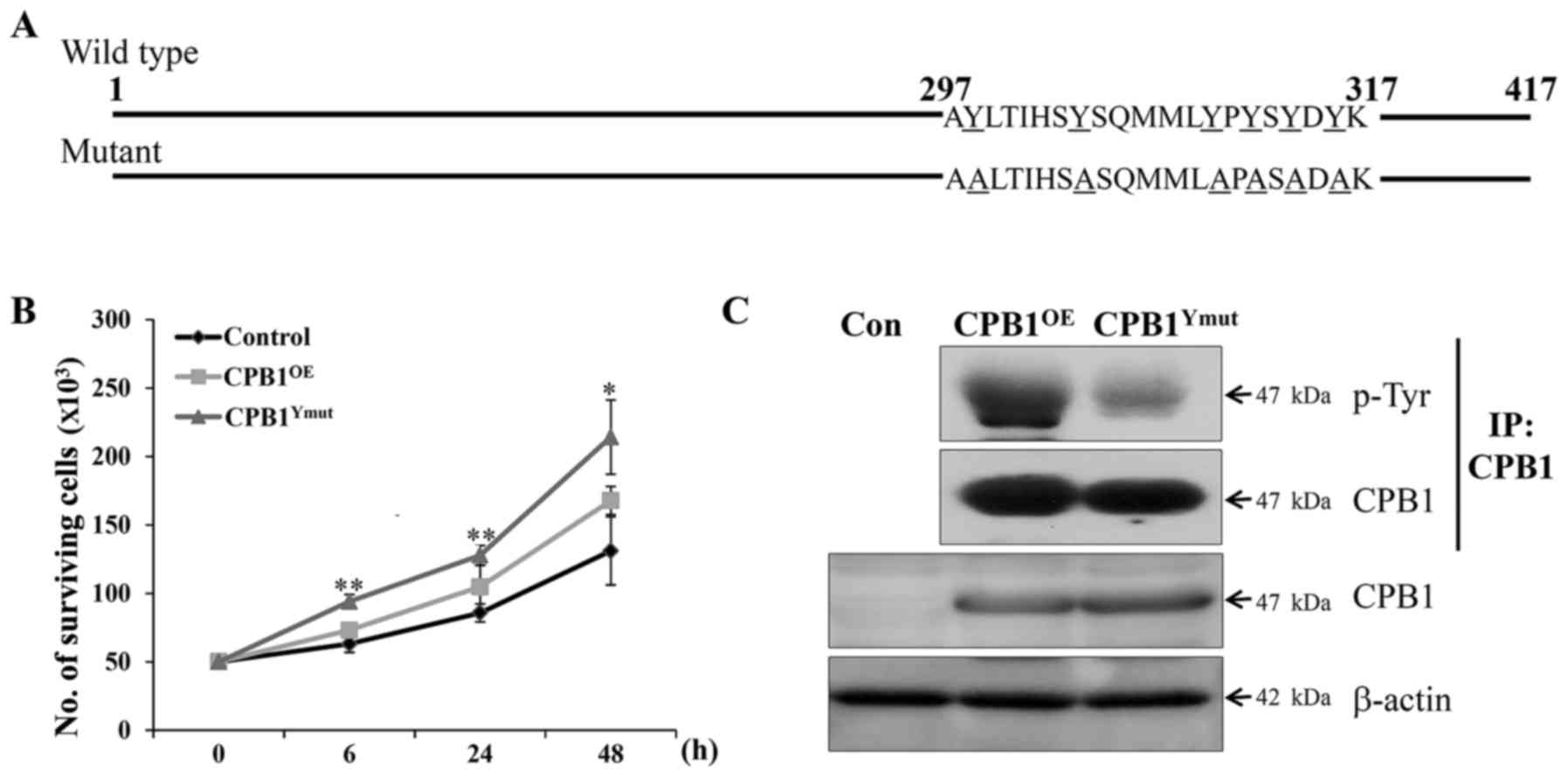
replication. First, the PTM region of the CPB1 protein was mutated
by phenylalanine-to-tyrosine substitution to reduce the
phosphorylation level (Fig. 3A).
Mutations at 6 sites (Y298F, Y304F, Y310F, Y312F, Y314F and Y316F)
were confirmed by sequencing.
Rat β-cell lines overexpressing wild-type or mutant
CPB1 were established. CPB1-overexpressing (CPB1OE) cell lines
showed higher proliferation than the control (Fig. 3B). In addition, reduced tyrosine
phosphorylation in the CPB1OE-mutY cell line resulted in increased
proliferation compared to that in CPB1OE cells. To confirm the
reduced tyrosine phosphorylation of CPB1, we performed
immunoprecipitation of the CPB1 antibody and detected
phosphorylated CPB1 by western blotting. Tyrosine phosphorylation
of CPB1 in CPB1OE-mutY cell lines decreased (Fig. 3C). Therefore, we clearly
demonstrated that the dephosphorylation of CPB1 was associated with
pancreatic regeneration.
Discussion
In the present study, we applied 2-DE analysis to
investigate PTMs of proteins related to pancreatic regeneration.
However, the present study has certain limitations, one of these
being the small sample size. Although the experimental group was
not sufficiently large enough for a comprehensive protein hunt, we
used it only to screen for proteins differentially expressed in
pancreatectomy, and we do not claim complete coverage. We observed
20 spots, including 4 CPB1 spots having different PIs, and we
confirmed the modification of CPB1 by MALDI-TOF/MS.
Carboxypeptidases have been examined for their functions in
digestion, and several studies on the synthesis of mature proteins
or their regulation of biological processes are underway. On the
basis of their active site mechanism, carboxypeptidases are divided
into three classes: metallocarboxypeptidases, serine
carboxypeptidases, and cysteine carboxypeptidases. CPB is a
metallocarboxypeptidase, and it has two isoforms: CPB1 and CPB2
(39). CPB was the first
pancreas-specific protein identified from proteins differentially
expressed between the normal pancreas and pancreatic carcinoma
(40). Moreover, CPB has been
suggested as a serum marker for acute pancreatitis and dysfunction
of pancreatic transplants (41–43).
CPB1 is secreted as a procarboxypeptidase form,
together with other pancreatic proenzymes, and is converted to the
active form through the action of trypsin (44–46). Only few PTMs of CPB1 have been
reported thus far. 3-Nitrotyrosine has been reported to be a type
of oxidative PTM of CPB1, and nitration of tyrosine plays an
important role in regulating enzyme activity. Sites of tyrosine
nitration are located at Try-198 and Tyr-248 in the catalytic site
of CPB1 (47,48). However, there is no report on the
association between PTMs of CPB1 and pancreatic regeneration. We
identified and confirmed PTMs of CPB1 by 2-DE and mass spectrometry
that may be associated with partial pancreatectomy. In islet cells
and β-cells, AMP-activated protein kinase (AMPK) regulates enzyme
activity by phosphorylation, and active AMPK inhibits insulin
secretion and proinsulin gene expression (49). Hussain et al suggested that
the phosphorylation of CREB-binding protein involved in the cyclic
AMP signaling pathway affects β-cell proliferation (50). Moreover, Rab-GTPase-activating
protein, having a critical role in glucose and lipid metabolism,
regulates phosphorylation by glucose in rat β-cells (51). Khoury et al reported a
statistical analysis of PTMs in the SWISS-PROT database;
phosphorylation is the most widely experimentally observed PTM
(52). Moreover, protein
phosphorylation plays important roles in activation, inactivation,
or modification of protein function. Therefore, we compared CPB1
phosphorylation levels between pancreases from sham-operated and
partially pancreatectomized mice using immunoprecipitation and
immunohistochemistry. We confirmed a reduction in CPB1
phosphorylation by partial pancreatectomy. To our knowledge, this
study is the first to identify the association between CPB1
dephosphorylation and pancreatic regeneration. As exact
phosphorylation sites of CPB1 have not been reported to date, we
substituted 6 anticipated tyrosines for phenylalanine and focused
on the PTMs of CPB1 and pancreatic regeneration. We found that CPB1
dephosphorylation affected β-cell proliferation. However, the
identification of the exact phosphorylation sites and mechanisms
warrants further studies. Glucagon-like peptide-1, cyclin D1,
parathyroid hormone-related protein (PTHrP), hepatocyte growth
factor/c-Met signaling, and double-stranded RNA-dependent protein
kinase have been identified as candidate proteins functioning in
pancreatic β-cell proliferation (53–57).
5′-N-ethylcarboxyamidoadenosine, an adenosine kinase
inhibitor, and domperidone, a dopamine D2 receptor antagonist, were
recently identified as small-molecule enhancers of β-cell
proliferation for the treatment of diabetes (58,59). In addition, injection of
amino-terminal peptides of PTHrP enhanced β-cell proliferation in
partially pancreatectomized mice (60). Therefore, as a next step, it needs
to be determined if peptides containing the dephosphorylation site
of CPB1 indicated in Fig. 3A can
enhance β-cell proliferation in vivo. Diabetes induced by a
reduction in pancreatic β-cells may be treatable by replenishing
the β-cell mass. However, pancreas or islet transplantation has
limited applicability owing to difficulties such as shortage of
organ donors and immunorejection of the transplanted tissue.
Therefore, the identification of β-cell proliferation mechanisms
forms the basis for developing efficient diabetes treatment
approaches. In this study, we provided evidence supporting the
relevance of CPB1 phosphorylation in β-cell proliferation.
Acknowledgments
This study was supported by the National Research
Foundation of Korea Grant funded by the Korean Government
(NRF-2012S1A2A1A0129168, NRF-2010-0007110 and
NRF-2015R1D1A3A01019948).
References
|
1
|
Seaberg RM, Smukler SR, Kieffer TJ,
Enikolopov G, Asghar Z, Wheeler MB, Korbutt G and van der Kooy D:
Clonal identification of multipotent precursors from adult mouse
pancreas that generate neural and pancreatic lineages. Nat
Biotechnol. 22:1115–1124. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
D'Amour KA, Bang AG, Eliazer S, Kelly OG,
Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK and Baetge
EE: Production of pancreatic hormone-expressing endocrine cells
from human embryonic stem cells. Nat Biotechnol. 24:1392–1401.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kroon E, Martinson LA, Kadoya K, Bang AG,
Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J,
et al: Pancreatic endoderm derived from human embryonic stem cells
generates glucose-responsive insulin-secreting cells in vivo. Nat
Biotechnol. 26:443–452. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lampeter EF, Gurniak M, Brocker U, Klemens
C, Tubes M, Friemann J and Kolb H: Regeneration of beta-cells in
response to islet inflammation. Exp Clin Endocrinol Diabetes.
103(Suppl 2): 74–78. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayashi KY, Tamaki H, Handa K, Takahashi
T, Kakita A and Yamashina S: Differentiation and proliferation of
endocrine cells in the regenerating rat pancreas after 90%
pancreatectomy. Arch Histol Cytol. 66:163–174. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kopp JL, Dubois CL, Schaffer AE, Hao E,
Shih HP, Seymour PA, Ma J and Sander M: Sox9+ ductal
cells are multipotent progenitors throughout development but do not
produce new endocrine cells in the normal or injured adult
pancreas. Development. 138:653–665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Criscimanna A, Coudriet GM, Gittes GK,
Piganelli JD and Esni F: Activated macrophages create
lineage-specific microenvironments for pancreatic acinar- and
β-cell regeneration in mice. Gastroenterology. 147:1106–1118. 2014.
View Article : Google Scholar
|
|
8
|
Dor Y, Brown J, Martinez OI and Melton DA:
Adult pancreatic beta-cells are formed by self-duplication rather
than stem-cell differentiation. Nature. 429:41–46. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thorel F, Népote V, Avril I, Kohno K,
Desgraz R, Chera S and Herrera PL: Conversion of adult pancreatic
alpha-cells to betacells after extreme beta-cell loss. Nature.
464:1149–1154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chera S, Baronnier D, Ghila L, Cigliola V,
Jensen JN, Gu G, Furuyama K, Thorel F, Gribble FM, Reimann F, et
al: Diabetes recovery by age-dependent conversion of pancreatic
δ-cells into insulin producers. Nature. 514:503–507. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Inada A, Nienaber C, Katsuta H, Fujitani
Y, Levine J, Morita R, Sharma A and Bonner-Weir S: Carbonic
anhydrase II-positive pancreatic cells are progenitors for both
endocrine and exocrine pancreas after birth. Proc Natl Acad Sci
USA. 105:19915–19919. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu X, D'Hoker J, Stangé G, Bonné S, De Leu
N, Xiao X, Van de Casteele M, Mellitzer G, Ling Z, Pipeleers D, et
al: Beta cells can be generated from endogenous progenitors in
injured adult mouse pancreas. Cell. 132:197–207. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lim HW, Lee JE, Shin SJ, Lee YE, Oh SH,
Park JY, Seong JK and Park JS: Identification of differentially
expressed mRNA during pancreas regeneration of rat by mRNA
differential display. Biochem Biophys Res Commun. 299:806–812.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shin JS, Lee JJ, Lee EJ, Kim YH, Chae KS
and Kim CW: Proteome analysis of rat pancreas induced by
pancreatectomy. Biochim Biophys Acta. 1749:23–32. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
De León DD, Farzad C, Crutchlow MF,
Brestelli J, Tobias J, Kaestner KH and Stoffers DA: Identification
of transcriptional targets during pancreatic growth after partial
pancreatectomy and exendin-4 treatment. Physiol Genomics.
24:133–143. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang M, Liu W, Wang CY, Liu T, Zhou F, Tao
J, Wang Y and Li MT: Proteomic analysis of differential protein
expression in early process of pancreatic regeneration in
pancreatectomized rats. Acta Pharmacol Sin. 27:568–578. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Choi JH, Lee MY, Kim Y, Shim JY, Han SM,
Lee KA, Choi YK, Jeon HM and Baek KH: Isolation of genes involved
in pancreas regeneration by subtractive hybridization. Biol Chem.
391:1019–1029. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi JH, Lee MY, Ramakrishna S, Kim Y,
Shim JY, Han SM, Kim JY, Lee DH, Choi YK and Baek KH: LCP1
up-regulated by partial pancreatectomy supports cell proliferation
and differentiation. Mol Biosyst. 7:3104–3111. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rukstalis JM and Habener JF: Neurogenin3:
A master regulator of pancreatic islet differentiation and
regeneration. Islets. 1:177–184. 2009. View Article : Google Scholar
|
|
20
|
Reichert M, Takano S, von Burstin J, Kim
SB, Lee JS, Ihida-Stansbury K, Hahn C, Heeg S, Schneider G, Rhim
AD, et al: The Prrx1 homeodomain transcription factor plays a
central role in pancreatic regeneration and carcinogenesis. Genes
Dev. 27:288–300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ahlgren U, Jonsson J, Jonsson L, Simu K
and Edlund H: beta-cell-specific inactivation of the mouse
Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and
maturity onset diabetes. Genes Dev. 12:1763–1768. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li S, Iakoucheva LM, Mooney SD and
Radivojac P: Loss of post-translational modification sites in
disease. Pacific Symposium on Biocomputing. Pac Symp Biocomput.
337–347. 2010.
|
|
23
|
Paulo JA, Kadiyala V, Brizard S, Banks PA,
Steen H and Conwell DL: Post-translational modifications of
pancreatic fluid proteins collected via the endoscopic pancreatic
function test (ePFT). J Proteomics. 92:216–227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Petersen HV, Peshavaria M, Pedersen AA,
Philippe J, Stein R, Madsen OD and Serup P: Glucose stimulates the
activation domain potential of the PDX-1 homeodomain transcription
factor. FEBS Lett. 431:362–366. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khoo S, Griffen SC, Xia Y, Baer RJ, German
MS and Cobb MH: Regulation of insulin gene transcription by ERK1
and ERK2 in pancreatic beta cells. J Biol Chem. 278:32969–32977.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lebrun P, Montminy MR and Van Obberghen E:
Regulation of the pancreatic duodenal homeobox-1 protein by
DNA-dependent protein kinase. J Biol Chem. 280:38203–38210. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boucher MJ, Selander L, Carlsson L and
Edlund H: Phosphorylation marks IPF1/PDX1 protein for degradation
by glycogen synthase kinase 3-dependent mechanisms. J Biol Chem.
281:6395–6403. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng R, Al-Quobaili F, Müller I, Götz C,
Thiel G and Montenarh M: CK2 phosphorylation of Pdx-1 regulates its
transcription factor activity. Cell Mol Life Sci. 67:2481–2489.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Frogne T, Sylvestersen KB, Kubicek S,
Nielsen ML and Hecksher-Sørensen J: Pdx1 is post-translationally
modified in vivo and serine 61 is the principal site of
phosphorylation. PLoS One. 7:e352332012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grijalva JL, Huizenga M, Mueller K,
Rodriguez S, Brazzo J, Camargo F, Sadri-Vakili G and Vakili K:
Dynamic alterations in Hippo signaling pathway and YAP activation
during liver regeneration. Am J Physiol Gastrointest Liver Physiol.
307:G196–G204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moles A, Butterworth JA, Sanchez A, Hunter
JE, Leslie J, Sellier H, Tiniakos D, Cockell SJ, Mann DA, Oakley F,
et al: A RelA(p65) Thr505 phospho-site mutation reveals an
important mechanism regulating NF-κB-dependent liver regeneration
and cancer. Oncogene. 35:4623–4632. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bonner-Weir S, Trent DF and Weir GC:
Partial pancreatectomy in the rat and subsequent defect in
glucose-induced insulin release. J Clin Invest. 71:1544–1553. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim HR, Kang JK, Yoon JT, Seong HH, Jung
JK, Lee HM, Sik Park C and Jin DI: Protein profiles of bovine
placenta derived from somatic cell nuclear transfer. Proteomics.
5:4264–4273. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Harding JD and Rutter WJ: Rat pancreatic
amylase mRNA. Tissue specificity and accumulation during embryonic
development. J Biol Chem. 253:8736–8740. 1978.PubMed/NCBI
|
|
35
|
Jelenik T and Roden M: Mitochondrial
plasticity in obesity and diabetes mellitus. Antioxid Redox Signal.
19:258–268. 2013. View Article : Google Scholar :
|
|
36
|
Dephoure N, Gould KL, Gygi SP and Kellogg
DR: Mapping and analysis of phosphorylation sites: A quick guide
for cell biologists. Mol Biol Cell. 24:535–542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ackermann AM and Gannon M: Molecular
regulation of pancreatic beta-cell mass development, maintenance,
and expansion. J Mol Endocrinol. 38:193–206. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li WC, Rukstalis JM, Nishimura W,
Tchipashvili V, Habener JF, Sharma A and Bonner-Weir S: Activation
of pancreaticduct-derived progenitor cells during pancreatic
regeneration in adult rats. J Cell Sci. 123:2792–2802. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Marinkovic DV, Marinkovic JN, Erdös EG and
Robinson CJ: Purification of carboxypeptidase B from human
pancreas. Biochem J. 163:253–260. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pousette A, Fernstad R, Sköldefors H and
Carlström K: Novel assay for pancreatic cellular damage: 1.
Characterization of protein profiles in human pancreatic cytosol
and purification and characterization of a pancreatic specific
protein. Pancreas. 3:421–426. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yamamoto KK, Pousette A, Chow P, Wilson H,
el Shami S and French CK: Isolation of a cDNA encoding a human
serum marker for acute pancreatitis. Identification of
pancreas-specific protein as pancreatic procarboxypeptidase B. J
Biol Chem. 267:2575–2581. 1992.PubMed/NCBI
|
|
42
|
Chen CC, Wang SS, Chao Y, Chen SJ, Lee SD,
Wu SL, Jeng FS and Lo KJ: Serum pancreas-specific protein in acute
pancreatitis. Its clinical utility in comparison with serum
amylase. Scand J G astroenterol. 29:87–90. 1994. View Article : Google Scholar
|
|
43
|
Printz H, Siegmund H, Wojte C, Schäfer C,
Hesse H, Rothmund M and Göke B: 'Human pancreas-specific protein'
(procarboxypeptidase B): A valuable marker in pancreatitis?
Pancreas. 10:222–230. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Burgos FJ, Salvà M, Villegas V, Soriano F,
Mendez E and Avilés FX: Analysis of the activation process of
porcine procarboxypeptidase B and determination of the sequence of
its activation segment. Biochemistry. 30:4082–4089. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Appelros S, Thim L and Borgström A:
Activation peptide of carboxypeptidase B in serum and urine in
acute pancreatitis. Gut. 42:97–102. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Müller CA, Appelros S, Uhl W, Büchler MW
and Borgström A: Serum levels of procarboxypeptidase B and its
activation peptide in patients with acute pancreatitis and
non-pancreatic diseases. Gut. 51:229–235. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sokolovsky M: Porcine carboxypeptidase B.
Nitration of the functional tyrosyl residue with tetranitromethane.
Eur J Biochem. 25:267–273. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chatterjee S, Lardinois O, Bonini MG,
Bhattacharjee S, Stadler K, Corbett J, Deterding LJ, Tomer KB,
Kadiiska M and Mason RP: Site-specific carboxypeptidase B1 tyrosine
nitration and pathophysiological implications following its
physical association with nitric oxide synthase-3 in experimental
sepsis. J Immunol. 183:4055–4066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
da Silva Xavier G, Leclerc I, Salt IP,
Doiron B, Hardie DG, Kahn A and Rutter GA: Role of AMP-activated
protein kinase in the regulation by glucose of islet beta cell gene
expression. Proc Natl Acad Sci USA. 97:4023–4028. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hussain MA, Porras DL, Rowe MH, West JR,
Song WJ, Schreiber WE and Wondisford FE: Increased pancreatic
beta-cell proliferation mediated by CREB binding protein gene
activation. Mol Cell Biol. 26:7747–7759. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rütti S, Arous C, Nica AC, Kanzaki M,
Halban PA and Bouzakri K: Expression, phosphorylation and function
of the Rab-GTPase activating protein TBC1D1 in pancreatic
beta-cells. FEBS Lett. 588:15–20. 2014. View Article : Google Scholar
|
|
52
|
Khoury GA, Baliban RC and Floudas CA:
Proteome-wide post-translational modification statistics: Frequency
analysis and curation of the swiss-prot database. Sci Rep. 1:12011.
View Article : Google Scholar
|
|
53
|
De León DD, Deng S, Madani R, Ahima RS,
Drucker DJ and Stoffers DA: Role of endogenous glucagon-like
peptide-1 in islet regeneration after partial pancreatectomy.
Diabetes. 52:365–371. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang X, Gaspard JP, Mizukami Y, Li J,
Graeme-Cook F and Chung DC: Overexpression of cyclin D1 in
pancreatic beta-cells in vivo results in islet hyperplasia without
hypoglycemia. Diabetes. 54:712–719. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Williams K, Abanquah D, Joshi-Gokhale S,
Otero A, Lin H, Guthalu NK, Zhang X, Mozar A, Bisello A, Stewart
AF, et al: Systemic and acute administration of parathyroid
hormone-related peptide (1-36) stimulates endogenous beta cell
proliferation while preserving function in adult mice.
Diabetologia. 54:2867–2877. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Alvarez-Perez JC, Ernst S, Demirci C,
Casinelli GP, Mellado-Gil JM, Rausell-Palamos F, Vasavada RC and
Garcia-Ocaña A: Hepatocyte growth factor/c-Met signaling is
required for β-cell regeneration. Diabetes. 63:216–223. 2014.
View Article : Google Scholar
|
|
57
|
Gao L, Tang W, Ding Z, Wang D, Qi X, Wu H
and Guo J: Proteinbinding function of RNA-dependent protein kinase
promotes proliferation through TRAF2/RIP1/NF-κB/c-Myc pathway in
pancreatic β cells. Mol Med. 21:154–166. 2015.PubMed/NCBI
|
|
58
|
Andersson O, Adams BA, Yoo D, Ellis GC,
Gut P, Anderson RM, German MS and Stainier DY: Adenosine signaling
promotes regeneration of pancreatic β cells in vivo. Cell Metab.
15:885–894. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sakano D, Choi S, Kataoka M, Shiraki N,
Uesugi M, Kume K and Kume S: Dopamine D2 receptor-mediated
regulation of pancreatic β cell mass. Stem Cell Reports. 7:95–109.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mozar A, Lin H, Williams K, Chin C, Li R,
Kondegowda NG, Stewart AF, Garcia-Ocaña A and Vasavada RC:
Parathyroid hormone-related peptide (1-36) enhances beta cell
regeneration and increases beta cell mass in a mouse model of
partial pancreatectomy. PLoS One. 11:e01584142016. View Article : Google Scholar : PubMed/NCBI
|