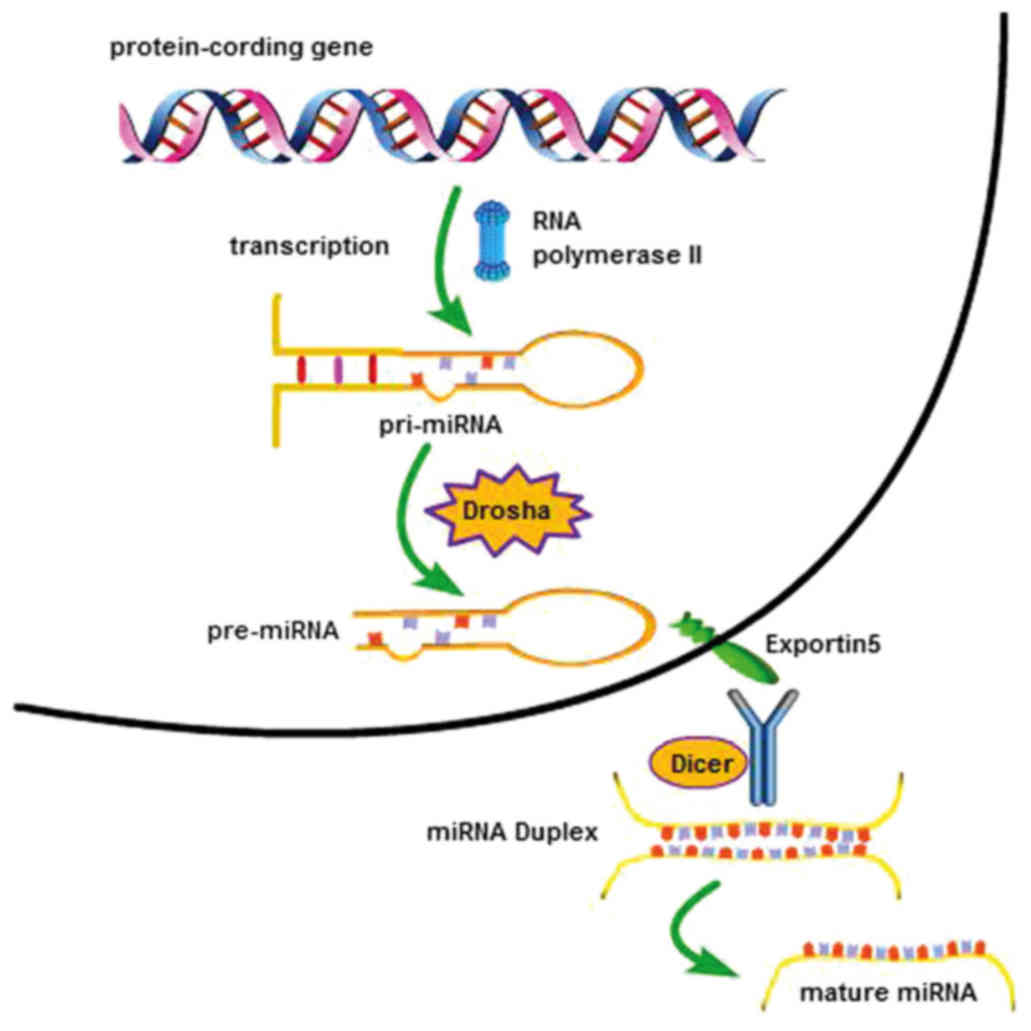
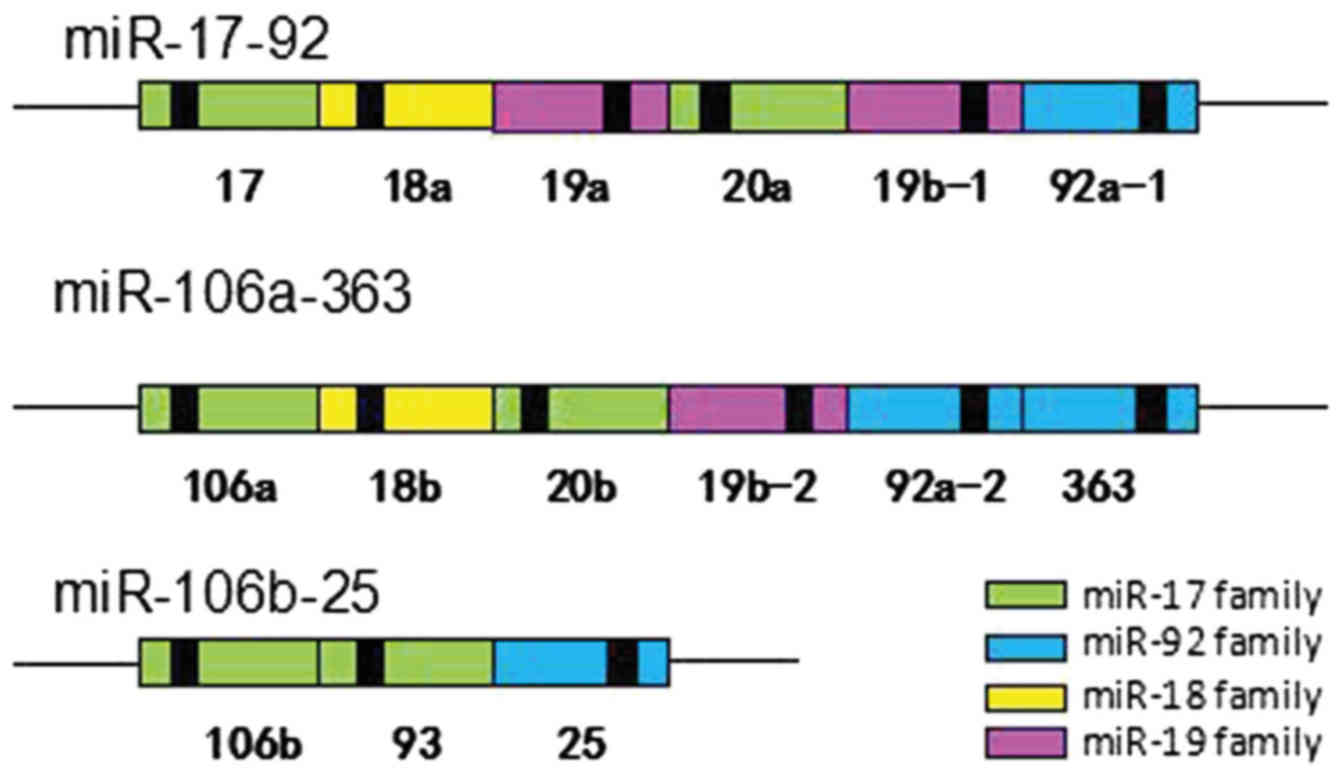
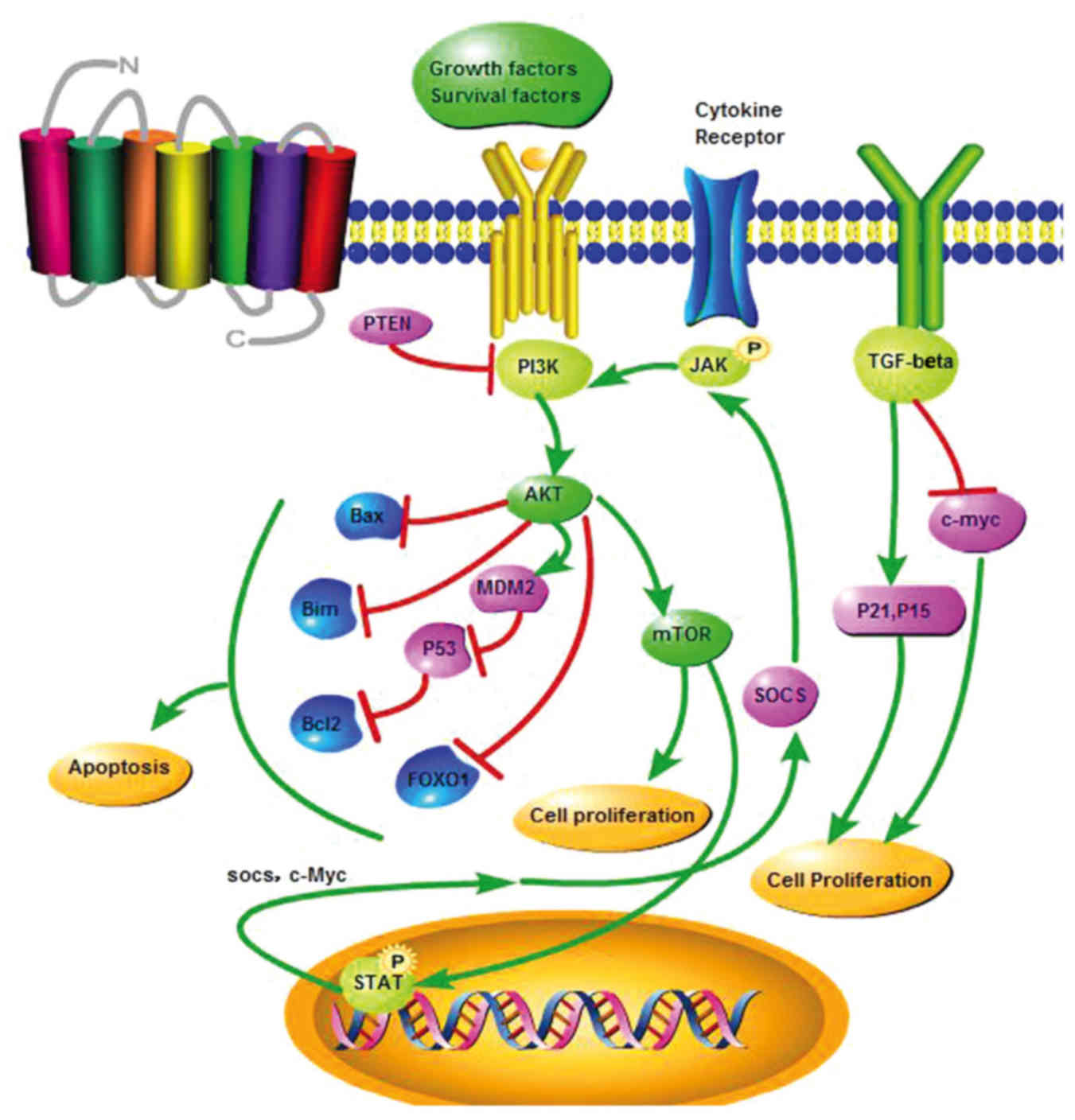
1. Introduction
MicroRNAs (miRNAs) are a class of single-stranded,
non-coding small RNAs, ~22 nucleotides (nt) in length, that are
able to bind to target mRNAs via partial or complete complementary
base pairing. miRNAs regulate gene expression and may inhibit
oncogenes or tumor-suppressor genes at the post-transcriptional
level (1). miRNAs take the RNA
induced silencing complex (RISC) to the target mRNA containing
complementary sequences, and induce its degradation (2). miRNAs are derived from the
transcription of a set of protein-coding genes, but are
structurally and functionally different from the mRNA transcribed
by the common gene. In particular, each miRNA originates from a
longer primary transcript, referred to as pri-miRNA, which is
transcribed in the nucleus from genomic DNA by the RNA polymerase
II. The pri-miRNA is then cleaved by the specific endonuclease
Drosha into a pre-miRNA hairpin consisting of ~70 nt and containing
the sequence complementary to the target mRNA (3). This pre-miRNA hairpin is transported
into the cytoplasm by the nuclear export protein exportin 5 and
cleaved by Dicer to form a short double-stranded molecule, in which
each strand is a mature miRNA (Fig.
1).
To date, ~8,000 genes coding for miRNAs have been
identified in various organisms, such as plants, viruses and
animals, including 1,000 human miRNAs that have been confirmed
(4). It has been demonstrated
that one single miRNA may regulate the expression of >200 target
genes, and the expression of certain target mRNAs may also be
regulated by several miRNAs. Overall, over one-third of structural
human genes were found to be regulated by miRNAs (5). miRNAs are involved in the regulation
of a plethora of biological processes, such as cell
differentiation, proliferation, metastasis, metabolism, apoptosis,
tumorigenesis, angiogenesis and others. Considering their roles, it
is not surprising that abnormal expression of miRNAs is associated
with several pathologies, thus making miRNAs useful clinical
biomarkers that may be used in the diagnosis, treatment and
prognosis of tumors (6). As a
consequence, numerous studies have demonstrated that miRNAs are
directly implicated in the occurrence and development of cancer,
thus attracting even more interest in the research field.
Approximately 60% of miRNAs may express
independently, 15% as a cluster, and 35% cannot express, being
located in introns. Clustered miRNAs have demonstrated a
cooperative function in regulating gene expression (7). In 2005, He et al first
discovered the miR-17-92 cluster, an oncogenic gene in human B-cell
lymphomas (8). The aim of the
present review was to summarize the functions and mechanisms
through which the miR-17-92 cluster is involved in cancer, thus
providing a theoretical basis to study the effect and molecular
mechanism of the miR-17-92 cluster in regulating the development of
prostate cancer cells.
2. Characteristics of the miRNA-17-92
cluster
miRNA clusters are mainly expressed in vertebrates
and mammals, and result from genome duplication (9). As a consequence, miRNAs were
classified as clusters due to their high sequence homology. The
miR-17-92 cluster is a typical highly conserved polycistronic miRNA
cluster, which is located in the human chromosome 13 open reading
frame 25 (C13orf25), encoding six mature miRNAs, including miR-17,
miR-18a, miR-19a, miR-19b, miR-20a and miR-92a (10). Both human miR-17 and miR-20a were
included in the miR-17 family due to their high sequence homology
(Fig. 2).
In detail, the miR-17-92 cluster has two paralogue
gene clusters named miR-106a-363 and miR-106b-25. The miR-106A-363
cluster encodes for miR-106a, miR-18b, miR-20b, miR-19b-2, miR92a-2
and miR-363; the miR-106b-25 cluster encodes for miR-106b, miR-93
and miR-25 (Fig. 2). According to
the homology of the seed-sequence, all these miRNAs have been
grouped into four families, namely the miR-17, miR-92, miR-18 and
miR-19 families (11). The miRNA
sequences of the three clusters miR-17-92, miR-106b-25 and
miR-106b-363 were found to be highly similar, with overlapping
functions (Fig. 2). Previous
studies have demonstrated that murine knockout models for the
miR-17-92 cluster died soon after birth due to lung function
insufficiency and ventricular septal defects. The simultaneous
deletion of both miR-17-92 and miR-106b-25 caused severe apoptosis
of fetal liver cells and central nervous system cells in mice.
However, the simultaneous or separated deletion of miR-106b-25 and
miR-106a-363 did not affect the individual development (12). These results indicated that there
are some overlapping roles in members of the miR-17 and miR-92
families within the miR-17-92 and miR-106b-25 clusters, while the
miR-18 and miR-19 families, only present in the miR-17-92 cluster,
play a critical role in developmental processes. A recent study
analyzed the expression of the miR-17-92 and miR-106b-25 clusters
in spermatogonial stem cells, demonstrating that the miR-106b-25
cluster may be upregulated in germ cells without affecting
spermatogonial development when the miR-17-92 cluster is deleted
(13). This indicated that the
miR-17-92 and miR-106b-25 clusters may synergistically regulate
reproductive development.
3. Expression and regulation of the
miRNA-17-92 cluster in tumor cells
Expression and functions of
miR-17/20a
The miRNA-17-92 cluster may be highly expressed in a
wide range of tumor cells and types of cancer, such as lung,
breast, pancreatic, prostate and thyroid cancer, as well as
lymphomas (7,14). Therefore, it is also referred to
as 'oncomiR1'. The majority of the previous studies have been aimed
at studying the potential carcinogenicity of the miR-17-92 cluster,
but this cluster also possesses antitumor properties. For example,
the miR-17 component acts as a tumor suppressor in breast and
prostate cancer by individually targeting AIB1 and PCAF (15,16). Of note, the development of
erythroleu-kemia induced by miR-92a was inhibited by the
co-expression of miR-92a and miR-17, indicating that the expression
of miR-17 may inhibit the carcinogenesis induced by miR-92a
(17). Moreover, it has been
demonstrated that miR-17-5p is able to induce prostate tumor growth
and invasion by regulating TIMP3 (18). In addition, miR-20a may have
different functions in various pathological processes, with a dual
behavior (acts as an oncogene or tumor suppressor). In fact,
several studies have demonstrated that miR-20a was found to be
upregulated in the serum of hepatitis C virus-infected individuals,
and in uveal melanoma, osteosarcoma, neuroglioma, undifferentiated
thyroid cancer, cervical, gastric and prostate cancer, while it was
down-regulated in breast, liver and pancreatic cancer cells
(19-26).
Expression and functions of
miR-19/miR-92a/miR-18a
Several studies reported miR-19, one of the major
oncogenes in the miR-17-92 cluster, to be highly expressed in
gastric and prostate cancer (27). In addition, miR-18a was found to
be highly expressed in breast, nasopharyngeal, prostate and
colorectal cancer (28). In
glioma, colorectal adenoma, renal clear cell carcinoma, small-cell
lung cancer, hepatocellular carcinoma, multiple myeloma and
non-Hodgkin lymphoma, the transcriptional level of miR-92a was
found to be higher compared with that of other miRNAs present in
the miR-17-92 cluster (29-31). However, the expression of the same
miRNA in breast cancer tissues was lower compared with normal
tissues. Further studies have also suggested that the expression
level of miR-92a may be associated with the size of the tumor and
lymph node metastasis.
Recent evidence demonstrated that the expression of
miR-17, miR18a and miR-19a increased during tumor angiogenesis,
displaying proangiogenic functions through the regulation of the
target protein kinase JAK1, while miR-92a decreased and inhibited
vascular network formation by regulating integrin α5 (ITGα5)
(32). In fact, the
overexpression of miR-17, miR-18a and miR-20a partially restored
the impaired endothelial network formation, but suppressed
angiogenic sprout formation in zebrafish (33). There have been no related studies
on human cells and tissues to date; however, this evidence suggests
that miR-92a is a negative regulator of angiogenesis.
The function of miRNAs as tumor suppressors is
similar to that of tumor suppressor genes: Their downregulation or
inactivation directly leads to the occurrence and development of
cancer. However, only few studies provided evidence enabling a
better understanding of the main function of the miR-17-92 cluster,
considering that it may promote carcinogenesis as well as act as a
tumor suppressor. Our study group aimed to investigate the
effectiveness of the miR-17-92 cluster as a diagnostic biomarker in
different stages of prostate cancer development, as well as the
molecular mechanism through which this cluster regulates prostate
cancer cell growth, migration and invasion, with the aim to
determine its potential use in clinical practice.
4. Mechanism of action of the miR-17-92
cluster in tumorigenesis and tumor development
Mechanism of action of the miR-17-92
cluster in tumori- genesis
Tumorigenesis is associated with a disorder of the
mechanism that maintains normal cell activity. It is associated
with multiple complex processes, such as excessive cell
proliferation, and interruptive apoptosis and differentiation,
among others. miRNAs play an important role in tumorigenesis and
tumor development, participating in various stages of these
processes. For example, a disorder in the regulation of the
interactions among miRNAs and target genes may lead to the
occurrence of a tumor. In cancer, the different regulation of
targeting genes allows miRNAs to have various biological functions,
as oncogenes or tumor suppressors.
The miR-17-92 cluster plays a crucial role in
tumorigenesis, mainly via the activation of oncogenes and the
inactivation of tumor suppressor genes. The expression of cell
cycle regulatory genes plays an important role in tumorigenesis.
For example, it has been demonstrated that the miR-17-92 cluster
can inhibit the expression of the tumor suppressor p21 and the
apoptotic gene Bim in lymphoma (34). Furthermore, miR-20a acts as an
oncogene and, through inhibition of the expression of early growth
response (EGR)2, it may promote cell proliferation and induce cell
cycle progression in osteosar-coma (21). Accordingly, miR-20a regulates
carcinogenesis in gastric cancer cells though the EGR2 signaling
pathway (25). By contrast,
miR-17-5p exerts an antitumor effect by inhibiting the expression
of AIB1 in breast cancer (15,35).
The miRNA-17-92 cluster promotes tumor cell
proliferation and apoptosis by regulating different target genes
and signaling pathways. Several transcription factors regulate the
miR-17-92 cluster, affecting its carcinogenic activity. The first
confirmed miR-17-92 transcription factor was MYC, which is involved
in multiple mechanisms regulating gene expression. Overexpressed in
approximately half of human cancers, MYC binds to specific genomic
sites directly activating miR-17-92 expression. MYC may also
inhibit specific target genes, such as Sin3b, Btg1 and the
apoptosis-regulating factor Bim (14,36). In neuroblastoma cells, the
miR-17-92 cluster is upregulated via MYCN (37). Along with MYC, p21 represents an
important target of the miRNA-17-92 cluster. The expression of p21
may be inhibited by c-Myc, which promotes tumor cell proliferation
and, thus, tumor growth. It has been demonstrated that miR-20
affects the regulatory factor CDKN1A/p21, which is activated by
transforming growth factor (TGF)-β, thus preventing the
antiproliferative effect induced by TGF-β in colorectal cancer
(38). In addition, the
transcription factors E2F1, E2F2 and E2F3, which are members of the
E2F family, have been identified as target genes of miR-17 and
miR-20a (39-41). In fact, the suppression of
miR-17-92 in cervical carcinoma led to the upregulation of E2F1
(24). Overall, it may be argued
that the regulation of gene expression by miRNAs may be implemented
through mechanisms similar to those of transcription factors.
The regulation of almost all cellular processes
occurs through several signaling pathways. The Janus kinase/signal
transducer and activator of transcription (JAK-STAT) pathway plays
a pivotal role in the mechanism of action of the miR-17-92 cluster.
In multiple myeloma, miR-17-92 enhances cell proliferation and
inhibits cell apoptosis by inhibiting the tumor suppressor gene
SOCS-1 and activating the JAK-STAT pathway (42). Recent studies have revealed that
phosphoinositide-3 kinase (PI3K)/AKT/mammalian target of rapamycin
is another important axis that regulates tumor development by
inhibiting apoptotic and activating anti-apoptotic factors, thus
promoting cell survival. Once activated, AKT regulates cell
proliferation, growth and survival by phosphorylating different
downstream targets, such as enzymes, kinases, transcription factors
and others. The activation of this pathway may downregulate the
expression of the tumor suppressor gene p53, thus inhibiting
apoptosis. Another tumor suppressor gene is phosphatase and tensin
homolog (PTEN), a phosphatase of phosphatidylinositol (3,4,5)-trisphosphate, which is the first to
be found in the tumor suppressor gene and, through downregulation
of the PI3K/AKT signaling pathway, promotes apoptosis, thus acting
as a tumor suppressor (Fig. 3).
The miR-17-92 cluster activates the AKT̸glycogen synthase kinase
pathway through downregulation of the expression of PTEN, and
promotes cell proliferation and angiogenesis throught the PI3K/AKT
pathway (43,44). The expression of the pro-apoptotic
factor Bim may be inhibited by the miR-17-92 cluster, thus blocking
apoptosis (34). Bim is
also a target of miR-92a, and it is associated with the
tumor malignancy in
colon adenoma (30).
Mechanism of action of the miR-17-92
cluster in tumor development Effect of the miR-17-92 cluster on
cancer stem cells
Cancer stem cells (CSCs), also referred to as
tumor-initiating cells, represent the origin of the primary tumor,
with their capacity of self-renewal and multiple differentiation
potential. CSCs play a crucial role in the occurrence, development,
metastasis and recurrence of tumors. However, there is currently
controversy regarding CSCs, although a growing volume of
experimental evidence (e.g., flow cytometry, sorting technologies
and animal models) support the CSC theory (45). CSCs maintain tumor cell viability
by self-renewal and their unlimited proliferation ability. Specific
miRNAs and long-chain non-coding RNAs regulate certain
characteristics of tumor stem cells, including asymmetric cell
division, high tumorigenicity, drug resistance, invasion and
metastasis (46). Zagorac et
al reported that the genetic targeting of DNMT1 through
epigenetic reactivation of the miR-17-92 cluster in reprogrammed
pancreatic cancer stem cells reduced their self-renewal (47). In addition, the overexpression of
miR-17-92 reduced CSC self-renewal capacity in vivo by
targeting multiple signaling cascade members, such as
NODAL/ACTIVIN/TGF-β1, and directly inhibiting the downstream
targets p21, p57 and TBX3. miR-17-92, thus, represents a potential
target for the prognosis of pancreatic cancer and may provide a
guide to diagnosis and treatment (48).
Therefore, the reduction or elimination of the
self-renewal ability of CSCs by miRNAs, such as the miR-17-92
cluster, may represent a promising new direction towards designing
novel cancer therapies.
Epithelial-to-mesenchymal transition
(EMT) and its role in tumor development
Epithelial and mesenchymal cells are the two main
types of cells in human tissues. Epithelial cells exhibit polarity,
and are connected to each other through adhesions, bridging and gap
junctions. Conversely, mesenchymal cells do not exhibit polarity,
lack intercellular junctions, and are able to migrate through the
extracellular matrix. EMT is a biological process during which
epithelial cells transform to cancer cells with mesenchymal
characteristics, such as the ability to invade and migrate under
physiological and pathological conditions. The expression of
E-cadherin and markers of mesenchymal cells (N-cadherin, vimentin
and fibronectin) represent the main characteristics of EMT, along
with decreased cell adhesion (49). EMT is critical for normal
embryonic development, wound healing, tissue regeneration, organ
fibrosis, and it also occurs during tumor development, invasion and
metastasis (50). A study on
colon and pancreatic cancer reported the presence of a mutual
feedback loop between members of the miR-200 family and ZEB1, which
is involved in the invasion and metastasis induced by EMT (51). The expression of miR-200 family
members was significantly associated with the expression of
E-cadherin, thus inhibiting the expression of ZEB1 and SIP1. By
contrast, ZEB1 binds directly with the promoter region of miR-200,
thus inhibiting the expression of genes and forming a double
negative feedback pathway. Increased expression of miR-19 is able
to trigger EMT in lung cancer cells, reduce cell adhesion, and
enhance cell migration and invasion through regulating epithelial
and mesenchymal proteins (52).
In colon cancer, miR-17 induces EMT consistently with the cancer
stem cell phenotype by regulating CYP7B1 expression (53). The expression of the miR-17-92
cluster is correlated with inhibition of EMT by reducing the
expression of mesenchymal markers, such as N-cadherin, vimentin,
Twist1, Slug and TCF8/ZEB1, and by promoting the expression of the
epithelial marker E-cadherin (54).
The miRNA-17-92 cluster facilitates tumor
cell migration, invasion and metastasis
The miR-17-92 cluster, acting as an oncogene,
induces tumor cell invasion and metastasis by regulating its target
genes. miR-19 may contribute to the development of c-Myc-induced
lymphoma, particularly by playing a key role in stimulating
lymphoma cell migration, invasion and metastasis (27). Similarly, miR-19a/b has been found
to be upregulated in metastatic gastric cancer, in which it
promotes cell migration, invasion and metastasis, by regulating the
tumor suppressor MXD1, a c-Myc antagonist (55). It has been demonstrated that
miR-92a directly targets the E-cadherin (CDH1) gene, which is
associated with human esophageal squamous cell carcinoma (56). In aggressive leukemia, such as
erythroleukemia caused by the activation of the Fli-1 gene by the
Friend virus, miR-92a may accelerate the development of the Friend
virus by regulating the p53 pathway (57). However, Ohyagi-Hara et al
confirmed that miR-92a can directly target ITGα5 and decrease the
expression of ITGα5 in ovarian cancer, thus inhibiting tumor cell
adhesion, metastasis and proliferation (58). mir-20a can promote cell invasion
and migration by targeting the ABL2 gene in prostate cancer, and
TIMP2 and ATG7 in glioma stem cells and ovarian cancer (22,26,59). miR-20a was found to be highly
expressed in undifferentiated thyroid carcinoma, and plays an
antitumor role in thyroid cancer. Its effect is mainly exerted
through the inhibition of the proliferation and invasion of thyroid
cancer cells by targeting the LIMK1 gene (23). Reversely, STAT-3 downregulation
inhibited malignant pleural mesothelioma cell invasion and tumor
migration by miR-17 (Fig. 4)
(60). All the abovementioned
studies demonstrated that the miR-17-92 cluster may play different
roles in several cancer tissues, but its mechanism of action
remains to be elucidated by further studies. However, these
findings provide novel insights to the treatment of different
cancers.
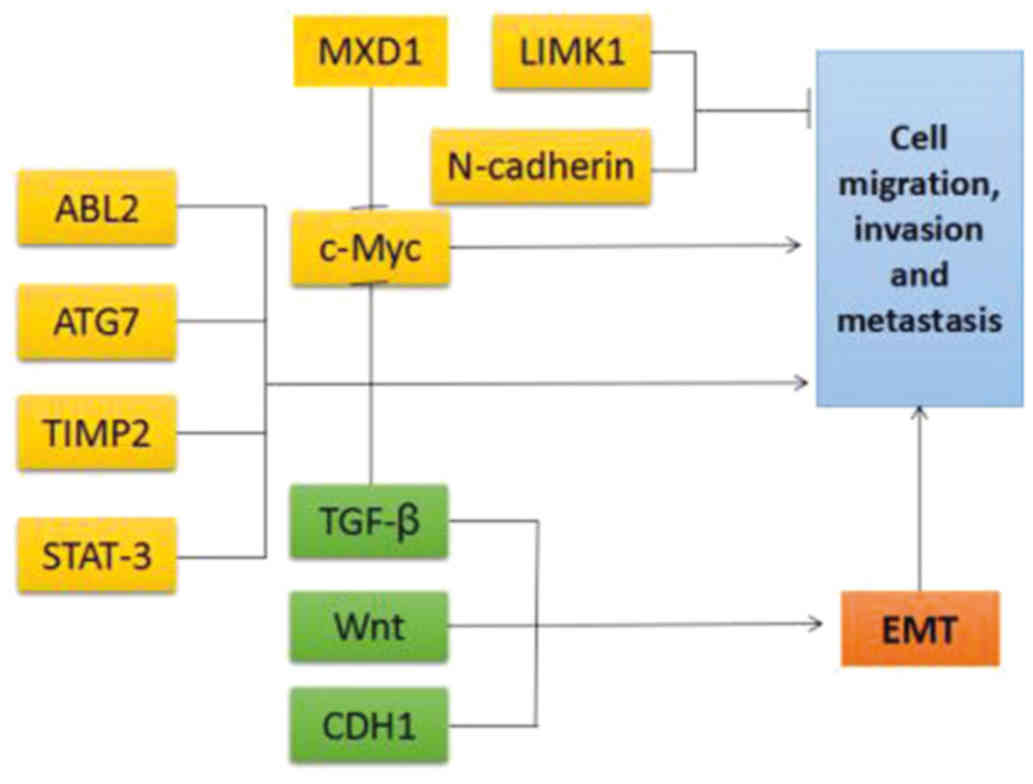 | Figure 4Target genes and signaling pathways
regulated by the miR-17-92 cluster that are involved in tumor cell
migration, invasion and metastasis. Green arrows, activating
effect; red lines, inhibitory effect. TGF, transforming growth
factor; STAT, signal transducer and activator of transcription;
CDH, cadherin; TIMP, tissue inhibitor of metalloproteinases; MXD1,
MAX dimerization protein 1; LIMK1, LIM domain kinase 1; EMT,
epithelial-to-mesenchymal transition. |
5. Clinical applications and perspectives
for the miRNA-17-92 cluster
miRNAs are key players in biological processes such
as cell proliferation, differentiation, tumorigenesis, immune
regulation and several others. A number of studies have
demonstrated specific expression of members of the miRNA-17-92
cluster in various diseases, particularly in different types of
cancer, suggesting that the miRNA-17-92 cluster may represent a new
direction for the diagnosis and treatment of cancer. Monitoring the
expression changes of the members of the miRNA-17-92 cluster under
specific tumor conditions may be a powerful tool for the early
detection of cancer. The miRNA-17-92 cluster is also predicted to
provide important supplementary tools for tumor classification,
determination of the treatment plan and analysis of prognosis by
clinicians. For example, the use of miR-17 antagonists represents a
novel therapeutic approach to the treatment of chronic lymphocytic
leukemia (61). In animal models,
the intravenous injection of anti-miR-17-92 may cure allograft
medulloblastoma by decreasing cell proliferation and suppressing
tumor growth (62). Experiments
on evaluating the effects of the overexpression and silencing of
the miRNA-17-92 cluster in the embryonic and postnatal mouse heart
demonstrated that this cluster may induce the proliferation of
cardiac muscle cells. This technology may become a therapeutic
target method for myocardial repair and regeneration (63). Recently, a close association was
demonstrated between miR-92a and lymphoma metastasis in colorectal
cancer, indicating that miR-92a may be a potential marker for
colorectal cancer (64). Overall,
the use of the miRNA-17-92 cluster in clinical practice represents
a promising tool, considering the accumulating evidence on its
specific functions. The study of miRNAs with clinical aims may pave
the way to major advances in cancer treatment in the near
future.
Acknowledgments
The authors are grateful to the staff members of the
Analytical Center for Spectral Measurement and Activity Test Center
for Antibacterial Activity of the Laboratory of Chemistry for
Natural Products of Guizhou Province and the Chinese Academy of
Sciences. The study was supported by the specific project of China
Postdoctoral Science Foundation (grant no. 2015T80106), the Main
Science Project of Guizhou Province [grant no. (2013)6012], the
Postdoctoral Science Foundation of Beijing (grant no. 2015-ZZ-45)
and the Natural Science Foundation of Guizhou Province [grant no.
(2014)2099].
References
|
1
|
Lai EC, Tomancak P, Williams RW and Rubin
GM: Computational identification of Drosophila microRNA genes.
Genome Biol. 4:R422003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Danielson LS, Reavie L, Coussens M,
Davalos V, Castillo-Martin M, Guijarro MV, Coffre M, Cordon-Cardo
C, Aifantis I, Ibrahim S, et al: Limited miR-17-92 overexpression
drives hematologic malignancies. Leuk Res. 39:335–341. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Visone R and Croce CM: miRNAs and cancer.
Am J Pathol. 174:1131–1138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mendes ND, Freitas AT and Sagot MF:
Current tools for the identification of miRNA genes and their
targets. Nucleic Acids Res. 37:2419–2433. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chhabra R, Dubey R and Saini N:
Cooperative and individualistic functions of the microRNAs in the
miR-23a-27a-24-2 cluster and its implication in human diseases. Mol
Cancer. 9:2322010. View Article : Google Scholar
|
|
8
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ, et al: A microRNA polycistron as a potential human
oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun J, Gao B, Zhou M, Wang ZZ, Zhang F,
Deng JE and Li X: Comparative genomic analysis reveals evolutionary
characteristics and patterns of microRNA clusters in vertebrates.
Gene. 512:383–391. 2013. View Article : Google Scholar
|
|
10
|
Ota A, Tagawa H, Karnan S, Tsuzuki S,
Karpas A, Kira S, Yoshida Y and Seto M: Identification and
characterization of a novel gene, C13orf25, as a target for
13q31-q32 amplification in malignant lymphoma. Cancer Res.
64:3087–3095. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mendell JT: miRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ventura A, Young AG, Winslow MM, Lintault
L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone
JR, et al: Targeted deletion reveals essential and overlapping
functions of the miR-17 through 92 family of miRNA clusters. Cell.
132:875–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tong MH, Mitchell DA, McGowan SD, Evanoff
R and Griswold MD: Two miRNA clusters, Mir-17-92 (Mirc1) and
Mir-106b-25 (Mirc3), are involved in the regulation of
spermato-gonial differentiation in mice. Biol Reprod. 86:722012.
View Article : Google Scholar
|
|
14
|
Mogilyansky E and Rigoutsos I: The
miR-17/92 cluster: A comprehensive update on its genomics,
genetics, functions and increasingly important and numerous roles
in health and disease. Cell Death Differ. 20:1603–1614. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hossain A, Kuo MT and Saunders GF:
Mir-17-5p regulates breast cancer cell proliferation by inhibiting
translation of AIB1 mRNA. Mol Cell Biol. 26:8191–8201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gong AY, Eischeid AN, Xiao J, Zhao J, Chen
D, Wang ZY, Young CY and Chen XM: miR-17 5p targets the
300/CBP-associated factor and modulates androgen receptor
transcriptional activity in cultured prostate cancer cells. BMC
Cancer. 12:4922012. View Article : Google Scholar
|
|
17
|
Yang X, Du WW, Li H, Liu F, Khorshidi A,
Rutnam ZJ and Yang BB: Both mature miR-17-5p and passenger strand
miR-17-3p target TIMP3 and induce prostate tumor growth and
invasion. Nucleic Acids Res. 41:9688–9704. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Vecchiarelli-Federico LM, Li YJ,
Egan SE, Spaner D, Hough MR and Ben-David Y: The miR-17-92 cluster
expands multipotent hematopoietic progenitors whereas imbalanced
expression of its individual oncogenic miRNAs promotes leukemia in
mice. Blood. 119:4486–4498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shrivastava S, Petrone J, Steele R, Lauer
GM, Di Bisceglie AM and Ray RB: Upregulation of circulating miR-20a
is correlated with hepatitis C virus-mediated liver disease
progression. Hepatology. 58:863–871. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou J, Jiang J, Wang S and Xia X:
Oncogenic role of microRNA 20a in human uveal melanoma. Mol Med
Rep. 14:1560–1566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhuo W, Ge W, Meng G, Jia S, Zhou X and
Liu J: MicroRNA 20a promotes the proliferation and cell cycle of
human osteosarcoma cells by suppressing early growth response 2
expression. Mol Med Rep. 12:4989–4994. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z, Wang B, Shi Y, Xu C, Xiao HL, Ma
LN, Xu SL, Yang L, Wang QL, Dang WQ, et al: Oncogenic miR-20a and
miR-106a enhance the invasiveness of human glioma stem cells by
directly targeting TIMP-2. Oncogene. 34:1407–1419. 2015. View Article : Google Scholar
|
|
23
|
Xiong Y, Zhang L and Kebebew E: miR-20a is
upregulated in anaplastic thyroid cancer and targets LIMK1. PLoS
One. 9:e961032014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Landais S, Landry S, Legault P and Rassart
E: Oncogenic potential of the miR-106-363 cluster and its
implication in human T-cell leukemia. Cancer Res. 67:5699–5707.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li X, Zhang Z, Yu M, Li L, Du G, Xiao W
and Yang H: Involvement of miR-20a in promoting gastric cancer
progression by targeting early growth response 2 (EGR2). Int J Mol
Sci. 14:16226–16239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qiang XF, Zhang ZW, Liu Q, Sun N, Pan LL,
Shen J, Li T, Yun C, Li H and Shi LH: miR-20a promotes prostate
cancer invasion and migration through targeting ABL2. J Cell
Biochem. 115:1269–1276. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Olive V, Bennett MJ, Walker JC, Ma C,
Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ and He L: miR-19
is a key oncogenic component of mir-17-92. Genes Dev. 23:2839–2849.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsu TI, Hsu CH, Lee KH, Lin JT, Chen CS,
Chang KC, Su CY, Hsiao M and Lu PJ: MicroRNA-18a is elevated in
prostate cancer and promotes tumorigenesis through suppressing STK4
in vitro and in vivo. Oncogenesis. 3:e992014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Niu H, Wang K, Zhang A, Yang S, Song Z,
Wang W, Qian C, Li X, Zhu Y and Wang Y: miR-92a is a critical
regulator of the apoptosis pathway in glioblastoma with inverse
expression of BCL2L11. Oncol Rep. 28:1771–1777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsuchida A, Ohno S, Wu W, Borjigin N,
Fujita K, Aoki T, Ueda S, Takanashi M and Kuroda M: miR-92 is a key
oncogenic component of the miR-17-92 cluster in colon cancer.
Cancer Sci. 102:2264–2271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H
and Hu C: Circulating microRNA-92a and microRNA-21 as novel
minimally invasive biomarkers for primary breast cancer. J Cancer
Res Clin Oncol. 139:223–229. 2013. View Article : Google Scholar :
|
|
32
|
Santoro MM and Nicoli S: miRNAs in
endothelial cell signaling: The endomiRNAs. Exp Cell Res.
319:1324–1330. 2013. View Article : Google Scholar :
|
|
33
|
Doebele C, Bonauer A, Fischer A, Scholz A,
Reiss Y, Urbich C, Hofmann WK, Zeiher AM and Dimmeler S: Members of
the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic
function in endothelial cells. Blood. 115:4944–4950. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Venturini L, Battmer K, Castoldi M,
Schultheis B, Hochhaus A, Muckenthaler MU, Ganser A, Eder M and
Scherr M: Expression of the miR-17-92 polycistron in chronic
myeloid leukemia (CML) CD34+ cells. Blood.
109:4399–4405. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shan SW, Lee DY, Deng Z, Shatseva T,
Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, et al:
MicroRNA miR-17 retards tissue growth and represses fibronectin
expression. Nat Cell Biol. 11:1031–1038. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Choi PS, Casey SC, Dill DL and
Felsher DW: MYC through miR-17-92 suppresses specific target genes
to maintain survival, autonomous proliferation, and a neoplastic
state. Cancer Cell. 26:262–272. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mestdagh P, Boström AK, Impens F, Fredlund
E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquière B,
Schulte S, Dews M, et al: The miR-17-92 microRNA cluster regulates
multiple components of the TGF-β pathway in neuroblastoma. Mol
Cell. 40:762–773. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sokolova V, Fiorino A, Zoni E, Crippa E,
Reid JF, Gariboldi M and Pierotti MA: The effects of miR-20a on
p21: Two mechanisms blocking growth arrest in TGFbeta responsive
colon carcinoma. J Cell Physiol. 230:3105–3114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Woods K, Thomson JM and Hammond SM: Direct
regulation of an oncogenic micro-RNA cluster by E2F transcription
factors. J Biol Chem. 282:2130–2134. 2007. View Article : Google Scholar
|
|
40
|
O'Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: c-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sylvestre Y, De Guire V, Querido E,
Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G and Chartrand P:
An E2F/miR-20a autoregulatory feedback loop. J Biol Chem.
282:2135–2143. 2007. View Article : Google Scholar
|
|
42
|
Pichiorri F, Suh SS, Ladetto M, Kuehl M,
Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, et
al: MicroRNAs regulate critical genes associated with multiple
myeloma pathogenesis. Proc Natl Acad Sci USA. 105:12885–12890.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou P, Ma L, Zhou J, Jiang M, Rao E, Zhao
Y and Guo F: miR-17-92 plays an oncogenic role and conveys
chemo-resistance to cisplatin in human prostate cancer cells. Int J
Oncol. 48:1737–1748. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dou L, Meng X, Sui X, Wang S, Shen T,
Huang X, Guo J, Fang W, Man Y, Xi J, et al: miR-19a regulates PTEN
expression to mediate glycogen synthesis in hepatocytes. Sci Rep.
5:116022015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Takahashi RU, Miyazaki H and Ochiya T: The
role of microRNAs in the regulation of cancer stem cells. Front
Genet. 4:2952014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hwang WL, Jiang JK, Yang SH, Huang TS, Lan
HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW, et al:
MicroRNA-146a directs the symmetric division of Snail-dominant
colorectal cancer stem cells. Nat Cell Biol. 16:268–280. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zagorac S, Alcala S, Fernandez Bayon G,
Bou Kheir T, Schoenhals M, González-Neira A, Fernandez Fraga M,
Aicher A, Heeschen C and Sainz B Jr: DNMT1 Inhibition reprograms
pancreatic cancer stem cells via upregulation of the miR-17-92
cluster. Cancer Res. 76:4546–4558. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cioffi M, Trabulo SM, Sanchez-Ripoll Y,
Miranda-Lorenzo I, Lonardo E, Dorado J, Reis Vieira C, Ramirez JC,
Hidalgo M, Aicher A, et al: The miR-17-92 cluster counteracts
quiescence and chemoresistance in a distinct subpopulation of
pancreatic cancer stem cells. Gut. 64:1936–1948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
He H and Magi-Galluzzi C:
Epithelial-to-mesenchymal transition in renal neoplasms. Adv Anat
Pathol. 21:174–180. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Burk U, Schubert J, Wellner U, Schmalhofer
O, Vincan E, Spaderna S and Brabletz T: A reciprocal repression
between ZEBl and member of the miR-200 family promotes EMT and
invasion in cancer cells. EMBO Rep. 9:521–522. 2008. View Article : Google Scholar
|
|
52
|
Li J, Yang S, Yan W, Yang J, Qin YJ, Lin
XL, Xie RY, Wang SC, Jin W, Gao F, et al: MicroRNA-19 triggers
epithelial-mesenchymal transition of lung cancer cells accompanied
by growth inhibition. Lab Invest. 95:1056–1070. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xi XP, Zhuang J, Teng MJ, Xia LJ, Yang MY,
Liu QG and Chen JB: MicroRNA-17 induces epithelial-mesenchymal
transition consistent with the cancer stem cell phenotype by
regulating CYP7B1 expression in colon cancer. Int J Mol Med.
38:499–506. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ottman R, Levy J, Grizzle WE and
Chakrabarti R: The other face of miR-17-92a cluster, exhibiting
tumor suppressor effects in prostate cancer. Oncotarget.
7:73739–73753. 2016.PubMed/NCBI
|
|
55
|
Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P,
Nie Y, Wu K, Shi Y and Fan D: miR-19a/b modulate the metastasis of
gastric cancer cells by targeting the tumour suppressor MXD1. Cell
Death Dis. 5:e11442014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z,
Sun J, Tan FW, Ding DP, Xu XH, Zhou F, et al: microRNA-92a promotes
lymph node metastasis of human esophageal squamous cell carcinoma
via E-cadherin. J Biol Chem. 286:10725–10734. 2011. View Article : Google Scholar :
|
|
57
|
Landskroner-Eiger S, Qiu C, Perrotta P,
Siragusa M, Lee MY, Ulrich V, Luciano AK, Zhuang ZW, Corti F,
Simons M, et al: Endothelial miR-17-92 cluster negatively regulates
arteriogenesis via miRNA-19 repression of WNT signaling. Proc Natl
Acad Sci USA. 112:12812–12817. 2015. View Article : Google Scholar
|
|
58
|
Ohyagi-Hara C, Sawada K, Kamiura S, Tomita
Y, Isobe A, Hashimoto K, Kinose Y, Mabuchi S, Hisamatsu T,
Takahashi T, et al: miR-92a inhibits peritoneal dissemination of
ovarian cancer cells by inhibiting integrin α5 expression. Am J
Pathol. 182:1876–1889. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhao S, Yao D, Chen J, Ding N and Ren F:
miR-20a promotes cervical cancer proliferation and metastasis in
vitro and in vivo. PLoS One. 10:e01209052015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang M, Liu Q, Mi S, Liang X, Zhang Z, Su
X, Liu J, Chen Y, Wang M, Zhang Y, et al: Both miR-17-5p and
miR-20a alleviate suppressive potential of myeloid-derived
suppressor cells by modulating STAT3 expression. J Immunol.
186:4716–4724. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Dereani S, Macor P, D'Agaro T, Mezzaroba
N, Dal-Bo M, Capolla S, Zucchetto A, Tissino E, Del Poeta G, Zorzet
S, et al: Potential therapeutic role of antagomiR17 for the
treatment of chronic lymphocytic leukemia. J Hematol Oncol.
7:792014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Murphy BL, Obad S, Bihannic L, Ayrault O,
Zindy F, Kauppinen S and Roussel MF: Silencing of the miR-17-92
cluster family inhibits medulloblastoma progression. Cancer Res.
73:7068–7078. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen J, Huang ZP, Seok HY, Ding J, Kataoka
M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, et al: mir-17-92 cluster
is required for and sufficient to induce cardiomyocyte
proliferation in postnatal and adult hearts. Circ Res.
112:1557–1566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou T, Zhang G, Liu Z, Xia S and Tian H:
Overexpression of miR-92a correlates with tumor metastasis and poor
prognosis in patients with colorectal cancer. Int J Colorectal Dis.
28:19–24. 2013. View Article : Google Scholar
|