Introduction
The cofilin 2 (CFL2) gene plays a key role in
muscle development in mammals. CFL plays a crucial role in the
regulation of actin by enhancing the turnover of actin filaments
(1,2). Mammalian CFL occurs as
non-muscle type (CFL1) or muscle-type (CFL2). In
embryos, CFL1 is predominantly expressed in non-myoblast
cells. The CFL2 protein appears in skeletal and cardiac myoblasts,
particularly in the I bands and Z lines of skeletal myoblasts.
During muscle development, CFL1 expression decreases, while
CFL2 expression increases and eventually becomes predominant
in mature mammalian skeletal myoblasts (3). The CFL2 transcript is spliced using
either exon1a or exon1b to form the CFL2a and CFL2b
transcripts, respectively. The CFL2a transcript is found in
various tissues, while the CFL2b transcript is mainly found
in mature skeletal muscle. CFL binds and polymerizes filamentous
F-actin and inhibits the polymerization of monomeric G-actin in a
pH-dependent manner, in part through interactions with tropomyosins
(4,5).
Four isoforms of myosin heavy chain (MyHC)
are expressed in skeletal muscle throughout the period of
development, and are determinants of muscle development (6): I, slow oxidative; IIa, fast
oxidative; IIb, fast glycolytic; and IIx/d, mixed (7). It was previously suggested that
MyHC composition affects meat quality in livestock (8). Meat characteristics, in particular
edibility, may be strongly affected by muscle fiber types, which
depend on MyHC mRNA expression (8). Indeed, the expression of
MyHC-I is associated with the pH value of the meat 24 h
after death (pH24h) and drip loss (a characteristic of
meat that represents its water-retaining capacity), while
MyHC-II expression is correlated with juiciness, off-flavor,
and tenderness of the meat (8).
Our previous study demonstrated that CFL2b overexpression
regulated the fast muscle fiber trait by increasing the expression
of MyHC-IIb and -IIx/d (9). However, the effects on MyHC
when CFL2 is disrupted have not been determined.
MyHC and CFL expression are associated
with four cellular signaling pathways, including AMP-activated
protein kinase (AMPK) (10,11), calmodulin (CaM), Rho, and
mitogen-activated protein kinase (MAPK) (12–14). Five important proteins in these
pathways, including extracellular signal-regulated kinase 2 (ERK2)
(15), p38 MAPK (16), myocyte enhancer factor 2C (MEF2C)
(17,18), cAMP-response element-binding
protein (CBP) (16) and AMPKα1
(19), may affect the expression
of MyHC after inhibiting CFL2 expression.
To identify the involvement of CFL2 in
skeletal muscle and the period during which CFL2 exerts its
effects, the present study investigated the effects of CFL2
on actin fibers by studying the CFL2 and MyHC genes
in undifferentiated and differentiated C2C12 cells transfected with
the CFL2 siRNA. In addition, the protein expression of
pathway-related genes, including ERK2, p38 MAPK, MEF2C, CBP and
AMPKα1, was determined.
Materials and methods
Cell culture and transfection
Mouse C2C12 myoblasts (GNM26) obtained from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China) were
grown in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% (v/v) fetal calf serum (both from Invitrogen; Thermo
Fisher Scientific, Carlsbad, CA, USA) at 37°C with 5%
CO2 in humidified chambers. Differentiation of C2C12
cells was induced by replacing the medium with 2% horse serum for 6
days.
Cells in 6-well plates were transfected with
CFL2 small interfering RNA (siRNA) using the liposome method
(FuGene HD transfection reagent; Roche, Shanghai, China) when they
reached 50% confluence, according to the manufacturer's
instructions (9). The RNA
interfering effects of siRNAs were confirmed by western blotting
and reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). The undifferentiated cells were then screened with 500
µg/ml of G418 (Roche) to obtain single cell clones.
siRNA for CFL2 gene
Two siRNA pairs (CFL2-1 and CFL2-2;
Table I) were selected for
targeting the murine CFL2 gene (NM_007688) using a web-based
siRNA design application (http://sirna.wi.mit.edu/home.php) (20). The selected siRNA pairs were then
synthesized by Genechem (Shanghai, China). Undifferentiated cells
were transfected with siRNA concentrations of 50, 100 and 150
nmol/l of CFL2-1, and 50 and 100 nmol/l of CFL2-2.
Cells transfected with scramble siRNA were used as controls. The
effects of CFL2 siRNA were identified by RT-PCR and western
blotting.
 | Table IsiRNA sequences for the target sites
in the CFL2 gene. |
Table I
siRNA sequences for the target sites
in the CFL2 gene.
| siRNA | Primer sequences
(5′-3′) | Target site |
|---|
| CFL2-1 | F:
CUGAAAGUGCACCGUUAAAdTdT | 315–337 |
| R:
UUUAACGGUGCACUUUCAGdTdT | |
| CFL2-2 | F:
GCUCUAAAGAUGCCAUUAAdTdT | 354–376 |
| R:
UUAAUGGCAUCUUUAGAGCdTdT | |
| Scramble |
CTTGAAGGAAAGCCACTAT | – |
Experimental groups
Cells were divided into the following groups:
Undifferentiated, C2C12 cells transfected with scramble siRNA
(UDN); undifferentiated, C2C12 cells transiently transfected with
CFL2 siRNA (UDT); differentiated, C2C12 cells transfected
with scramble siRNA (DN); and differentiated, C2C12 cells
transiently transfected with CFL2 siRNA (DT).
RT-PCR
RNA was extracted from C2C12 cells using an RNeasy
mini kit (Qiagen, Hilden, Germany), and first-strand cDNA was
synthesized from purified total RNA with a reverse transcription
kit (Takara, Dalian, China). RT-PCR was performed according to
standard protocols using the primers indicated in Table II. The final volumes of each
reaction were 20 µl, including 10 pmol primers, 2 mM
MgCl2, 2.5 mM dNTP, and 1 U TaqDNA polymerase (Takara).
The samples underwent PCR as follows: Denaturation for 5 min at
94°C; 30 cycles of 30 sec at 94°C, 30 sec at 56.7°C, and 45 sec at
72°C; and extension for 10 min at 72°C. Following amplification, 10
µl PCR product was analyzed using 1% agarose gel
electrophoresis and visualized using ethidium bromide and UV
light.
 | Table IIRT-PCR primers for the target
genes. |
Table II
RT-PCR primers for the target
genes.
| Target genes | Primer sequences
(5′-3′) | PCR product
size |
|---|
| CFL2 | F:
ATCTTGGTGGGTGACATTGG | 322 bp |
| R:
CAAGGGAAACTACAACACTGC | |
|
GAPDHa | F:
GCAGTGGCAAAGTGGAGATT | 790 bp |
| R:
TGAAGTCGCAGGAGACAACC | |
|
GAPDHb | F:
GCGAGACCCCACTAACATC | 171 bp |
| R:
TTCACACCCATCACAAACA | |
RT-qPCR
RT-qPCR was performed to detect mRNA expression of
MyHC-I, MyHC-IIa, MyHC-IIb, MyHC-IIx
and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in UDN,
UDT, DN and DT. The cDNA template was prepared from mRNA directly
harvested from CFL2-transfected cells at 1×106
cells/well. First-strand reverse transcription was performed using
100 ng total RNA with a reverse transcription kit (Takara); the
RT-qPCR primers were also from Takara (Table III). SYBR-Green RT-qPCR assay
for the target genes was performed in optical 96-well plates using
the SYBR® Premix EX Taq™ II (DRR081A; Takara) and the
7500 Fast Real-Time PCR system (Applied Biosystems, Foster City,
MA, USA), according to the manufacturer's instructions. Results
were detected quantitatively in real-time by relative quantitation
of two standard curves. The process consisted of 41 cycles,
including 1 cycle of 10 sec at 95°C and 40 cycles of 5 sec at 95°C
and 20 sec at 60°C. All experiments were performed three times,
each time in triplicate.
 | Table IIIOligonucleotide primers for
RT-qPCR. |
Table III
Oligonucleotide primers for
RT-qPCR.
| Genes | Primer sequences
(5′-3′) | PCR product
size |
|---|
| MyHC-I | F:
ATGAGCTGGAGGCTGAGCA | 124 bp |
| R:
TGCAGCCGCAGTAGGTTCTT | |
|
MyHC-IIa | F:
ATTCTCAGGCTTCAGGATTTGGTG | 114 bp |
| R:
CTTGCGGAACTTGGATAGATTTGTG | |
|
MyHC-IIb | F:
GAGTTCATTGACTTCGGGATGG | 143 bp |
| R:
TGCTGCTCATACAGCTTGTTCTTG | |
|
MyHC-IIx | F:
AAGGGTCTGCGCAAACATGA | 173 bp |
| R:
TTGGCCAGGTTGACATTGGA | |
| GAPDH | F:
GCGAGACCCCACTAACATC | 171 bp |
| R:
TTCACACCCATCACAAACA | |
Western blotting
Western blotting for CFL2, p38, ERK2, CBP, AMPKα1,
MEF2C and GAPDH was performed in UDN and UDT. The cells were
washed, harvested, lysed with a lysis buffer [20 mM Tris-Cl, pH
7.5, 150 mM NaCl, 1% sodium dodecyl sulfate (SDS), 1% Triton X-100,
10 µg/ml leupeptin, 1 mM aprotinin and 1 mM
phenylmethanesulfonyl fluoride] on ice for 30 min, and centrifuged
at 14,000 × g at 4°C for 15 min. Proteins were resolved by
SDS-polyacrylamide gel electrophoresis, transferred onto
polyvinylidene difluoride membranes, blocked at room temperature
for 1 h in 5% bovine serum albumin (BSA), and then incubated
overnight at 4°C on a rotator with the primary antibodies: CFL2
(1:1,000; goat, SAB2500255; Sigma-Aldrich; Merck KGaA, St. Louis,
MO, USA), p38 (1:1,000; mouse, ab31828), ERK2 (1:1,000; rabbit,
ab32081), CBP (1:1,000; mouse, ab50702), AMPKα1 (1:1,000; mouse,
ab80039) (all from Abcam, Cambridge, MA, USA), MEF2C (1:1,000;
rabbit, SAB2103534; Sigma-Aldrich; Merck KGaA) and GAPDH (1:2,000;
mouse, ab8245; Abcam). The membranes were washed four times with
TBST and incubated for 1 h with an appropriate horseradish
peroxidase (HRP)-conjugated secondary antibody (1:3,000).
Immunoreactivity was visualized with an enhanced chemiluminescence
detection reagent (ECL; Pierce, Rockford, IL, USA). All the
experiments were performed three times, each time in
triplicate.
CFL2 and F-actin analysis by fluorescence
microscopy
CFL2 siRNA was transfected into cells.
CFL2 and F-actin were labeled with fluorescein
isothiocyanate (FITC) and tetramethylrhodamine (TRITC)-labeled
phalloidin (Sigma-Aldrich; Merck KGaA) to differentiate between UDN
and UDT. In terms of TRITC-labeled phalloidin methodology, UDN and
UDT were grown on glass coverslips until they reached the
logarithmic growth stage. Specimens were then washed three times in
phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde
at room temperature for 30 min. After washing three times with PBS,
the specimens were permeabilized with 0.2% Triton X-100 for 10 min,
washed three times with PBS, and blocked with 1% BSA for 30 min.
The coverslips were incubated for 10 min with diluted phalloidin
(1:200, protected from light) and washed three times in PBS. The
coverslips were mounted and observed with 90% glycerol in PBS.
Notably, the FITC methodology is equivalent to the TRITC-labeled
phalloidin approach, but additionally requires an overnight
antibody incubation step prior to hybridization with diluted FITC
(1:100, protected from light). UDN were used as controls. The cells
were examined using a Zeiss epifluorescence microscope (Carl Zeiss
AG, Oberkochen, Germany), as described in Table IV.
 | Table IVFluorescent staining of CFL2
and F-actin in C2C12 cells. |
Table IV
Fluorescent staining of CFL2
and F-actin in C2C12 cells.
| CFL2 expression
levels | Fluorescent
staining method
|
|---|
| CFL2 | F-actin |
|---|
| Low | IgG antibody
labeling with FITC | TRITC-labeled
phalloidin |
| Normal | IgG antibody
labeling with FITC | TRITC-labeled
phalloidin |
Statistical analyses
The results were analyzed using SPSS 17.0 software
(IBM, Armonk, NY, USA). Data are presented as means ± standard
deviation. Experimental and control groups were compared using
analysis of variance followed by Least Significant Difference post
hoc analysis. P-values <0.05 were considered to indicate
statistically significant differences.
Results
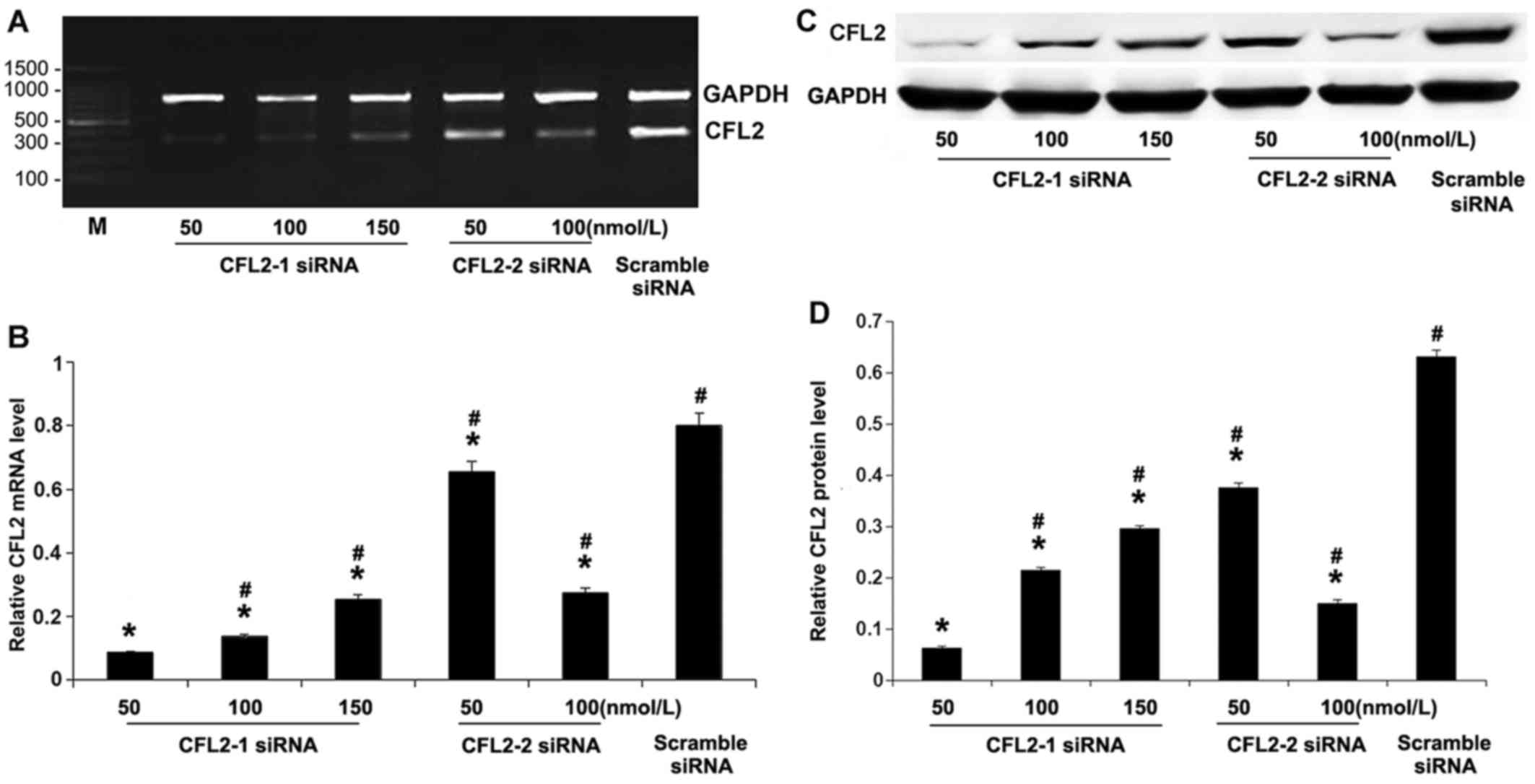
Knockdown of CFL2 by CFL2 siRNA
We first validated the RNA interference (RNAi)
effects of CFL2 siRNAs. RT-PCR amplification of CFL2
and GAPDH was analyzed (Fig.
1A). The results revealed that CFL2-1 siRNA at 50 nM exerted
the strongest inhibitory effect of all five experimental groups
(P<0.05; Fig. 1B), whereas, as
expected, the scramble siRNA exerted no effect on CFL2
expression. The results of western blotting were consistent with
the RT-PCR results (Fig. 1C and
D). Therefore, the most effective siRNA sequence for
CFL2 was CFL2-1 siRNA at a concentration of 50
nM.
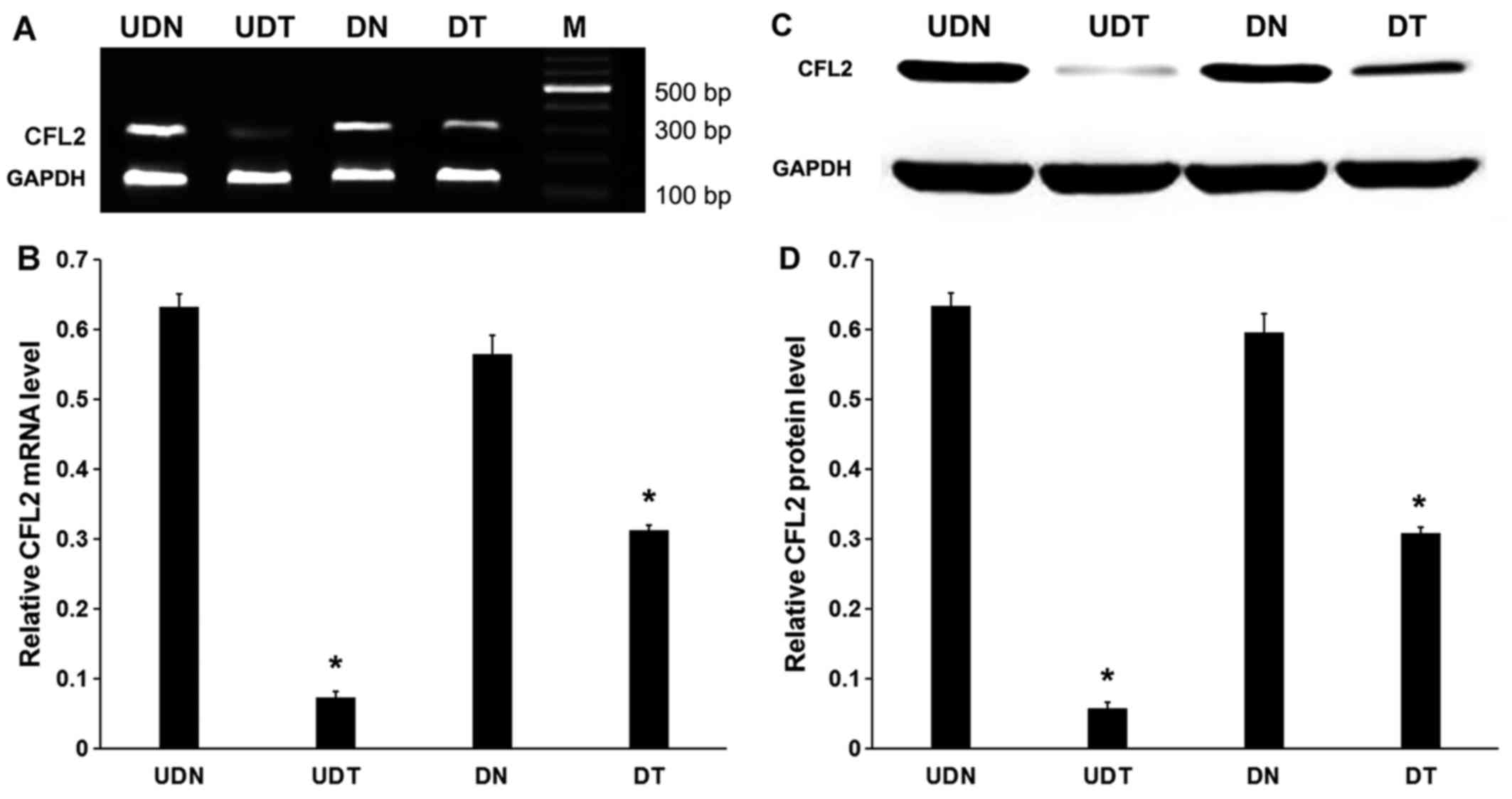
MyHC expression is downregulated in
undifferentiated C2C12 cells treated with CFL2 RNAi
RNAi significantly inhibited CFL2 in
undifferentiated cells (P<0.05) and in DT compared with DN
(P<0.05; Fig. 2A and B).
Therefore, regardless of cellular differentiation, RNAi
significantly inhibited CFL2 mRNA expression in C2C12 cells.
Western blot analyses were also consistent with the RT-PCR results
(P<0.05; Fig. 2C and D).
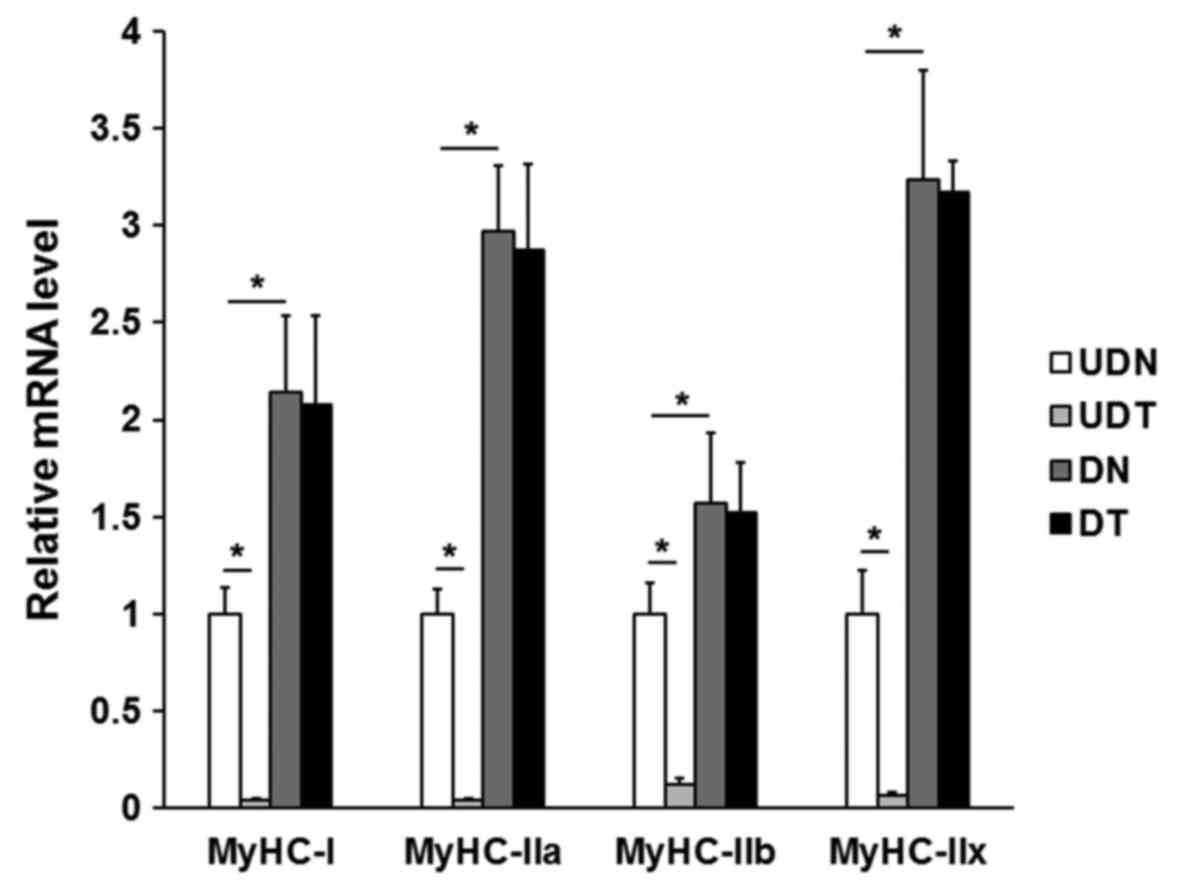
RT-qPCR amplification was used to determine the
expression of MyHC genes in differentiated and
undifferentiated C2C12 cells following CFL2 siRNA
transfection. The MyHC/GAPDH expression in
undifferentiated and differentiated cells is shown in Fig. 3. MyHC-I, MyHC-IIa,
MyHC-IIb and MyHC-IIx gene expressions were
significantly decreased in UDT (P<0.05). However, in DT, RNAi
had almost no effect on MyHC expression, suggesting that
MyHC expression is generally proportional to that of
CFL2 prior to differentiation of cells into muscle fibers.
These results suggest that MyHC mRNA expression was elevated
after differentiation (P<0.05), and that MyHC genes were
not significantly affected by downregulation of CFL2 only in
undifferentiated cells. With this in mind, subsequent experiments
were only performed in the UDT and UDN groups.
Muscle fiber signaling pathway-related
factors are altered by CFL2 RNAi in undifferentiated C2C12
cells
The expression of key signaling molecules in the
AMPK, CaM, Rho and MAPK pathways (including p38 MAPK, ERK2, CBP,
AMPKα1 and MEF2C) was measured by western blotting. The expressions
of p38 MAPK, CBP, AMPKα1 and MEF2C proteins were significantly
decreased in UDT (P<0.05). By contrast, ERK2 protein expression
was significantly increased compared with UDN (P<0.05; Fig. 4).
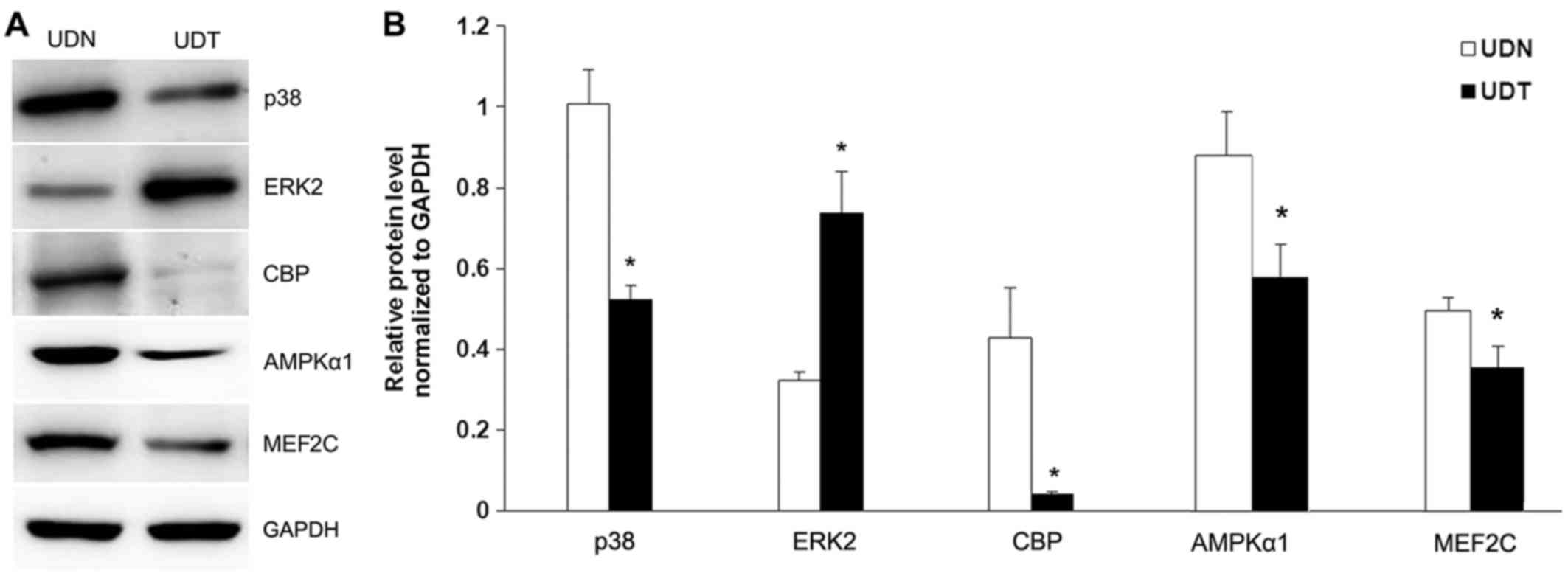 | Figure 4p38, ERK2, CBP, AMPKα1 and MEF2C
protein expression in UDN and UDT. (A) Representative image of
western blotting of p38, ERK2, CBP, AMPKα1 and MEF2C protein
expression in UDN and UDT. (B) Statistical data. Glyceraldehyde
3-phosphate dehydrogenase (GAPDH) was used as an internal control.
All experiments were performed three times, each time in
triplicate. *P<0.05 vs. UDN. UDN, undifferentiated
C2C12 cells transfected with scramble siRNA; UDT, undifferentiated
C2C12 cells transiently transfected with CFL2 siRNA; ERK2,
extracellular signal-regulated kinase 2; CBP, cAMP-response
element-binding protein; AMPK, AMP-activated protein kinase; MEF2C,
myocyte enhancer factor 2C. |
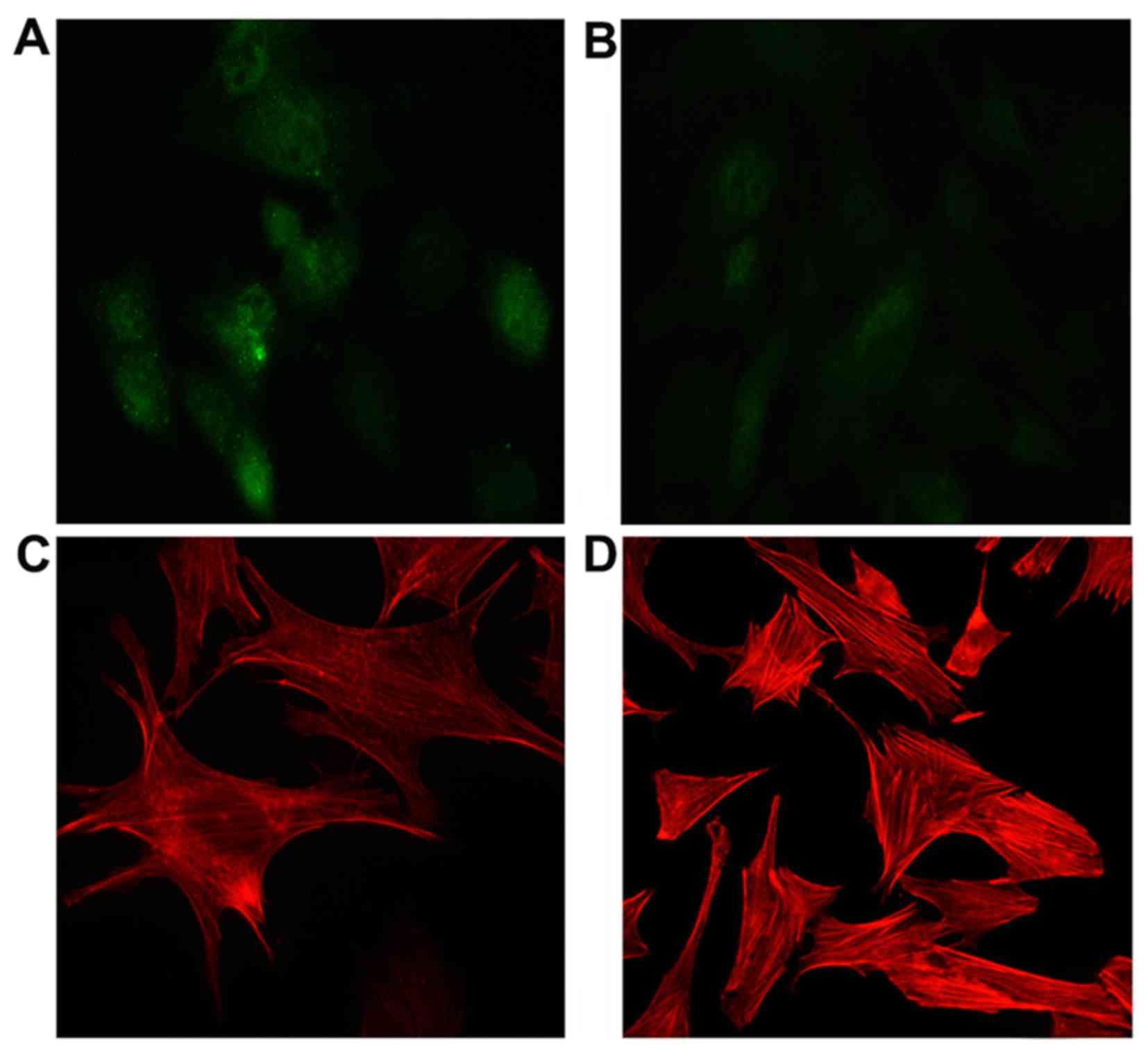
CFL2 alters F-actin formulation in C2C12
cells
In UDN, CFL2 (green fluorescence) was
diffusely distributed in the cytoplasm and nucleus of the cells
(Fig. 5A). UDT exhibited a
similar diffuse CFL2 distribution pattern (Fig. 5B).
F-actin structures were not significantly different
in UDT compared to UDN with CFL2 expression. The F-actin
bundles, however, exhibited a tendency to form amorphous diffuse
patterns (Fig. 5C and D).
Discussion
The aim of the present study was to demonstrate the
effects of the mouse myosin-binding protein CFL2 on the expression
of MyHC, which may play a pivotal role in the cytoskeleton
in undifferentiated C2C12 cells. The functions of CFL2 in
undifferentiated and differentiated C2C12 cells were investigated
and the results suggested that, in undifferentiated cells, the
expression of MyHC genes decreased significantly after
CFL2 RNAi; however, there was no obvious change in
differentiated cells.
Effects of CFL2 on muscle fiber
types
Four MyHC isoforms (I, IIa, IIx and IIb) are
expressed in adult skeletal muscles (6). MyHCs are the primary
molecular markers for distinguishing muscle fiber types and
studying their characteristics. When CFL2 gene expression in
C2C12 myoblasts is suppressed, the expression of MyHC-I,
MyHC-IIa, MyHC-IIb and MyHC-IIx is
significantly downregulated.
In addition, 6 days after C2C12 differentiation was
induced using 2% horse serum, the CFL2 mRNA and protein
expressions in DN were not significantly different from those in
undifferentiated cells. The expression of MyHC-I,
MyHC-IIa, MyHC-IIb and MyHC-IIx increased,
indicating that MyHC is associated with myoblast
differentiation. However, RNAi no longer affected the types of
MyHC. This may indicate that interactions between
CFL2 and MyHCs only occur in undifferentiated
cells.
Molecular mechanisms of MyHC in C2C12
cells and CFL2 involvement
Within the four key cellular signaling pathways
involved in muscle fiber regulation, there are five important
signaling factors: p38 MAPK, ERK2, CBP, MEF2C and AMPKα1. The fast
muscle fibers (MyHC-IIb/IIx) are preferentially affected by
MAPK signaling. The dual-specific mitogen-activated protein kinase
kinase 3 (MKK3) and the constitutively active MKK6 mutant upstream
of p38 MAPK are involved in the maintenance of the fast muscle
fiber phenotype (21); they may
also be involved in the regulation of MyHC-IIx promoter
activity in myotubes (12,22).
The ERK pathway is also a major pathway that affects fast fibers,
as demonstrated by increased MyHC-IIx and MyHC-IIb
transcripts and decreased MyHC-I expression following
ablation of the ERK1/2 pathway in cultured rat fetal myocytes
(23). This mechanism is also
consistent with the downregulation of CFL2 genes as a means
to decrease p38 MAPK, MyHC-IIb and MyHC-IIx
expression and to increase ERK2 expression.
It was previously reported that ERK1/2 activity is
more than 2-fold higher in fast muscle fibers compared with that in
slow muscle fibers (15),
suggesting that the ERK1/2 pathway may play a key role in
maintaining the fast fiber phenotype. When p38 MAPK activity is
inhibited, the promoter activities of fast-type MyHC-IIb and
MyHC-IIx are downregulated. Moreover, p38α/β MAPK is known
to mediate MyHC-IIb and MyHC-IIx expression via CBP
and the MEF2C/D heterodimer (16). When combined with other
transcriptional factors (17),
MEF2C may affect MyHC isoform expression in mouse
skeletal muscle (18). These
prior findings are highly consistent with the results of the
present study, in which MEF2C and CBP expression was
decreased by CFL2 RNAi. Furthermore, a decrease in
MEF2C expression has been associated with a decrease in
MyHC-I muscle fibers (24). Expression of the MEF2C
subunits may also be involved in activation of the p38 MAPK
pathway. Thus, MEF2C and p38 synergistically regulate and
promote type I muscle fibers.
Previously, the authors observed that skeletal
muscle in wild-type mice commonly undergoes an AMPK-mediated shift
from MyHC-IIb to MyHC-IIa and MyHC-IIx during
exercise training (10). These
prior results indicated that AMPKα1 and AMPKα2 may
also alter skeletal muscle composition. This is consistent with the
present results, which demonstrated down-regulation of
AMPKα1, MyHC-IIa, MyHC-IIb and MyHC-IIx
following CFL2 RNAi.
Notably, MyHC gene expression was altered in
UDT due to RNAi, resulting in downregulation of MyHC-I,
MyHC-IIa, MyHC-IIb and MyHC-IIx expression.
Glycolytic fibers (MyHC-IIb) are the most common among these
four muscle fiber types. According to the MyHC conversion
rules (25–27), MyHC-I primarily transforms
into MyHC-IIa. Additional studies are required to confirm
the findings of the present study, in part owing to the complexity
of MyHC conversion due to overlapping signaling pathways.
Additionally, the MyHC isoforms that play a key role in the
regulation of fast fibers may play a less important role in the
regulation of slow fibers under certain physiological conditions.
Although MyHC-I expression was also found to be decreased in
the present study, other isoforms may be able to convert
MyHCs. Future studies are required to elucidate these
isoforms and their roles in the mechanism of MyHC
conversion.
In conclusion, the findings of the present study
indicate that mouse CFL2 may regulate MyHC. In
addition, the expression of four MyHC isoforms in
undifferentiated cells with CFL2 RNAi was found to be
significantly decreased compared with that in differentiated cells.
However, further investigations on the exact association of
CFL2 with signaling pathway-related factors are required to
fully elucidate the role of CFL2 in the regulation of
MyHC.
Acknowledgments
The present study was supported by the National
Natural Science Foundations of China (grant no. 31272415/C170102),
the Natural Science Foundations of Liaoning Province (grant no.
2015020765) and the Training Programs of Innovation and
Entrepreneurship for Undergraduates of Liaoning Province (grant no.
201610160051).
References
|
1
|
Chhabra D and dos Remedios CG: Cofilin,
actin and their complex observed in vivo using fluorescence
resonance energy transfer. Biophys J. 89:1902–1908. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Papalouka V, Arvanitis DA, Vafiadaki E,
Mavroidis M, Papadodima SA, Spiliopoulou CA, Kremastinos DT,
Kranias EG and Sanoudou D: Muscle LIM protein interacts with
cofilin 2 and regulates F-actin dynamics in cardiac and skeletal
muscle. Mol Cell Biol. 29:6046–6058. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mohri K, Takano-Ohmuro H, Nakashima H,
Hayakawa K, Endo T, Hanaoka K and Obinata T: Expression of cofilin
isoforms during development of mouse striated muscles. J Muscle Res
Cell Motil. 21:49–57. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gillett GT, Fox MF, Rowe PS, Casimir CM
and Povey S: Mapping of human non-muscle type cofilin (CFL1) to
chromosome 11q13 and muscle-type cofilin (CFL2) to chromosome 14.
Ann Hum Genet. 60:201–211. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ono S and Ono K: Tropomyosin inhibits
ADF/cofilin-dependent actin filament dynamics. J Cell Biol.
156:1065–1076. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asaduzzaman M, Kinoshita S, Siddique BS,
Asakawa S and Watabe S: Multiple cis-elements in the 5′-flanking
region of embryonic/larval fast-type of the myosin heavy chain gene
of torafugu, MYH(M743-2), function in the transcriptional
regulation of its expression. Gene. 489:41–54. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Solomon MB, West RL and Carpenter JW:
Fiber types in the longissimus muscle from water buffalo and
selected domestic beef breeds. Meat Sci. 13:129–135. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang YK, Choi YM, Lee SH, Choe JH, Hong KC
and Kim BC: Effects of myosin heavy chain isoforms on meat quality,
fatty acid composition, and sensory evaluation in Berkshire pigs.
Meat Sci. 89:384–389. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao W, Su YH, Su RJ, Ba CF, Zeng RX and
Song HJ: The full length cloning of a novel porcine gene CFL2b and
its influence on the MyHC expression. Mol Biol Rep. 36:2191–2199.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Röckl KS, Hirshman MF, Brandauer J, Fujii
N, Witters LA and Goodyear LJ: Skeletal muscle adaptation to
exercise training: AMP-activated protein kinase mediates muscle
fiber type shift. Diabetes. 56:2062–2069. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miranda L, Carpentier S, Platek A, Hussain
N, Gueuning MA, Vertommen D, Ozkan Y, Sid B, Hue L, Courtoy PJ, et
al: AMP-activated protein kinase induces actin cytoskeleton
reorganization in epithelial cells. Biochem Biophys Res Commun.
396:656–661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Long YC, Widegren U and Zierath JR:
Exercise-induced mitogen-activated protein kinase signalling in
skeletal muscle. Proc Nutr Soc. 63:227–232. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Madak-Erdogan Z, Ventrella R, Petry L and
Katzenellenbogen BS: Novel roles for ERK5 and cofilin as critical
mediators linking ERα-driven transcription, actin reorganization,
and invasiveness in breast cancer. Mol Cancer Res. 12:714–727.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Won KJ, Park SH, Park T, Lee CK, Lee HM,
Choi WS, Kim SJ, Park PJ, Jang HK, Kim SH, et al: Cofilin
phosphorylation mediates proliferation in response to
platelet-derived growth factor-BB in rat aortic smooth muscle
cells. J Pharmacol Sci. 108:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang SH, Sharrocks AD and Whitmarsh AJ:
MAP kinase signalling cascades and transcriptional regulation.
Gene. 513:1–13. 2013. View Article : Google Scholar
|
|
16
|
Meissner JD, Chang KC, Kubis HP, Nebreda
AR, Gros G and Scheibe RJ: The p38alpha/beta mitogen-activated
protein kinases mediate recruitment of CREB-binding protein to
preserve fast myosin heavy chain IId/x gene activity in myotubes. J
Biol Chem. 282:7265–7275. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Al Madhoun AS, Mehta V, Li G, Figeys D,
Wiper-Bergeron N and Skerjanc IS: Skeletal myosin light chain
kinase regulates skeletal myogenesis by phosphorylation of MEF2C.
EMBO J. 30:2477–2489. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu J, McKinsey TA, Nicol RL and Olson EN:
Signal-dependent activation of the MEF2 transcription factor by
dissociation from histone deacetylases. Proc Natl Acad Sci USA.
97:4070–4075. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wright DC, Hucker KA, Holloszy JO and Han
DH: Ca2+ and AMPK both mediate stimulation of glucose
transport by muscle contractions. Diabetes. 53:330–335. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reynolds A, Leake D, Boese Q, Scaringe S,
Marshall WS and Khvorova A: Rational siRNA design for RNA
interference. Nat Biotechnol. 22:326–330. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Delling U, Tureckova J, Lim HW, De Windt
LJ, Rotwein P and Molkentin JD: A calcineurin-NFATc3-dependent
pathway regulates skeletal muscle differentiation and slow myosin
heavy-chain expression. Mol Cell Biol. 20:6600–6611. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kramer HF and Goodyear LJ: Exercise, MAPK,
and NF-kappaB signaling in skeletal muscle. J Appl Physiol (1985).
103:388–395. 2007. View Article : Google Scholar
|
|
23
|
Shi H, Scheffler JM, Pleitner JM, Zeng C,
Park S, Hannon KM, Grant AL and Gerrard DE: Modulation of skeletal
muscle fiber type by mitogen-activated protein kinase signaling.
FASEB J. 22:2990–3000. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bassel-Duby R and Olson EN: Signaling
pathways in skeletal muscle remodeling. Annu Rev Biochem. 75:19–37.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weiss A, McDonough D, Wertman B,
Acakpo-Satchivi L, Montgomery K, Kucherlapati R, Leinwand L and
Krauter K: Organization of human and mouse skeletal myosin heavy
chain gene clusters is highly conserved. Proc Natl Acad Sci USA.
96:2958–2963. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Whitmer JD, Koslovsky JS, Bähler M and
Mercer JA: Chromosomal location of three unconventional myosin
heavy chain genes in the mouse. Genomics. 38:235–237. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Knotts S, Rindt H, Neumann J and Robbins
J: In vivo regulation of the mouse beta myosin heavy chain gene. J
Biol Chem. 269:31275–31282. 1994.PubMed/NCBI
|