|
1
|
Chi ZL, Akahori M, Obazawa M, Minami M,
Noda T, Nakaya N, Tomarev S, Kawase K, Yamamoto T, Noda S, et al:
Overexpression of optineurin E50K disrupts Rab8 interaction and
leads to a progressive retinal degeneration in mice. Hum Mol Genet.
19:2606–2615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rezaie T, Child A, Hitchings R, Brice G,
Miller L, Coca-Prados M, Héon E, Krupin T, Ritch R, Kreutzer D, et
al: Adult-onset primary open-angle glaucoma caused by mutations in
optineurin. Science. 295:1077–1079. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allingham RR, Liu Y and Rhee DJ: The
genetics of primary open-angle glaucoma: a review. Exp Eye Res.
88:837–844. 2009. View Article : Google Scholar
|
|
4
|
Gleason CE, Ordureau A, Gourlay R, Arthur
JSC and Cohen P: Polyubiquitin binding to optineurin is required
for optimal activation of TANK-binding kinase 1 and production of
interferon β. J Biol Chem. 286:35663–35674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hattula K and Peränen J: FIP-2, a
coiled-coil protein, links huntingtin to Rab8 and modulates
cellular morphogenesis. Curr Biol. 10:1603–1606. 2000. View Article : Google Scholar
|
|
6
|
del Toro D, Alberch J, Lázaro-Diéguez F,
Martín-Ibáñez R, Xifró X, Egea G and Canals JM: Mutant huntingtin
impairs post-Golgi trafficking to lysosomes by delocalizing
optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell.
20:1478–1492. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagabhushana A, Chalasani ML, Jain N,
Radha V, Rangaraj N, Balasubramanian D and Swarup G: Regulation of
endocytic trafficking of transferrin receptor by optineurin and its
impairment by a glaucoma-associated mutant. BMC Cell Biol.
11:42010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al
RIKEN Genome Exploration Research Group and Genome Science Group
(Genome Network Project Core Group): The transcriptional landscape
of the mammalian genome. Science. 309:1559–1563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mercer TR, Dinger ME, Sunkin SM, Mehler MF
and Mattick JS: Specific expression of long noncoding RNAs in the
mouse brain. Proc Natl Acad Sci USA. 105:716–721. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Amaral PP and Mattick JS: Noncoding RNA in
development. Mamm Genome. 19:454–492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ng SY, Lin L, Soh BS and Stanton LW: Long
noncoding RNAs in development and disease of the central nervous
system. Trends Genet. 29:461–468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johnson R: Long non-coding RNAs in
Huntington's disease neurodegeneration. Neurobiol Dis. 46:245–254.
2012. View Article : Google Scholar
|
|
14
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Jin Y, Zheng X, Wu Y and Fu H: The long noncoding RNA expression
profile of hepatocellular carcinoma identified by microarray
analysis. PLoS One. 9:e1017072014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maruyama H, Morino H, Ito H, Izumi Y, Kato
H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al:
Mutations of optineurin in amyotrophic lateral sclerosis. Nature.
465:223–226. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moran VA, Perera RJ and Khalil AM:
Emerging functional and mechanistic paradigms of mammalian long
non-coding RNAs. Nucleic Acids Res. 40:6391–6400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weisschuh N, Alavi MV, Bonin M and
Wissinger B: Identification of genes that are linked with
optineurin expression using a combined RNAi - microarray approach.
Exp Eye Res. 85:450–461. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liao Q, Xiao H, Bu D, Xie C, Miao R, Luo
H, Zhao G, Yu K, Zhao H, Skogerbø G, et al: ncFANs: A web server
for functional annotation of long non-coding RNAs. Nucleic Acids
Res. 39:W118–W124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
23
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schlitt T, Palin K, Rung J, Dietmann S,
Lappe M, Ukkonen E and Brazma A: From gene networks to gene
function. Genome Res. 13:2568–2576. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yan B, Tao ZF, Li XM, Zhang H, Yao J and
Jiang Q: Aberrant expression of long noncoding RNAs in early
diabetic retinopathy. Invest Ophthalmol Vis Sci. 55:941–951. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Albagha OM, Visconti MR, Alonso N,
Langston AL, Cundy T, Dargie R, Dunlop MG, Fraser WD, Hooper MJ,
Isaia G, et al: Genome-wide association study identifies variants
at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget's
disease of bone. Nat Genet. 42:520–524. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Soto I, Oglesby E, Buckingham BP, Son JL,
Roberson ED, Steele MR, Inman DM, Vetter ML, Horner PJ and
Marsh-Armstrong N: Retinal ganglion cells downregulate gene
expression and lose their axons within the optic nerve head in a
mouse glaucoma model. J Neurosci. 28:548–561. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chodroff RA, Goodstadt L, Sirey TM, Oliver
PL, Davies KE, Green ED, Molnár Z and Ponting CP: Long noncoding
RNA genes: conservation of sequence and brain expression among
diverse amniotes. Genome Biol. 11:R722010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kapitein LC and Hoogenraad CC: Building
the neuronal microtubule cytoskeleton. Neuron. 87:492–506. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Espinet C, Gonzalo H, Fleitas C, Menal MJ
and Egea J: Oxidative stress and neurodegenerative diseases: A
neurotrophic approach. Curr Drug Targets. 16:20–30. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wakatsuki S, Furuno A, Ohshima M and Araki
T: Oxidative stress-dependent phosphorylation activates ZNRF1 to
induce neuronal/axonal degeneration. J Cell Biol. 211:881–896.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Herrup K, Neve R, Ackerman SL and Copani
A: Divide and die: Cell cycle events as triggers of nerve cell
death. J Neurosci. 24:9232–9239. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dobolyi A, Vincze C, Pál G and Lovas G:
The neuroprotective functions of transforming growth factor beta
proteins. Int J Mol Sci. 13:8219–8258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang SP, Wang ZH, Peng DY, Li SM, Wang H
and Wang XH: Therapeutic effect of mesenchymal stem cells in rats
with intracerebral hemorrhage: Reduced apoptosis and enhanced
neuroprotection. Mol Med Rep. 6:848–854. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu G, Wang X, Shao G and Liu Q:
Genetically modified Schwann cells producing glial cell
line-derived neurotrophic factor inhibit neuronal apoptosis in rat
spinal cord injury. Mol Med Rep. 9:1305–1312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
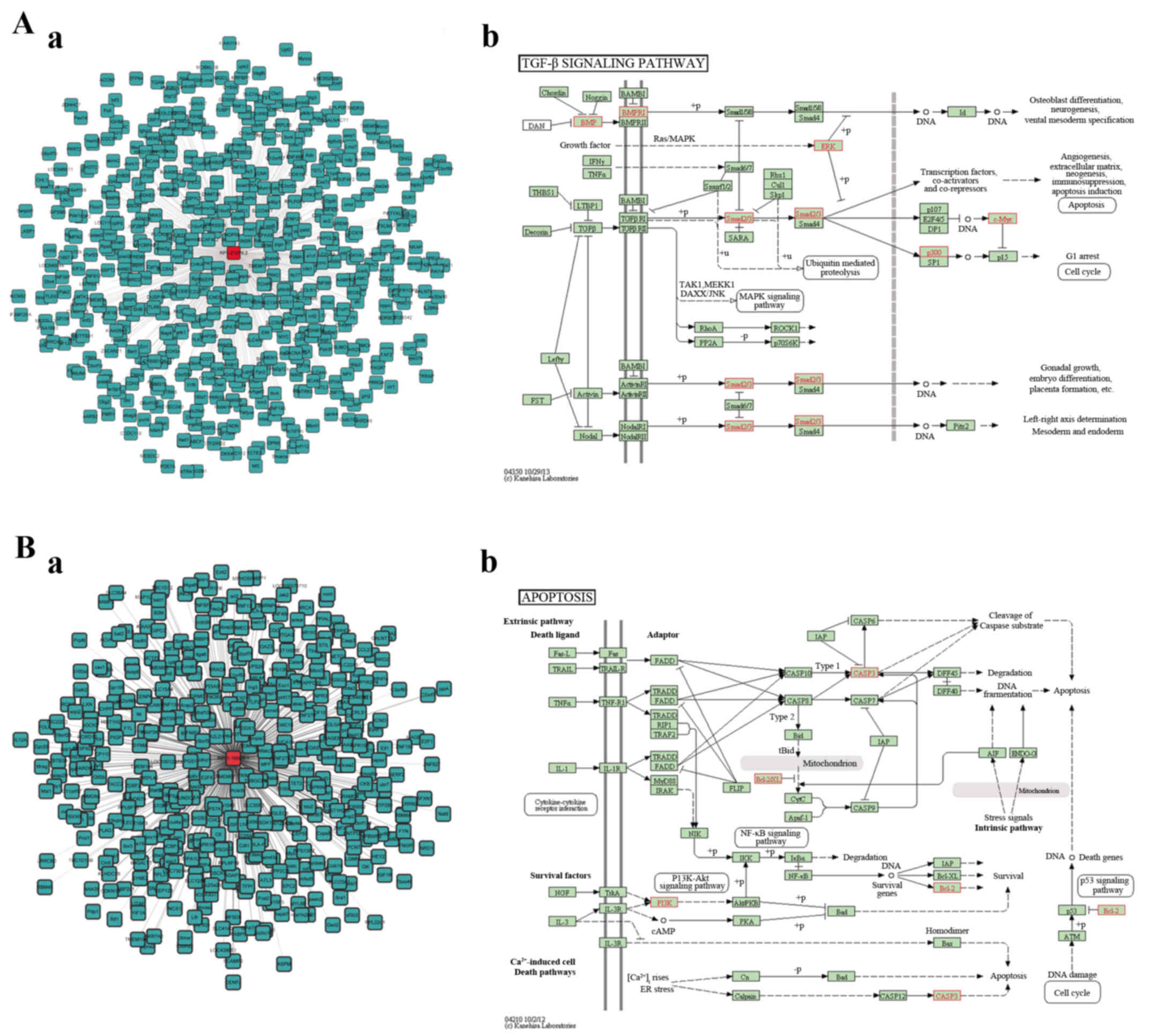
Zhao B and Chen YG: Regulation of TGF-β
signal transduction. Scientifica (Cairo). 2014:8740652014.
|
|
37
|
Wordinger RJ, Clark AF, Agarwal R, Lambert
W and Wilson SE: Expression of alternatively spliced growth factor
receptor isoforms in the human trabecular meshwork. Invest
Ophthalmol Vis Sci. 40:242–247. 1999.PubMed/NCBI
|
|
38
|
McDowell CM, Tebow HE, Wordinger RJ and
Clark AF: Smad3 is necessary for transforming growth factor-beta2
induced ocular hypertension in mice. Exp Eye Res. 116:419–423.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zode GS, Clark AF and Wordinger RJ: Bone
morphogenetic protein 4 inhibits TGF-β2 stimulation of
extracellular matrix proteins in optic nerve head cells: role of
gremlin in ECM modulation. Glia. 57:755–766. 2009. View Article : Google Scholar
|
|
40
|
Sethi A, Wordinger RJ and Clark AF:
Gremlin utilizes canonical and non-canonical TGFβ signaling to
induce lysyl oxidase (LOX) genes in human trabecular meshwork
cells. Exp Eye Res. 113:117–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kuehn MH, Fingert JH and Kwon YH: Retinal
ganglion cell death in glaucoma: mechanisms and neuroprotective
strategies. Development. 1:32005.
|
|
42
|
Levkovitch-Verbin H: Retinal ganglion cell
apoptotic pathway in glaucoma: Initiating and downstream
mechanisms. Prog Brain Res. 220:37–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sánchez Y and Huarte M: Long non-coding
RNAs: Challenges for diagnosis and therapies. Nucleic Acid Ther.
23:15–20. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gao W, Chan JY and Wong TS: Differential
expression of long noncoding RNA in primary and recurrent
nasopharyngeal carcinoma. Biomed Res Int. 2014:4045672014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Michelhaugh SK, Lipovich L, Blythe J, Jia
H, Kapatos G and Bannon MJ: Mining Affymetrix microarray data for
long non-coding RNAs: Altered expression in the nucleus accumbens
of heroin abusers. J Neurochem. 116:459–466. 2011. View Article : Google Scholar
|