Introduction
Chinese herbal medicine has become a popular field
of study. For instance, artemisinins inhibit the development of a
broader age range of parasites than quinine and other antimalarial
drugs (1). Myricetin has a
negative effect on proliferation and induces apoptosis in gastric
cancer cells (2). Evodiamine
(Evo) is a quinozole alkaloid constituent of Evodia
rutaecarpa extracted from the fruit of Evodia fructus (dried
fruits of Evodia rutaecarpa Bentham, Rutaceae) (3,4).
Evodia fruits, known as 'Wu-Chu-Yu', have been prescribed for the
treatment of headaches, thoracicoabdominal pain and vomiting caused
by a cold or a cold constitution in both traditional Chinese and
Japanese medicine. The Chinese literature refers to Evodia fruits
as a 'hot nature' herb, which is the same category chilli peppers
belong to. Previous studies have reported that Evo exhibits
antitumor, anti-inflammatory and vasorelaxant properties (5–7).
Furthermore, was demonstrated to Evo activate extracellular
signal-regulated kinase (ERK) signalling, which inhibited
adipogenesis (8). However, at
present, the effect of Evo on myogenesis has not been reported.
Skeletal muscle accounts for 40% of the total human
body mass. Skeletal muscle has extensive metabolic and functional
plasticity and a robust regenerative capacity (9). During myogenesis, there are two
phases of proliferation and differentiation, which are precisely
regulated by an ordered set of cellular events. The MyoD family, a
basic helix-loop-helix, includes four members [myogenic
differentiation 1 (Myod), myogenic factor 5,
Myogenin (MyoG) and myogenic factor 6], are
the major molecular mediators regulating skeletal myogenesis
(10,11). C2C12 cells are regarded as a
bioassay system in vitro. At present, this model has been
used extensively in studies of the effects of muscle growth factors
on muscular atrophy and hypertrophy, cell replication and apoptosis
(12,13).
Wnt/β-catenin signalling has a vital role in the
development of embryonic muscle and proliferation of satellite
cells during skeletal muscle regeneration (14,15). β-catenin translocation to nucleus
activates Wnt signalling pathway, and WNT3a enhances the myogenic
differentiation of C2C12 muscle cells (16). The association of Axin and
glycogen synthases kinase (GSK) 3β disrupts the β-catenin complex,
which results in the accumulation of β-catenin in the nucleus and
activation of target genes through displacing transcriptional
repressors. (17).
In normal mammalian cells, ERK1/ERK2 has an
important central role in the regulation of cell proliferation and
is required for cells to move from G0 through G1 and into S phase
(18). In 3T3-L1 preadipocytes,
Evo activates ERK phosphorylation, which in turn inhibits adipocyte
differentiation (8).
Given that Evo regulates the proliferation,
differentiation and apoptosis of multiple cell types and that
Wnt/β-catenin signalling is essential for myogenesis, we
hypothesize that Evo may affect C2C12 myoblast differentiation
through the Wnt/β-catenin signalling pathway. In the present study,
it was demonstrated that Evo promotes the differentiation of C2C12
cells and inhibits proliferation.
Materials and methods
Cell culture
C2C12 mouse myoblast cell line (American Type
Culture Collection, Manassas, VA, USA) was cultured in Dulbecco's
modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% (v/v)
foetal bovine serum (HyClone; GE Healthcare Life Sciences, Logan,
UT, USA), 2 mM glutamine and 1% (v/v) penicillin/streptomycin
[growth medium, (GM)]. Cells were cultured at 37°C with 5%
CO2. When the cell density reached 90% confluence, the
GM was replaced with a differentiation medium (DM), which was DMEM
supplemented with 2% (v/v) horse serum (HyClone; GE Healthcare Life
Sciences), 2 mM L-glutamine, and 1% (v/v) penicillin/streptomycin
solution. The media was changed every 2 days. Evo (purity
>98.0%; Xi'an Guanyu Bio-Tech Co., Ltd., Xi'an, China) was
diluted to 50, 100 and 200 nM using dimethyl sulfoxide (DMSO).
Evo supplementation and DMSO negative
control
To determine the effect of Evo on the
differentiation of C2C12, there were two Evo treatments. Initially,
different concentrations (50, 100 and 200 nM) of Evo were added to
the DM on differentiation day 0, and then the experiments were
performed on differentiation day 2 or 4. Subsequently, following
differentiation for 2 days, differentiation concentrations (100 and
200 nM) of Evo were added to the DM, and experiment were performed
on differentiation day 4 or 6. To determine the effect of Evo on
the proliferation of C2C12, GM was supplemented with Evo when the
cell density reached 30–40%.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using a TRIzol®
reagent (Life Technologies; Thermo Fisher Scientific, Inc.). The
RNA quality and concentration were determined using agarose gel
electrophoresis and NanoDrop 2.0 (NanoDrop Technologies; Thermo
Fisher Scientific, Inc., Wilmington, DE, USA), respectively. Then,
1 µg of the total RNA was processed into single-stranded
cDNA using an RT kit (Takara Biotechnology Co., Ltd., Dalian,
Chin). RNA (1 µg) was reverse transcribed using PrimeScript
RT reagent kit with gDNA Eraser. The RT program include two steps
as follows: 42°C for 2 min, followed by 15 min at 37°C and 85°C for
5 sec. qPCR was performed using a SYBR®-Green kit
(Takara Biotechnology Co., Ltd.) in triplicate on a Bio-Rad iQTM5
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). The qPCR program
was as follows: 95°C for 30 sec, followed by 40 cycles of 5 sec at
95°C and 30 sec at 60°C. The relative mRNA expression level was
normalized to GAPDH. The 2−ΔΔCq algorithm was employed
to estimate the relative expression level of each gene (19). Primer sequences are listed in
Table I.
 | Table ISpecific primers for reverse
transcription-quantitative polymerase chain reaction. |
Table I
Specific primers for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Sequences
(5′-3′) |
|---|
| MyoD | F:
CATTCCAACCCACAGAACCT |
| R:
CAAGCCCTGAGAGTCGTCTT |
| MyoG | F:
CAATGCACTGGAGTTCGGT |
| R:
CTGGGAAGGCAACAGACAT |
| MyHC | F:
CGCAAGAATGTTCTCAGGCT |
| R:
GCCAGGTTGACATTGGATTG |
| Axin2 | F:
GGTTCCGGCTATGTCTTTGC |
| R:
CAGTGCGTCGCTGGATAACTC |
|
β-catenin | F:
CTCTTGTACGCACCGTCCTT |
| R:
AAAGGCGCATGATTTGCTGG |
| Cyclin
B | F:
AACTTCAGCCTGGGTCG |
| R:
CAGGGAGTCTTCACTGTAGGA |
| Cyclin
D | F:
TAGGCCCTCAGCCTCACTC |
| R:
CCACCCCTGGGATAAAGCAC |
| Cyclin
E | F:
CAGACCAGCGAGCAGGAGC |
| R:
GCAGCTGCTTCCACACCACT |
| c-Myc | F:
CCCTATTTCATCTGCGACGAG |
| R:
GAGAAGGACGTAGCGACCG |
| p21 | F:
AGTGTGCCGTTGTCTCTTCG |
| R:
ACACCAGAGTGCAAGACAGC |
| GAPDH | F:
AACTTTGGCATTGTGGAAGG |
| R:
ACACATTGGGGGTAGGAACA |
Flow cytometry
C2C12 cells were seeded in 60 mm dishes
(1.6×105 cells per dish). At 24 h after supplementation
with Evo, when the cell density reached 80% confluence,
proliferating myoblasts were washed three times with cold
phosphate-buffered saline (PBS) and fixed in 70% ethanol at −20°C
overnight. Following centrifugation at 476 × g, at 4°C for 5 min,
the supernatants were discarded, and the cells were washed with PBS
twice and resuspended in 0.5 ml 50 µg/ml propidium iodide
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan) solution
containing 0.2% (v/v) Triton X-100 and 100 µg/ml RNase A for
30 min at 4°C. The cells were then tested with a flow cytometry
instrument (FACSCalibur; BD Biosciences, Franklin Lakes, NJ, USA),
and data were analysed with the ModFit software (LT4.1; Verity
Software House Inc., Topsham, ME, USA). The proliferative index
indicates the proportion of mitotic cells from a total of 20,000
cells examined.
Proliferation assay
Labelling with 5-ethynyl-2′-deoxyuridine (EdU) is
regarded as a marker that can detect DNA synthesis. At 24 and 48 h
after supplementation with Evo, 10 µM EdU (Guangzhou RiboBio
Co., Ltd., Guangzhou, China) was added into the GM and incubated
for 4 h. All the procedures were performed according to the
manufacturer's protocol. Cell nuclei were stained with Hoechst
33342 (Invitrogen; Thermo Fisher Scientific, Inc.) at a
concentration of 5 µg/ml for 10 min. Subsequently,
EdU-positive cells were visualized under a fluorescence microscope
(Nikon Corporation, Tokyo, Japan). The percentage of positive cells
(EdU-staining cells/total number of cells) was then calculated.
Western blot analysis
C2C12 cells were rinsed twice with ice-cold PBS.
Total and nuclear proteins of C2C12 cells were extracted using
ice-cold lysis buffer (radioimmunoprecipitation assay buffer;
Beyotime Institute of Biotechnology, Haimen, China) including a
protease inhibitor (Pierce; Thermo Fisher Scientific, Inc.). The
lysates were centrifuged (5,712 × g) at 4°C for 15 min, and then 5X
protein loading buffer was added to the lysates prior to full
denaturation in boiling water for 10 min. A total of 25 µg
of protein (quantified using the BCA Protein Assay kit; cat. no.
CW0014; CWBIO, Beijing, China) were electrophoresed on a 12%
SDS-polyacrylamide gel and transferred to a polyvinylidene
difluoride membrane (Cell Signalling Technology, Inc., Danvers, MA,
USA). The membrane was blocked in 5% (w/v) skim milk at room
temperature for 2 h and then incubated at 4°C overnight with
primary antibodies, including anti-MyoG (ab124800; Abcam,
Cambridge, UK), muscle myosin heavy chain (MyHC; AF2387; R&D
Systems, Minneapolis, MN, USA), β-catenin (bs-1165R; Bioss,
Beijing, Chin), lamin B1 (ab16048; Abcam), ERK1/2 (8544; Cell
Signalling Technology, Inc.), anti-p-ERK1/2 (9101; Cell Signalling
Technology, Inc.), p21 (BA0272; Wuhan Boster Biological Technology,
Ltd., Wuhan, China) and GAPDH (A00227; Wuhan Boster Biological
Technology, Ltd.). The next day, the membranes were washed with
TBST (20% Tween) and incubated with goat anti-mouse or anti-rabbit
secondary antibodies [HRP-conjugated goat anti-mouse IgG (BA1050)
and HRP-conjugated goat anti-rabbit IgG (BA1054); Wuhan Boster
Biological Technology, Ltd.] at room temperature for 2 h. The
membranes were washed three times with TBST and then exposed using
a ChemiDoc XRS imaging system (Bio-Rad Laboratories, Inc.).
Immobilon Western Chemiluminescent HRP substrate (WBKLS0500;
Millipore Corporation, Billerica, MA, USA) was used to produce
signal to detect western blot results. The captured images were
analysed by Image Lab software (Bio-Rad Laboratories, Inc.). The
protein bands were quantified using the Image J program (version
1.4.3.67; National Institutes of Health, Bethesda, MA, USA). The
protein level of whole cell lysates was normalized against the
expression of GAPDH and β-tubulin (KM9003T; Sungene Biotech,
Tianjing, China), whereas the protein level of nuclear lysates was
normalized against the expression of lamin B1.
Cell counting kit-8 (CCK-8)
detection
C2C12 were seeded in 96-well plates at
2×103 cells per well in 100 µl GM. At 24 and 48 h
after supplementation with Evo, 10 µl CCK-8 (Beyotime
Institute of Biotechnology) was used to measure the cell
proliferation index according to the manufacturer's instructions.
The absorbance was quantified on a microplate reader (PerkinElmer
Inc., Waltham, MA, USA). This experiment was independently repeated
at least three times.
Immunocytofluorescence analysis
At 4 days post-myogenic differe ntiation, C2C12
cells were washed three times with cold PBS and fixed with 4% (w/v)
paraformaldehyde for 30 min at room temperature. 0.5% (v/v) Triton™
X-100 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used for
permeabilization for 20 min at room temperature. The cells were
then blocked in 5% (w/v) bovine serum albumin diluted in PBS. After
blocking for 1 h at room temperature, the cells were incubated with
anti-MyHC antibody (1:200 in TBST; MAB4470; R&D Systems China
Co., Ltd., Shanghai, China) at 37°C for 2 h, followed by incubation
with Alexa Fluor® 568-labelled secondary antibody (cat.
no. a-11079; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C
for 1 h. The nuclei were labelled with DAPI (1:1,000 in PBS; Roche
Diagnostics, Basel, Switzerland) for 10 min at room temperature.
Images were captured using a Nikon TE2000 microscope (Nikon
Corporation). The differentiation index was determined as the
percentage of MyHC-positive nuclei among total nuclei. The numbers
of myotubes with 1–5 and 5 nuclei were counted and averaged among
three images per treatment.
Statistical analysis
Each experiment was repeated three times
independently, and each independent repeat was performed in
triplicate. All experimental data are presented as the mean ±
standard error and all figures were produced with GraphPad Prism 5
(GraphPad Software, Inc., La Jolla, CA, USA) One-way analy sis of
variance followed by LSD and Duncan post hoc tesst in SPSS 18.0
software was used to perform the variance analysis and significance
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Evo promotes C2C12 myogenic
differentiation
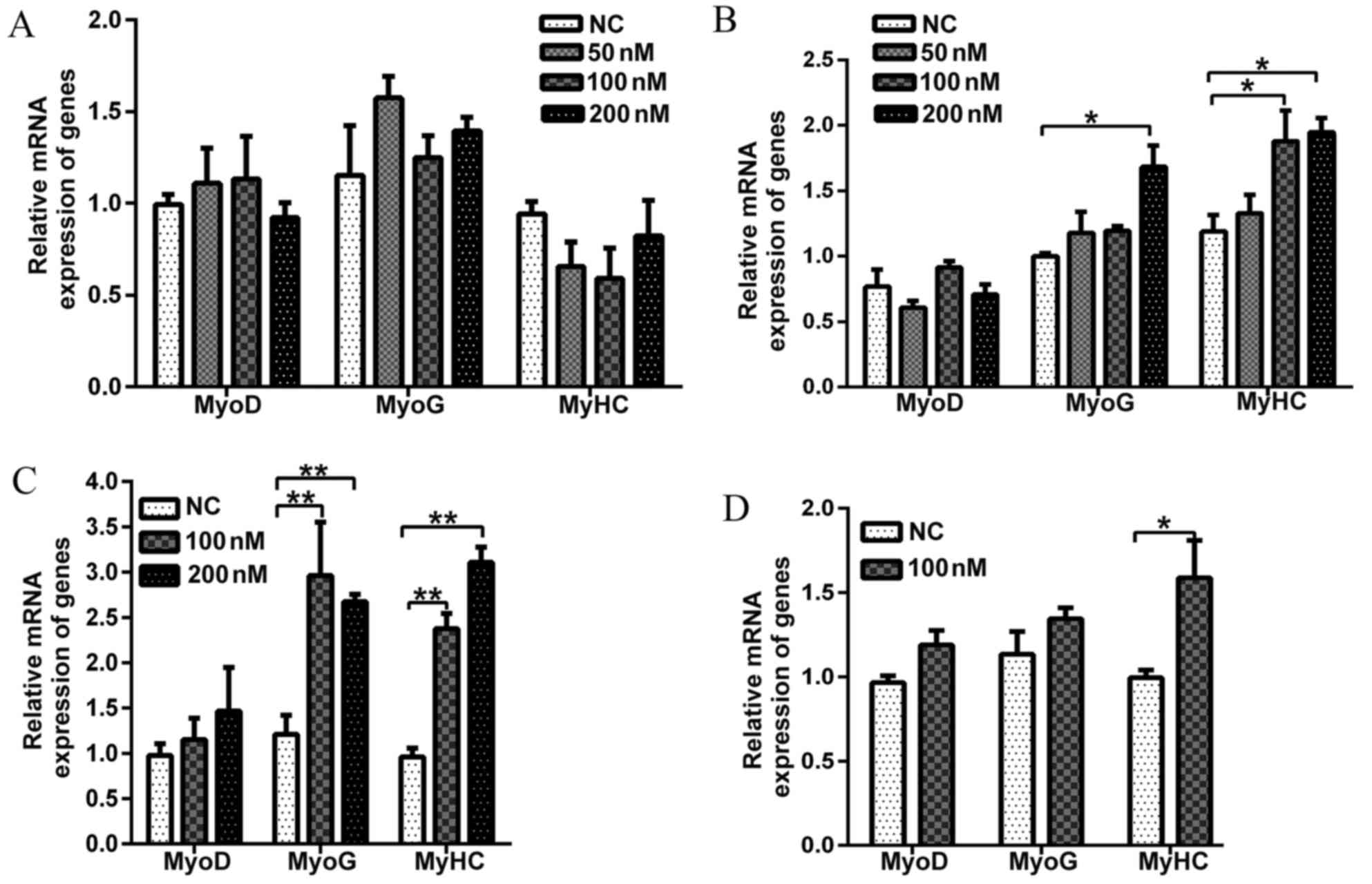
To determine the effect of Evo during the early
phase of C2C12 differentiation, Evo was added to the DM on
differentiation day 0. Fig. 1A
demonstrates that the different concentrations of Evo did not
significantly influence myogenic differentiation on day 2
post-differentiation. In order to determine the effect of long term
treatment on C2C12 differentiation, Evo was added to DM for 4 days.
Fig. 1B demonstrates that 50 nM
Evo did not alter the mRNA expression of myogenic marker genes
(MyoD, MyoG and MyHC). However, compared with
the control treatment, 100 nM Evo significantly increased the
expression of MyHC, a marker of late stage of muscle
differentiation. Compared with the control, 200 nM Evo
significantly increased the mRNA expression of MyoG and
MyHC (Fig. 1B). The
expression of MyoD, a well-known marker of early myogenic
stage, was not changed by Evo addition. Therefore, Evo may
positively impact the middle and late stages of C2C12
differentiation.
Subsequently, Evo was added either at 100 or 200 nM
to DM on differentiation day 2. The mRNA expression levels of two
myogenic marker genes (MyoG and MyHC) were increased
>2.5-fold, compared with those of the negative control (Fig. 1C). Similarly, the same treatment
was extended to differentiation day 6 (Fig. 1D), and 100 nM Evo increased mRNA
expression of MyHC but did not alter the expression of MyoD
and MyoG compared with the control group(Fig. 1D).
These results indicated that Evo treatment increases
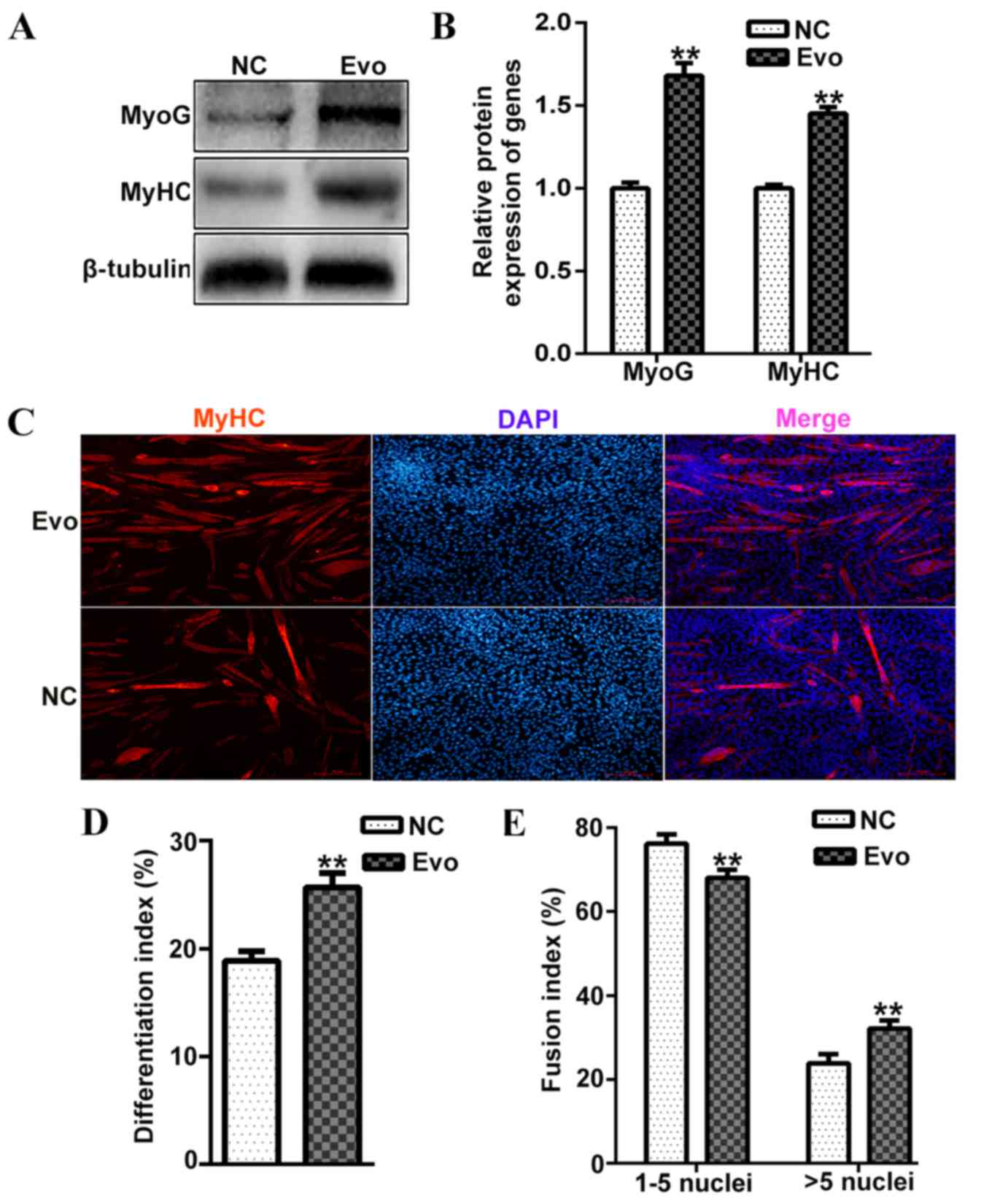
the mRNA expression of MyoG and MyHC. Additionally,
the protein levels of MyoG and MyHC were significantly increased
compared with the control on differentiation day 4 (Fig. 2A and B). The population of
myotubes was also markedly increased by Evo compared with the
control (Fig. 2C). Furthermore,
the index of differentiation (Fig.
2D) and fusion (Fig. 2E) were
also increased in cells treated with 100 nM Evo compared with the
negative control. Thus, 100 nM Evo was selected as an appropriate
concentration for further experiments. These results indicated that
Evo had a positive effect on C2C12 differentiation at the middle
and late stage.
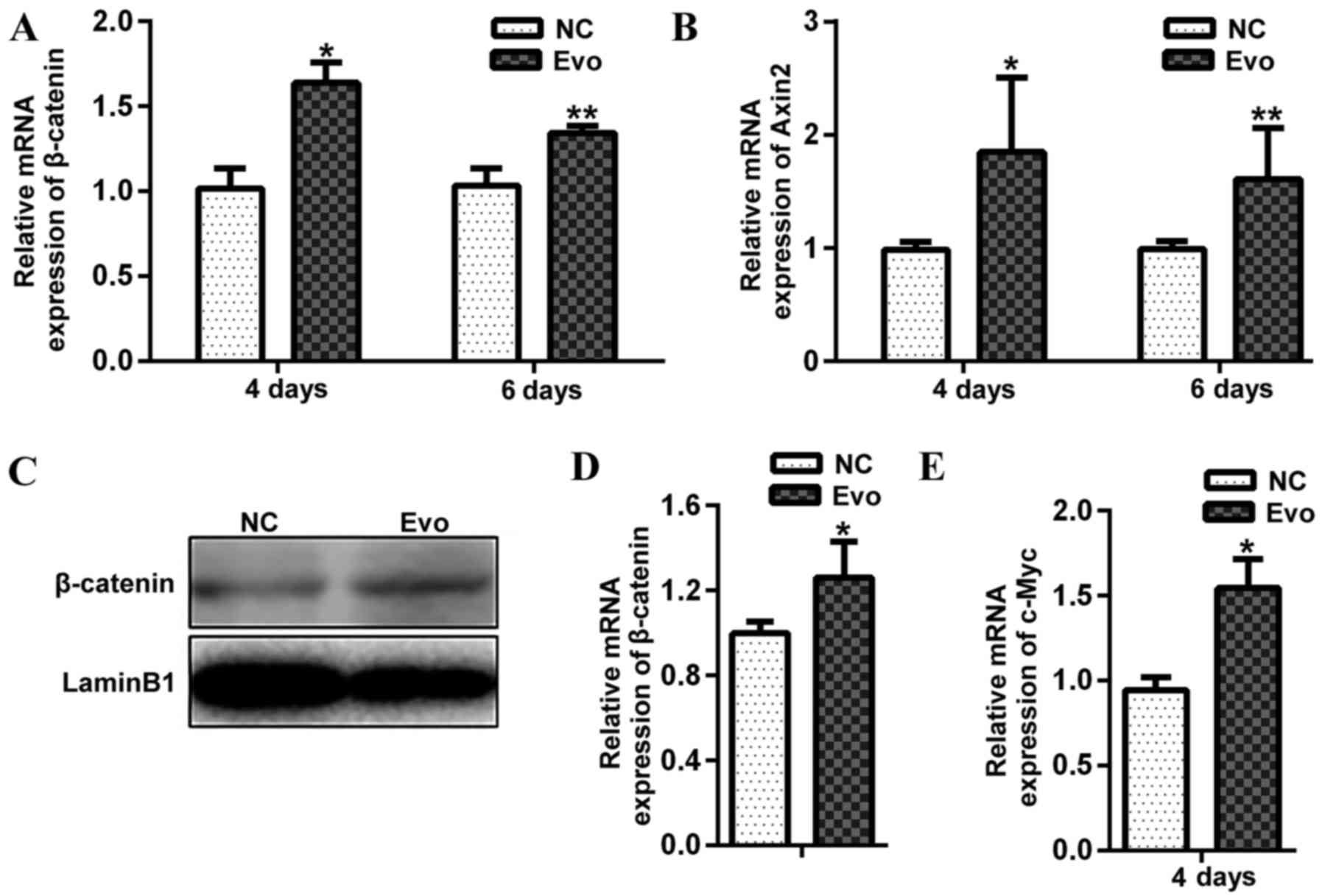
Effect of on Wnt/β-catenin signalling in
differentiating C2C12
To determine whether Evo promotes myogenesis via
Wnt/β-catenin signalling Evo was added to DM on differentiation day
2. On differentiation day 4 and 6 Axin2 transcription and
the nuclear translocation of β-catenin were determined, which are
two indicators of Wnt/β-catenin activity. Evo treatment
significantly increased the mRNA expression of β-catenin and
Axin2 at day 4 and 6 of differentiation compared with the
negative control (Fig. 3A and B).
Evo treatment significantly increased the level of nuclear
β-catenin (Fig. 3C and D). In
addition, the mRNA expression of c-Myc, which is a known
target of the canonical Wnt/β-catenin signalling pathway, was
significantly upregulated in cells treated with Evo compared with
the control (Fig. 3E). These
results suggested that Evo promotes C2C12 differentiation through
activation of the Wnt/β-catenin signalling pathway.
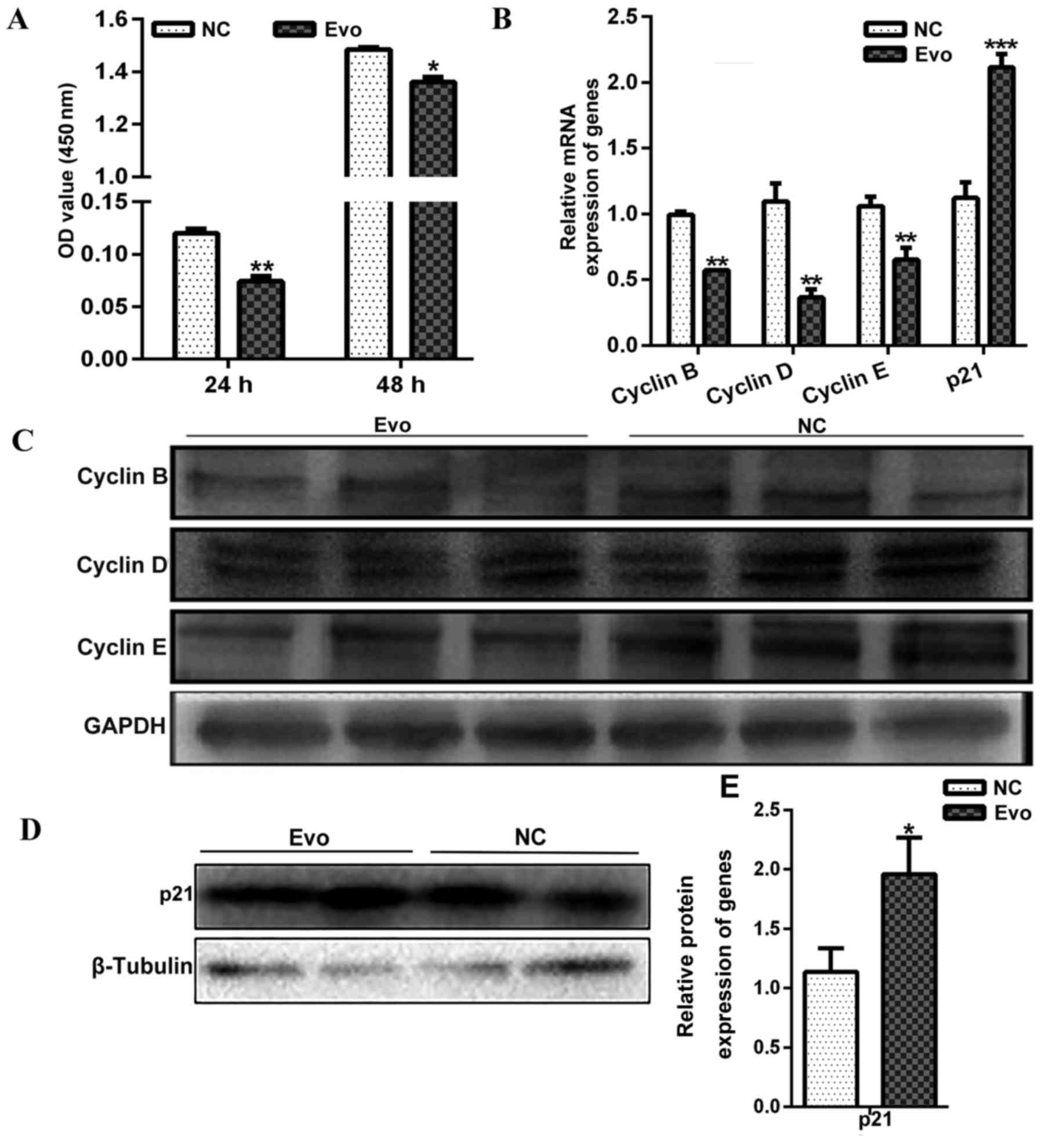
Proliferation of C2C12 myoblasts is
repressed by Evo treatment
To investigate the effect of Evo on C2C12 myoblast
proliferation, proliferation assays were performed in C2C12 cells
by adding 100 nM Evo at different times. CCK-8 kit assays indicated
that Evo decreased C2C12 cell proliferation after 24 and 48 h
treatment compared with the control (Fig. 4A).
Additionally, mRNA expression analyses of genes
associated with proliferation revealed that mRNA expression of
Cyclin B (sc-166152; Santa Cruz Biotechnology, Santa Cruz,
CA, USA), Cyclin D (BM4272; Wuhan Boster Biological
Technology, Ltd.) and Cyclin E (sc-377100; Santa Cruz
Biotechnology) were significantly decreased compared with the
control after 24 h treatment (Fig.
4B). However, the mRNA expression of p21, a well-known negative
regulator of the cell cycle, was significantly increased by Evo
(Fig. 4B). The protein levels of
the above genes were also detected by western blotting, and the
results with consistent with the level of mRNA expression (Fig. 4C–E).
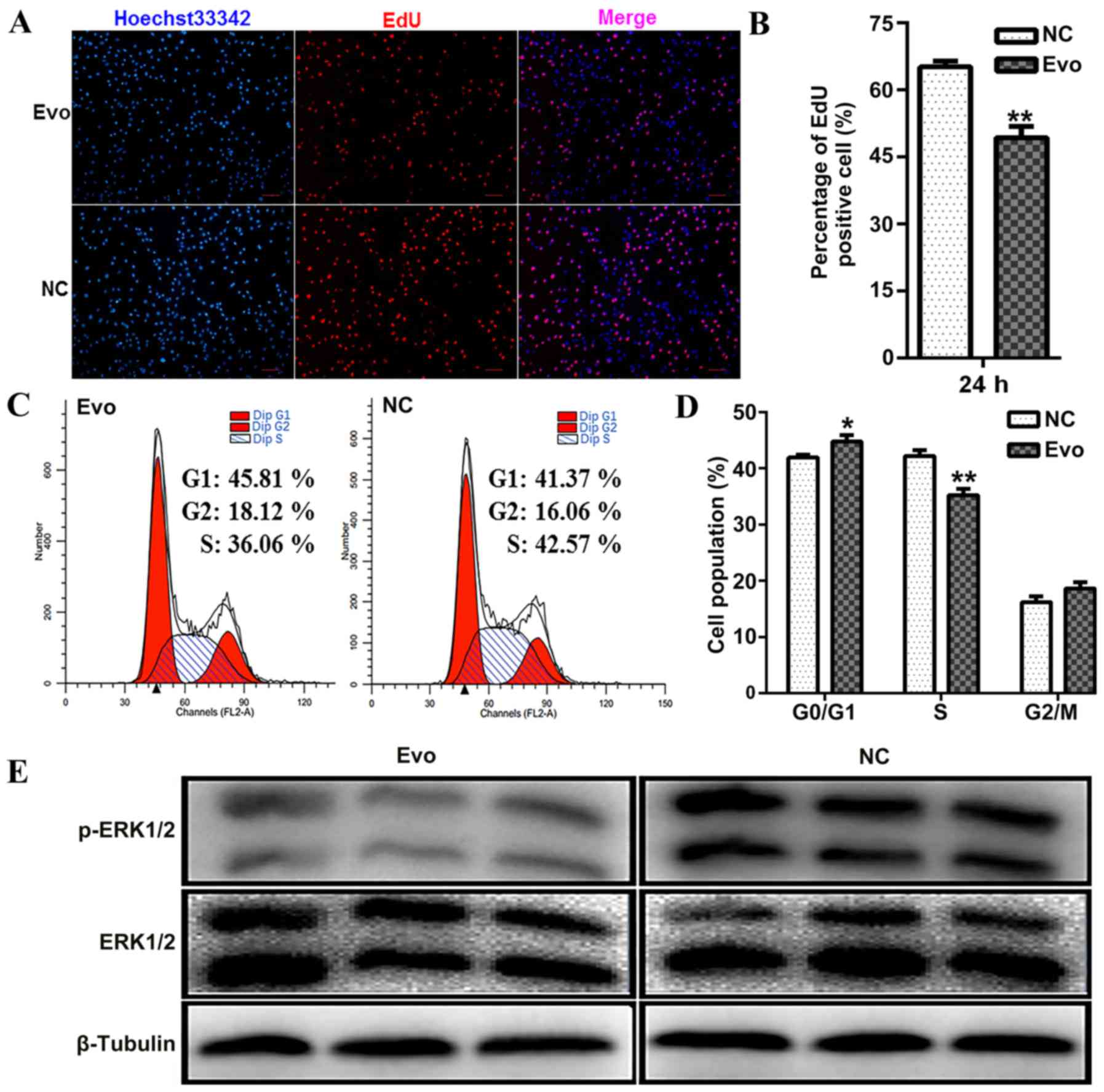
To further examine the direct effect of Evo on C2C12
proliferation, EdU incorporation was detected as an estimate of
cell proliferation at 24 h. EdU is a thymidine analogue that can be
inserted in nascent DNA molecules at the cell proliferation stage.
After 24 h of treatment with Evo, the percentage of EdU-positive
cells was significantly lower in the Evo-treated group compared
with the control group (Fig. 5A and
B). Additionally, proliferation analysis was performed using
flow cytometry, which indicated that Evo led to cell-cycle delay
with the fraction of S-phase cells decreased by Evo and the G0/G1
phase proportion increased by Evo (Fig. 5C and D).
Finally, whether Evo could affect ERK1/2
phosphorylation in proliferating C2C12 cells was determined.
Western blotting demonstrated that Evo markedly inhibited
phosphorylation of ERK1/2 (Fig.
5E). Therefore, we hypothesize that Evo may partially function
by inhibiting the activity of the ERK pathway to repress C2C12
myoblast proliferation.
Discussion
In the present study, it was aimed to investigate
the effects of Evo on C2C12 myoblasts. Three different
concentrations of Evo (50, 100 and 200 nM) were used initially. On
day 2 of differentiation, the mRNA expression level of MyoD,
MyoG and MyHC was examined. However, Evo had no effect
on the mRNA expression of the markers gene. This finding suggested
that Evo does not affect early myogenic differentiation. On day 4
of differentiation, the high concentration of Evo significantly
increased the mRNA expression of MyoG and MyHC, which
are markers of middle and later differentiation stages in C2C12
myoblasts, respectively. Based on these findings, Evo does not have
any influence on the mRNA expression of MyoD, a skeletal
muscle-specific early transcription factor that has a
transcriptional regulation effect on the myogenic program (20). Therefore, we hypothesized that Evo
may use other mechanisms to initiate myogenic differentiation. At 2
days post-differentiation of C2C12 cells, 100 or 200 nM Evo was
added to the DM. Consistent with expectations, the mRNA expression
and protein levels of MyoG and MyHC were dramatically
increased compared with the negative control at 4 days of
differentiation, although the effect was not concentration
dependent. Further confirmation of the above findings was provided
by immunocytofluorescence analysis of MyHC expression in
myotubes at 4 days of differentiation. Evo treatment markedly
induced the expression of MyoG and MyHC, and
increased the formation of myotubes. Analysis was continued to 6
days, and MyHC expression was remained significantly
increased at this time point. Day 6 is the late period of
differentiation for C2C12 cells, MyoD and MyoG
expression was not significantly increased by Evo at this stage.
These results indicate that Evo has an important effect on C2C12
myogenic differentiation.
The Wnt family of signalling molecules consists of
21 secreted glycoproteins. Wnt signalling includes canonical,
planar-cell-polarity and calcium-dependent pathways (21). Wnt/β-catenin signalling initiates
myogenic differentiation of satellite cells by replacing Notch
signalling, which is critical for self-renewal of satellite cells
through the inhibition of GSK-3 (22). In gastric cancer stem cells, Evo
treatment suppressed the Wnt/β catenin signalling pathway to
inhibit proliferation (7).
Notably, the results of the current study indicated that Evo
increased the nuclear translocation and accumulation of β-catenin,
and activated the transcription of downstream genes, Axin2
and c-Myc, which are two indicators of Wnt/β-catenin
signalling activity (23,24). The differing results may be due to
the different cell types used.
The role of Evo has already been extensively
investigated in various cell types. In 3T3-L1 preadipocytes, Evo
activated the epidermal growth factor receptor-protein kinase
Cα-ERK signalling pathway to inhibit adipogenesis (8). However, the data of the current
study indicated that Evo inhibited the phosphorylation of ERK. The
opposing result that was reported previously could be due to the
use of different cell types. In hyperlipidemic HepG2 cells, Evo
exhibited anti-lipogenic and anti-gluconeogenic effects through
activation of human constitutive androstane receptor (25). In tumour cells, Evo promotes
anti-proliferative effects and induces cell apoptosis (26–28). Evo inhibited the growth of DU145
and PC3 prostate cancer cell lines via accumulation at the G2/M
phase and an induction of apoptosis (29). Evo has a negative effect on
angiogenesis by inhibiting vascular endothelial growth factor
expression and ERK phosphorylation (30). These results support that Evo
typically has a negative effect on cell proliferation and that the
ERK pathway may be involved in this process.
In conclusion, the findings of the current study
demonstrate that Evo can positively regulates differentiation and
suppresses the proliferation of C2C12 myoblasts. Evo may be a novel
therapeutic agent for enhancing muscle growth.
Acknowledgments
The present study was granted by the Fundamental
Research Funds for the Central Universities (grant nos. 2452015278
and 2452017079) and the National Science and Technology Major
Project of China (grant no. 2016ZX08006003).
References
|
1
|
White NJ: Qinghaosu (artemisinin): The
price of success. Science. 320:330–334. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feng J, Chen X, Wang Y, Du Y, Sun Q, Zang
W and Zhao G: Myricetin inhibits proliferation and induces
apoptosis and cell cycle arrest in gastric cancer cells. Mol Cell
Biochem. 408:163–170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pearce LV, Petukhov PA, Szabo T, Kedei N,
Bizik F, Kozikowski AP and Blumberg PM: Evodiamine functions as an
agonist for the vanilloid receptor TRPV1. Org Biomol Chem.
2:2281–2286. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kobayashi Y, Nakano Y, Kizaki M, Hoshikuma
K, Yokoo Y and Kamiya T: Capsaicin-like anti-obese activities of
evodiamine from fruits of Evodia rutaecarpa, a vanilloid receptor
agonist. Planta Med. 67:628–633. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takada Y, Kobayashi Y and Aggarwal BB:
Evodiamine abolishes constitutive and inducible NF-kappaB
activation by inhibiting IkappaBalpha kinase activation, thereby
suppressing NF-kappaB-regulated antiapoptotic and metastatic gene
expression, up-regulating apoptosis, and inhibiting invasion. J
Biol Chem. 280:17203–17212. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heo SK, Yun HJ, Yi HS, Noh EK and Park SD:
Evodiamine and rutaecarpine inhibit migration by LIGHT via
suppression of NADPH oxidase activation. J Cell Biochem.
107:123–133. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wen Z, Feng S, Wei L, Wang Z, Hong D and
Wang Q: Evodiamine, a novel inhibitor of the Wnt pathway, inhibits
the self-renewal of gastric cancer stem cells. Int J Mol Med.
36:1657–1663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang T, Wang Y and Yamashita H: Evodiamine
inhibits adipogenesis via the EGFR-PKCalpha-ERK signaling pathway.
FEBS Lett. 583:3655–3659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shan T, Zhang P, Bi P and Kuang S: Lkb1
deletion promotes ectopic lipid accumulation in muscle progenitor
cells and mature muscles. J Cell Physiol. 230:1033–1041. 2015.
View Article : Google Scholar :
|
|
10
|
Di Marcantonio D, Galli D, Carubbi C,
Gobbi G, Queirolo V, Martini S, Merighi S, Vaccarezza M, Maffulli
N, Sykes SM, et al: PKCε as a novel promoter of skeletal muscle
differentiation and regeneration. Exp Cell Res. 339:10–19. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He K, Hu J, Yu H, Wang L, Tang F, Gu J, Ge
L, Wang H, Li S, Hu P and Jin Y: Serine/threonine kinase 40 (Stk40)
functions as a novel regulator of skeletal muscle differentiation.
J Biol Chem. 292:351–360. 2017. View Article : Google Scholar :
|
|
12
|
Taylor WE, Bhasin S, Artaza J, Byhowe F,
Azam M, Willard DH Jr, Kull FC Jr and Gonzalez-Cadavid N: Myostatin
inhibits cell proliferation and protein synthesis in C2C12 muscle
cells. Am J Physiol Endocrinol Metab. 280:E221–E228. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Q, Shi XE, Song C, Sun S, Yang G and
Li X: BAMBI promotes C2C12 myogenic differentiation by enhancing
Wnt/β-catenin signaling. Int J Mol Sci. 16:17734–17745. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Suzuki A, Scruggs A and Iwata J: The
temporal specific role of WNT/β-catenin signaling during
myogenesis. J Nat Sci. 1:e1432015.
|
|
15
|
Armstrong DD and Esser KA:
Wnt/beta-catenin signaling activates growth-control genes during
overload-induced skeletal muscle hypertrophy. Am J Physiol Cell
Physiol. 289:C853–C859. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang J, Mo C, Bonewald L and Brotto M:
Wnt3a and Wnt1 enhance myogenesis of C2C12 myoblasts–potential
mechanisms of osteocyte to muscle cell signaling. The FASEB
Journal. 29(1 Supplement): 947.132015.
|
|
17
|
Zhang L, Shi S, Zhang J, Zhou F and ten
Dijke P: Wnt/β-catenin signaling changes C2C12 myoblast
proliferation and differentiation by inducing Id3 expression.
Biochem Biophys Res Commun. 419:83–88. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meloche S and Pouysségur J: The ERK1/2
mitogen-activated protein kinase pathway as a master regulator of
the G1- to S-phase transition. Oncogene. 26:3227–3239. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Andrews JL, Zhang X, McCarthy JJ,
McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S,
Reid MB, Walker JR, et al: CLOCK and BMAL1 regulate MyoD and are
necessary for maintenance of skeletal muscle phenotype and
function. Proc Natl Acad Sci USA. 107:19090–19095. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cadigan KM and Nusse R: Wnt signaling: A
common theme in animal development. Genes Dev. 15:3286–3305. 1997.
View Article : Google Scholar
|
|
22
|
Brack AS, Conboy IM, Conboy MJ, Shen J and
Rando TA: A temporal switch from notch to Wnt signaling in muscle
stem cells is necessary for normal adult myogenesis. Cell Stem
Cell. 2:50–59. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jho EH, Zhang T, Domon C, Joo CK, Freund
JN and Costantini F: Wnt/beta-catenin/Tcf signaling induces the
transcription of Axin2, a negative regulator of the signaling
pathway. Mol Cell Biol. 22:1172–1183. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick
R, Hanash S, Cho KR and Fearon ER: Activation of AXIN2 expression
by beta-catenin-T cell factor. A feedback repressor pathway
regulating Wnt signaling. J Biol Chem. 277:21657–21665. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu L, Wang Z, Huang M, Li Y, Zeng K, Lei
J, Hu H, Chen B, Lu J, Xie W and Zeng S: Evodia alkaloids suppress
gluconeogenesis and lipogenesis by activating the constitutive
androstane receptor. Biochim Biophys Acta. 1859:1100–1111. 2016.
View Article : Google Scholar
|
|
26
|
Fei XF, Wang BX, Li TJ, Tashiro S, Minami
M, Xing DJ and Ikejima T: Evodiamine, a constituent of Evodiae
Fructus, induces anti-proliferating effects in tumor cells. Cancer
Sci. 94:92–98. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang ZG, Chen AQ and Liu B:
Antiproliferation and apoptosis induced by evodiamine in human
colorectal carcinoma cells (COLO-205). Chem Biodivers. 6:924–933.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen MC, Yu CH, Wang SW, Pu HF, Kan SF,
Lin LC, Chi CW, Ho LLT, Lee CH and Wang PS: Anti-proliferative
effects of evodiamine on human thyroid cancer cell line ARO. J Cell
Biochem. 110:1495–1503. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kan SF, Yu CH, Pu HF, Hsu JM, Chen MJ and
Wang PS: Anti-proliferative effects of evodiamine on human prostate
cancer cell lines DU145 and PC3. J Cell Biochem. 101:44–56. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shyu KG, Lin S, Lee CC, Chen E, Lin LC,
Wang BW and Tsai SC: Evodiamine inhibits in vitro angiogenesis:
Implication for antitumorgenicity. Life Sci. 78:2234–2243. 2006.
View Article : Google Scholar
|