Introduction
Joint arthroplasty is an effective treatment for
severe trauma and arthritic joint disorders, in that it provides
reliable long-term improvements in joint function, pain and quality
of life (1). However, chronic
wear on these prostheses can generate metallic, polyethylene or
ceramic debris, which is released into the joint space and becomes
embedded into synovial tissues (2). The deposition of the debris induces
inflammatory cytokine production that directly or indirectly
initiates unexpected bone erosion (osteolysis) resulting from the
differentiation and activation of osteoclasts (3–5).
Several studies have reported that aseptic loosening due to
periprosthetic osteolysis has become the major cause of failure
following prosthesis implantation, and has a prevalence of >10%
(6–8). Unfortunately, the only treatment
currently available for periprosthetic osteolysis is revision
surgery; however, this method is rendered less effective by its
greater rate of morbidity and poorer functional outcomes. Given the
crucial role that prosthesis implantation has in treating patients
with joint disorders, it is important to identify key modulators
involved in the osteolytic process if the clinical outcomes of
patients receiving joint arthroplasty are to be improved. The
development, remodeling and repair of bone tissue are critically
dependent on angiogenesis (9).
Osteoclasts and chondroclasts first appear in conjunction with
blood vessel invasion. Furthermore, the formation of new
capillaries and resorption of mineralized matrices are essential
events for bone morphogenesis and growth (10). Therefore, cytokines that have
determinant roles in the angiogenesis process also have key
functions in maintaining the skeletal system. Vascular endothelial
growth factor (VEGF) is the most important mediator of angiogenesis
(11). VEGF is produced by
various types of cells, and is expressed in osteoclasts and
chondroclasts in skeletal tissue (12,13). Nakagawa et al (9) reported that VEGF directly increases
osteoclastic bone resorption and the survival time of mature
osteoclasts. Henriksen et al (14) reported that VEGF is capable of
inducing osteoclast differentiation and the functioning of receptor
activator of nuclear factor-κB ligand (RANKL). Taken together,
these findings indicate that VEGF promotes the onset of osteolysis.
Thus, modulation of VEGF activity may be a strategy for alleviating
bone erosion following joint arthroplasty.
Mammalian cells produce various non-coding RNA
molecules, including small non-coding RNAs such as microRNAs (miRs)
and long non-coding RNAs (lncRNAs). The use of miRs to regulate
gene expression in treatment of various diseases has been
previously reported (15).
Furthermore, lncRNAs are increasingly recognized as important
modulators of diverse cellular processes, including cell
proliferation, cell-cycle progression, apoptosis and cell growth
(16). Several miRs have been
reported to inhibit the transcription of factors involved in VEGF
regulation (17–19). One such miR is miR-22, which was
reported to suppress VEGF activity in colon cancer (20). This infers that upregulation of
miR-22 during the osteogenic differentiation of human adipose
tissue-derived mesenchymal stem cells may have a promoting effect
(21), which is paradoxical to
its suppressive effect on VEGF activity. Thus, it is necessary to
investigate the association between miR-22 and VEGF during the
osteolytic process. Aside from miRs, lncRNAs also affect
development of the skeletal system. Che et al (22) reported that metastasis-associated
lung adenocarcinoma transcript 1 (MALAT1) knockdown reduced growth
inhibition and cell cycle arrest in RANKL-induced cells.
Additionally, MALAT1 is able to regulate miR-22 activity via a
competitive endogenous RNA mechanism (23). Taken together, these findings
suggest that MALAT1, miR-22, VEGF and RANKL all interact with each
other during the osteolytic process that reduces the benefits of
joint arthroplasty.
Therefore, the present study was performed to reveal
the molecular regulatory mechanism of these various factors
(MALAT1, miR-22, VEGF and RANKL) in an osteolysis model using hFOB
1.19 osteoblast cells. The expression levels of osteolysis-related
indicators were examined in clinical interface membrane samples and
synovial tissues, and MALAT1 knockdown hFOB 1.19 cells. The
findings suggest that during the osteolytic process, MALAT1
increases RANKL activity and indirectly activates the VEGF pathway
by suppressing miR-22-5p activity.
Materials and methods
Chemicals
Antibodies against VEGF (BA0407), osteoprotegerin
(OPG), RANKL (M00363) and GAPDH (A00227) were all purchased from
Wuhan Boster Biological Technology, Ltd. (Wuhan, China). Mimics
(5′-AGUUCUUCAGUGGCAAGCUUUA-3′), inhibitors
(5′-UAAAGCUUGCCACUGAAGAACU-3′), negative control (NC;
5′-UUCUCCGAACGUGUCACGUTT-3′) miR-22-5p, specific MALAT1 small
interfering RNA (siRNA), and siVEGF (5′-UUCCUCUGGUGGCCAGGGGCA-3′)
and scrambled NC MALAT1 siRNA (siMALAT1,
5′-AAGAAAAAUAAAAGCUUUCCU-3′ and siNC, 5′-ACGUGACACGUUCGGAGAATT-3′)
were all obtained from Shanghai GenePharma Co., Ltd. (Shanghai,
China). Fragments of the 3′ untranslated region (3′UTR) of
MALAT1/VEGF and the mutant 3′UTR of MALAT1/VEGF were amplified from
human hFOB 1.19 cells by PCR and inserted into a psiCHECK-2 vector
(vector containing firefly luciferase under control of the SV40
promoter; Promega Corporation, Madison, WI, USA) to create
different wild type (WT) and mutant (MUT) versions of plasmids. The
ultra-high molecular weight polyethylene (UHMWPE) particles
(Clariant, Gersthofen, Germany) were utilized for establishing
in vitro osteolysis model and the size of UHMWPE used was
0.05–11.6 μm (mean, 1.74±1.43 μm). Prior to
injection, the particles were tested in a quantitative limulus
amebocyte lysate assay to ensure their endotoxin level was <0.25
EU/ml. Following testing, the particles were sterilized in 99.5%
ethanol at room temperature for 24 h; after which, they were dried
and suspended in fetal calf serum (cat. no. 10099133; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Cell culture
Human hFOB 1.19 fetal osteoblastic cell line was
obtained from Bioleaf Biotech Co., Ltd. (Shanghai, China). The
cells were cultured in a 1:1 mixture of Ham's F12 medium and
Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc.)
without phenol red, but supplemented with 10% fetal bovine serum
(Thermo Fisher Scientific, Inc.), 50 μg/ml penicillin, and
50 mg/l gentamicin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
at pH 7.2 in a humidified atmosphere containing 5%
CO2.
Clinical sample collection
From January 2015 to September 2016, 8 samples of
interface membrane tissue were collected from patients with
prosthetic aseptic loosening at Xiangya Hospital (Changsha, China),
and used to investigate the expression status of molecules involved
the osteolytic process that occurred after implantation of a
prosthesis (10 samples). Eight samples of synovial tissue collected
from patients that received a hip operation were used as normal
control samples. All enrolled patients, with 9/18 males, were
provided with detailed information concerning the clinical,
pathological and prognostic aspects of their disease, and were
diagnosed by a micro-computed tomography scan (data not shown). The
study protocol was approved by the Research Ethics Committee of the
Xiangya Hospital of Central South University (Changsha, China). The
Hospital's Ethics Committee approved the study-associated
screening, inspection and data collection procedures, and all
subjects signed a written informed consent document. Information
concerning the enrolled patients is presented in Table I. All study procedures complied
with provisions in the Declaration of Helsinki.
 | Table IClinical features of patients
involved in the study. |
Table I
Clinical features of patients
involved in the study.
| Patient no. | Gender | Age (years) | Diagnosis | Operation |
|---|
| 1 | Female | 77 | Prosthetic aseptic
loosening (left hip) | Revision (left
hip) |
| 2 | Female | 61 | Prosthetic aseptic
loosening (left hip) | Revision (left
hip) |
| 3 | Male | NR | Prosthetic aseptic
loosening (right hip) | Revision (right
hip) |
| 4 | Male | 47 | Prosthetic aseptic
loosening (left hip) | Revision (left
hip) |
| 5 | Male | 66 | Prosthetic aseptic
loosening (right hip) | Revision (right
hip) |
| 6 | Female | NR | – | – |
| 7 | Female | 40 | Femoral neck
fracture (left) | Total hip
replacement (left hip) |
| 8 | Male | 86 | Femoral neck
fracture (left) | Total hip
replacement (left hip) |
| 9 | Male | 75 | Femoral neck
fracture (left) | Total hip
replacement (left hip) |
| 10 | Female | 57 | Femoral neck
fracture (left) | Total hip
replacement (left hip) |
| 11 | Male | 40 | Femoral neck
fracture (bilateral) | Total hip
replacement (bilateral) |
| 12 | Male | 48 | Prosthetic aseptic
loosening (left hip) | Revision (left
hip) |
| 13 | Male | 40 | Femoral neck
fracture (bilateral) | Total hip
replacement (bilateral) |
| 14 | Male | 68 | Prosthetic aseptic
loosening (right hip) | Revision (right
hip) |
| 15 | Female | 79 | Femoral neck
fracture (left) | Revision (left
hip) |
| 16 | Female | 69 | Prosthetic aseptic
loosening (right hip) | Revision (right
hip) |
| 17 | Female | 67 | Femoral neck
fracture (left) | Total hip
replacement (left hip) |
| 18 | Female | NR | Femoral neck
fracture (left) | NR |
Cell seeding and particle treatment
Human hFOB 1.19 osteoblast cells (1×104
cells/well)were transferred into 96-well plates, and incubated at
37°C in an atmosphere containing 5% CO2. Fresh medium
containing UHMWPE particles was added to each well after 24 h of
culture. The inherent hydrophobicity of polyethylene was used to
ensure that sufficient contact occurred between the cells and
particles. The cell-particle ratio was 1:500, and the particle
treatment method was previously described by Kauther et al
(24).
Experimental design and transfection
Transfection with mimics or plasmids was performed
using Lipofectamine® 2000 (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
instructions. Details of the experimental designs are provided
below.
The following two groups of cells (i and ii) were
used in assays designed to determine how MALAT1 functions during
the osteolytic process: i) NC group (UHMWPE-treated hFOB cells
transfected with NC siRNA, 100 pmol per 6 well plates); and ii)
siMALAT1 group (UHMWPE-treated hFOB cells transfected with specific
MALAT1 siRNA, 100 pmol per 6 well plates).
Cells used to explore the role of miR-22-5p in bone
erosion were divided into the following four groups: i) mimics NC
group (UHMWPE-treated hFOB cells transfected with NC miR-22-5p
mimic, 100 pmol per 6 well plates); ii) mimics group
(UHMWPE-treated hFOB cells transfected with miR-22-5p mimic, 100
pmol per 6 well plates); iii) inhibitor NC group (UHMWPE-treated
hFOB cells transfected with a NC miR-22-5p inhibitor, 100 pmol per
6 well plates); and iv) inhibitor group (UHMWPE-tre ated hFOB cells
transfected with a miR-22-5p inhibitor, 100 pmol per 6 well
plates).
Cells used to detect interactions which occurred
between MALAT1 and miR-22-5p were divided into the following four
groups: i) blank group (UHMWPE-treated hFOB cells); ii) NC group
(UHMWPE-treated hFOB cells transfected with pcDNA vector, 3
μg per 6 well plates); iii) mimics group (UHMWPE-treated
hFOB cells transfected with miR-22-5p mimic, 3 μg per 6 well
plates); and iv) MALAT1 + mimics group (UHMWPE-treated hFOB cells
transfected with miR-22-5p mimic, 3 μg per 6 well plates,
and pcDNA-MALAT1 vector, 2 μg per 6 well plates).
Cells used to detect whether VEGF intervention could
recapitulate the effects of miR-22-5p overexpression were divided
into the following three groups: i) blank group (UHMWPE-treated
hFOB cells); ii) NC group (UHMWPE-treated hFOB cells transfected
with NC siRNA, 100 pmol per 6 well plates); and iii) siVEGF group
(UHMWPE-treated hFOB cells transfected with specific VEGF siRNA,
100 pmol per 6 well plates).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from different clinical
tissue and cell samples using an RNA Purified Total RNA Extraction
kit according to the manufacturer's instructions (cat. no. RP1201;
BioTeke Corporation, Beijing, China). GAPDH was selected as the
reference gene. cDNA templates were constructed by reverse transc
ription of RNA using Super M-MLV reverse transcriptase (cat. no.
PR6502; BioTeke Corporation). The mixture was incubated at 70°C for
5 min and then incubated at 37°C for 5 min. Each 20 μl
reaction mixture contained 10 μl of SYBR Premix Ex Taq II
(Clontech Laboratories, Inc., Mountainview, CA, USA) and 0.5
μl of each primer. RT-qPCR was performed as follows:
pre-denaturation at 95°C for 10 sec, followed by 40 cycles of
denaturation at 95°C for 10 sec and elongation at 60°C for 30 sec.
Relative expression levels were calculated using DataAssist
software, version 3.0 (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and the formula:
2−[Cq(Gene) − Cq(GAPDH)]. Each assay was performed three
times. The primers used in this study are presented in Table II.
 | Table IIPrimer sequences used in quantitative
polymerase chain reaction. |
Table II
Primer sequences used in quantitative
polymerase chain reaction.
| Genes | Sequences
(5′-3′) |
|---|
| OPG | F:
GCTGCTCAGTTTGTGGCG |
| R:
TGGACCTGGTTACCTATCATTTCT |
| RANK | F:
CATGTTTACTTGCCCGGTTTA |
| R:
AGCTGTGAGTGCTTTCCCTTT |
| RANKL | F:
TGCCAACATTTGCTTTCG |
| R:
TTCCTCCTTTCATCAGGGTAT |
| MALAT1 | F:
GCAGGGAGAATTGCGTCATT |
| R:
TTCTTCGCCTTCCCGTACTT |
| VEGF | F:
TGTGTATACTCGCGCTACCT |
| R:
GATCTGCATCCGGACTTGGT |
| miR-22-5p | F:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAAGAA |
| R:
TACACTCCAGCTGGGATTTCGAACGGTGACT |
| U6 | F:
CTCGCTTCGGCAGCACA |
| R:
AACGCTTCACGAATTTGCGT |
| β-actin | F:
ATCGTGCGTGACATTAAGGAGAAG |
| R:
AGGAAGGAAGGCTGGAAGAGTG |
Western blot analyses
The total proteins from different samples were
extracted using a Total Protein Extraction kit according to the
manufacturer's instructions (cat. no. WLA019; Wanleibo Co., Ltd.,
Beijing, China). GAPDH was used as an internal reference protein.
The protein concentration of each sample was determined using the
bicinchoninic acid method. An aliquot of protein (40 μg)
from each sample subjected to 10% SDS-PAGE performed at 80 V for
2.5 h. The separated proteins were transferred onto polyvinylidene
difluoride membranes, which were then washed with TBS-Tween for 5
min, and subsequently incubated with a powdered skim milk solution
for 1 h. Subsequently, the membranes were incubated with primary
antibodies against OPG (1:1,000), RANKL (1:500), VEGF (1:500) or
GAPDH (1:500) at 4°C overnight. Following incubation, the membranes
were washed four more times with TBS-Tween, and then incubated with
secondary IgG-horseradish peroxidase antibodies [1:5,000; goat
anti-mouse IgG (BA1050) and goat anti-rabbit IgG (BA1054), Wuhan
Boster Biological Technology, Ltd.] for 45 min at 37°C. Following
incubation, the membranes were washed six times with TBS-Tween, and
the blots were developed using Beyo ECL Plus reagent (Thermo Fisher
Scientific, Inc.). The images were recorded using a gel imaging
system.
Cell Counting Kit-8 assay (CCK-8)
CCK-8 assays were performed to detect cell
proliferation. Approximately 1×104 hFOB cells were
seeded into each well of a 96-well plate and cultured for 72 h. The
numbers of viable cells were determined using CCK-8 (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) at 0, 24, 48, and 72
h after seeding. Briefly, after treatment with CCK-8 at 37°C for 1
h, the absorbance of the hFOB cells at 450 nm was measured using a
microplate reader (Rayto Life and Analytical Sciences Co., Ltd.,
München, Germany).
Cell viability assay
Cell viability further assessed using the
5-ethynyl-2′-deoxyuridine (EdU) assay as performed with a
Cell-Light™ EdU kit (Guangzhou RiboBio Co., Ltd., Guangzhou,
China). The viability of cells subjected to diffe rent treatments
was assessed using a flow cytometer (Accuri C6; BD Biosciences,
Franklin Lakes, NJ, USA) as described below.
Flow cytometry
The percentage of apoptotic cells in each sample was
determined by using flow cytometry in conjunction with an Annexin
V-FITC/PI Apoptosis Detection kit (cat. no. BB-4101-2; BestBio Co.,
Shanghai, China; http://www.beibokit.com/goods.php?id=53) as described
in instructions provided by the manufacturer. Briefly, cultured
cells from different time-points were incubated with 5 μl
Annexin V for 10 min at room temperature, and then resuspended in
binding buffer supplemented with 5 μl propidium iodide. The
percentage of apoptotic cells was determined by flow cytometry.
Following staining, the cells were gated into quadrants, and the
rate of apoptosis was calculated as follows: Upper right (UR)
quadrant + lower right (LR) quadrant indicated the percentage of
apoptotic cells, which equaled the sum of the late apoptotic rate
(UR) and the early apoptotic rate (LR).
miRNA targets mRNA prediction
Targets mRNA of miRNA were identified using the
following websites: microRNA.org
and targetscan.org.
Dual luciferase assay
The concentration of hFOB cells was adjusted to
1×104 cells/ml, and the cells were then incubated for 24
h prior to co-transfection with different combinations of
vectors.
Cells used to determine interactions between VEGF
and miR-22-5p were divided into the following four groups: i) WT +
NC group (cells were co-transfected with WT VEGF plasmid, 0.2
μg, and NC miR-22-5p mimics, 5 pmol); ii) WT + mimics group
(cells were co-transfected with WT VEGF plasmid, 0.2 μg, and
miR-22-5p mimics, 5 pmol); iii) MUT + NC group (cells were
co-transfected with MUT VEGF plasmid, 0.2 μg, and NC
miR-22-5p mimics, 5 pmol); iv) MUT + mimics group (cells were
co-transfected with MUT VEGF plasmid, 0.2 μg, and miR-22-5p
mimics, 5 pmol). Following transfection, the luciferase activity
was measured using the Dual-Luciferase Reporter Assay kit (cat. no.
E1910; Promega Corporation). Results were normalized to Renilla
luciferase activity. The same method of grouping cells was used
when assessing interactions between MATAL1 and miR-22-5p.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Student's t-test, analysis of variance, and Duncan's
post hoc multiple comparisons test were performed using GraphPad
Prism 6 software (GraphPad Software, Inc., San Diego, CA, USA).
P≤0.05 was considered to indicate a statistically significant
difference.
Results
MALAT1 expression in upregulated in
samples with bone erosion
The expression of MALAT1 and molecules associated
with skeletal development was comprehensively investigated in
clinical samples. Demographic and clinical characteristics of the
patients that donated the samples are presented in Table I. The RANKL/RANK/OPG system is
considered as an important signal transduction pathway in
osteolysis. We found that the expression of RANK was lower in loose
tissue samples comparing with normal synovial tissues (Fig. 1). By contrast, molecules that
promote osteolysis showed significantly enhanced levels of
expression (Fig. 1) in samples of
loose tissue compared with normal control samples. Furthermore,
MALAT1 RNA levels were higher in loose samples compared with normal
samples.
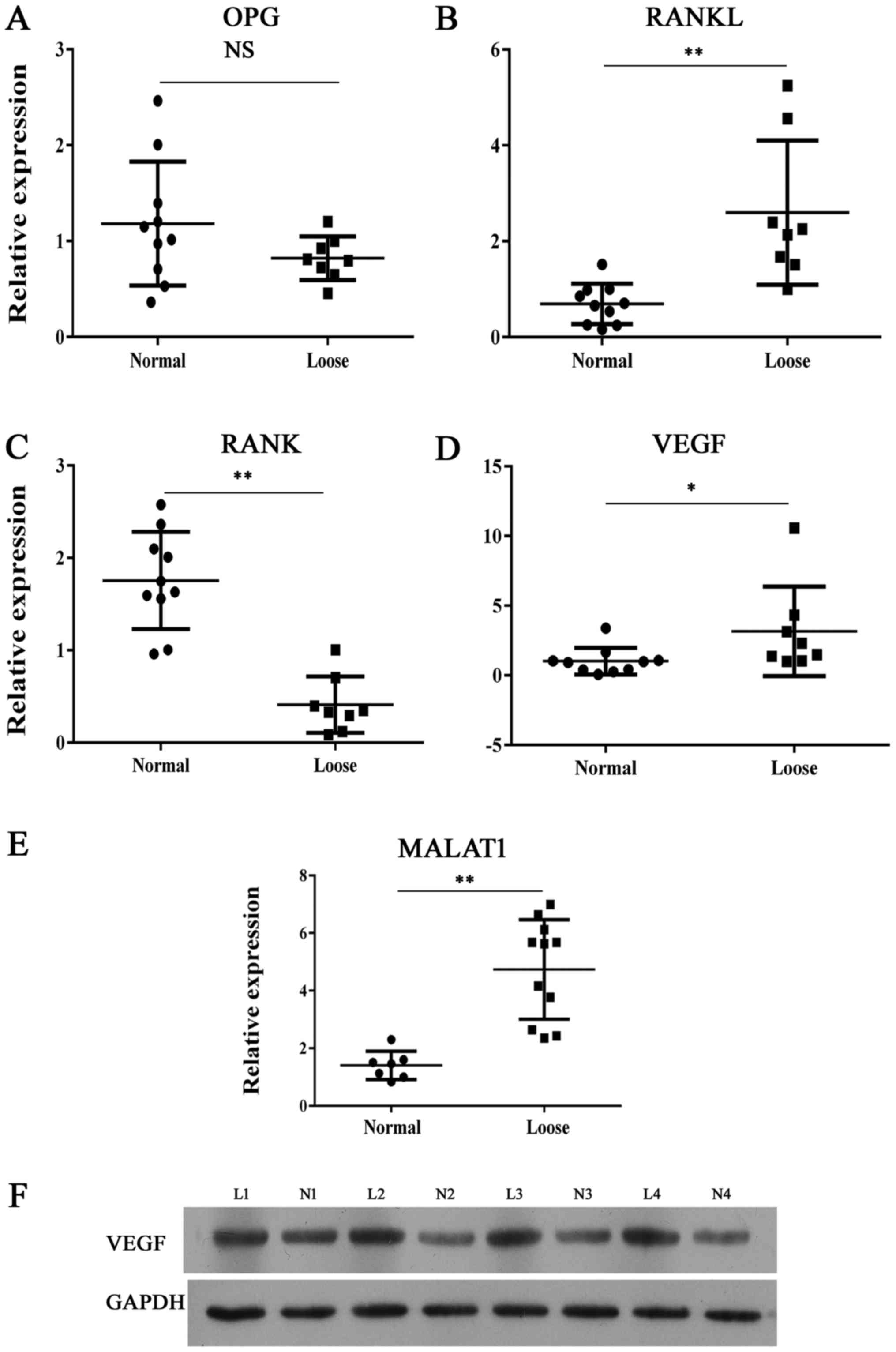 | Figure 1Expression of bone metabolism
indicators and MALAT1. Reverse transcription-quantitative
polymerase chain reaction analysis of (A) OPG, (B) RANKL, (C) RANK,
(D) VEGF and (E) MALAT1 expression in clinical interface membrane
tissues and normal tissues. (F) Representative images from western
blot analysis of VEGF protein expression in interface membrane
tissues showed enhanced expression of VEGF. *P<0.05,
**P<0.01 vs. normal sample. OPG, osteoprotegerin; NS,
not significant; RANK, receptor activator of nuclear factor-κB;
RANKL, RANK ligand; MALAT1, metastasis-associated lung
adenocarcinoma transcript 1; VEGF, vascular endothelial growth
factor; L, loose; N, normal. |
Knockdown of MALAT1 decreases cell
viability and induces cell apoptosis in hFOB 1.19 cells
The human hFOB 1.19 fetal osteoblastic cell line has
been used as an in vitro osteolysis model in numerous
studies. Thus, in the present study, MALAT1 gene expression was
altered in the cell line to assess its role in the osteolytic
process. Following silencing the MALAT1 gene using siRNA, the
viability of UHMWPE-treated hFOB 1.19 cells was significantly
reduced compared with the NC siRNA group (Fig. 2A and B). Results of CCK-8 assays
indicated that the MALAT1 knockdown UHMWPE-treated hFOB 1.19 cells
were significantly less viable than UHMWPE-treated hFOB 1.19 cells
from 48 h of the assay (Fig. 2A).
These results were further validated in the EdU assay, in which the
numbers of EdU-positive cells was reduced in the siMALAT1 group
compared with the NC group (Fig.
2B). Furthermore, the percentage of apoptotic cells in the
siMALAT1 group increased compared with the NC group, in conjunction
with their decreased viability (Fig.
2C), indicating that MALAT1 knockdown decreased the survival
rates of UHMWPE-treated hFOB 1.19 cells. At the molecular level,
knockdown of MALAT1 reduced the protein expression levels of RANKL
and VEGF, and enhanced the levels of OPG (Fig. 2D).
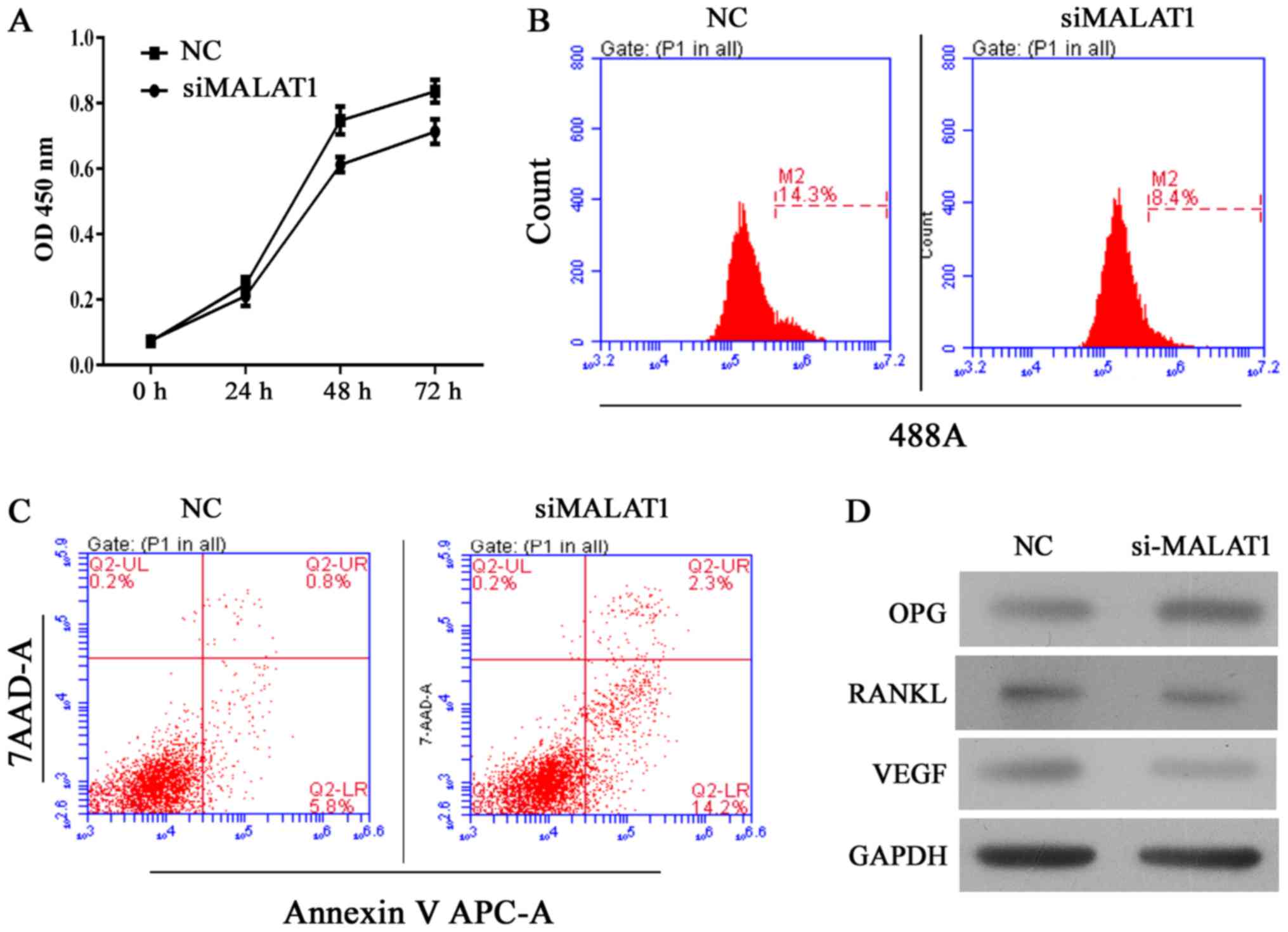 | Figure 2Knockdown of MALAT1 inhibits cell
growth and induces apoptosis in ultra-high molecular weight
polyethylene-treated hFOB 1.19 cells, and the effect was associated
is upregulation of OPG, and downregulation of RANKL and VEGF. (A)
Quantitative analysis of CCK-8 assay results at 24, 48 and 72 h
after siRNA transfection. (B) Representative images of
5-ethynyl-2′-deoxyuridine assay results after 48 h of siRNA
transfection. (C) Representative images of apoptosis rates as
detected by flow cytometry after 48 h of siRNA transfection. (D)
Representative images from western blot analyses of OPG, RANKL and
VEGF protein expression after 48 h of siRNA transfection. All
experiments were repeated at least 3 times. OD, optical density;
NC, negative control; si, small interfering RNA; MALAT1,
metastasis-associated lung adenocarcinoma transcript 1; 7-AAD,
7-aminoactinomycin D; APC, allophycocyanin; OPG, osteoprotegerin;
RANKL, receptor activator of nuclear factor-κB ligand; VEGF,
vascular endothelial growth factor. |
Knockdown of VEGF recapitulated the
biological effects observed following miR-22-5p overexpression
To further corroborate the pathway of miR-22-5p
targeting VEGF, which in turn regulated RANKL and OPG,
UHMWPE-treated hFOB 1.19 cells were treated with siVEGF to
determine whether this intervention induces the same effects as
miR-22-5p overexpression. The EdU and CCK-8 assays demonstrated
that the viability of UHMWPE-treated hFOB 1.19 cells was decreased
by siVEGF transfection (Fig. 3A and
B). Furthermore, the apoptosis of UHMWPE-treated hFOB 1.19
cells was induced by siVEGF (Fig.
3D). Western blot results (Fig.
3C) demonstrated that inhibited VEGF expression increased OPG
expression, while RANKL was decreased in hFOB 1.19 cells. These
results indicated that knockdown of VEGF induces the same
biological effects observed following miR-22-5p overexpression.
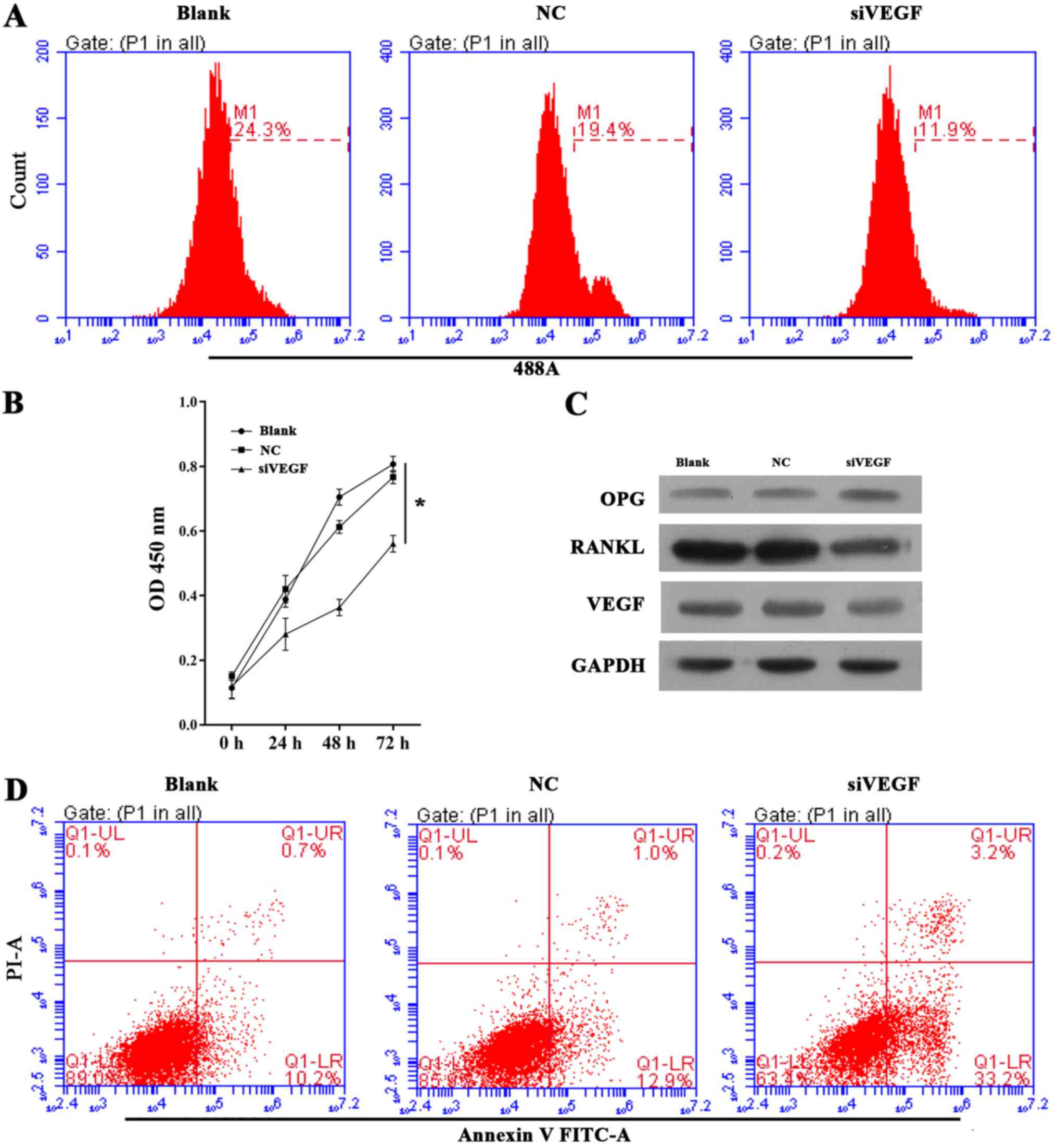 | Figure 3Effect of VEGF knockdown on cell
growth, apoptosis and OPG, RANKL and VEGF expression in ultra-high
molecular weight polyethylene-treated hFOB 1.19 cells. (A)
Representative images from 5-ethynyl-2′-deoxyuridine assays after
48 h of siRNA transfection. (B) Quantitative analysis of results of
CCK-8 assays at 24, 48 and 72 h after siRNA transfection. (C)
Representative images from western blot analyses of OPG, RANKL and
VEGF expression after 48 h of siRNA transfection. (D)
Representative images showing apoptosis rates detected by flow
cytometry after 48 h of siRNA transfection. All experiments were
repeated at least 3 times. NC, negative control; si, small
interfering RNA; VEGF, vascular endothelial growth factor; OD,
optical density; OPG, osteoprotegerin; RANKL, receptor activator of
nuclear factor-κB ligand; PI, propidium iodide; FITC, fluorescein
isothiocyanate. |
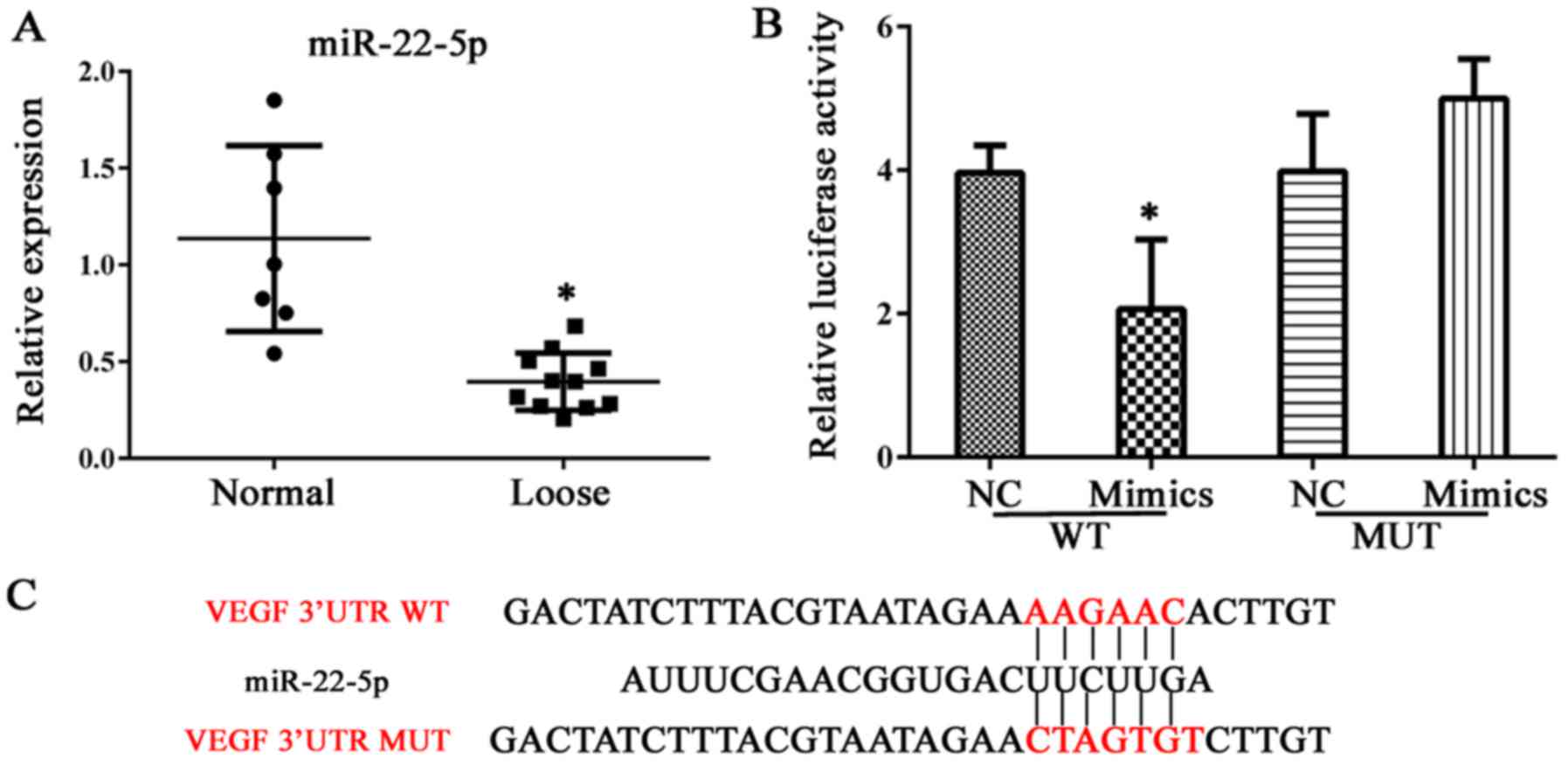
miR-22-5p decreases cell viability and
induces apoptosis in hFOB 1.19 cells, and reduces VEGF
expression
Expression of miR-22-5p was investigated in clinical
samples of abnormal bone tissue. As demonstrated in Fig. 4A, bone erosion was associated with
decreased miR-22-5p production; thus, the role of miR-22-5p in
osteolysis was examined further. VEGF was demonstrated to be a
direct target of miR-22-5p and this conclusion was validated in a
dual luciferase assay, in which transfection of miR-22-5p inhibited
luciferase activity from the plasmid containing the 3′UTR of VEGF
(Fig. 4B and C). Subsequently,
UHMWPE-treated hFOB 1.19 cells were transfected with mimics or an
inhibitor of miR-22-5p. The viability of UHMWPE-treated hFOB 1.19
cells was decreased by miR-22-5p mimics and miR-22-5p inhibitor
(Fig. 5A and B). Western blot
analysis demonstrated that the effect of miR-22-5p on the viability
of UHMWPE-treated hFOB 1.19 cells was associated changes in the
levels of OPG, RANKL and VEGF expression. Consistent with results
of the dual luciferase assay, miR-22-5p mimics reduced VEGF protein
expression (Fig. 5C).
Furthermore, the expression of RANKL, which is reported to function
in conjunction with VEGF during osteoclast differentiation
(25), was also reduced by
miR-22-5p mimics (Fig. 5C). In
contrast to the effects on VEGF and RANKL, expression of the
anti-osteolysis factor OPG was induced by miR-22-5p mimics and
suppressed by miR-22-5p inhibitors (Fig. 5C). Furthermore, apoptosis of
UHMWPE-treated hFOB 1.19 cells was induced by miR-22-5p mimics and
suppressed by an miR-22-5p inhibitor (Fig. 5D). These results demonstrated that
in contrast to the effect of MALAT1, miR-22-5p inhibited the
osteolytic process.
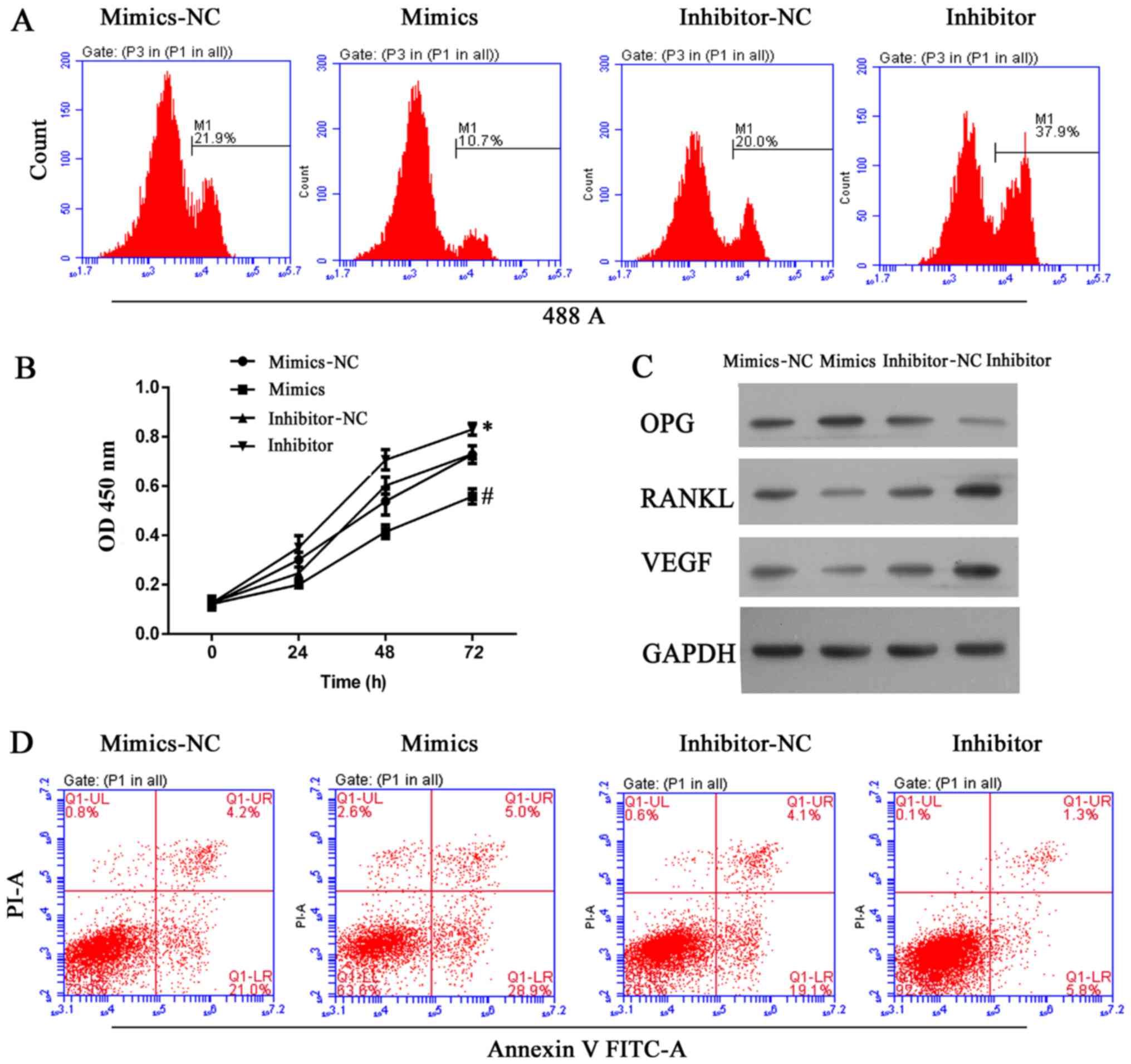 | Figure 5miR-22-5p antagonized osteolysis by
inhibiting cell growth and inducing apoptosis in ultra-high
molecular weight polyethylene-treated hFOB 1.19 cells, and the
effect was associated with upregulation of OPG and downregulation
of RANKL and VEGF. (A) Representative images from
5-ethynyl-2′-deoxyuridine assays after 48 h of transfection. (B)
Quantitative analysis of results from CCK-8 assays at 24, 48 and 72
h after transfection. (C) Representative images from western blot
analyses of OPG, RANKL and VEGF expression after 48 h of
transfection. (D) Representative images showing apoptosis rates as
detected by flow cytometry at 48 h after transfection. All
experiments were repeated at least 3 times. NC, negative control;
OD, optical density; OPG, osteoprotegerin; RANKL, RANKL, receptor
activator of nuclear factor-κB ligand; VEGF, vascular endothelial
growth factor; PI, propidium iodide; FITC, fluorescein
isothiocyanate. |
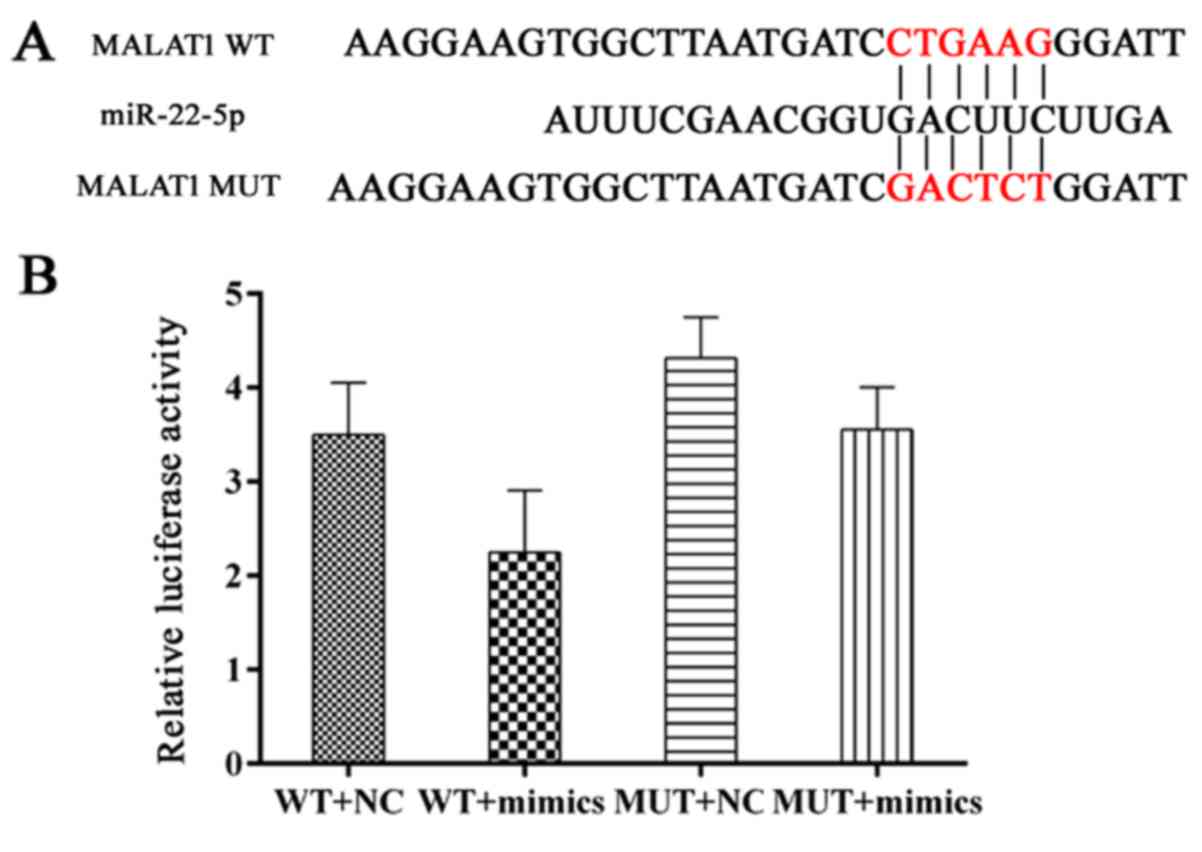
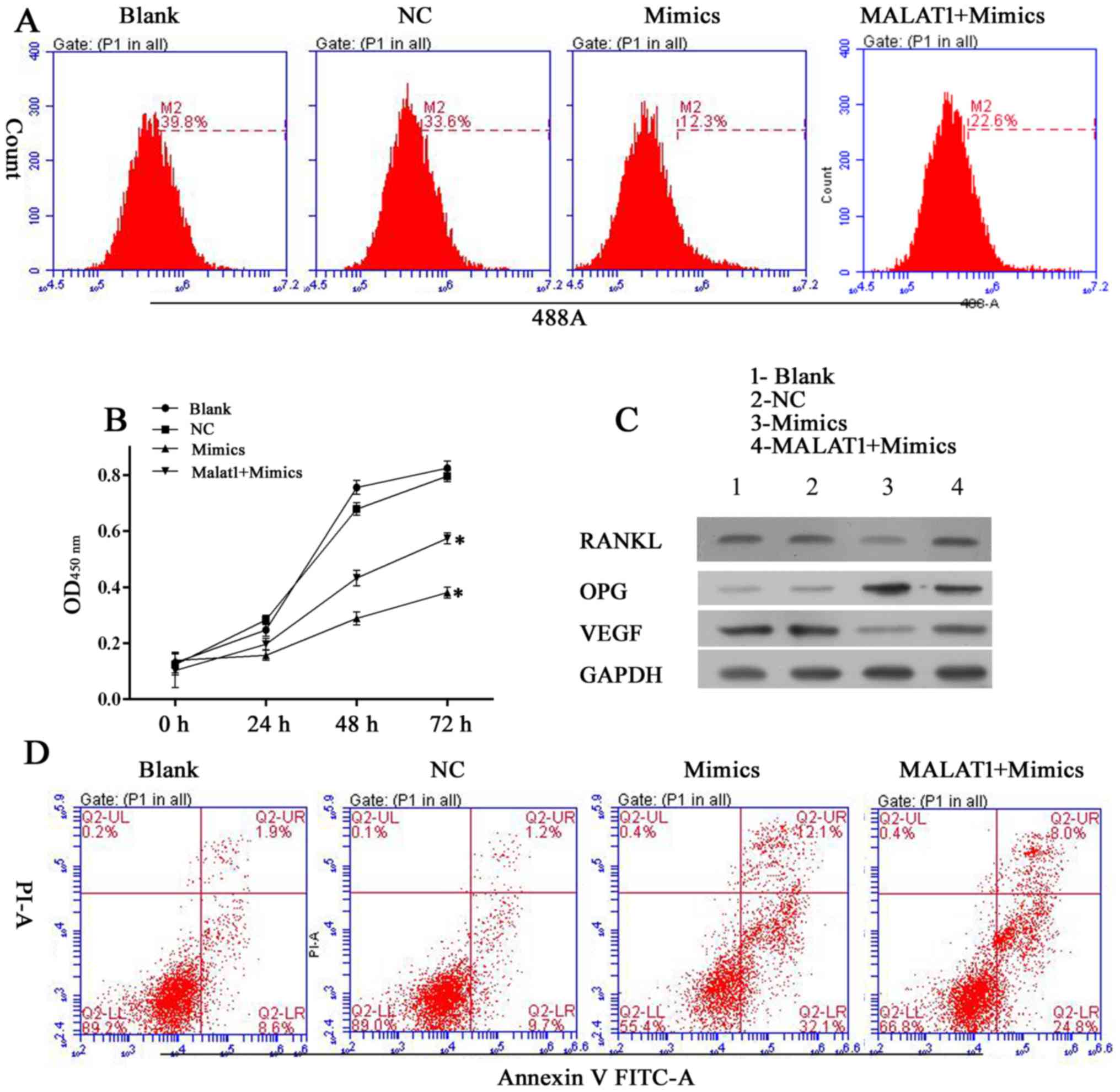
MALAT1 induces osteolysis by upregulating
the expression of VEGF and RANKL
MALAT1 was expressed at relatively high levels in
samples of loose interface membrane tissue compared with normal
samples (Fig. 1E). As
Demonstrated in Fig. 6A and B,
miR-22-5p targets MALAT1 due to its matched sequence and
interaction between MALAT1 and miR-22-5p was validated in a dual
luciferase assay. To further examine the modulating sequence of the
two RNA members, hFOB 1.19 cells were transfected with different
combinations of MALAT1 expression vector and miR-22-5p mimics. It
was observed that when co-transfected, the pro-survival effect of
MALAT1 on hFOB 1.19 cells reduced the anti-survival effect of
miR-22-5p. Furthermore, when tested in the EdU and CCK-8 assays,
cells transfected with pcDNA-MALAT1 and miR-22-5p mimics exhibited
increased viability compared with cells transfected with miR-22-5p
mimics (Fig. 7A and B). The
antagonistic effect of MATAL1 reversed the decreased levels of
RANKL and VEGF expression induced by miR-22-5p mimics (Fig. 7C). The above results indicate that
MALAT1 induces osteolysis, potentially by inhibiting miR-22-5p
expression, and thus further initiating pro-osteolysis signaling
processes.
Discussion
Homeostasis of bone metabolism depends on
maintaining a balance between osteoblastic bone formation and
osteoclastic bone resorption (22). It is commonly recognized that the
transition between osteoblastic and osteoclastic processes is
regulated by the relative levels of OPG and RANKL (26,27). Therefore, upstream regulators of
these two indicators assist in modulating bone metabolism, and
represent potential targets for treating bone erosion associated
with an implanted prosthesis. In the present study, MALAT1 lncRNA
induced the osteoclastic process in UHMWPE-treated hFOB 1.19 cells
by suppressing miR-22-5p activity. This suppression may inhibit
osteolysis by blocking VEGF signaling and increasing RANKL
activity.
MALAT1 is a typical member of the lncRNA family,
which is a novel class of mRNA-like transcripts that have numerous
structural and functional roles cells. Previous studies have
reported that overexpression of MALAT1 modulates alternative
splicing, and is associated with metastasis and a poor prognosis in
patients with lung cancer (28,29). Furthermore, a deficiency of MALAT1
led to enhanced sprouting and migration of endothelial cells in a
sphere model (30). Che et
al (22) reported that MALAT1
knockdown reversed RANKL-induced cell growth inhibition and cell
cycle arrest in normal hFOB 1.19 cells. Additionally, the previous
study also demonstrated that MALAT1 knockdown by siRNA
significantly downregulated the levels of OPG protein in hFOB 1.19
cells, suggesting the modulation of OPG by MALAT1 in osteoblastic
cells (22). In the present
study, it was initially validated that MALAT1 was overexpressed in
clinical samples, and overexpression was associated with a
downregulation of OPG and upregulation of RANKL. In healthy bone
tissue, OPG acts as a soluble receptor antagonist that neutralizes
RANKL, and thus blocks RANKL-RANK interaction. This blocking effect
maintains a balance between osteoblastic and osteoclastic metabolic
processes (26,31,32). To further determine the role of
MALAT1 in osteolysis, hFOB 1.19 cells were pre-treated with UHMWPE,
which induced the osteoclastic process in those cells. The cells
were then transfected with MALAT1 specific siRNA, and cell growth
was inhibited by MALAT1 knockdown. Furthermore, the decrease in
cell growth was accompanied by increased OPG expression, and
suppression of RANKL and VEGF expression. UHMWPE-treated hFOB 1.19
cells were characterized by osteoclastic features; therefore, the
inhibitory effect of MALAT1 knockdown on the osteoclastic process
in those cells confirmed that MALAT1 has a role in the onset of
osteolysis. Accordingly, MALAT1 exerted its function via the
OPG/RANKL pathway. It was also demonstrated that other molecules
have crucial roles in bone metabolism, as VEGF expression (33–35) was also modulated by MALAT1
knockdown.
Results of previous studies suggest that the
potential connection between MALAT1 and VEGF may be modulated by
two separate pathways. Yamakuchi et al (20) reported that VEGF protein
expression was reduced in cells that overexpressed miR-22 (20), and Tang et al (23) reported that MALAT1 protects the
endothelium from oxidized low-density lipoprotein-induced
endothelial dysfunction by competing with miR-22 (23). Those findings justified performing
a comprehensive investigation of the regulating sequence among
those three molecules. In the present study, a dual luciferase
assay was used to validate that VEGF and MALAT1 directly interact
with miR-22-5p. Transfection with miR-22-5p inhibited the growth of
UHMWPE-treated hFOB 1.19 cells, and reduced VEGF expression.
However, it was also found that overexpression of MALAT1 in
UHMWPE-treated hFOB 1.19 cells reversed the inhibiting effect of
miR-22-5p mimics on the cells, and increased VEGF protein levels.
The potential regulatory effect of MALAT1 on miR-22-5p can be
explained by 'competitive endogenous RNA (ceRNA)' theory. That
theory suggests that miRNAs function as gene-regulating non-coding
RNAs by directing the RNA-induced silencing complex towards
miRNA-response elements (MREs) (36,37). These MREs are located on the
3′UTR, coding sequence and 5′UTR of mRNA, or non-protein coding
transcripts such as lncRNAs (38). The vast majority of RNA molecules
harbor several MREs, and are thus repressed by different miRNAs.
This target multiplicity underlies the hypothesis that different
RNAs (either pseudo-targets or legitimate targets) compete for
limited pools of miRNAs (39,40), and thus act as ceRNAs. This
suggests that any regulatory effect of MALAT1 on VEGF may be due to
inhibition of miR-22-5p, and further supports the notion that
MALAT1 induces osteolysis. In addition to the MALAT1/miR-22-5p
mechanism for regulating VEGF, MALAT1 may also induce VEGF activity
via a RANKL-dependent mechanism. Zhang et al (41) reported that VEGF-C is a target
gene of RANKL, and that VEGF-C expression was upregulated by RANKL
in osteoclasts (41). Although
such a mechanism was not explicitly validated in the present study,
the suppressed VEGF levels found after MALAT1 knockdown may
partially reflect participation of a MALAT1/RANKL/VEGF pathway in
the initiation of osteolysis.
In conclusion, the findings of the current study
demonstrate that MALAT1 has a key role in the onset of osteolysis.
MALAT1 was upregulated in interface membrane samples from patients
receiving a knee replacement operation compared with normal
controls. Furthermore, knockdown of MALAT1 inhibited the growth of
UHMWPE-treated hFOB 1.19 cells. The promoting effect of MALAT1 on
the osteoclastic process was exerted via three pathways: i) MALAT1
initiated RANKL signaling by decreasing the OPG level; ii) MALAT1
increased VEGF levels via a mechanism that inhibited miR-22-5p; and
iii) MALAT1 increased VEGF levels in a RANKL-dependent manner.
Thus, MALAT1 is a potential target for treating osteolysis
associated with an implanted prosthesis. However, additional
comprehensive studies are required prior to developing treatment
strategies based on MALAT1.
Acknowledgments
This study was supported by the Fundamental Research
Funds for Central Universities (grant no. 2012QNZT119) and the
National Natural Science Foundation of China (grant no.
8170090962).
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Kandahari AM, Yang X, Laroche KA, Dighe
AS, Pan D and Cui Q: A review of UHMWPE wear-induced osteolysis:
the role for early detection of the immune response. Bone Res.
4:160142016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harris WH: Wear and periprosthetic
osteolysis: the problem. Clin Orthop Relat Res. 393:66–70. 2001.
View Article : Google Scholar
|
|
3
|
Fu C, Xie J, Hu N, Liang X, Chen R, Wang
C, Chen C, Xu C, Huang W and Paul Sung KL: Titanium particles
up-regulate the activity of matrix metalloproteinase-2 in human
synovial cells. Int Orthop. 38:1091–1098. 2014. View Article : Google Scholar :
|
|
4
|
Katsuyama E, Miyamoto H, Kobayashi T, Sato
Y, Hao W, Kanagawa H, Fujie A, Tando T, Watanabe R, Morita M, et
al: Interleukin-1 receptor-associated kinase-4 (IRAK4) promotes
inflammatory osteolysis by activating osteoclasts and inhibiting
formation of foreign body giant cells. J Biol Chem. 290:716–726.
2015. View Article : Google Scholar :
|
|
5
|
Vallés G, Pérez C, Boré A, Martín-Saavedra
F, Saldaña L and Vilaboa N: Simvastatin prevents the induction of
interleukin-6 gene expression by titanium particles in human
osteoblastic cells. Acta Biomater. 9:4916–4925. 2013. View Article : Google Scholar
|
|
6
|
Beck RT, Illingworth KD and Saleh KJ:
Review of periprosthetic osteolysis in total joint arthroplasty: an
emphasis on host factors and future directions. J Orthop Res.
30:541–546. 2012. View Article : Google Scholar
|
|
7
|
Wooley PH and Schwarz EM: Aseptic
loosening. Gene Ther. 11:402–407. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang Y, Jia T, Wooley PH and Yang SY:
Current research in the pathogenesis of aseptic implant loosening
associated with particulate wear debris. Acta Orthop Belg. 79:1–9.
2013.PubMed/NCBI
|
|
9
|
Nakagawa M, Kaneda T, Arakawa T, Morita S,
Sato T, Yomada T, Hanada K, Kumegawa M and Hakeda Y: Vascular
endothelial growth factor (VEGF) directly enhances osteoclastic
bone resorption and survival of mature osteoclasts. FEBS Lett.
473:161–164. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Favus MJ: Primer on the Metabolic Bone
Diseases and Disorders of Mineral Metabolism. Rittenhouse Book
Distributors. 2006.
|
|
11
|
Leung DW, Cachianes G, Kuang WJ, Goeddel
DV and Ferrara N: Vascular endothelial growth factor is a secreted
angiogenic mitogen. Science. 246:1306–1309. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goad DL, Rubin J, Wang H, Tashjian AH Jr
and Patterson C: Enhanced expression of vascular endothelial growth
factor in human SaOS-2 osteoblast-like cells and murine osteoblasts
induced by insulin-like growth factor I. Endocrinology.
137:2262–2268. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gerber HP, Vu TH, Ryan AM, Kowalski J,
Werb Z and Ferrara N: VEGF couples hypertrophic cartilage
remodeling, ossification and angiogenesis during endochondral bone
formation. Nat Med. 5:623–628. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Henriksen K, Karsdal M, Delaisse JM and
Engsig MT: RANKL and vascular endothelial growth factor (VEGF)
induce osteoclast chemotaxis through an ERK1/2-dependent mechanism.
J Biol Chem. 278:48745–48753. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gibb EA, Vucic EA, Enfield KS, Stewart GL,
Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S,
Brown CJ, et al: Human cancer long non-coding RNA transcriptomes.
PLoS One. 6:e259152011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gutschner T and Diederichs S: The
hallmarks of cancer: a long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu B, Peng XC, Zheng XL, Wang J and Qin
YW: miR-126 restoration down-regulate VEGF and inhibit the growth
of lung cancer cell lines in vitro and in vivo. Lung Cancer.
66:169–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cascio S, D'Andrea A, Ferla R, Surmacz E,
Gulotta E, Amodeo V, Bazan V, Gebbia N and Russo A: miR-20b
modulates VEGF expression by targeting HIF-1 alpha and STAT3 in
MCF-7 breast cancer cells. J Cell Physiol. 224:242–249.
2010.PubMed/NCBI
|
|
19
|
Lei Z, Li B, Yang Z, Fang H, Zhang GM,
Feng ZH and Huang B: Regulation of HIF-1alpha and VEGF by miR-20b
tunes tumor cells to adapt to the alteration of oxygen
concentration. PLoS One. 4:e76292009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamakuchi M, Yagi S, Ito T and Lowenstein
CJ: MicroRNA-22 regulates hypoxia signaling in colon cancer cells.
PLoS One. 6:e202912011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang S, Wang S, Bian C, Yang Z, Zhou H,
Zeng Y, Li H, Han Q and Zhao RC: Upregulation of miR-22 promotes
osteogenic differentiation and inhibits adipogenic differentiation
of human adipose tissue-derived mesenchymal stem cells by
repressing HDAC6 protein expression. Stem Cells Dev. 21:2531–2540.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Che W, Dong Y and Quan HB: RANKL inhibits
cell proliferation by regulating MALAT1 expression in a human
osteoblastic cell line hFOB 1.19. Cell Mol Biol. 61:7–14.
2015.PubMed/NCBI
|
|
23
|
Tang Y, Jin X, Xiang Y, Chen Y, Shen CX,
Zhang YC and Li YG: The lncRNA MALAT1 protects the endothelium
against ox-LDL-induced dysfunction via upregulating the expression
of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett.
589:3189–3196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kauther MD, Xu J and Wedemeyer C:
Alpha-calcitonin gene-related peptide can reverse the catabolic
influence of UHMWPE particles on RANKL expression in primary human
osteoblasts. Int J Biol Sci. 6:525–536. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao S, Liu D, Pan F and Wise GE: Effect of
vascular endothelial growth factor on RANK gene expression in
osteoclast precursors and on osteoclastogenesis. Arch Oral Biol.
51:596–602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lacey DL, Timms E, Tan HL, Kelley MJ,
Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S,
et al: Osteoprotegerin ligand is a cytokine that regulates
osteoclast differentiation and activation. Cell. 93:165–176. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Simonet WS, Lacey DL, Dunstan CR, Kelley
M, Chang MS, Lüthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, et
al: Osteoprotegerin: a novel secreted protein involved in the
regulation of bone density. Cell. 89:309–319. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thum T and Fiedler J: LINCing MALAT1 and
angiogenesis. Circ Res. 114:1366–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yasuda H, Shima N, Nakagawa N, Yamaguchi
K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A,
et al: Osteoclast differentiation factor is a ligand for
osteoprotegerin/osteoclastogenesis-inhibitory factor and is
identical to TRANCE/RANKL. Proc Natl Acad Sci USA. 95:3597–3602.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takahashi N, Udagawa N and Suda T: A new
member of tumor necrosis factor ligand family,
ODF/OPGL/TRANCE/RANKL, regulates osteoclast differentiation and
function. Biochem Biophys Res Commun. 256:449–455. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y, Berendsen AD, Jia S, Lotinun S,
Baron R, Ferrara N and Olsen BR: Intracellular VEGF regulates the
balance between osteoblast and adipocyte differentiation. J Clin
Invest. 122:3101–3113. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu K and Olsen BR: Osteoblast-derived VEGF
regulates osteoblast differentiation and bone formation during bone
repair. J Clin Invest. 126:509–526. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tombran-Tink J and Barnstable CJ:
Osteoblasts and osteoclasts express PEDF, VEGF-A isoforms, and VEGF
receptors: possible mediators of angiogenesis and matrix remodeling
in the bone. Biochem Biophys Res Commun. 316:573–579. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hendrickson DG, Hogan DJ, McCullough HL,
Myers JW, Herschlag D, Ferrell JE and Brown PO: Concordant
regulation of translation and mRNA abundance for hundreds of
targets of a human microRNA. PLoS Biol. 7:e10002382009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM and Hatzigeorgiou
AG: DIANA-LncBase: experimentally verified and computationally
predicted microRNA targets on long non-coding RNAs. Nucleic Acids
Res. 41:D239–D245. 2013. View Article : Google Scholar :
|
|
39
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta Stone of a hidden RNA
language. Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Seitz H: Redefining microRNA targets. Curr
Biol. 19:870–873. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Q, Guo R, Lu Y, Zhao L, Zhou Q,
Schwarz EM, Huang J, Chen D, Jin ZG, Boyce BF, et al: VEGF-C, a
lymphatic growth factor, is a RANKL target gene in osteoclasts that
enhances osteoclastic bone resorption through an autocrine
mechanism. J Biol Chem. 283:13491–13499. 2008. View Article : Google Scholar : PubMed/NCBI
|