Introduction
Type 2 diabetes mellitus (T2DM) is one of the most
prevalent metabolic diseases, which is characterized by high levels
of blood glucose as a result of insulin resistance and relative
insulin deficiency. Diabetes is often associated with impaired bone
metabolism and calcium and phosphorus metabolism disorders, causing
secondary osteopenia, osteoporosis and other diabetic bone
diseases. Diabetes interferes with bone formation, increases the
risk of fractures and impedes fracture healing. Jaw bones of DM
patients often display loss of alveolar bone and osteoporosis,
resulting in tooth loosening or even loss (1,2).
Dental implantation is an important method for the repair of
missing teeth. However, diabetes-induced skeletal complications and
an abnormal metabolic environment cause pathological changes that
greatly reduce the success rate of dental implantation (3).
Unlike skeletal limbs, which originate from the
mesoderm, the jaw develops from the neural crest of the embryo. Jaw
injury is repaired by bone marrow mesenchymal stem cells (BMSCs) of
neural crest origin, while limb bone injuries are repaired by
mesoderm-derived BMSCs. Studies that examined the BMSCs of the jaw
and the BMSCs of the tibia indicated that BMSCs of different
sources had different biological characteristics. BMSCs from the
two abovementioned sources exhibited differences in their ability
of bone formation and proliferation, with the osteogenic abilities
of neural crest-derived BMSCs being greater due to the differences
in embryonic origin and HOX gene expression in the process of bone
regeneration (4). Furthermore,
the jaw and skeletal limbs have different patterns of bone
formation, which proceed via intramembrane osteogenesis in the jaw
and via endochondral osteogenesis in skeletal limbs. Gene
expression, including mRNA expression, during bone formation and
the regulatory mechanisms under any stimulation may be different in
bone tissues of these two origins. Previous studies on diabetic
osteopathia have mainly focused on cells or tissues derived from
skeletal limbs. Even fewer studies have focused on the mechanism of
diabetes-associated pathological changes in the jaw, in which teeth
are anchored and with which dental implants are combined.
MicroRNAs (miRNAs/miRs) are noncoding
single-stranded RNA molecules consisting of ~22 nucleotides. miRNAs
are widely distributed in all types of organisms. By binding to the
3′-untranslated region (3′-UTR) of the mRNA of a target gene,
miRNAs may cause their degradation or translational repression
(5,6) and post-transcriptionally regulate
the process of gene expression, which is associated with individual
growth, development and diseases. Various miRNAs have important
roles in numerous human diseases, including diabetes, and are
aberrantly expressed in affected patients/tissues. Studies have
indicated that certain miRNAs are important in the maintenance of
bone development and metabolism. They regulate osteogenesis by
modulating osteogenic transcription factors and their associated
factors. A group of miRNAs, including miR-133, miR-23a and miR-30c,
have been reported to inhibit osteogenesis by targeting the key
transcription factor Runt-related protein 2 (Runx2) (7–12).
Another group of miRNAs, including miR-145, miR-214 and miR-31,
inhibit osteogenesis by targeting Osterix (13–18). Distal-less homeobox 5, special
AT-rich sequence-binding protein 2 and other associated factors
have been reported to be the targets of miRNAs (19–21). miRNA also regulates
osteogenesis-associated signaling pathways to regulate bone
formation. Bone morphogenetic protein (BMP)/transforming growth
factor (TGF)-β and Wnt signaling pathways have been widely reported
to be regulated by a group of miRNAs. miR-29b, miR-135a and
miR-199a have been reported to inhibit osteogenesis by targeting
factors of the BMP/TGF-β pathway (8,22-25), while miR-335-5p, miR-29a and
miR-27a regulate osteogenesis by targeting factors associated with
the Wnt pathway (26–29). While numerous factors and pathways
associated with osteogenesis that have been identified to be
regulated by miRNAs (30), only
few studies have assessed miRNA expression in the jaw bone, and it
has therefore remained elusive whether miRNAs are involved in loss
of alveolar bone and osteoporosis of the jaw in diabetes.
The present study intended to i) determined whether
miRNAs have a role in the regulation of bone loss in the jaw bone
in T2DM, and ii) investigate the underlying mechanisms. The results
indicated that the expression of a group of miRNAs in the mandibles
of diabetic rats is different from that in the mandibles of normal
rats. For the first time, to the best of our knowledge, miR-203-3p
was identified to be upregulated in the diabetic group and to
participate in osteogenesis inhibition. Subsequently, miR-203-3p
was indicated to target the 3′-UTR of Smad1 mRNA, the protein of
which is an important mediator of the BMP/Smad pathway, and to
thereby inhibit Smad1 expression and degrade Smad1 mRNA. Therefore,
the BMP/Smad pathway was repressed by miR-203-3p. The present study
provided a basis for a gene therapeutic approach by inhibition of
miR-203-3p in diabetic jaw bones, which may ameliorate diabetic
osteoporosis and improve fracture healing, tooth stability and
implant osseointegration.
Materials and methods
Experimental animals
A total of 6 male Sprague Dawley (SD) rats (age, 10
weeks; weight, 290–310 g) were purchased from the Experimental
Animal Center of Chongqing Medical University (Chongqing, China).
The rats were maintained in the same room under a 12-h light/dark
cycle (lights on at 08:00 a.m.; 150 lux) with 60±10% relative
humidity and an ambient temperature of 25±2°C in barrier
conditions. The rats had access to sterile chow and water ad
libitum in a specific pathogen-free facility in Chongqing Key
Laboratory of Oral Diseases and Biomedical Sciences (Chongqing,
China). The rats were randomly assigned to a diabetic group and
control group. The diabetic group was treated by intraperitoneal
injection of streptozotocin (STZ; 30 mg/kg body weight;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and was fed a
high-fat diet, which was composed of 66.5% basic feed, 20% sucrose,
10% lard, 2.5% cholesterol and 1% cholic acid salt, for 8 weeks.
Rats were considered to be diabetic when their blood glucose levels
exceeded 16.7 mmol/l at 8 weeks after injection. The control group
was injected with saline instead of STZ and was fed a normal diet.
Another 3 male SD rats (age, 8 weeks; weight, 270–280 g) were used
for isolation of rat (r) BMSCs. The study was approved by the
Ethics Committee of Chongqing Medical University (Chongqing,
China).
Bone collection
Three rats of each group were sacrificed after 8
weeks. Left and right mandibles of every rat were isolated within 5
min and washed with PBS (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). After the teeth were extracted, the mandibles
were placed into liquid nitrogen and transferred into cryogenic
vials (Corning Inc., Corning, NY, USA) and stored at −80°C.
Cell culture and differentiation
rBMSCs were isolated from 3 male SD rats (age, 8
weeks). The bone marrow of the mandibles was flushed out with
α-Modified Eagle's Medium (α-MEM; Gibco; Thermo Fisher Scientific,
Inc.) containing 20% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and seeded in 60-mm dishes. After 72 h of
incubation, the medium was removed and the non-adherent cells were
removed by washing with PBS (Gibco; Thermo Fisher Scientific,
Inc.). The adherent cells were further cultured with α-MEM
containing 10% FBS and passaged twice a week at a 1:3 ratio, and
cells at passage 3–4 were used in the subsequent experiments.
C3H101/2 clone 8 cells from the American Type Culture Collection
(Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) containing 10%
FBS. rBMSCs or C3H101/2 cells were seeded in 24- or 6-well plates
and induced using osteogenic medium with 5.5 or 25 mM glucose plus
50 µg/ml ascorbic acid, 10 nM dexamethasone and 10 mM
β-glycerophosphate. After 7 days, the RNA or protein was extracted
at the indicated times, and the cells were fixed in 4%
paraformaldehyde for histochemical detection of alkaline
phosphatase (ALP) (31).
RNA preparation
The mandibles were ground in liquid nitrogen. The
total RNA of mandibles and cells was isolated with TRIzol reagent
(Ambion; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The quality of the RNA samples for
high-throughput deep sequencing was determined with a Bioanalyzer
2100 (Agilent Technologies, Inc., Santa Clara, CA, USA), and the
purity of the RNA samples for reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) was determined by measuring the
absorbance at 260/280 nm with a Nanodrop spectrophotometer
(NanoDrop; Thermo Fisher Scientific, Inc.).
High-throughput small RNA sequencing
After the left sides of the mandibles of 3 rats of
each group were ground into powder in liquid nitrogen, the RNA was
isolated with TRIzol reagent (Ambion; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The RNA was
subjected to high-throughput small RNA sequencing, which was
performed by the Beijing Genomics Institute (Shenzhen, China). RNA
(18–30 nt) was isolated by PAGE and sequentially ligated to 5′ and
3′ small RNA adapters. The 5′ adapter ligation reaction was
performed at 20°C for 6 h, and RNA (40–60 nt) was isolated by PAGE
and recycled. Following this, the 3′ adapter ligation reaction was
performed at 20°C for 6 h, and RNA (60–800 nt) was isolated by PAGE
and recycled. Complementary DNA (cDNA) was then synthesized and
amplified using proprietary RT primers and amplification primers
(Illumina, Inc., San Diego, CA, USA). The reverse transcription
reaction was performed as follows: 65°C for 10 min, 48°C for 3 min,
42°C for 1 h and 70°C for 15 min with First Strand Master Mix
(Invitrogen; Thermo Fisher Scientific, Inc.) and SuperScript™ II
(Invitrogen; Thermo Fisher Scientific, Inc.). Several rounds of PCR
amplification were performed to enrich the cDNA fragments as
follows: 98°C for 30 sec, 12–15 cycles of 98°C for 10 sec and 72°C
for 15 sec, followed by 72°C for 10 min and 4°C hold. Subsequently,
PCR-amplified fragments were extracted and purified using PAGE. The
cDNA libraries were qualified and quantified with an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc.) and an ABI StepOnePlus
Real-Time PCR machine (Thermo Fisher Scientific, Inc.). The 49-nt
sequence tags from Hiseq sequencing were subjected to data cleaning
analysis to obtain credible clean tags. The difference in
expression between the diabetic group and the control group was
analyzed. miRNA expression levels were visualized by heatmaps
generated by R 3.1.2. TargetScan (http://www.TargetScan.org), miRanda (http://www.microRNA.org) and Pictar (http://pictar.mdc-berlin.de) were used to search for
predicted miRNA targets.
RT-qPCR assay
Total RNA isolated from the right sides of the
mandibles was used for RT-qPCR analysis. The indicated miRNA RT
primer was designed for each selected miRNA, which was
differentially expressed in the diabetic group and control group
detected by high-throughput small RNA sequencing (Table I). The cDNA of miRNA was obtained
according to the instructions of the GoScript™ Reverse
Transcription system (Promega Corp., Madison, WI, USA). Equal
amounts of cDNA were used in duplicate and amplified with
GoTaq® qPCR Master Mix (Promega Corp.). U6 small nuclear
RNA was used as an internal reference. The primers used for qPCR
are displayed in Table II.
 | Table ISequences of reverse transcription
primers for miRs. |
Table I
Sequences of reverse transcription
primers for miRs.
| Gene name | Sequence
(5′-3′) |
|---|
| miR-203a-3p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTAGTG |
| miR-23a-3p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGAAAT |
| miR-96-5p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCAAA |
| miR-30c-5p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTGAG |
| miR-17-5p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT |
| miR-320-3p |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCGCCC |
 | Table IISequences of primers used for
polymerase chain reaction analysis of miRs. |
Table II
Sequences of primers used for
polymerase chain reaction analysis of miRs.
| Gene name | Sequence
(5′-3′) |
|---|
| miR-203a-3p
(F) |
GCGCGTGAAATGTTTAGGAC |
| miR-23a-3p (F) |
TGCATCACATTGCCAGGG |
| miR-96-5p (F) |
TCGTTTGGCACTAGCACATT |
| miR-30c-5p (F) |
GCGCTGTAAACATCCTACACT |
| miR-17-5p (F) |
CGCAAAGTGCTTACAGTGC |
| miR-320-3p (F) |
CGCAAAAGCTGGGTTGAGA |
| miRNA (R) |
GTGCAGGGTCCGAGGT |
| U6 (F) |
CTCGCTTCGGCAGCACA |
| U6 (R) |
AACGCTTCACGAATTTGCGT |
The RNA isolated from mandibles or cells was also
processed using the GoScript™ Reverse Transcription system (Promega
Corp.) with OligodT as the primer. The subsequent qPCR was
performed according to the instructions of the GoTaq®
qPCR Master Mix. β-actin was used as an internal reference. The
forward and reverse PCR primers are displayed in Table III.
 | Table IIISequences of primers used for
polymerase chain reaction analysis of mRNAs. |
Table III
Sequences of primers used for
polymerase chain reaction analysis of mRNAs.
| Gene name | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| β-actin (m) |
CAGCCTTCCTTCTTGGGTAT |
TGGCATAGAGGTCTTTACGG |
| Runx2 (m) |
GGGAACCAAGAAGGCACAGA |
GGATGAGGAATGCGCCCTAA |
| Smad1 (m) |
CAGAGGAGATGTTCAGGCAGTT |
CTGTGAAACCATCCACCAGC |
| ALP (m) |
TCGGAACAACCTGACTGACC |
GTCAATCCTGCCTCCTTCCA |
| Osterix (m) |
TGCTTGAGGAAGAAGCTCACTA |
TGAAAGGTCAGCGTATGGCT |
| OC (m) |
ACCTCACAGATGCCAAGCC |
GCCGGAGTCTGTTCACTACC |
| β-actin (r) |
CCCGCGAGTACAACCTTCTTG |
GTCATCCATGGCGAACTGGTG |
| Col1a1 (r) |
AAGTCTCAAGATGGTGGCCG |
TACTCTCCGCTCTTCCAGTCA |
| Runx2 (r) |
CCGAGACCAACCGAGTCATTTA |
AAGAGGCTGTTTGACGCCAT |
| Smad1 (r) |
TCAATAGAGGAGATGTTCAAGCAGT |
GAAACCATCCACCAACACGC |
| ALP (r) |
CCTGCAGGATCGGAACGTCAATTA |
TGAGTTGGTAAGGCAGGGTCC |
| Osterix (r) |
GCCAGTAATCTTCGTGCCAG |
TAGTGAGCTTCTTCCTGGGGA |
| OC (r) |
CTCAACAATGGACTTGGAGCC |
GGCAACACATGCCCTAAACG |
All PCRs were performed with a Bio-Rad CFX Manager
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). The cycling
profile consisted of an initial denaturation at 95°C for 10 min,
followed by 40 cycles of 95°C for 15 sec, 60°C for 60 sec and 95°C
for 15 sec, followed by melt curve analysis. RNA fold changes
between different groups were calculated by the 2−∆∆Cq
method (32).
Western blot analysis
Cells were washed with pre-cooled PBS twice and
treated with radioimmunoprecipitation assay buffer (RIPA; Beyotime
Institute of Biotechnology, Haimen, China) containing 1 mM
phenylmethane sulfonyl fluoride and phosphatase inhibitor (Beyotime
Institute of Biotechnology) for 5 min, and the cells and the
lysates were collected and centrifuged at 4°C for 10 min at 13,400
× g. The supernatant was collected in fresh tubes, and the
concentration was detected with a BCA protein assay kit (Beyotime
Institute of Biotechnology) according to the manufacturer's
instructions. After the concentration of every sample was adjusted
with RIPA, the samples were added to loading buffer (WB0091;
Beijing Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China)
and boiled for 5 min. The treated samples were stored at −80°C for
further analysis. For western blot analysis, total protein (50
µg per lane) was separated using 10% SDS-PAGE according to
the instructions of the SDS-PAGE preparation gel kit (Beijing
Dingguo Changsheng Biotechnology Co., Ltd.), followed by transfer
onto polyvinylidene difluoride membranes (EMD Millipore, Billerica,
MA, USA). The membranes were blocked using Tris/HCl-buffered salt
solution containing 0.1% Tween-20 and 5% bovine serum albumin
(Sigma-Aldrich; Merck KGaA) for 2 h at room temperature. The
membranes were incubated with specific antibodies to Runx2
(1:1,000; cat. no. sc-10758; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), β-actin (1:1,000; cat. no. 13E5), Smad1 (1:1,000;
cat. no. D59D7), phosphorylated (p)-Smad1 (1:1,000; cat. no. D40B7)
and p-Smad1/5/8 (1:1,000; cat. no. D5B10; Cell Signaling
Technology, Inc., Danvers, MA, USA) overnight at 4°C. Following
this, the membranes were washed three times in 0.1% TBST and
incubated for 1.5 h with the secondary antibody (1:2,000; cat. no.
A21020-1; Abkine, Wuhan, HB, China) at room temperature. Blots were
developed using an enhanced chemiluminescence (ECL) kit (BeyoECL
Plus; Beyotime Institute of Biotechnology), and images were
captured using a Bio-Rad Moleculer Imager® ChemiDoc™ XRS
Imaging system (Bio-Rad Laboratories, Inc.). Densitometry was
performed with Quantity One® Software v. 4.6.9 (Bio-Rad
Laboratories, Inc.).
Plasmid construction
Plasmid construction was performed by Lengshuiquan
Biotechnology Co., Ltd. (Chengdu, China). The RNA of mouse livers
was extracted and reverse transcribed into cDNA. The wild-type
3′-UTR of Smad1 and a mutated sequence were amplified by PCR using
primers with XbaI and FseI restriction sites. The
wild-type primer sequences were
5′-TGTTCTAGAAGACCTGTGGCTTCCGTCTCTTG-3′ (forward) and
5′-AATGGCCGGCCTTATTTAAAAATTTCCAGTAGAG-3′ (reverse), and the mutant
primer sequences were 5′-TGTTCTAGAAGACCTGTGGCTTCCGTCTCTTG-3′
(forward) and 5′-ATAGAGAGATCTTTAATCCTGCGAATAATGAAC-3′ (reverse).
The PCR product was purified and then inserted into the
XbaI-FseI site of the pGL3-control vector (Promega,
Madison, WI, USA), and it was identified by DNA sequencing. The
wild-type plasmid containing the 3′-UTR of Smad1 with the
complementary sequence of miR-203-3p (pGL3-Smad1 3′-UTR WT) and the
mutant plasmid with the mutation sequence containing the 3′-UTR of
Smad1 without the complementary sequence of miR-203-3p (pGL3-Smad1
3′-UTR mut) were created.
To construct a Smad1 expression vector,
cytomegaloviral vector (pCMV)-Smad1, the Smad1 coding sequence was
RT-PCR-amplified from mouse mRNA using the primer sequences
5′-GTGTAGAATTCGACCAGCCGCTATGAATGT-3′ (forward) and
5′-TTTGCTCGAGACGGAAGCCACAGGTC-3′ (reverse). The PCR product was
digested with EcoRI-XbaI and sub-cloned into the
similarly digested pCMV vector (Invitrogen; Thermo Fisher
Scientific, Inc.). The plasmids were sequenced to ensure
authenticity.
miRNA and plasmid transfection
Prior to transfection, cells were seeded into 6-well
or 24-well plates and were transfected at 50–70% confluence.
Agomir-203-3p, Agomir-NC, Antagomir-203-3p and Antagomir-NC were
purchased from GenePharma Co., Ltd. (Shanghai, China) and were
trans-fected into indicated cells at a concentration of 100 nM with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in Opti-MEM® (Gibco; Thermo Fisher
Scientific, Inc.). Smad1 overexpression plasmids were transfected
into cells at 1 µg per well for 24-well plates or 4
µg per well for 6-well plates. After 6 h of transfection,
the Opti-MEM® was replaced by medium with 10% FBS.
Luciferase reporter assay
C3H10T1/2 cells were seeded in 24-well plates,
co-transfected with three luciferase constructs containing
pGL3-Smad1 3′-UTR wild or mut, phRL-null (Renilla plasmid
for normalization) and 100 nm Agomir-203-3p or Agomir-NC.
Luciferase assays were performed using the Promega Dual Luciferase
Assay system (cat. no. E1910; Promega Corp.) according to the
manufacturer's instructions. Three independent experiments were
performed for 24 h and assayed in quadruplicate per group. Relative
luciferase activity (firefly/Renilla) was detected with a
Promega Glomax (Promega Corp.).
Statistical analysis
GraphPad Prism 5 software (GraphPad Software, Inc.,
La Jolla, CA, USA) was used for all statistical tests. Values are
expressed as the mean ± standard deviation. For comparisons between
2 groups, a two-tailed Student's t-test was used. For comparisons
between multiple groups, one-way analysis of variance followed by
Tukey's test was applied. RT-qPCR assays, western blot analysis and
luciferase reporter assays were repeated at least three times and
representative results are presented. P<0.05 was considered to
indicate a statistically significant difference.
Results
Osteogenesis is downregulated in the
mandibles of diabetic rats and high-glucose-treated rBMSCs
The mRNA expression of osteogenic genes in the
mandibles of SD rats with high-fat diet- and STZ-induced T2DM as
well as in normal rats was detected by RT-qPCR. The osteogenic
genes were significantly down-regulated in the mandibles of
diabetic rats (Fig. 1A), which
was consistent with the results of another study (33). rBMSCs were osteogenically induced
in high-glucose or normal medium, and the histochemical detection
of ALP on day 7 indicated that ALP expression was downregulated in
cells grown in high-glucose medium (Fig. 1B). The protein expression of the
osteogenic key transcriptional factor Runx2 in cells cultured in
high-glucose medium was downregulated on days 1, 4, 7, 10, 14 and
21 (Fig. 1C), and the mRNA
expression of osteogenic genes was also downregulated during
osteogenesis; however, towards the end of the in vitro
experiment there was no significant difference for Runx2 and Alp
(Fig. 1D–G). These results
suggested that diabetes and high glucose levels repress
osteogenesis.
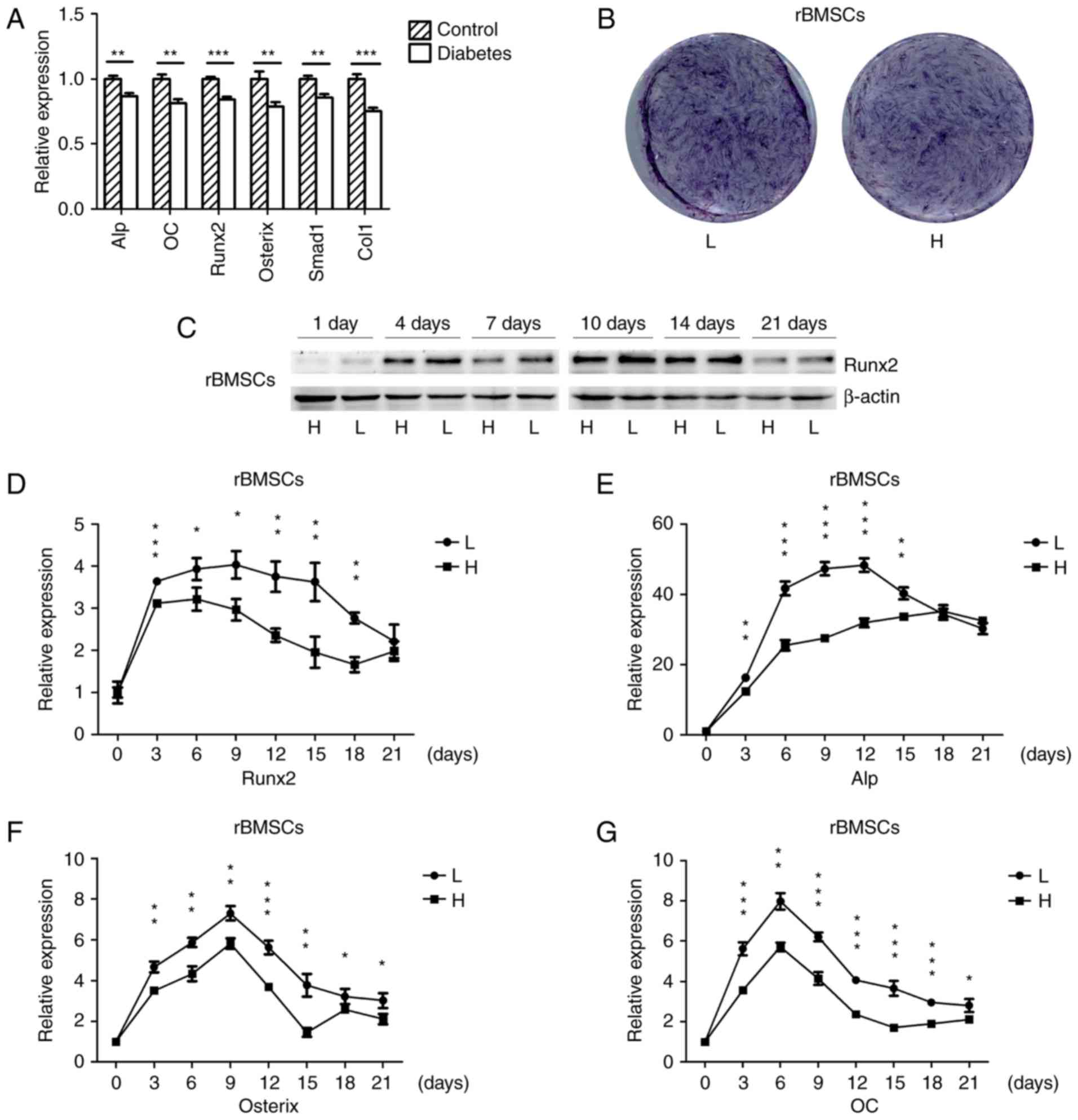 | Figure 1Osteogenesis is downregulated in the
mandibles of diabetic rats and high-glucose-treated rBMSCs. (A)
RT-qPCR analysis of the mRNA expression of ALP, OC, Runx2, Osterix,
Smad1 and Col1 in the mandibles of high-fat diet- and STZ-induced
type 2 diabetic, as well as in normal rats. (B) Histochemical
detection of ALP of osteogenically-induced rBMSCs in high-glucose
or normal medium on day 7 of culture. (C) Protein expression of
Runx2 in rBMSCs osteogenically induced for 1, 4, 7, 10, 14 and 21
days was detected by western blot analysis. (D–G) RT-qPCR analysis
of mRNA expression of Runx2, ALP, Osterix and OC in rBMSCs
osteogenically induced in high-glucose and low-glucose medium for
0, 3, 6, 9, 12, 15, 18 and 21 days. Values are expressed as the
mean ± standard deviation (n=3). *P<0.05,
**P<0.01, ***P<0.001. RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; L,
low-glucose medium; H, high-glucose medium; rBMSCs, rat bone marrow
mesenchymal stem cells; ALP, alkaline phosphatase; Runx2,
runt-related transcription factor 2; Col1, collagen type 1 α 1; OC,
osteocalcin. |
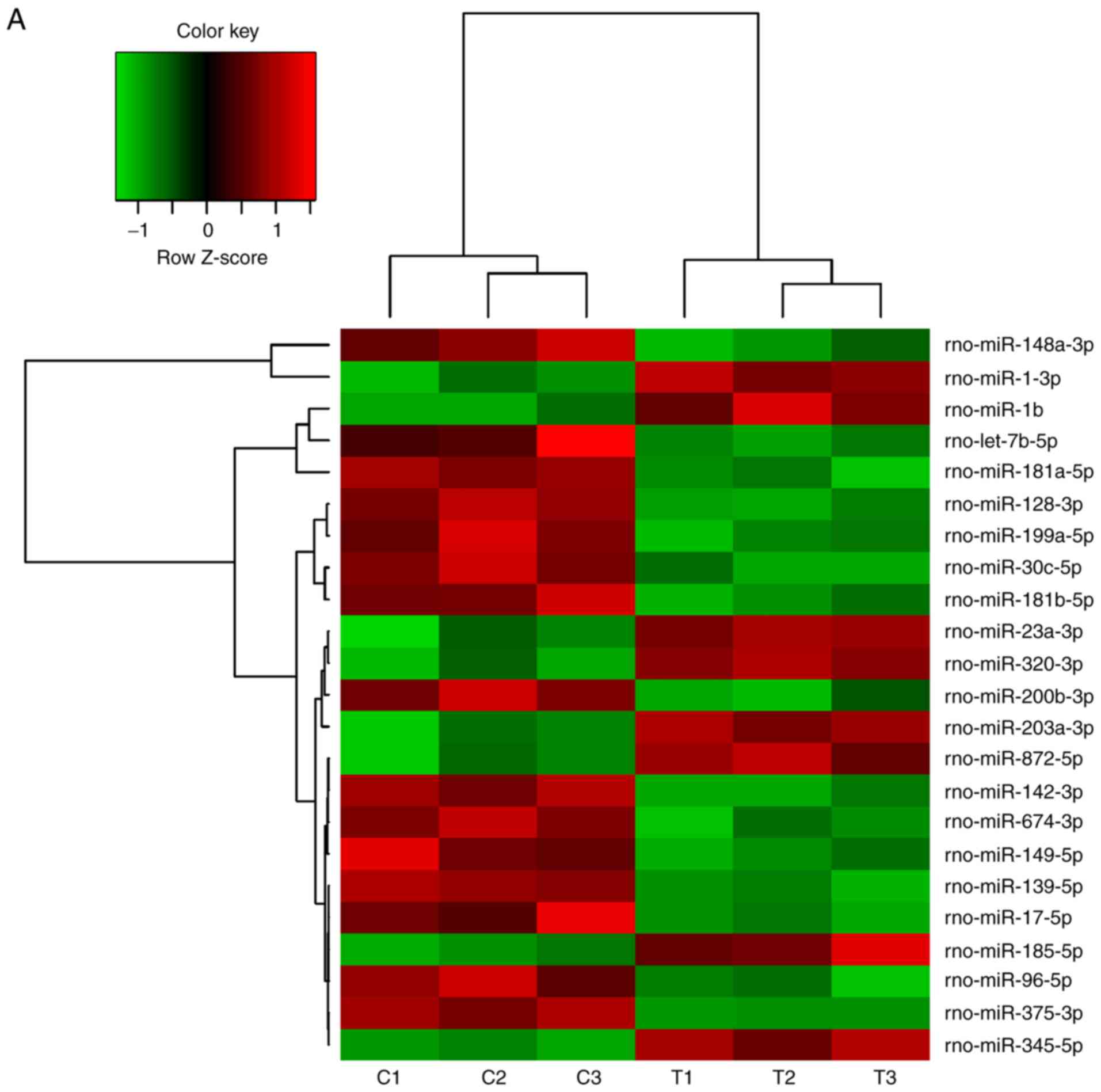
Changes of miRNA expression in mandibles
of diabetic rats
A number of miRNAs has been verified to regulate
osteogenesis in various studies (30). In a previous study, the miR-17~92
cluster was reported to critically regulate osteoblast
differentiation (26). Therefore,
the present study used high-throughput small RNA sequencing to
detect any differences in miRNA expression between experimental
T2DM rats and normal rats. The heat map indicates that 126 miRNAs
were differently expressed between the two groups of rats (Fig. 2A). RT-qPCR was performed to verify
these results on the miRNAs from the high-throughput sequencing.
miR-30c-5p, miR-96-5p and miR-17-5p were down-regulated in the
diabetic group, while miR-203a-3p, miR-23a-3p and miR-320-3p were
upregulated (Fig. 2B–G), which
was consistent with the results of the high-throughput
sequencing.
Gain- and loss-of-function analyses
Agomir-203-3p, antagomir-203-3p and their negative
control were transfected into C3H10T1/2 cells or rBMSCs (Fig. 3A). Cells were osteogenically
induced and fixed in 4% paraformaldehyde for histochemical
detection of ALP on day 7. Histological staining for ALP, an early
marker of bone formation, revealed that miR-203-3p reduced ALP and
inhibited osteogenesis (Fig. 3B).
The protein expression of the key transcriptional factor Runx2 was
inhibited by miR-203-3p and promoted by the inhibitor of miR-203-3p
on days 4, 7 and 14 of osteogenesis (Fig. 3C). The mRNA expression of several
osteogenic genes was detected by RT-qPCR on days 1, 4, 7 and 14
(Fig. 3D–G). The expression of
Runx2 and ALP was inhibited by miR-203-3p and promoted by its
inhibitor (Fig. 3D and E);
similarly, the expression of Osterix and osteocalcin (OC) was
retarded by miR-203-3p and promoted by its inhibitor (Fig. 3F and G), which suggested that
miR-203-3p inhibits osteogenesis in C3H10T1/2 cells and rBMSCs.
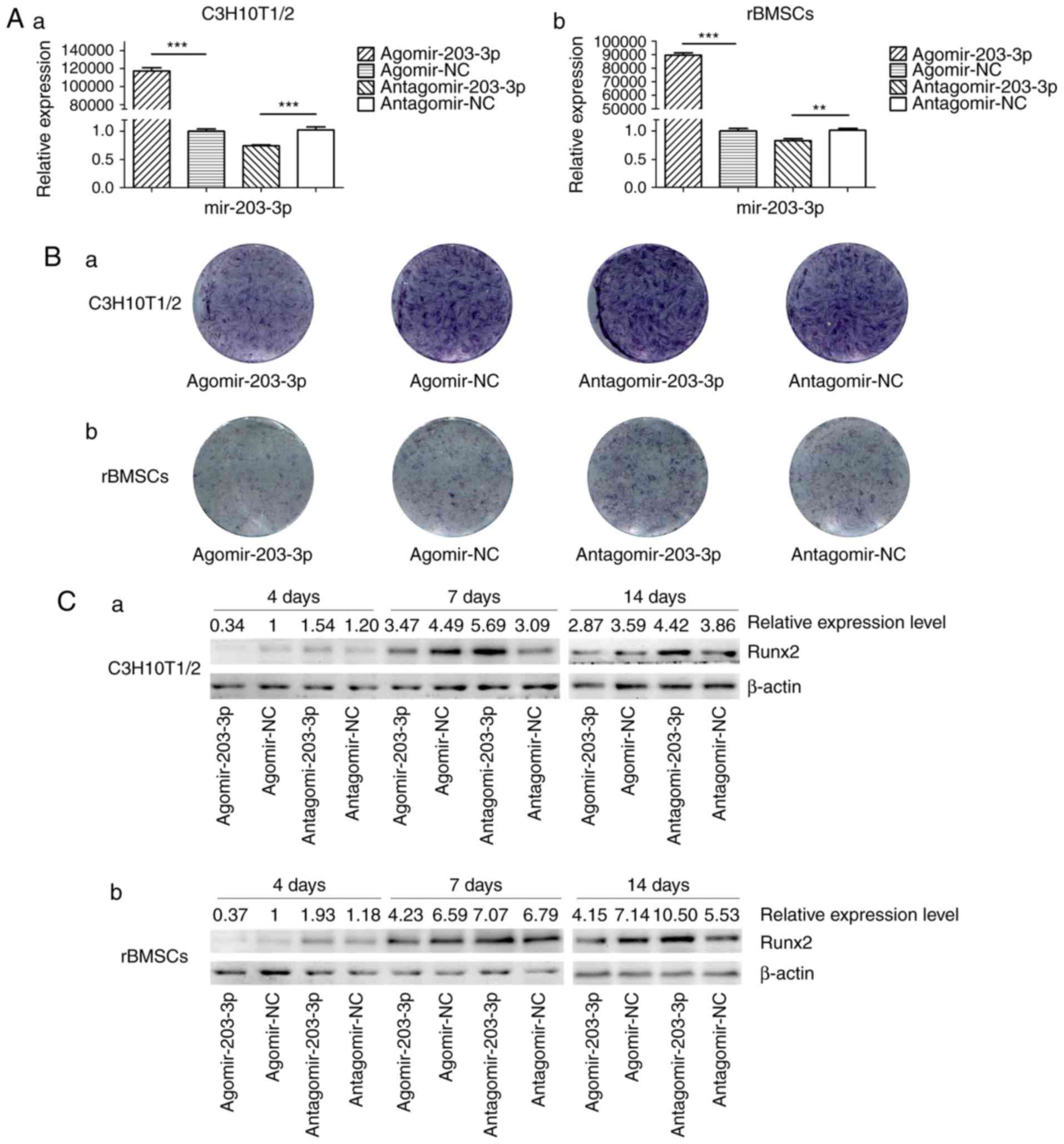 | Figure 3Gain- and loss-of-function
experiments. (A) RT-qPCR analysis of levels of miR-203-3p
expression in (a) C3H10T1/2 cells and (b) rBMSCs trans-fected with
agomir-203-3p, agomir-NC, antagomir-203-3p and antagomir-NC. (B)
Cells were osteogenically induced and fixed in 4% paraformaldehyde
for histochemical detection of ALP after 7 days of culture.
Representative histological staining images (magnification, ×1.4)
for ALP (ALP staining) of (a) C3H10T1/2 cells and (b) rBMSCs
transfected with agomir-203-3p, agomir-NC, antagomir-203-3p and
antagomir-NC are displayed. (C) Protein expression of Runx2 in (a)
C3H10T1/2 cells and (b) rBMSCs transfected with agomir-203-3p,
agomir-NC, antagomir-203-3p and antagomir-NC after 4, 7 and 14 days
of osteogenic induction detected by western blot analysis,
revealing that miR-203-3p inhibited the expression of Runx2, while
the inhibitor of miR-203-3p promoted its expression. (D-G) The mRNA
expression of (D) Runx2, (E) ALP, (F) Osterix and (G) OC in (a)
C3H10T1/2 cells and (b) rBMSCs transfected with agomir-203-3p,
agomir-NC, antagomir-203-3p and antagomir-NC after 1, 4, 7 and 14
days of osteogenic induction was detected by RT-qPCR. The
expression of Runx2 and ALP was inhibited by miR-203-3p and
promoted by its inhibitor, while the expression of Osterix and OC
was retarded by miR-203-3p and promoted by its inhibitor. Values
are expressed as the mean ± standard deviation (n=3).
*P<0.05, **P<0.01,
***P<0.001. miR, microRNA; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; rBMSCs, rat
bone marrow mesenchymal stem cells; ALP, alkaline phosphatase;
Runx2, runt-related transcription factor 2; OC, osteocalcin; NC,
negative control; Agomir, miR agonist; Antagomir, miR
antagonist. |
miR-203-3p directly targets Smad1
Using TargetScan software, a putative binding site
for miR-203-3p was identified in the 3′-UTR of Smad1 mRNA, which is
highly conserved in mammals (Fig.
4A). To determine whether Smad1 is the direct target of
miR-203-3p, agomir-203-3p, antagomir-203-3p and their negative
control were transfected into in C3H10T1/2 cells or rBMSCs
(Fig. 4B), and after 48 h of
osteogenesis, the expression of Smad1 was assessed by western blot
analysis. The results demonstrated that the protein levels of Smad1
were repressed following enhancement of miR-203-3p and increased
following inhibition of miR-203-3p (Fig. 4Ca). The protein levels of further
Smads were detected in C3H10T1/2 cells or rBMSCs. Western blot
analysis indicated that enhancement of miR-203-3p obvi ously
repressed the protein levels of Smad1, p-Smad1 and p-Smad1/5/8 in
C3H10T1/2 cells or rBMSCs (Fig.
4Cb). Furthermore, the mRNA expression of Smad1 was depressed
by overexpression of miR-203-3p in C3H10T1/2 cells and rBMSCs
following 24 and 48 h of osteogenesis (Fig. 4D). Furthermore, enhancement of
miR-203-3p suppressed the expression of Smad1 in C3H10T1/2 cells
and rBMSCs, while inhibition of miR-203-3p resulted in the
promotion of Smad1 at various time-points of osteogenesis. Agomir
delayed the expression of Smad1 so that, in the osteogenic process,
the Agomir group demonstrated the highest expression levels of
Smad1 later than the Agomir-NC group. At day 14, Smad1 appeared to
be increased in the Agomir group compared with that observed in the
Agomir-NC group (Fig. 4E). To
demonstrate the direct interaction between miR-203-3p and Smad1
mRNA, a pGL3 luciferase reporter system containing a binding site
(Smad1-3′-UTR WT) or a mutated site (Smad1-3′-UTR mut) located in
the downstream region of the luciferase reporter gene was
constructed. C3H10T1/2 cells were co-transfected with pGL3-Smad1-WT
or pGL3-Smad1-mut, phRL-null and agomir-203-3p or agomir-NC. The
luciferase activity of the pGL3-Smad1 vector in the agomir-203-3p
group was decreased by 37.3% (P<0.01) compared with that in the
agomir-NC group, while agomir-203-3p did not affect the luciferase
activity of the pGL3-Smad1 mut vector (Fig. 4F). These results were consistent
with the bioinformatics prediction that the 3′-UTR of Smad1 mRNA is
a direct target of miR-203-3p (Fig.
4A).
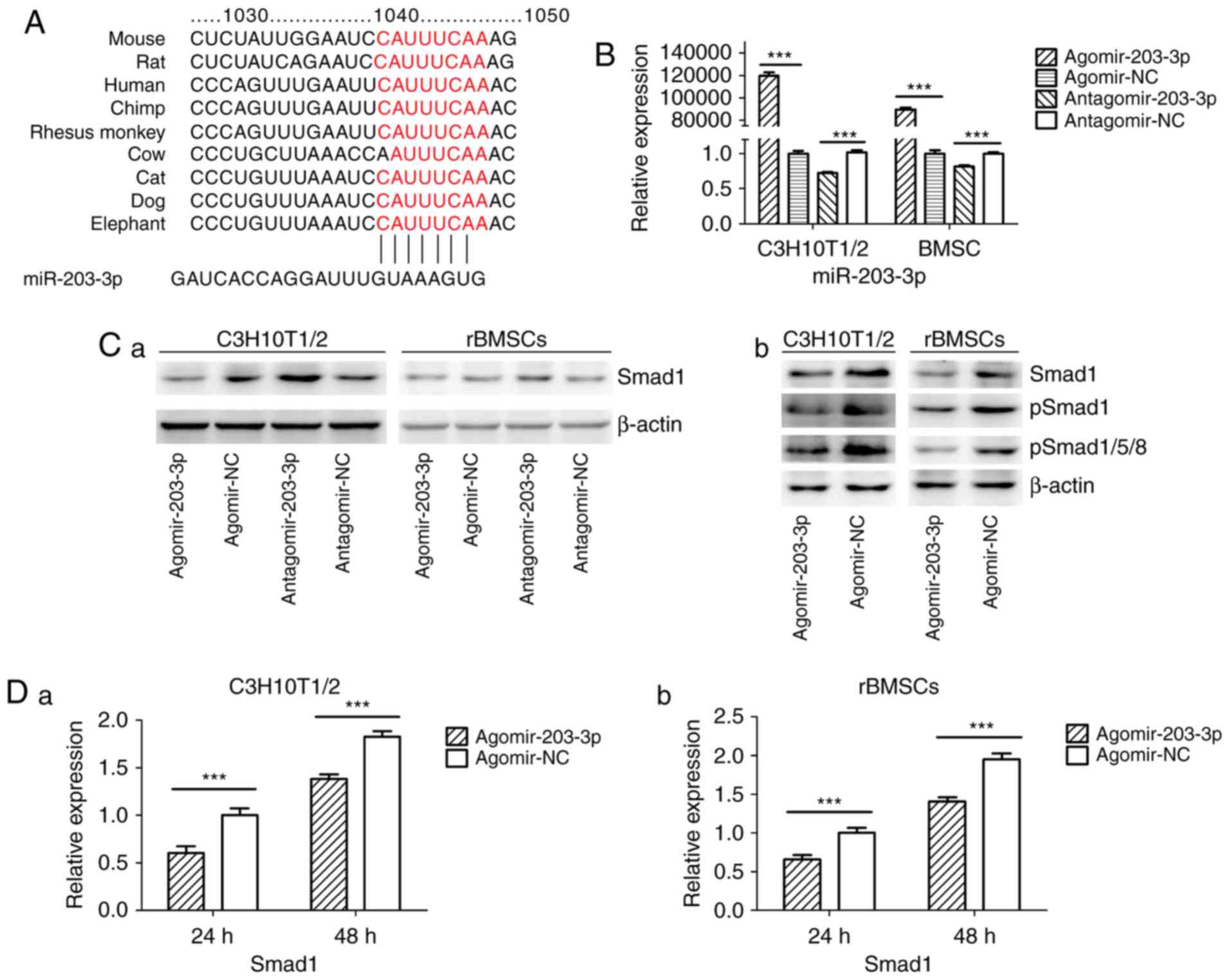 | Figure 4miR-203-3p targeting the Smad1 gene.
(A) A putative target site of miR-203-3p, which is highly conserved
in mammals, was predicted to be located in the 3′-UTR of Smad1
mRNA, by using TargetScan software. The numbers represent the
position of the 'seed region' matching miR-203-3p within the 3′-UTR
sequences. (B) Reverse transcription-quantitative polymerase chain
reaction analysis of levels of miR-203-3p expression in C3H10T1/2
cells and rBMSCs transfected with agomir-203-3p, agomir-NC,
antagomir-203-3p and antagomir-NC. (C) Protein was extracted at 48
h after transfection. (a) Western blot analysis demonstrated that
the protein levels of Smad1 were repressed by overexpression of
miR-203-3p and promoted by inhibition of miR-203-3p. (b) Expression
of bone morphogenetic protein/Smad pathway-associated proteins in
C3H10T1/2 cells and rBMSCs. Representative western blot images
demonstrate that Smad1, p-Smad1 and p-Smad1/5/8 levels are
repressed by overexpression of miR-203-3p. (D) mRNA was extracted
at 24 and 48 h after transfection. The mRNA expression of Smad1 was
depressed by overexpression of miR-203-3p in (a) C3H10T1/2 cells
and (b) rBMSCs. (E) Smad1 protein levels in (a) C3H10T1/2 cells and
(b) rBMSCs transfected with agomir-203-3p, agomir-NC,
antagomir-203-3p and antagomir-NC on the days 4, 7 and 14 of
osteogenic induction was detected by western blot analysis,
revealing that overexpression of miR-203-3p caused the expression
of Smad1 to be (a) depressed and retarded in C3H10T1/2 cells and
(b) to be depressed in rBMSCs. Overexpression of miR-203-3p
suppressed the expression of Smad1 in (a) C3H10T1/2 cells and (b)
rBMSCs, while inhibition of miR-203-3p resulted in the promotion of
Smad1. Agomir delayed the expression of Smad1 so that, in the
osteogenic process, the Agomir group demonstrated the highest
expression levels of Smad1 later than the Agomir-NC group. At day
14, Smad1 appeared to be increased in the Agomir group compared
with that observed in the Agomir-NC group. (F-a) A luciferase
reporter system containing a binding site (Smad1-3′-UTR-WT) or a
mutated site (Smad1-3′-UTR-mut) located in the downstream region of
the pGL3 luciferase reporter gene was constructed. C3H10T1/2 cells
were co-transfected with pGL3-Smad1 or pGL3-Smad1-mut, phRL-null
and Agomir-203-3p or Agomir-NC. (b) Compared with the negative
control, the luciferase activity in the group co-transfected with
Agomir-203-3p and pGL3-Smad1-WT was decreased by 37.3%, while
Agomir-203-3p did not affect the luciferase activity of the
pGL3-Smad1-mut vector. Values are expressed as the mean ± standard
deviation (n=3). ***P<0.001. miR, microRNA; NC,
negative control; Agomir, miR agonist; Antagomir, miR antagonist;
UTR, untranslated region; mut/Mt, mutant; WT, wild-type; rBMSCs,
rat bone marrow mesenchymal stem cells; pSmad, phosphorylated Smad;
Luc, luciferase. |
Ectopic overexpression of Smad1 abrogates
the suppressive effect of miR-203-3p on osteogenesis
To further investigate the association between
miR-203-3p and Smad1, a Smad1 overexpression plasmid was
constructed and transfected into C3H10T1/2 cells (Fig. 5A); in addition, the Smad1
overexpression plasmid was co-transfected with agomir-203-3p or
agomir-NC into C3H10T1/2 cells and overexpression of Smad1 protein
was confirmed by western blot analysis (Fig. 5B). Ectopic expression of Smad1
appeared to attenuate the miR-203-3p-mediated suppression of
osteogenic differentiation (Fig. 5C
and D). These results indicated that miR-203-3p inhibited
osteogenic differentiation, in part via the downregulation of Smad1
expression.
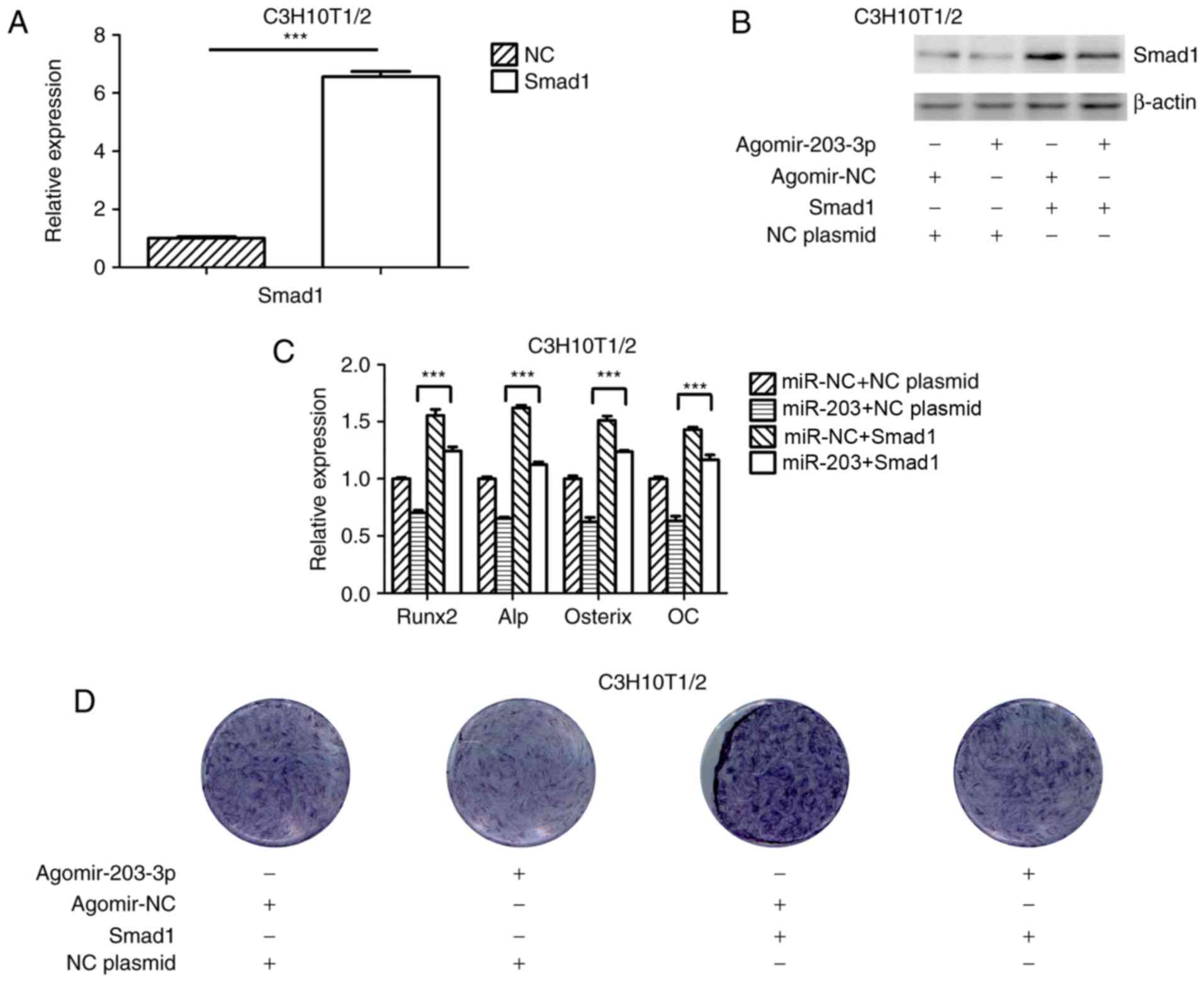 | Figure 5Overexpression of Smad1 rescues the
repression of osteogenesis by miR-203-3p. (A) RT-qPCR analysis of
levels of Smad1 expression in C3H10T1/2 cells transfected with
Smad1 overexpression plasmid and NC plasmid. (B) Smad1 protein
expression in C3H10T1/2 cells transfected with Smad1 overexpression
plasmid or NC plasmid and agomir-203-3p or agomir-NC was detected
by western blot analysis. (C and D) Smad1 overexpression plasmid or
NC plasmid and agomir-203-3p or agomir-NC were co-transfected into
C3H10T1/2 cells, followed by osteogenic induction for 7 days. (C)
RT-qPCR was used to analyze the expression levels of Runx2, Alp,
Osterix and OC. (D) Histochemical detection of ALP in C3H10T1/2
cells (magnification, ×1.4). Overexpression of Smad1 appeared to
attenuate the miR-203-3p-mediated suppression of osteogenic
differentiation. Values are expressed as the mean ± standard
deviation (n=3). ***P<0.001. miR, microRNA; NC,
negative control; Agomir, miR agonist; Antagomir, miR antagonist;
ALP, alkaline phosphatase; Runx2, runt-related transcription factor
2; OC, osteocalcin; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction. |
Discussion
Diabetes mellitus is a metabolic disorder that
affects bone metabolism in the entire body, which, in turn,
inevitably affects the bone metabolism and bone mass in the jaw
(1,2). Unlike skeletal limbs, which
originate from the mesoderm, the jaw is derived from the neural
crest of the embryo. Jaw injury is repaired by BMSCs of neural
crest origin, while limb injury is repaired by BMSCs derived from
the mesoderm. Studies that examined BMSCs of the jaw and of the
tibia demonstrated that BMSCs of different sources had different
biological characteristics (4).
To date, studies on the effects of diabetes on osteogenesis have
mostly focused on skeletal limbs and cells cultured in
vitro, while few studies have focused on the jaw bone (34–36). Therefore, to determine the
potential tissue-specific factors that affect the stability of
teeth and dental implants, DM-associated changes in jaw bones were
assessed. In the present study, a rat model of T2DM was generated
and the osteogenesis-associated gene expression in mandibles was
assessed, revealing that osteogenesis was inhibited in the diabetic
group, as were the cells cultured with high-glucose medium in
vitro, which was consistent with the results of previous
studies (34–36).
miRNAs may act as developmental regulators and
modulate physiological and pathological processes by
post-transcriptionally inhibiting gene expression (37). Previous studies have discovered
that multiple miRNAs are involved in osteogenic differentiation. In
the present study, it was hypothesized that certain miRNAs
participate in inhibition of osteogenesis in diabetic jaw bones. To
assess this, high-throughput small RNA sequencing was performed,
which determined that the expression of a number of miRNAs in the
mandibles of diabetic rats was different from that in the mandibles
of normal rats, which was verified by RT-qPCR. Among them, the
expression of certain miRNAs was consistent with the results
obtained by screening BMSCs cultured under different glucose
concentrations in vitro, including miR-181a-5p, miR-345-5p
and miR-872-5p, while other miRNAs were not, including miR-149-5p,
miR-185-5p and miR-674-3p (38).
Therefore, there are differences between the in vivo and
in vitro results, indicating that high-glucose culture may
not completely simulate diabetic conditions, and better models need
to be established. A literature review did not provide any studies
that had performed miRNA screening of bone in diabetic conditions,
not even of skeletal limbs. In the diabetic group, the present
study revealed and upregulation of miR-203-3p, which has been
reported to regulate the inflammatory response, inhibit the
progression of several types of tumor and participate in the
regulation of pathological processes of diabetes (39-43). It was identified that osteogenesis
was repressed by overexpression of miR-203-3p and promoted when
miR-203-3p was inhibited, which indicated that miR-203-3p
participates in the repression of osteogenesis. In addition, to the
best of our knowledge, the present study was the first to
demonstrate that miR-203-3p suppresses osteogenesis.
To further study the mechanisms of osteogenesis
repression by miR-203-3p, its target mRNAs were predicted with
TargetScan, miRanda and Pictar, and the common results provided a
pool of candidate genes. Among them, the predicted target site of
Smad1 was evolutionarily conserved among vertebrates. The
experimental results demonstrated that Smad1 expression was lower
in the mandibles of diabetic rats as compared with that in the
mandibles of normal rats. RT-qPCR and western blot analyses
indicated that miR-203-3p markedly decreased Smad1 expression
levels, while inhibition of miR-203-3p resulted in the upregulation
of Smad1 at the mRNA and protein level, respectively. Using a miRNA
target luciferase reporter assay, it was revealed that miR-203-3p
overexpression decreased the luciferase activity of the reporter
vector driven by the Smad1 3′-UTR, which indicated that miR-203-3p
inhibits Smad1 expression by directly binding to its 3′-UTR and
thereby degrading Smad1 mRNA or inhibiting its translation. To
determine whether miR-203-3p modulates osteogenesis by repressing
Smad1, cells were transfected with agomir-203-3p, resulting in
attenuation of the protein expression of Smad1, as indicated by
western blot analysis. Finally, repressed osteogenesis by
miR-203-3p was attenuated by ectopic overexpression of Smad1, which
indicated that miR-203-3p represses osteogenesis partially through
downregulation of Smad1. The BMP/Smad1/5/8 pathway was demonstrated
to be repressed by miR-203-3p.
Smad1 is an immediate downstream transducer
mediating the BMP receptor in BMP signaling (44). Mice with osteoblast-specific Smad1
gene knockout displayed impaired postnatal bone formation (45). TGF-β superfamily members have a
variety of biological functions, as Smads may combine with a
variety of transcription factor-targeting genes.
Supra-physiological glucose and insulin levels of T2DM inhibit the
maturation of primary human osteoblasts, and human immortalized
BMSCs treated with sera from T2DM patients exhibited significantly
increased TGF-β signaling, while inhibition of this signaling
effectively restored osteo-blast maturation in the T2DM group
(46). Smad proteins are
classified into three subtypes: Receptor-regulated Smads (R-Smads),
common-partner Smads (Co-Smads) and inhibitory Smads. Among them,
Smad2 and Smad3 serve as R-Smads transducing TGF-β/activin-like
signals, whereas Smad1, Smad5 and Smad8 act as R-Smads transducing
BMP-like signals. R-Smads that are activated by BMP and Runx2
induce osteogenic differentiation. Smad4 is the only Co-Smad in
mammals and is shared by the TGF-β/activin and BMP pathways
(47). TGF-β has been described
as the central factor in bone homeostasis, as it induces the
recruitment and proliferation of osteoblasts (48), and several murine studies suggest
that TGF-β may also negatively influence the bone structure
(49,50). Smad1/5/8 and Smad2/3 compete for
combination with Smad4 for signal transduction (51). In addition, miR-203-3p was
reported to target Runx2 (52,53), which, as a specific transcription
factor, has an important role in the formation and reconstruction
of bone tissue. Runx2 interacts with Smads to enhance the
expression of osteogenesis-specific genes (54). Using RT-qPCR, the present study
demonstrated that miR-203-3p markedly decreased Smad1 mRNA
expression, and western blot analysis indicated that miR-203-3p
markedly decreased Smad1 protein expression, while the inhibition
of miR-203-3p resulted in the upregulation of Smad1 mRNA and
protein. It may therefore be hypothesized that high expression of
miR-203-3p prevails in T2DM, which represses osteogenesis by
targeting Smad1 to directly attenuate the BMP/Smads pathway or
promote the TGF-β/activin pathway, which has an inhibitory effect
on osteoblast differentiation by reducing Smad1 and Smad2/3
competition for Smad4 indirectly, and these two pathways acting
synergistically, as well as by targeting Runx2 to attenuate the
synergistic effect on the BMP/Smads pathways or downregulating the
downstream transcription of osteogenic genes (Fig. 6). Of course, further experiments
are necessary to verify these mechanisms.
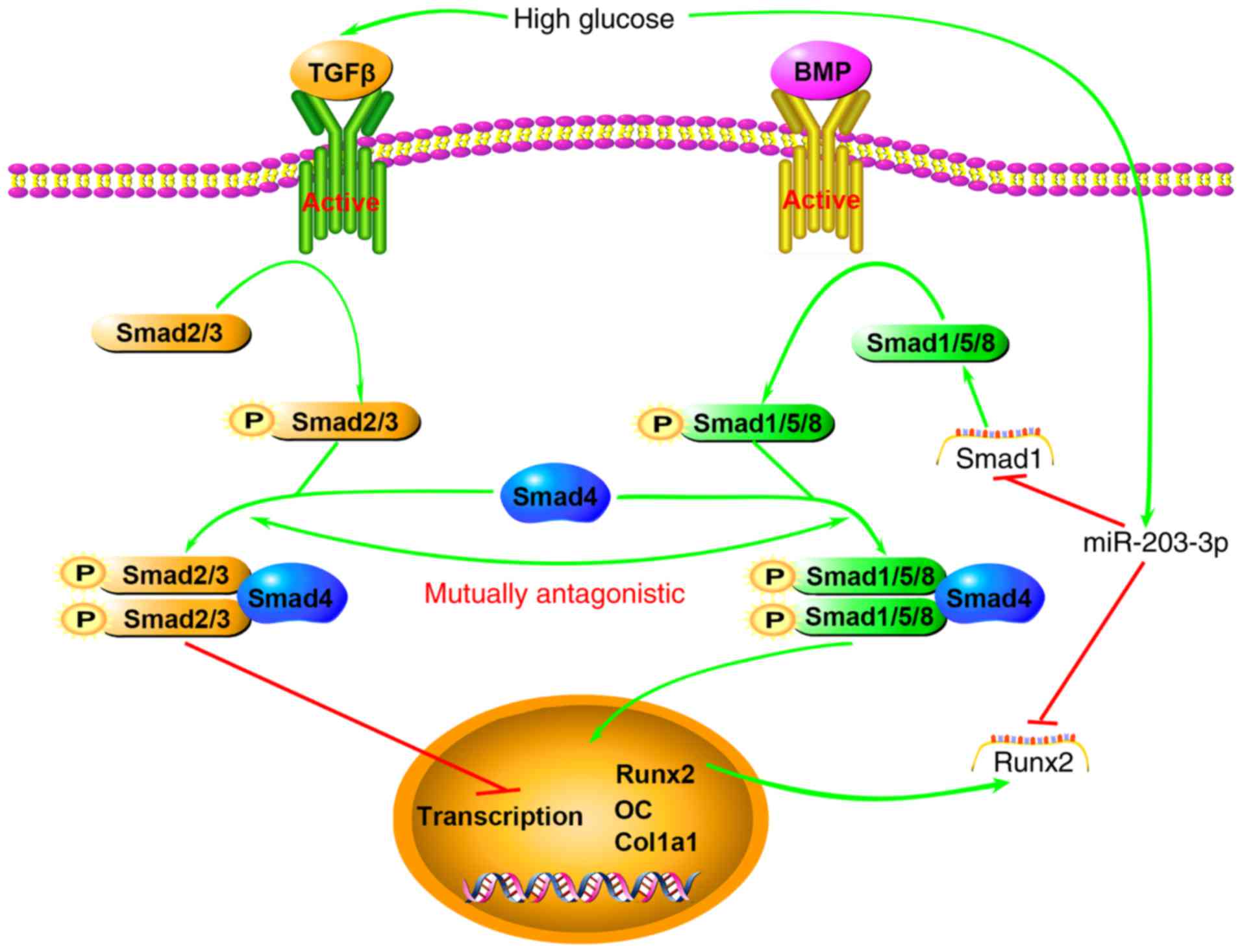 | Figure 6Schematic illustrating the
association between high glucose, miR-203-3p and the BMP/Smad1
pathway. High glucose results in high expression of miR-203-3p,
which represses osteogenesis by i) targeting Smad1 to directly
attenuate the BMP/Smads pathway or promote the TGF-β/activin
pathway, which has an inhibitory role in osteoblast
differentiation, by indirectly reducing Smad1 and Smad2/3
competition for Smad4 and the two pathways act synergistically; ii)
targeting Runx2 to attenuate the synergistic effect on the
BMP/Smads pathway or downregulating the downstream transcription of
osteogenic genes. BMP, bone morphogenetic protein; TGF,
transforming growth factor; miR, microRNA; Runx2, runt-related
transcription factor 2; OC, osteocalcin; P, phosphate; Col1a1,
collagen type 1 α 1. |
In conclusion, the present study suggested that
miR-203-3p has a role in the inhibition of osteogenesis in the jaw
bones of diabetic rats, and overexpression of miR-203-3p in the jaw
bones of diabetic rats or cells cultured in high-glucose medium
inhibits osteogenesis by targeting Smad1, which is an important
mediator of the BMP/Smad pathway. Therefore, the BMP/Smad pathway
is attenuated, and the TGF-β/activin pathway may be promoted
through the reduction of Smad1. The present study provides a basis
for gene therapy by inhibition of miR-203-3p in diabetic jaw bones,
which may ameliorate diabetic osteoporosis and improve fracture
healing, tooth stability and implant osseointegration.
Acknowledgments
The present study was supported by the Program for
the Natural Science Foundation of China (grant no. 81470772), the
Natural Science Foundation of Chongqing (grant nos.
cstc2015jcyjA10028 and cstc2016jcyjA0238), the Medical Scientific
Research Project of Chongqing (grant nos. 20141013 and
2015HBRC009), and the Program for Innovation Team Building at
Institutions of Higher Education in Chongqing in 2016 and the
Chongqing Municipal Key Laboratory of Oral Biomedical Engineering
of Higher Education in 2014.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Clementini M, Rossetti PH, Penarrocha D,
Micarelli C, Bonachela WC and Canullo L: Systemic risk factors for
peri-implant bone loss: A systematic review and meta-analysis. Int
J Oral Maxillofac Surg. 43:323–334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nemtoi A, Ladunca O, Dragan E, Budacu C,
Mihai C and Haba D: Quantitative and qualitative bone assessment of
the posterior mandible in patients with diabetes mellitus: A cone
beam computed tomography study. Rev Med Chir Soc Med Nat Iasi.
117:1002–1008. 2013.
|
|
3
|
de Morais JA, Trindade-Suedam IK, Pepato
MT, Marcantonio E Jr, Wenzel A and Scaf G: Effect of diabetes
mellitus and insulin therapy on bone density around osseointegrated
dental implants: A digital subtraction radiography study in rats.
Clin Oral Implants Res. 20:796–801. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dobreva G, Chahrour M, Dautzenberg M,
Chirivella L, Kanzler B, Fariñas I, Karsenty G and Grosschedl R:
SATB2 is a multifunctional determinant of craniofacial patterning
and osteoblast differentiation. Cell. 125:971–986. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu Q and Paroo Z: Biochemical principles
of small RNA pathways. Annu Rev Biochem. 79:295–319. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Xie RL, Gordon J, LeBlanc K,
Stein JL, Lian JB, van Wijnen AJ and Stein GS: Control of
mesenchymal lineage progression by microRNAs targeting skeletal
gene regulators Trps1 and Runx2. J Biol Chem. 287:21926–21935.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Z, Hassan MQ, Volinia S, van Wijnen AJ,
Stein JL, Croce CM, Lian JB and Stein GS: A microRNA signature for
a BMP2-induced osteoblast lineage commitment program. Proc Natl
Acad Sci USA. 105:13906–13911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu H, Sun Q, Wan C, Li L, Zhang L and
Chen Z: MicroRNA-338-3p regulates osteogenic differentiation of
mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2.
J Cell Physiol. 229:1494–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim EJ, Kang IH, Lee JW, Jang WG and Koh
JT: MiR-433 mediates ERRγ-suppressed osteoblast differentiation via
direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci.
92:562–568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zuo B, Zhu J, Li J, Wang C, Zhao X, Cai G,
Li Z, Peng J, Wang P, Shen C, et al: microRNA-103a functions as a
mechanosensitive microRNA to inhibit bone formation through
targeting Runx2. J Bone Miner Res. 30:330–345. 2015. View Article : Google Scholar
|
|
12
|
Tomé M, López-Romero P, Albo C, Sepúlveda
JC, Fernández- Gutiérrez B, Dopazo A, Bernad A and González MA:
miR-335 orchestrates cell proliferation, migration and
differentiation in human mesenchymal stem cells. Cell Death Differ.
18:985–995. 2011. View Article : Google Scholar :
|
|
13
|
Jia J, Tian Q, Ling S, Liu Y, Yang S and
Shao Z: miR-145 suppresses osteogenic differentiation by targeting
Sp7. FEBS Lett. 587:3027–3031. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi K, Lu J, Zhao Y, Wang L, Li J, Qi B,
Li H and Ma C: MicroRNA-214 suppresses osteogenic differentiation
of C2C12 myoblast cells by targeting Osterix. Bone. 55:487–494.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baglìo SR, Devescovi V, Granchi D and
Baldini N: MicroRNA expression profiling of human bone marrow
mesenchymal stem cells during osteogenic differentiation reveals
Osterix regulation by miR-31. Gene. 527:321–331. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goettsch C, Rauner M, Pacyna N, Hempel U,
Bornstein SR and Hofbauer LC: miR-125b regulates calcification of
vascular smooth muscle cells. Am J Pathol. 179:1594–1600. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen S, Yang L, Jie Q, Lin YS, Meng GL,
Fan JZ, Zhang JK, Fan J, Luo ZJ and Liu J: MicroRNA-125b suppresses
the proliferation and osteogenic differentiation of human bone
marrow-derived mesenchymal stem cells. Mol Med Rep. 9:1820–1826.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang JF, Fu WM, He ML, Wang H, Wang WM,
Yu SC, Bian XW, Zhou J, Lin MC, Lu G, et al: miR-637 maintains the
balance between adipocytes and osteoblasts by directly targeting
Osterix. Mol Biol Cell. 22:3955–3961. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Itoh T, Nozawa Y and Akao Y: MicroRNA-141
and -200a are involved in bone morphogenetic protein-2-induced
mouse pre-osteoblast differentiation by targeting distal-less
homeobox 5. J Biol Chem. 284:19272–19279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie Q, Wang Z, Bi X, Zhou H, Wang Y, Gu P
and Fan X: Effects of miR-31 on the osteogenesis of human
mesenchymal stem cells. Biochem Biophys Res Commun. 446:98–104.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Dong J, Zhang ZH, Zhang DC, You XY,
Zhong Y, Chen MS and Liu SM: miR-10a restores human mesenchymal
stem cell differentiation by repressing KLF4. J Cell Physiol.
228:2324–2336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Z, Hassan MQ, Jafferji M, Aqeilan RI,
Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS and Lian JB:
Biological functions of miR-29b contribute to positive regulation
of osteoblast differentiation. J Biol Chem. 284:15676–15684. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin EA, Kong L, Bai XH, Luan Y and Liu CJ:
miR-199a, a bone morphogenic protein 2-responsive MicroRNA,
regulates chondrogenesis via direct targeting to Smad1. J Biol
Chem. 284:11326–11335. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kureel J, Dixit M, Tyagi AM, Mansoori MN,
Srivastava K, Raghuvanshi A, Maurya R, Trivedi R, Goel A and Singh
D: miR-542-3p suppresses osteoblast cell proliferation and
differentiation, targets BMP-7 signaling and inhibits bone
formation. Cell Death Dis. 5:e10502014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luzi E, Marini F, Sala SC, Tognarini I,
Galli G and Brandi ML: Osteogenic differentiation of human adipose
tissue-derived stem cells is modulated by the miR-26a targeting of
the SMAD1 transcription factor. J Bone Miner Res. 23:287–295. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zheng L, Tu Q, Meng S, Zhang L, Yu L, Song
J, Hu Y, Sui L, Zhang J, Dard M, et al: Runx2/DICER/miRNA pathway
in regulating osteogenesis. J Cell Physiol. 232:182–191. 2017.
View Article : Google Scholar
|
|
27
|
Ko JY, Chuang PC, Chen MW, Ke HC, Wu SL,
Chang YH, Chen YS and Wang FS: MicroRNA-29a ameliorates
gluco-corticoid-induced suppression of osteoblast differentiation
by regulating β-catenin acetylation. Bone. 57:468–475. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang T and Xu Z: miR-27 promotes
osteoblast differentiation by modulating Wnt signaling. Biochem
Biophys Res Commun. 402:186–189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang
W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL,
et al: miR-218 directs a Wnt signaling circuit to promote
differentiation of osteoblasts and osteomimicry of metastatic
cancer cells. J Biol Chem. 287:42084–42092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Clark EA, Kalomoiris S, Nolta JA and
Fierro FA: Concise review: MicroRNA function in multipotent
mesenchymal stromal cells. Stem cells. 32:1074–1082. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gaur T, Hussain S, Mudhasani R, Parulkar
I, Colby JL, Frederick D, Kream BE, van Wijnen AJ, Stein JL, Stein
GS, et al: Dicer inactivation in osteoprogenitor cells compromises
fetal survival and bone formation, while excision in differentiated
osteoblasts increases bone mass in the adult mouse. Dev Biol.
340:10–21. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu H, Kraut D, Gerstenfeld LC and Graves
DT: Diabetes interferes with the bone formation by affecting the
expression of transcription factors that regulate osteoblast
differentiation. Endocrinology. 144:346–352. 2003. View Article : Google Scholar
|
|
34
|
García-Hernández A, Arzate H,
Gil-Chavarria I, Rojo R and Moreno-Fierros L: High glucose
concentrations alter the biomineralization process in human
osteoblastic cells. Bone. 50:276–288. 2012. View Article : Google Scholar
|
|
35
|
Ogawa N, Yamaguchi T, Yano S, Yamauchi M,
Yamamoto M and Sugimoto T: The combination of high glucose and
advanced glycation end-products (AGEs) inhibits the mineralization
of osteoblastic MC3T3-E1 cells through glucose-induced increase in
the receptor for AGEs. Horm Metab Res. 39:871–875. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gopalakrishnan V, Vignesh RC, Arunakaran
J, Aruldhas MM and Srinivasan N: Effects of glucose and its
modulation by insulin and estradiol on BMSC differentiation into
osteoblastic lineages. Biochem Cell Biol. 84:93–101. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
van Rooij E: The art of microRNA research.
Circ Res. 108:219–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang JC: miR-467f modulates osteogenic
differentiation of mice bone mesenchymal stem cells in high glucose
environment. Military Medical School; Beijing: 2013, In
Chinese.
|
|
39
|
Yang Z, Zhong L, Zhong S, Xian R and Yuan
B: miR-203 protects microglia mediated brain injury by regulating
inflammatory responses via feedback to MyD88 in ischemia. Mol
Immunol. 65:293–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Okumura T, Shimada Y, Moriyama M, Takei Y,
Omura T, Sekine S, Nagata T, Shimizu K and Tsukada K: MicroRNA-203
inhibits the progression of esophageal squamous cell carcinoma with
restored epithelial tissue architectur in vivo. Int J Oncol.
44:1923–1932. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu X, Jiang X, Li H, Guo L, Jiang W and Lu
SH: miR-203 inhibits the proliferation and self-renewal of
esophageal cancer stem-like cells by suppressing stem renewal
factor Bmi-1. Stem Cells Dev. 23:576–585. 2014. View Article : Google Scholar
|
|
42
|
Delic D, Eisele C, Schmid R, Luippold G,
Mayoux E and Grempler R: Characterization of Micro-RNA changes
during the progression of type 2 diabetes in Zucker diabetic fatty
rats. Int J Mol Sci. 17:pii: E665. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu J, Xu Y, Shu B, Wang P, Tang J, Chen
L, Qi S, Liu X and Xie J: Quantification of the differential
expression levels of microRNA-203 in different degrees of diabetic
foot. Int J Clin Exp Pathol. 8:13416–13420. 2015.
|
|
44
|
Hoodless PA, Haerry T, Abdollah S,
Stapleton M, O'Connor MB, Attisano L and Wrana JL: MADR1, a
MAD-related protein that functions in BMP2 signaling pathways.
Cell. 85:489–500. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang M, Jin H, Tang D, Huang S, Zuscik MJ
and Chen D: Smad1 plays an essential role in bone development and
postnatal bone formation. Osteoarthritis Cartilage. 19:751–762.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ehnert S, Freude T, Ihle C, Mayer L, Braun
B, Graeser J, Flesch I, Stöckle U, Nussler AK and Pscherer S:
Factors circulating in the blood of type 2 diabetes mellitus
patients affect osteoblast maturation-description of a novel in
vitro model. Exp Cell Res. 332:247–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Miyazono K, Maeda S and Imamura T:
Coordinate regulation of cell growth and differentiation by
TGF-beta superfamily and Runx proteins. Oncogene. 23:4232–4237.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pfeilschifter J, Bonewald L and Mundy GR:
Characterization of the latent transforming growth factor beta
complex in bone. J Bone Miner Res. 5:49–58. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Edwards JR, Nyman JS, Lwin ST, Moore MM,
Esparza J, O'Quinn EC, Hart AJ, Biswas S, Patil CA, Lonning S, et
al: Inhibition of TGF-β signaling by 1D11 antibody treatment
increases bone mass and quality in vivo. J Bone Miner Res.
25:2419–2426. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mohammad KS, Chen CG, Balooch G, Stebbins
E, McKenna CR, Davis H, Niewolna M, Peng XH, Nguyen DH,
Ionova-Martin SS, et al: Pharmacologic inhibition of the TGF-beta
type I receptor kinase has anabolic and anti-catabolic effects on
bone. PLoS One. 4:e52752009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Matsumoto Y, Otsuka F, Hino J, Miyoshi T,
Takano M, Miyazato M, Makino H and Kangawa K: Bone morphogenetic
protein-3b (BMP-3b) inhibits osteoblast differentiation via Smad2/3
pathway by counteracting Smad1/5/8 signaling. Mol Cell Endocrinol.
350:78–86. 2012. View Article : Google Scholar
|
|
52
|
Taipaleenmaki H, Browne G, Akech J, Zustin
J, van Wijnen AJ, Stein JL, Hesse E, Stein GS and Lian JB:
Targeting of Runx2 by miR-135 and miR-203 impairs progression of
breast cancer and metastatic bone disease. Cancer Res.
75:1433–1444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Saini S, Majid S, Yamamura S, Tabatabai L,
Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y and Dahiya R:
Regulatory Role of mir-203 in prostate cancer progression and
metastasis. Clin Cancer Res. 17:5287–5298. 2011. View Article : Google Scholar
|
|
54
|
Lee MH, Kwon TG, Park HS, Wozney JM and
Ryoo HM: BMP-2-induced Osterix expression is mediated by Dlx5 but
is independent of Runx2. Biochem Biophys Res Commun. 309:689–694.
2003. View Article : Google Scholar : PubMed/NCBI
|