Introduction
Retinoblastoma (RB) is a common intraocular
malignant tumor in infants that develops from embryonic retinal
nervous tissue. RB frequently occurs in children aged ≤3 years, and
several patients often succumb to the disease as a result of its
high-grade malignancy and systemic metastases (1,2).
At present, among the therapeutic schedules for RB, including
photocoagulation, condensation, radiotherapy and chemotherapy,
chemotherapy is the standard treatment. However, the presently
available chemotherapeutics, such as vincristine (VCR), etoposide
(ETO) and carboplatin (CBP), often result in drug resistance, which
is associated with a low success rate or a high recurrence rate.
Therefore, investigating the mechanism underlying the development
of drug resistance in RB and overcoming this obstacle will greatly
improve the survival rate and prognosis of the patients (3,4).
High-mobility group protein B1 (HMGB1) is a nuclear
chromatin protein with strong mobility. HMGB1 is able to move
freely and rapidly among various proteins and chromatins and
selectively binds to specific sites. HMGB1 has been found to be
closely associated with cancer drug resistance. When exposed to
chemotherapeutics, cancer cells, such as osteosarcoma, colon
cancer, endometrial cancer, lung cancer and leukemia cells, release
higher amounts of HMGB1 and, subsequently, HMGB1 reduces the
sensitivity of cancer cells to chemotherapeutics through enhancing
autophagy and suppressing apoptosis (5–8).
The molecular mechanisms linked to HMGB1 have been
previously investigated (9–12).
HMGB1 affects cell behavior, including inflammation, proliferation,
metastasis and invasion, by binding to the receptor for advanced
glycation endproducts (RAGE), Toll-like receptor (TLR)-2, TLR-4 and
TLR-9, and then regulating downstream signaling pathways. Moreover,
regardless of the type of receptor HMGB1 binds to, it affects
cancer cell apoptosis and autophagy via finally transducing the
signals to nuclear factor-κB (NF-κB) (9–12).
Therefore, it was hypothesized that the HMGB1̸NF-κB pathway may
also play a crucial role in the drug resistance of RB cells. We
herein performed a series of experiments, including gene silencing,
cell viability, apoptosis, autophagy and western blot analysis, in
order to confirm this hypothesis.
Materials and methods
Materials and cell culture
Ammonium pyrrolidinedithiocarbamate (PDTC),
bicinchoninic acid (BCA) kit, Hoechst staining kit, methyl
thiazolyl tetrazolium (MTT), and horseradish peroxidase-labeled
goat anti-rabbit IgG (H+L; A0206) were purchased from Beyotime
Institute of Biotechnology (Nantong, China). Lipofectamine™ 2000
was purchased from Invitrogen; Thermo Fisher Scientific (Carlsbad,
CA, USA). Rabbit anti-Beclin 1 (ab62557), anti-p62 (ab56416),
anti-cleaved-caspase-3 (ab2302), anti-cleaved-poly(ADP-ribose)
polymerase (PARP; ab32064), anti-NF-κB (ab16502), anti-LC3
(ab48394), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH;
ab8245) and anti-HMGB1 (ab79823) polyclonal antibodies were from
Abcam (Cambridge, MA, USA). VCR, ETO and CBP were purchased from
Sigma-Aldrich; Merck KGaA (St. Louis, MO, USA). siRNA HMGB1 (sense,
5′-CCC GUU AUG AAA GAG AAA UTT-3′ and antisense, 5′-AUU UCU CUU UCA
UAA CGG GTT-3′), the negative control (siRNA NC, sense, 5′-UUC UCC
GAA CGU GUC ACG UTT-3′ and antisense, 5′-ACG UGA CAC GUU CGG AGA
ATT-3′), and FAM-labeled siRNA were prepared by GenePharma
(Shanghai, China).
The human RB cell lines Weri-Rb-1 and Y79 were
obtained from the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China) and cultured in DMEM media containing
10% (v/v) fetal bovine serum (FBS) (both from HyClone, Logan, UT,
USA).
HMGB1 levels after drug treatment
Weri-Rb-1 and Y79 cells were seeded into 6-well
plates. After 48 h, 10 µM VCR, 5 µM ETO, and 1
µM CBP was added to each well. The cells were then incubated
for 48 h, collected, and subjected to western blot analysis as
described below.
Cells transfection
siRNA HMGB1, siRNA NC and FAM-labeled siRNA were
transfected into cells using the Lipofectamine™ 2000 kit, according
to the manufacturer's instructions. When the confluence of Y79
cells reached 50–70%, the mixture of siRNA and Lipofectamine™ 2000
was added to the cells. After 6 h, the medium was replaced by DMEM
supplemented with 10% (v/v) FBS. After incubation for 48 h at 37°C
and 5% CO2, FAM-labeled cells were photographed under a
fluorescence microscope and the transfection efficiency was
calculated by semi-quantitatively analyzing fluorescence intensity
with Image-Pro Plus software. Other cells were submitted to western
blot analysis for HMGB1 determination.
Cell viability
Y79 cells were seeded into 96-well plates and
transfected with siRNA HMGB1 and siRNA NC, as described above.
After 48 h, the cells were treated with 0.1, 0.3, 1, 3, 10, 30, 100
and 300 µM VCR. After a further 48 h, 20 µl MTT (5
mg/ml) was added to each well. After incubation for 4 h, the medium
was discarded and 150 µl dimethyl sulfoxide was added to
each well. After vibration for 10 min, the plates were subjected to
absorbance determination at 560 nm using a microplate reader
(Infinite F200/M200; Tecan, Männedorf, Switzerland). Cells treated
with VCR and without siRNA transfection served as control. The
relative cell viability and the IC50 value of VCR to Y79
were calculated.
Cell apoptosis
Y79 cells were seeded into 96-well plates and were
transfected with siRNA HMGB1 and siRNA NC, as described above.
Additional cells were exposed to 10 µM PDTC. After
incubation for 48 h, 10 µM VCR was added into each well.
After a further 48 h, the cells were treated with the Hoechst
staining kit according to the manufacturer's instructions. The
cells were then visualized under a microscope (TS100; Nikon, Tokyo,
Japan).
Western blot analysis
Y79 cells were seeded into 96-well plates and
transfected with siRNA HMGB1 and siRNA NC, as described above.
Additional cells were exposed to 10 µM PDTC. After
incubation for 48 h, VCR was added to each well. After a further 48
h, the cells were collected and lysed in RIPA buffer for 30 min.
The cell lysate was then centrifuged for 10 min at 7,000 × g and
4°C. The supernatant was carefully collected and total protein
concentration was analyzed with BCA kits. Subsequently, protein was
denatured and loaded quantitatively to perform sodium dodecyl
sulfate-polyacrylamide gel electrophoresis. Membrane transfer was
performed with the wet transfer method for 30–50 min. The membranes
were incubated with primary antibody buffer (1:100; rabbit
anti-HMGB1, anti-beclin 1, anti-p62, anti-cleaved caspase-3,
anti-cleaved PARP, anti-NF-κB and anti-GAPDH polyclonal antibody)
overnight at 4°C, followed by rinsing and incubation with secondary
antibody buffer for 1–2 h at room temperature. After rinsing again,
enhanced chemiluminescent (ECL) solution was dripped on the
membrane and the membrane was exposed to a gel imaging system
(ChemiDoc™ XRS; Bio-Rad, Hercules, CA, USA). 'Quantity One'
software (Bio-Rad) was utilized to analyze the gray values of the
protein strips.
Immunofluorescence assay
Y79 cells were seeded on glass slides and
transfected with siRNA HMGB1 and siRNA NC, as described above.
After incubation with PDTC and VCR, the cells were fixed with
precooled 4% (w/v) paraformaldehyde for 10 min and rinsed with
phosphate-buffered saline (PBS) three times, followed by incubation
with serum containing 0.1% (w/v) Triton X-100 for 30 min and then
with rabbit anti-LC3 polyclonal antibody at 4°C overnight. After
washing with PBS, secondary antibody was added at room temperature
for 1 h. The cells were again washed with PBS and stained with
DAPI. Finally, the slides were mounted and observed under a
confocal microscope. The average LC3 puncta were calculated by
semi-quantitatively analyzing fluorescence intensity with Image-Pro
Plus software based on at least 100 cells, which represents cell
autophagy levels.
Statistical analysis
All data are expressed as mean ± standard deviation
and were analyzed with Student's t-test using SPSS 17.0 software
(SPSS Inc., Chicago, IL, USA). Statistical significance was set at
P<0.05
Results
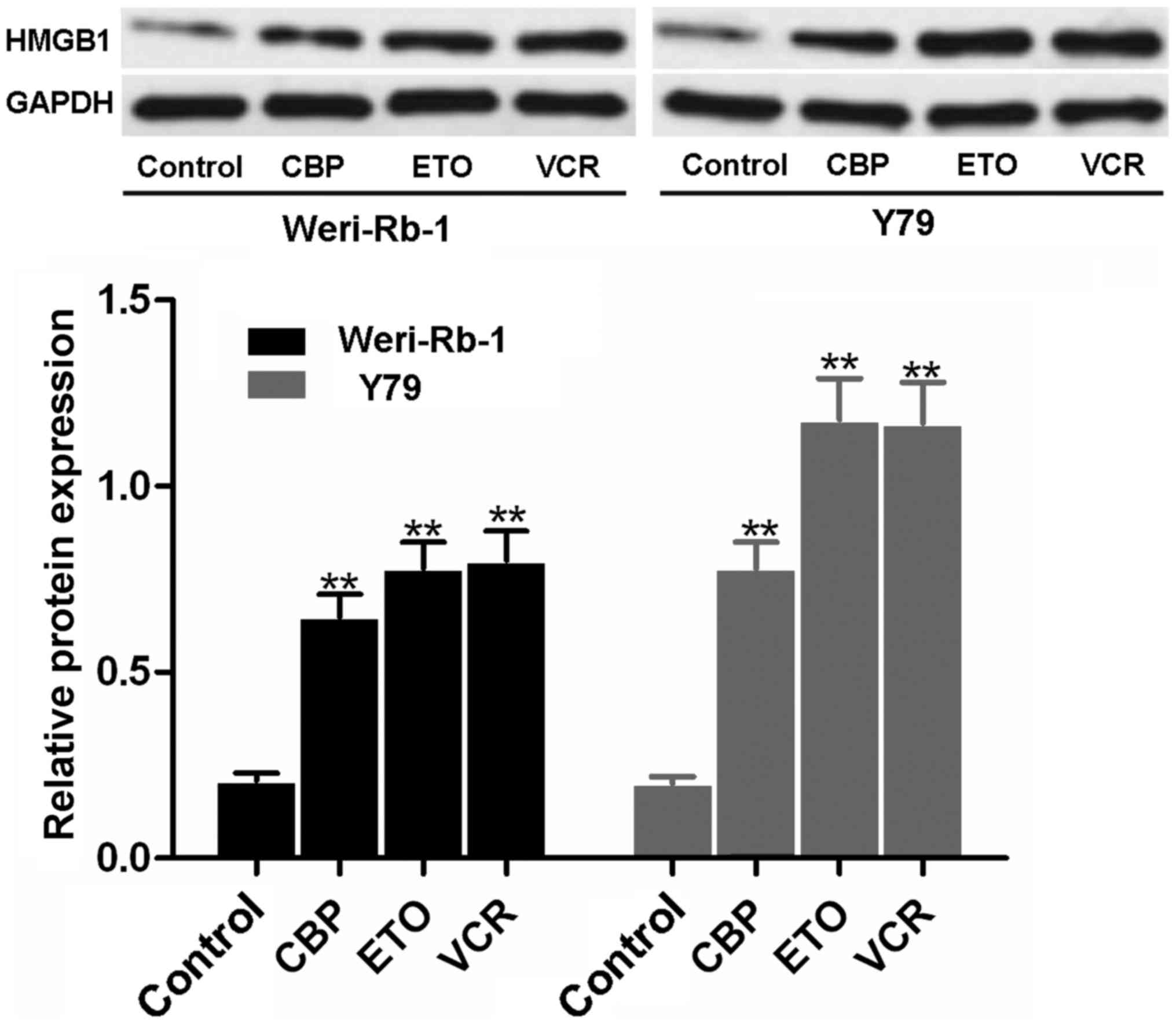
Effects of chemotherapeutics on HMGB1
expression
As shown in Fig.
1, a faint blot representing a small quantity of HMGB1 was
observed in the untreated control Weri-Rb-1 and Y79 cells. However,
following VCR, ETO or CBP treatment, HMGB1 expression was found to
be significantly upregulated in both types of RB cells (all
P<0.01).
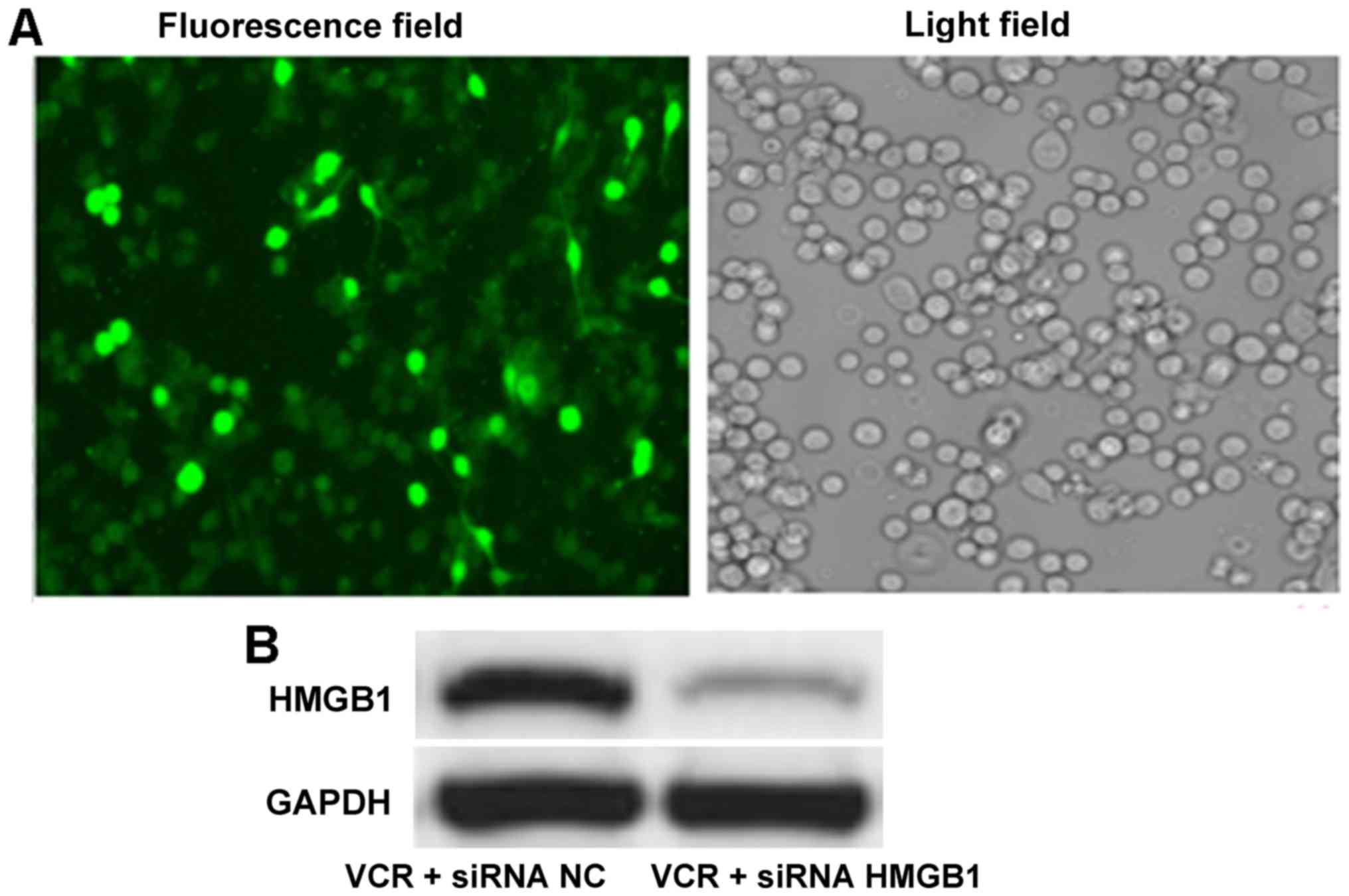
siRNA HMGB1 induces downregulation of
HMGB1 expression in Y79 cells
After FAM-labeled siRNA HMGB1 transfection by
Lipofectamine™ 2000, the percentage of positive Y79 cells reached
90%, suggesting successful transfection into RB cells (Fig. 2A). Western blot analysis revealed
that, following siRNA HMGB1 transfection, the HMGB1 band was
notably fainter compared with siRNA NC transfection (Fig. 2B). Quantitative results
demonstrated that the gray value of HMGB1 in the siRNA HMGB1 group
(0.09±0.01) was significantly smaller compared with that in the
siRNA NC group (0.49±0.04; P<0.01), indicating that siRNA HMGB1
induced downregulation of HMGB1 expression in RB cells.
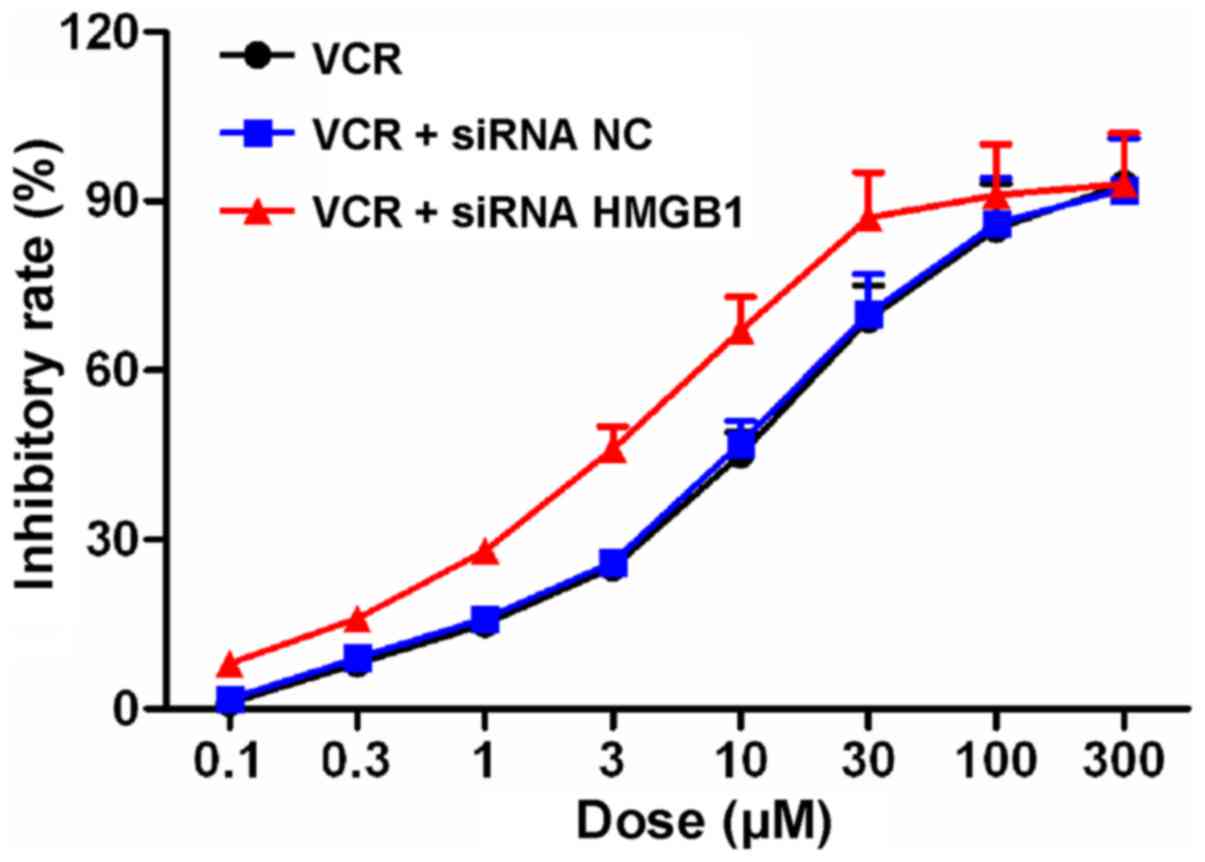
siRNA HMGB1 suppresses the proliferation
of Y79 cells
The effects of siRNA NC and siRNA HMGB1 on the
inhibitory profile of VCR in Y79 cells was determined with the MTT
assay (Fig. 3). Following VCR
treatment at various concentrations for 48 h, the IC50
value of VCR in Y79 cells was 10.59±1.28. However, following siRNA
NC and siRNA HMGB1 transfection, the IC50 values were
10.50±1.21 and 3.60±0.30, respectively, suggesting that siRNA HMGB1
enhanced the sensitivity of Y79 cells to VCR and reduced the
effective concentration of VCR.
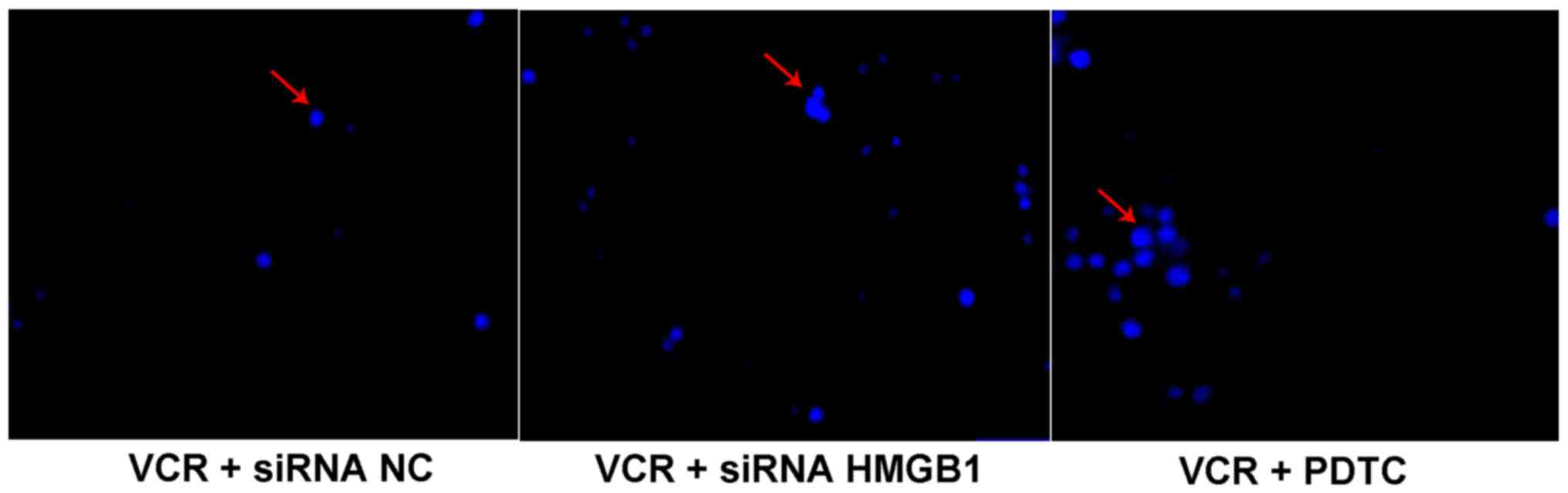
siRNA HMGB1 promotes apoptosis of Y79
cells
The apoptosis of Y79 cells was evaluated with
Hoechst staining and the apoptotic cells were identified as light
blue spots (Fig. 4), whereas
non-apoptotic cells were not stained. Compared with siRNA NC, siRNA
HMGB1 and PDTC observably promoted the apoptosis of Y79 cells
following VCR treatment.
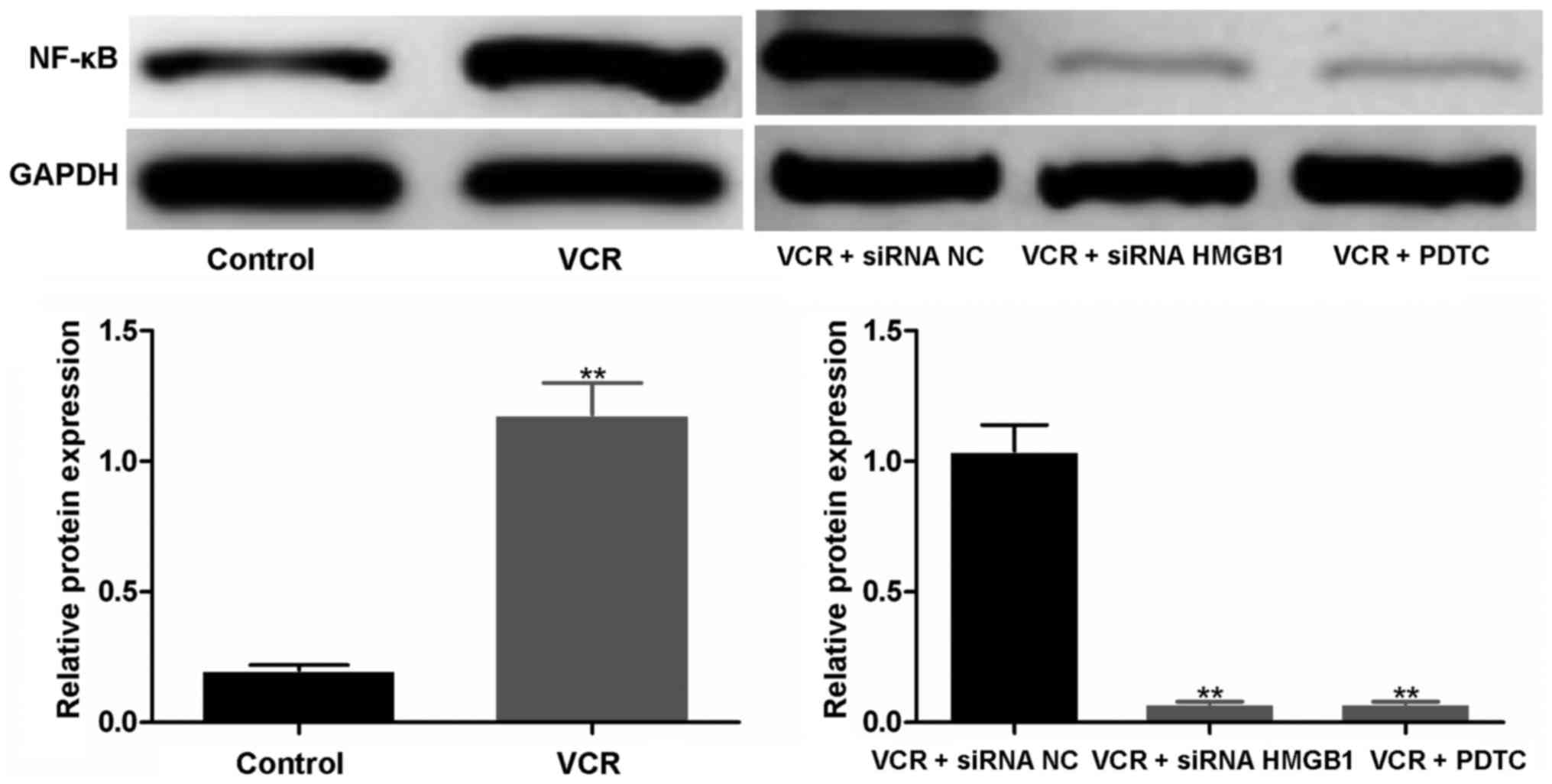
siRNA HMGB1 and PDTC downregulate the
expression of NF-κB
As shown in Fig.
5, compared with the untreated control, the expression of NF-κB
was markedly elevated in VCR-treated cells; the gray values in the
control and VCR groups were 0.13±0.01 and 0.88±0.08, respectively
(P<0.01). When siRNA NC and siRNA HMGB1 were transfected into
Y79 cells prior to VCR treatment, a dark band remained in the siRNA
NC group, while the band in the siRNA HMGB1 group was almost
invisible. The gray value in the siRNA NC group was significantly
higher compared with that in the siRNA HMGB1 group (P<0.01).
Similarly, the expression of NF-κB was markedly inhibited by
PDTC.
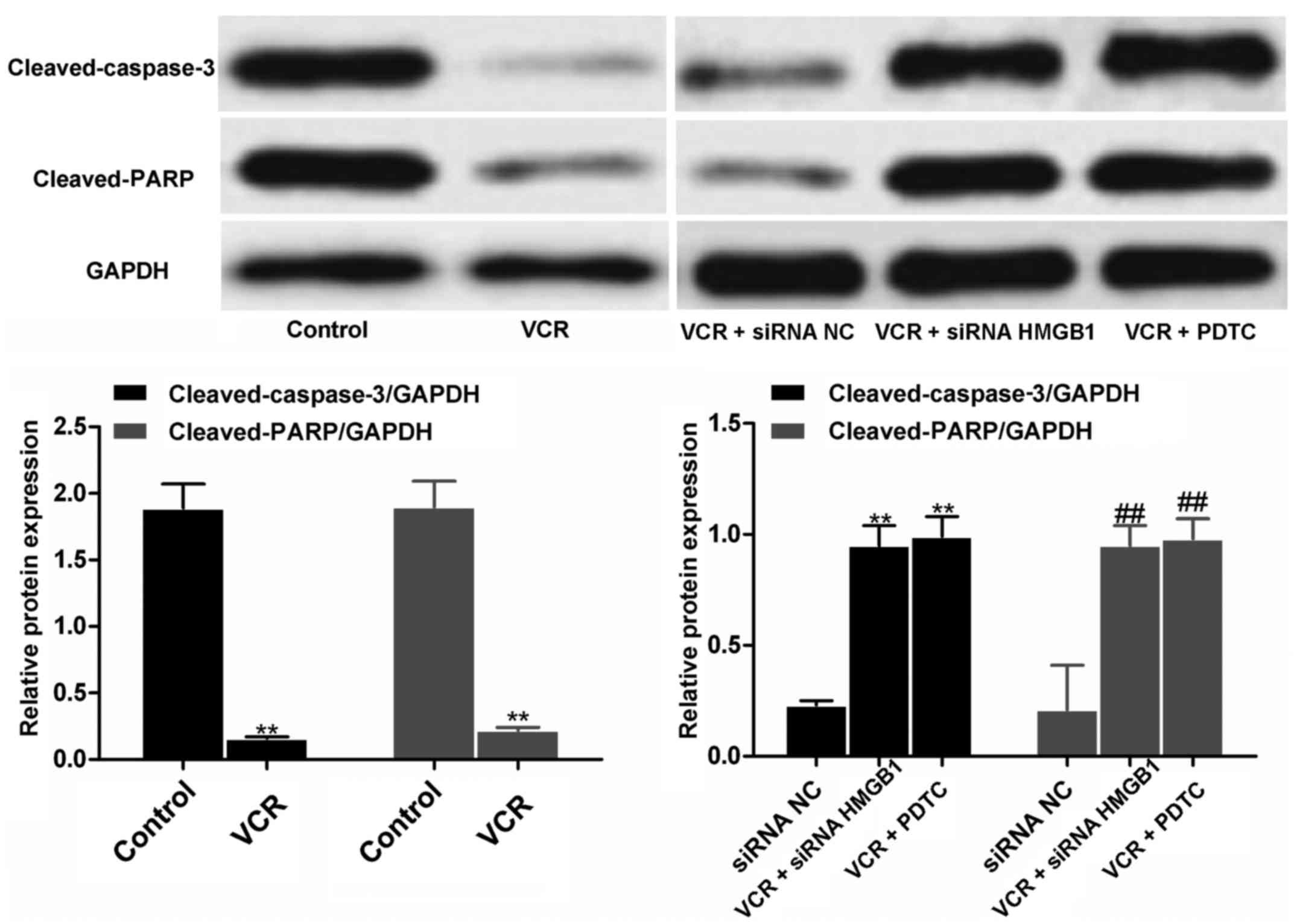
siRNA HMGB1 and PDTC upregulate the
expression of cleaved caspase-3 and cleaved PARP
Qualitative and quantitative western blot analysis
revealed that, after VCR treatment, the expression of cleaved
caspase-3 and cleaved PARP in Y79 cells was significantly reduced
(both P<0.01; Fig. 6).
However, siRNA HMGB1 and PDTC treatment significantly increased the
expressions in VCR-treated cells compared with siRNA NC (all
P<0.01), indicating that siRNA HMGB1 and PDTC were able to
reverse the effects of VCR on cleaved caspase 3 and cleaved PARP
expression in RB cells.
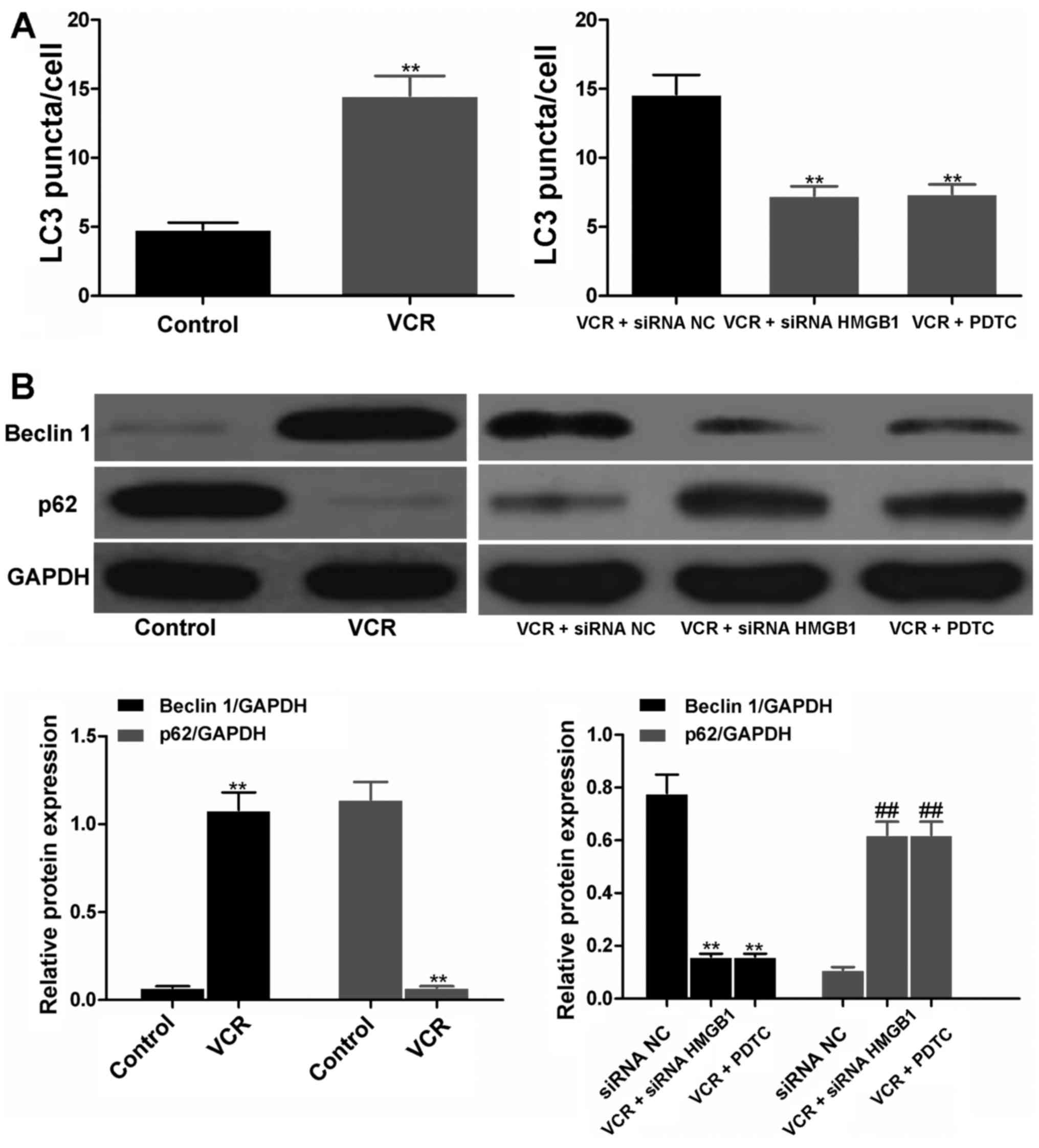
siRNA HMGB1 and PDTC suppressed autophagy
by regulating the expression of beclin 1 and p62
The LC3 puncta in cells represented autophagy. As
shown in Fig. 7A, the LC3 puncta
were notably increased in VCR- and siRNA NC-treated cells. However,
in the siRNA HMGB1 and PDTC groups, the LC3 puncta were
significantly decreased (both P<0.01), suggesting the effective
inhibition of siRNA HMGB1 and PDTC on autophagy of Y79 cells.
Furthermore, Y79 cells treated with VCR for 48 h produced a
significantly higher amount of beclin 1 and lower amount of p62
compared with the control (both P<0.01; Fig. 7B). When cells were treated with
siRNA NC, siRNA HMGB1 and PDTC prior to VCR treatment, cells in
both the siRNA HMGB1 and PDTC groups released a significantly lower
amount of beclin 1 and higher amount of p62 compared with the siRNA
NC group (all P<0.01), suggesting that siRNA HMGB1 and PDTC were
able to reverse the effects of VCR on beclin 1 and p62 expression
in RB cells.
Discussion
HMG proteins were first identified by Goodwin in
thymic cell in 1973 (13) and
gained their name due to their high migration rates in PAGE. HMG
proteins are composed of classic HMG proteins and HMG motif
proteins. We herein investigated the classic HMG proteins that are
abundant in eucaryotes, namely HMGB, HMGA and HMGN, with HMGB being
the most abundant. HMGB contains a special structural domain
HMG-box and comprises three proteins, HMGB1, HMGB2 and HMGB3. The
HMGB1 gene is located on chromosome 13q12 and its protein sequence
is highly conserved, with two homologous HMG-boxes. HMGB1 has been
found to affect the expression of downstream proteins, cell
apoptosis, cell proliferation and autophagy through binding to
endonuclear DNA and then regulating multiple gene transcriptions
and gene recombination (14,15).
HMGB1 had been reported to be overexpressed in
several cancers, including RB, and it is closely correlated with
tumorigenesis and tumor development. Singh et al found that
HMGB1 was expressed in 55.07% (38/69) primary RB cases through
immunohistochemistry, and HMGB1 mRNA expression was observed in
77.41% (24/31) fresh tumor tissue samples via reverse
transcription-polymerase chain reaction analysis (16). Moreover, the expression of HMGB1
was higher in tumors with poor differentiation and optic nerve
invasion (16). In addition,
numerous studies demonstrated that HMGB1 was associated with cancer
resistance to chemotherapy. Luo et al reported that
doxorubicin promoted the expression of HMGB1 in CT27 colon cancer
cells (9). Liu et al also
demonstrated that chemotherapeutic drugs upregulated the expression
of HMGB1 in leukemia cells (17).
Zhang et al reported that doxorubicin, cisplatin and
methotrexate stimulated the upregulation of HMGB1 expression in
A549 lung cancer cells, and the expression of HMGB1 in A549/DDP
drug-resistant cells was significantly higher compared with that in
A549 cells (18). Therefore, in
the present study, the expression of HMGB1 in the RB cell lines
Weri-Rb-1 and Y79 after treatment with 10 µM VCR, 5
µM ETO and 1 µM CBP for 48 h was investigated through
western blot analysis, and the results demonstrated that all three
drugs markedly elevated the expression of HMGB1 in RB cells. VCR
and Y79 cells were employed as the model drug and model RB cells in
the following experiments, respectively.
In addition, shRNA HMGB1 reduced the IC50
value of cisplatin to A549/DDP cells and enhanced the
susceptibility of A549/DDP cells to cisplatin (18). HMGB1-neutralizing antibody may
improve the sensitivity of leukemia cells to chemotherapeutic drugs
(17). Similarly, HMGB1 gene
expression was silenced in Y79 cells by siRNA HMGB1 transfection.
Based on the cell viability analysis by MTT assay, it was observed
that siRNA HMGB1 significantly reduced the IC50 value of
VCR to RB cells and enhanced the inhibitory effect of VCR on RB
cells. These results suggested that HMGB1 was closely associated
with the chemotherapeutics of RB cells and the downregulation of
HMGB1 expression was able to notably intensify the susceptibility
of RB cells to chemotherapeutic drugs.
Due to having two homologous box structural domains,
the HMGB1 gene can bind to the corresponding receptors RAGE, TLR-2
and TLR-4. These binding complexes then activate multiple signaling
pathways, including NF-κB, mitogen-activated protein kinase,
phosphatidylinositol 3-kinase/AKT, and Janus kinase/signal
transducer and activator of transcription, consequently regulating
cell proliferation, differentiation and autophagy. Among those
pathways, NF-κB was the most common pathway of HMGB1 receptors RAGE
and TLR-4, and HMGB1 significantly affected cell behavior via
activating the NF-κB pathway (9–12).
Following activation by upstream signals, NF-κB with special DNA
sequences shifts into the nucleus, binds to promoter or enhancer of
downstream target genes, and then regulates the expression of
apoptosis-related genes and inflammatory factors. In addition,
NF-κB was reported to be involved in the chemotherapy of several
types of cancer (19,20). Therefore, in the present study,
the expression of NF-κB in Y79 cells treated by VCR for 48 h was
evaluated through western blot analysis and it was revealed that,
although VCR treatment obviously elevated the expression of NF-κB,
after siRNA HMGB1 transfection and PDTC (specific NF-κB inhibitor)
treatment, the expression of NF-κB was markedly dowregulated. These
findings suggest that siRNA HMGB1 promoted death and apoptosis of
RB cells through markedly decreasing the high level of NF-κB
induced by chemotherapeutic drugs.
Multiple factors and processes are involved in the
resistance of cancer cells to chemotherapeutic drugs, mainly
including membrane glycoprotein-mediated drug efflux, DNA repair
and apoptosis pathway abnormalities. The inhibition of cancer cell
apoptosis is a major cause of cancer drug resistance and
chemotherapy failure; thus, it was hypothesized that the induction
of cancer cell apoptosis may effectively enhance the sensitivity of
cancer cells to chemotherapeutic drugs and suppress the development
of cancer drug resistance (21,22). Therefore, in the present study,
the apoptosis of Y79 cells following treatment with siRNA HMGB1,
PDTC (a specific NF-κB inhibitor) and VCR was investigated. The
results of Hoechst staining revealed that both siRNA HMGB1 and
NF-κB inhibitor obviously aggravated the apoptosis of RB cells
induced by chemotherapy. In addition, cleaved caspase-3 and cleaved
PARP are markers of cell apoptosis, whereas caspase is the executor
of cell apoptosis. As it is in the middle and downstream of the
caspase cascade reactions, caspase-3 is the principal effector
molecule of the apoptotic process and the intersection point of the
apoptosis pathway. As a DNA repairase in eukaryocytes, PARP is the
substrate of the caspase family. The cleavage of PARP reflects the
activation of the caspase family. In the present study, the
expression of cleaved caspase-3 and cleaved PARP in Y79 cells after
treatment with siRNA HMGB1, PDTC and VCR was determined; the
results of qualitative and quantitative western blot analysis
demonstrated that siRNA HMGB1 and PDTC markedly upregulated their
expression in RB cells after chemotherapy. This result was in
agreement with previous studies (18,23). Zhang et al revealed that
shRNA HMGB1 promoted cell apoptosis after cisplatin induction
through upregulating the expression of cleaved caspase-3 (18). Liu et al demonstrated that
the downregulation of HMGB1 in chemotherapeutic drug-treated Y79
cells may induce oxidative stress injury and DNA damage, increase
the activity of caspase-3, and upregulate the expression of cleaved
caspase-3 and cleaved PARP1 (23). Accordingly, it was indicated that
siRNA HMGB1 intensified chemotherapy-induced apoptosis in RB cells
and promoted the susceptibility of RB cells to chemotherapeutic
drugs through downregulating the expression of NF-κB and
subsequently upregulating the expression of cleaved caspase-3 and
cleaved PARP.
Furthermore, HMGB1 may alleviate the resistance of
cancer cells to drugs by restraining autophagy and facilitating
cell apoptosis (17,18). As a self-rescue mode under
conditions of stress or alimentary deficiency, autophagy not only
eliminates aging or damaged organelles, but also repairs DNA damage
and protein misfolding. The chemotherapeutic drug resistance in a
number of cancer cells was enhanced along with increased activity
of autophagy; thus, the development of drug resistance in cancer
cells may be inhibited and the sensitivity of cancer cells to drugs
may be increased through suppressing the increase in cancer cell
autophagy (24). In the present
study, the autophagy protein markers beclin 1 and p62 in
VCR-treated Y79 cells were determined after siRNA HNGB1 and PDTC
treatment using western blot analysis. It was observed that, after
HMGB1 gene silencing by siRNA or NF-κB inhibition by PDTC, beclin 1
expression was downregulated and p62 expression was upregulated. It
was suggested that the chemotherapeutic drug-induced autophagy in
RB cells was suppressed by regulating beclin 1 and p62. Similarly,
exogenous HMGB1 reduced the sensitivity of leukemia cells to
chemotherapeutic drugs and enhanced cell autophagy (17). HMGB1 silencing by shRNA may
promote cell apoptosis induced by cisplatin and inhibit the
expression of autophagy-related proteins (18).
In conclusion, VCR, ETO and CBP markedly upregulated
the expression of HMGB1 in RB cells. However, after HMGB1 silencing
by siRNA, the susceptibility of RB cells to chemotherapeutic drugs
and the apoptosis of RB cells with chemotherapeutic drug treatment
were notably improved. In addition, siRNA HMGB1 inhibited the
expression of NF-κB and, similar to the NF-κB inhibitor PDTC, siRNA
HMGB1 treatment significantly increased the expression of cleaved
caspase-3, cleaved PARP and p62, and reduced the expression of
beclin 1. Consequently, siRNA HMGB1 promoted apoptosis and
suppressed autophagy of RB cells through downregulating NF-κB.
Therefore, the downregulation of the HMGB1̸NF-κB pathway was able
to overcome the resistance of RB cells to chemotherapy. These
results may contribute to the molecular diagnosis and targeted
therapy of RB based on HMGB1.
References
|
1
|
Zheng M, Wu H, Wang J, Jin G, Jiang W and
Sha O: Progress of retinoblastoma pathogenesis. Anat Res.
37:223–228. 2015.In Chinese.
|
|
2
|
Li J and Bo L: Progress in therapy and
research in retinoblastoma. J Clin Ophthalmol. 22:185–188. 2014.In
Chinese.
|
|
3
|
Liu J and Zhu Y: Recent advances in
retinoblastoma treatment. Recent Adv Ophthalmol. 33:91–94. 2013.In
Chinese.
|
|
4
|
Manjandavida FP and Shields CL: The role
of intravitreal chemotherapy for retinoblastoma. Indian J
Ophthalmol. 63:141–145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang J, Liu K, Song D, Ding M, Wang J,
Jin Q and Ni J: Krüppel-like factor 4 promotes high-mobility group
box 1-induced chemotherapy resistance in osteosarcoma cells. Cancer
Sci. 107:242–249. 2016. View Article : Google Scholar :
|
|
6
|
Li X, Wang S, Chen Y, Liu G and Yang X:
miR-22 targets the 3′UTR of HMGB1 and inhibits the HMGB1-associated
autophagy in osteosarcoma cells during chemotherapy. Tumour Biol.
35:6021–6028. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Naumnik W, Nilklińska W, Ossolińska M and
Chyczewska E: Serum levels of HMGB1, survivin, and VEGF in patients
with advanced non-small cell lung cancer during chemotherapy. Folia
Histochem Cytobiol. 47:703–709. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ran X, Yang J, Liu C, Zhou P, Xiao L and
Zhang K: MiR-218 inhibits HMGB1-mediated autophagy in endometrial
carcinoma cells during chemotherapy. Int J Clin Exp Pathol.
8:6617–6626. 2015.PubMed/NCBI
|
|
9
|
Luo Y, Chihara Y, Fujimoto K, Sasahira T,
Kuwada M, Fujiwara R, Fujii K, Ohmori H and Kuniyasu H: High
mobility group box 1 released from necrotic cells enhances regrowth
and metastasis of cancer cells that have survived chemotherapy. Eur
J Cancer. 49:741–751. 2013. View Article : Google Scholar
|
|
10
|
Hu Y, Yang L and Zhang C: HMGB1-a as
potential target for therapy of hematological malignancies-review.
J Exp Hematol. 24:560–564. 2014.In Chinese.
|
|
11
|
Cao Q, Liu Y and Ling C: Research progress
of the correlation between HMGB1 and tumor. Chin Clin Oncol.
17:282–285. 2012.In Chinese.
|
|
12
|
Stoetzer OJ, Fersching DM, Salat C,
Steinkohl O, Gabka CJ, Hamann U, Braun M, Feller AM, Heinemann V,
Siegele B, et al: Circulating immunogenic cell death biomarkers
HMGB1 and RAGE in breast cancer patients during neoadjuvant
chemotherapy. Tumour Biol. 34:81–90. 2013. View Article : Google Scholar
|
|
13
|
Goodwin GH, Sanders C and Johns EW: A new
group of chromatin-associated proteins with a high content of
acidic and basic amino acids. Eur J Biochem. 38:14–19. 1973.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang X, Xiang L, Li H, Chen P, Feng Y,
Zhang J, Yang N, Li F, Wang Y, Zhang Q, et al: The role of HMGB1
signaling pathway in the development and progression of
hepatocellular carcinoma: A review. Int J Mol Sci. 16:22527–22540.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Erlandsson Harris H and Andersson U:
Mini-review: The nuclear protein HMGB1 as a proinflammatory
mediator. Eur J Immunol. 34:1503–1512. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Singh MK, Singh L, Pushker N, Sen S,
Sharma A, Chauhan FA and Kashyap S: Correlation of high mobility
group box-1 protein (HMGB1) with clinicopathological parameters in
primary retinoblastoma. Pathol Oncol Res. 21:1237–1242. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu
Y, Xie M, Yin X, Livesey KM, Lotze MT, et al: HMGB1-induced
autophagy promotes chemotherapy resistance in leukemia cells.
Leukemia. 25:23–31. 2011. View Article : Google Scholar
|
|
18
|
Zhang R, Li Y, Wang Z, Chen L, Dong X and
Nie X: Interference with HMGB1 increases the sensitivity to
chemotherapy drugs by inhibiting HMGB1-mediated cell autophagy and
inducing cell apoptosis. Tumour Biol. 36:8585–8592. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Prajoko YW and Aryandono T: Expression of
nuclear factor kappa B (NF-κB) as a predictor of poor pathologic
response to chemotherapy in patients with locally advanced breast
cancer. Asian Pac J Cancer Prev. 15:595–598. 2014. View Article : Google Scholar
|
|
20
|
Endo F, Nishizuka SS, Kume K, Ishida K,
Katagiri H, Ishida K, Sato K, Iwaya T, Koeda K and Wakabayashi G: A
compensatory role of NF-κB to p53 in response to 5-FU-based
chemotherapy for gastric cancer cell lines. PLoS One. 9:e901552014.
View Article : Google Scholar
|
|
21
|
Wu Y, Fang W and Li Y: Mechanisms and
reversing drugs of cancer multidrug resistance. Pharma Clin Res.
24:43–47. 2016.In Chinese.
|
|
22
|
Pan G and Yan L: Research advancement on
mechanism of tumor multidrug resistance. Med Recapitulate.
15:1162–1164. 2009.In Chinese.
|
|
23
|
Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang
R, Cao L, Tang D and Duan X: MIR34A regulates autophagy and
apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy.
10:442–452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang Y and Zhang J and Zhang J:
Progresses on autophagy and drug resistance of tumor. Life Sci Res.
19:62–67. 2015.In Chinese.
|