Introduction
Colorectal cancer (CRC), the third most common
malignancy worldwide (1), has
been treated using combination therapy, including surgery,
radiotherapy, chemotherapy and targeted drugs. However, a high
proportion of CRC patients were diagnosed in the advanced stage
(2), which resulted in poor
prognosis and recurrence or metastasis even after treatment.
Multiple genes are responsible for this process (3–8);
however, the molecular mechanism remains unclear. Therefore,
identifying novel genes is considered to be vital for analyzing
pathophysiological variations, evaluating medical conditions and
defining novel targets in CRC.
Annexins are a group of Ca2+-dependent
phospholipid-binding proteins, of which the family members include
A, B, C, D and E subgroups. In particular, numerous members of the
Annexin A (ANXA) subgroup are closely associated with cancer
development (9–11). ANXA9 is a family member of ANXA. A
Japanese study demonstrated that expression of ANXA9 mRNA in CRC
was associated with a poor prognosis (12), indicating that ANXA9 may be
associated with CRC development. However, there are few studies
regarding the molecular mechanism of ANXA9 in CRC. The aim of the
present study was to investigate the value of ANXA9 protein
detection in CRC evaluation and understand the mechanism of ANXA9
in CRC cells. Therefore, in the present study, expression levels of
the ANXA9 protein were detected in clinical samples obtained from
patients with CRC and the correlation between the ANXA9 protein and
clinicopathologic features was analyzed. In addition, the prognosis
of the CRC patients was recorded. Furthermore, the variations in
CRC cell activity, invasion and migration were investigated under
RNA interference by inhibiting ANXA9 expression in HCT116 cells and
the alteration of associated genes [ADAM metallopeptidase domain 17
(ADAM17), matrix metallopeptidase 9 (MMP-9), tissue inhibitors of
metalloproteinases-1 (TIMP-1), E-cadherin and N-cadherin] was also
detected.
Materials and methods
Ethical approval
The study protocol was approved by the Medical
Ethics Committee of the Third Hospital of Hebei Medical University
(Shijiazhuang, China). All methods used in the present study were
performed according to the International Ethical Guidelines for
Biomedical Research Involving Human Subjects, and informed consent
was obtained from all participants prior to the study.
Participant enrollment
A total of 105 CRC patients whose cancer was removed
at The Third Hospital of Hebei Medical University were recruited
between January and December 2010. The mean age was 56.36±9.19 year
(range, 38–78 years) with 30 females and 75 males. All participants
were first diagnosed with CRC without any other malignancies and
the diagnosis was conformed by pathological examination. No
participants had received radio- or chemotherapy or targeted
therapy prior to surgery. Complete clinicopathological data and
follow-ups were recorded. TNM Classification of Malignant Tumors
was performed according to Union for International Cancer
Control/American Joint Committee on Cancer gastric cancer staging
system (13). The follow-ups
ended in December 2015. Paraffin-embedded samples of tumor tissues
and adjacent mucosa (≥2 cm from the edge of the caner with no
cancer cells verified) were obtained for the detection of ANXA9
protein expression. Additional fresh samples of tumor tissues and
adjacent mucosa were collected from 20 participants who had
undergone surgery at The Third Hospital of Hebei Medical University
(Shijiazhuang, China) between March and October 2016. These samples
were maintained at −80°C and the expression level of ANXA9 proteins
were detected by western blotting.
Cell lines and reagents
Human CRC cell lines, Caco-2, HCT116, SW620, SW480
and LOVO were purchased from the Cell
Resource Center of Life Sciences (Shanghai, China). Cells from the
HT-29 cell line were preserved and incubated at the Scientific
Research Center of The Third Hospital of Hebei Medical University
(Shijiazhuang, China). Lipofectamine™ 2000 Reagent (Invitrogen;
Thermo Fisher Scientific, Inc., USA), Gibco Dulbecco's modified
Eagle's medium (DMEM) and Gibco fetal bovine serum (FBS; Thermo
Fisher Scientific, Inc.) were applied. In addition, Thiazolyl Blue
Tetrazolium Bromide (MTT), TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.), and the quantitative polymerase chain reaction
(qPCR) and protein extraction kits (cat. no. PROTTOT) were obtained
from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). The PCR
primers and ANXA9-siRNA were designed and synthesized by Sangon
Biotech, Co., Ltd. (Shanghai, China). ANXA9 (cat. no. sc-373934),
ADAM17 (cat. no. sc-390859), MMP-9 (cat. no. sc-12759), TIMP-1
(cat. no. sc-365905), E-cadherin (cat. no. sc-71008), N-cadherin
(cat. no. sc-59987) and β-actin (cat. no. sc-8432) antibodies were
obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
IHC staining assay
Paraffin specimens were deparaffinized and
rehydrated, and immunohistochemical staining of surfactant proteins
(S–P) (cat. no. sc-80621) was performed according to the
manufacturer's instruction (Santa Cruz Biotechnology, Inc.) Five
random visual fields (magnification, ×400; 100 cells per field) for
each section were evaluated by pathologists. Expression levels of
the ANXA9 protein were determined as positive if yellow or brown
plasmids were observed in the cytoplasm or on the cell membrane.
Expression positivity was scored as follows: i) Darkness of
staining (transparent, 0; light yellow, 1; brownish-yellow, 2; and
brown, 3; ii) ratio of positive to negative cells (positivity of
0%, 0; ≤10%, 1; 11–50%, 2; 51–75%, 3; and >75%). The two scores
were added and ≤2 was considered to be negative (−) and >2 was
considered to be positive (+).
Western blot assay
Tissue and cell lysates were prepared with lysis
buffer [1% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl (pH 7.4), 1 mM
ethylene diamine tetraacetic acid (EDTA), 1 mM ethylene
glycol-bis(β-aminoethylether)tetraacetic acid (pH 8.0), 0.2 mM
Na3VO4, 0.2 mM phenylmethylsulfonyl fluoride,
and 0.5% NP-40]. The samples were rinsed in ice-cold lysis buffer
for 20 min followed by centrifugation for 10 min at 7,104 × g at
4°C. The supernatant was collected and the bicinchoninic acid assay
was performed for quantitation of protein. Equal quantities of
protein (60 µg) from each sample were separated in 10% dodecyl
sulfate, sodium salt-polyacrylamide gel electrophoresis gels and
electrotransferred to polyvinylidene difluoride membranes (100 V, 2
h). The membranes were blocked in 5% non-fat milk for 2 h at room
temperature, followed by incubation in diluted antibodies at 4°C
overnight. The following antibodies were used: Mouse anti-ANXA9
(1:200, cat. no. sc-373934), mouse anti-ADAM17 (1:200, cat. no.
sc-390859), mouse anti-MMP-9 (1:400, cat. no. sc-12759), mouse
anti-TIMP-1 (1:200, cat. no. sc-365905), mouse anti-E-cadherin
(1:200, cat. no. sc-71008), mouse anti-N-cadherin (1:800, cat. no.
sc-59987), mouse anti-β-actin (1:200, cat. no. sc-8432) all Santa
Cruz Biotechnology, Inc. Following three rinses with Tris-Hcl, NaCl
and Tween-20 (TBST), blots were incubated with
peroxidase-conjugated donkey anti-mouse antibody (1:2,000; cat. no.
AB10085; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA,
USA) for 2 h at room temperature. After three rinses with TBST and
one with TBS, the optical density (OD) of the bands was detected
using an enhanced chemiluminescence detection system. The
concentration of proteins in the samples was then determined by
comparing the OD of the samples to the standard curve.
Cell culture
All cell lines were cultured in DMEM supplemented
with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin. Cells
were maintained at 37°C in an incubator saturated with 5%
CO2. Cells were dissociated with 0.25% trypsin
containing 0.02% EDTA and were passaged. Cells in the exponential
growth phase were used for the experiments.
ANXA9-siRNA transfection
Design and synthesis of the sequence of siRNA
targeting ANXA9 was performed as below: siRNA,
5′-GCAGUCUACAAACACAAUUtt-3′ and non-specific control siRNA
(NS-siRNA), 5′-UUCUCCGAACGUGUCACGUtt-3′. HCT116 cells were
transplanted into 6-well plates 24 h prior to transfection (density
of 1×106 cells/ml). Plasmid transfection was performed
using Lipofectamine™ 2000 according to the manufacturer's
instructions after samples were washed with serum- and
antibody-free DMEM. Efficiency of transfection and ANXA9
suppression was detected 24 h after transfection. Experimental
samples were divided into three groups according to the
transfection status, which were the Lipofectamine™ 2000 transfected
group (blank group), NS-siRNA transfected group (NS-siRNA group)
and the ANXA9-siRNA transfected group (ANXA9-siRNA group).
RNA extraction and qPCR
Total cellular RNA in the tissue specimens and cells
of different groups was extracted using TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
qPCR was performed in a total volume of 20 µl containing 1 µl
reverse transcription product as a template for PCR, 2X UltraSYBR
mixture (10 µl; Applied Biosystems; Thermo Fisher Scientific,
Inc.), 10 µmol/l per 1 µl primer, 8 µl DNase/RNase-Free water. The
primer sequences used in PCR are presented in Table I. PCR was performed over 35 cycles
as follows: Initial denaturation at 95°C for 5 min, denaturation at
95°C for 30 sec, annealing at 60°C for 30 sec and elongation at
72°C for 30 sec. Fluorescence was detected at the end of each
cycle. The specificity of the products was confirmed by melting
curve analysis. GAPDH served as an endogenous reference to
standardize relative expression levels for data analysis to
calculate the expression levels.
 | Table IPrimer sequences for quantitative
polymerase chain reaction. |
Table I
Primer sequences for quantitative
polymerase chain reaction.
| Gene | Forward primer (5′ to
3′) | Reverse primer (5′ to
3′) |
|---|
| Annexin A9 |
TGAGCCCAATTACCAAGTCC |
GTTCAGCCAAACACGGAAAT |
| ADAM metallopeptidase
domain 17 |
ATCAAACCCTTTCCTGCG |
CAAACCCATCCTCGTCCA |
| Matrix
metallopeptidase 9 |
AGAACCAATCTCACCGACAGG |
CGACTCTCCACGCATCTCT |
| Tissue inhibitors
of metalloproteinases-1 |
ACTTCCACAGGTCCCACAAC |
GCATTCCTCACAGCCAACAG |
| GAPDH |
GACCCCTTCATTGACCTCAACC |
GCTCCTGGAAGATGGTGAT |
MTT assay
Cells were incubated in 96-well plates at a density
of 1×105 cells/ml. When the cell density reached 70–80%
confluence, ANXA9-siRNA or NS-siRNA was transfected. The cells were
plated in 6 replicate wells per cell density. Following incubation
at 37°C for 20 h, 20 µl (5 mg/ml) MTT was added for another 4-h
incubation and discarded, followed by the addition of 150 µl DMSO
in each well and gentle shaking at room temperature for 15 min. The
OD value was measured at a wavelength of 490 nm using a microplate
spectrophotometer. Each treatment was performed in triplicate.
Wound healing assay
HCT116 cells were formed into mono-layer suspension
(density, 1×106 cell/ml) and seeded in each well of
6-well culture plates. The cells were transfected with ANXA9-siRNA
or NS-siRNA at 60–70% confluence and cultured until 100%. The
culture medium was removed and the cells were rinsed with
phosphate-buffered saline (PBS). This monolayer was then scored
using a sterile pipette tip to form scratches and was rinsed with
PBS again to remove any dislodged cells. Wound closure was
visualized using a microscope. The procedures were performed in
triplicate.
Transwell assay
Each well in the upper chamber was coated with 100
µl Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) under
ultraviolet light. HCT116 cells were suspended and plated at
1×106 cells/ml in 6-well plates. These cells were
cultured until 60–70% confluence for transfection. After a 24-h
incubation at 37°C, 200 µl cells were extracted from each group and
plated in the upper chamber of a Transwell, while DMEM was added to
the lower chamber. After removing any excess Matrigel and
non-invading cells from the upper chamber, the Transwell membranes
were fixed in methanol for 10 min and stained with crystal violet
for 30 min at 37°C. Cells on the underside of the membranes that
had invaded the Matrigel were counted under an inverted microscope
(Carl Zeiss AG, Oberkochen, Germany). The treatment was repeated
three times.
Statistical analysis
All of the data was analyzed by SPSS 26.0
statistical software (IBM Corp., Armonk, NY, USA). Quantitative
data was represented as the mean ± standard deviation, and the
deviation between groups was analyzed using one way analysis and
Dunnett t-test and variation analysis. Categorical data were
expressed as percentages and analyzed using the χ2 test.
Kaplan-Meier analysis and COX's proportional hazard regression
model were utilized to investigate the prognostic factors of the
ANXA9 protein. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of ANXA9 in CRC tissues
and adjacent mucosa
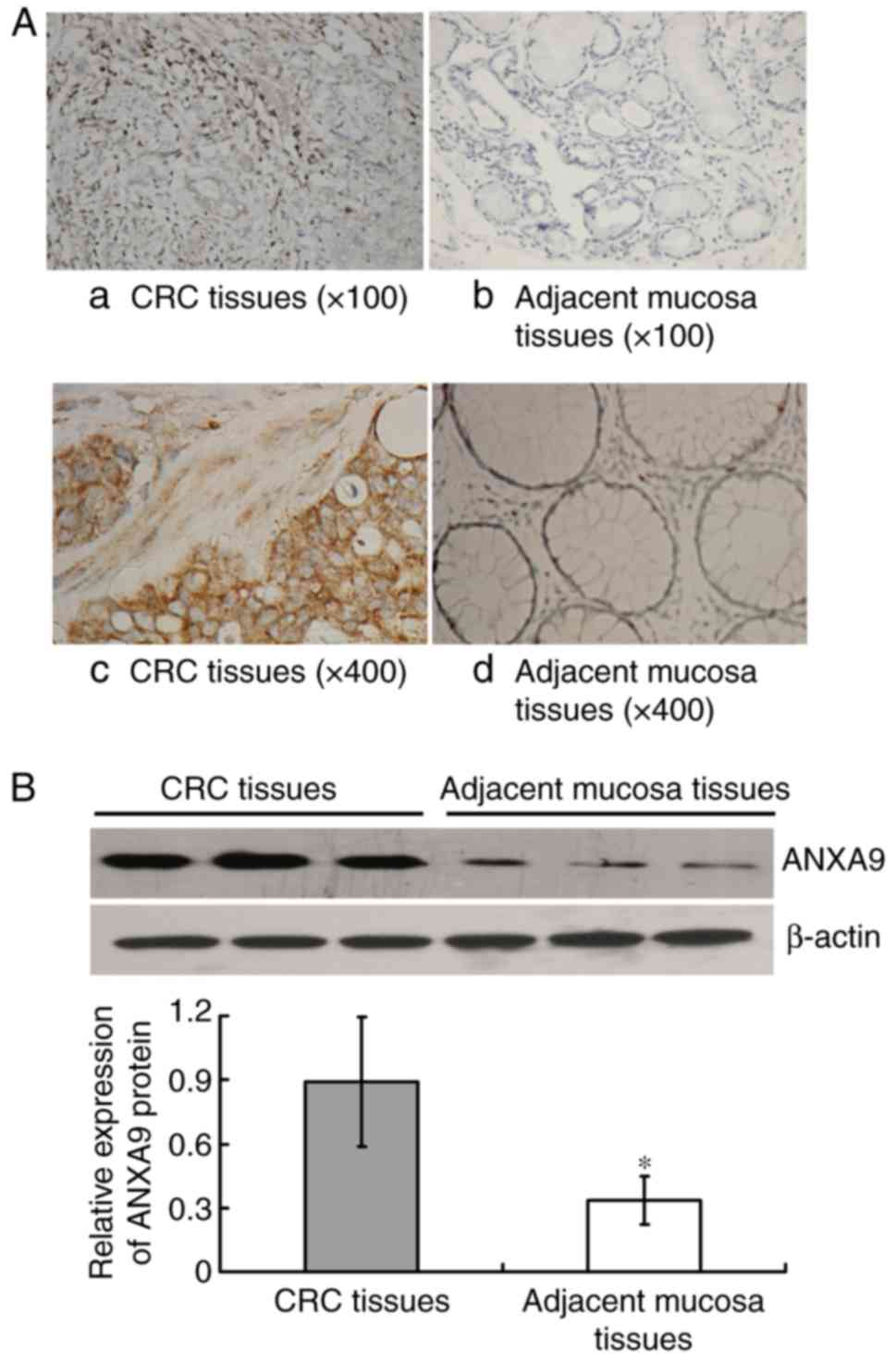
The IHC result demonstrated that the positive rate
of ANXA9 protein expression in CRC tissue samples was higher than
that in the adjacent mucosa with 76.19% (80/105), and 16.19%
(17/105), respectively (χ2=76.041; P<0.001), as
illustrated in Fig. 1A.
Similarly, the result of western blot analysis demonstrated that
the ANXA9 expression level was higher in CRC tissues compared with
the adjacent mucosa (P<0.001) (Fig. 1B).
Association between expression levels of
ANXA9 in CRC tissue and clinicopathological characteristics with
CRC patients
The result demonstrated that a higher ANXA9 positive
rate presented deeper-infiltration and lymphatic metastasis in the
CRC tissue samples (P<0.05), and no significant correlation was
identified between ANXA9 and other clinicopathological parameters
(P>0.05). The results were illustrated in Table II.
 | Table IIAssociation between ANXA9 protein and
clinicopathological parameters in CRC patients (n=105). |
Table II
Association between ANXA9 protein and
clinicopathological parameters in CRC patients (n=105).
| Clinicopathological
parameter | Positive
(n=80) | Negative
(n=25) | χ2 | P-value |
|---|
| Sex | | | | |
| Male | 59 | 16 | 0.887 | 0.346 |
| Female | 21 | 9 | | |
| Age (years) | | | | |
| ≥60 | 28 | 6 | 1.053 | 0.305 |
| <60 | 52 | 19 | | |
| Tumor
differentiation | | | | |
| Well
differentiated | 53 | 19 | 0.840 | 0.359 |
| Poorly
differentiated | 27 | 6 | | |
| Depth of
invasion | | | | |
| With serosal
infiltration | 56 | 11 | 5.576 | 0.018 |
| Without serosal
infiltration | 24 | 14 | | |
| Lymphatic
metastasis | | | | |
| Positive | 51 | 10 | 4.413 | 0.036 |
| Negative | 29 | 15 | | |
| Nerve/vessel
invaded | | | | |
| Invaded | 34 | 13 | 0.695 | 0.404 |
| Not invaded | 46 | 12 | | |
| TNM stages | | | | |
| I/II | 35 | 10 | 0.109 | 0.751 |
| III/IV | 45 | 15 | | |
| Distant
metastasis | | | | |
| Positive | 6 | 3 | 0.492 | 0.483 |
| Negative | 74 | 22 | | |
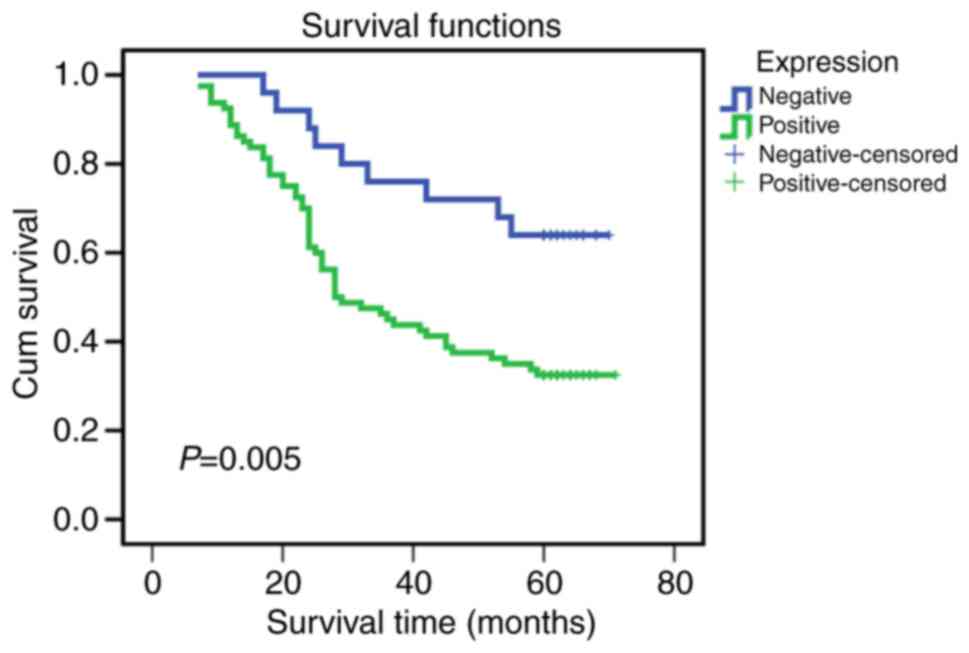
Prognostic value of ANXA9 detection for
CRC patients
The association between ANXA9 expression levels and
prognosis was analyzed and presented using a Kaplan-Meier survival
curve (Fig. 2). The data
demonstrates that the overall survival rate was lower in the
patients with positive ANXA9 expression compared with those with
negative ANXA9 expression (P=0.005). According to Cox's
proportional hazards regression model presented in Table III, the present study
illustrated that ANXA9 expression level was an independent risk
factor in CRC prognosis (P=0.022), and other independent risk
factors, including lymphatic metastasis, differentiation and
distant metastasis (P=0.017, 0.021 and 0.026, respectively).
 | Table IIIAnalysis of COX proportional hazards
model results in the colorectal cancer patients. |
Table III
Analysis of COX proportional hazards
model results in the colorectal cancer patients.
| Variable | B | SE | Wald | df | Sig | Exp(B) | 95% CI for
Exp(B)
|
|---|
| Lower | Upper |
|---|
| ANXA9
expression | 0.992 | 0.432 | 5.273 | 1 | 0.022 | 2.696 | 1.156 | 6.284 |
| Lymphatic
metastasis | 0.879 | 0.367 | 5.730 | 1 | 0.017 | 2.409 | 1.173 | 4.950 |
| TNM stages | −1.289 | 0.903 | 2.039 | 1 | 0.153 | 0.276 | 0.047 | 1.617 |
| Invasion | −.066 | 0.862 | 0.006 | 1 | 0.939 | 0.936 | 0.173 | 5.068 |
| Sex | 0.189 | 0.288 | 0.430 | 1 | 0.512 | 1.207 | 0.687 | 2.122 |
| Age (years) | −0.010 | 0.015 | 0.486 | 1 | 0.486 | 0.990 | 0.961 | 1.019 |
|
Differentiation | 0.624 | 0.272 | 5.286 | 1 | 0.021 | 1.867 | 1.097 | 3.179 |
| Nerve/vessel | −0.101 | 0.272 | 0.139 | 1 | 0.709 | 0.904 | 0.531 | 1.539 |
| Distant
metastasis | 1.000 | 0.450 | 4.949 | 1 | 0.026 | 2.719 | 1.126 | 6.563 |
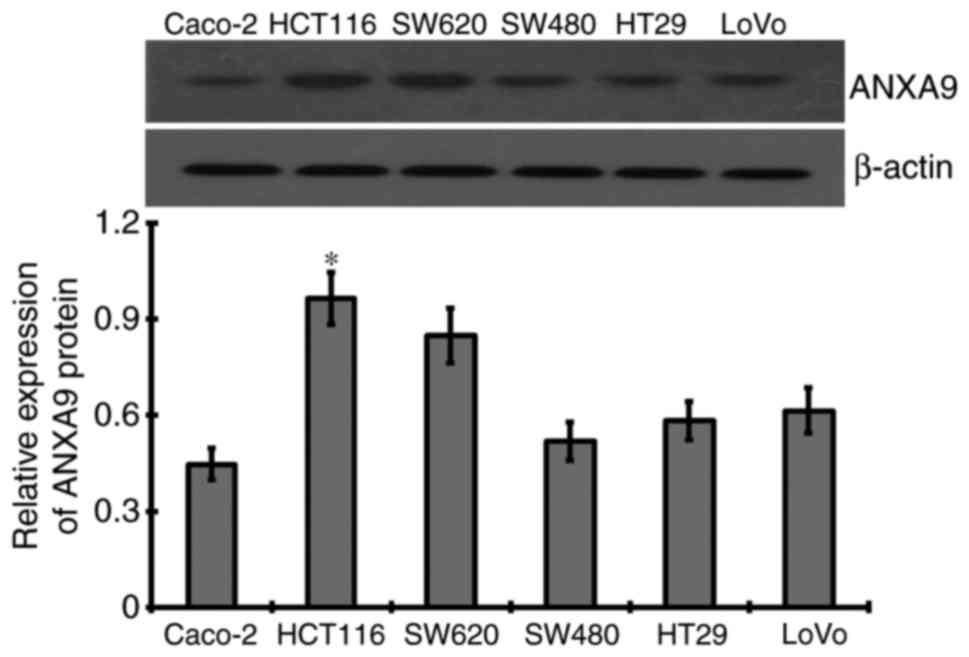
ANXA9 expression levels in CRC cell
lines
As a result of western blotting, different levels of
ANXA9 protein were detected in six CRC cell lines, among which the
highest expression level of ANXA9 protein was demonstrated in
HCT116 cells, and thus was selected for subsequent experiments
(Fig. 3).
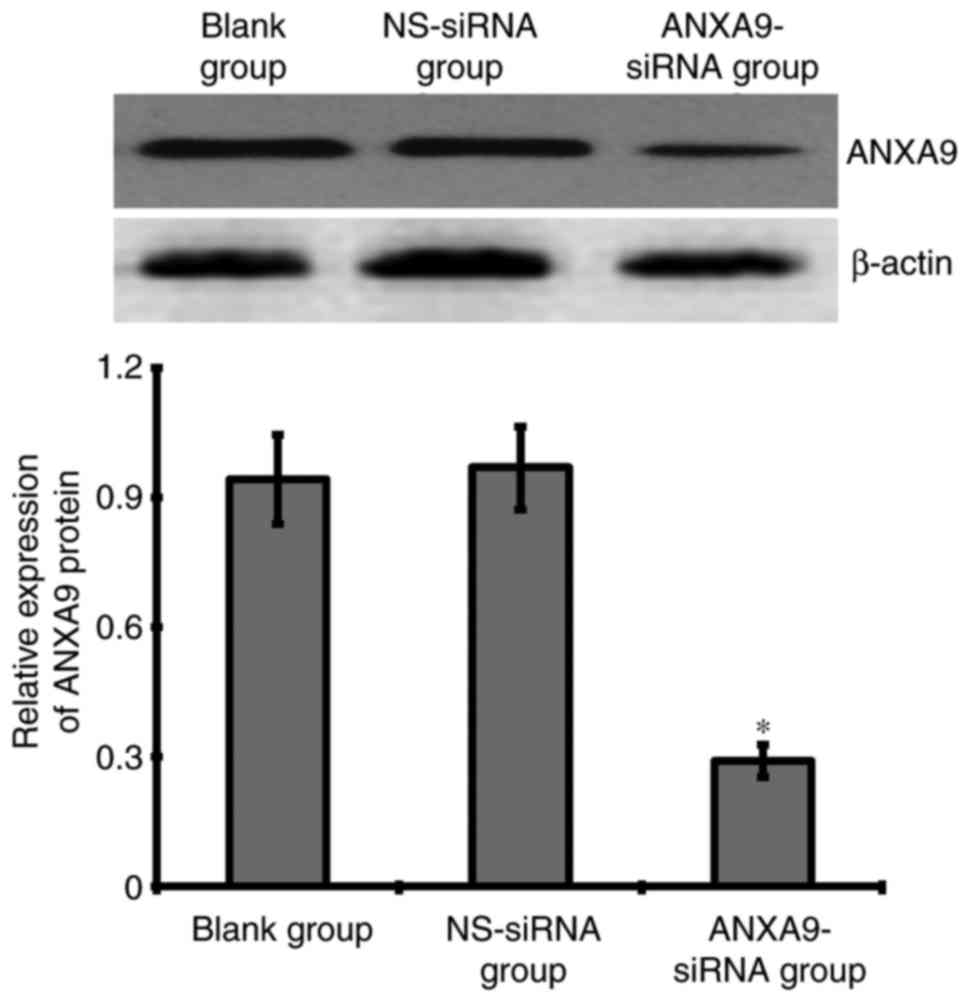
Effect of ANXA9-siRNA on ANXA9 protein in
HCT116 cells
The result of western blotting demonstrated that
after a 48-h transfection with 20 µmol/l ANXA9-siRNA, the ANXA9
expression level was downregulated more significantly when compared
with the NS-siRNA and blank groups (Fig. 4).
Impact of ANXA9-siRNA on activity of
HCT116 cells
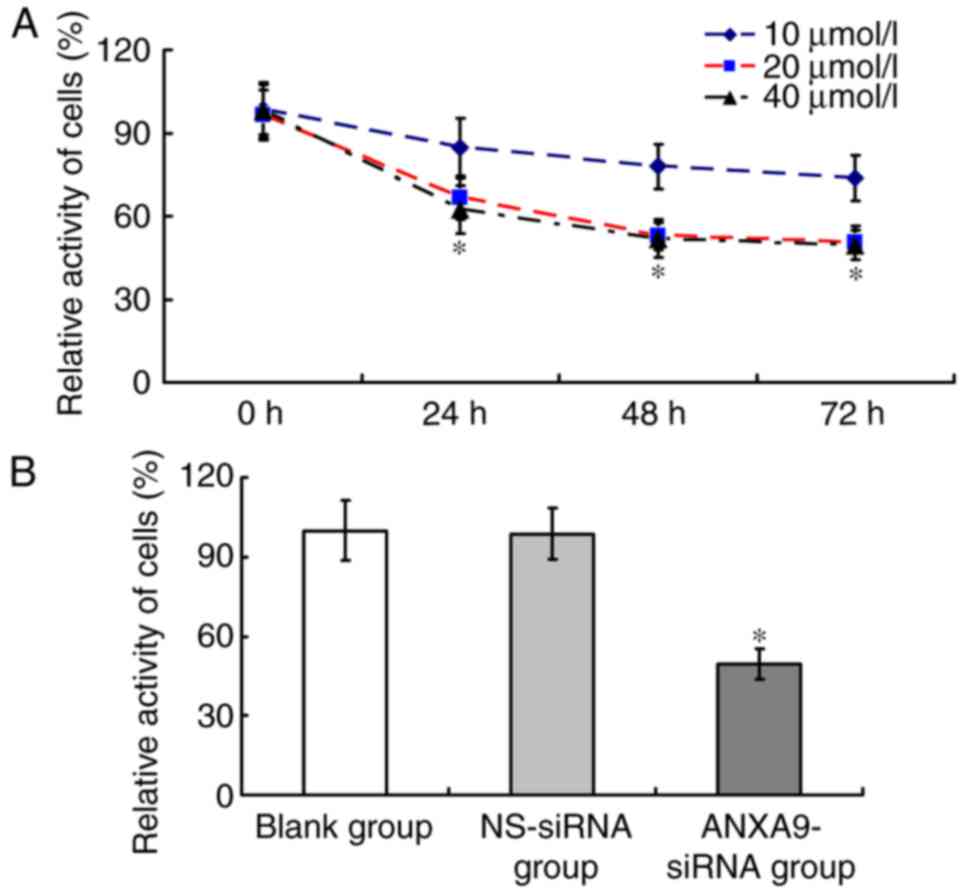
The cell activity of the ANXA9-siRNA group varied
with different concentrations and durations (Fig. 5A). Following transfection with
ANXA9-siRNA (20 µmol/l) for 48 h, the cell activity of the
ANXA9-siRNA group (49.64±5.82%) was significant lower than that in
the NS-siRNA group (98.62±9.69%) and the blank group (100±11.24%;
P<0.05), as demonstrated in Fig.
5B.
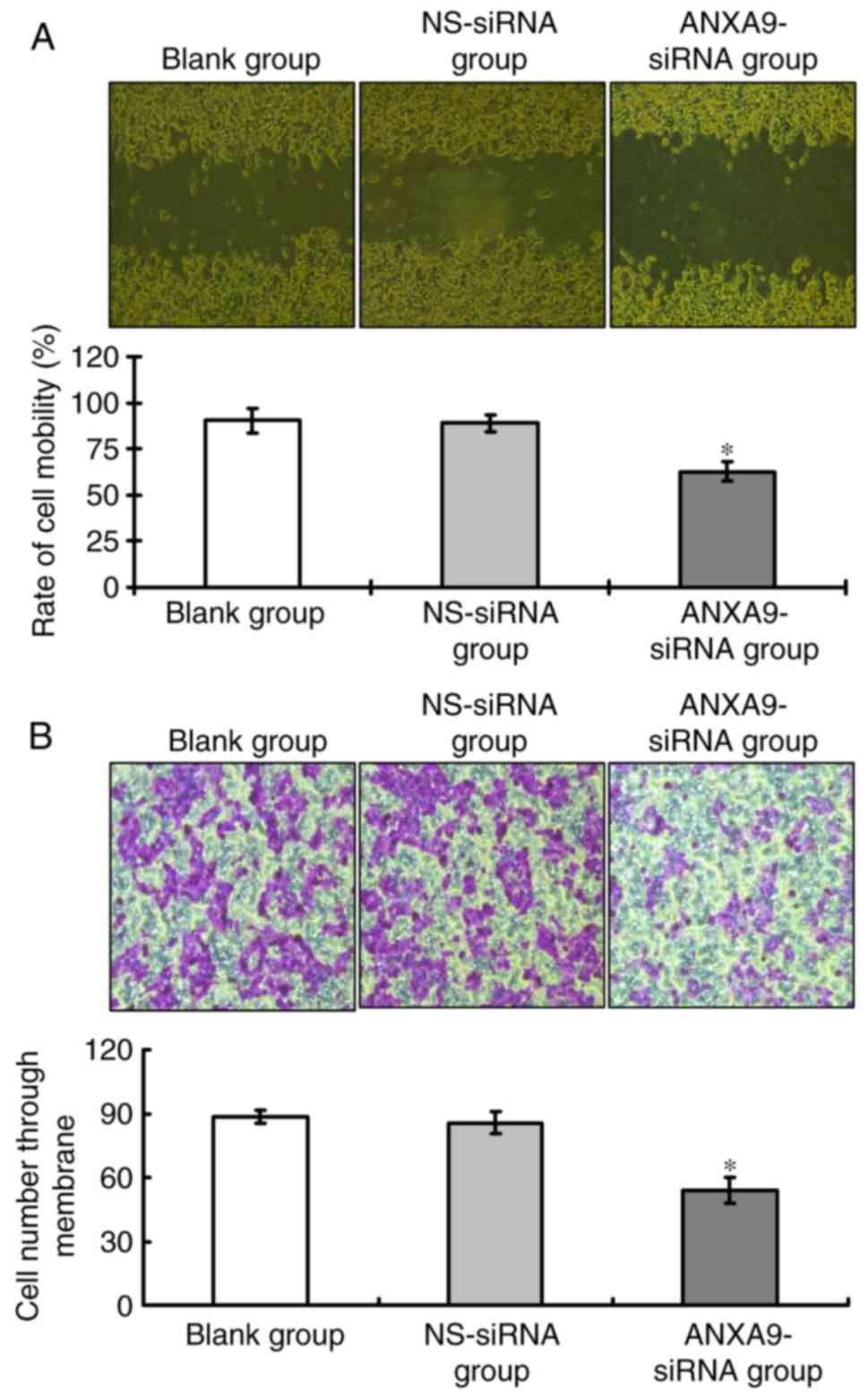
Effect of ANXA9-siRNA on migration and
invasion activities in HCT116 cells
Results of the wound healing assay (Fig. 6A) and Transwell assay (Fig. 6B) demonstrate that, following
ANXA9-siRNA transfection, the migration and invasion of HCT116
cells treated with ANXA9-siRNA were significantly decreased when
compared with the NS-siRNA group and the blank group (P<0.05),
as shown in Fig. 6.
Effect of ANXA9-siRNA on expression
levels of ADAM17, MMP-9, TIMP-1, E-cadherin and N-cadherin in
HCT116 cells
Expression levels of ADAM17 and MMP-9 mRNA and
proteins were significantly downregulated, while TIMP-1 and
E-cadherin mRNA and protein expression levels were significantly
upregulated in HCT116 cells following ANXA9-siRNA transfection
(P<0.05), and no obvious variation was observed in N-cadherin
following ANXA9-siRNA transfection (P>0.05; Fig. 7).
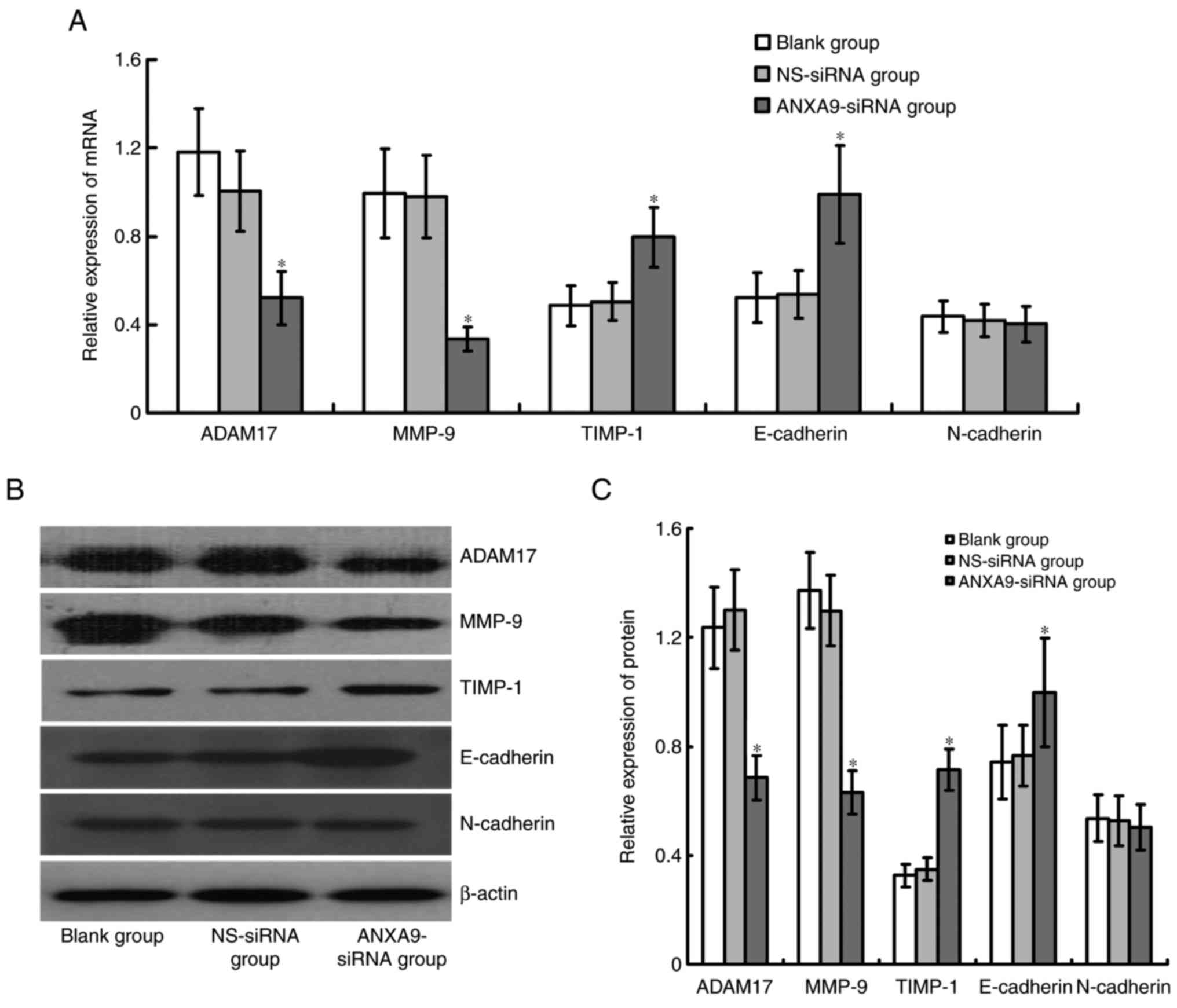 | Figure 7Effect of ANXA9-siRNA on the
expression levels of ADAM17, MMP-9 and TIMP-1 genes in HCT116
cells. (A) HCT116 cells were transfected with ANXA9-siRNA, and
subjected to quantitative polymerase chain reaction to detect the
mRNA expression levels of ADAM17, MMP-9 and TIMP-1. (B and C)
HCT116 cells were transfected with ANXA9-siRNA, and subjected to
western blot analysis to detect the proteins of ADAM17, MMP-9 and
TIMP-1. The expression levels of ADAM17 and MMP-9 decreased, while
TIMP-1 expression increased in the HCT116 cells.
*P<0.05 vs. blank group or NS-siRNA group. ANXA9,
Annexin A9; non-specific control siRNA; ADAM17, ADAM
metallopeptidase domain 17; MMP-9, matrix metallopeptidase 9;
TIMP-1, tissue inhibitors of metalloproteinases-1. |
Discussion
The incidence of CRC has increased in recent years
(14) and has unsatisfactory
treatment outcomes. Although certain risk factors of CRC have been
identified in terms of diet (15,16), environment (17) and genetics (18), factors that determine the risk of
disease remain poorly understood. In the early stages, the symptoms
of CRC are often insidious; therefore patients with CRC are
typically diagnosed at the advanced stage with a relatively rapid
progression and metastases. One of the reasons for the rapid
progression of CRC is the strong ability of the cancerous cells to
invade and metastasize (3,4).
Therefore, suppression of invasion and metastasis of CRC cells may
contribute to improvement of the treatment of this illness.
Multiple genes and signaling pathways are important in the
progression of CRC (5–8,19,20), including various members of the
ANXA family. Zhang et al (21) demonstrated that non-steroid
anti-inflammatory drugs affect the activity of the nuclear
factor-κB signaling pathway resulting in ANXA1 inhibition, which
may lead to growth suppression of CRC cells. In the study by Yang
et al (22), ANXA2 was
verified to be correlated with the clinicopathological
characteristics of CRC. Miyoshi et al (12) reported that a high expression
level of ANXA9 mRNA was a marker of poor prognosis for CRC. These
studies indicate that ANXAs are significantly associated with CRC
development. However, to the best of our knowledge, the association
between the ANXA9 gene and CRC has only been examined in one study
and only the mRNA expression level of clinical value was reported
(12).
In the present study, the clinical value of ANXA9
expression in patients with CRC was investigated in cancer and
adjacent mucosa tissue samples (obtained from 105 patients) using
IHC. The results demonstrated that a positive ANXA9 expression rate
in the cancer tissue samples was higher than that in the mucosal
tissue samples. Furthermore, the western blot result was consistent
with the IHC result, indicating that ANXA9 may be involved in
carcinogenesis development and progression. In addition, further
analysis demonstrated that ANXA9 was associated with tumor
infiltration depth and lymphatic metastasis, implying that ANXA9
may contribute to CRC invasion and migration. Furthermore,
prognostic analysis demonstrated a lower survival rate in the
patients with positive ANXA9 protein expression, which was also an
independent risk factor for patient survival. These results
indicated that ANXA9 protein may be significant in prognostic
evaluation, as well as being a marker of poor prognosis with
positive expression.
The ANXA9 gene (size, 8,233 bp) is located in human
chromosome 1q21, contains 14 exons and encodes 345 amino acid
chains (23,24). The association between ANXA9, and
CRC cell invasion and migration has not yet been defined. Our
further aim is therefore to analyze the function of ANXA9 in CRC
invasion and metastasis using RNA interference technology to
suppress ANXA9 expression of HCT116 cells in CRC. The present study
demonstrated that ANXA9 inhibition resulted in a significant
decrease in HCT116 cell proliferation, as well as decreased ability
of invasion and migration. In order to understand the molecular
mechanism of regulation by ANXA9, the changes of ADAM17, MMP-9 and
TIMP-1 expression levels were detected in HCT116 cells folloing
inhibition of ANXA9 expression. ADAM17 is a family members of
disintegrin and metalloprotease, of which the upregulated
expression in CRC participates in tumor progression (25). Furthermore, a previous study
demonstrated that suppression of ADAM17 expression in the CRC cell,
MC38CEA results in inhibition of activity and migration (26). MMP-9, an important member of the
MMP family, is significant in CRC progression (27,28), whereas TIMP-1 (an MMP-9
suppressor) inhibits MMP-9 and therefore decreases the ability of
cancerous cells to invade and migrate (29,30). The present study demonstrated that
inhibitation of ANXA9 expression resulted in reduction of ADAM17
expression levels, whereas the level of MMP-9 expression increased
in TIMP-1. In addition, ANXA9 was identified to be correlated with
ADAM17, where MMP-9 is downregulated by mediation of ADAM17
(31). These results indicate
that inhibition of ANXA9 expression levels in HCT116 cells may
induce suppression of ADMA17 and MMP-9 expression levels, but
increase the TIMP-1 expression level contributing to the weakness
of tumor cells in invasion and migration (26,32). Epithelial-mesenchymal transition
(EMT) contributes to the invasion and migration of CRC (33). E-cadherin and N-cadherin are
important in EMT of CRC (34,35). The results of the current study
demonstrate that E-cadherin was upregulated after ANXA9 inhibition,
whereas no obvious variation was verified in N-cadherin. To better
understand the association between ANXA9 and CRC, further studies
at the molecular level are required.
In conclusion, the present study demonstrates that
ANXA9 may be a novel marker of poor prognosis. Inhibition of ANXA9
expression may suppress the activity, invasion and metastasis of
CRC cells by regulating ADAM17, MMP-9, TIMP-1 and E-cadherin. This
indicates that ANXA9 may be associated with invasion and metastasis
of CRC. Thus, the current study may provide evidence for further
research into CRC development, prognostic markers and gene targeted
therapeutic strategies.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Schreuders EH, Ruco A, Rabeneck L, Schoen
RE, Sung JJ, Young GP and Kuipers EJ: Colorectal cancer screening:
A global overview of existing programmes. Gut. 64:1637–1649. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mol L, Ottevanger PB, Koopman M and Punt
CJ: The prognostic value of WHO performance status in relation to
quality of life in advanced colorectal cancer patients. Eur J
Cancer. 66:138–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Wyk HC, Roxburgh CS, Horgan PG, Foulis
AF and McMillan DC: The detection and role of lymphatic and blood
vessel invasion in predicting survival in pa'tients with node
negative operable primary colorectal cancer. Crit Rev Oncol
Hematol. 90:77–90. 2014. View Article : Google Scholar
|
|
4
|
Ning Y and Lenz HJ: Targeting IL-8 in
colorectal cancer. Expert Opin Ther Targets. 16:491–497. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu M, Wang J, Tang W, Zhan X, Li Y, Peng
Y, Huang X, Bai Y, Zhao J, Li A, et al: FOXK1 interaction with FHL2
promotes proliferation, invasion and metastasis in colorectal
cancer. Oncogenesis. 5:e2712016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeong SH, Jeon YJ and Park SJ: Inhibitory
effects of dieckol on hypoxia-induced epithelial-mesenchymal
transition of HT29 human colorectal cancer cells. Mol Med Rep.
14:5148–5154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim BR, Kang MH, Kim JL, Na YJ, Park SH,
Lee SI, Kang S, Joung SY, Lee SY, Lee DH, et al: RUNX3 inhibits the
metastasis and angiogenesis of colorectal cancer. Oncol Rep.
36:2601–2608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi W, Ye Z, Zhuang L, Li Y, Shuai W, Zuo
Z, Mao X, Liu R, Wu J, Chen S and Huang W: Olfactomedin 1
negatively regulates NF-κB signalling and suppresses the growth and
metastasis of colorectal cancer cells. J Pathol. 240:352–365. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boudhraa Z, Bouchon B, Viallard C, D'Incan
M and Degoul F: Annexin A1 localization and its relevance to
cancer. Clin Sci. 130:205–220. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qi H, Liu S, Guo C, Wang J, Greenaway FT
and Sun MZ: Role of Annexin A6 in cancer. Oncol Lett. 10:1947–1952.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lauritzen SP, Boye TL and Nylandsted J:
Annexins are instrumental for efficient plasma membrane repair in
cancer cells. Semin Cell Dev Biol. 45:32–38. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miyoshi N, Yamamoto H, Mimori K, Yamashita
S, Miyazaki S, Nakagawa S, Ishii H, Noura S, Ohue M, Yano M, et al:
ANXA9 gene expression in colorectal cancer: A novel marker for
prognosis. Oncol Lett. 8:2313–2317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bode AM, Dong Z and Wang H: Cancer
prevention and control: Alarming challenges in China. Natl Sci Rev.
3:117–127. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu J, Zhou Q, Xu J, Wang J and Zhang Y:
Detection of EGFR expression in patients with colorectal cancer and
the therapeutic effect of cetuximab. J BUON. 21:95–100.
2016.PubMed/NCBI
|
|
16
|
Bhopal RS: Diet and colorectal cancer
incidence. JAMA Intern Med. 175:1726–1727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marley AR and Nan H: Epidemiology of
colorectal cancer. Int J Mol Epidemiol Genet. 7:105–114.
2016.PubMed/NCBI
|
|
18
|
Cai Z, Han S, Li Z, He L, Zhou J, Huang W
and Xu Y: A genome-wide assessment of variations of primary
colorectal cancer maintained in metastases. Gene. 595:18–24. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang H, Xi Q and Wu G: Fatty acid synthase
regulates invasion and metastasis of colorectal cancer via Wnt
signaling pathway. Cancer Med. 5:1599–1606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zou F, Mao R, Yang L, Lin S, Lei K, Zheng
Y, Ding Y, Zhang P, Cai G, Liang X and Liu J: Targeted deletion of
miR-139-5p activates MAPK, NF-κB and STAT3 signaling and promotes
intestinal inflammation and colorectal cancer. FEBS J.
283:1438–1452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Z, Huang L, Zhao W and Rigas B:
Annexin 1 induced by anti-inflammatory drugs binds to NF-kappaB and
inhibits its activation: Anticancer effects in vitro and in vivo.
Cancer Res. 70:2379–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang T, Peng H, Wang J, Yang J, Nice EC,
Xie K and Huang C: Prognostic and diagnostic significance of
Annexin A2 in colorectal cancer. Colorectal Dis. 15:e373–e381.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boczonadi V and Määttä A: Annexin A9 is a
periplakin interacting partner in membrane-targeted cytoskeletal
linker protein complexes. FEBS Lett. 586:3090–3096. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goebeler V, Ruhe D, Gerke V and Rescher U:
Atypical properties displayed by annexin A9, a novel member of the
annexin family of Ca2+ and lipid binding proteins. FEBS
Lett. 546:359–364. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Blanchot-Jossic F, Jarry A, Masson D,
Bach-Ngohou K, Paineau J, Denis MG, Laboisse CL and Mosnier JF:
Up-regulated expression of ADAM17 in human colon carcinoma:
Co-expression with EGFR in neoplastic and endothelial cells. J
Pathol. 207:156–163. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Das S, Czarnek M, Bzowska M, Mężyk-Kopeć
R, Stalińska K, Wyroba B, Sroka J, Jucha J, Deneka D, Stokłosa P,
et al: ADAM17 silencing in mouse colon carcinoma cells: The effect
on tumoricidal cytokines and angiogenesis. PLoS One. 7:e507912012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lima AI, Mota J, Monteiro SA and Ferreira
RM: Legume seeds and colorectal cancer revisited: Protease
inhibitors reduce MMP-9 activity and colon cancer cell migration.
Food Chem. 197:30–38. 2016. View Article : Google Scholar
|
|
28
|
Liu F, Zhang T, Zou S, Jiang B and Hua D:
B7-H3 promotes cell migration and invasion through the
Jak2/Stat3/MMP9 signaling pathway in colorectal cancer. Mol Med
Rep. 12:5455–5460. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Weidle UH, Birzele F and Krüger A:
Molecular targets and pathways involved in liver metastasis of
colorectal cancer. Clin Exp Metastasis. 32:623–635. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Christensen IJ, Brünner N, Dowell B, Davis
G, Nielsen HJ, Newstead G and King D: Plasma TIMP-1 and CEA as
markers for detection of primary colorectal cancer: A prospective
validation study including symptomatic and non-symptomatic
individuals. Anticancer Res. 35:4935–4941. 2015.PubMed/NCBI
|
|
31
|
Nakayama H, Fukuda S, Inoue H,
Nishida-Fukuda H, Shirakata Y, Hashimoto K and Higashiyama S: Cell
surface annexins regulate ADAM-mediated ectodomain shedding of
proamphiregulin. Mol Biol Cell. 23:1964–1975. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao LJ, Lin P, Lin F, Liu X, Qin W, Zou
HF, Guo L, Liu W, Wang SJ and Yu XG: ADAM17 targets MMP-2 and MMP-9
via EGFR-MEK-ERK pathway activation to promote prostate cancer cell
invasion. Int J Oncol. 40:1714–1724. 2012.
|
|
33
|
Li Q, Wang Y, Lai Y, Xu P and Yang Z:
HspB5 correlates with poor prognosis in colorectal cancer and
prompts epithelial-mesenchymal transition through ERK signaling.
PLoS One. 12:e01825882017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iseki Y, Shibutani M, Maeda K, Nagahara H,
Ikeya T and Hirakawa K: Significance of E-cadherin and CD44
expression in patients with unresectble metastatic colorectal
cancer. Oncol Lett. 14:1025–1034. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yan X, Yan L, Liu S, Shan Z, Tian Y and
Jin Z: N-cadherin, a novel prognostic biomarker, drives malignant
progression of colorectal cancer. Mol Med Rep. 12:2999–3006. 2015.
View Article : Google Scholar : PubMed/NCBI
|