Introduction
Osteoarthritis (OA), a degenerative joint disease,
can give rise to articular cartilage loss, bone sclerosis,
osteophyte formation and synovial inflammation (1,2).
As the target tissue of OA, articular cartilage, which is
characterized by >90% surrounding matrix, <10%
progenitor/stem cells in cell volume and an avascular structure,
has low potential in the tissue repair and regenerative process
(3). The articular cartilage
consists of surface, middle and deep zones, with the articular
surface is crucial in regulating the appositional growth of the
tissue (4). As previously
described (5), ulcerated
cartilage, when destroyed, is never recovered. Expanding on this,
stem cell research has led to substantial advances in cell therapy
and tissue regeneration (6–9).
Embryonic stem cells, adult stem cells and induced pluripotent stem
cells have all been used for cartilage reconstruction in
vitro (10–15). Similarly, cartilage
stem/progenitor cells (CSPCs) exist extensively within human,
equine, bovine and chicken articular cartilage. Initially stated in
2003, Barbero et al reported that human articular
chondrocytes enhanced cartilage repair capacity (16). In 2004, the existence of
multipotential mesenchymal progenitor cells derived from human
articular cartilage was identified by virtue of the expression of
the cell surface markers (CD105 and CD166) (17). In 2007, isolated stem cells, which
existed in the superficial region of bovine articular cartilage,
were determined by fluorescent cell sorting and immunoblot assays
(18). Worthley et al also
stated that bone and cartilage can develop from a dedicated and
committed postnatal progenitor population (19).
To date, numerous references to the use of CSPCs in
investigations have been reported, however, the growth mechanisms
and functions of articular cartilage remain to be elucidated.
Several problems remain unresolved for CSPCs, including origin,
efficient isolation method, lack of definitive surface markers, and
application potential. The present study aimed to isolate a
population of progenitor cells from the surface zone of fetal sheep
articular cartilage, and examine their biological characteristics
with regard to growth kinetics, karyotype, immunophenotype,
specific markers and differentiation potential.
Materials and methods
Experimental animals
The 45-day-old fetal sheep embryos (12 in total, 6
males and 6 females) were provided by the Chinese Academy of
Agricultural Sciences farm. All animal procedures in the present
study were approved by the Institutional Animal Care and Use
Committee of the Chinese Academy of Agricultural Sciences (Beijing,
China). Unless otherwise stated, all experimental reagents were
purchased from Sigma-Aldrich; Merck Millipore (Darmstadt,
Germany).
CSPC isolation and culture
The method of CSPC isolation was as previously
described (20,21). All procedures were performed under
sterile conditions. Fresh joints were exposed and mechanically
peeled off from the articular cartilages of the fetal sheep, and
were washed six times with phosphate-buffered saline (PBS). The
samples were diced and the enzymatic digestion reaction was
performed with 0.2% collagenase II for 2 h at 37°C. When filtered
through a 70-μm nylon cell strainer, the cell suspension was
transferred into 15-ml conical centrifuge tubes and centrifuged at
200 × g for 8 min at room temperature. Subsequently, the cell
pellet was gently resuspended in growth medium [DMEM/F12, 10% (v/v)
FBS, 2 mM L-glutamine and 104 IU/ml
penicillin/streptomycin] and seeded in Petri dishes, which were
coated with 10 μg/ml fibronectin, at
5×104/cm2 (22). At ~24 h post-seeding at 37°C/5%
CO2, the supernatants were collected into a new cell
culture dish to remove other cells. On reaching 80-90% confluence,
the separated and purified CSPCs were disaggregated using 0.25%
Trypsin-EDTA at a ratio of 1:1 to further expansion.
Cell population dynamics assay
For the growth kinetics assay of CSPCs, the adherent
cells (1.0×104/well) at passages 5, 15 and 25 were
trypsinized, and then collected into a 24-well plate. Subsequently,
the cells from three randomly selected wells were counted each day
for 7 days. Growth curves were drawn in accordance with the mean
cell numbers, and the population doubling time (PDT) was calculated
based on the following formula: PDT = (t − t0)
lg2/(lgNt − lgN0), where t0 is the
starting time of culture; t is the time of culture termination;
N0 is the initial cell number in the culture;
Nt is the ultimate cell number in the culture.
Karyotype analysis
Chromosome spreads of the CSPCs were processed as
previously described (23,24).
Briefly, following the addition of the 0.1 μg/ml colcemid
for 4 h, the 2×106 cells were dissociated with 0.25%
trypsin-EDTA when they reached 80-90% confluence, and then treated
with 0.075 M KCL. After 30 min at 37°C, the CSPCs were fixed with
methanol/glacial acetic acid (3:1) and metaphase chromosome spreads
were observed under BX41 light microscope (magnification, ×1,000;
Olympus, Tokyo, Japan) using Giemsa staining.
Immunofluorescent detection
Cells from passage six that had reached ~60-70%
confluence, were processed by use of paraformaldehyde fixation.
Following extensive washing with cold PBS, 0.2% Triton X-100 was
used for the experiments, and the samples were transferred to PBS
containing 10% (v/v) normal goat serum (BIOSS, Beijing, China) for
30 min. Following incubation with the primary antibodies for 1 h,
the appropriate FITC-labeled goat anti rabbit IgG antibodies
(1:100; BIOSS) were subsequently used for incubation of the cells
for 1 h in the dark. All experimental procedures mentioned above
were performed at room temperature. The primary antibodies used in
the present study were as follows: Rabbit anti-CD29 (cat. no.
bs-20631R; 1:100), rabbit anti-CD166 (cat. no. bs-1251R; 1:100),
rabbit anti-collagen type I (cat. no. bs-7158R; 1:100), rabbit
anti-fibroblast growth factor receptor 3 (FGFR3; cat. no. bs-1301R;
1:100) and rabbit anti-SRY-Box 9 (SOX9; cat. no. bs-4177R; 1:100),
all purchased from BIOSS. Finally, the cells were thoroughly
counterstained with 1 μg/ml DAPI and visualized with a Nikon
TE-2000-E confocal microscope (magnification, ×200; Nikon
Corporation, Tokyo, Japan).
RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR) assays
A 2-μg sample of total RNA, which was
isolated from the cultured CSPCs (P5, P15 and P25) and
differentiation-induced cells with TRIzol reagent (Ambion; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), was subjected to RT-PCR
analysis as previously described. The extracted RNA was reverse
transcribed to cDNA using an RNA PCR kit (version 3.0; Takara Bio,
Inc., Otsu, Japan). The PCR analysis was performed using the PCR
Master Mix kit (Takara Bio, Inc.) in a 20-μl mixture
containing 1 μl template cDNA, 0.5 μl each of forward
and reverse primers, 8 μl ddH2O and 10 μl
2X PCR Mix. A thermal cycler was programmed for 35 cycles as
follows: 1 cycle at 94°C for 5 min; 35 cycles including a
denaturation step at 94°C for 30 sec, annealing step at 50-60°C for
30 sec and elongation step at 72°C for 30 sec; final single cycle
at 72°C for 8 min. Subsequently, the product sizes were assessed by
the application of 2.5% agarose gel electrophoresis. The specific
primers sequences are indicated in Table I, which were designed by NCBI
Primer-BLAST software (Primer3 and Blast; https://www.ncbi.nlm.nih.gov/tools/primer-blast/)
(25). RT-quantitative PCR
(RT-qPCR) was performed using the SYBR® Premix Ex Taq™
II kit (Takara Bio, Inc.) on an Applied Biosystems QuantStudio™ 6
Flex thermocycler (Thermo Fisher Scientific, Inc.), and the
relative gene expression was normalized to GAPDH expression, and
was calculated using the 2−ΔΔCq relative quantification
method (26). Statistical
analysis of each group included three independent samples and the
intergroup difference was analyzed via Student's t-test (27).
 | Table IPrimer sequences used in reverse
transcription-polymerase chain reaction assays. |
Table I
Primer sequences used in reverse
transcription-polymerase chain reaction assays.
| Gene | Primer
sequence | Product length
(bp) | Temperature
(°C) |
|---|
| GADPH | F:
5′-AGATGGTGAAGGTCGGAGTG-3′ | 154 | 50 |
| R:
5′-TGGGTGGAATCATACTGGAAC-3′ | | |
| ITGB1 | F:
5′-GCTAAACTCAGGAACCCTTGC-3′ | 160 | 57 |
| R:
5′-CATCAAAGCCACCTTCTGGT-3′ | | |
| ALCAM | F:
5′-TTCAGCAGCCATCACAGTTC-3′ | 135 | 60 |
| R:
5′-TTCATCCACACCACAGTTGC-3′ | | |
| FGFR3 | F:
5′-CTGGCTGAAGAACGGCAAGGA-3′ | 97 | 58 |
| R:
5′-CACCACGCTCTCCATGACCA-3′ | | |
| COL1A2 | F:
5′-GGTCATCACGGCGATCAAGGT-3′ | 106 | 60 |
| R:
5′-GCTGTCCAGTGCGACCATCTT-3′ | | |
| SOX9 | F:
5′-ACCGCCTTGTCGTTAGACTG-3′ | 116 | 60 |
| R:
5′-GAATCTCCATCGTCCTCCAC-3′ | | |
| OPN | F:
5′-AGGTGATAGTGTGGCTTATG-3′ | 233 | 58 |
| R:
5′-GATTGGAATGCTTGCTCTC-3′ | | |
| COLL1 | F:
5′-CAGAATGGAGCAGTGGTT-3′ | 305 | 58 |
| R:
5′-GCAATGGTAGGTGATGTTC-3′ | | |
| LPL | F:
5′-TGAAGACTCGTTCTCAGATG-3′ | 218 | 57 |
| R:
5′-CAATTCTCCAATATCCACCTC-3′ | | |
| PPAR-γ | F:
5′-ATCAAGTTCAAGCACATCAG-3′ | 154 | 58 |
| R:
5′-CATTCAAGTCAAGGTTCACA-3′ | | |
Flow cytometric analysis
Flow cytometry was used to positively identify the
CSPCs. Briefly, cells of P6 in the logarithmic phase were collected
into the FACS tubes and processed with cold 70% ethanol fixation at
4°C overnight. Subsequently, 1% BSA (cat. no. A7030; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) in PBS was added to block
nonspecific reacting. The expression levels of specific markers of
the culture-expanded cells were evaluated by incubation with
fluorescence-conjugated rabbit anti-CD29, rabbit anti-CD166, rabbit
anti-collagen type I, rabbit anti-FGFR3, rabbit anti-SOX9
monoclonal antibodies at 1:100 at room temperature for 1 h using
flow cytometry.
In vitro differentiation of CSPCs
For the assessment of differentiation potential, the
present study aimed to determine whether the CSPCs of the fetal
sheep were able to differentiate into adipogenic, osteogenic or
chondrogenic lineages. To examine adipogenic differentiation
potential, the CSPCs (P4) were incubated under adipogenic induction
conditions comprising DMEM/F12 supplemented with 10% FBS, 1 mM
dexamethasone, 0.5 mM IBMX, 10 mg/ml insulin and 60 mM
indomethacin. Following one week, adipogenic differentiation was
evaluated using intracellular lipid accumulation by Oil Red O
staining solution for 30 min at room temperature. To identify the
osteogenic differentiation potential, the cells were cultured under
osteogenic induction conditions, which comprised 10% FBS, 0.5 mM
dexamethasone, 50 μg/ml vitamin C and 10 mM
β-glycerophosphate. Subsequent to two weeks, the osteogenic
differentiation potential was determined by the calcium salt
deposition by Alizarin Red staining solution for 30 min at room
temperature. Chondrogenic differentiation was processed in 2-D
monolayer cultures and in pellet mass cultures under chondrogenic
induction (CID) conditions. CSPCs (P4) in 2-D monolayer cultures
were treated with 5% FBS, 1% insulin-transferrin-selenium, 50
μg/ml L-proline, 0.1 μM dexamethasone, 0.9 mM sodium
pyruvate, 50 μg/ml vitamin C and 10 ng/ml transforming
growth factor-β3 for 21 days. The differentiation induction medium
was refreshed twice weekly. For the pellet mass cultures, the cells
were resuspended and transferred to 1.5 ml conical polypropylene
tubes. The samples, which consisted of 2.5×105 cells in
chondrogenic medium, were centrifuged at 200 × g for 5 min at room
temperature to form a pellet. Following induction for 21 days, the
pellets were fixed and then embedded into paraffin wax.
Subsequently, the samples were subjected to histochemical Alcian
Blue and Toluidine Blue staining. The normal goat serum (BIOSS) was
used to block nonspecific reactions. The samples were then stained
with rabbit anti-collagen type I (cat. no. bs-7158R; 1:100; BIOSS)
and rabbit anti-collagen type II (cat. no. bs-10589R; 1:100; BIOSS)
overnight at 4°C, respectively. The subsequent day, the sections
were treated with FITC-conjugated goat anti-rabbit antibody (cat.
no. ZF-0311; 1:100; OriGene Technologies, Inc., Rockville, MD, USA)
for 1 h at room temperature and were then processed using 1
μg/ml DAPI for nuclear staining. Finally, the
differentiation capacities of the induced cells were visualized by
confocal microscopy and RT-PCR analysis, respectively. RNA samples
of CSPCs prior to differentiation were included as negative control
groups.
Statistical analysis
All data are presented as the mean ± standard
deviation, and an unpaired two-tailed t-test was used to determine
differences between two groups (for example, induction, vs.
control). All statistical analyses were performed using GraphPad
Prism 7.0 software (GraphPad Software, Inc., La Jolla, CA,
USA).
Results
Morphology, proliferation and karyotype
analyses of CSPCs
The freshly isolated primary cells were observed to
adhere to the fibronectin-coated 6-well culture plates under the
inverted microscope by use of differential adhesion (Fig. 1Aa). Following initial seeding, the
CSPCs continued to proliferate rapidly and formed colonies. After 5
days, the colonies were expanded gradually in a monolayer culture
(Fig. 1Ab). The passaged cells at
P1, P2, P3, P5, P9, P13, P23, P25 and P28 grew rapidly, and the
morphological structure of the cells was long fusiform or polygonal
(Fig. 1Ac–k). The cells were
passaged every 2 days to P32, following which the majority of cells
exhibited cell senescence (Fig.
1Al). The growth curves of the CSPC cultures at different
passages (P5, P15 and P25), as shown in Fig. 1B, were all typically sigmoidal in
shape, indicating similar proliferative potential. The PDTs were
determined as 39.45, 42.35 and 45.72 h for the cells at P5, P15 and
P25, respectively. Karyotype analysis was used for analyzing the
reproducibly diploid CSPC cell line (Fig. 1C). The results confirmed the
frequencies of cells in P5, P15 and P25 with 2n=54 were 94.4, 93.8
and 91.6%, respectively, indicating no cross contamination.
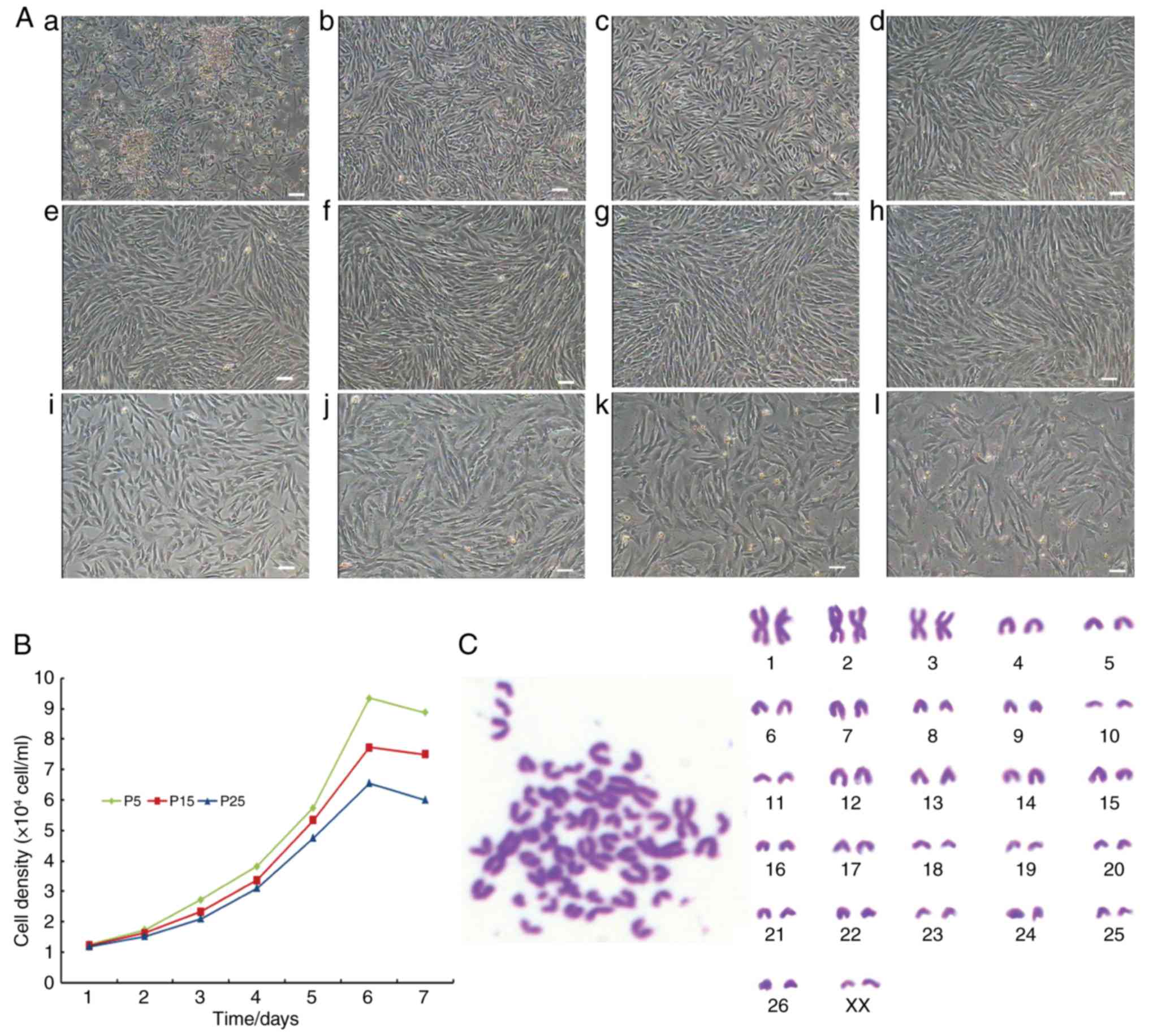 | Figure 1Morphology, growth curves and
karyotype of CSPCs. (A) Morphology of CSPCs on day (b) 1 and (b) 5
of primary culture. (c–l) Cell morphology analysis of subcultured
CSPCs at (c) P1, (d) P2, (e) P3, (f) P5, (g) P9, (h) P13, (i) P23,
(j) P25, (k) P28 and (l) P32. Scale bar=50 μm. (B) Typically
sigmoidal growth curves of CSPCs at P5, P15 and P25. (C) Karyotype
analysis of CSPCs of female (XX) type. CSPCs, cartilage
stem/progenitor cells; P, passage. |
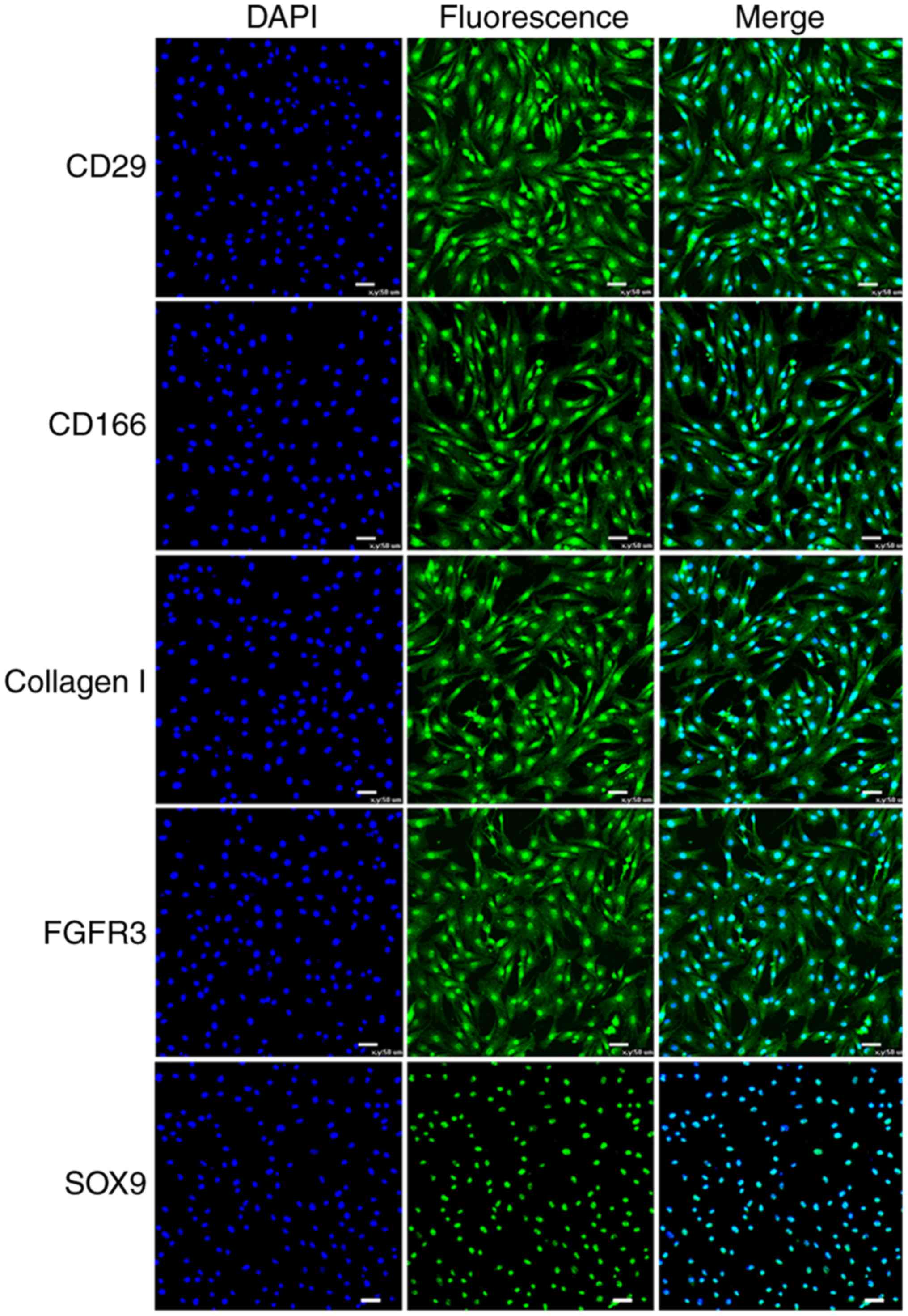
Detection of stem cell markers
The surface antigen expression was assessed using
RT-PCR analysis. Similar to human CSPCs, the CSPCs of the fetal
sheep were positive for specific marker genes integrin β1 (ITGB1),
activated leukocyte cell adhesion molecule (ALCAM), FGFR3, collagen
type I α2 (COL1A2) and SOX9 (Fig. 2A
and B). The analysis of CSPCs for FGFR3 and SOX9 by
immunofluorescence was consistent with the results mentioned above
(Fig. 3). Additionally, the
expression of markers CD29, CD166 and COL1A2 was verified by
immunohistochemical staining analysis under the confocal microscope
(Fig. 3).
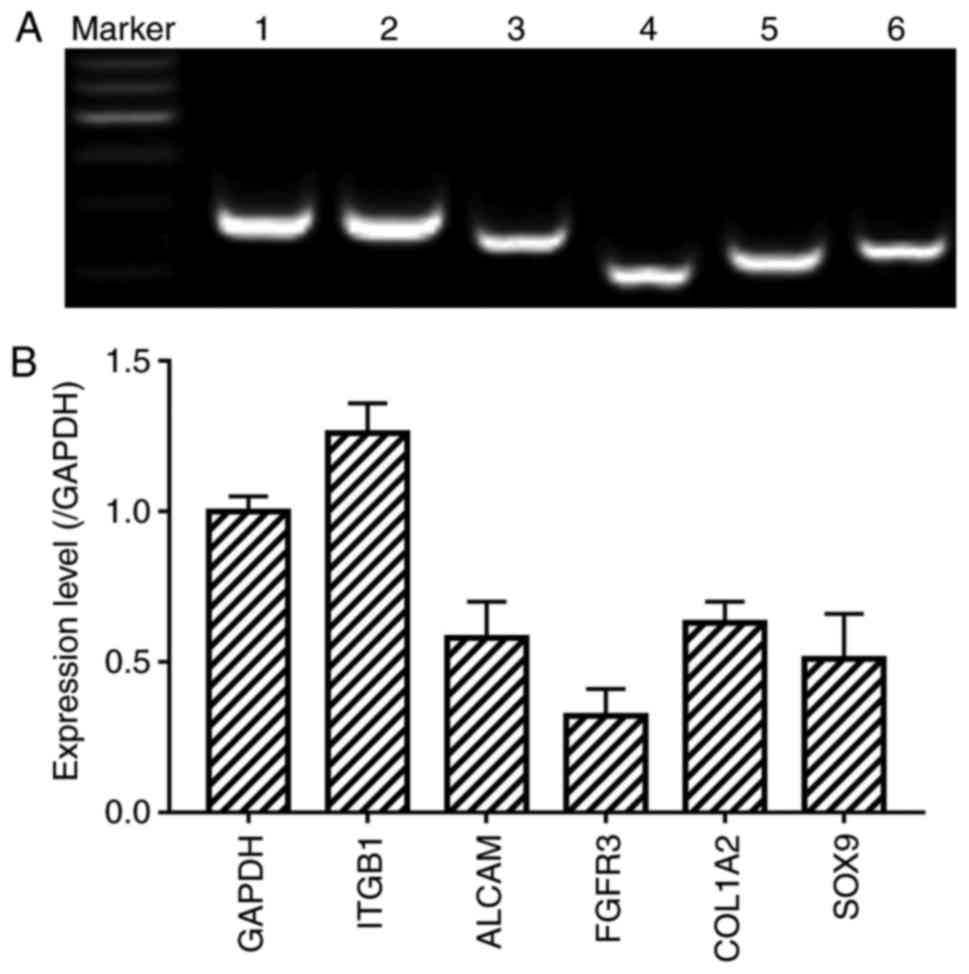 | Figure 2Detection of CSPC markers. (A)
Expression of GADPH, ITGB1, ALCAM, FGFR3, COL1A2 and SOX9 was
examined by RT-PCR analysis. Lane 1, GADPH as an internal control;
Lane 2, ITGB1; lane 3, ALCAM; lane 4, FGFR3; lane 5, COL1A2; lane
6, SOX9. Marker=600 bp; (B) Relative mRNA expression of specific
marker genes were measured by RT-PCR analysis. RT-PCR, reverse
transcription-polymerase chain reaction; ITGB1, integrin β1; ALCAM,
activated leukocyte cell adhesion molecule; FGFR3, fibroblast
growth factor receptor 3; COL1A2, collagen type I α2; SOX9, SRY
Box-9. |
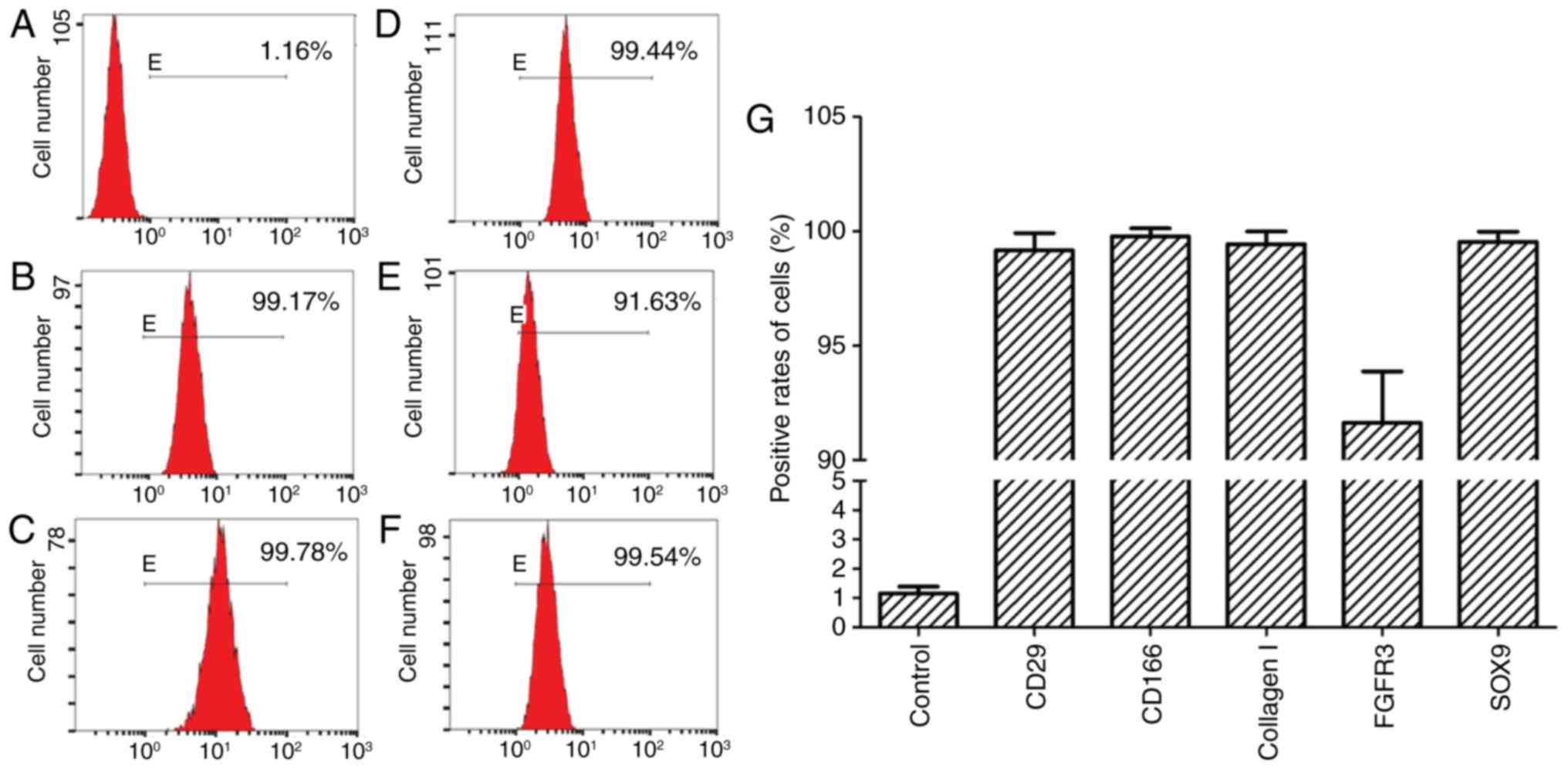
FACS analysis
Using FACS analysis, the majority of CSPCs (>90%)
expressed high levels of the above-mentioned surface markers in the
viable cell population (Fig.
4A–G), which was in accordance with the phenotype of human
CSPCs.
Differentiation of CSPCs
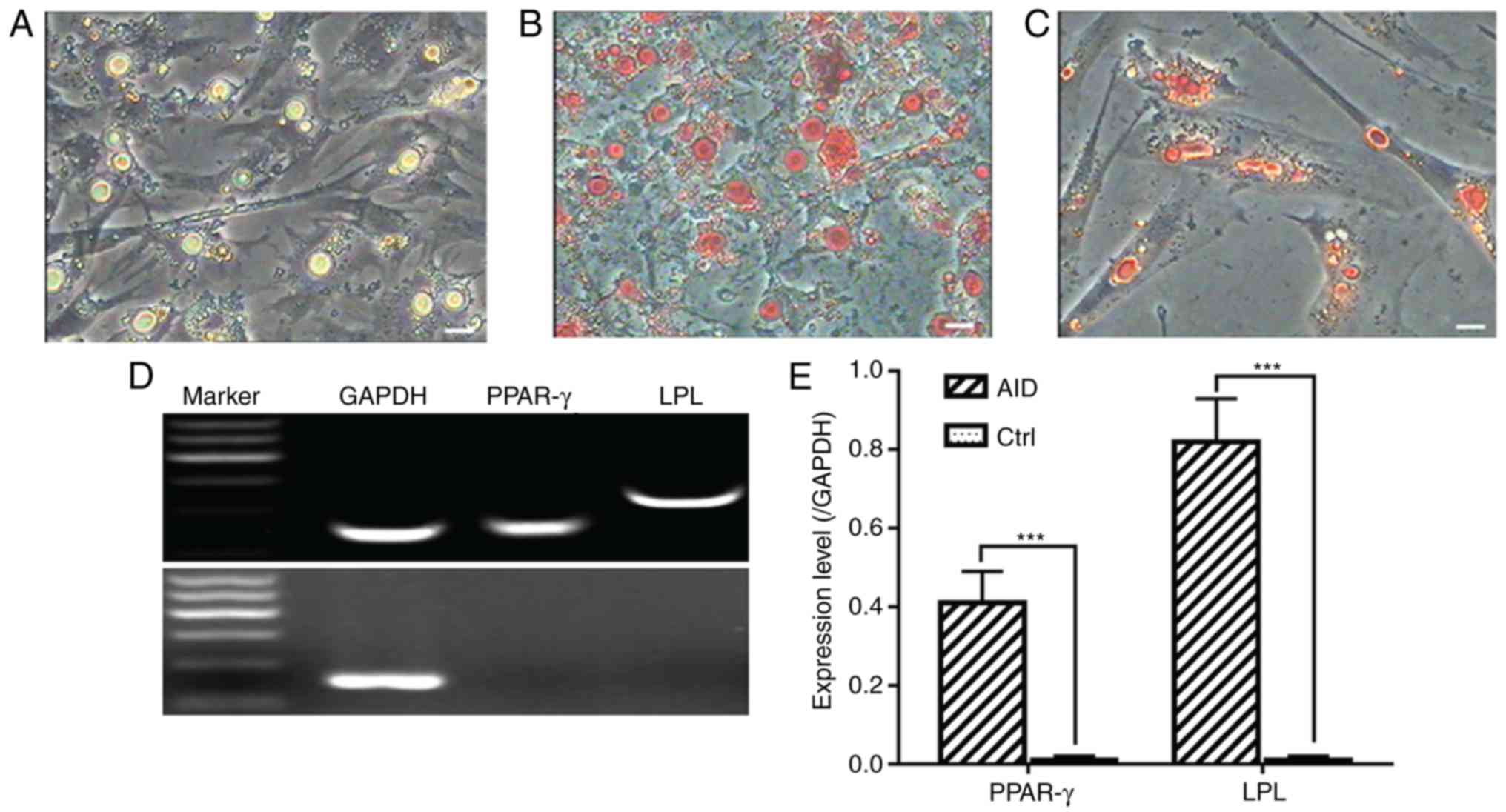
Adipogenic differentiation
For adipogenic differentiation, the CSPCs were
plated and precultured at confluence in complete culture medium,
followed by exposure to adipogenic medium for 7 days, at which time
larger droplets were generated (Fig.
5A). Positive Oil Red O staining revealed time-dependent
increases in the size and number of lipid droplets (Fig. 5B and C). The cells were cultured
for an additional 7 days, and RNA isolation of the induced cells
was performed to examine the expression of adipocyte-specific genes
peroxisome proliferator-activated receptor-γ (PPAR-γ) and
lipoprotein lipase (LPL) during adipogenesis of the CSPCs by
RT-qPCR analysis (Fig. 5D and
E).
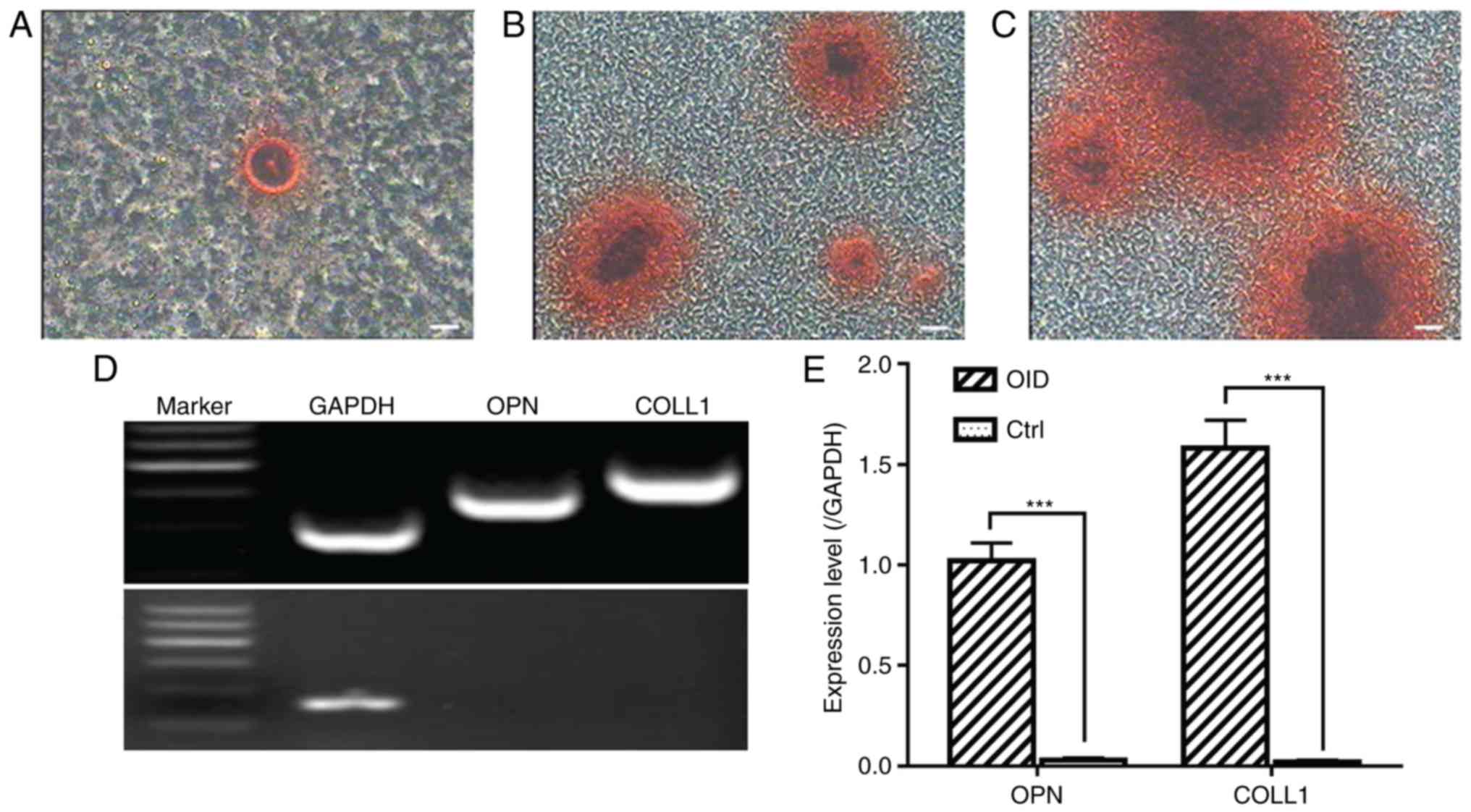
Osteogenic differentiation
The CSPCs were found to be capable of osteogenic
differentiation. Following induction in osteo-genic differentiation
medium for 7 days, the morphology of the CSPCs was altered and
mineralization appeared (Fig.
6A). As the time increased, the nodules increased in size and
number. Following incubation in osteogenic medium for 2 weeks,
Alizarin Red staining was used to reveal the formation of
mineralized bone nodules (Fig. 6B and
C). The expression of osteopontin (OPN) and COLL1 were detected
in the induced cells by RT-qPCR analysis (Fig. 6D and E).
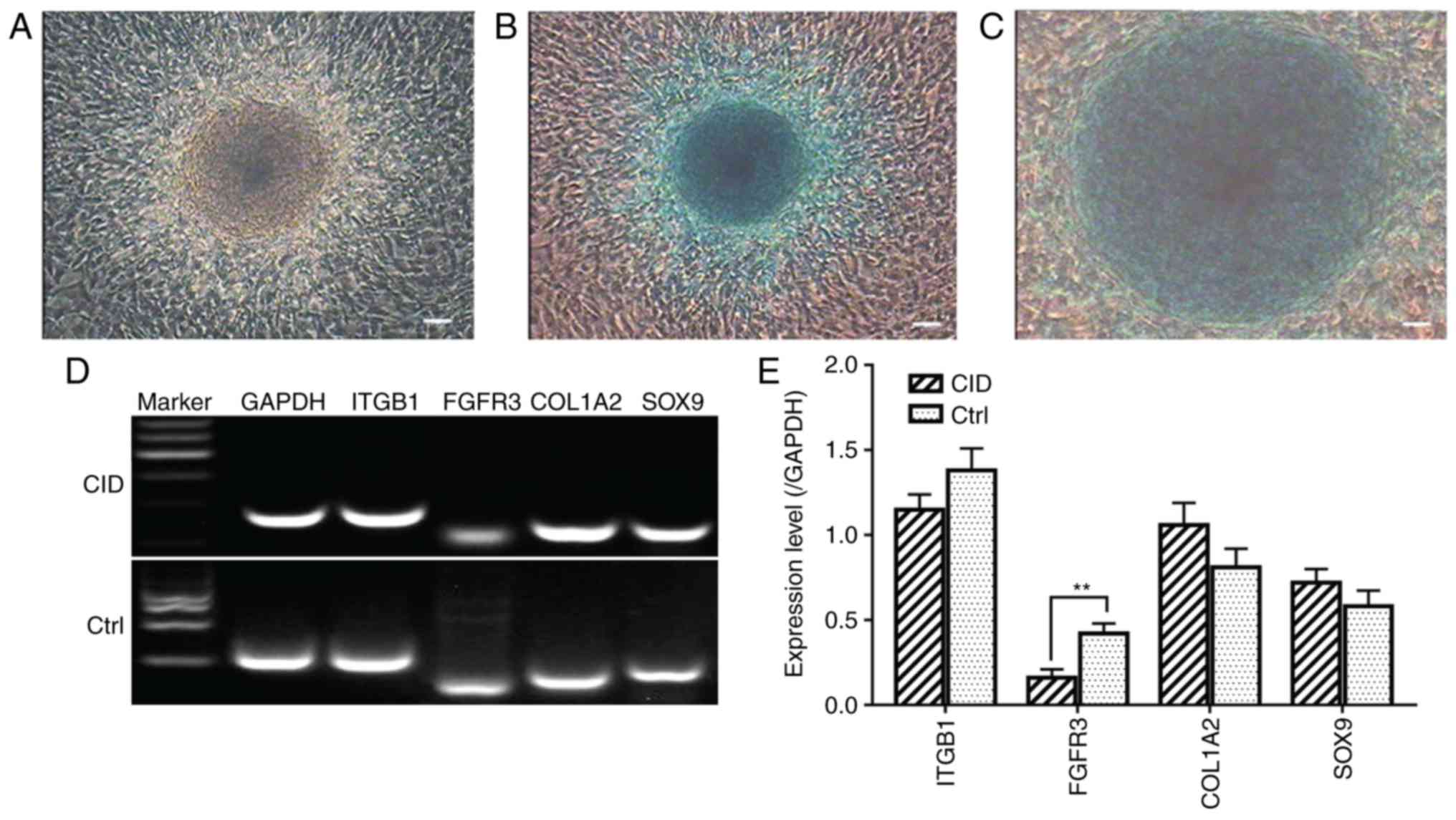
Chondrogenic differentiation
The induced CSPCs in 2-D monolayer cultures were
incubated for 21 days using chondrogenesis medium and exhibited a
notable blue color following staining by Alcian Blue (Fig. 7A–C). The total RNA of the induced
cells was isolated and RT-qPCR analysis was performed. The specific
genes detected were unchanged, with the exception of FGFR3
(Fig. 7D and E). The induced
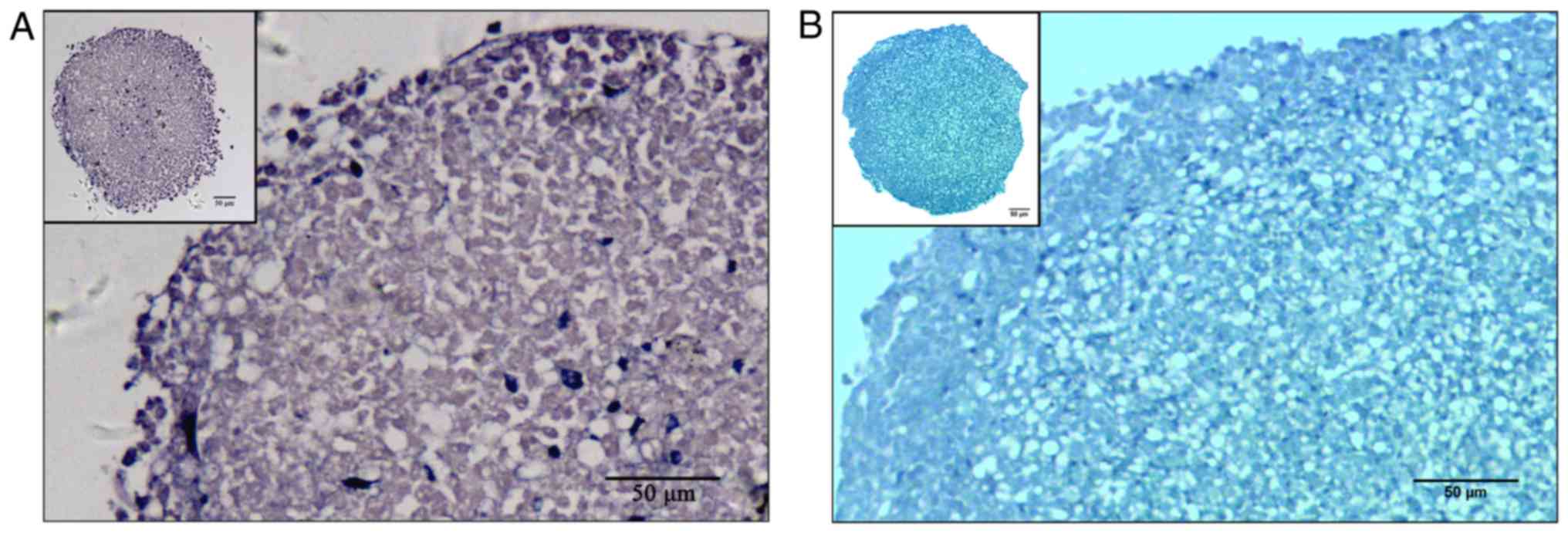
CSPCs in 3-D monolayer cultures were smooth and iridescent in
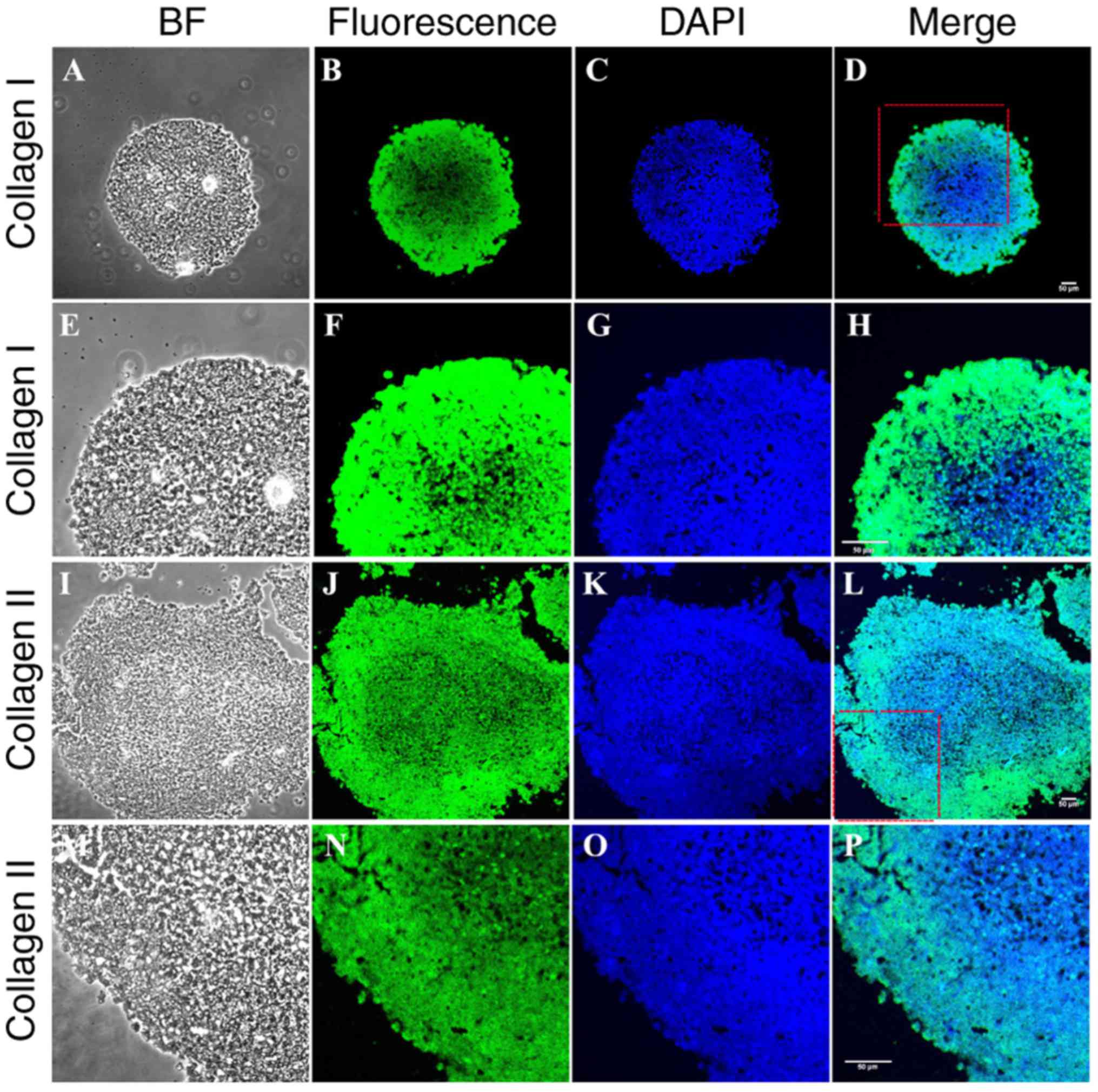
appearance, and stained positively for Toluidine Blue (Fig. 8A) and Alcian Blue (Fig. 8B). Immunohistochemistry revealed
positive labeling for type I (Fig.
9A–H) and type II (Fig. 9I–P)
collagen.
Discussion
The multipotency of stem cell populations, together
with their self-renewal ability, proliferation and
nontumorigenicity characteristics, in the culture process make them
promising candidate cells for cell-based therapies and tissue
engineering applications (28–31). In the present study, CSPCs were
successfully isolated from the articular cartilages of fetal sheep
with a fibronectin adhesion assay. The results showed that the
biological characteristics of the newly isolated stem cells were
stable. Chondrocytes and certain fibroblasts were present together,
however, the cell type was homogeneous through purification for 2-3
passages. Due to the high activity of stem cells from younger
embryos, 45-day-old Small-tailed Han sheep embryos were selected
for experimental materials. Therefore, the CSPCs were cultured
successfully in vitro and maintained high activity for at
least 32 passages. The CSPC growth dynamics were detected by
drawing of a growth curve. Growth curves are a conventional method
to detect cell growth rhythm, which has been widely used. The
results showed that the growth curve had a typical 'S' shape and
had a normal population doubling time. Karyotype analysis is an
important method for distinguishing normal cells from variants. The
CSPCs cultured in the present study were all normal diploid cells
and the genetic properties of the cells remained stable during the
sequential passaging.
At present, CSPCs have not been well defined due to
a lack of stable markers for tracing their lineage. Quintin et
al stated that isolated cells were capable of multilineage
differentiation from human fetal femurs at weeks 14-16 of
embryogenesis (32). Wu et
al used microdissection to identify different subpopulations of
cartilage-forming cells in human embryonic limb buds. In the latter
study, two cell subpopulations present at weeks 5-6 were of
particular interest: CD166low/−CD73+CD146
low/− cells, with the capacity to undergo multilineage
differentiation, including chondrogenesis; and
CD166low/−CD73+CD146low/−LIN–CD44low
cells, which were able to undergo chondrogenesis only (33). In the present study, the
distribution of cell surface markers of the fetal sheep CSPCs was
verified, including positivity for collagen I, collagen II and
relevant markers, though the use of immunofluorescence and RT-PCR
analysis. The expression rate of the fetal sheep CSPCs was also
detected by FACS analysis.
There is increasing interest in investigations
associated with the multilineage differentiation potentiality of
stem cells. In vitro, under the action of specific inducing
factors, the expression of certain key genes in the signaling
pathway associated with stem cell differentiation can change
(34,35). In the present study, fetal sheep
CSPCs were successfully induced to differentiate into chondrogenic,
osteogenic and adipogenic lineages. In addition, the expression of
type I collagen and type II collagen In CSPC pellets under CID were
consistent with the results reported by McCarthy et al
(34) and Williams et al
(36,37), suggesting that CSPCs retained
their normal phenotype characteristics in 3-D monolayer cultures.
CSPCs may be crucial in investigations targeting cartilage repair
in human and veterinary medicine following extended expansion.
Therefore, further investigations are required with regard to using
these cells for future studies and therapy.
In conclusion, a Small-tailed Han sheep embryo CSPC
line was successfully established in the present study. Cell
morphology, surface markers and biological characteristics were
observed and detected. In addition, the multipotency of CSPCs was
confirmed, with cells having the ability to be induced to
differentiate into chondrocytes, adipocytes and osteoblasts. These
findings suggested that CSPCs may be superior cells in producing
cartilage capable of functional repair.
References
|
1
|
Jo CH, Chai JW, Jeong EC, Oh S, Shin JS,
Shim H and Yoon KS: Intra-articular Injection of mesenchymal stem
cells for the treatment of osteoarthritis of the knee: A 2-year
follow-up study. Am J Sports Med. 45:2774–2783. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scotece M and Mobasheri A: Leptin in
osteoarthritis: Focus on articular cartilage and chondrocytes. Life
Sci. 140:75–78. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hayes AJ, MacPherson S, Morrison H,
Dowthwaite G and Archer CW: The development of articular cartilage:
Evidence for an appositional growth mechanism. Anat Embryol.
203:469–479. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dowthwaite GP, Bishop JC, Redman SN, Khan
IM, Rooney P, Evans DJ, Haughton L, Bayram Z, Boyer S, Thomson B,
et al: The surface of articular cartilage contains a progenitor
cell population. J Cell Sci. 117:889–897. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schmidt KJ, Tirico LE, McCauley JC and
Bugbee WD: Fresh osteochondral allograft transplantation: Is graft
storage time associated with clinical outcomes and graft
survivorship? Am J Sports Med. 45:2260–2266. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Seol D, McCabe DJ, Choe H, Zheng H, Yu Y,
Jang K, Walter MW, Lehman AD, Ding L, Buckwalter JA, et al:
Chondrogenic progenitor cells respond to cartilage injury.
Arthritis Rheum. 64:3626–3637. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alizadeh A, Moztarzadeh F, Ostad SN, Azami
M, Geramizadeh B, Hatam G, Bizari D, Tavangar SM, Vasei M and Ai J:
Synthesis of calcium phosphate-zirconia scaffold and human
endometrial adult stem cells for bone tissue engineering. Artif
Cells Nanomed Biotechnol. 44:66–73. 2016. View Article : Google Scholar
|
|
8
|
Guo DL, Wang ZG, Xiong LK, Pan LY, Zhu Q,
Yuan YF and Liu ZS: Hepatogenic differentiation from human
adipose-derived stem cells and application for mouse acute liver
injury. Artif Cells Nanomed Biotechnol. 45:224–232. 2017.
View Article : Google Scholar
|
|
9
|
Zhang Z, Pu Y, Pan Q, Xu X and Yan X:
Influences of keratinocyte growth factor-mesenchymal stem cells on
chronic liver injury in rats. Artif Cells Nanomed Biotechnol.
44:1810–1817. 2016. View Article : Google Scholar
|
|
10
|
Yamashita A, Liu S, Woltjen K, Thomas B,
Meng G, Hotta A, Takahashi K, Ellis J, Yamanaka S and Rancourt DE:
Cartilage tissue engineering identifies abnormal human induced
pluripotent stem cells. Sci Rep. 3:19782013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oakes BW, Handley CJ, Lisner F and Lowther
DA: An ultrastructural and biochemical study of high density
primary cultures of embryonic chick chondrocytes. J Embryol Exp
Morphol. 38:239–263. 1977.PubMed/NCBI
|
|
12
|
Okita K, Ichisaka T and Yamanaka S:
Generation of germ-line-competent induced pluripotent stem cells.
Nature. 448:313–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Saito T, Yano F, Mori D, Ohba S, Hojo H,
Otsu M, Eto K, Nakauchi H, Tanaka S, Chung UI and Kawaguchi H:
Generation of Col2a1-EGFP iPS cells for monitoring chondrogenic
differentiation. PLoS One. 8:e741372013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bai C, Li X, Gao Y, Yuan Z, Hu P, Wang H,
Liu C, Guan W and Ma Y: Melatonin improves reprogramming efficiency
and proliferation of bovine-induced pluripotent stem cells. J
Pineal Res. 61:154–167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang M, Yuan Z, Ma N, Hao C, Guo W, Zou G,
Zhang Y, Chen M, Gao S, Peng J, et al: Advances and prospects in
stem cells for cartilage regeneration. Stem Cells Int.
2017:41306072017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barbero A, Ploegert S, Heberer M and
Martin I: Plasticity of clonal populations of dedifferentiated
adult human articular chondrocytes. Arthritis Rheum. 48:1315–1325.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alsalameh S, Amin R, Gemba T and Lotz M:
Identification of mesenchymal progenitor cells in normal and
osteoarthritic human articular cartilage. Arthritis Rheum.
50:1522–1532. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hattori S, Oxford C and Reddi AH:
Identification of superficial zone articular chondrocyte
stem/progenitor cells. Biochem Biophys Res Commun. 358:99–103.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Worthley DL, Churchill M, Compton JT,
Tailor Y, Rao M, Si Y, Levin D, Schwartz MG, Uygur A, Hayakawa Y,
et al: Gremlin 1 identifies a skeletal stem cell with bone,
cartilage, and reticular stromal potential. Cell. 160:269–284.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li L, Ma Y, Li X, Li X, Bai C, Ji M, Zhang
S, Guan W and Li J: Isolation, culture, and characterization of
chicken cartilage stem/progenitor cells. Biomed Res Int.
2015:5862902015.PubMed/NCBI
|
|
21
|
Archer CW, McDowell J, Bayliss MT,
Stephens MD and Bentley G: Phenotypic modulation in sub-populations
of human articular chondrocytes in vitro. J Cell Sci. 97:361–371.
1990.PubMed/NCBI
|
|
22
|
Qu Z, Balkir L, van Deutekom JC, Robbins
PD, Pruchnic R and Huard J: Development of approaches to improve
cell survival in myoblast transfer therapy. J Cell Biol.
142:1257–1267. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma C, Liu C, Li X, Lu T, Bai C, Fan Y,
Guan W and Guo Y: Cryopreservation and multipotential
characteristics evaluation of a novel type of mesenchymal stem
cells derived from small tailed han sheep fetal lung tissue.
Cryobiology. 75:7–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vasa M, Fichtlscherer S, Aicher A, Adler
K, Urbich C, Martin H, Zeiher AM and Dimmeler S: Number and
migratory activity of circulating endothelial progenitor cells
inversely correlate with risk factors for coronary artery disease.
Circ Res. 89:E1–E7. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ye J, Coulouris G, Zaretskaya I,
Cutcutache I, Rozen S and Madden TL: Primer-BLAST: A tool to design
target-specific primers for polymerase chain reaction. BMC
Bioinformatics. 13:1342012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar
|
|
27
|
Lee H, Im J, Won H, Kim JY, Kim HK, Kwon
JT, Kim YO, Lee S, Cho IH, Lee SW and Kim HJ: Antinociceptive
effect of Valeriana fauriei regulates BDNF signaling in an animal
model of fibromyalgia. Int J Mol Med. 41:485–492. 2018.
|
|
28
|
Bai C, Li X, Gao Y, Wang K, Fan Y, Zhang
S, Ma Y and Guan W: Role of microRNA-21 in the formation of
insulin-producing cells from pancreatic progenitor cells. Biochim
Biophys Acta. 1859:280–293. 2016. View Article : Google Scholar
|
|
29
|
Gao Y, Bai C, Zheng D, Li C, Zhang W, Li
M, Guan W and Ma Y: Combination of melatonin and Wnt-4 promotes
neural cell differentiation in bovine amniotic epithelial cells and
recovery from spinal cord injury. J Pineal Res. 60:303–312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kariminekoo S, Movassaghpour A, Rahimzadeh
A, Talebi M, Shamsasenjan K and Akbarzadeh A: Implications of
mesenchymal stem cells in regenerative medicine. Artif Cells
Nanomed Biotechnol. 44:749–757. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mohammadian M, Abasi E and Akbarzadeh A:
Mesenchymal stem cell-based gene therapy: A promising therapeutic
strategy. Artif Cells Nanomed Biotechnol. 44:1206–1211. 2016.
|
|
32
|
Quintin A, Schizas C, Scaletta C, Jaccoud
S, Applegate LA and Pioletti DP: Plasticity of fetal cartilaginous
cells. Cell Transplant. 19:1349–1357. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu L, Bluguermann C, Kyupelyan L, Latour
B, Gonzalez S, Shah S, Galic Z, Ge S, Zhu Y, Petrigliano FA, et al:
Human developmental chondrogenesis as a basis for engineering
chondrocytes from pluripotent stem cells. Stem Cell Reports.
1:575–589. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Morgado AL, Rodrigues CM and Solá S:
MicroRNA-145 regulates neural stem cell differentiation through the
Sox2-Lin28/let-7 signaling pathway. Stem Cells. 34:1386–1395. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luo S, Shi Q, Zha Z, Yao P, Lin H, Liu N,
Wu H and Sun S: Inactivation of Wnt/β-catenin signaling in human
adipose-derived stem cells is necessary for chondrogenic
differentiation and maintenance. Biomed Pharmacother. 67:819–824.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McCarthy HE, Bara JJ, Brakspear K,
Singhrao SK and Archer CW: The comparison of equine articular
cartilage progenitor cells and bone marrow-derived stromal cells as
potential cell sources for cartilage repair in the horse. Vet J.
192:345–351. 2012. View Article : Google Scholar
|
|
37
|
Williams R, Khan IM, Richardson K, Nelson
L, McCarthy HE, Analbelsi T, Singhrao SK, Dowthwaite GP, Jones RE,
Baird DM, et al: Identification and clonal characterisation of a
progenitor cell sub-population in normal human articular cartilage.
PLoS One. 5:e132462010. View Article : Google Scholar : PubMed/NCBI
|