Introduction
The PRDI-BFI and RIZ homology domain containing
(PRDM) family contains 17 orthologs in primates, and 16 orthologs
in rodents, birds and amphibians, the regulation of which has been
associated with cancer.
PRDM2 was the first tumor suppressor identified
within the PRDMs whose expression causes apoptosis and/or cell
cycle arrest in cancer cell lines, and suppresses tumor growth
in vivo (1,2). PRDM1 was identified as a tumor
suppressor of diffuse large B-cell lymphoma in two mouse models of
PRDM1 deficiency in the B cell compartment (3). PRDM3 and PRDM16 have been identified
as different isoforms associated with leukemia (4,5),
and PRDM5 is a potential tumor suppressor involved in cell adhesion
and extracellular matrix formation along with several genes in the
Wnt pathway (6–8). Therefore, several members of the
PRDM family can suppress tumors.
Following Basic Local Alignment Search Tool searches
of the National Center for Biology Information database, the
present study focused on one of the PRDMs, PRDM13, which is
expressed in the nervous system. PRDM13 encodes a protein
containing an N-terminal PR domain and four zinc finger repeats,
which mediate sequence-specific DNA binding and protein-protein
interactions. However, to the best of our knowledge, neither the
expression nor the role of PRDM13 in glioma has been reported.
Glioma is a common central nervous system tumor, which is
frequently life threatening, despite optimized treatment including
neurosurgery, radiotherapy and chemotherapy. Therefore, it is
necessary to understand the molecular mechanisms underlying the
proliferation and migration of gliomas. In the present study, it
was identified that the expression of PRDM13 was low in U87 cells,
therefore, it was hypothesized that the upregulation of PRDM13 may
be important for glioma development. Subsequently, the effects of
PRDM13 on cell proliferation, migration and invasion were
investigated in the U87 human glioma cell line. The results
demonstrated that PRDM13 may upregulate anti-oncogenes to inhibit
glioma progression. These findings suggested that the
overexpression of PRDM13 may be a useful molecular target for
treating human glioma.
Materials and methods
Cell lines and cell culture
The U87 cell line was purchased from the Cell Bank
of the Chinese Academy of Sciences (Shanghai, China; http://www.biovector.net/search?wd=U87).
The cells were routinely cultured in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA);
all media contained 10% fetal calf serum (Biochrom, Ltd.,
Cambridge, UK) and 1% penicillin/streptomycin (Invitrogen; Thermo
Fisher Scientific, Inc.), and the cells were cultured at 37°C in an
atmosphere containing 5% CO2.
Lentivirus construction and
transfection
The PRDM13 lentivirus construct was designed by
Shanghai GeneChem Co., Ltd. (Shanghai, China) and a GFP lentivirus
was used as the negative control (NC). PRDM13 was inserted into the
Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin lentiviral vector. The U87
cells were plated in a six-well plate and the cells were
transfected with PRDM13 lentivirus and NC lentivirus (LV control)
at a multiplicity of infection of 10 with polybrene (Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany). Non-transfected cells were
used as controls. The cells were cultured in a 5% CO2
incubator at 37°C for a further 7 days. The expression of GFP was
observed by fluorescence microscopy.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted using the RNA spin Mini RNA
isolation kit (Axygen Scientific, Inc., Union City, CA, USA). cDNA
was synthesized with the Promega Reverse Transcriptase system
(Promega Corporation, Madison, WI, USA). The PCR amplification was
performed using Taq Master mix (TransGen Biotech Co., Ltd.,
Beijing, China). Thirty-one primer pairs of different genes were
designed and tested. The PCR cycling conditions were as follows:
94°C for 30 sec, followed by 40 repeats of 94°C for 5 sec and 60°C
for 30 sec. The relative expression levels of the genes of interest
were normalized to the levels of GAPDH. The results were quantified
as target gene/GAPDH using the 2−ΔΔCq method (9). The primers used are shown in
Table I. All measurements were
performed in triplicate.
 | Table IPrimer sequences for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I
Primer sequences for reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Sense (5′-3′) | Antisense
(5′-3′) |
|---|
| RORA |
AACGGGCTGACGGAACTTCA |
GGGAAGGCTGTATGTCCAGGT |
| EGR1 |
TTCCCTGAGCCACAAAGCCA |
GGAACCTGGAAGCCACCCTT |
| WNT7B |
GTGGACGCTCGGGAGATCAA |
TCCTGCCGGCCTCATTGTTA |
| BHLHE41 |
GAGAACGACACGGACACCGA |
GGAGGCTCCTGCTTGATGGT |
| ROS1 |
CTCCCAGCTGCACCCTCTG |
TGAGAGCCAAACAGCACCGA |
| PLEKHG5 |
AGAGCTCTGCCTGGCTGTTC |
AGCAGGGTGGTCCTGATTCG |
| SNAI2 |
GCACTGCGATGCCCAGTCTA |
TGCCGCAGATCTTGCAAACA |
| SEMA5A |
CCTTGCCTTGGCCCATCTCT |
GAGCAAGAGCGGGTCCTCAT |
| TWIST1 |
CCCTCGGACAAGCTGAGCAA |
TCGCTCTGGAGGACCTGGTA |
| ASAP3 |
GTGAAGAGCAGGCCTGGGAA |
GAGTGACTGCATGCGCGAAA |
| ATOH8 |
GCTGTCCAAACTGGCCATCC |
TTGCTGTGGTCGGCACTGTA |
| GADD45B |
GAGGCCCGAGACCTGCATT |
CACCAAGCCGTGGCTCTTC |
| PIM1 |
GTGCCCATGGAAGTGGTCCT |
AACTGTCGGGCCTCTCGAAC |
| ARHGAP30 |
TGGGATGGGCTACCTGGAG |
TGCCTCATCCAGAGACAGGT |
| INCA1 |
AAGTGTTCCAGGGTGGTCAG |
GGTGGGCCTTTGATTAAGGT |
| GRPR |
GACAGTGCTGGTGTTTGTGG |
TGACAAAGTGGAGCATGGAG |
| IL24 |
GCCAGCAAGCTCAGGATAAC |
ATAGCAGAAACCGCCTGTGT |
| ADAMTS12 |
GTGGGAAACAGTGGCAAGAT |
CAAGGATTGGGAAGTGAGGA |
| DIRAS3 |
GCCTTCATGGAGATTTCAGC |
GCATCTGGGATTTCTTCTCG |
| SLIT2 |
GATGTGGCCATTCAGGACTT |
TTGTTGCTACATCGGACGAC |
| TFPI2 |
CGGATTGAGAACAGGTTTCC |
ATTGTCATTCCCTCCACAGC |
| CTNNBIP1 |
ATGGGATCAAACCTGACAGC |
ATCACCACGTCCTCTGCAC |
| EPHB1 |
CCTCCCTAATGTCCCAGGAT |
CCTCAGACCAGAAGGCTGAC |
| CXCL14 |
CTACAGCGACGTGAAGAAGC |
CGCTCTTGGTGGTGATGATA |
| COL4A6 |
CTGGCTTTCTTGGCATCAAT |
CCAACAGCTCCTTGAGTTCC |
| ADAMTS6 |
AAGTCTGCCCTTCAACAACG |
ATGTGTCATTCAGCCACCAA |
| DLC1 |
CCAGCAACTTGGCAGGCAAT |
GCTTCAGGCTCTCCATCCGT |
| ITGBL1 |
GTGGTCGCTGTGTGTTTGTGAG |
CGAGCACAATATGCCATCTG |
| WNT10B |
CATCCTCAAGCGCGGTTTCC |
CTCAGCCGATCCTGCTCACC |
| SOX7 |
ATGGTTTGGGCCAAGGACGA |
CAGCGCCTTCCACGACTTTC |
| GAPDH |
AGAAGGCTGGGGCTCATTTG |
AGGGGCCATCCACAGTCTTC |
Western blot analysis
The cells were washed twice with cold PBS, and then
lysed in cold lysis buffer. The mix solution was centrifuged at
29,040 g for 50 min at 4°C. Following collection of the
supernatant, the protein concentration was determined with the BCA
method (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China),
according to the manufacturer's protocol. A total of 50 μg
of protein was loaded and separated by 8–10% SDS-PAGE, according to
molecular weight, and then transferred onto PVDF membranes.
Subsequently, the membranes were incubated with antibodies at 4°C
overnight. The antibodies used were as follows: PRDM13 (1:1,000;
cat. no. ab40542), Flag (1:1,000; cat. no. ab192404), and deleted
in liver cancer 1 (DLC1; 1:1,000; cat. no. ab1220) from Abcam
(Cambridge, MA, USA), Rho GTPase activating protein 30 (ARHGAP30;
1:500; cat. no. AP4996b; Abgent Biotech Co., Ltd., Wuhan, China), A
disintegrin and metalloproteinase with thrombospondin motifs 12
(ADAMTS12; 1:500; cat. no. 24934-1-AP; ProteinTech Group, Inc.,
Wuhan, China), inhibitor of cyclin-dependent kinase (CDK)
interacting with cyclin A1 (INCA1; 1:500; cat. no. bsm-7899R;
BIOSS, Beijing, China), and GAPDH (1:6,000; cat. no. 60004-1-Ig;
ProteinTech Group, Inc.). The membranes were washed and incubated
with the secondary antibodies (1:10,000) at room temperature for 90
min, comprising goat anti-rabbit IgG (cat. no. bsm-0295G) and goat
anti-mouse IgG (cat. no. bsm-0296G) from BIOSS. All antibodies were
diluted in 5% skim milk (BD Difco, cat. no. 2332100, SBJBio,
Nanjing, China). Finally, the membranes were developed and
quantified using Odyssey CLx Infrared Imaging system software,
Image Studio Version 4.0 (LI-COR Biosciences, Lincoln, NE,
USA).
Cell proliferation assay
Following transfection with lenti-viruses expressing
PRDM13 or the scramble control using polybrene, the U87 cells were
cultured for 24 h. The cells were collected and seeded in 96-well
plates at a density of 2,000 cells/well. Cell proliferation rates
were determined once per day for 5 days using a Cell Counting Kit-8
kit (TransGen Biotech Co., Ltd.). The samples were assayed in
triplicate and OD values were detected at 450 nm.
Flow cytometric analysis of cell
cycle
The U87 cells were transfected with lentiviruses
expressing PRDM13 or the scramble control. Following incubation for
72 h, the cells were washed twice with PBS solution, trypsinized
and fixed with pre-cooled 70% alcohol at 4°C overnight. The cells
were washed with PBS and stained with propidium iodide (Cell Cycle
Detection kit; KeyGen Biotech Co., Ltd.) at 4°C for 30 min. Flow
cytometry was performed using a FACSCalibur (BD Biosciences,
Franklin Lakes, NJ, USA).
Wound-healing assay
The U87 cells were seeded in 60-mm cell culture
plates and incubated at 37°C overnight in DMEM with 10% FBS. Upon
reaching 90% confluence, the cells were cultured in serum-free
medium for 24 h. A 200-μl sterile pipette tip was used to
create a '+' wound. The cells were washed twice with PBS to remove
cell debris and grown in FBS-free medium. Images of the wounds were
captured under a microscope and repeated three times in
duplicate.
Transwell invasion assay
Cell invasion was measured using BD BioCoat™
Matrigel™ Invasion Chambers (BD Biosciences). Transwell chambers
were coated with Matrigel at a final concentration of 1 mg/ml. A
total of 1×104 cells/ml were seeded into the upper
chamber with 100 μl serum-free DMEM, and 500 μl DMEM
with 10% FBS medium was added into the lower chamber. The cells
were incubated in a humidified environment with 5% CO2
at 37°C for 24 h. The cells on the top surface of the membrane were
removed with a cotton swab, and the migratory cells on the bottom
surface of the membrane were fixed in 90% alcohol for 30 min at
room temperature. The chambers were washed with PBS and stained
with 5% crystal violet for 30 min. Migratory cells in the bottom
surface of the membrane were counted by light microscopy. A total
of six high-power microscopic fields (magnification, ×200) were
randomly selected and images captured, and the cells were
counted.
Transcriptome sequencing
The total RNA of cells with lenti-viruses expressing
either PRDM13 or the scramble control was extracted using TRIzol
reagent. The integrity of total RNA was examined using a NanoDrop
2000 spectrophotometer (Thermo Fisher Scientific, Inc.).
Transcriptome sequencing was performed by Vazyme Biotech Co., Ltd.
(Nanjing, China). Significant differentially expressed genes and
the gene ontology (GO) function enrichment were obtained between
two groups.
Validation of RNA-sequencing (RNA-Seq)
analysis by RT-qPCR and western blot analyses
According to the transcriptome sequencing results,
genes of interest were selected relative to the proliferation and
migration function with the following criteria: P<0.05 and
absolute fold change >2. RT-qPCR and western blot analyses were
performed to validate the RNA-Seq analysis. The primers used are
listed in Table I, and the
methods for RT-qPCR and western blot analyses were as described
above.
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assay was performed using the Pierce™
Agarose ChIP kit (cat. no. 26156; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Immunoprecipitated DNA
enrichment was normalized relative to the input. The enrichment of
each target sequence was determined by RT-qPCR analysis. Flag
antibody (1:1,000; cat. no. ab192404, Abcam) was used to perform
ChIP. The primers used were as follows: DLC1 ChIP, forward 1
5′-CTCTCCGGAGGTCCTTTAGC-3′; DLC1 ChIP, reverse 1
5′-CCAGCCGTCTGTCTCTAGTGT-3′; DLC1 ChIP, forward 2
5′-CTTTGAAAACGCGAGGTCAC-3′; and DLC1 ChIP, reverse 2
5′-TGCAACCCAATGACTCACCT-3′. All samples were assessed in
triplicate.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Statistical analyses were performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Comparisons between the NC
group and the PRDM13 group were made using Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Efficient transfection of Lenti-PRDM13
into the U87 cell line
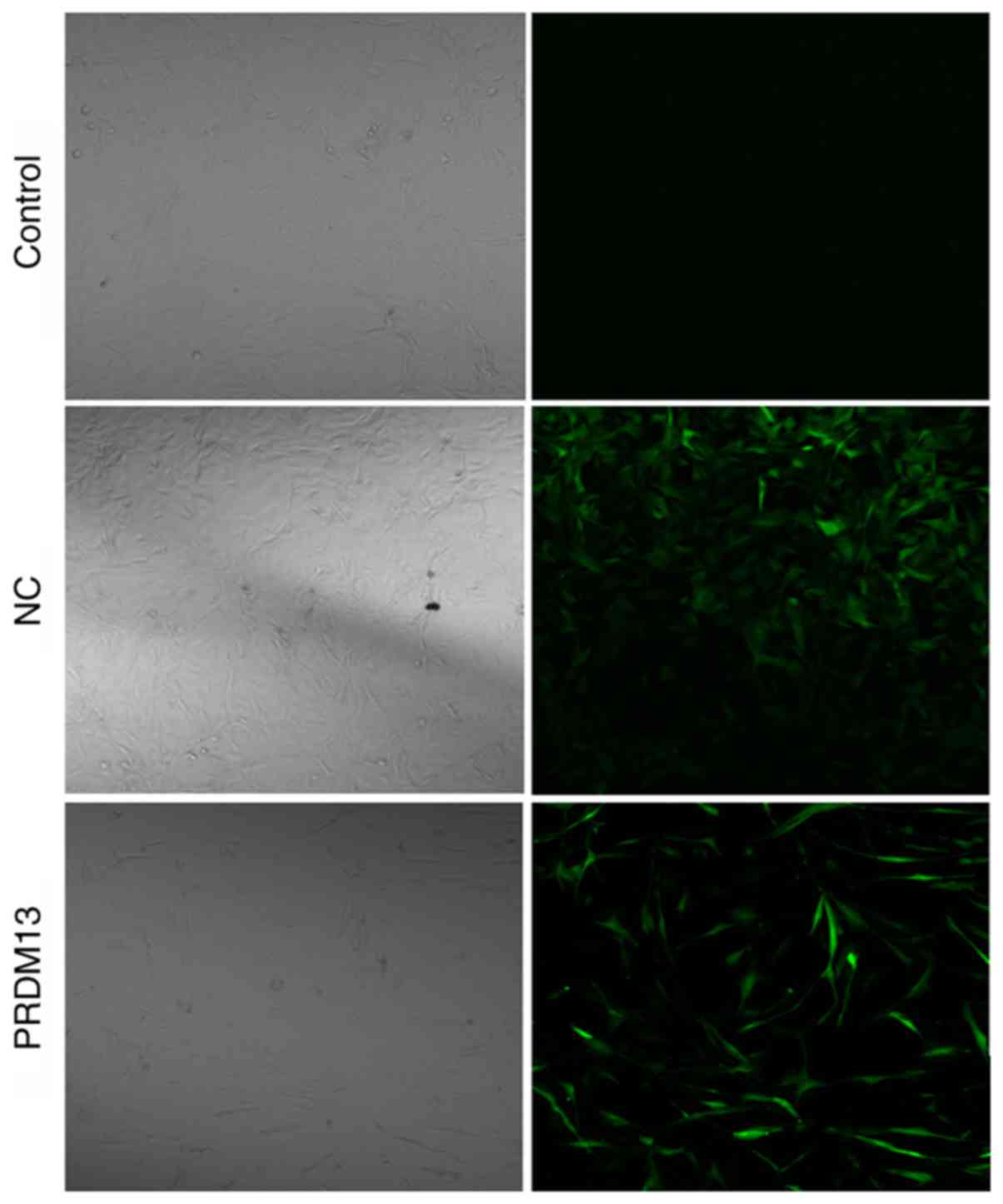
The lower expression of PRDM13 in U87 human glioma
cells suggested that PRDM13 may be involved in glioma development.
To examine the role of PRDM13 in gliomas, the Lenti-PRDM13 and
Lenti-Con (NC) lentiviruses were transfected into U87 cells. The
successful transfection of Lenti-PRDM13 or Lenti-Con into the U87
cells was confirmed by microscopic green fluorescence detection. As
shown in Fig. 1, the transfection
efficiency was >80% by 3 days post-infection with lentivirus.
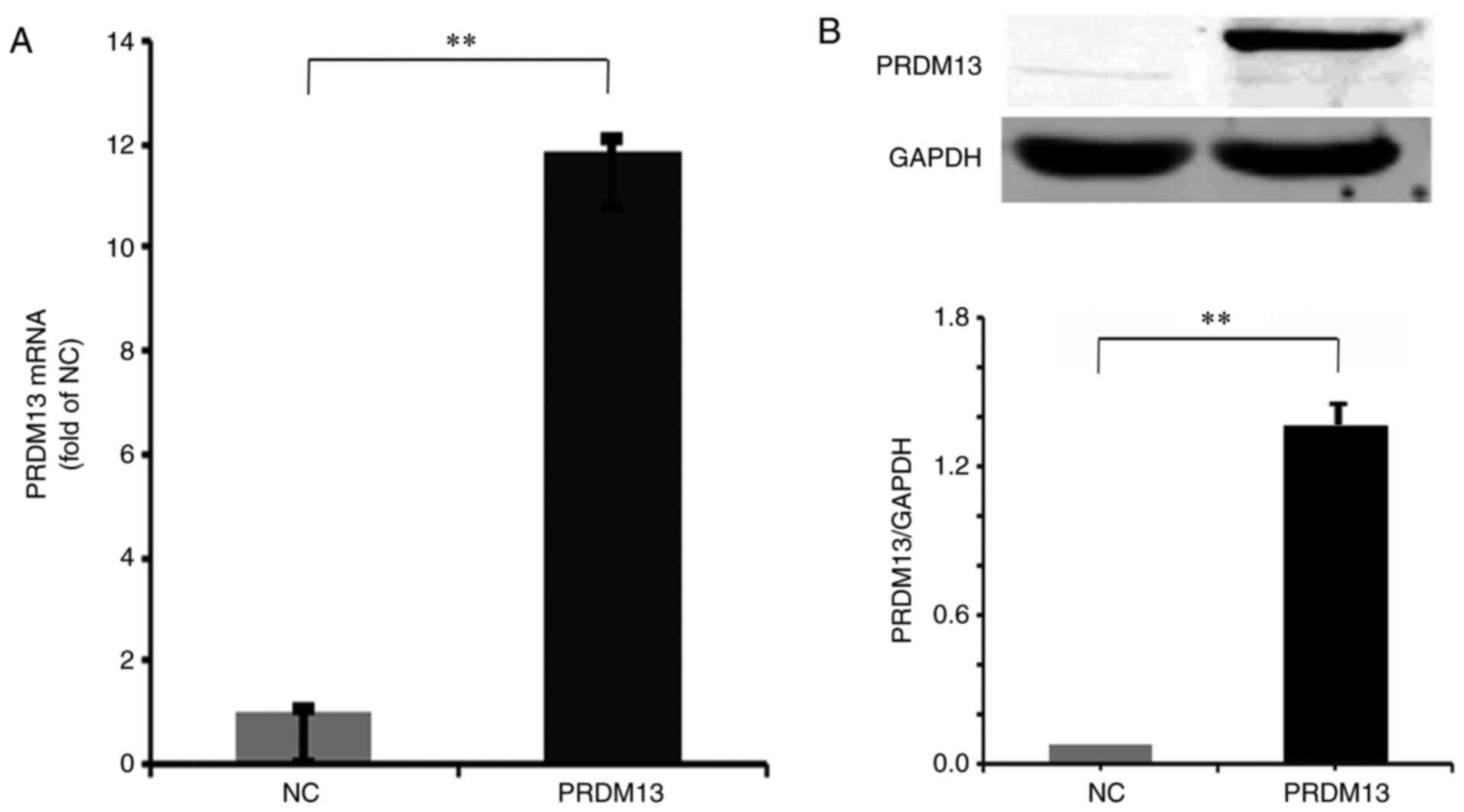
The RT-qPCR data demonstrated that the mRNA expression level of
PRDM13 was significantly increased in the Lenti-PRDM13-infected
cells (P<0.01; Fig. 2A). In
addition, the protein expression level of Flag-PRDM13, detected by
western blot analysis, was markedly increased in the
Lenti-PRDM13-infected cells, compared with that in the NC cells
(P<0.01; Fig. 2B). These data
indicated that the PRDM13 gene was effectively overexpressed at the
mRNA and protein levels.
Effects of the overexpression of PRDM13
on U87 cells
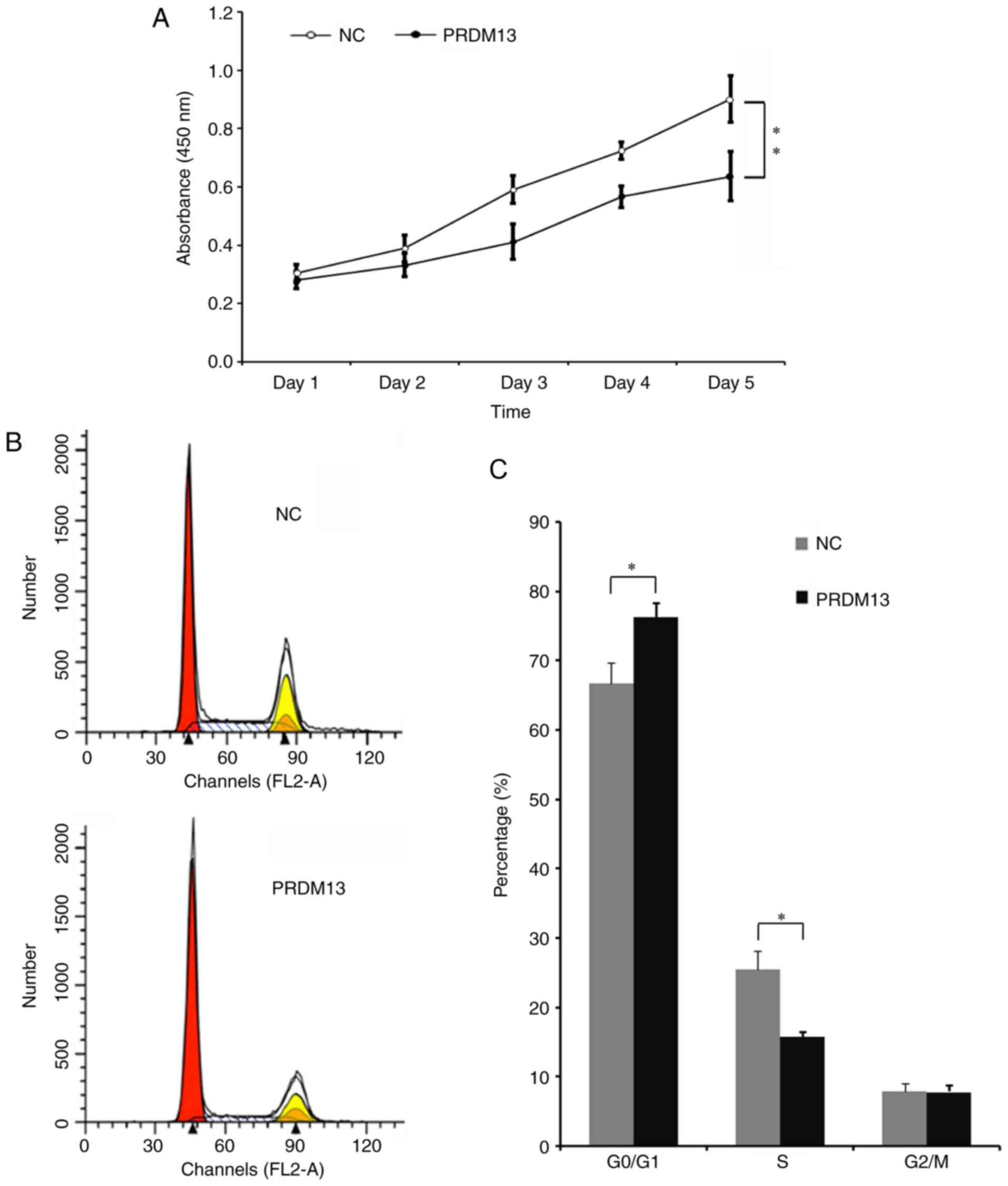
Tumor cell proliferation is a vital characteristic
of tumors. Therefore, it was necessary to examine the impact of the
overexpression of PRDM13 on cell proliferation in the U87 cell
line. It was demonstrated that cell proliferation was decreased
from the third day in the cells infected with Lenti-PRDM13,
compared with that in the NC cells (Fig. 3A). The role of PRDM13 in the cell
cycle of the U87 cells was analyzed by flow cytometry (Fig. 3B). It was demonstrated that the
cell percentage in the G0/G1 phase was
increased from 66.66% in the NC group to 77.34% in the
Lenti-PRDM13-infected cells, and the cell percentage in the S phase
was decreased to 15.81% from 25.83% (P<0.05). These results
indicated that PRDM13 inhibited DNA duplication in the U87 human
glioma cells.
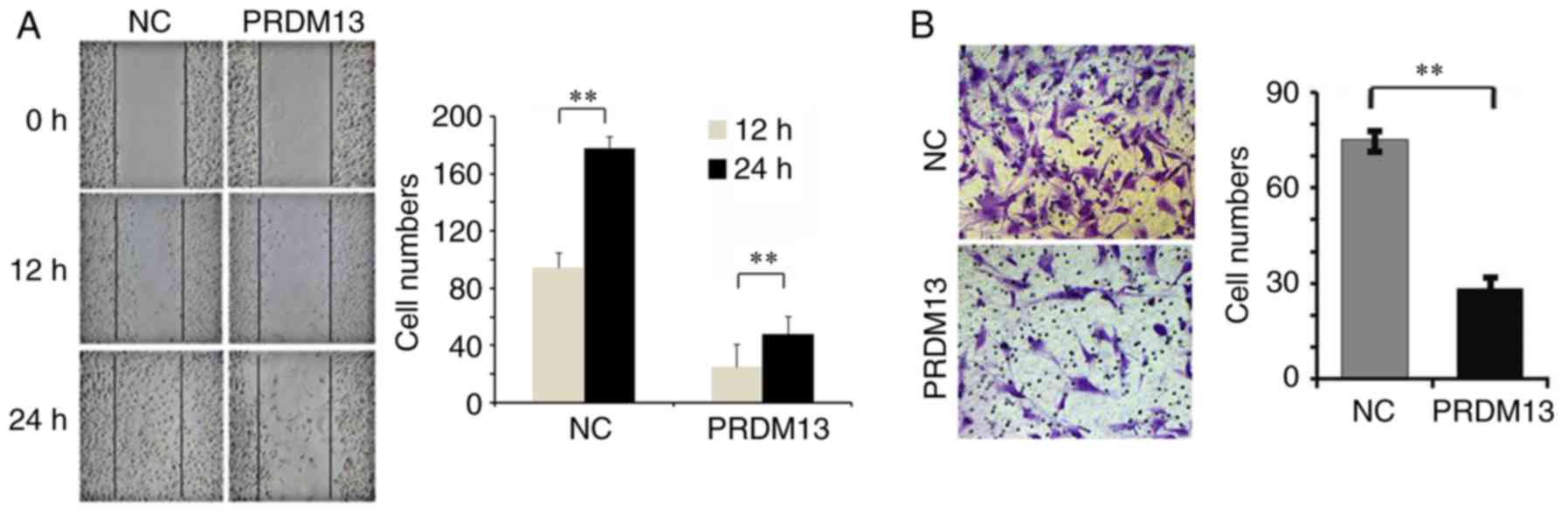
The results of the wound-healing assay demonstrated
that the migration of U87 cells was lower in the PRDM13
over-expression group than in the NC group (Fig. 4A). The results from the Transwell
invasion assay demonstrated that the overexpression of PRDM13
suppressed the invasion of U87 cells (Fig. 4B). The average invasive cell
number was 28 in the PRDM13 overexpression group, and 75 in the NC
group (P<0.01; Fig. 4B). These
data showed that the overexpression of PRDM13 may suppress the
migration and invasion of U87 cells.
Analysis of transcriptome sequencing
data
To further investigate the mechanisms underlying the
effect of the overexpression of PRDM13 in U87 glioma cells,
transcrip-tome sequencing data were analyzed in order to reveal the
molecular mechanisms of U87 cells infected with Lenti-PRDM13 or
Lenti-control. The volcano plot revealed 760 differentially
expressed genes between the two groups, including 222 upregulated
genes and 538 downregulated genes (P<0.05; Fig. 5A). GO analysis revealed three
differentially expressed gene functional enrichments: Molecular
function, cellular component and biological process. The genes were
primarily involved in 'receptor binding', 'molecular function
regulator', 'extracellular region part', 'cell periphery', 'system
development', 'single-multicellular organism process', 'regulation
of multicellular organismal process', 'anatomical structure
development' and 'regulation of cell migration' (FDR<0.05;
Fig. 5B).
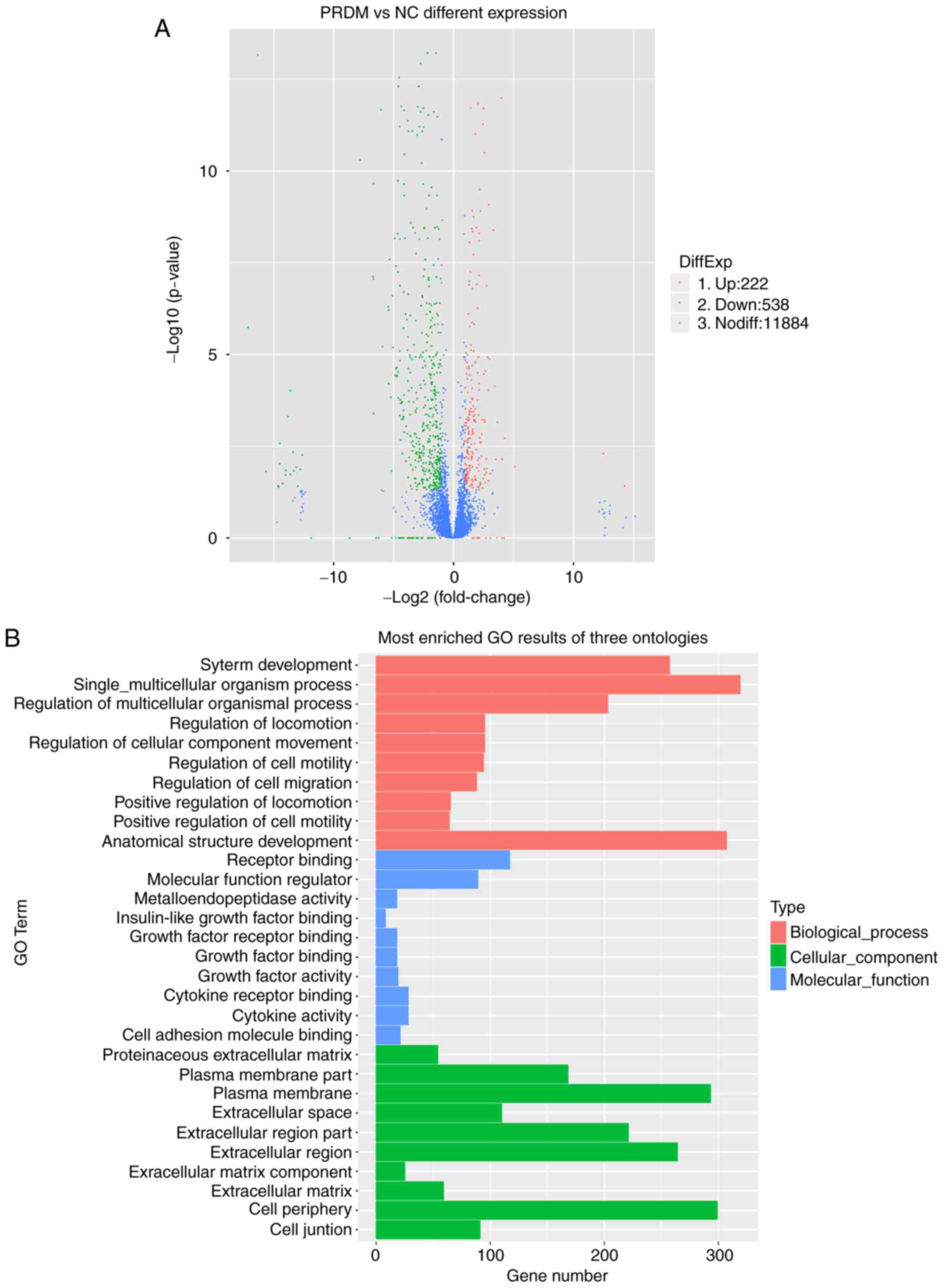 | Figure 5Analysis of gene expression changes in
U87 cells with overexpression of PRDM13 by transcriptome
sequencing. (A) Volcano plot shows differentially expressed genes
in the U87 human malignant glioma cell line infected with a
lentivirus expressing either PRDM13 or Lenti-con under the criteria
of |log2Ratio| ≥1 and P≤0.05. The genes are denoted by different
colored dots. The fold change is shown at the bottom in the image
(red represents upregulated genes, green represents downregulated
genes, and blue represents no difference). (B) Functional
enrichment was analyzed via GO. The x-axis represents the number of
significantly different genes and the y-axis represents the GO
term. Different colors represent different categories of genes (red
represents biological process, blue represents cellular component
and green represents molecular function). PRDM13, PRDI-BFI and RIZ
homology domain containing 13; NC, negative control; GO, gene
ontology; DiffExp, differential expression; up, upregulated; down,
downregulated; nodiff, no difference. |
Significant differential expression in genes between
the two groups was analyzed. Following consultation of the relevant
literature, it was identified that a number of these genes were
involved in tumorigenesis, proliferation and migration, including
SRY-Box 7 (SOX7), tumor necrosis factor receptor superfamily member
8 (TNFRSF8), RAR-related orphan receptor α (RORA), early growth
response 1 (EGR1), basic helix-loop-helix family, member e41
(BHLHE41), Snail family transcriptional repressor 2 (SNAI2), ArfGAP
with SH3 domain, ankyrin repeat and PH domain 3 (ASAP3), Growth
arrest and DNA-damage-inducible β (GADD45B), PIM1, ADAMTS12,
integrin β-like 1 (ITGBL1), DLC1 and ADAMTS6. These genes have been
demonstrated to have a promoting or inhibitory effect on tumors. A
number of known inflammatory and immune-associated genes, including
interleukin (IL)24, chemokine (C-X-C motif) ligand 14 and
interleukin 1 receptor like 1, were also identified. The results of
the analyses of these genes is likely to facilitate investigations
into the associated mechanisms.
Anti-oncogenes are involved in the
PRDM13-mediated progression of glioma cells
According to the results of transcriptome sequencing
data, genes associated with the regulation of cell proliferation
and migration were identified from GO sources. A total of 15
upregulated genes and 15 downregulated genes were validated by
RT-qPCR analysis. All genes coincided with the results of the
transcriptome sequencing data (Fig.
6A). Of note, the anti-oncogenes DLC1, ARHGAP30, INCA1 and
ADAMTS12 were all upregulated at the protein level (Fig. 6B). The ChIP assay demonstrated
that PRDM13 may bind to the DLC1 promoter to regulate its
expression (Fig. 6C). As
summarized in the diagram in Fig.
6D, PRDM13 may upregulate DLC1 and ARHGAP30, suppressing U87
cell migration, upregulating INCA1 and ADAMTS12, and causing a
decrease in U87 cell proliferation.
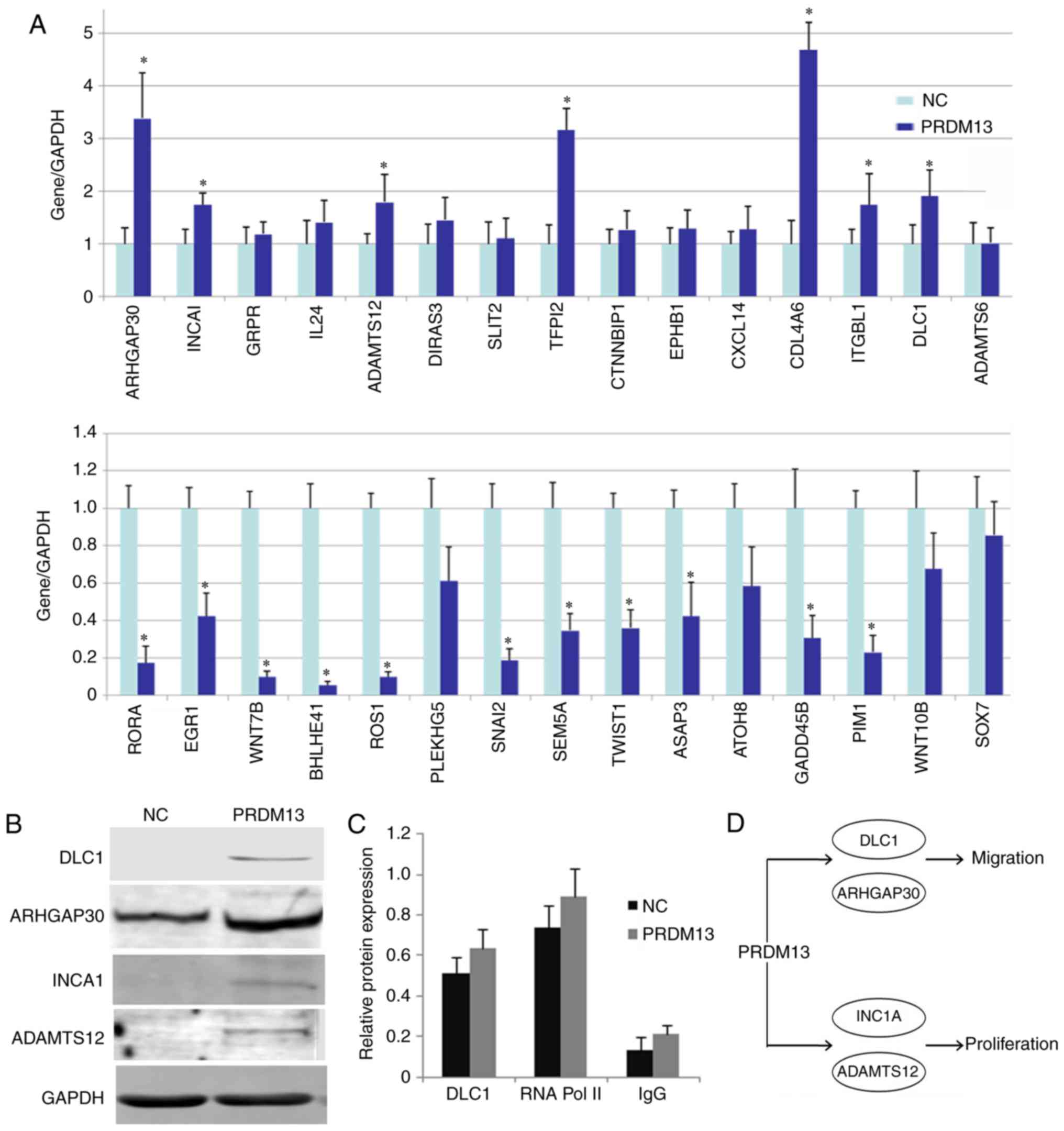 | Figure 6Proliferation and migration-associated
genes involved in U87 cells infected with Lenti-PRDM13 and
Lenti-Con. (A) Total RNA was extracted and reverse
transcription-quantitative polymerase chain reaction analysis was
performed to quantify the expression of different proliferation-
and migration-associated genes at the mRNA level. The mRNA level
was normalized to that of GAPDH. *P<0.05 (B) Protein
expression levels of DLC1, ARHGAP30, INCA1 and ADAMTS12 were
analyzed by western blot analysis in U87 cells. GAPDH was used as
an internal control. (C) Chromatin immuno- precipitation assay in
U87 cells exhibited a direct association between PRDM13 and the
DLC1 promoter. (D) Diagram summarizing the conclusion that PRDM13
may upregulate DLC1 and ARHGAP30, which suppress U87 cell migration
and upregulate INCA1 and ADAMTS12, causing a decrease in U87 cell
proliferation. PRDM13, PRDI-BFI and RIZ homology domain containing
13; DLC1, deleted in liver cancer 1; ARHGAP30, Rho GTPase
activating protein 30; ADAMTS12, A disintegrin and
metalloproteinase with thrombospondin motifs 12; INCA1, inhibitor
of cyclin-dependent kinase interacting with cyclin A1; NC, negative
control. |
Discussion
The PRDM family members have been reported to
correlate closely with cancer, and the majority are tumor
suppressors. PRDM13 is a member of the PRDM family that contains a
PR domain and a zinc finger motif (10). However, the biological function of
PRDM13 in glioma has not been elucidated previously. Previous
studies have demonstrated that PRDM13 is a transcriptional
repressor, which regulates the excitatory/inhibitory balance
(11). In the present study,
lower expression levels of PRDM13 were identified in glioma cells
and glioma tissues, therefore, the role of PRDM13 in glioma cells
was examined. The results demonstrated that the over-expression of
PRDM13 inhibited the proliferation, migration and invasion of U87
cells, and decreased the percentage of cells in the S phase of the
cell cycle, suggesting that PRDM13 may influence DNA
replication.
Dysregulation of the cell cycle is one of the
important reasons behind the malignant transformation and
uncontrolled proliferation of cells and, ultimately, the formation
of malignant tumors. INCA and CDK inhibitor may inhibit the
activity of CDK2 and cell proliferation. This inhibition depends on
cell cycle protein interactions. Mitogen and oncogenic signaling
may inhibit INCA1, which is caused by cell cycle arrest. Studies
have identified that the expression of novel CDK inhibitor INCA1
was decreased in acute leukemia myeloid and lymphoid cells. This
gene is important for cell survival, including genetic damage
detection and repair (12). The
expression of ADAMTS12 has been reported to be associated with a
tumor cell anti-proliferation effect by modulating the
extracellular signal-regulated kinase signaling pathway. It has
also been identified as a novel antitumor protease. It
simultaneously induces interstitial cells that may act as part of
normal tissues in controlling cancer progression (13). In the absence of ADAMTS12,
angiogenesis and tumor invasion in host tissues increase. El Hour
et al (14) reported that
ADAMTS12 exhibited anti-angiogenic properties and promoted host
defenses against tumors. The expression of ADAMTS12 is associated
with tumor histological type, tumor invasion and lymph node
metastasis. If the expression of ADAMTS12 is low or missing in the
extracellular matrix, the survival rate of the patient is
relatively poor. ADASTM12 is important in tumor development; its
expression may be a good prognostic indicator in colon cancer
(15). The PRDM13-mediated
inhibition of proliferation in glioma is associated with the above
proteins.
Among tumor characteristics, invasion and growth are
vital for glioma tumors (16).
The results of the present study demonstrated that the
overexpression of PRDM13 inhibited the migration and invasion of
glioma cells. In order to obtain further insights into the
mechanism of PRDM13-associated glioma progression, RNA-Seq analysis
for U87 cells over-expressing PRDM13 was performed, and hundreds of
genes exhibited significant differential expression. Among them,
the gene expression levels of Rho protein and GTP enzyme activation
protein were examined in U87 cells. The overexpression of PRDM13 in
U87 cells induced upregulation of the expression of DLC1 and
ARHGAP30. ChIP was performed to show that PRDM13 was involved with
the transcription start site to upregulate the expression of DLC1.
These results indicated that PRDM13 inhibited the migration of
glioma cells through the upregulation of Rho family proteins.
The Rho family proteins function as important
regulators in the organization of the actin cytoskeleton, and
dysregulation of the Rho GTPase-associated signaling pathways has
been reported in tumors (17).
Low expression levels of ARHGAP30, a Rho GTPase-activating protein,
have been reported to be associated with poor survival rates in
patients with colorectal cancer (CRC) (18). ARHGAP30 is required for p53
acetylation and functional activation in CRC, and the expression of
ARHGAP30 is a potential prognostic marker for CRC (18). DLC1, which is associated with
human HCC, is also termed ARHGAP7 and is a Rho GTPase-activating
protein. DLC1 has been found to be downregulated in a number of
cancer tissues and cells. It has been reported that DLC1 is a tumor
suppressor gene in liver cancer in addition to other types of
cancer (19,20). The DLC-1 gene, deleted in liver
cancer at chromosome 8p21-22, is altered primarily by genomic
deletion or aberrant promoter methylation in a large number of
types of human cancer, including breast, liver, colon and prostate
cancer, and is known to have an inhibitory effect on breast and
liver tumor cell growth (21). A
feature of many advanced tumors is the activation of the
Rho-GTPases RhoA and RhoC. These small GTPases, which are
Ras-related proteins, may contribute to various parameters of
abnormal cell growth, including viability, migration, invasion,
proliferation and anchorage-independent growth (22–24). GTPase-activating protein
expression is frequently downregulated or silenced in a variety of
human malignancies. The observations in the present study revealed
that the overexpression of PRDM13 is likely to be associated with
the GTPase-activating proteins, inhibiting glioma cell migration.
In the future, it may be worthwhile to investigate the mechanism
underlying PRDM1-mediated Rho protein expression.
In conclusion, the present study demonstrated that
PRDM13 was important in glioma cell malignant progression in
vitro, as the overexpression of PRDM13 prevented proliferation,
migration and invasion. Verhaak et al (25) reported the gene expression-based
molecular classification of GBM into proneural, neural, classical,
and mesenchymal subtypes. Aberrations and gene expression of
epidermal growth factor receptor, neurofibromin 1, and platelet
derived growth factor receptor α/isocitrate dehydrogenase 1 each
define the different subtypes, Their findings were from brain
tissues of patients. The findings of the present study are
significant, however, the role of PRDM13 on glioma cells was
discussed using the U87 cell line in vitro only. This was a
limitation of the present study and future investigations into the
detailed molecular mechanisms of the role of PRDM13 in glioma are
required. These results provide the biological basis for a novel
therapeutic approach in glioma.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 301460300)
and West China Top Class Discipline Project in Basic Medical
Sciences, Ningxia Medical University (grant no. NXYLXK2017B07).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ, YW and QH designed the study. LZ, HC, TH, JY and
HT performed the research. LZ, HC, TH and QH analyzed the data. LZ,
YW and QH wrote and revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chadwick RB, Jiang GL, Bennington GA, Yuan
B, Johnson CK, Stevens MW, Niemann TH, Peltomaki P, Huang S and de
la Chapelle A: Candidate tumor suppressor RIZ is frequently
involved in colorectal carcinogenesis. Proc Natl Acad Sci USA.
97:2662–2667. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang GL and Huang S: Adenovirus
expressing RIZ1 in tumor suppressor gene therapy of
microsatellite-unstable colorectal cancers. Cancer Res.
61:1796–1798. 2001.PubMed/NCBI
|
|
3
|
Mandelbaum J, Bhagat G, Tang H, Mo T,
Brahmachary M, Shen Q, Chadburn A, Rajewsky K, Tarakhovsky A,
Pasqualucci L and Dalla-Favera R: BLIMP1 is a tumor suppressor gene
frequently disrupted in activated B cell-like diffuse large B cell
lymphoma. Cancer Cell. 18:568–579. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wieser R: The oncogene and developmental
regulator EVI1: Expression, biochemical properties, and biological
functions. Gene. 396:346–357. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shing DC, Trubia M, Marchesi F, Radaelli
E, Belloni E, Tapinassi C, Scanziani E, Mecucci C, Crescenzi B,
Lahortiga I, et al: Overexpression of sPRDM16 coupled with loss of
p53 induces myeloid leukemias in mice. J Clin Invest.
117:3696–3707. 2007.PubMed/NCBI
|
|
6
|
Deng Q and Huang S: PRDM5 is silenced in
human cancers and has growth suppressive activities. Oncogene.
23:4903–4910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meani N, Pexximenti F, Deflorian G, Mione
M and Alcalay M: The tumor suppressor PRDM5 regulates Wnt signaling
at early stages of zebrafish development. PLoS One. 4:e42732009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Burkitt Wright EMM, Spencer HL, Daly SB,
Manson FDC, Zeef LAH, Urquhart J, Zoppi N, Bonshek R, Tosounidis I,
et al: Mutations in PRDM5 in brittle cornea syndrome identify a
pathway regulating extracellular matrix development and
maintenance. Am J Hum Genet. 88:767–777. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
10
|
Hohenauer T and Moore AW: The Prdm family:
Expanding roles in stem cells and development. Development.
139:2267–2282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang JC, Meredith DM, Mayer PR, Borromeo
MD, Lai HC, Ou YH and Johnson JE: Prdm13 mediates the balance of
inhibitory and excitatory neurons in somatosensory circuits. Dev
Cell. 25:182–195. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bäumer N, Tickenbrock L, Tschanter P,
Lohmeyer L, Diederichs S, Bäumer S, Skryabin BV, Zhang F,
Agrawal-Singh S, Köhler G, et al: Inhibitor of cyclin-dependent
kinase (CDK) interacting with cyclin A1 (INCA1) regulates
proliferation and is repressed by oncogenic signaling. J Biol Chem.
286:28210–28222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moncada-Pazos A, Obaya AJ, Fraga MF,
Viloria CG, Capellá G, Gausachs M, Esteller M, López-Otín C and Cal
S: The ADAMTS12 metalloprotease gene is epigenetically silenced in
tumor cells and transcriptionally activated in the stroma during
progression of colon cancer. J Cell Sci. 122:2906–2913. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
El Hour M, Moncada-Pazos A, Blacher S,
Masset A, Cal S, Berndt S, Detilleux J, Host L, Obaya AJ, Maillard
C, et al: Higher sensitivity of Adamts12-deficient mice to tumor
growth and angiogenesis. Oncogene. 29:3025–3032. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang D, Zhu T, Zhang FB and He C:
Expression of ADAMTS12 in colorectal cancer-associated stroma
prevents cancer development and is a good prognostic indicator of
colorectal cancer. Dig Dis Sci. 56:3281–3287. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang H, Zhu W, Su X, Wu S, Lin Y, Li J,
Wang Y, Chen J, Zhou Y, Qiu P, et al: Triprolide inhibits
proliferation and invasion of malignant glioma cells. J Neurooncol.
109:53–62. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmitz AA, Govek EE, Böttner B and Van
Aelst L: Rho GTPases: Signaling, migration, and invasion. Exp Cell
Res. 261:1–12. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Qian J, Hu Y, Kong X, Chen H, Shi
Q, Jiang L, Wu C, Zou W, Chen Y, et al: ArhGAP30 promotes p53
acetylation and function in colorectal cancer. Nat Commun.
5:47352014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan BZ, Miller MJ, Keck CL, Zimonjic DB,
Thorgeirsson SS and Popescu NC: Cloning, characterization, and
chromosomal localization of a gene frequently deleted in human
liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res.
58:2196–2199. 1998.PubMed/NCBI
|
|
20
|
Qian X1, Li G, Asmussen HK, Asnaghi L,
Vass WC, Braverman R, Yamada KM, Popescu NC, Papageorge AG and Lowy
DR: Oncogenic inhibition by a deleted in liver cancer gene requires
cooperation between tensin binding and Rho-specific
GTPase-activating protein activities. Proc Natl Acad Sci USA.
104:9012–9017. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan BZ, Jefferson AM, Baldwin KT,
Thorgeirsson SS, Popescu NC and Reynolds SH: DLC-1 operates as
tumor suppressor gene in human non-small cell lung carcinomas.
Oncogene. 23:1405–1411. 2004. View Article : Google Scholar
|
|
22
|
Karnoub AE, Symons M, Campbell SL and Der
CJ: Molecular basis for Rho GTPase signaling specificity. Breast
Cancer Res Treat. 84:61–71. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ridley AJ: Rho proteins and cancer. Breast
Cancer Res Treat. 84:13–19. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gómez del Pulgar T, Benitah SA, Valerón
PF, Espina C and Lacal JC: Rho GTPase expression in tumourigenesis:
Evidence for a significant link. Bioessays. 27:602–613. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Verhaak RG, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Integrated genomic analysis identifies clinically relevant subtypes
of glioblastoma characterized by abnormalities in PDGFRA, IDH1,
EGFR, and NF1. Cancer Cell. 17:98–110. 2010. View Article : Google Scholar : PubMed/NCBI
|