Introduction
Oral squamous cell carcinoma (OSCC) is the sixth
most common malignant cancer worldwide (1). Each year, ~500,000 new cases of OSCC
are diagnosed worldwide (2,3).
Although there have been many recent achievements, the 5-year
survival rate for patients with OSCC has not significantly improved
because of regional recurrence or distant metastases causing poor
prognosis (4,5). Chemotherapy has traditionally been
used as the standard treatment for OSCC, but its toxic effects and
the potential for chemo-resistance impair its useful application
(6,7). Therefore, novel strategies should be
developed to address this disease, including more sensitive, but
less toxic, therapeutic molecular targets. Gene therapy has offered
a promising strategy for the treatment of OSCC (8,9).
Tumor necrosis factor (TNF)-related
apoptosis-inducing ligand (TRAIL), which is a member of the TNF
superfamily, can induce tumor cell apoptosis while exerting no
effect on normal human cells (10,11). Mounting evidence has demonstrated
that TRAIL exerts anti-tumor effects via suppression of tumor
proliferation and promotion of tumor apoptosis in multiple types of
cancer both in vivo and in vitro; examples include:
Colorectal cancer (12), breast
cancer (13), esophageal cancer
(14), lung cancer (15), gastric cancer (16) and hepatocellular carcinoma (HCC)
(17). A treatment strategy for
cancer based on TRAIL has previously been identified as a promising
option (18). However, malignant
tumors may become resistant to TRAIL-induced apoptosis.
Human telomerase reverse transcriptase (hTERT), a
catalytic subunit of telomerase, is overexpressed in ~90% of
malignant tumors (19). These
tumor cells have been reported to obtain immortalized features by
maintaining their telomere lengths with the help of hTERT (20). Suppression of hTERT gene activity
can impair tumor proliferation by shortening their replicative life
span, further arresting tumor growth (21). Therefore, hTERT is another
potential molecular target for tumor treatment.
Recently, combination treatment has become the
prevailing approach in tumor therapy as an effective way to address
cancer. For instance, the combination of cisplatin and paclitaxel
has been demonstrated to induce antitumor effects of varying
degrees (22,23). However, a lack of specificity,
off-target activity, chemo-resistance and normal cell damage due to
toxicity, markedly impaired efficacy of this treatment. A more
targeted combination therapeutic strategy involving multiple genes
may be a more promising solution.
In the present study, the synergistic antitumor
effects of adeno-associated virus (AAV)-mediated TRAIL
overexpression combined with lentivirus vector-mediated hTERT,
which induces tumor apoptosis and inhibits tumor proliferation
in vitro and in vivo, were investigated. The findings
suggested that AAV-mediated TRAIL overexpression combined with
lentivirus vector-mediated hTERT may be an effective therapy for
OSCC.
Materials and methods
Ethical approval and informed
consent
The present study was approved by the Ethics
Committee at Sun Yat-sen Memorial Hospital (Sun Yat-sen University,
Guangzhou, China). Animal experiments were approved by the Ethics
Committee of Sun Yat-Sen University and were conducted following
the official instructions of the laboratory animal center at Sun
Yat-Sen University.
Cell lines and cell culture
Two human tongue squamous cell carcinoma cell lines,
SCC25 and UM1, were used in the present study. SCC25 cell line was
purchased from American Type Culture Collection (ATCC; Manassas,
VA, USA) and UM1 cell line was kindly supplied by Professor Wang
from The First Affiliated Hospital of Sun Yat-sen University. Both
cell lines were cultured in RPMI-1640 medium (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS and
1% penicillin/streptomycin. 293T cells were purchased from ATCC and
maintained in Dulbecco’s modified Eagle’s medium supplemented with
10% FBS. Cells were cultured at 37°C in a humidified atmosphere
containing 5% CO2.
AAV-TRAIL viral preparation
Recombinant AAV2 vectors were produced using a
triple transient transfection method. The plasmid carrying TRAIL
cDNA (pAAV-CMV-EGFP-TRAIL; Clontech Laboratories, Inc.,
Mountainview, CA, USA), pAAV-RC plasmid expressing rep/cap gene and
the pHelper plasmid (Applied Viromics, LLC. Fremont, CA, USA) were
mixed at a respective ratio of 15:10:10 µg and
co-transfected into 293T cells with polyethylenimine (Polysciences,
Inc., Warrington, PA, USA) for 72 h according to the manufacturer’s
protocol. The viral supernatant was collected following
centrifuging at 4°C at 500 × g for 10 min and 293T cells containing
packaged particles were collected, purified and concentrated by
using HiTrap Heparin HP and Amicon Ultra 4 ml Filters (GE
Healthcare, Chicago, IL, USA) to obtain concentrated recombinant
AAV vectors. The particle titers were determined by quantitative
polymerase chain reaction (qPCR), as detailed below, and are
presented as genome particles/ml. To evaluate the efficiency of the
recombinant AAV system on TRAIL upregulation, enhanced green
fluorescence protein (EGFP) gene was inserted as a reporter gene
and its expression was analyzed using Nikon Eclipse Ti fluorescence
microscopy (magnification, ×400; Nikon Corporation, Tokyo,
Japan).
Lentivirus preparation and gene
silencing
The lentivirus vector was constructed using the
LV-008 plasmid (Forevergen Biosciences Co., Ltd., Guangzhou, China)
with a verified short hairpin RNA against TERT (shTERT) inserted,
the pMD2.G plasmid (cat. no. 12259; Addgene, Inc., Cambridge, MA,
USA) expressing the vesicular stomatitis virus glycoprotein gene
and psPAX2 plasmid (cat. no. 12260; Addgene, Inc.) carrying the
gag/pol-gene. For viral packaging, 8 µg shRNA plasmid, 2
µg pMD2.G plasmid and 4 µg psPAX2 plasmid were
co-transfected using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) into 293T cells plated in 100-mm
dishes. Transfected lentiviruses were collected 72 h following
transfection and concentrated via ultra-centrifugation at 87,000 ×
g for 180 min at 4°C using a Beckman SW28 rotor (Beckman Coulter,
Inc., Brea, CA, USA). SCC25 and UM1 cells were cultured with
RPMI-1640 and seeded in a six-well plate. When cells reached 50–70%
confluence, cells were infected with hTERT-lentivirus or a negative
control (NC)-shRNA lentivirus, respectively, in the presence of 5
µg/ml polybrene (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany). At 5 days following transfection, the knockdown
efficiency was examined by reverse transcription (RT)-qPCR and
western blotting. To select for stably silenced cells, puromycin (2
µg/ml; Sigma-Aldrich; Merck KGaA) was added to the
medium.
RNA extraction and RT-qPCR
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and RT was
performed to synthesize cDNA using a Prime Script™ RT reagent kit
according to the manufacturer’s protocol (Takara Biotechnology Co.,
Ltd., Dalian, China). qPCR was performed with a SYBR Green PCR
Master Mix (Takara Biotechnology Co., Ltd.). The reaction mixture
consisted of a volume of 20 µl, containing 2 µl cDNA,
10 µl SYBR-Green Mix, 4 µl primer mix and 4 µl
ddH2O. The PCR cycling conditions were 95°C for 5 min,
followed by 30 cycles of 95°C for 30 sec and 50°C for 30 sec.
β-actin was used as reference gene and the primers used are
presented in Table I. Relative
gene expression was calculated using the 2−ΔΔCq method
(24).
 | Table IReverse transcription-quantitative
polymerase chain reaction primers. |
Table I
Reverse transcription-quantitative
polymerase chain reaction primers.
| Target gene | Primer
sequence |
|---|
| TRAIL | Forward:
5′-GTTGGCACATGCCTGTAGTCC-3′ |
| Reverse:
5′-AAACCAAGTCTCGCTCTGTCG-3′ |
| Caspase-3 | Forward:
5'-GAGCTGCCTGTAACTTG-3' |
| Reverse:
5'-ACCTTTAGAACATTTCCACT-3' |
| Caspase-8 | Forward:
5'-CTGCTGGGGATGGCCACTGTG-3' |
| Reverse:
5'-TCGCCTCGAGGACATCGCTCTC-3' |
| Caspase-9 | Forward:
5'-TTCCCAGGTTTTGTTTCCTG-3' |
| Reverse:
5'-CCTTTCACCGAAACAGCATT-3' |
| Bcl-2 | Forward:
5'-CATGTGTGTGGAGAGCGTCAA-3' |
| Reverse:
5'-GCCGGTTCAGGTACTCAGTCA-3' |
| hTERT | Forward:
5'-CGTCCAGACTCCGC TTCATC-3' |
| Reverse:
5'-GAGACGCTCGGCCCTCTT-3' |
| β-actin | Forward:
5'-AGGGGCCGGACTCGTCATACT-3' |
| Reverse:
5'-GGCGGCACCACCATGTACCCT-3' |
Western blotting
Cells were lysed in sample lysis solution (50 mM
Tris-HCl, pH 8.0; 1% SDS; 1 mM EDTA; 5 mM DTT; 10 mM
phenylmethylsulfonyl fluoride; 1 mM NaF; 1 mM
Na3VO4; and protease inhibitor cocktail
(Roche Diagnostics GmbH, Mannheim, Germany), and protein
concentration was determined with a bicinchoninic acid assay kit
(Abcam, Cambridge, UK). Equal amounts of protein (20 µg)
were separated by 10% SDS-PAGE and transferred onto polyvinylidene
fluoride membranes (GE Healthcare). Membranes were probed with
antibodies against TRAIL (cat. no. 3219), caspase-3 (cat. no.
9662), caspase-8 (cat. no. 9746), caspase-9 (cat. no. 9504;
1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA),
β-actin (cat. no. 3700; 1:2,000; Cell Signaling Technology, Inc.),
B cell lymphoma-2 (Bcl-2; cat. no. ab196495) and hTERT (cat. no.
ab191523; 1:1,000; Abcam) overnight at 4°C, and subsequently with
horseradish peroxidase (HRP)-conjugated secondary anti-mouse (cat.
no. 7076; 1:3,000,) and anti-rabbit (cat. no. 7074; 1:3,000)
antibodies (both Cell Signaling Technology, Inc.). Protein signals
were visualized using an enhanced chemiluminescence kit (GE
Healthcare) according to the manufacturer’s instructions.
Anti-β-actin was used as a control sample for protein loading.
Protein expression was analyzed using ImageJ software version 14.8
(National Institutes of Health, Bethesda, MD, USA).
Immunocytochemistry
The streptavidin-peroxidase conjugated method was
used to detect TRAIL, caspase-8, caspase-9, caspase-3 and Bcl-2
expression in cells. SCC25 cells were grown and fixed on coverslips
in 4% paraformaldehyde (PFA) and blocked with normal goat serum
(cat. no. 566380; Sigma-Aldrich; Merck KGaA) containing 0.25%
Triton for 15 min at room temperature. Slides were incubated with
relative primary antibodies against TRAIL (cat. no. 3219),
caspase-3 (cat. no. 9662), caspase-8 (cat. no. 9746), caspase-9
(cat. no. 9504; 1:400; Cell Signaling Technology, Inc., Danvers,
MA, USA) and B cell lymphoma-2 (Bcl-2; cat. no. ab196495; 1:500;
Abcam), respectively, for 1 h at 37°C. The slides were then rinsed
with PBS three times and subsequently incubated with a
biotin-labeled anti-rabbit/mouse secondary antibody (cat. no.
SP-9001/9002; 1:500; OriGene Technologies, Inc., Rockville, MD,
USA) for 30 min at 37°C. HRP-conjugated streptavidin (Dako; Agilent
Technologies, Inc., Santa Clara, CA, USA) was added to the slides
before visualization of the antigen-antibody complex with 3,3′
diaminobenzidine (DAB) for 15 min at room temperature.
Apoptosis assay and cell cycle assay
Results of the apoptosis assay were observed via
flow cytometry performed using an Annexin V-fluorescein
isothiocyanate (FITC) kit (BD Biosciences, San Jose, CA, USA)
according to the manufacturer’s protocol. Cells were harvested,
washed and resuspended with binding buffer. Cells were subsequently
incubated with Annexin V-FITC then washed with cold PBS and
resuspended. Finally, propidium iodide (PI) was added to mark
apoptotic cells, which were then analyzed using a flow
cytometer.
Cell cycle staging was also analyzed using flow
cytometry. The cell suspension was fixed with 70% cold ethanol
overnight at 4°C, then washed with cold PBS three times and stained
with PI solution for 30 min in the dark at 37°C prior to being
analyzed with a flow cytometer, to estimate the frequency of cells
in the sub-G0/G1 phase. Each measurement was
repeated in triplicate.
Cell proliferation and viability
assay
To evaluate the contribution of TRAIL and hTERT to
OSCC cell proliferation, an MTT assay of cell proliferation and
cell viability was performed. Following infection with AAV-TRAIL or
shTERT virus, OSCC cells were treated with MTT following the
manufacturer’s procedures. The reaction was terminated using
dimethylsulfoxide. Subsequently, absorbance was measured at 570 nm
using a microplate reader (Molecular Devices, LLC, Sunnyvale, CA,
USA). Each transfection was measured every day in triplicate, and
the percentage of viable cells were calculated.
In vivo mice xenograft assay
BALB/c nude mice (age, 4 weeks; weight, 12–16
g) were used for tumor implantation. A total of 25 male mice were
purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai,
China). All animal experiments complied with the Regulations for
the Administration of Affairs Concerning Experimental Animals
(25). The mice were housed in
controlled conditions of 20–22°C, relatively humidity of 50–55%,
and a 12-h light-dark cycle. All mice had free access to food and
water.
To generate the tumor implantation mouse model,
SCC25 cells were infected with empty vector, AAV-TRAIL, shTERT and
AAV-TRAIL + shTERT virus. The negative control (NC) group was not
infected. Then, 1×106 cells in 0.1 ml were
subcutaneously injected into the axilla of each mouse (n=5 mice per
group). Following injection, tumor appearance in mice was inspected
each week by observation and palpation for 6 weeks. The greatest
longitudinal diameter (a) and the greatest transverse diameter (b)
were measured weekly using a digital caliper. All mice were
euthanized with CO2 at the end of 6 weeks following
implantation. Tumor volume was calculated using the following
formula: Tumor volume=1/2 ab2. Tumors were harvested,
weighed and kept at −80°C for further analysis.
Immunohistochemistry
Immunohistochemistry was performed to evaluate
TRAIL, caspase-3, caspase-8, caspase-9, Bcl-2 and hTERT expression
in the tumors of each xenograft mice group. Tissue samples were
fixed in 4% PFA at 4°C overnight, embedded in paraffin and cut into
~4-µm thick sections. The slides were heated at 65°C for 30
min, deparaffinized with xylene and rehydrated with a gradient
series of alcohol and rinsed in PBS, then subjected to antigen
retrieval in citrate buffer (pH 6.0; 10 mM citric acid) at 95°C for
15 min in a microwave oven (Oriental Rotor Ltd., Tokyo, Japan). The
slides were subsequently quenched in PBS containing 3% hydrogen
peroxide at room temperature for 10 min to block endogenous
peroxidase activity. Non-specific binding was blocked with PBS
containing 5% BSA for 5 min at room temperature. The sections were
incubated at 4°C overnight with primary antibodies against: TRAIL,
caspase-9, (1:50; Cell Signalling Technology, Inc.), caspase-3
(1:300; Cell Signalling Technology, Inc.), Bcl-2 (cat. no. AM43;
1:300; Merck KGaA), caspase-8 (cat. no. MA1-41280; 1:50; Thermo
Fisher Scientific, Inc.), Ki67 (cat. no. ab16667; 1:100; Abcam),
poly (ADP-ribose) polymerase (PARP; cat. no. 5625; 1:100; Cell
Signalling Technology, Inc.) and hTERT (cat. no. PC563; 1:50; Merck
KGaA). Slides were rinsed in PBS three times, and incubated with
HRP-conjugated goat and anti-rabbit/mouse secondary antibodies with
a Dako Real™ EnVision™ detection system kit (cat. no. K5007; 1:100;
Dako; Agilent Technologies, Inc.). Reaction was visualized with DAB
(Dako) for 5 min at room temperature and hematoxylin was used to
counterstain the slides at room temperature for 1 min. The slides
were observed under a light microscope (Nikon Corporation;
magnification, ×100).
Terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling
(TUNEL) assay
TUNEL staining was used for the in situ
analysis of apoptotic cells. Tumor tissues were treated in the
aforementioned manner to obtain specimen slides from each group.
TUNEL staining was performed with an In Situ Cell Death
Detection kit (Roche Diagnostics GmbH) according to the
manufacturer’s instructions. In brief, formalin-fixed,
paraffin-embedded tumor tissues were cut into 5-µm thick
sections. Following heating at 65°C for 30 min, the sections were
deparaffinized in xylene and rehydrated through graded ethanol. The
tissue sections were rinsed in PBS and antigen retrieval was
performed in sodium citrate solutions. Sections were further
incubated with TUNEL reaction mixture at 37°C for 1 h in humid
conditions. Coverslips were then washed in PBS and counterstained
with DAPI (Sigma-Aldrich; Merck KGaA) at room temperature for 1
min. The slides were observed under the Nikon E600 fluorescence
microscope (Nikon Corporation; magnification, ×100).
Statistical analysis
All data are expressed as the mean ± standard
deviation, and the differences between groups were analyzed by
one-way analysis of variance with Tukey’s post hoc test using SPSS
17.0 (SPSS, Inc., Chicago, IL, USA). Pearson’s χ2 test
was also employed to compare difference among groups in univariate
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
AAV-mediated TRAIL overexpression induces
OSCC expression of caspase-3, caspase-8, and caspase-9, as well as
Bcl-2 suppression
As presented in Fig.
1A, a high level of EGFP expression was detected in SCC25
cells, indicating that the virus vectors constructed for the
present study effectively infected cells. To confirm the effect of
TRAIL overexpression on the AAV system, RT-qPCR was carried out.
Results demonstrated that in the AAV-TRAIL group, the TRAIL,
caspase-3, caspase-8 and caspase-9 mRNA were increased, whereas
BCL-2 mRNA was decreased, compared with the NC and vector groups
(Fig. 1B). TRAIL has been well
characterized as inducing tumor cells apoptosis (8,9).
The expression level of several genes closely associated with
apoptosis including caspase-3, caspase-8, caspase-9 and Bcl-2, were
monitored by western blotting and immunocytochemistry assays. As
presented in Fig. 1C,
overexpression of TRAIL resulted in marked upregulation of
caspase-3, caspase-8 and caspase-9, and downregulation of Bcl-2, an
apoptosis suppressor gene, in comparison with the NC and vector
groups. This phenomenon was also confirmed by immunocytochemistry
assay (Fig. 1D).
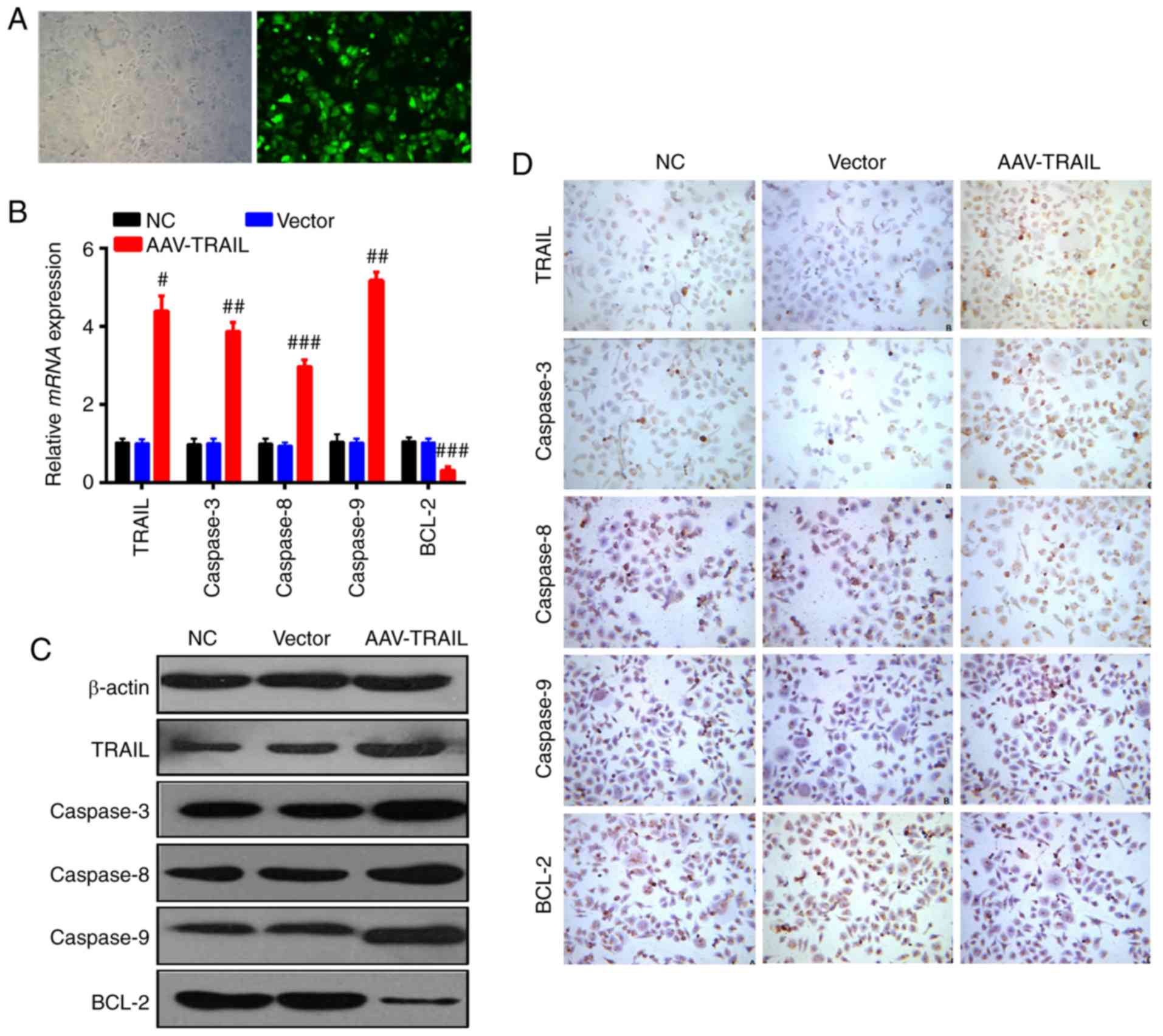 | Figure 1AAV-mediated TRAIL overexpression
induced increased expression of caspase-3, caspase-8, and
caspase-9, and reduced expression of Bcl-2 in OSCC cells. (A)
Immunofluorescence assay was carried out to detect the infection
efficiency of the recombinant AAV system in the SCC25 OSCC cell
line (magnification, ×400). The left panel presents light
microscopy and the right field presents fluorescence microscopy.
(B) Reverse transcription-quantitative polymerase chain reaction,
(C) western blotting and (D) immunocytochemistry analysis
(magnification, ×200) were performed to measure apoptosis-related
genes expression changes in SCC25 cells with upregulation of TRAIL.
Data are presented as the mean ± standard deviation. Each assay was
performed independently at least three times.
#P<0.05, ##P<0.01,
###P<0.001 vs. NC. AAV, adeno-associated virus;
TRAIL, tumor necrosis factor-related apoptosis-inducing ligand;
Bcl-2, B cell lymphoma-2; OSCC, oral squamous cell carcinoma; NC,
negative control. |
AAV-mediated TRAIL upregulation promotes
tumor cell apoptosis and suppresses cell proliferation and cell
cycle arrest
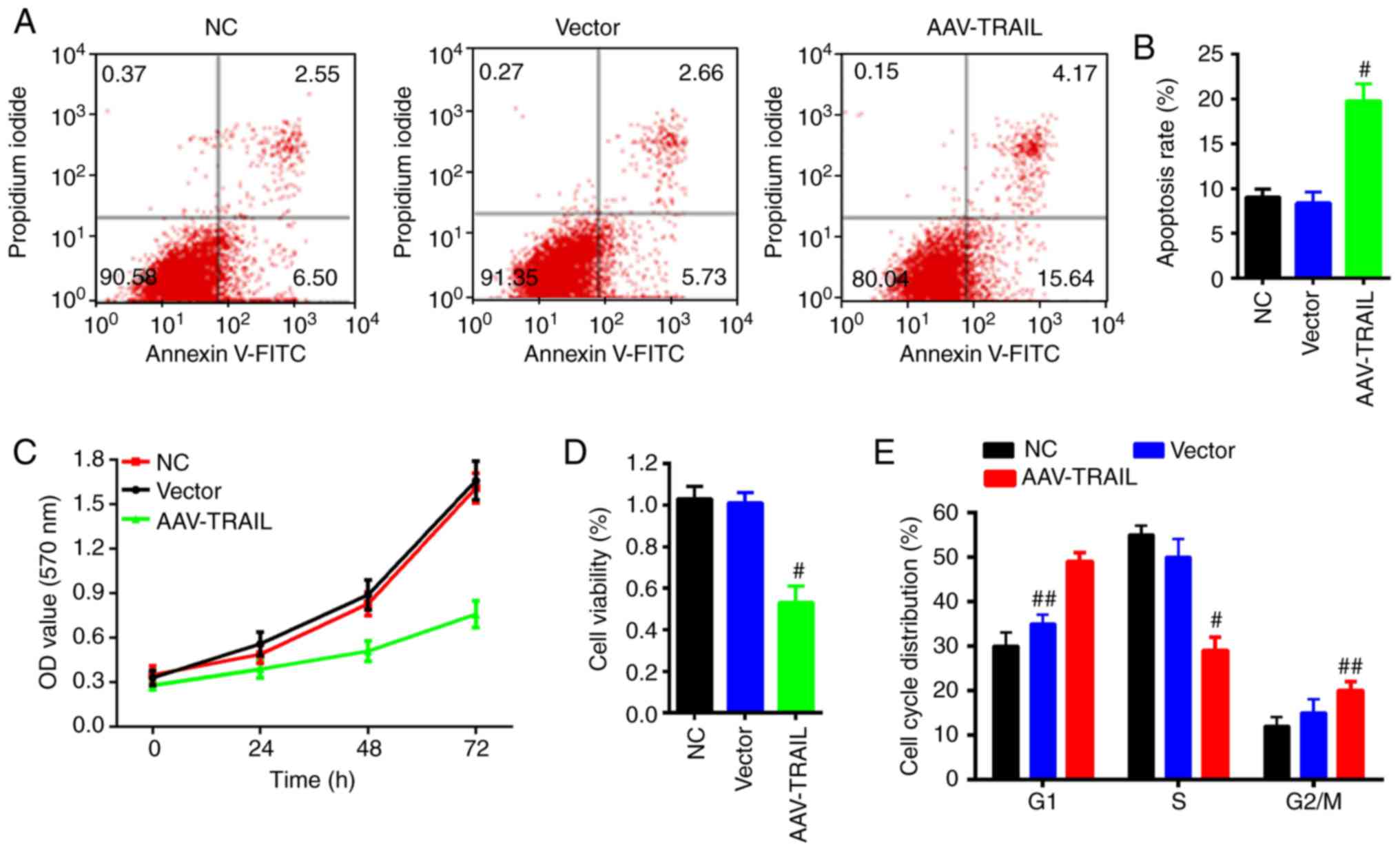
To further explore the function of TRAIL in OSCC
cells, an apoptosis assay was performed on SCC25 cells by flow
cytometry. The apoptosis rate of the cells infected with AAV-TRAIL
was significantly increased compared with the NC group (Fig. 2A and B). In addition, cell
proliferation ability and viability of SCC-25 cells transduced with
AAV-TRAIL were evaluated using the MTT assay. The results
demonstrated that the cell proliferation and viability were
prohibited with exogenous overexpression of TRAIL by the AAV
system, compared with the NC group (Fig. 2C and D).
Cell cycle distribution was also analyzed to assess
the effect of TRAIL on cell cycle. The results demonstrated that
the proportion of cells in G1 phase in the AAV-TRAIL
group was higher than that of the NC and vector groups; whereas the
proportion of cells in the S phase in the AAV-TRAIL group was
significantly lower than that of the NC group (Fig. 2E). These results suggest that
overexpression of TRAIL promotes OSCC cell apoptosis, inhibits
proliferation and arrests the cell cycle.
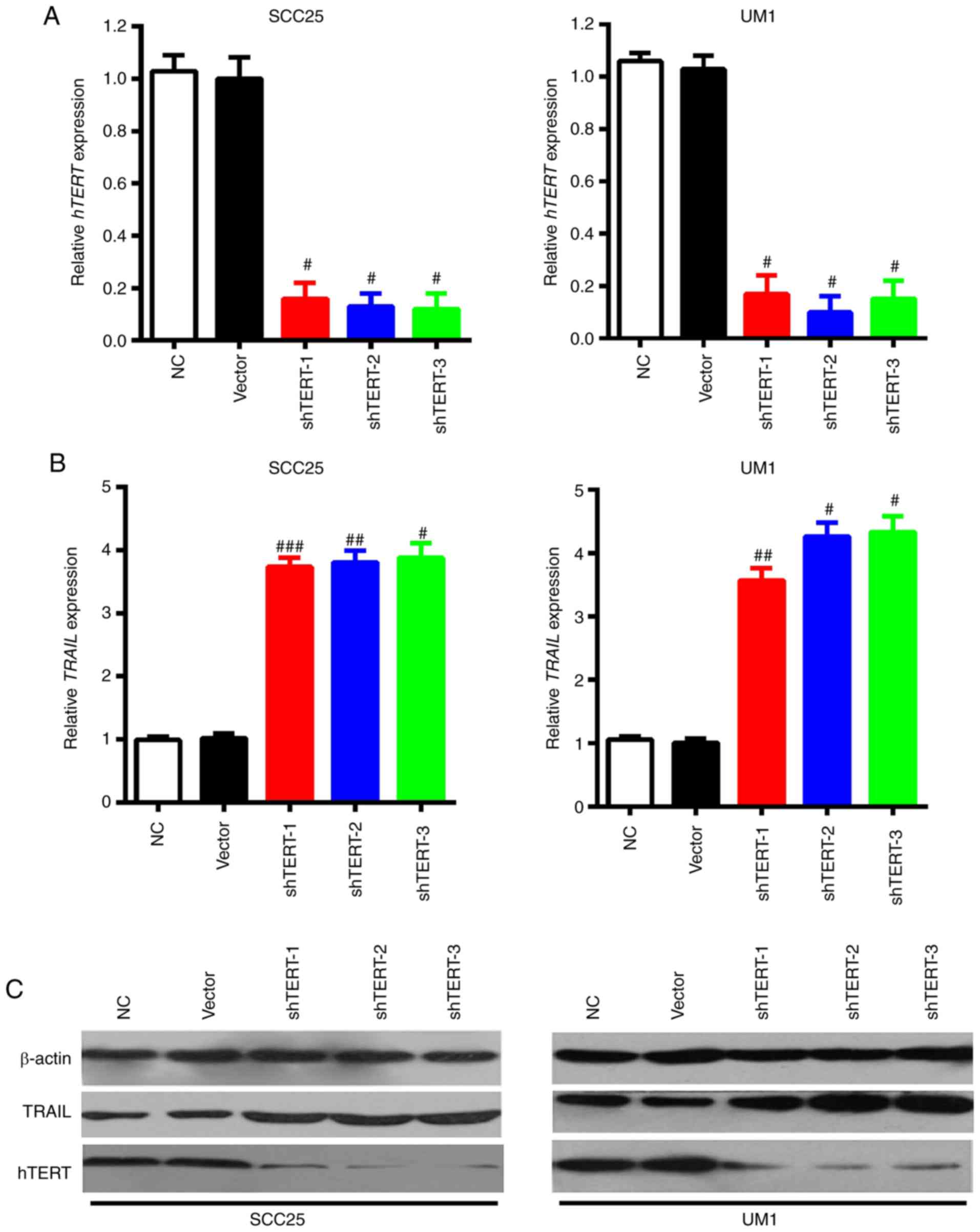
Lentivirus-mediated hTERT interference
upregulates TRAIL expression
It has been reported previously that the aberrant
expression of hTERT can promote cancer progression; there is a
negative correlation between TRAIL and hTERT in multiple cancer
types (26). As such, the present
study aimed to clarify the association between TRAIL and hTERT in
OSCC. Initially, three lentivirus vectors targeted to different
fragments of hTERT were constructed to interfere with hTERT gene
expression. Then, the interference efficiencies were measured in
SCC25 (epithelial) and UM1 (mesenchymal) cells, via RT-qPCR and
western blotting. Results of these experiments revealed that all
three shRNAs significantly inhibited hTERT expression, when
compared with the NC group, in SCC25 and UM1 cells (Fig. 3A). TRAIL expression changes were
further detected via silencing hTERT through shRNAs with RT-qPCR
and western blotting. The results demonstrated that mRNA and
protein expression of TRAIL was elevated following knocking down
hTERT expression in both SCC25 and UM1 cells (Fig. 3B and C). These results suggest
that attenuation of hTERT expression upregulates TRAIL
expression.
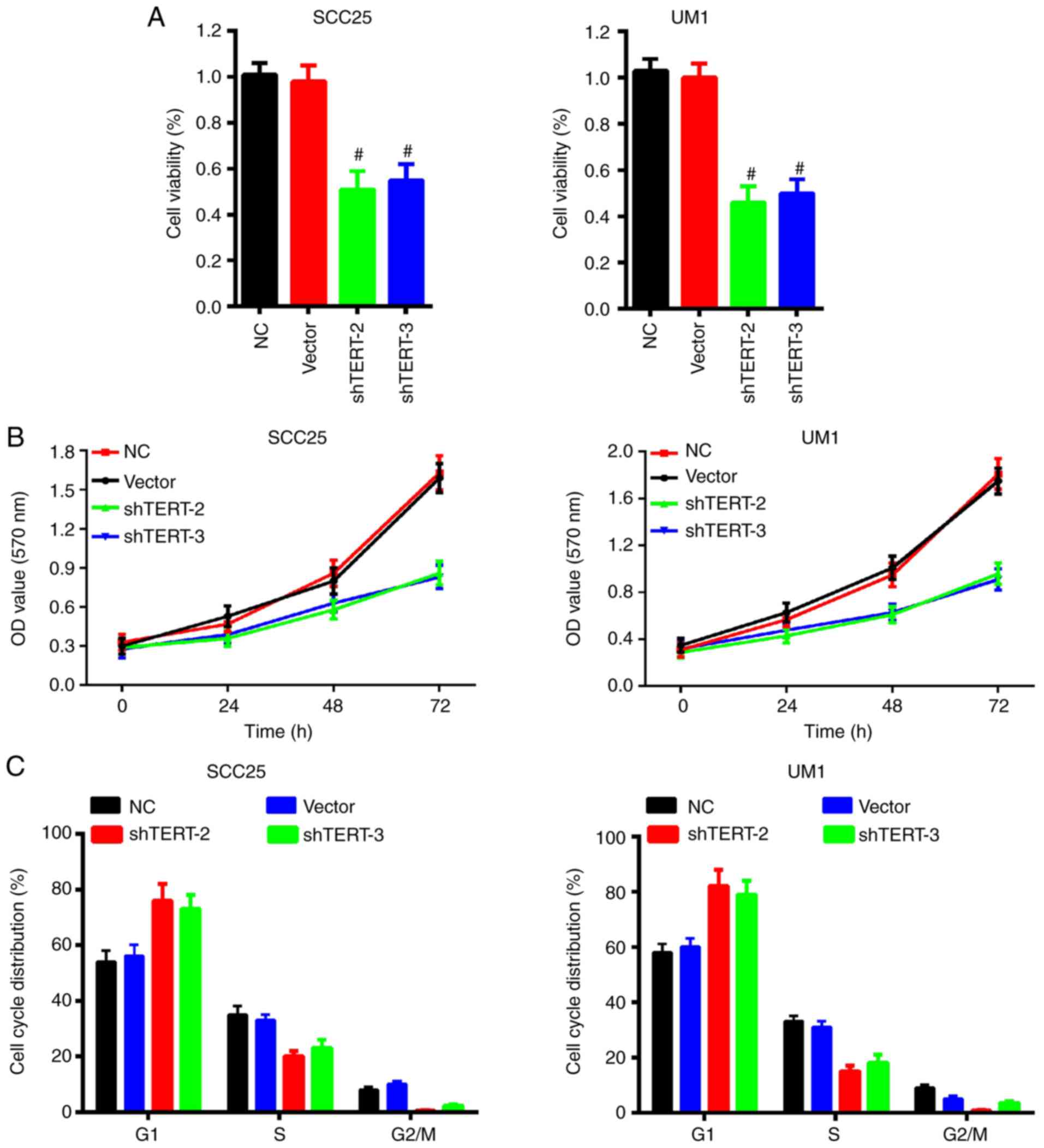
Silencing of hTERT expression impairs
OSCC cell viability, proliferation and cell cycle arrest
We further investigated cell viability and
proliferation ability with lentivirus-mediated hTERT silencing
using flow cytometry and MTT assay. The results indicated that
silencing hTERT significantly impaired cell viability in SCC25 and
UM1 cells (Fig. 4A). The MTT
assay also demonstrated that knockdown of hTERT expression
prohibited OSCC cell proliferation in SCC25 and UM1 cells (Fig. 4B). Furthermore, the cell cycle was
also analyzed to evaluate the hTERT silencing effect. The results
demonstrated that knockdown of hTERT markedly increased the
proportion of cells in the G1 phase, but decreased the
proportion of cells in the S phase in SCC25 and UM1 cells (Fig. 4C). These results indicate that
inhibition of hTERT activity regulates cell cycle
distribution, inhibits the proliferation of tumor cells and
promotes apoptosis.
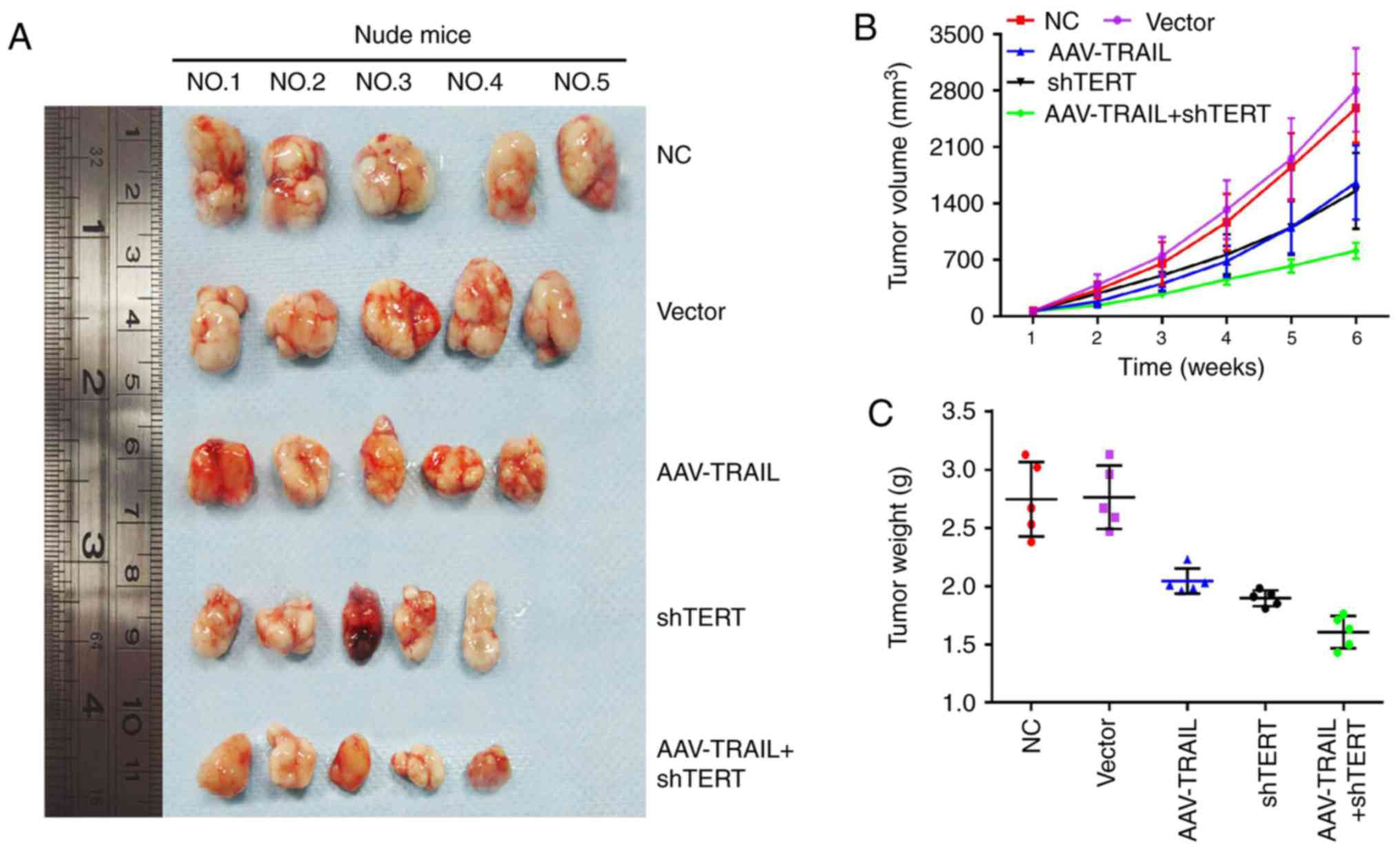
Combination of AAV-TRAIL and hTERT-shRNA
suppresses OSCC xenograft growth in BALB/c nude mice
A BALB/c nude mouse xenograft model was used as an
in vivo model to verify the tumor suppression effects of
AAV-mediated TRAIL overexpression combined with lentivirus
vector-mediated hTERT silencing in OSCC. The five groups of SCC25
cell lines (NC, vector, AAV-TRAIL, shTERT and AAV-TRAIL+shTERT)
were subcutaneously implanted in the axilla of the mice. Xenograft
growth curves were plotted according to the tumor volume changes
(Fig. 5A and B). As presented on
the respective plots, there was a marked reduction in tumor size
and tumor weight in the AAV-TRAIL and shTERT group, compared with
the NC and vector groups (Fig. 5B and
C). Notably, there was a marked suppression of tumor growth in
the AAV-TRAIL+shTERT combination group, compared with the NC and
vector groups (Fig. 5).
Combination of AAV-TRAIL and hTERT-shRNA
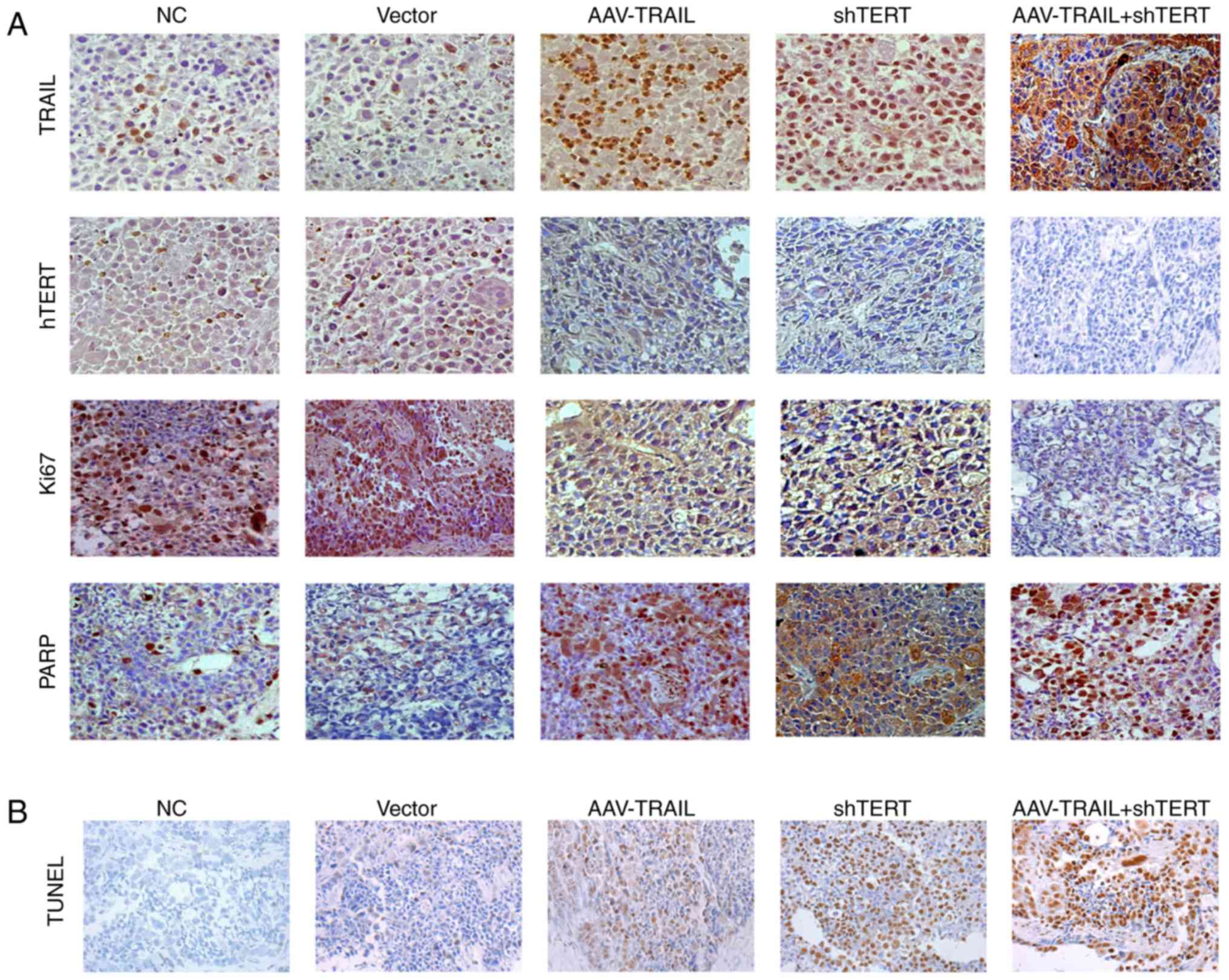
promotes OSCC xenograft apoptosis in BALB/c nude mice
To confirm these in vivo results,
immunohistochemistry was used to detect the combination effect on
apoptosis in mouse tumor tissues from each subgroup. The results
demonstrated that compared with the NC and vector groups, AAV-TRAIL
group has a higher level of TRAIL and PARP expression, as well as
lower expression of hTERT and Ki67. The shTERT group also exhibited
higher expression of TRAIL and PARP, and lower expression of hTERT
and Ki67, compared with the negative control and vector groups.
Notably, the TRAIL+hTERT group exhibited the highest expression of
TRAIL and PARP, and the lowest expression of hTERT and Ki67. These
results suggest that silencing hTERT upregulates TRAIL expression
and results in promotion of apoptosis and inhibition of
proliferation in OSCC. It is also clear that shTERT has a
synergistic effect on TRAIL-induced OSCC apoptosis enhancement and
proliferation suppression in a trial of combined AAV-TRAIL and
shTERT treatment (Fig. 6A).
Furthermore, this effect was further verified via TUNEL assay in
xenograft tissues (Fig. 6B).
The present findings indicate that
lentivirus-mediated TERT inhibition synergistically promotes
TRAIL-induced OSCC apoptosis enhancement and proliferation
suppression in vitro and in vivo.
Discussion
In the present study, TRAIL expression was
upregulated and hTERT expression was inhibited experimentally using
a stable AAV-mediated system and a lentivirus-mediated interfering
system in OSCC cells. The upregulation of TRAIL activated
apoptosis, suppressed proliferation and induced cell cycle arrest
in OSCC cells in vitro. Additionally, silencing hTERT
expression triggered elevation of TRAIL expression and displayed
the same effects. Finally, the upregulation of TRAIL and
suppression of hTERT potently inhibited the growth of xenograft
OSCC tumors in BALB/c nude mice; with a combination of TRAIL
overexpression and hTERT silencing exhibiting the most potent
suppressive effect on tumor growth.
Dysregulation of apoptosis-related genes is a
pivotal event during carcinogenesis and has been identified as a
critical mechanism in cancer development (27). The TRAIL gene efficiently induces
apoptosis in multiple types of tumor cells while sparing normal
cells, making TRAIL an ideal candidate for cancer gene therapy
(28,29). TRAIL induces malignant tumor cell
apoptosis by binding to the TRAIL receptors, such as death
receptors DR4 and DR5 (15,30). This interaction activates the
signal pathway for apoptosis, including Bcl-2, PARP and the caspase
cascade (31). Caspase-8
activation, initiated via cleaving of pro-apoptotic protein,
induces a downstream expression cascade caspase-9 and caspase-3.
Caspase-3 is the ultimate effector protein that directly triggers
apoptotic process (32). The
downstream signal of the Bcl-2 pathway accompanied with
upregulation of cleaved PARP activates caspase-3 to directly induce
apoptosis in certain types of malignant tumors, such as HCC
(33,34). In the present study, the
expression of TRAIL triggered the apoptosis process and pathway,
including the upregulation of caspase-8, caspase-9 and caspase-3,
as well as the inhibition of Bcl-2.
The catalytic subunit of telomerase, hTERT, can
reverse the telomere shortening process. The abnormal elevation of
hTERT expression and activation is a common event that causes
chemoresistance and increased proliferative ability in tumor cells
(35–37). Previous large studies have
demonstrated that hTERT has a pivotal role in cancer tumorigenesis,
growth, migration and invasion (38,39). The upregulation of hTERT is
positively correlated with metastasis and poor prognosis in many
types of malignant tumors, including HCC, ovarian cancer and
colorectal cancer (37,40,41). Therefore, hTERT is a potential
therapeutic target for cancer treatment, and elucidating the
underlying regulatory mechanisms of hTERT is essential.
In previous studies, TRAIL has been administered in
combination with chemotherapy drugs, such as doxorubicin or
flavonoids (42–44). Several studies have suggested as
association between hTERT and TRAIL, which has demonstrated that
downregulation of hTERT by RNA interference (RNAi) technology
significantly improves the sensitivity of HCC cells to TRAIL
treatment, including TRAIL-resistant cells (35,45–47). An underlying mechanism is that
knockdown of hTERT can increase cell apoptosis in TRAIL-treatment
cells via the mitochondrial type II apoptosis pathway and
telomerase-dependent pathway (47,48). In the present study, a regulatory
association between hTERT and TRAIL was demonstrated; that
lentivirus-mediated interference of hTERT expression can
significantly upregulate the mRNA and protein expression of TRAIL.
In addition, the present study provided, to the best of our
knowledge, the first evidence that downregulated hTERT and
overexpressed TRAIL cells exhibit reduced cell viability and
increased cell apoptosis in vivo and in vitro,
indicating that interfering with hTERT expression may benefit the
therapeutic effect of TRAIL in OSCC. Another previous study
regarding the association between hTERT and TRAIL revealed that
AAV-mediated TRAIL gene expression driven by hTERT promoter has an
inhibitory role in HCC growth in mice, which revealed the clinical
application of hTERT promoter-driven TRAIL gene therapy (26). In contrast with this therapeutic
strategy, the present study suggested a combination use of hTERT
knockdown and TRAIL overexpression to enhance the effect of OSCC
treatment.
Consistent with previous results, the present
results also confirmed that silencing of hTERT increases TRAIL
expression and promotes the anti-tumor effect induced by TRAIL in
OSCC cells (49). This effect was
also verified via in vivo xenograft nude mice
experimentation. The combination of AAV-TRAIL and shTERT
demonstrated a synergistic effect in the suppression of tumor
growth and overall tumor apoptosis compared with groups treated
with a single gene therapy target.
The present results also provide a rationale for a
combined virotherapy strategy to treat OSCC. The downregulation of
induced myeloid leukemia cell differentiation protein Mcl-1 has
been demonstrated to sensitize hTERT-adenovirus in TRAIL-mediated
apoptosis in HCC (50). Another
previous study reported that hTERT RNAi overcomes TRAIL resistance
via the mitochondrial type II apoptosis pathway and the
telomerase-dependent pathway (47,48). Therefore, the main limitation of
the present study is that the interaction between TRAIL and hTERT
was not evaluated, and the molecular mechanism that regulates OSCC
cell apoptosis promotion and growth inhibition was not elucidated.
Accordingly, further investigation must be performed to clarify the
underlying interaction mechanisms between TRAIL and hTERT that
synergistically regulate OSCC progression in vitro and in
vivo.
Together, the present results indicate that
AAV-mediated TRAIL expression and lentivirus-mediated hTERT
suppression display anti-tumor effects induced by TRAIL, including
apoptosis promotion, proliferation inhibition and cell cycle arrest
in OSCC both in vitro and in vivo. In addition, hTERT
interference was demonstrated to effectively upregulate TRAIL
expression. Furthermore, treatment with a combination of TRAIL
overexpression and hTERT interference has enhanced anti-tumor
effects, inhibiting tumor cell proliferation and increasing
apoptosis in a xenograft model. This suggests that there is a
synergistic effect between TRAIL and hTERT. The present results
indicate that the delivery of AAV-TRAIL and shTERT in combination
may be an effective treatment strategy for OSCC gene therapy.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Nature Science Foundation of China (grant no. 30872891); the
Natural Science Foundation of Guangdong Province, China (grant no.
2016A030313196); and the Science and Technology Planning Project of
Guangzhou City (Guangdong, China) (grant no. 201704020130).
Availability of data and materials
All data generated or analysed during the present
study are included in this published article.
Authors’ contributions
CZ and JS carried out the viral vector construction
and data analysis. XinZ and ZL performed animal experiments. SZ
carried out the reverse transcription-quantitative polymerase chain
reaction and western blotting analyses. XinZ wrote the manuscript.
JW and XiaZ conceived the present study and helped to draft the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee at Sun Yat-sen Memorial Hospital (Sun Yat-sen University,
Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lo Muzio L, Sartini D, Santarelli A,
Rocchetti R, Morganti S, Pozzi V, Rubini C, Bambini F and Emanuelli
M: Expression and prognostic significance of apoptotic genes in
oral squamous cell carcinoma. Mol Carcinog. 53:264–271. 2014.
View Article : Google Scholar
|
|
5
|
Yanamoto S, Yamada S, Takahashi H,
Yoshitomi I, Kawasaki G, Ikeda H, Minamizato T, Shiraishi T, Fujita
S, Ikeda T, et al: Clinicopathological risk factors for local
recurrence in oral squamous cell carcinoma. Int J Oral Maxillofac
Surg. 41:1195–1200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhong LP, Zhang CP, Ren GX, Guo W, William
WN Jr, Sun J, Zhu HG, Tu WY, Li J, Cai YL, et al: Randomized phase
III trial of induction chemotherapy with docetaxel, cisplatin, and
fluorouracil followed by surgery versus up-front surgery in locally
advanced resectable oral squamous cell carcinoma. J Clin Oncol.
31:744–751. 2013. View Article : Google Scholar
|
|
7
|
Yajima T, Ochiai H, Uchiyama T, Takano N,
Shibahara T and Azuma T: Resistance to cytotoxic
chemotherapy-induced apoptosis in side population cells of human
oral squamous cell carcinoma cell line Ho-1-N-1. Int J Oncol.
35:273–280. 2009.PubMed/NCBI
|
|
8
|
Jiang G, Li J, Zeng Z and Xian L:
Lentivirus-mediated gene therapy by suppressing survivin in BALB/c
nude mice bearing oral squamous cell carcinoma. Cancer Biol Ther.
5:435–440. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang L, Wang H, Zhou L and Yu D:
Akt-FOXO3a signaling axis dysregulation in human oral squamous cell
carcinoma and potent efficacy of FOXO3a-targeted gene therapy. Oral
Oncol. 47:16–21. 2011. View Article : Google Scholar
|
|
10
|
Wajant H, Pfizenmaier K and Scheurich P:
TNF-related apoptosis inducing ligand (TRAIL) and its receptors in
tumor surveillance and cancer therapy. Apoptosis. 7:449–459. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang S: The promise of cancer therapeutics
targeting the TNF-related apoptosis-inducing ligand and TRAIL
receptor pathway. Oncogene. 27:6207–6215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McLornan DP, Barrett HL, Cummins R,
McDermott U, McDowell C, Conlon SJ, Coyle VM, Van Schaeybroeck S,
Wilson R, Kay EW, et al: Prognostic significance of TRAIL signaling
molecules in stage II and III colorectal cancer. Clin Cancer Res.
16:3442–3451. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Polanski R, Vincent J, Polanska UM,
Petreus T and Tang EK: Caspase-8 activation by TRAIL monotherapy
predicts responses to IAPi and TRAIL combination treatment in
breast cancer cell lines. Cell Death Dis. 6:e18932015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim K, Nakagawa H, Fei P, Rustgi AK and
El-Deiry WS: Targeting Bcl-xL in esophageal squamous cancer to
sensitize to chemotherapy plus TRAIL-induced apoptosis while normal
epithelial cells are protected by blockade of caspase 9. Cell Death
Differ. 11:583–587. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen M, Wang X, Zha D, Cai F, Zhang W, He
Y, Huang Q, Zhuang H and Hua ZC: Apigenin potentiates TRAIL therapy
of non-small cell lung cancer via upregulating DR4/DR5 expression
in a p53-dependent manner. Sci Rep. 6:354682016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Wen XZ, Bu ZD, Cheng XJ, Xing XF,
Wang XH, Zhang LH, Guo T, Du H, Hu Y, et al: Paclitaxel enhances
tumoricidal potential of TRAIL via inhibition of MAPK in resistant
gastric cancer cells. Oncol Rep. 35:3009–3017. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Piras-Straub K, Khairzada K, Trippler M,
Baba HA, Kaiser GM, Paul A, Canbay A, Weber F, Gerken G and Herzer
K: TRAIL expression levels in human hepatocellular carcinoma have
implications for tumor growth, recurrence and survival. Int J
Cancer. 136:E154–E160. 2015. View Article : Google Scholar
|
|
18
|
Holoch PA and Griffith TS: TNF-related
apoptosis-inducing ligand (TRAIL): A new path to anti-cancer
therapies. Eur J Pharmacol. 625:63–72. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu ST, Chen L, Wang HJ, Tang XD, Fang DC
and Yang SM: hTERT promotes the invasion of telomerase-negative
tumor cells in vitro. Int J Oncol. 35:329–336. 2009.PubMed/NCBI
|
|
20
|
Xi L and Cech TR: Inventory of telomerase
components in human cells reveals multiple subpopulations of hTR
and hTERT. Nucleic Acids Res. 42:8565–8577. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lü MH, Liao ZL, Zhao XY, Fan YH, Lin XL,
Fang DC, Guo H and Yang SM: hTERT-based therapy: A universal
anticancer approach (Review). Oncol Rep. 28:1945–1952. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu L, Yin S, Banerjee S, Sarkar F and
Reddy KB: Enhanced anticancer effect of the combination of
cisplatin and TRAIL in triple-negative breast tumor cells. Mol
Cancer Ther. 10:550–557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang WB, Zhou YL, Heng DF, Miao CH and Cao
YL: Combination of tumor necrosis factor-related apoptosis-inducing
ligand (TRAIL) and canstatin gene suppression therapy on breast
tumor xenograft growth in mice. Breast Cancer Res Treat.
110:283–295. 2008. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
O’Neill PM, Amewu RK, Charman SA, Sabbani
S, Gnädig NF, Straimer J, Fidock DA, Shore ER, Roberts NL, Wong MH,
et al: A tetraoxane-based antimalarial drug candidate that
overcomes PfK13-C580Y dependent artemisinin resistance. Nat Commun.
8:151592017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Ma H, Zhang J, Liu S, Liu Y and
Zheng D: AAV-mediated TRAIL gene expression driven by hTERT
promoter suppressed human hepatocellular carcinoma growth in mice.
Life Sci. 82:1154–1161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kuo WT, Lee TC, Yang HY, Chen CY, Au YC,
Lu YZ, Wu LL, Wei SC, Ni YH, Lin BR, et al: LPS receptor subunits
have antagonistic roles in epithelial apoptosis and colonic
carcinogenesis. Cell Death Differ. 22:1590–1604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang K, Kievit FM, Jeon M, Silber JR,
Ellenbogen RG and Zhang M: Nanoparticle-mediated target delivery of
TRAIL as gene therapy for glioblastoma. Adv Healthc Mater.
4:2719–2726. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Li L, Shao N, Hu Z, Chen H, Xu L,
Wang C, Cheng Y and Xiao J: Triazine-modified dendrimer for
efficient TRAIL gene therapy in osteosarcoma. Acta Biomater.
17:115–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Abe K, Kurakin A, Mohseni-Maybodi M, Kay B
and Khosravi-Far R: The complexity of TNF-related
apoptosis-inducing ligand. Ann N Y Acad Sci. 926:52–63. 2000.
View Article : Google Scholar
|
|
31
|
Kojima Y, Nakayama M, Nishina T, Nakano H,
Koyanagi M, Takeda K, Okumura K and Yagita H: Importin β1
protein-mediated nuclear localization of death receptor 5 (DR5)
limits DR5/tumor necrosis factor (TNF)-related apoptosis-inducing
ligand (TRAIL)-induced cell death of human tumor cells. J Biol
Chem. 286:43383–43393. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Valley CC, Lewis AK, Mudaliar DJ,
Perlmutter JD, Braun AR, Karim CB, Thomas DD, Brody JR and Sachs
JN: Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)
induces death receptor 5 networks that are highly organized. J Biol
Chem. 287:21265–21278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vizetto-Duarte C, Custódio L, Gangadhar
KN, Lago JH, Dias C, Matos AM, Neng N, Nogueira JM, Barreira L,
Albericio F, et al: Isololiolide, a carotenoid metabolite isolated
from the brown alga Cystoseira tamariscifolia, is cytotoxic and
able to induce apoptosis in hepatocarcinoma cells through caspase-3
activation, decreased Bcl-2 levels, increased p53 expression and
PARP cleavage. Phytomedicine. 23:550–557. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu P, Cai X, Zhang W, Li Y, Qiu P, Lu D
and He X: Flavonoids of Rosa roxburghii Tratt exhibit
radioprotection and anti-apoptosis properties via the
Bcl-2(Ca(2+))/Caspase-3/PARP-1 pathway. Apoptosis. 21:1125–1143.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dudognon C, Pendino F, Hillion J, Saumet
A, Lanotte M and Ségal-Bendirdjian E: Death receptor signaling
regulatory function for telomerase: hTERT abolishes TRAIL-induced
apoptosis, independently of telomere maintenance. Oncogene.
23:7469–7474. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luiten RM, Pene J, Yssel H and Spits H:
Ectopic hTERT expression extends the life span of human CD4+ helper
and regulatory T-cell clones and confers resistance to oxidative
stress-induced apoptosis. Blood. 101:4512–4519. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dômont J, Pawlik TM, Boige V, Rose M,
Weber JC, Hoff PM, Brown TD, Zorzi D, Morat L, Pignon JP, et al:
Catalytic subunit of human telomerase reverse transcriptase is an
independent predictor of survival in patients undergoing curative
resection of hepatic colorectal metastases: A multicenter analysis.
J Clin Oncol. 23:3086–3093. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Z, Li Q, Li K, Chen L, Li W, Hou M,
Liu T, Yang J, Lindvall C, Björkholm M, et al: Telomerase reverse
transcriptase promotes epithelial-mesenchymal transition and stem
cell-like traits in cancer cells. Oncogene. 32:4203–4213. 2013.
View Article : Google Scholar
|
|
39
|
Zhao T, Hu F, Qiao B, Chen Z and Tao Q:
Telomerase reverse transcriptase potentially promotes the
progression of oral squamous cell carcinoma through induction of
epithelial-mesenchymal transition. Int J Oncol. 46:2205–2215. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Choi MJ, Cho KH, Lee S, Bae YJ, Jeong KJ,
Rha SY, Choi EJ, Park JH, Kim JM, Lee JS, et al: hTERT mediates
norepinephrine-induced Slug expression and ovarian cancer
aggressiveness. Oncogene. 34:3402–3412. 2015. View Article : Google Scholar
|
|
41
|
Qin YZ, Xie XC, Liu HZ, Lai H, Qiu H and
Ge LY: Screening and preliminary validation of miRNAs with the
regulation of hTERT in colorectal cancer. Oncol Rep. 33:2728–2736.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wennerberg E, Sarhan D, Carlsten M,
Kaminskyy VO, D’Arcy P, Zhivotovsky B, Childs R and Lundqvist A:
Doxorubicin sensitizes human tumor cells to NK cell- and
T-cell-mediated killing by augmented TRAIL receptor signaling. Int
J Cancer. 133:1643–1652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kahana S, Finniss S, Cazacu S, Xiang C,
Lee HK, Brodie S, Goldstein RS, Roitman V, Slavin S, Mikkelsen T
and Brodie C: Proteasome inhibitors sensitize glioma cells and
glioma stem cells to TRAIL-induced apoptosis by
PKCepsilon-dependent downregulation of AKT and XIAP expressions.
Cell Signal. 23:1348–1357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dhandapani L, Yue P, Ramalingam SS, Khuri
FR and Sun SY: Retinoic acid enhances TRAIL-induced apoptosis in
cancer cells by upregulating TRAIL receptor 1 expression. Cancer
Res. 71:5245–5254. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin T, Huang X, Gu J, Zhang L, Roth JA,
Xiong M, Curley SA, Yu Y, Hunt KK and Fang B: Long-term tumor-free
survival from treatment with the GFP-TRAIL fusion gene expressed
from the hTERT promoter in breast cancer cells. Oncogene.
21:8020–8028. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jacob D, Bahra M, Schumacher G, Neuhaus P
and Fang B: Gene therapy in colon cancer cells with a
fiber-modified adenovector expressing the TRAIL gene driven by the
hTERT promoter. Anticancer Res. 24:3075–3079. 2004.PubMed/NCBI
|
|
47
|
Zhang RG, Zhao JJ, Yang LQ, Yang SM, Wang
RQ, Chen WS, Peng GY and Fang DC: RNA interference-mediated hTERT
inhibition enhances TRAIL-induced apoptosis in resistant
hepatocellular carcinoma cells. Oncol Rep. 23:1013–1019.
2010.PubMed/NCBI
|
|
48
|
Chen L, Lü MH, Zhang D, Hao NB, Fan YH, Wu
YY, Wang SM, Xie R, Fang DC, Zhang H, et al: miR-1207 5p and
miR-1266 suppress gastric cancer growth and invasion by targeting
telomerase reverse transcriptase. Cell Death Dis. 5:e10342014.
View Article : Google Scholar
|
|
49
|
Zang G, Miao L, Mu Y, Qiao C, Liu J, Ke X,
Zheng C and Sun H: Adenoviral mediated transduction of adenoid
cystic carcinoma by human TRAIL gene driven with hTERT tumor
specific promoter induces apoptosis. Cancer Biol Ther. 8:966–972.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wirth T, Kühnel F, Fleischmann-Mundt B,
Woller N, Djojosubroto M, Rudolph KL, Manns M, Zender L and Kubicka
S: Telomerase-dependent virotherapy overcomes resistance of
hepatocellular carcinomas against chemotherapy and tumor necrosis
factor-related apoptosis-inducing ligand by elimination of Mcl-1.
Cancer Res. 65:7393–7402. 2005. View Article : Google Scholar : PubMed/NCBI
|