Introduction
Mesenchymal stem cells (MSCs) are adult stem cells
that have the ability to self-renew and differentiate into various
mesodermal cells, including osteoblasts, chondrocytes and
adipocytes (1). Populations of
MSCs are present in almost every tissue in the body, including bone
marrow, adipose tissue, dental pulp and synovial tissue (2-4).
Adipose-derived stem cells (ASCs) are MSCs that are present within
the adipose tissue, as first described by Zuk et al
(5). It has been reported that
ASCs are easier to isolate and acquire compared with other resident
stem cell populations, and the cell yield is much higher than that
of bone marrow-derived MSCs (BMSCs) (6). ASCs have garnered attention in the
scientific and medical fields due to their potential clinical
applications (7). Numerous
ASCs-based clinical trials have been performed over recent years,
and it has been suggested that ASCs possess therapeutic potential
for the future treatment of various diseases (7). To fully exploit the therapeutic
value of ASCs in clinical application, an in-depth understanding of
the molecular pathways by which ASCs proliferate and differentiate
is essential.
Yes-associated protein (YAP; gene symbol, YAP1) is a
key transcriptional co-factor that is regulated by the Hippo
signaling pathway (8). YAP acts
as a transcriptional co-activator of the TEA domain-containing
sequence-specific transcription factor, which regulates the
expression of several 'stemness' genes (9). Core components of the Hippo pathway
include the kinases MST and LATS (10). Upon activation of the Hippo
pathway, MST phosphorylates and activates LATS, which subsequently
phosphorylates and inhibits YAP. YAP phosphorylation leads to
cytoplasmic retention and degradation by proteasomes (10). Conversely, inhibition of the Hippo
pathway results in YAP nuclear retention and activation of
transcriptional activity (11).
It has previously been reported that sustained YAP expression is
associated with liver enlargement and eventual tumorigenesis, thus
suggesting an important role for YAP in cell proliferation and
tumor formation (12).
The upstream signaling mechanisms that regulate the
Hippo signaling pathway remain elusive. A previous study
demonstrated that the mechanical properties of the extracellular
matrix (ECM), along with cell matrix attachment, may regulate the
localization and activity of YAP via a process involving the actin
cytoskeleton (13). Furthermore,
G protein-coupled receptors and their agonists, including
lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P), have
been revealed to regulate YAP activity via modulating the actin
cytoskeleton (14).
YAP and its paralog, transcriptional co-activator
with the PDZ binding motif (TAZ), are the main downstream
regulators of the Hippo signaling pathway (15). TAZ has been demonstrated to
co-activate genes dependent on Runt-related transcription factor 2
(RUNX2), which is the transcriptional regulator of the osteoblastic
lineage, while suppressing the transcription of genes dependent on
peroxisome proliferator-activated receptor γ (PPARγ), which is the
master regulator of the adipogenic lineage, in MSCs. Our previous
study demonstrated that the phytomolecule icariin may promote the
proliferation and osteogenic differentiation of rASCs via the Ras
homolog gene family, member A-TAZ signaling pathway (16). YAP and TAZ are often considered to
be orthologs of Drosophila Yorkie; however, it has been
reported that the differentiation-regulating functions of YAP are
not the same as those of TAZ (17).
The present study aimed to evaluate the upstream
factors that affect YAP expression and subcellular distribution in
rASCs, as well as the role of YAP in rASC proliferation and
osteogenic/adipogenic differentiation.
Materials and methods
Isolation and culture of rASCs
Male Sprague-Dawley rats (age, 6-8 weeks; weight,
200–250 g, n=24) used in the present study were purchased from the
Laboratory Animal Center of the Tongji Medical College (Wuhan,
China). All rats were kept in ventilated filter-top cages under
standard laboratory conditions: 12-h light/dark cycle and a
constant temperature of 24°C with 60% humidity. Rats were given
ad libitu access to conventional rodent chow and water. All
experimental animals were sacrificed via cervical dislocation.
Prior to cervical dislocation, rats were anesthetized by
intra-peritoneal injection of 2% pentobarbital sodium (35 mg/kg
body weight). Animal death was confirmed by monitoring the
heartbeat and body temperature. The present study was approved by
the Experimental Animal Ethics Committee of Tongji Medical College.
Briefly, rASCs were isolated, cultured and characterized as
described in our previous study (16). Adipose tissues from the epididymis
of male Sprague-Dawley rats were harvested, finely minced and
digested with 0.1% type I collagenase (Wuhan Boster Biological
Technology, Ltd., Wuhan, China) at a 1:1 volume ratio at 37°C for 1
h, followed by centrifugation at 100 × g for 10 min at 37°C. The
supernatant layer was discarded and the remaining cells were
collected and resuspended in Dulbecco's modified Eagle's
medium/Ham's F-12 (DMEM/F12; Hyclone; GE Healthcare Life Sciences,
Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Undigested debris was removed by filtering through a sterile 75-mm
nylon mesh. Finally, cells were cultured with DMEM/F12 supplemented
with 10% FBS and 1% penicillin-streptomycin at 37°C in an
atmosphere containing 5% CO2. Cells at passage 3 were
used for subsequent experiments.
Osteogenic and adipogenic differentiation
of rASCs
For osteogenic and adipogenic differentiation, ASCs
at passage 3 were plated into 6-well plates at a density of
1×104 cells/cm2 and cultured at 37°C in an
atmosphere containing 5% CO2. After 24 h, the cell
culture medium was removed and replaced with adipogenic or
osteogenic differentiation medium. The adipogenic differentiation
medium comprised DMEM supplemented with 10% FBS, 1 µM
dexamethasone (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 10
mg/ml insulin (Invitrogen; Thermo Fisher Scientific, Inc.), 0.5 mM
methyl-isobutyl-xanthine (Sigma-Aldrich; Merck KGaA) and 100 mM
indomethacin (Sigma Aldrich; Merck KGaA). The osteogenic
differentiation medium consisted of DMEM supplemented with 10% FBS,
10 mM β-glycerophosphate (Sigma-Aldrich; Merck KGaA), 0.1 µM
dexamethasone and 0.1 mM ascorbate-2-phosphate (Sigma-Aldrich;
Merck KGaA).
Cell treatment
rASCs at passage 3 were seeded at varying densities
(1,500 or 12,000 cells/cm2) and cultured for 2 days in
an atmosphere containing 5% CO2 at 37°C with DMEM/F12
supplemented with 10% FBS. Cells were harvested and the gene
expression of CTGF and Ankrd1 was evaluated via reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). To
evaluate the role of the actin cytoskeleton in YAP activity, cells
were seeded at a density of 1,500 cells/cm2 and cultured
for 2 days in an atmosphere containing 5% CO2 at 37°C
with DMEM/F12 supplemented with 10% FBS. After treatment with 1
µg/ml Latrunculin B for 30 min, cells were harvested and the
expression levels of CTGF and Ankrd1 were evaluated by RT-qPCR.
rASCs were seeded in 6-cm dishes at a density of
1,500 cells/cm2 and incubated in serum-free DMEM/F12
medium for 24 h at 37°C prior to treatment with various reagents.
LPA at 10 µmol/l and S1P at 10 µmol/l were used in
the present study. Latrunculin B (LatB; 1 µg/ml) was used to
disrupt F-actin fiber organization. All reagents were purchased
from Sigma-Aldrich (Merck KGaA).
Immunofluorescence staining
To evaluate the effects of contact-inhibited
proliferation on YAP subcellular localization, rASCs were plated on
coverslips at varying densities (1,500 or 12,000
cells/cm2) and cultured for 2 days in an atmosphere
containing 5% CO2 at 37°C with DMEM/F12 supplemented
with 10% FBS. To evaluate the effects of the actin cytoskeleton on
YAP subcellular localization, cells were seeded on coverslips at a
density of 1,500 cells/cm2 with DMEM/F12 supplemented
with 10% FBS and treated with 1 µg/ml Latrunculin B at 37°C
for 30 min. Cells were subsequently fixed with 4% paraformaldehyde
for 15 min and permeabilized with 0.1% Triton X-100 for 10 min at
room temperature. After blocking with 5% bovine serum albumin (BSA;
Wuhan Boster Biological Technology, Ltd.) for 1 h at 37°C, slides
were incubated with YAP primary antibody (cat. no. 4912; Cell
Signaling Technology, Inc., Danvers, MA, USA) diluted with 5% BSA
(dilution 1:200) at 4°C overnight. After washing three times with
PBS, slides were incubated with Alexa Fluor® 594-labeled
donkey anti-rabbit secondary antibodies (cat. no. R37119;
Invitrogen; Thermo Fisher Scientific, Inc.; dilution 1:1,000) for 1
h at 37°C. For staining of F-actin, cells were incubated with
fluorescein isothiocyanate-conjugated phalloidin (cat. no. P5282;
Sigma-Aldrich; Merck KGaA) 1 h after blocking at 37°C. After
washing with PBS, cell nuclei were visualized with DAPI for 5 min.
To reduce background fluorescence, slides were washed with PBS.
Finally, cells were observed under a fluorescence microscope
(FV500; Olympus Corporation, Tokyo, Japan).
Western blot analysis
Cells were washed with PBS and treated with lysis
buffer containing 50 mM Tris-HCl, 0.1 mM EDTA, 0.1% Triton X-100
and 1 mM phenylmethylsulfonyl fluoride for 30 min at 37°C. The
proteins were prepared using commercial bicinchoninic acid kits
(Beyotime Institute of Biotechnology, Jiangsu, China) according to
the manufacturer's protocols. Aliquots containing equal amounts of
protein (25 µg/lane) were separated by SDS-PAGE (7–15%
gradient cross-linked polyacrylamide gels) and were then
transferred to polyvinylidene fluoride membranes (EMD Millipore,
Billerica, MA, USA). After transfer, membranes were probed
overnight at 4°C with anti-YAP rabbit polyclonal antibodies (cat.
no. 4912; Cell Signaling Technology, Inc.; dilution 1:1,000),
anti-RUNX2 rabbit polyclonal antibodies (cat. no. ab23981; Abcam,
Cambridge, UK; dilution 1:200), anti-PPARγ rabbit polyclonal
antibodies (cat. no. ab59256 Abcam; dilution 1:200) and anti-GAPDH
rabbit polyclonal antibodies (cat. no. BA2913; Wuhan Boster
Biological Technology, Ltd.; dilution 1:200) followed by
horseradish peroxidase-conjugated secondary antibodies for 1 h at
37°C (cat. no. BA1054; Wuhan Boster Biological Technology, Ltd.;
dilution 1:10,000). Blots were developed by enhanced
chemiluminescence (ECL) using Western ECL Substrate kit (Pierce;
Thermo Fisher Scientific, Inc.). The intensity of each band was
semi-quantified using digital image analysis software (Quantity
One, version 4.6; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
GAPDH was selected as a protein loading control.
RT-qPCR analysis
For relative mRNA expression analysis, cells in each
group were washed with PBS and treated with TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) to extract total RNA,
according to the manufacturer's protocol. RNA samples were then
purified and reverse-transcribed to cDNA using the 1st Strand cDNA
Synthesis kit (Toyobo Life Science, Osaka, Japan), as per the
manufacturer's protocol. The cDNA samples were then subject to qPCR
to determine the expression of various genes. qPCR was performed
with cDNA samples in triplicate under the following conditions:
95°C for 5 sec, followed by 40 cycles at 94°C for 20 sec, 60°C for
20 sec and extension at 72°C for 10 sec. GAPDH was used as an
internal control. Primers used in the present study were as
follows: Connective tissue growth factor (CTGF), 5′-CGT TAG CCT CGC
CTT GGT G-3′ (sense) and 5′-GGG AGC CGA AGT CGC AGA-3′ (antisense);
ankyrin repeat domain 1 (Ankrd1), 5′-CGG CTC TTG ATG ACC TTC G-3′
(sense) and 5′-GCA TTC TCC TTG AGG CTG TC-3′ (antisense); YAP,
5′-CCC AAG GCT TGA CCC TCG T-3′ (sense) and 5′-CGT ATT GCC TGC CGA
AAT AAC T-3′ (antisense); proliferating cell nuclear antigen
(PCNA), 5′-AAG GGC TGA AGA TAA TGC TGA TAC-3′ (sense) and 5′-CAT
ATA CGT GCA AAT TCA CCA GAT-3′ (antisense); RUNX2, 5′-TGG TAC TTC
GTC AGC GTC CTA TC-3′ (sense) and 5′-GCT TCC ATC AGC GTC AAC ACC-3′
(antisense); PPARγ, 5′-ACA TCA GTG GGA ATT AAG GCA-3′ (sense) and
5′-TCA AAG GAA TGG GAGTGG TC-3′ (antisense); and GAPDH, 5′-GGC AAG
TTC AAC GGC ACA G-3′ (sense) and 5′-CGC CAG TAG ACT CCA CGA CAT-3′
(antisense).
Cell Counting kit (CCK)-8 cell
proliferation assay
Cells were cultured under various conditions and
subsequently seeded into 96-well plates at a density of
2×103 cells/well. Cells were divided into three groups:
Serum starvation (SS) group (DMEM/F12 medium only), SS + LPA group
(DMEM/F12 medium supplemented with 10 µmol/l LPA) and SS +
short hairpin (sh) RNA-targeting YAP (shYAP) + LPA group (cells
transduced with shYAP lentivirus and cultured in DMEM/F12 medium
supplemented with 10 µmol/l LPA). Cells were cultured for 1,
2 or 3 days at 37°C and each sample was assessed for cellular
proliferation. A total of 10 µl CCK-8 solution (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) was added to each
well and incubated at 37°C in the dark for 2 h. Absorbance was read
on a microplate spectrophotometer (Bio-Rad Laboratories, Inc.) at a
wavelength of 490 nm.
Cell cycle distribution analysis
rASCs were seeded in 6-well plates at a density of
1,500 cells/cm2 with serum-free DMEM/F12 medium at 37°C.
Cell cycle distribution and DNA content were analyzed by flow
cytometry. Cells were divided into the following groups: SS group,
SS + LPA group, SS + S1P group (DMEM/F12 medium supplemented with
10 µmol/l S1P), SS + shYAP + LPA group and SS + shYAP + S1P
group (cells transduced with shYAP lentivirus cultured in DMEM/F12
medium supplemented with 10 µmol/l S1P). After 3 days
treatment, cultured cells were rinsed with PBS twice and fixed with
70% cold ethanol at -20°C for 2 h. Fixed cells were treated with
RNase A (50 µg/ml; cat. no. R4875; Sigma-Aldrich; Merck
KGaA) at 37°C for 30 min and then stained with propidium iodide
(PI; 65 µg/ml; Sigma-Aldrich; Merck KGaA) in the dark for 30
min at 37°C. PI fluorescence of individual nuclei was measured
using a flow cytometer (FACSort; BD Biosciences, Franklin Lakes,
NJ, USA).
Alkaline phosphatase (ALP) staining
rASCs were seeded in 6-well plates at a density of
1×104 cells/cm2 and cultured at 37°C in an
atmosphere containing 5% CO2 in osteogenic medium. Cells
were divided into four groups: Control group (DMEM/F12 supplemented
with 10% FBS), Osteo group (cells cultured in osteogenic
differentiation medium), Osteo + control shRNA (shControl) group
(cells transduced with control lentivirus cultured in osteogenic
differentiation medium) and Osteo + shYAP group (cells transduced
with shYAP lentivirus cultured in osteogenic differentiation
medium). Following 5 days of culture, cells were rinsed with PBS
three times and fixed with 4% paraformaldehyde for 15 min at 4°C.
Cells were rinsed again with deionized water and stained with
naphthol AS-MX phosphate and fast blue RR salt (Sigma-Aldrich;
Merck KGaA) for 30 min at 37°C in the dark. Excess dye was removed
with PBS and photomicrographs were captured under a light
microscope.
Oil Red O staining
rASCs were seeded in 6-well plates at a density of
1×104 cells/cm2 and cultured at 37°C in an
atmosphere containing 5% CO2 in adipogenic medium. Cells
were divided into four groups: Control group, Adipo group (cells
cultured in adipogenic differentiation medium), Adipo + shControl
group (cells transduced with control lentivirus cultured in
adipogenic differentiation medium) and Adipo + shYAP group (cells
transduced with shYAP lentivirus cultured in adipogenic
differentiation medium). Following 5 days of culture, cells were
rinsed with PBS three times and fixed with 4% paraformaldehyde for
15 min at 4°C. Cells were rinsed again with deionized water and
stained with Oil Red O for 30 min at 37°C. Excess dye was removed
with PBS and photomicrographs were captured under a light
microscope.
Construction of recombinant lentiviral
vectors
Specific restriction sites of YAP (NM_001034002;
Shanghai GeneChem Co., Ltd., Shanghai, China) were inserted into a
GV118 plasmid (Shanghai GeneChem Co., Ltd.). The target of shYAP#1
was GGC AAT ACG GAA TAT CAA T, whereas that of shYAP#2 was GAC AGT
CTT CCT TTG AGA T. Linked products were identified by double
digestion, and DNA sequencing confirmed accurate insertion. The
lentiviral vector system included three parts: pGC-LV vector,
pHelper 1.0 vector and pHelper 2.0 vector. pHelper 1.0 plasmid DNA
(15 µg), pHelper 2.0 plasmid DNA (10 µg) and pGC-LV
shYAP were co-transfected into subconfluent 293T cells (80%
confluence) at 37°C for 8 h using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Supernatants were
collected by ultracentrifugation at 4,000 × g for 10 min at room
temperature and the viral pellet was resuspended in Hanks' balanced
salt solution. The vector titers were determined by serial
dilution. Lentiviral null vectors with scrambled shRNA (sequence:
sense, 5′-UUC UCC GAA CGU GUC ACG UTT-3′ and antisense 5′-ACG UGA
CAC GUU CGG AGA ATT-3′) (Shanghai GeneChem Co., Ltd.) were used as
the shControl group.
Transduction of rASCs with lentiviral
vectors
rASCs at the third passage were seeded at the
density of 3×104 cells/ml in 24-well tissue culture
plates and cultured until cells reached 30% confluence. Following
removal of the cell culture medium, rASCs were incubated with
DMEM/F12 containing lentivirus (shYAP or shControl) and 5
µg/ml polybrene (Sigma-Aldrich; Merck KGaA) for 5 h at 37°C.
Multiplicity of infection (MOI) ranges between 10 and 200 were
calculated. Following lentiviral vector infection, the medium was
replaced with normal growth medium comprised of DMEM/F12 and 10%
FBS. A total of 1 day post-transduction, reporter gene expression
[green fluorescent protein (GFP)] was examined with fluorescence
microscopy. ASCs were transduced with lentiviruses at a MOI of 100
and passaged for further study.
Statistical analysis
All data are presented as the means ± standard
deviation. SPSS 13.0 was used for general statistical analysis
(SPSS, Inc., Chicago, IL, USA). Significant differences in
numerical data between two groups were determined using a Student's
t-test. For multiple group comparisons, one-way analysis of
variance was used with the Bonferroni post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
YAP subcellular distribution and
expression in rASCs is regulated by cell density and the actin
cytoskeleton
YAP is a transcriptional co-activator, which has
been reported to serve critical roles in cell proliferation and
differentiation; however, the functions of YAP and its upstream
regulating factors in rASCs have yet to be elucidated. The present
study demonstrated that the subcellular distribution of YAP in
rASCs was regulated by cell density and F-actin integrity. rASCs
were seeded at varying densities in tissue culture plates (1,500
and 12,000 cells/cm2). When cultured at 1,500
cells/cm2 (low density), cells showed no contact with
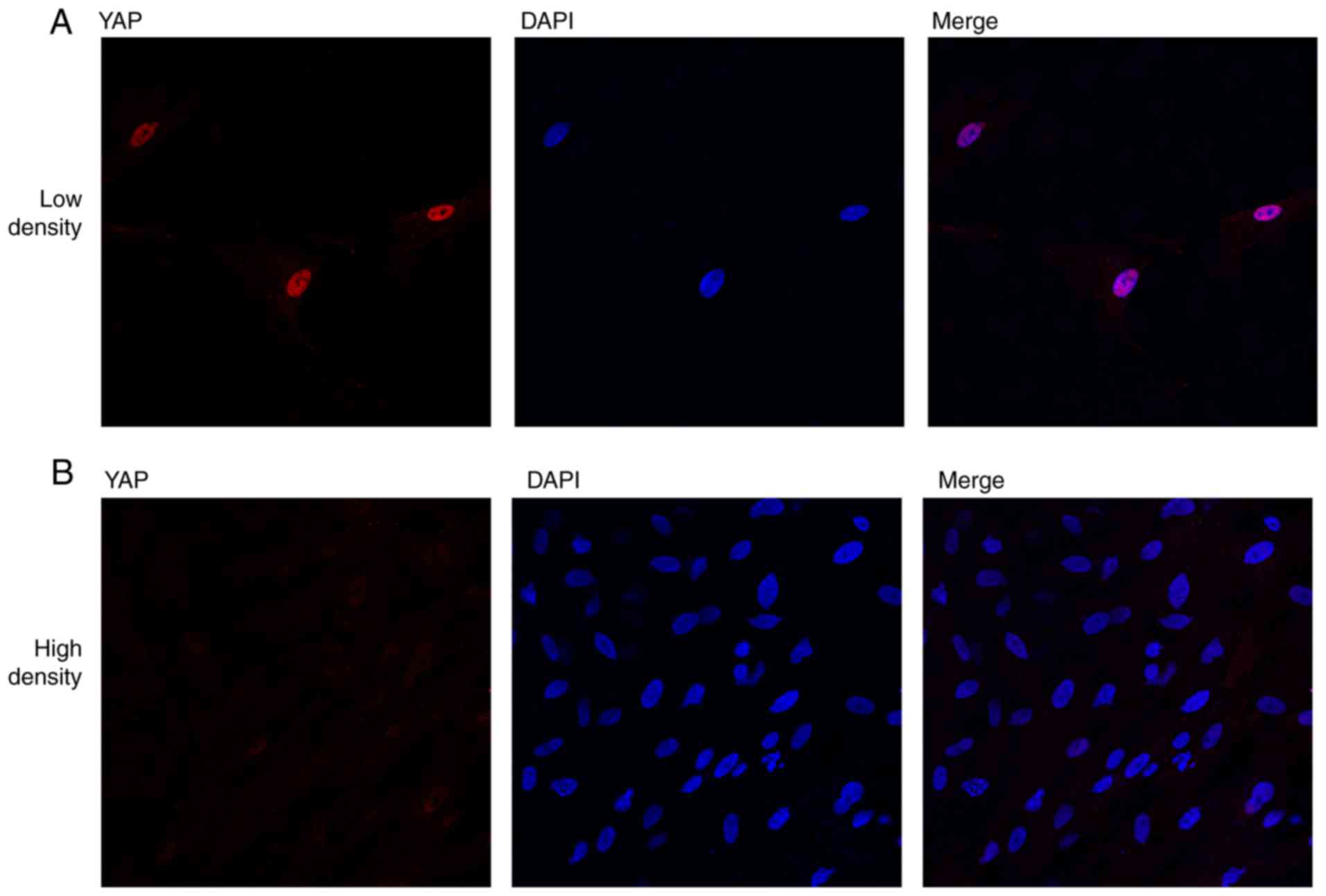
neighboring cells (Fig. 1A). When
cultured at a higher density of 12,000 cells/cm2 (high
density), a marked amount of contact was observed with neighboring
cells (Fig. 1B). In addition, in
the low density group, YAP expression was mainly localized to the
nuclei of rASCs (Fig. 1A).
Conversely, nuclear YAP expression was markedly decreased when
rASCs were cultured at a higher density (Fig. 1B). In addition, the effects of the
actin cytoskeleton on YAP subcellular distribution were
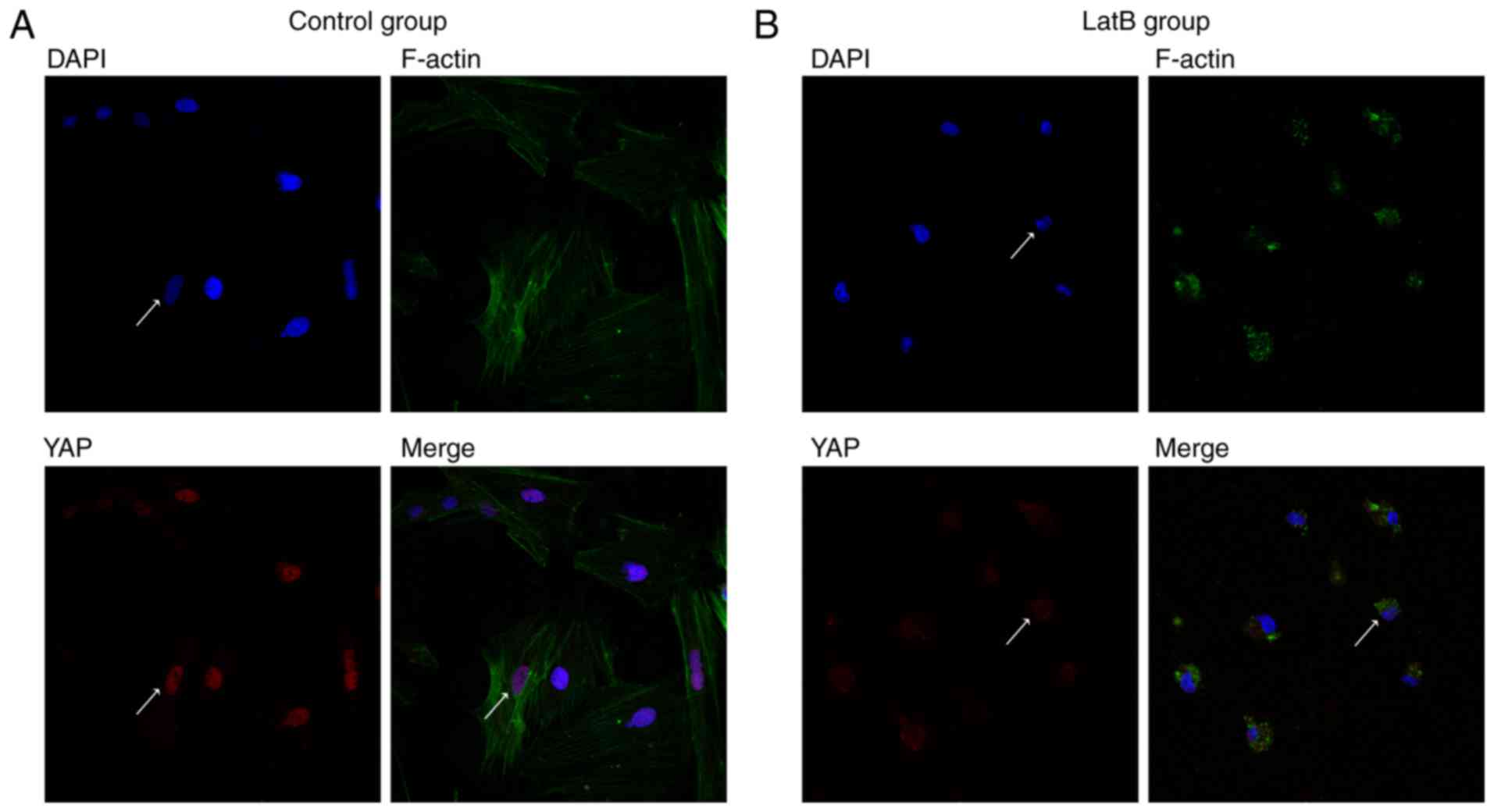
investigated. The results demonstrated that under normal
conditions, YAP was mainly localized in the nuclei of rASCs
(Fig. 2A); however, disruption of
the actin cytoskeleton with 1 µg/ml LatB for 30 min induced
marked YAP cytoplasmic translocation (Fig. 2B).
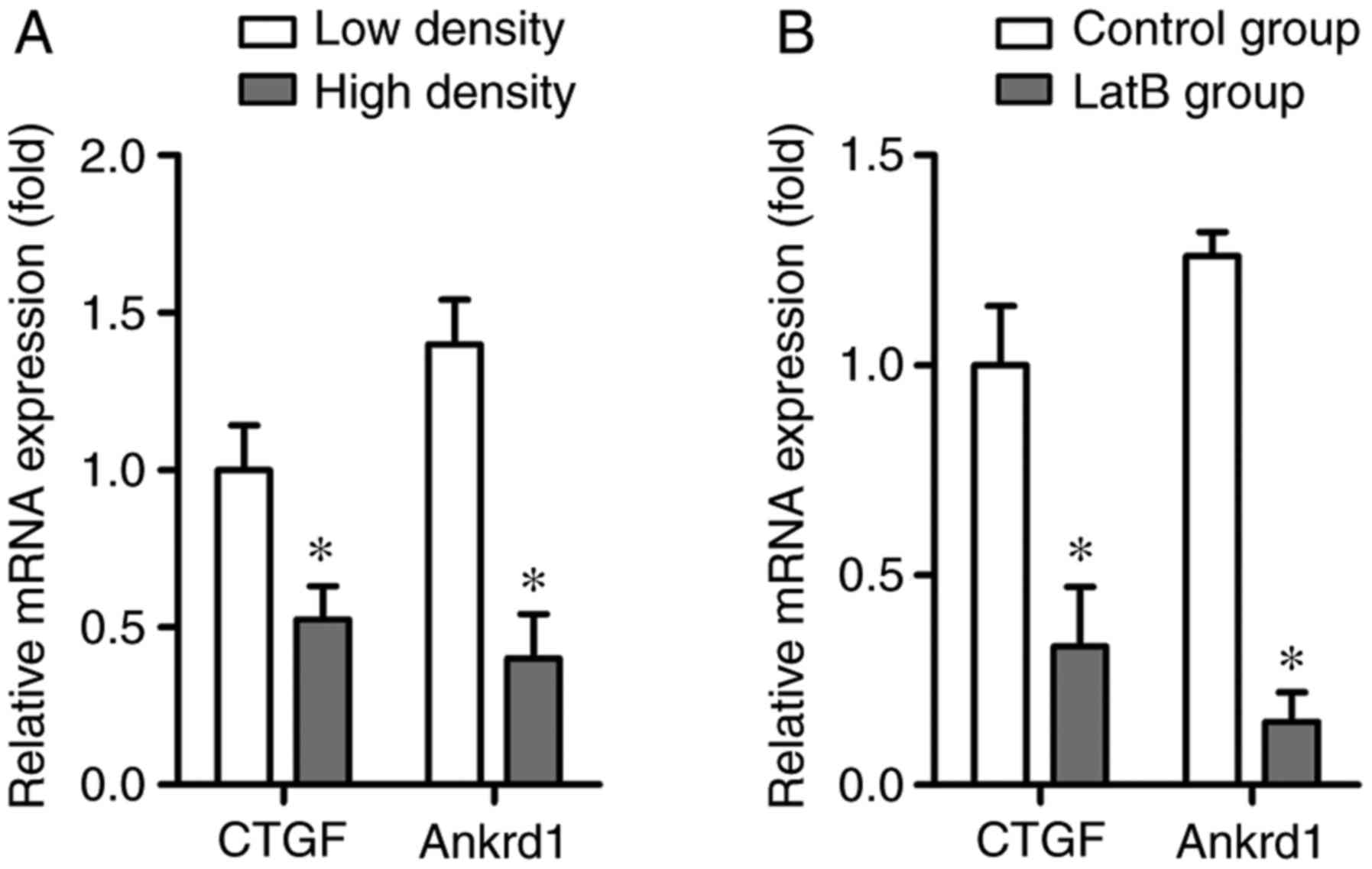
RT-qPCR was conducted to analyze the expression
levels of the YAP target genes, CTGF and Ankrd1. The results
indicated that the mRNA expression levels of CTGF and Ankrd1 were
significantly decreased in the high density group compared with in
the low density group, which was consistent with decreased YAP
nuclear localization (Fig. 3A).
Furthermore, CTGF and Ankrd1 mRNA expression was markedly inhibited
by LatB treatment compared with in the control group, thus
suggesting that the integrity of the actin cytoskeleton may be
closely associated with YAP activity (Fig. 3B).
LPA promotes YAP protein expression in
rASCs
It has previously been reported that LPA activates
YAP expression in epithelial cells, such as MCF10A cells (14). However, to the best of our
knowledge, the effects of LPA on YAP protein expression in rASCs
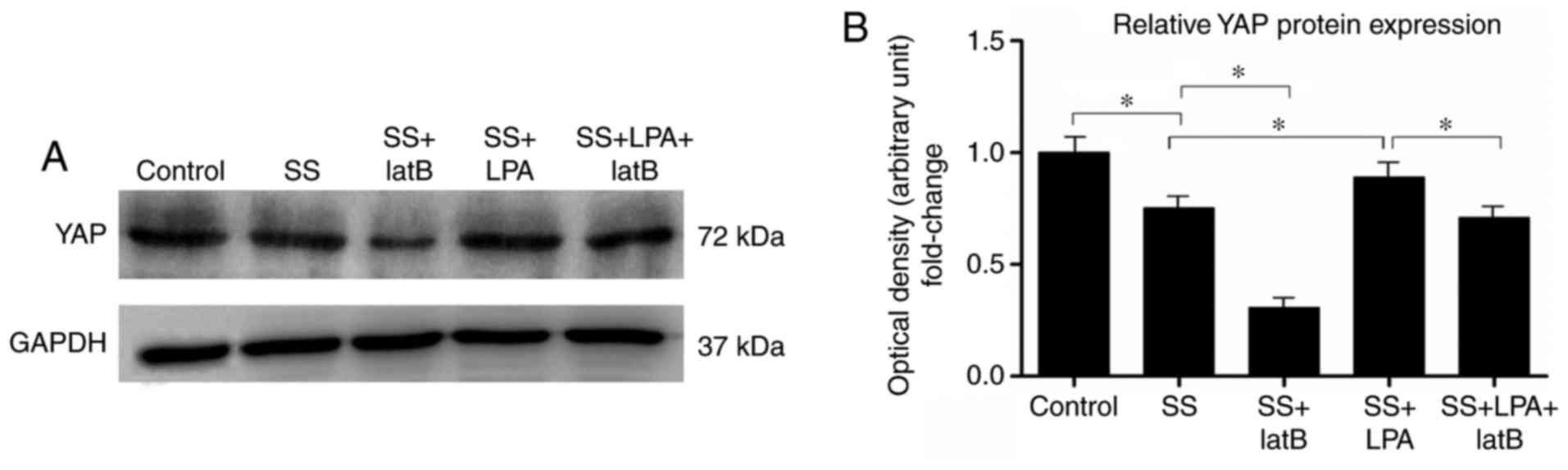
have yet to be determined. The present study demonstrated that SS
of rASCs for 24 h markedly decreased YAP protein expression
(Fig. 4). However, when cells
were treated with LPA (10 µmol/l) for 1 h after 24 h SS, YAP
protein expression was significantly increased (Fig. 4). When cells were treated with the
actin cytoskeleton disruption agent, LatB, for 1 h after 24 h SS
treatment, YAP protein expression was further decreased compared
with in the SS group (Fig. 4).
Notably, the LPA-induced upregulation of YAP protein expression in
rASCs was inhibited in the SS + LPA + LatB group compared with in
the SS + LPA group (Fig. 4).
These results suggested that LPA may promote YAP protein expression
in rASCs, and that integrity of the actin cytoskeleton is critical
for the regulation of YAP protein by LPA.
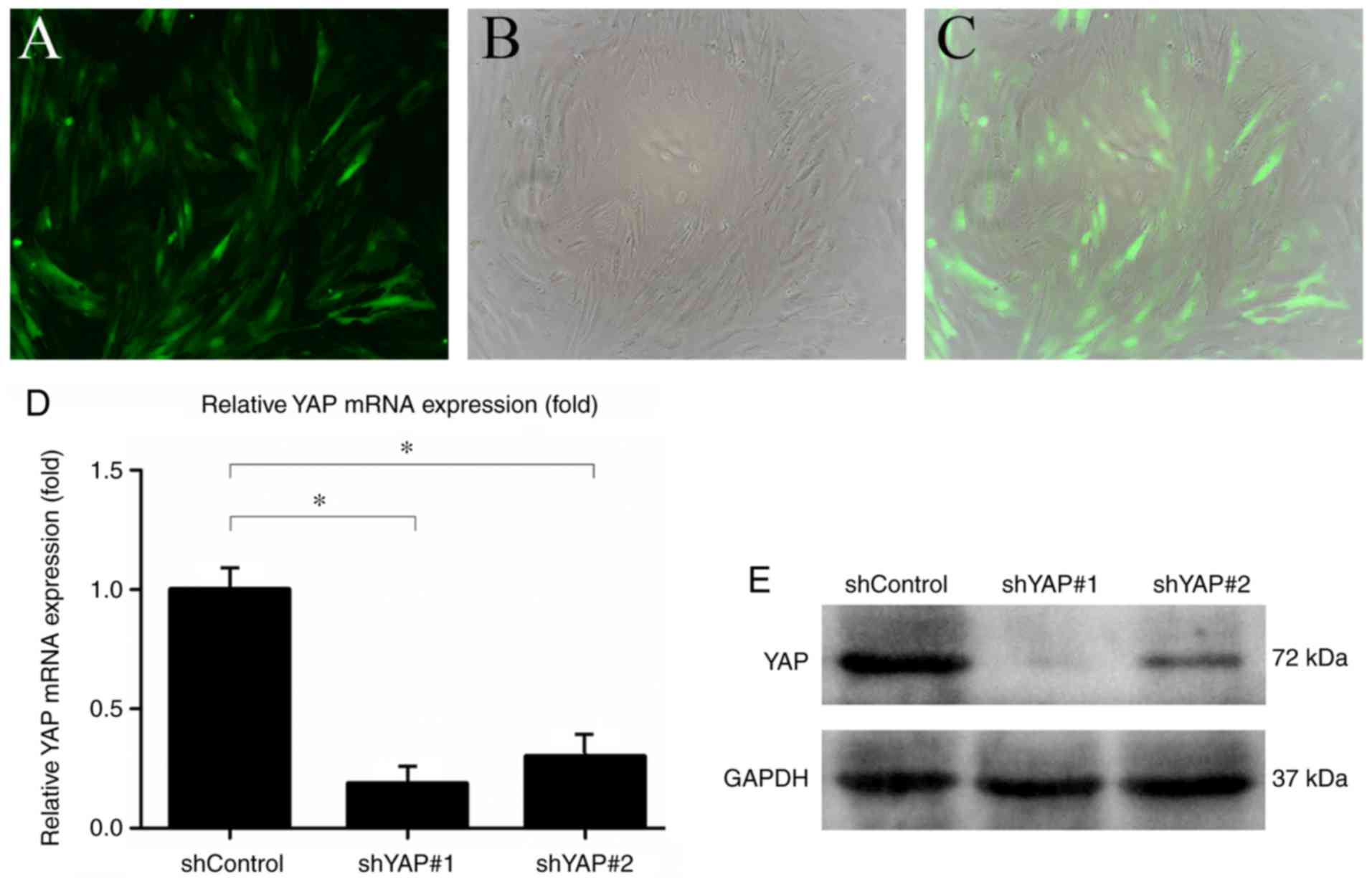
Transduction of rASCs with the shYAP
lentiviral system
rASCs at passage three were infected with
lentiviruses carrying shYAP-GFP or shControl-GFP. Fluorescence
microscopy was used to detect GFP expression in transduced rASCs.
GFP-expressing rASCs were observed by fluorescence microscopy 24 h
post-transduction (Fig. 5A–C). To
verify that the lentiviral system had been successfully infected
into rASCs, the mRNA expression levels of YAP were measured by
RT-qPCR 5 days post-transduction. The results revealed that YAP
mRNA expression in the shYAP#1 and shYAP#2 groups was significantly
lower than in the shControl group (Fig. 5D). Furthermore, YAP protein
expression in the shYAP#1 and shYAP#2 groups was markedly lower
compared with in the shControl group 7 days post-transduction
(Fig. 5E). The shYAP#1 lentiviral
system was used for subsequent experiments.
LPA and S1P promote rASC proliferation by
activating YAP
YAP is a transcriptional coactivator that promotes
the expression of various downstream genes, including CTGF and
Ankrd1 (14). The present results
demonstrated that CTGF, Ankrd1 and YAP mRNA expression levels were
significantly increased when rASCs were treated with LPA (10
µmol/l) or S1P (10 µmol/l) for 1 h compared with in
the SS control group (Fig. 6A–C).
However, rASCs transduced with shYAP lentiviruses exhibited no
increase in CTGF, Ankrd1 and YAP mRNA expression when treated with
LPA or S1P (Fig. 6A–C).
Furthermore, the expression levels of PCNA in rASCs were evaluated
following treatment with LPA and S1P for 3 days. The results
demonstrated that LPA and S1P significantly increased PCNA mRNA
expression in rASCs (Fig. 6D).
However, when rASCs were transduced with a shYAP lentivirus, the
effects of LPA and S1P on PCNA expression were abrogated (Fig. 6D).
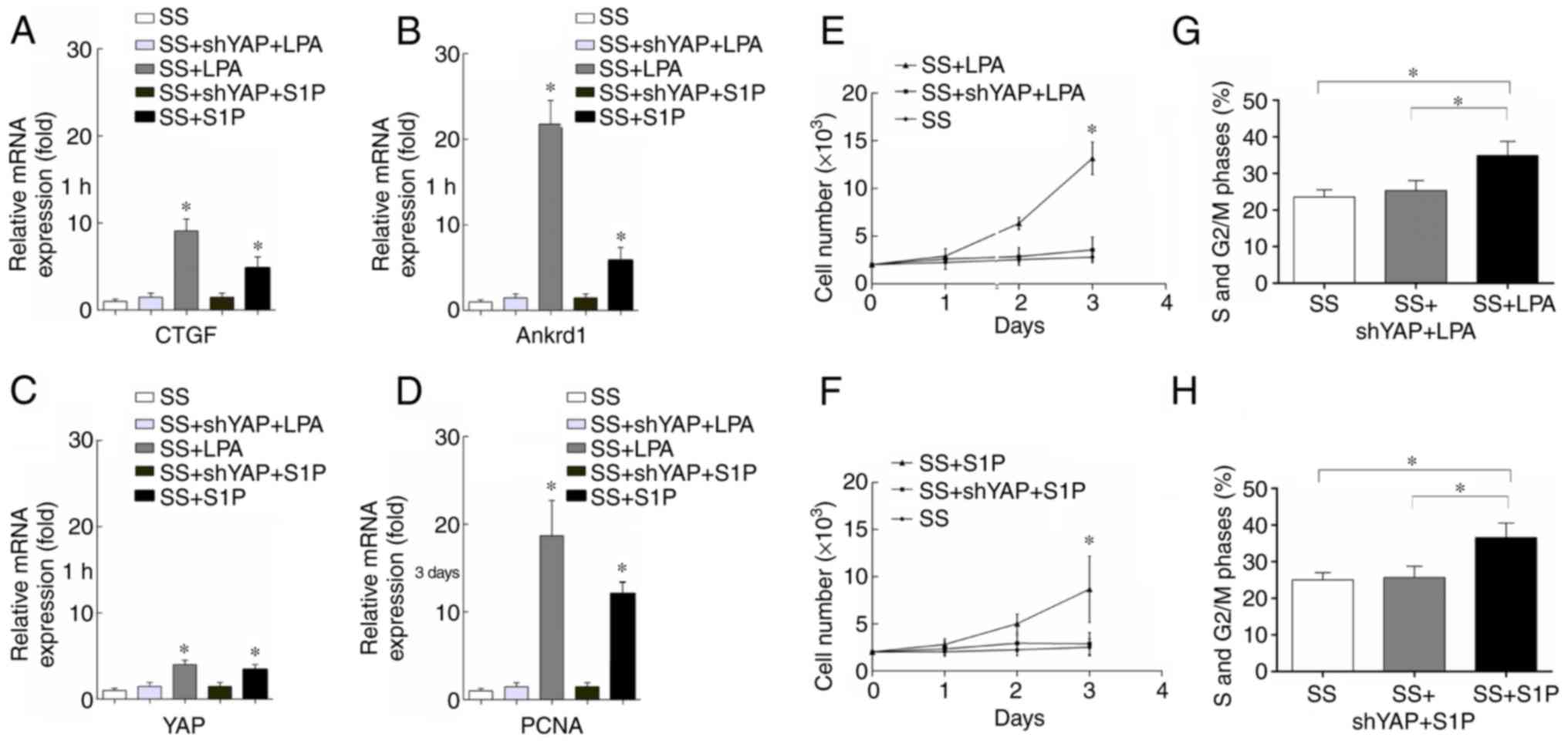 | Figure 6YAP activation promotes the
proliferation of rASCs. (A–C) CTGF, Ankrd1 and YAP mRNA expression
was assessed 1 h after LPA or S1P treatment using RT-qPCR. Data are
presented as the means ± standard deviation, n=3.
*P<0.05, SS + LPA vs. SS + shYAP + LPA or SS + S1P
vs. SS + shYAP + S1P. (D) PCNA expression was determined 3 days
after LPA or S1P treatment using RT-qPCR. Data are presented as the
means ± standard deviation, n=3. *P<0.05, SS + LPA
vs. SS + shYAP + LPA or SS + S1P vs. SS + shYAP + S1P. (E and F)
Cell Counting kit-8 cell proliferation assay. Data are presented as
the means ± standard deviation, n=6. *P<0.05, SS +
LPA or SS + S1P vs. SS. (G and H) Percentage of rASCs in S and
G2/M phases. Data are presented as the means ± standard
deviation, n=3. *P<0.05. Ankrd1, ankyrin repeat
domain 1; CTGF, connective tissue growth factor; LPA,
lysophosphatidic acid; PCNA, proliferating cell nuclear antigen;
rASCs, rat adipose-derived stem cells; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; sh, short
hairpin RNA; SS, serum starvation; YAP, yes-associated protein. |
rASC proliferation was investigated using CCK-8 cell
proliferation assays and flow cytometric analysis. CCK-8 results
revealed that LPA and S1P significantly increased rASC
proliferation compared with in the SS control group 2 and 3 days
after treatment (Fig. 6E and F).
Infection with a shYAP lentivirus significantly inhibited LPA and
S1P-induced cell proliferation (Fig.
6E and F). Cell cycle distribution of rASCs was also
investigated using flow cytometry, and the representative flow
cytometric profile is presented. Flow cytometric analysis revealed
that treating rASCs with LPA and S1P for 3 days significantly
increased the percentage of rASCs in S and G2/M phases
compared with in the SS control group (Fig. 6G and H). However, when rASCs were
infected with a shYAP lentivirus, no significant difference was
observed in the percentage of rASCs in the S and G2/M phases
compared with in the SS control group (Fig. 6G and H). In conclusion, these
results suggested that LPA and S1P may induce rASC proliferation by
activating YAP expression.
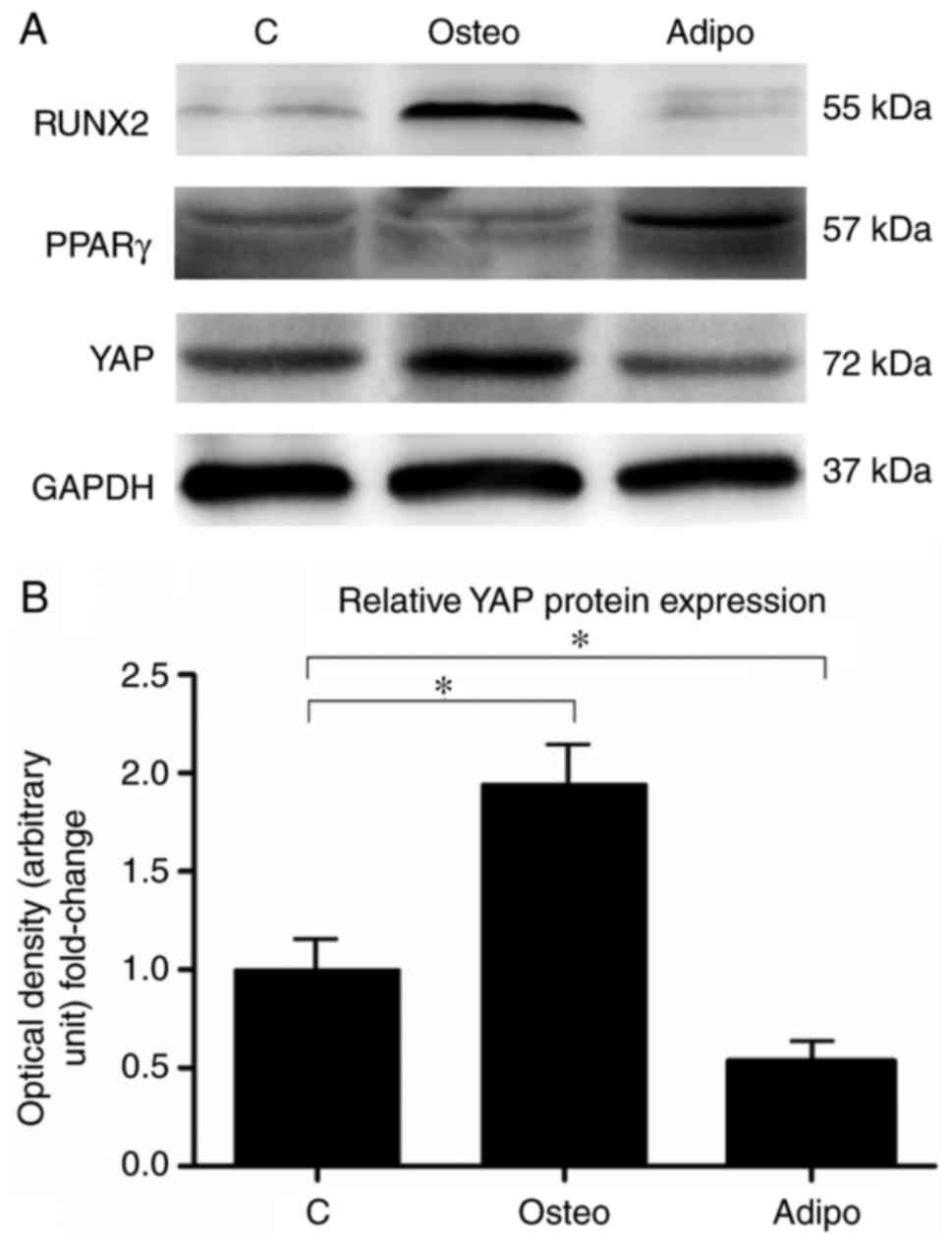
YAP inactivation promotes osteogenesis
and inhibits adipogenesis of rASCs
The protein expression levels of YAP were
investigated 7 days after osteogenic and adipogenic differentiation
of rASCs. The results revealed that YAP protein expression was
increased during osteogenic differentiation of rASCs (Fig. 7A and B). Conversely, the protein
expression of YAP was significantly decreased during adipogenic
differentiation (Fig. 7A and
B).
The present study also investigated the effects of
YAP depletion on osteogenic and adipogenic differentiation of
rASCs. YAP knockdown following transduction with shYAP led to a
marked increase in osteogenic differentiation, as demonstrated by
increased ALP staining in the Osteo + shYAP group compared with in
the Osteo and Osteo + shControl groups 5 days after cell culture
(Fig. 8A). In accordance with
this, the expression levels of the osteogenic
differentiation-related gene RUNX2 were also increased in the Osteo
+ shYAP group compared with in the Osteo and Osteo + shControl
groups 5 days after cell culture (Fig. 8B). The effects of YAP knockdown on
adipogenic differentiation of rASCs were also investigated. The
results suggested that adipogenic differentiation of rASCs was
reduced following YAP inhibition, as adipocyte formation was
markedly decreased 5 days after cell culture in the Adipo + shYAP
group compared with in the Adipo and Adipo + shControl groups
(Fig. 8C). The adipogenic
differentiation-related gene PPARγ was also decreased by YAP
knockdown 5 days after cell culture compared with in the control
groups (Fig. 8D). In conclusion,
these results suggested that YAP may serve an important role in
rASCs osteogenic and adipogenic differentiation.
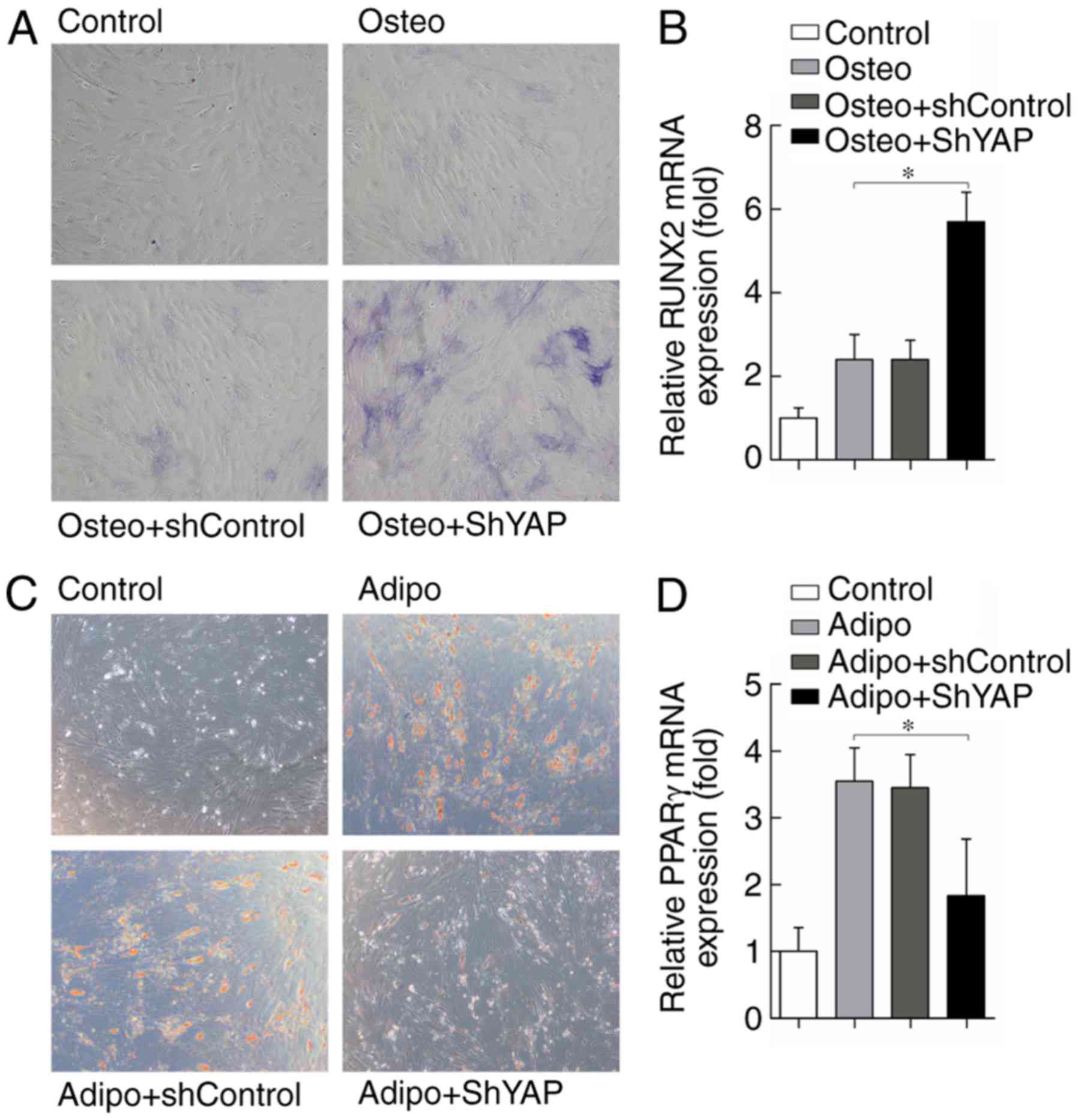 | Figure 8YAP inactivation promotes
osteogenesis and inhibits adipogenesis of rASCs. (A) Alkaline
phosphatase staining of rASCs 5 days after cell culture.
Magnification, ×100. (B) Relative expression of RUNX2 mRNA, as
determined by RT-qPCR 5 days after cell culture. (C) Oil Red O
staining of rASCs 5 days after cell culture. Magnification, ×100.
(D) Relative expression of PPARγ mRNA, as determined by RT-qPCR 5
days after cell culture. Data are presented as the means ± standard
deviation, n=3. *P<0.05. PPARγ, peroxisome
proliferator-activated receptor γ; rASCs, rat adipose-derived stem
cells; RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; RUNX2, Runt-related transcription factor 2; sh, short
hairpin RNA; YAP, yes-associated protein. |
Discussion
MSCs are promising cells in the field of
regenerative medicine. BMSCs and ASCs are two distinct lineages of
MSCs, which are currently being investigated due to their potential
clinical applications (18). ASCs
represent an abundant and easily accessible source of adult stem
cells that can differentiate along numerous lineage pathways
(19,20). ASC-based therapies for the
treatment of musculoskeletal disorders are currently being
developed (21); however, the
factors that mediate ASC proliferation and differentiation remain
poorly understood. Increasing understanding of the molecular
mechanisms governing proliferation and differentiation of ASCs may
reveal their potential clinical applications.
The regulation of YAP subcellular localization has
not been studied extensively. A study by Zhao et al in
epithelial cells revealed that YAP localization and phosphorylation
are dependent on cell density (22). At low cellular densities, YAP is
predominantly localized in the nuclei, translocating to the
cytoplasm when cell density is increased (22). Furthermore, Dupont et al
reported that YAP/TAZ subcellular localization and activity are
regulated by ECM stiffness and cell geometry (23). In addition, the activity of
YAP/TAZ in mammary epithelial cells grown on stiff hydrogels has
been reported to be comparable to that of cells grown on tissue
culture plastic, whereas cells cultured on soft matrices exhibit
decreased YAP/TAZ activity (23).
It has also been reported that inhibition of the actin cytoskeleton
decreases YAP/TAZ nuclear accumulation and transcriptional activity
in mammary epithelial cells (23).
The present study demonstrated that the subcellular
distribution of YAP in rASCs was regulated by cell density and the
actin cytoskeleton. In the low density group, YAP was predominantly
localized to the nucleus, whereas in the high density group, YAP
expression was markedly decreased in rASC nuclei; however, no
significant cytoplasmic localization was detected. These results
are not in agreement with those for epithelial cells (22,23). It may be hypothesized that this
difference is due to the stem-like characteristics of rASCs and
their primordial state, although more thorough investigations are
required to confirm this. Consistent with a previous study
(23), the present study also
indicated that disruption of the actin cytoskeleton with LatB may
induce YAP cytoplasmic translocation. The present results suggested
that the expression of YAP target genes, CTGF and Ankrd1, was
markedly decreased in the high density and LatB-treated groups
compared with in the corresponding control groups.
It has previously been reported that LPA acts
through G12/13-coupled receptors to inhibit LATS kinase in the
Hippo signaling pathway, thereby activating transcriptional
co-activators YAP/TAZ (14). In
the present study, the effects of LPA on YAP protein expression
were detected in rASCs. LPA is a glycerophospholipid-signaling
molecule present in all tissues that binds to receptors, such as
LPA1-6, to initiate intracellular signaling cascades (24). Because serum contains LPA, in the
present study, rASCs were serum starved for 24 h, in order to avoid
the effects of LPA in serum on YAP expression. YAP expression was
decreased in rASCs cultured in DMEM/F12 (SS without LPA), whereas
treatment of rASCs with LatB further decreased YAP expression.
Treatment with LPA (10 µmol/l) significantly increased YAP
expression, whereas LatB treatment partly abolished the effects of
LPA on rASCs. These results suggested that LPA may stimulate YAP
expression in rASCs by modulating the actin cytoskeleton.
Previous studies have revealed that YAP
overexpression increases liver and heart size by increasing cell
number (25–27). YAP overexpression in other
tissues, such as the skin and intestines, results in an enlargement
of the stem cell pool but no overall organ enlargement (28,29). The general conclusion from
previous genetic analyses is that YAP induces cell proliferation
and tissue development. In the present study, the effects of YAP
activation on rASC proliferation were detected. S1P has been
reported to possess overlapping effects with LPA (30). The present results revealed that
LPA and S1P promoted rASC proliferation, potentially by increasing
YAP expression. CTGF and Ankrd1 are both well-characterized YAP
target genes. Treatment with LPA and S1P increased CTGF, Ankrd1 and
YAP gene expression in rASCs. Furthermore, treatment with LPA or
S1P increased the mRNA expression levels of PCNA in rASCs.
Conversely, YAP knockdown significantly abrogated the proliferative
effects of LPA and S1P on rASCs, thus suggesting that LPA and S1P
promote rASC proliferation by activating YAP.
Coordinated proliferation and differentiation of
adult stem cells is important for the regeneration and homeostasis
of adult tissues. Balanced proliferation and differentiation of
muscle satellite stem cells, which express myogenic regulator
factor 5, are critical for the innate regeneration response of
adult skeletal muscles (31). It
has previously been reported that YAP is mainly localized in the
nuclei of mouse myoblasts (32),
and upon differentiation, YAP is translocated to the cytoplasm and
phosphorylated (32). These
findings suggested that YAP localization and activity serve a role
in the regulation of cell differentiation. The osteogenic
differentiation-related master gene RUNX2 has been revealed to
interact with YAP and TAZ (33–35). It has also been demonstrated that
recruitment of TAZ to RUNX2 target genes significantly promotes
osteogenic differentiation of MSCs, whereas TAZ knockdown in MSCs
results in reduced osteogenic differentiation (35). To the best of our knowledge, the
role of YAP in the osteogenic or adipogenic differentiation of
rASCs has yet to be fully elucidated. The present study
investigated the effects of YAP inactivation on the osteogenesis
and adipogenesis of rASCs. Initially, YAP protein expression was
examined during osteogenic and adipogenic differentiation in rASCs;
YAP protein expression was increased during osteogenesis and
decreased during adipogenesis. Furthermore, YAP knockdown increased
osteogenic differentiation and inhibited adipogenic
differentiation. Since Wnt signaling is believed to be a major
signaling pathway that controls MSCs osteogenic differentiation, it
may be hypothesized that YAP blocks osteogenic differentiation
induced by Wnt signaling by binding β-catenin or inducing negative
regulators of Wnt signaling. As such, YAP knockdown may enhance
osteogenic differentiation, while inhibiting adipogenic
differentiation of rASCs. These results suggested that YAP is a
critical regulator of rASC differentiation; however, to the best of
our knowledge, it has only been reported in the present study that
YAP protein expression was increased during osteogenesis and
decreased during adipogenesis, whereas YAP knockdown increased
osteogenic differentiation and inhibited adipogenic
defferentiation, thus further studies are required to confirm the
detailed molecular mechanisms.
In conclusion, the present study demonstrated that
YAP subcellular localization in rASCs was regulated by cell density
and the actin cytoskeleton. Furthermore, it was revealed that YAP
expression in rASCs may be regulated, in part, by LPA and the actin
cytoskeleton. YAP activation was demonstrated to promote rASC
proliferation, whereas YAP knockdown promoted osteogenesis and
inhibited adipogenesis. Therefore, targeted modulation of YAP in
rASCs may be an effective novel strategy to control rASC
proliferation and differentiation for the treatment of various
musculoskeletal system diseases. However, the present study is
potentially limited by the use of only rat-derived ASCs, and
further studies are required using human ASCs, in order to
translate these findings into humans.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81702155
and 81371915).
Availability of data and materials
The data and materials used and/or analyzed during
the current study are available from the corresponding author on
reasonable request.
Authors' contributions
XJ, YY and FG conceived and designed the
experiments. XJ, WY, JW and WZ performed the experiments. WZ and YY
analyzed the data. ZF and GL contributed
reagents/materials/analysis tools and performed the experiments. XJ
and YY wrote the paper. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Experimental
Animal Ethics Committee of Tongji Medical College (Wuhan,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Murphy MB, Moncivais K and Caplan AI:
Mesenchymal stem cells: Environmentally responsive therapeutics for
regenerative medicine. Exp Mol Med. 45:e542013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Davies OG, Cooper PR, Shelton RM, Smith AJ
and Scheven BA: A comparison of the in vitro mineralisation and
dentinogenic potential of mesenchymal stem cells derived from
adipose tissue, bone marrow and dental pulp. J Bone Miner Metab.
33:371–382. 2015. View Article : Google Scholar
|
|
3
|
Ye Y, Du Y, Guo F, Gong C, Yang K and Qin
L: Comparative study of the osteogenic differentiation capacity of
human bone marrow- and human adipose-derived stem cells under
cyclic tensile stretch using quantitative analysis. Int J Mol Med.
30:1327–1334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qi J, Chen A, You H, Li K, Zhang D and Guo
F: Proliferation and chondrogenic differentiation of CD105-positive
enriched rat synovium-derived mesenchymal stem cells in
three-dimensional porous scaffolds. Biomed Mater. 6:0150062011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zuk PA, Zhu M, Ashjian P, De Ugarte DA,
Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P and Hedrick
MH: Human adipose tissue is a source of multipotent stem cells. Mol
Biol Cell. 13:4279–4295. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Ugarte DA, Morizono K, Elbarbary A,
Alfonso Z, Zuk PA, Zhu M, Dragoo JL, Ashjian P, Thomas B, Benhaim
P, et al: Comparison of multi-lineage cells from human adipose
tissue and bone marrow. Cells Tissues Organs. 174:101–109. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lim MH, Ong WK and Sugii S: The current
landscape of adipose-derived stem cells in clinical applications.
Expert Rev Mol Med. 16:e82014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Halder G and Johnson RL: Hippo signaling:
Growth control and beyond. Development. 138:9–22. 2011. View Article : Google Scholar :
|
|
9
|
Lian I, Kim J, Okazawa H, Zhao J, Zhao B,
Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al:
The role of YAP transcription coactivator in regulating stem cell
self-renewal and differentiation. Genes Dev. 24:1106–1118. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu FX, Zhang Y, Park HW, Jewell JL, Chen
Q, Deng Y, Pan D, Taylor SS, Lai ZC and Guan KL: Protein kinase A
activates the Hippo pathway to modulate cell proliferation and
differentiation. Genes Dev. 27:1223–1232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao B, Tumaneng K and Guan KL: The Hippo
pathway in organ size control, tissue regeneration and stem cell
self-renewal. Nat Cell Biol. 13:877–883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong J, Feldmann G, Huang J, Wu S, Zhang
N, Comerford SA, Gayyed MF, Anders RA, Maitra A and Pan D:
Elucidation of a universal size-control mechanism in Drosophila and
mammals. Cell. 130:1120–1133. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao B, Li L, Wang L, Wang CY, Yu J and
Guan KL: Cell detachment activates the Hippo pathway via
cytoskeleton reorganization to induce anoikis. Genes Dev. 26:54–68.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu FX, Zhao B, Panupinthu N, Jewell JL,
Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al:
Regulation of the Hippo-YAP pathway by G-protein-coupled receptor
signaling. Cell. 150:780–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan D: Hippo signaling in organ size
control. Genes Dev. 21:886–897. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye Y, Jing X, Li N, Wu Y, Li B and Xu T:
Icariin promotes proliferation and osteogenic differentiation of
rat adipose-derived stem cells by activating the RhoA-TAZ signaling
pathway. Biomed Pharmacother. 88:384–394. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seo E, Basu-Roy U, Gunaratne PH, Coarfa C,
Lim DS, Basilico C and Mansukhani A: SOX2 regulates YAP1 to
maintain stemness and determine cell fate in the osteoadipo
lineage. Cell Rep. 3:2075–2087. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Izadpanah R, Trygg C, Patel B, Kriedt C,
Dufour J, Gimble JM and Bunnell BA: Biologic properties of
mesenchymal stem cells derived from bone marrow and adipose tissue.
J Cell Biochem. 99:1285–1297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gimble JM, Katz AJ and Bunnell BA:
Adipose-derived stem cells for regenerative medicine. Circ Res.
100:1249–1260. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Ye Y, Tian H, Yang S, Jin X, Tong
W and Zhang Y: In vitro osteogenesis of human adipose-derived stem
cells by coculture with human umbilical vein endothelial cells.
Biochem Biophys Res Commun. 412:143–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Steinert AF, Rackwitz L, Gilbert F, Nöth U
and Tuan RS: Concise review: The clinical application of
mesenchymal stem cells for musculoskeletal regeneration: Gurrent
status and perspectives. Stem Cells Transl Med. 1:237–247. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim
J, Xie J, Ikenoue T, Yu J, Li L, et al: Inactivation of YAP
oncoprotein by the Hippo pathway is involved in cell contact
inhibition and tissue growth control. Genes Dev. 21:2747–2761.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dupont S, Morsut L, Aragona M, Enzo E,
Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M,
Bicciato S, et al: Role of YAP/TAZ in mechanotransduction. Nature.
474:179–183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Choi JW, Herr DR, Noguchi K, Yung YC, Lee
CW, Mutoh T, Lin ME, Teo ST, Park KE, Mosley AN, et al: LPA
receptors: Subtypes and biological actions. Annu Rev Pharmacol
Toxicol. 50:157–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu
Q, Wang Y, Halder G, Finegold MJ, Lee JS and Johnson RL: Hippo
signaling is a potent in vivo growth and tumor suppressor pathway
in the mammalian liver. Proc Natl Acad Sci USA. 107:1437–1442.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee KP, Lee JH, Kim TS, Kim TH, Park HD,
Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, et al: The
Hippo-Salvador pathway restrains hepatic oval cell proliferation,
liver size, and liver tumorigenesis. Proc Natl Acad Sci USA.
107:8248–8253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xin M, Kim Y, Sutherland LB, Qi X,
McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R and Olson EN:
Regulation of insulin-like growth factor signaling by YAP governs
cardiomyocyte proliferation and embryonic heart size. Sci Signal.
4:ra702011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Camargo FD, Gokhale S, Johnnidis JB, Fu D,
Bell GW, Jaenisch R and Brummelkamp TR: YAP1 increases organ size
and expands undifferentiated progenitor cells. Curr Biol.
17:2054–2060. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang H, Pasolli HA and Fuchs E:
Yes-associated protein (YAP) transcriptional coactivator functions
in balancing growth and differentiation in skin. Proc Natl Acad Sci
U S A. 108:2270–2275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rosen H, Gonzalez-Cabrera PJ, Sanna MG and
Brown S: Sphingosine 1-phosphate receptor signaling. Annu Rev
Biochem. 78:743–768. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tedesco FS, Dellavalle A, Diaz-Manera J,
Messina G and Cossu G: Repairing skeletal muscle: Regenerative
potential of skeletal muscle stem cells. J Clin Invest. 120:11–19.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Watt KI, Judson R, Medlow P, Reid K, Kurth
TB, Burniston JG, Ratkevicius A, De Bari C and Wackerhage H: Yap is
a novel regulator of C2C12 myogenesis. Biochem Biophys Res Commun.
393:619–624. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yagi R, Chen LF, Shigesada K, Murakami Y
and Ito Y: A WW domain-containing Yes-Associated Protein (YAP) is a
novel transcriptional co-activator. EMBO J. 18:2551–2562. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui CB, Cooper LF, Yang X, Karsenty G and
Aukhil I: Transcriptional coactivation of bone-specific
transcription factor Cbfa1 by TAZ. Mol Cell Biol. 23:1004–1013.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hong JH, Hwang ES, McManus MT, Amsterdam
A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp
PA, et al: TAZ, a transcriptional modulator of mesenchymal stem
cell differentiation. Science. 309:1074–1078. 2005. View Article : Google Scholar : PubMed/NCBI
|