Introduction
Endometriosis is a benign gynecological disease,
which is characterized by the presence of functional endometrial
glands and stroma outside the uterine cavity. The characteristics
of endometriosis, for example invasion and metastasis ability, are
similar to the malignant tumors, and endometriosis affects up to
20% of women of reproductive age who suffer from dysmenorrheal,
chronic pelvic pain, subfertility and dyspareunia (1). It decreases the life quality of the
affected women, and the clinical prognosis of endometriosis is
still poor due to the high recurrence rate. Present medications
only ameliorate symptoms and cause significant side effects such as
osteoporosis at the same time. Therefore, novel therapeutic agents
that suppress endometriotic lesion establishment and growth to
reduce the high burden conferred by this disease are required
(2). The pathogenesis of
endometriosis is not fully characterized. Sampson's theory of
retrograde menstruation, first described in 1927, remains the most
accepted theory for the origin of this disease (3). According to Sampson's theory,
endometrial stromal cells (ESCs) could escape from the uterine
cavity and adhere to the peritoneal surface to mediate the invasion
and metastasis to the target organs (4).
In the last decade, we found that various cellular
physiological and pathological processes (including cell cycle,
metabolism, cell-cell communication, cell survival and apoptosis,
immune responses and oncogenesis) were induced or promoted due to
dozens of genes targeted by a single miRNA through the perfect or
partial base pairing with the 3′-untranslated region (3′-UTR) of
the target mRNAs (5,6). Additionally, studies suggest that
miRNAs play an important role in endometriosis (7–9).
For example, miR-210 can promote the proliferation, resistance to
apoptosis and VEGF production of endometriosis through STAT3
activation (6). It can be seen
that miRNAs have important roles in pathological processes of
endometriosis, though the mechanisms responsible for endometriosis
remain largely unknown.
miRNAs belong to a large class of small,
single-stranded, non-coding RNAs, molecules approximately 18 to 25
nucleotides in length. They can bind to mRNA, preventing
translation and accelerating mRNA deadenylation and subsequent
degradation, thus having a gene silencing effect (10). In a previous study (ref.
?), miR-363 was found significantly downregulated in
endometriosis tissues than in normal tissues by microRNA chip
technology. We supposed that miR-363 plays a significant role in
endometriosis mediating target genes. However, the mechanism of
miR-363 in endometriosis process remains largely unexplored.
In this study, we sought to determine the expression
pattern of miR-363 targeted mRNAs by sequence-specific base pairing
on the 3′-UTR, and explored their potential functions and pathways,
which are involved in the biological behavior of human
endometriosis. This study may afford new clues for understanding
the pathogenesis of endometriosis, and miR-363 may serve as a
therapeutic option in endometriosis treatment.
Materials and methods
Cell lines and cell culture
The human ESC line was obtained from the American
Type Culture Collection (ATCC; Manassas, VA, USA). Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% (v/v) fetal bovine serum (FBS), 100 U/ml penicillin and
100 U/ml streptomycin (all from Gibco Life Technologies, Carlsbad,
CA, USA) at 37°C in a humidified atmosphere containing 5%
CO2.
Lentiviral transfection of ESC cells
ESC cells were seeded into 6-well plates and
incubated overnight, reached 60–80% confluence before transfection.
Transient transfection of ESC with mimics for hsa-miR-363 or
counterpart negative control used lentiviral vector (LV-3-GFP-PURO;
Shanghai GenePharma Co., Ltd., Shanghai, China) according to the
manufacturer's instructions. The experiments were classified into
two groups, hsa-miR-363 (or miR-363 mimics) group, and negative
control (NC) group. After 24 h post-transfection, the medium was
removed and the fresh culture medium with 10% FBS was injected. The
transfection efficiency of lentiviral vector was observed by
fluorescence microscope at 48 and 72 h after transfection.
Wound migration assay
A density of 1×105 ESC cells/well were
seeded into 6-well plates. When they were approximately 80%
confluent, ESC cells were transfected with hsa-miR-363 or negative
control vector for 24 h. The monolayer was scratched with a sterile
10 µl pipette tip, and rinsed with phosphate-buffered saline
(PBS) to remove cellular debris. Subsequently, fresh medium was
added and cells were incubated for 48 h to allow time for migration
into the cell-free area. The images of the migratory cells were
captured at times 0, 24 and 48 h post-wounding using an Olympus
IX70 microscope equipped with digital camera (Olympus America Inc.,
Melville, Ny, USA). Images were analyzed by the TScratch software
(CSE, Switzerland). Experiments were run in triple independent
repeats.
Gene microarray analysis
Standard gene microarray was performed to analyze
the gene expression characteristics after miRNA-363 overexpression
(11). After ESC cells were
transfected with hsa-miR-363 mimics or counterpart negative control
lentivirus vector for 72 h, respectively, total RNA was isolated
using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA,
USA) according to the manufacturer's instructions. Total RNA was
extracted using Qiagen RNeasy Mini kit and RNase-free DNase I (both
from Qiagen Gmbh, hilden, Germany) according to the manufacturer's
instructions. The purity and concentration of total RNA were
measured by the UV absorbance at 260 and 280 nm (260/280 nm).
Global gene expression was detected using Affymetrix GeneChip human
Gene 1.0 ST array (Affymetrix, Inc., Santa Clara, CA, USA). About
500 ng of total RNA were employed in each experiment. Briefly,
total RNA of each sample was amplified using the Ambion WT
Expression kit (Applied Biosystems, Foster City, CA, USA) according
to the manufacturer's instructions. Approximately 5.5 µg of
generated cDNA was fragmented and labeled using GeneChip3 WT
Terminal Labeling kit and Control kit (both from Affymetrix, Inc.).
Labeled cDNA target was hybridized to the Affymetrix GeneChip human
Gene 1.0 ST array using GeneChip3 hybridization, Wash, and Stain
kit (Affymetrix, Inc.) at 45°C for 16 h, then hybridized arrays
were washed and stained on the Affymetrix GeneChip Command Console
(AGCC; Affymetrix, Inc.), and scanned on a GeneChip2 Scanner 3000
7G (Affymetrix, Inc.). The acquired array images were analyzed by
Affymetrix GeneChip Operating Software (GCOS). Affymetrix
expression console software was used to perform quantile
normalization and subsequent data processing. Data transformation
was applied to set all negative raw values at 1.0, followed by
quantile normalization. A filter on low gene expression was used to
keep only the probes expressed in at least one sample.
Differentially regulated genes were identified via fold change
filtering.
Real-time PCR quantification of miR-363
and mRNAs
Total RNA was isolated from cultured cells using
TRIzol reagent (Invitrogen Life Technologies) according to the
manufacturer's instructions. miR-363 was reverse transcribed to
cDNA by using TaqMan MicroRNA Reverse Transcription kit with
specific probes according to the TaqMan MicroRNA assay protocol
(both from Applied Biosystems), and human snRNA U6 as an endogenous
control. qRT-PCR was performed using a ABI 7900 PCR system (Applied
Biosystems). The 20 µl reaction volume included 100 ng RT
product, 10 µl 2× TaqMan Universal Master mix II and 1
µl primers, and was performed for 2 min at 50°C, 10 min at
95°C, 15 sec at 95°C and 1 min at 60°C for 40 cycles followed by
the thermal denaturation protocol. All reactions were done in
triplicate. The expression of miR-363 was defined based on the
threshold cycle (Ct), and relative expression levels were
calculated as 2−ΔΔCt after normalization with reference
to expression of snRNA U6.
For quantification of mRNA, the purified total RNA
(2 µg) was reverse transcribed using the PrimeScript RT
Master mix (Perfect Real Time) (cat. no. RR036A; Takara Bio, Inc.,
Otsu, Japan), and qPCR was performed on an ABI 7900 PCR system
using Power SyBR-Green PCR Master mix (2×) (both from Applied
Biosystems). GAPDH was used as a normalization control. Primers for
amplification are listed in Table
I. Experiments were performed in triplicates in three
independent experiments. Errors are reported as mean ± SD. The
alteration in mRNA expression was calculated as the fold change
relative to control. Student's t-tests were used for statistical
analyses with two-tailed distributions and two-sample unequal
variance parameters.
 | Table. IPrimers for amplification. |
Table. I
Primers for amplification.
| Target genes | Forward
primers | Reverse
primers |
|---|
| ITGA5 |
AGGACAGATGCCACACAAGG |
CTGCCTGGATTAGGTGCCAA |
| ITGA6 |
GTGGCTATTCTCGCTGGGAT |
AGAGCGTTTAAAGAATCCACACT |
| FHL2 |
GGGGAGACTGGTTGCTGAAA |
AGGGTCTCAAAGCACACCAC |
| PRKCB |
GGGTGTCCTGTCCAGAAAGT |
CTTCCTGAGAGCATGTCCAA |
| SNORD15A |
GATGTTCTCTTTGCCCAGGT |
TTCAACCAGGGCTCTTTAAACT |
| GGT7 |
CCACTCCTGCCTGTCTATGA |
GCCACAATGAAGTCATCAGG |
| SERPINA9 |
AGAACTTCCCATTCCTGGTG |
ACGGCATCTCCCTTGTAATC |
| SERPINB6 |
AGGAGAACACCGAGGAGAGA |
TTCCTTGCCAACATATGGAA |
| ABHD14B |
ATATGACCCTTCGCTTGAGG |
GTGTGGAGGCTCATGTATGC |
| ADAM19 |
CAAGGGCCAACACCTTATTT |
AGGTCGCTTCTTGGTCTGTT |
| GAS7 |
ATCTGGCTTTCTCAGCCACT |
GCCGAATGAGGTAGTTCCAT |
| FAM198A |
CAGATGGACCCAGTGTTCTG |
TCGTCCCTGAAGAGTGTGAG |
| EGR2 |
GAGTTGGGTCTCCAGGTTGT |
CACCGGGTAGATGTTGTCAG |
Gene Ontology (GO) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) analysis
GO function and KEGG enrichment for differentially
expressed genes were used to identify the significantly enriched
biological terms and pathways (11). The Database for Annotation,
Visualization and Integrated Discovery (DAVID) (12) was applied to perform GO function
and KEGG enrichment for differentially expressed genes. Putative
genes that were downregulated more than 2-fold with a P-value
<0.05 after miR-363 overexpression compared to miR-363 NC group
were considered as differentially expressed and selected to submit
to DAVID Bioinformatics Resources 6.7, NIAID/NIH (http://david.abcc.ncifcrf.gov/). The overall
functions regulated by these miR-363 modulated genes were
identified by functional annotation clustering and ranked by
enrichment scores.
Statistical analysis
Each experiment was done at least in triplicate. All
statistical analyses were carried out by two-tailed Student's
t-test. Numerical data were presented as means ± standard deviation
(SD). A P-value <0.05 was considered statistically significant.
Computer-based calculations were conducted using SPSS version 20
(SPSS, Inc., Chicago, IL, USA).
Results
The efficiency of lentiviral transfection
in ESC
To determine the efficiency of lentiviral
transfection and the miR-363 expression level in ESC, fluorescence
microscope and qRT-PCR were used at 96 h after transfection. The
results showed that over 90% of cells expressed green fluorescent
protein (GFP) (Fig. 1A). Compared
to ESC miR-363 NC cell group, the expression levels of miR-363 in
ESC miR-363 mimics cell group were significantly upregulated
3,264.58-fold (Fig. 1B), and in a
lentivirus dose-dependent manner. These results confirmed the
transfection efficiency of lentiviral vector with miR-363 in
ESC.
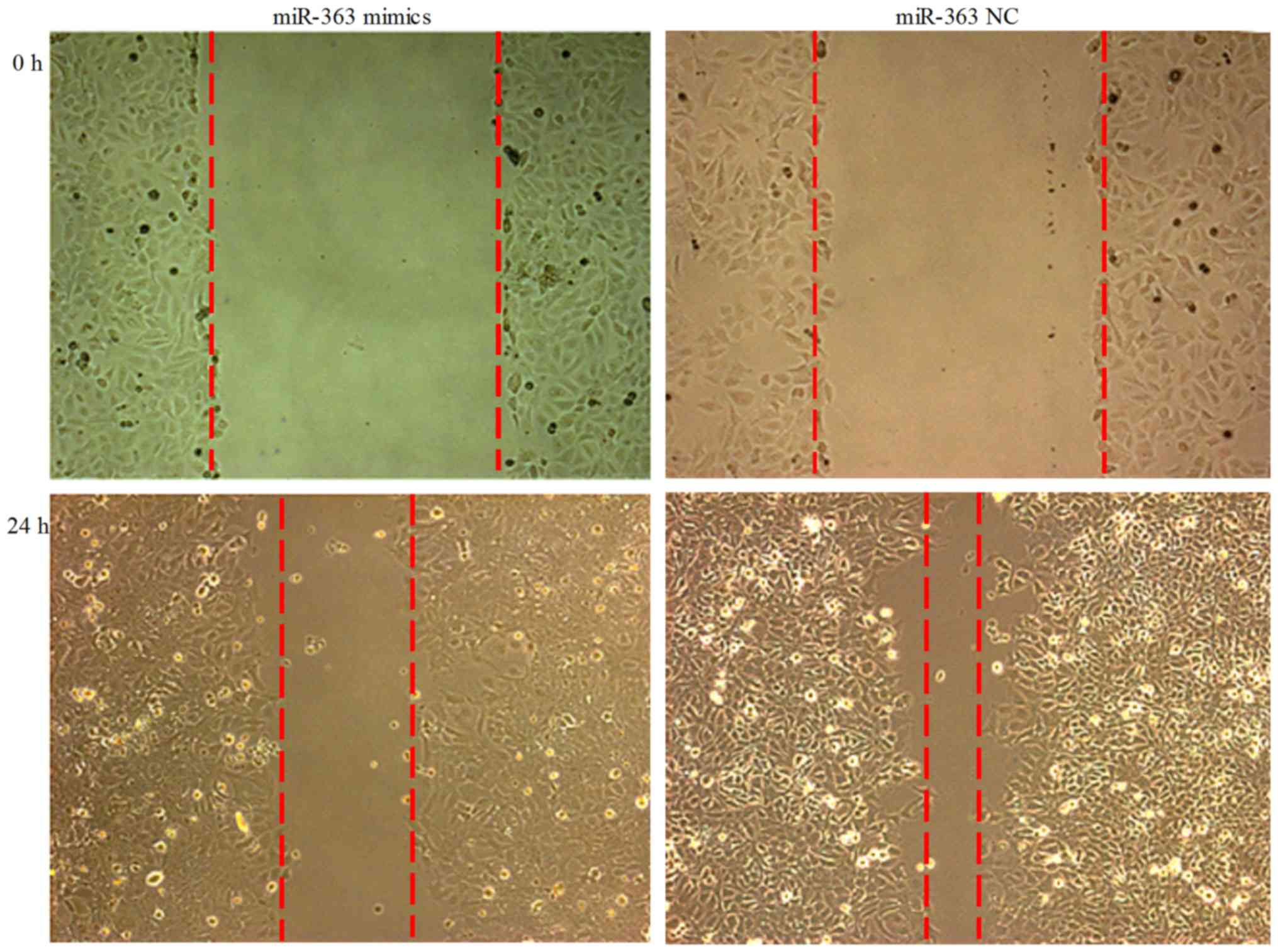
Wound migration assay
ESC cells were transfected with hsa-miR-363 or NC
vector for 24 h. Subsequently, scratch wound migration assay was
performed. There was an observable trend of decrease of the cell
migration potential at 24 h after transfection (Fig. 2).
Differentially expressed genes
In order to explore the potential mechanism of
miR-363 in human breast cancer, gene microarray was carried out to
detect the whole coding gene expression pattern after miR-363
overexpression. The gene expression profiles showed that, compared
to ESC miR-363 NC cell group, 249 genes were upregulated in ESC
miR-363 mimics group, and 139 genes were downregulated (fold change
threshold was ≥2.0) (Fig. 3A).
Although we used a 2-fold change as our threshold in gene
expression, only some of these genes exhibited >2.0-fold change
in gene expression and these differences reached statistical
significance (P<0.05) (Fig.
3B). To assess the validity of the microarray data, real-time
quantitative PCR was performed to compare gene expression levels
between the two cell groups. We selected 10 genes from array
dataset (raw intensity of ESC miR-363 NC was ≥100) and 3 genes
which were not downregulated >2.0-fold in the dataset, but had
great connections to the endometriosis through bioinformatics
analysis of miR-363 to validate the microarray data, all of which
were downregulated in ESC miR-363 mimics cell group. RT-PCR data
was consistent with the results of the microarray, suggesting that
the dataset obtained from the microarray analysis accurately
reflects gene expression differences between ESC miR-363 NC cells
and ESC miR-363 mimics cells (Fig.
3C).
 | Figure 3Differentially expressed genes in
endometrial stromal cells (ESC) miR-363 mimics cells and ESC NC
cells. (A) Heat map shows regulated genes by miR-363 that
significantly increased or decreased after lentiviral transfection.
Dot plot overview of up and downregulated genes by miR-363 after
lentiviral transfection are compared to miR-363 NC group control.
(B) Green lines represent unchanged (middle), 2-fold upregulated
(upper-left) or 2-fold down-regulated (lower right) genes. To
confirm the validity of the microarray data, qRT-PCR was used to
compared gene expression levels between the two cell groups.
Compared to ESC NC cells, 13 genes (ITGA5, ITGA6, FHL2, PRKCB,
SNORD15A, GGT7, SERPINA9, SERPINB6, ABHD14B, ADAM19, GAS7, FAM198A
and EGR2) were significantly downregulated in ESC miR-363 mimics
cell group (calculated as 2−ΔΔCt). (C) The results of
qRT-PCR were consistent with the results of the microarray.
*P<0.05, compared to NC. |
GO and pathways mapping analysis
We performed GO and KEGG analysis to identify the
biological pathways of these differentially expressed genes in the
dataset. GO analysis is a functional analysis associating
differentially expressed genes with GO categories. We employed a GO
approach using DAVID tools as an unbiased method for identifying
the predominant biological process and pathways representing the
target genes. GO analysis result showed that the genes were
enriched in the following biological processes: regulation of cell
death, mitotic cell cycle, regulation of apoptosis, cell cycle,
mitosis, negative regulation of gene expression, regulation of cell
development, regulation of cell motion, regulation of growth, and
regulation of translation (Fig.
4A); the gene production mainly composed of cytoskeleton,
mitochondrion, nucleolus, centrosome, spindle, nuclear body,
transcriptional repressor complex, actin cytoskeleton,
transcription factor complex and adherens junction (Fig. 4B); the molecular functions of
these genes included nucleotide binding, ATP binding,
ribonucleotide binding, DNA binding, nucleoside-triphosphatase
regulator activity, cytoskeletal protein binding, transcription
factor binding, transcription activator activity, actin binding,
and transcription regulator activity (Fig. 4C). Then, we used the KEGG pathway
mapping tools to identify the biological pathways of these genes,
included p53 signaling pathway, pathways in cancer, MAPK signaling
pathway, regulation of actin cytoskeleton, focal adhesion, adherens
junction, RNA polymerase, mTOR signaling pathway, apoptosis, amino
sugar and nucleotide sugar metabolism, Wnt signaling pathway, Notch
signaling pathway and GnRh signaling pathway (Fig. 4D).
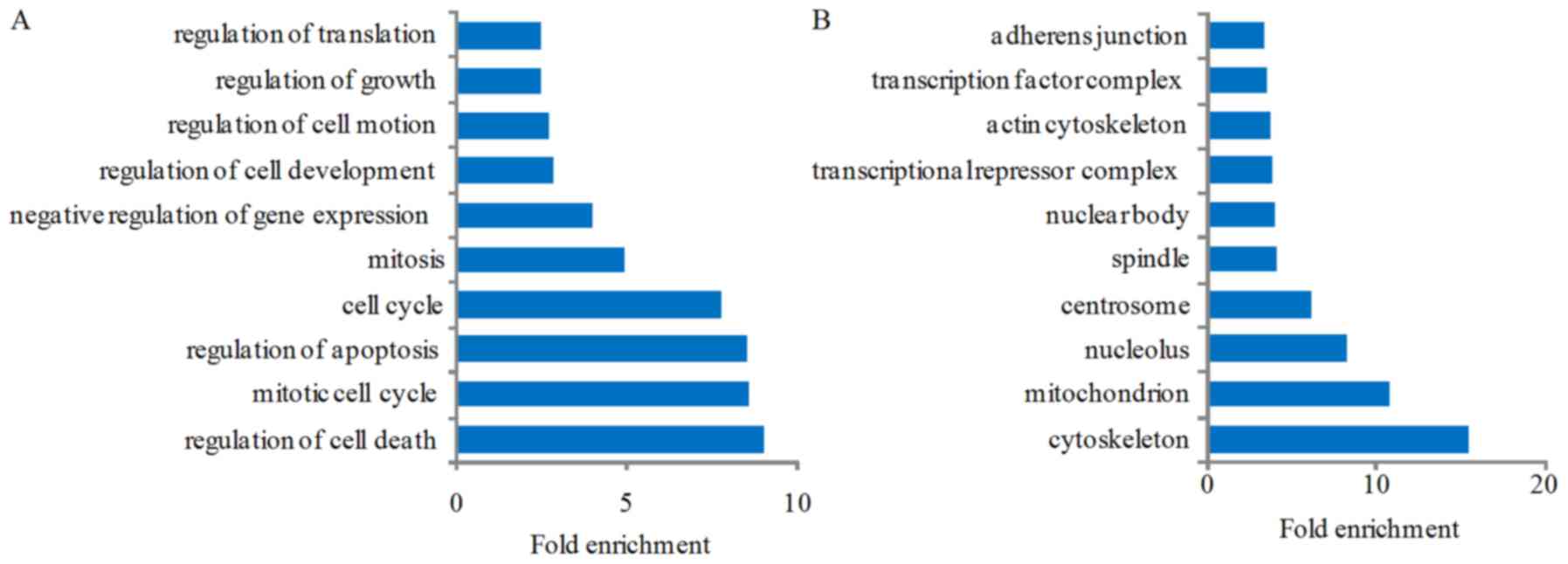 | Figure 4Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analysis of the
differentially expressed genes after miR-363 overexpression. GO
analysis result show that these genes were enriched in the
following biological processes: (A) regulation of cell death,
mitotic cell cycle, regulation of apoptosis, cell cycle, mitosis,
negative regulation of gene expression, regulation of cell
development, regulation of cell motion, regulation of growth, and
regulation of translation; (B) these genes' production mainly
composed into cytoskeleton, mitochondrion, nucleolus, centrosome,
spindle, nuclear body, transcriptional repressor complex, actin
cytoskeleton, transcription factor complex and adherens junction;
(C) the molecular functions of these genes included nucleotide
binding, ATP binding, ribonucleotide binding, DNA binding,
nucleoside-triphosphatase regulator activity, cytoskeletal protein
binding, transcription factor binding, transcription activator
activity, actin binding, and transcription regulator activity; (D)
the biological pathways of these genes, included p53 signaling
pathway, pathways in cancer, MAPK signaling pathway, regulation of
actin cytoskeleton, focal adhesion, adherens junction, RNA
polymerase, mTOR signaling pathway, apoptosis, amino sugar and
nucleotide sugar metabolism, Wnt signaling pathway, Notch signaling
pathway, and GnRH signaling pathway. |
Discussion
The invasion and metastasis characteristics play
important roles in endometriosis. Several mechanisms have been
proved to connect to the invasion and metastasis characteristics
and the alterations to miRNA expression levels is a new point in
endometriosis. In our study, the endometriosis cell line ESC was
widely used as an in vitro model for study of endometriosis.
The invasion and metastasis ability of ESC was mainly adjusted by
its own and local microenvironment, including multiple hormones,
cytokines, growth factors, extracellular matrix glycoprotein and
various kinds of transcription factors, and involving a large
amount of signaling pathways, such as MAPK, PI3K/Akt signaling
pathways (13–16).
miR-363 has been found to participate in regulating
cancer biological progress. Several studies have shown that miR-363
can regulate the invasion and metastasis of tumors (17,18). Whereas, there is no study
exploring the function of miR-363 in endometriosis. In our GO and
KEGG analysis, we found that the target genes of miR-363
participated in many significant biological processes, cellular
components, molecular functions, and signaling pathways. All of the
above were involved in migration, cell adhesion and invasion,
proliferation, apoptosis, alteration of endometrial cells.
The target gene ITGA5 is a member of the cell
adhesion molecule family that acts on both cell-cell and
cell-substratum adhesion. They can promote cell attachment to
proteins within ECM and enhance cell migration and invasion
(1). Vernet-Tomás et al
found integrins could be mediated endometrial cells and peritoneal
mesothelial cell adhesion, causing pelvic adhesion extensively
(19). According to Giannelli
et al, this family can also activate the ERK and JNK
signaling pathway in MAPK signaling pathway, promote the adhesion
and motion of endometrial cells (20).
In the last few years, aberrant activation of MAPK
was found to play important roles in pathological processes of
endometriosis progression (21).
Such as, aberrant activation of ERK was shown to be involved in
interleukin (IL)-1β and macrophage migration inhibitory
factor-induced cyclooxygenase-2 expression, prostaglandin
E2 (PGE2)-induced fibroblast growth factor 9
(FGF9) expression (21–25). In addition, activation of Akt
independently can regulate cell invasion, metastasis, angiogenesis,
and survival (26,27). Recently, associated studies
reported PI3K/AKT signaling pathway was activated in endometriosis.
Especially, p-AKT was highly expressed in endometriotic stromal
cells, leading to the reduced decidualization and cell survival
(26,28–30).
The mechanism of regulating cellular proliferation
in endometriosis is considered to be generally the same as normal
endometrium, as a result of the interaction between gender steroids
and their corresponding receptors. As an immunohistochemical marker
of cell proliferation, the expression of proliferating cell nuclear
antigen in endometriotic lesions is much higher than eutopic and
normal endometrium, indicating a higher proliferative activity. In
addition, the cyclical changes in cellular proliferation observed
in eutopic and normal endometrium are not discovered in
endometriotic lesions (1).
Apoptosis is a complex process, regulated by several
multifunctional mediators. As a programmed cell death it kills
unwanted cells without inducing an immune response or inflammation
due to these multifunctional mediators. In normal human
endometrium, apoptosis has been reported to appear in the mid
secretory phase, increasing in the late secretory phase, and is
maximal during the menstrual phase. Because of programmed cell
death, endometrial cells from healthy women during menstruation do
not survive in ectopic locations (1,31–33).
It has been demonstrated that apoptotic events
seemed to be decreased in endometriotic lesions compared with
corresponding eutopic and normal endometrium, on the basis of
endometriosis. Impaired sensitivity of the endometrial tissues to
spontaneous apoptosis leads to abnormal implantation and growth of
endometrial tissues in ectopic places (1). It has been suggested that the
concordant endometrial expression of many genes that trigger the
activation of apoptotic and cell-cycle signaling were identified.
The activation of antiapoptotic genes and the inhibition of
proapoptotic ones may contribute to the resistance of endometriotic
cells to apoptosis (1,31–34).
Many miRNAs have been predicted to regulate the
expression of genes with functions in apoptosis and cell cycle
regulation of several cancer cells. However, limited evidence
exists to support the regulatory functions of miRNAs in
proliferative response of normal, differentiated quiescent cells
(31,35–37). Several studies have suggested that
remodeling of the actin cytoskeleton is required for the cell
adhesion and movement. Via the restructuring of the cytoskeleton,
the cyclic architectural modifications observed in the endometrium
are achieved. It is confirmed that cytoskeletal protein is related
to the endometriotic cell proliferation, adhesion, invasion and
apoptosis (1).
Xu et al found that cytoskeletal protein
confilin 1 (CFL1) was overexpressed in endometriotic lesions and
eutopic endometrium with endometriosis. Silencing the CFL1
gene in endometriotic cells can attenuate their proliferation,
adhesion and invasion. On the contrary, once the CFL1 gene
is upregulated, the cell proliferation, adhesion and invasion
increased (1,38). As endometriosis is considered to
be a multigenic disease with complex genetic traits, differences in
hormonal regulation of the endometria is inferred as a result of an
abnormal gene expression pattern and may be responsible for
adhesion and invasion properties of the shed endometrial fragments
(1). Absenger et al
confirmed a highly regulated cysteine-rich 61 (Cyr61) gene
in endometrium from women with endometriosis, from the results of
microarray technology (39).
MicroRNAs are small non-coding RNAs of 18–25
nucleotides, which post-transcriptionally regulate gene expression
and can control a broad spectrum of normal and pathological
cellular functions via binding to their target mRNAs. Numerous
diseases, including benign and malignant gynecological disorders,
are associated with abnormal miRNA expression. Recently,
researchers identified that miRNAs relevant to molecular pathways
were associated with pathogenesis of endometriosis via conducting
the miRNA expression profiles of endometrium from women with
endometriosis (9,8,40).
In conclusion, we found that miR-363 was associated
with migration, cell adhesion and invasion, proliferation,
apoptosis, alteration of endometrial cells in endometriosis and
there were 139 potential target genes of miR-363 via microarray
analysis. Through GO and KEGG pathways analysis, we also identified
the biological processes and signaling pathways involved in
endometriosis of ESC miR-363 mimics cell line. These results
provided us a systematic understanding and global view of the
function of overexpression miR-363 related to the process of
endometriosis. Future investigation should be targeted to the
biological processes and signaling pathways that have been
identified. miR-363 may be a novel biomarker for the endometriosis
prediction and prognosis, while more efforts should be done to
identify the mechanism of miR-363 regulation of endometriosis.
Funding
This study was financially supported by the National
Natural Science Foundation of China (nos. 81401182, 81202007 and
81302304) and the Science and Technology Development Foundation of
Nanjing Medical University (no. 2012NJMU185).
Availability of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WL performed gene microarray analysis, and was a
major contributor in writing the manuscript. XF performed the ESC
cell culture and lentiviral transfection. MZ performed the wound
migration assay and real-time PCR. LH participated in gene
microarray analysis and the manuscript modification. SL
participated in gene microarray analysis and the manuscript
modification. LW performed the related literature collection. YW
analyzed the experiment data. CD participated in the ESC cell
culture and lentiviral transfection. JX participated in the
manuscript modification and the experiment data analysis. PX and ZF
performed gene ontology and pathways mapping analysis. XJ performed
the management of the experiment and foundation. XS performed
personnel assignment and the manuscript modification. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
Authors declare they have no competing interest.
Acknowledgments
Not applicable.
References
|
1
|
Jiang QY and Wu RJ: Growth mechanisms of
endometriotic cells in implanted places: a review. Gynecol
Endocrinol. 28:562–567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gao X, Outley J, Botteman M, Spalding J,
Simon JA and Pashos CL: Economic burden of endometriosis. Fertil
Steril. 86:1561–1572. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sampson J: Peritoneal endometriosis due to
menstrual dissemination of endometrial tissue into the peritoneal
cavity. Am J Obstet Gynecol. 14:422–469. 1927. View Article : Google Scholar
|
|
4
|
Nair AS, Nair HB, Lucidi RS, Kirchner AJ,
Schenken RS, Tekmal RR and Witz CA: Modeling the early
endometriotic lesion: mesothelium-endometrial cell co-culture
increases endometrial invasion and alters mesothelial and
endometrial gene transcription. Fertil Steril. 90(Suppl 4):
1487–1495. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Okamoto M, Nasu K, Abe W, Aoyagi Y, Kawano
Y, Kai K, Moriyama M and Narahara H: Enhanced miR-210 expression
promotes the pathogenesis of endometriosis through activation of
signal transducer and activator of transcription 3. Hum Reprod.
30:632–641. 2015. View Article : Google Scholar
|
|
7
|
Teague EM, Print CG and Hull ML: The role
of microRNAs in endometriosis and associated reproductive
conditions. Hum Reprod Update. 16:142–165. 2010. View Article : Google Scholar
|
|
8
|
Ohlsson Teague EM, Van der Hoek KH, Van
der hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG and
Hull LM: MicroRNA-regulated pathways associated with endometriosis.
Mol Endocrinol. 23:265–275. 2009. View Article : Google Scholar
|
|
9
|
Pan Q, Luo X, Toloubeydokhti T and Chegini
N: The expression profile of micro-RNA in endometrium and
endometriosis and the influence of ovarian steroids on their
expression. Mol Hum Reprod. 13:797–806. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lv J, Fu Z, Shi M, Xia K, Ji C, Xu P, Lv
M, Pan B, Dai L and Xie H: Systematic analysis of gene expression
pattern in has-miR-760 overexpressed resistance of the MCF-7 human
breast cancer cell to doxorubicin. Biomed Pharmacother. 69:162–169.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
13
|
Yu J, Wang Y, Zhou WH, Wang L, He YY and
Li DJ: Combination of estrogen and dioxin is involved in the
pathogenesis of endometriosis by promoting chemokine secretion and
invasion of endometrial stromal cells. Hum Reprod. 23:1614–1626.
2008. View Article : Google Scholar
|
|
14
|
De La Garza EM, Binkley PA, Ganapathy M,
Krishnegowda NK, Tekmal RR, Schenken RS and Kirma NB: Raf-1, a
potential therapeutic target, mediates early steps in endometriosis
lesion development by endometrial epithelial and stromal cells.
Endocrinology. 153:3911–3921. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li MQ, Li HP, Meng YH, Wang XQ, Zhu XY,
Mei J and Li DJ: Chemokine CCL2 enhances survival and invasiveness
of endometrial stromal cells in an autocrine manner by activating
Akt and MAPK/Erk1/2 signal pathway. Fertil Steril. 97:919–929.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tapia-Pizarro A, Argandoña F, Palomino WA
and Devoto L: Human chorionic gonadotropin (HCG) modulation of
TIMP1 secretion by human endometrial stromal cells facilitates
extravillous trophoblast invasion in vitro. Hum Reprod.
28:2215–2227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun Q, Zhang J, Cao W, Wang X, Xu Q, Yan
M, Wu X and Chen W: Dysregulated miR-363 affects head and neck
cancer invasion and metastasis by targeting podoplanin. Int J
Biochem Cell Biol. 45:513–520. 2013. View Article : Google Scholar
|
|
18
|
Qiao J, Lee S, Paul P, Theiss L, Tiao J,
Qiao L, Kong A and Chung DH: miR-335 and miR-363 regulation of
neuroblastoma tumorigenesis and metastasis. Surgery. 154:226–233.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vernet-Tomás MM, Pérez-Ares CT, Verdú N,
Fernández-Figueras MT, Molinero JL and Carreras R: The depolarized
expression of the alpha-6 integrin subunit in the endometria of
women with endometriosis. J Soc Gynecol Investig. 13:292–296. 2006.
View Article : Google Scholar
|
|
20
|
Giannelli G, Sgarra C, Di Naro E, Lavopa
C, Angelotti U, Tartagni M, Simone O, Trerotoli P, Antonaci S and
Loverro G: Endometriosis is characterized by an impaired
localization of laminin-5 and alpha3beta1 integrin receptor. Int J
Gynecol Cancer. 17:242–247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin SC, Wang CC, Wu MH, Yang SH, Li YH and
Tsai SJ: Hypoxia-induced microRNA-20a expression increases ERK
phosphorylation and angiogenic gene expression in endometriotic
stromal cells. J Clin Endocrinol Metab. 97:E1515–E1523. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu MH, Wang CA, Lin CC, Chen LC, Chang WC
and Tsai SJ: Distinct regulation of cyclooxygenase-2 by
interleukin-1beta in normal and endometriotic stromal cells. J Clin
Endocrinol Metab. 90:286–295. 2005. View Article : Google Scholar
|
|
23
|
Carli C, Metz CN, Al-Abed Y, Naccache PH
and Akoum A: Up-regulation of cyclooxygenase-2 expression and
prostaglandin E2 production in human endometriotic cells
by macrophage migration inhibitory factor: involvement of novel
kinase signaling pathways. Endocrinology. 150:3128–3137. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tamura M, Sebastian S, yang S, Gurates B,
Fang Z and Bulun SE: Interleukin-1beta elevates cyclooxygenase-2
protein level and enzyme activity via increasing its mRNA stability
in human endometrial stromal cells: an effect mediated by
extracellularly regulated kinases 1 and 2. J Clin Endocrinol Metab.
87:3263–3273. 2002.PubMed/NCBI
|
|
25
|
Chuang PC, Sun HS, Chen TM and Tsai SJ:
Prostaglandin E2 induces fibroblast growth factor 9 via
EP3-dependent protein kinase Cdelta and Elk-1 signaling. Mol Cell
Biol. 26:8281–8292. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu X, Zheng Q, Zhang Z, Zhang X, Liu R and
Liu P: Periostin enhances migration, invasion, and adhesion of
human endometrial stromal cells through integrin-linked kinase
1/Akt signaling pathway. Reprod Sci. 22:1098–1106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Zhang H, Wu J, Guan H, Yuan J, Huang
Z and Li M: Prognostic significance of integrin-linked kinase1
overexpression in astrocytoma. Int J Cancer. 126:1436–1444.
2010.
|
|
28
|
Yin X, Pavone ME, Lu Z, Wei J and Kim JJ:
Increased activation of the PI3K/AKT pathway compromises
decidualization of stromal cells from endometriosis. J Clin
Endocrinol Metab. 97:E35–E43. 2012. View Article : Google Scholar :
|
|
29
|
Samartzis EP, Noske A, Dedes KJ, Fink D
and Imesch P: ARID1A mutations and PI3K/AKT pathway alterations in
endometriosis and endometriosis-associated ovarian carcinomas. Int
J Mol Sci. 14:18824–18849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Franco-Murillo Y, Miranda-Rodríguez JA,
Rendón-Huerta E, Montaño LF, Cornejo GV, Gómez LP, Valdez-Morales
FJ, Gonzalez-Sanchez I and Cerbón M: Unremitting cell proliferation
in the secretory phase of eutopic endometriosis: involvement of
pAkt and pGSK3β. Reprod Sci. 22:502–510. 2015. View Article : Google Scholar
|
|
31
|
Pan Q and Chegini N: MicroRNA signature
and regulatory functions in the endometrium during normal and
disease states. Semin Reprod Med. 26:479–493. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Harada T, Taniguchi F, Izawa M, Ohama Y,
Takenaka Y, Tagashira Y, Ikeda A, Watanabe A, Iwabe T and Terakawa
N: Apoptosis and endometriosis. Front Biosci. 12:3140–3151. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brosens JJ and Gellersen B: Death or
survival - progesterone-dependent cell fate decisions in the human
endometrial stroma. J Mol Endocrinol. 36:389–398. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kao LC, Tulac S, Lobo S, Imani B, Yang JP,
Germeyer A, Osteen K, Taylor RN, Lessey BA and Giudice LC: Global
gene profiling in human endometrium during the window of
implantation. Endocrinology. 143:2119–2138. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Matsubara H, Takeuchi T, Nishikawa E,
Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M,
Nimura Y, et al: Apoptosis induction by antisense oligonucleotides
against miR-17-5p and miR-20a in lung cancers overexpressing
miR-17-92. Oncogene. 26:6099–6105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mertens-Talcott SU, Chintharlapalli S, Li
X and Safe S: The oncogenic microRNA-27a targets genes that
regulate specificity protein transcription factors and the G2-M
checkpoint in MDA-MB-231 breast cancer cells. Cancer Res.
67:11001–11011. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu YL, Wang DB, Liu QF, Chen YH and Yang
Z: Silencing of cofilin-1 gene attenuates biological behaviours of
stromal cells derived from eutopic endometria of women with
endometriosis. Hum Reprod. 25:2480–2488. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Absenger Y, Hess-Stumpp H, Kreft B,
Krätzschmar J, Haendler B, Schütze N, Regidor PA and Winterhager E:
Cyr61, a deregulated gene in endometriosis. Mol Hum Reprod.
10:399–407. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hull ML, Escareno CR, Godsland JM, Doig
JR, Johnson CM, Phillips SC, Smith SK, Tavaré S, Print CG and
Charnock-Jones DS: Endometrial-peritoneal interactions during
endometriotic lesion establishment. Am J Pathol. 173:700–715. 2008.
View Article : Google Scholar : PubMed/NCBI
|