Introduction
MicroRNAs (miRNAs/miRs) are a class of RNAs
measuring <25 bp in length that do not have protein-coding
functions (1). Previous studies
have demonstrated that miRNAs may regulate cell proliferation,
invasion, immune response and metabolism in physiological and
pathophysiological processes by binding to the 3′ untranslated
region (3′-UTR) of targeting mRNAs to decrease their expression
(2).
Hepatocellular carcinoma (HCC) is a serious health
burden worldwide, particularly in Asia and Africa (3). Although advanced therapeutic methods
have been implemented in previous years, clinical data indicate
that patients suffering with metastasis usually have poor prognoses
(4). Therefore, the control of
tumor metastasis is an important strategy to improve the prognosis
of HCC. At present, there is increasing evidence that miRNAs serve
critical roles in HCC metastasis (5,6).
For example, the downregulation of miR-1271 in HCC tissues was
associated with venous infiltration, and ectopic expression of
miR-1271 may inhibit invasion and lung metastatic nodules formation
(7).
miR-552, a novel cancer-associated miRNA, has been
studied primarily in colorectal cancer (CRC) (8). Functionally, miR-552 was
demonstrated to increase CRC cell migration and invasion abilities
(9). However, the functions of
miR-552 in the metastasis of HCC and detailed mechanisms associated
with epithelial-mesenchymal transition (EMT), which is an important
mechanism for cell migration and invasion (10), remains unclear.
The present study identified that miR-552 was
upregulated in HCC. High expression of miR-552 predicted poor
overall survival and disease-free survival in a 3-year follow-up
period. In vitro, miR-552 was demonstrated to increase
migration, invasion and EMT by targeting Wnt inhibitory factor 1
(WIF1). Additionally, glycogen synthase kinase 3β (GSK3β)/β-catenin
signaling activation was confirmed as an important mechanism for
the oncogenic functions of miR-552.
Materials and methods
Ethical review
The use of clinical samples was approved by the
Ethics Committee of First Affiliated Hospital of Xi’an Jiaotong
University (Xi’an, China) according to the 1975 Declaration of
Helsinki. Written informed consent was obtained and signed by each
patient enrolled in the present study.
Clinical samples and cell lines
A total of 76 HCC tissues and matched tumor-adjacent
tissues were collected from 53 male patients and 23 female patients
who received surgery from the Department of Hepatobiliary Surgery,
First Affiliated Hospital of Xi’an Jiaotong University between
January 2010 and January 2014. Tissues were stored in liquid
nitrogen. Human immortalized normal hepatocyte cell line (LO2) and
HCC cell lines (Huh-7, MHCC97-L, Hep3B, MHCC97-H) were purchased
from the Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences (Shanghai, China) and stored at the Centre for
Translational Medicine at the First Affiliated Hospital of Xi’an
Jiaotong University. Cells were cultured with Dulbecco’s modified
Eagle’s medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 1% V/V
penicillin/streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) in a humidified atmosphere containing 5% CO2 at
37°C.
Cell transfection
miR-552 mimics (miR10003215-1-5; miR-552), miR-552
inhibitor (miR20003215-1-5; miR-552-inhi), negative control mimics
(miR01201-1-5; miR-ctrl), negative control inhibitor (miR02201-1-5;
miR-ctrl-inhi), WIF1 small interfering (si)RNA (stB0009885;
si-WIF1) and negative control siRNA (siN05815122147-1-5; si-ctrl)
were purchased from RiboBio Co. (Guangzhou, China). Negative
control vectors (Vec-ctrl) and WIF1 plasmids (SC119176) were
obtained from OriGene (Rockville, MD, USA). A total of 10 μg
mimics, inhibitors, siRNAs or plasmids were used to transfect HCC
cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer’s protocol. After
48 h transfection, cells were harvested for further
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to isolate total RNA from tissues
and cultured cells. RT-PCR and qPCR were performed using the
Superscript III Reverse Transcriptase kit (Invitrogen; Thermo
Fisher Scientific, Inc.) and iTaq Universal SYBR-Green Supermix kit
(Bio-Rad Laboratories, Inc., Hercules, CA, USA), respectively. The
Bulge-Loop miR-552 RT-qPCR Primer Set (Guangzhou RiboBio, Co.,
Ltd., Guangzhou, China) was applied to measure the miR-552
expression. The primer sequences were as follows: WIF1 forward, 5′-
GTGTGAAATCAGCAAATGCC-3′ and reverse, 5′- GTCTTCCATGCCAACCTTCT-3′;
and GAPDH forward, 5′-CCAGGGCTGCTTTTAACTCT-3′ and reverse,
5′-GGACTCCACGACGTACTCA-3′). The thermocycling conditions were as
follows: Initial denaturation at 50°C for 2 min and enzyme
denaturation at 95°C for 10 min, followed by 40 cycles:
Denaturation at 95°C for 15 sec, annealing at 60°C for 30 sec,
elongation at 70°C for 1 min, and a final extension at 72°C for 10
min. The 2-ΔΔCq method (11) was used to calculate the relative
gene expression normalized by GAPDH.
Western blot analysis
Cells were lysed with radioimmunoprecipitation assay
lysis buffer (Xi’an Hut Biotechnology Co., Ltd., Xian, China) and
quantified with a BCA protein assay kit II (cat. no. 5000002;
Bio-Rad Laboratories, Inc.). Protein samples (40 μg) were
separated by 10% SDS-PAGE gel and transferred onto a nitrocellulose
membrane (Invitrogen; Thermo Fisher Scientific, Inc.) using Bio-Rad
tank blotting system (Bio-Rad Laboratories, Inc.). Membranes were
blocked with 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA)
with 1X TBST (TBS with 0.1% Tween-20) for 2 h at room temperature.
The membranes were incubated with the appropriate primary
antibodies for a 1:1,000 dilution overnight at 4°C. Horseradish
peroxidase (HRP)-conjugated secondary antibodies at a 1:2,000
dilution were used to incubate the membranes for 1 h at room
temperature subsequent to washing them with TBST (TBS with 0.1%
Tween-20) 3 times for 10 min. The targeting proteins on the
membrane were visualized with enhanced chemiluminescent (ECL)
reagents (EMD Millipore; Billerica, MA, USA). Membranes incubated
with ECL reagents were wrapped in plastic wrap box and exposed to
x-ray film. Exposed x-ray films were then scanned to a
black-and-white figure by a scanner (LiDE 220, Canon, Inc., Tokyo,
Japan). The intensity of every protein band was calculated using
ImageJ software [V1.8.0_112; National Institutes of Health (NIH),
Bethesda, MD, USA]. The corresponding GAPDH intensity was used to
calculate the relative intensity of the bands.
The primary antibodies used for the western blot
analysis were as follows: WIF1 (cat. no. ab155101) was purchased
from Abcam (Cambridge, MA, USA); epithelial cadherin (E-cadherin;
cat. no. 3195), vimentin (cat. no. 3932), GSK3β (cat. no. 12456)
and phosphorylated GSK3β (cat. no. 5558) antibodies were purchased
from Cell Signaling Technology, Inc. (Danvers, MA, USA); and matrix
metalloproteinase (MMP)-2 (cat. no. sc-13594) and GAPDH (cat. no.
sc-47724) antibodies were purchased from Santa Cruz Biotechnology,
Inc. (Dallas, Texas, USA).
Immunohistochemical staining
Paraformaldehyde-fixed paraffin-embedded HCC tissue
sections were prepared by Xi’an Hut Biotechnology Co., Ltd. and
used for immunohistochemical staining. WIF1 primary antibodies were
diluted in PBS at 1:100 to label the WIF1 at 4°C overnight.
Biotinylated goat anti-rabbit secondary antibodies (cat. no.
SP-9001; ZSGB-BIO; OriGene Technologies, Inc., Beijing, China) were
used to detect the primary antibodies. Complexes were detected by
HRP-streptavidin conjugates (ZSGB-BIO; OriGene Technologies, Inc.)
and visualized with DAB (ZSGB-BIO; OriGene Technologies, Inc.). The
final immunohistochemical staining scores were calculated as
described previously (12).
Wound healing assay
Following transfection, cells were cultured in
6-well plates and grown to 80-90% confluence. A wound across the
middle of the well was created by a 200 μl sterile pipette
tip. Subsequently, cells were cultured with serum-reduced (2% FBS
v/v) DMEM in a humidified 5% CO2 incubator at 37°C for 0
and 48 h. Images were captured with a phase-contrast microscope at
a magnification of ×40 and analyzed by Image J software
(V1.8.0_112; NIH).
Transwell assay
Transwell inserts (Nalge Nunc, Penfield, NY, USA)
were coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA)
at 1 mg/ml on the upper layer and incubated at 37°C for 30 min.
Briefly, 3×104 cells were seeded into the upper chamber
with serum-reduced (2% FBS v/v) DMEM. A total of 750 μl DMEM
containing 10% FBS was added into the lower chamber. Cells were
then incubated in a humidified 5% CO2 incubator at 37°C
for 24 h. Chambers were fixed in 4% paraformaldehyde for 5 min at
room temperature and then stained with 0.3% crystal violet dye for
10 min. Non-invading cells were removed by a cotton swab. Invaded
cells were counted under a light microscope at a magnification of
×200.
Dual-luciferase reporter assay
TargetScan (http://www.targetscan.org/) and MiRanda (http://www.microrna.org) were used to predict the
target of miR-552. The predicted wild-type WIF1 3′-UTR sequence
corresponding with miR-552 and the mutant 3′-UTR sequence were
synthesized and inserted into the pEZX-MT06 plasmid vector
(GeneCopoeia, Inc., Rockville, MD, USA). Recombinant constructs,
miR-552 mimics or miR-552 inhibitors were co-transfected into the
Huh-7 and Hep3B cells using Lipofectamine 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer’s protocol.
After 48 h, the cells were harvested, and luciferase activity was
measured using the Luc-Pair Duo-Luciferase Assay kit 2.0
(GeneCopoeia, Inc.). Firefly luciferase activity was normalized to
Renilla luciferase activity.
XAV939 cell treatment
XAV939 (cat. no. X3004) was purchased from
Sigma-Aldrich; Merck KGaA, and dissolved in dimethyl sulfoxide
(DMSO; Sigma-Aldrich; Merck KGaA). A total of 20×104
cells were seeded into 6-well plate and cultured at 37°C overnight.
XAV939 was used to treat cells at a final concentration of 5
μM for 24 h. DMSO was used to treat the cells as a
control.
Statistical analysis
Continuous variables are presented as the mean ±
standard deviation. Statistical analysis was performed using SPSS
version 21.0 software (SPSS Inc.; IBM Corp., Armonk, NY, USA) or
GraphPad PRISM 5 software (GraphPad Software, Inc., La Jolla, CA,
USA). Correlations between miR-552 and clinical features were
analyzed by Pearson’s χ2 test. The correlation between
miR-552 and WIF1 expression was analyzed by Spearman correlation
analysis. The difference between two groups was analyzed using an
unpaired Student t-test. One-way analysis of variance was used to
compare the data among multiple groups, and the Bonferroni method
was used for the post-hoc test. Survival analysis was performed
using a Kaplan-Meier curve and log-rank test. P<0.05 was
considered to indicated a statistically significant difference.
Results
Expression of miR-552 is upregulated in
HCC tissues and predicts poor prognosis
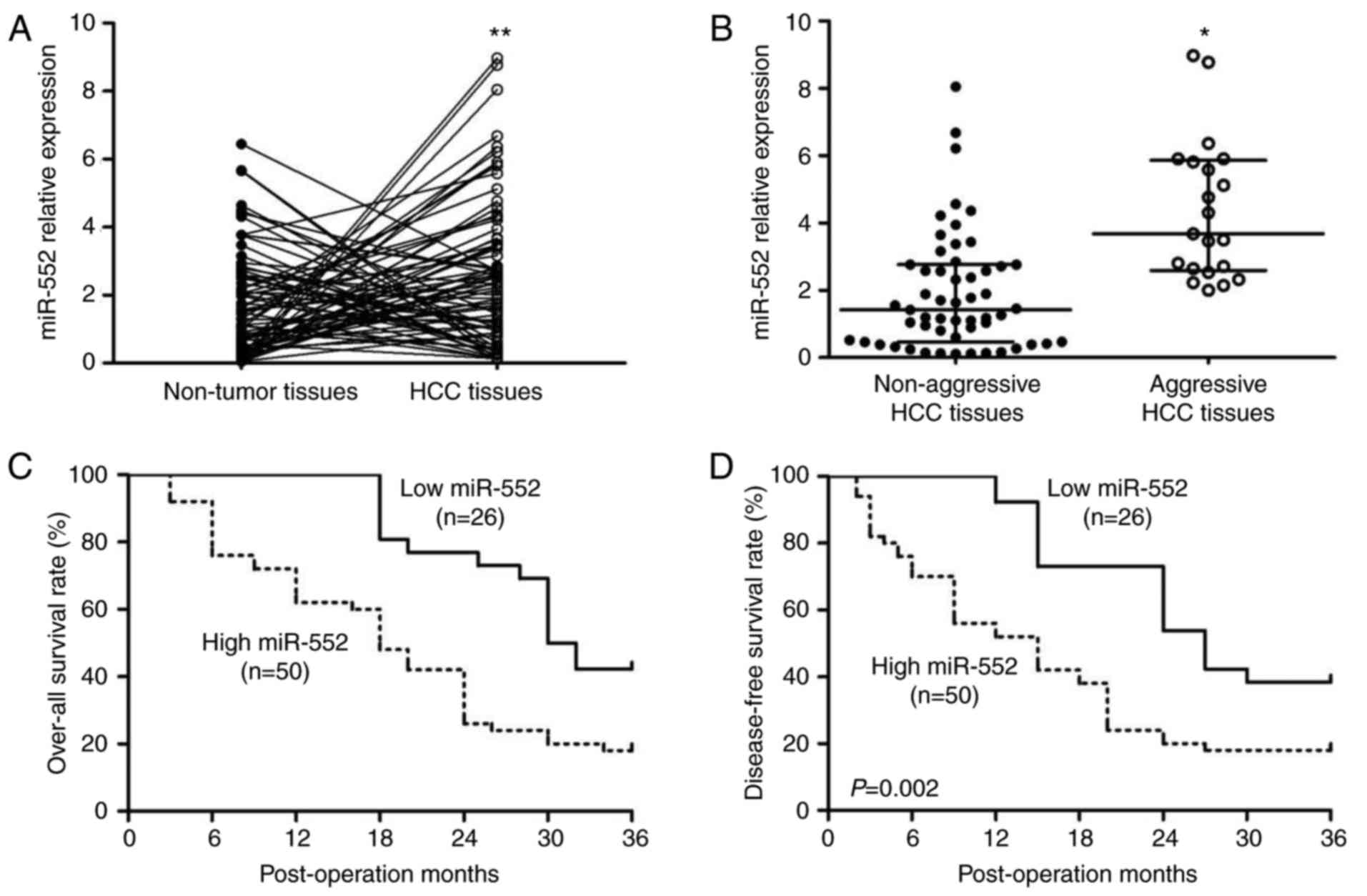
The expression of miR-552 in human tissues was
analyzed by RT-qPCR. A significant increase in miR-552 in HCC
tissues compared with the adjacent non-tumor tissues was identified
(Fig. 1A; P<0.01). The
clinical significance of miR-552 in patients with HCC was then
investigated. A total of two subgroups, miR-552 high expression
group (≥mean, n=50) and miR-552 low expression group (<mean,
n=26), in the cohort were divided according to the mean level of
miR-552. In Table I, the results
from Pearson’s χ2 test indicated that high expression of
miR-552 was associated with venous invasion (P=0.035) and advanced
Tumor Node Metastasis (13) stage
(III+IV; P=0.024), indicating that tumors with high miR-552
expression levels may be more aggressive. Therefore, the difference
in miR-552 expression between aggressive tissues (tumors with
intrahepatic metastasis, bile duct invasion or venous infiltration)
and non-aggressive HCC tissues was determined. As hypothesized,
compared with non-aggressive HCC tissues, miR-552 was significantly
upregulated in aggressive HCC tissues (Fig. 1B; P<0.05).
 | Table IClinical significance of miR-552 in
hepatocellular carcinoma. |
Table I
Clinical significance of miR-552 in
hepatocellular carcinoma.
| Clinical
features | miR-552 expression
| χ2 | P-value |
|---|
High
(n=50) | Low
(n=26) |
|---|
| Sex | | | 1.259 | 0.262 |
| Male | 37 | 16 | | |
| Female | 13 | 10 | | |
| Age, years | | | 1.284 | 0.257 |
| <60 | 22 | 15 | | |
| ≥60 | 28 | 11 | | |
| Hepatitis B
infection | | | 1.404 | 0.236 |
| Absent | 11 | 9 | | |
| Present | 39 | 17 | | |
| Tumor size, cm | | | 2.606 | 0.107 |
| <5 | 27 | 19 | | |
| ≥5 | 23 | 7 | | |
| Liver
cirrhosis | | | 0.866 | 0.352 |
| Absent | 14 | 10 | | |
| Present | 36 | 16 | | |
| Serum α fetoprotein
level, ng/ml | | | 2.155 | 0.142 |
| <400 | 20 | 15 | | |
| ≥400 | 30 | 11 | | |
| Venous
invasion | | | 4.451 | 0.035 |
| Absent | 33 | 23 | | |
| Present | 17 | 3 | | |
| Edmondson-Steiner
grading | | | 1.748 | 0.186 |
| I+II | 19 | 14 | | |
| III+IV | 31 | 12 | | |
| Tumor Node
Metastasis stage | | | 5.077 | 0.024 |
| I+II | 21 | 18 | | |
| III+IV | 29 | 8 | | |
Furthermore, Kaplan-Meier survival curves were used
to describe the prognostic value of miR-552 in HCC. Patients with
high miR-552 expression exhibited a decrease in overall survival
[Fig. 1C, hazard ratio
(HR)=2.541; 95% confidence interval (CI) 1.442-4.478; P=0.001] and
disease-free survival (Fig. 1D;
HR=2.435, 95% CI 1.390-4.263; P=0.002). These data suggest that
miR-552 is a potential biomarker for the clinical outcome of
patients with HCC.
miR-552 promotes the migration, invasion
and EMT of HCC cells
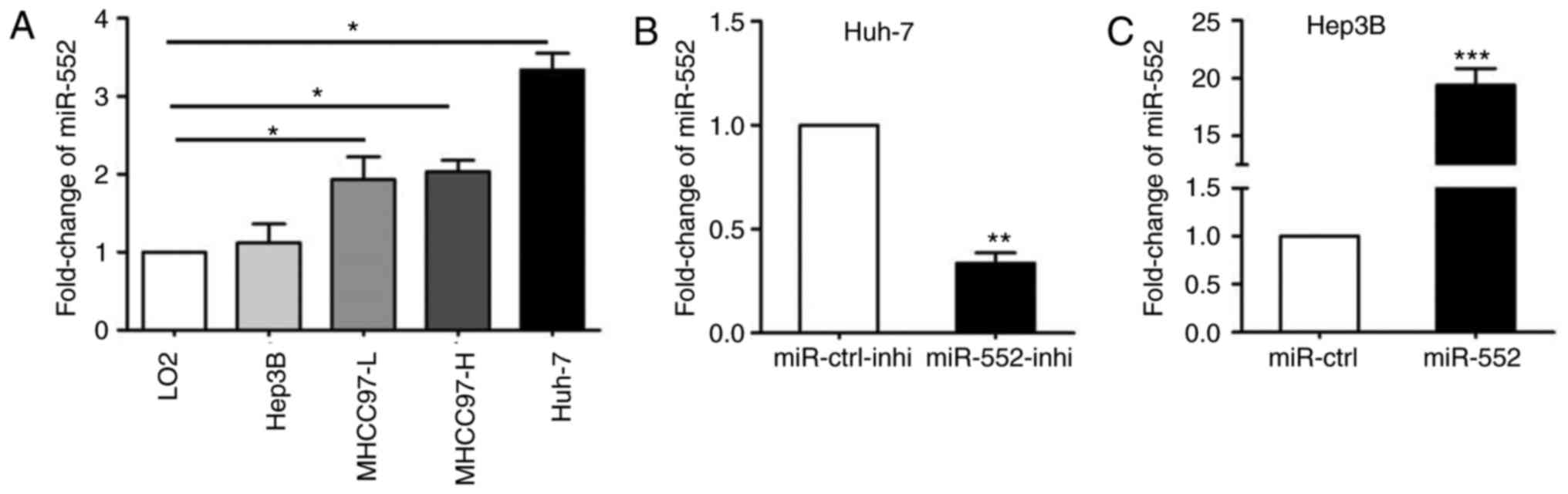
Next, qPCR was used to demonstrate that miR-552 was
upregulated in all HCC cell lines with the exception of Hep3B
(Fig. 2A; P<0.05). miR-552
expression was then silenced in Huh-7 cells (Fig. 2B; P<0.01) and overexpressed in
Hep3B cells (Fig. 2C;
P<0.001).
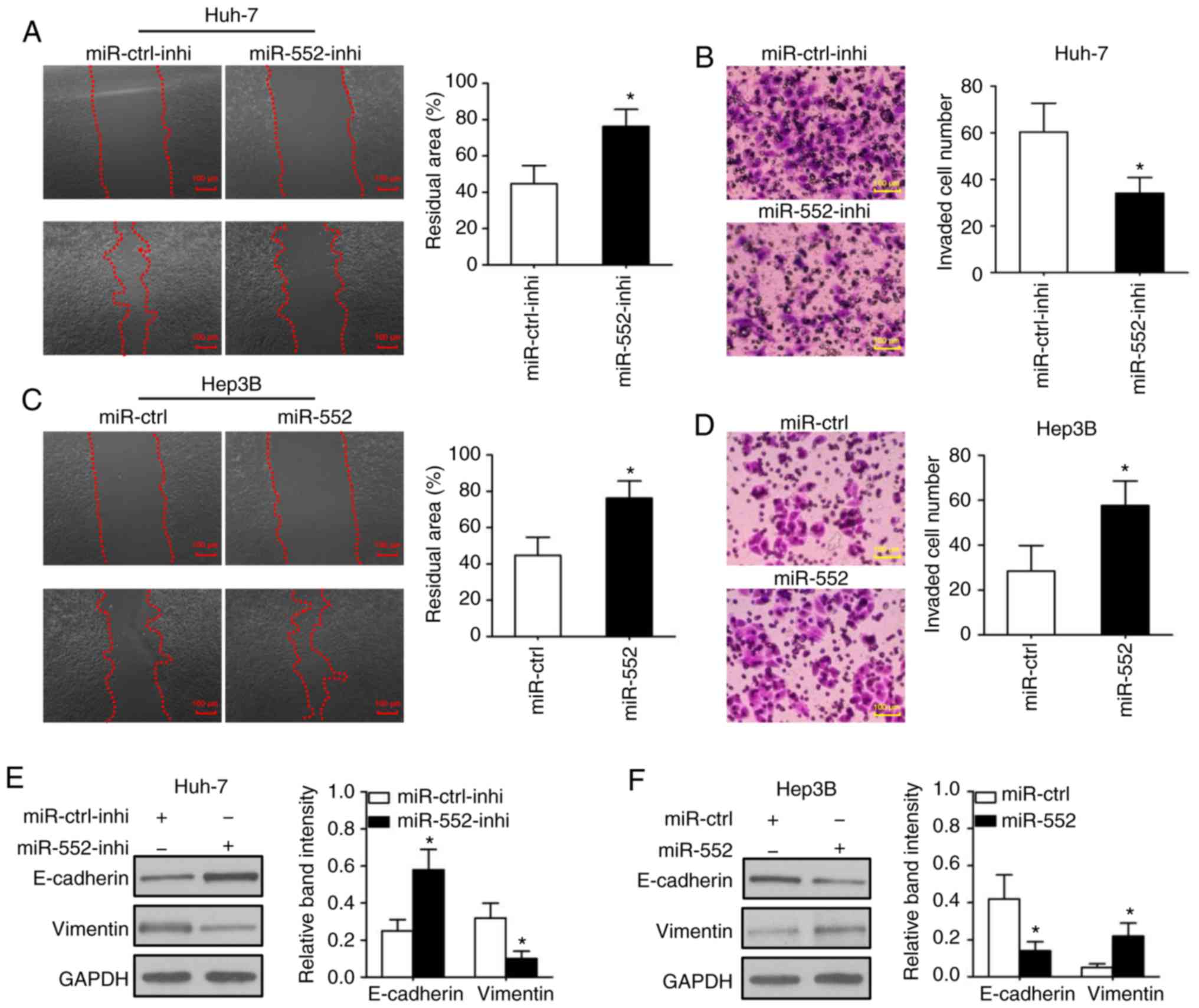
Wound-healing and Transwell assays were used to
determine the migration and invasion, respectively, in
vitro. In was observed that downregulation of miR-552 decreased
cell migration (Fig. 3A;
P<0.05) and invasion (Fig. 3B;
P<0.05) in Huh-7 cells. Conversely, the overexpression of
miR-552 significantly promoted cell migration (Fig. 3C; P<0.05) and invasion
(Fig. 3D, P<0.05) in Hep3B
cells. This result indicated that miR-552 may be associated with
tumor metastasis in HCC.
Tumor migration and invasion are complex processes
and are regulated by several cellular processes, including a loss
of cell-cell adhesion that is frequently accompanied by the
downregulation of E-cadherin and upregulation of vimentin. The
western blot analysis results of the present study indicated that
knockdown of miR-552 increased E-cadherin expression and decreased
vimentin expression in Huh-7 cells (Fig. 3E; P<0.05), whereas upregulation
of miR-552 inhibited E-cadherin, but induced vimentin expression in
Hep3B cells (Fig. 3F; P<0.05).
These results indicate that miR-552 possesses marked oncogenic
functions in HCC cells.
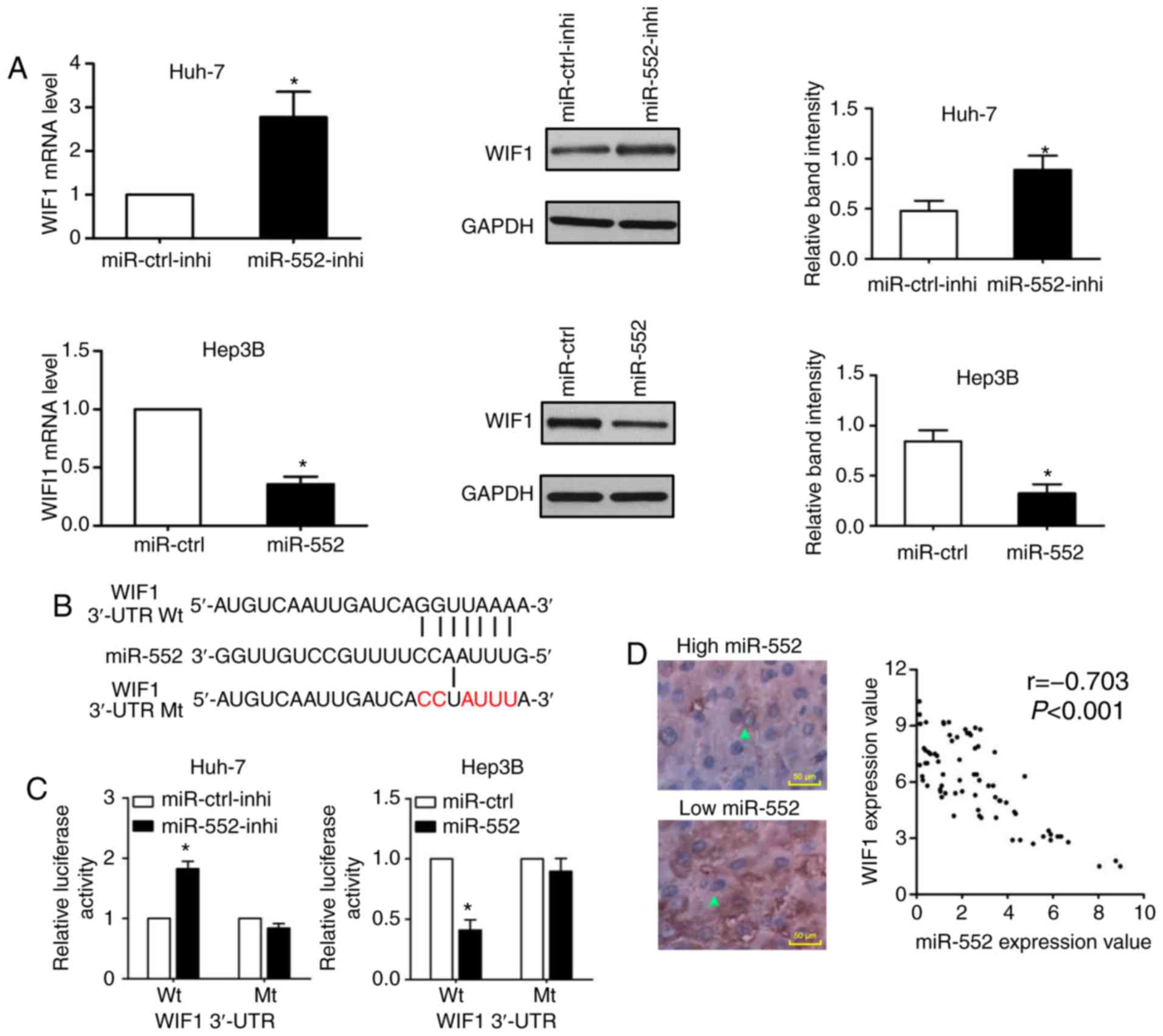
WIF1 is a downstream target of miR-552 in
HCC
miRNAs may bind to the 3′-UTR and act as inhibitors
to decrease certain mRNAs expression. A total of 2 online MicroRNA
targets prediction tools, TargetScan (http://www.targetscan.org/) and MiRanda (http://www.microrna.org), were used to predict WIF1 as
a downstream target of miR-552. As indicated in Fig. 4A, knockdown of miR-552 increased
WIF1 mRNA and protein expression in Huh-7 cells (P<0.05);
conversely, miR-552 overexpression decreased WIF1 expression in
Hep3B cells (P<0.05). Additionally, the wild-type and
mutant-type dual-luciferase reporters were constructed according
the binding sequence between miR-552 and WIF1 3′-UTR (Fig. 4B). As demonstrated in Fig. 4C, the dual-luciferase reporter
assay demonstrated that the downregulation of miR-552 increased the
luciferase activity of wild-type WIF1 3′-UTR (P<0.05), whereas
the upregulation of miR-552 decreased the luciferase activity of
the wild-type WIF1 3′-UTR (P<0.05). In either miR-552 knockdown
or induction, there was no significant change on the luciferase
activity of mutant WIF1 3′-UTR. Furthermore, a negative correlation
between miR-552 and WIF1 expression was confirmed by Spearman
correlation analysis (Fig. 4D;
r=-0.703; P<0.001). These data provided convincing evidence to
suggest that WIF1 is a direct target of miR-552 in HCC.
Oncogenic functions of miR-552 in HCC
cells are mediated by WIF1
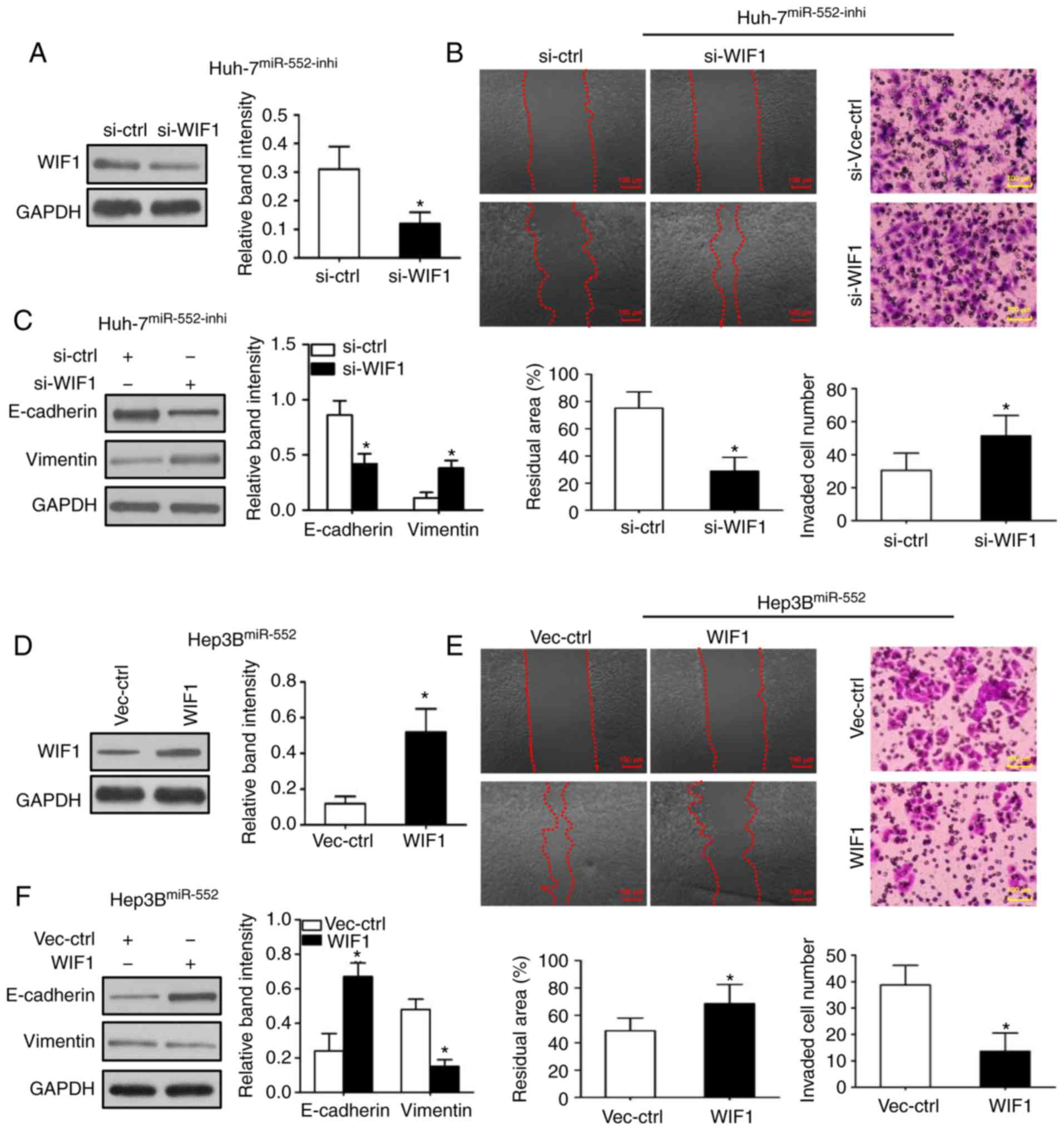
To additionally investigate the underlying
mechanisms of miR-552/WIF1 axis on tumor progression, WIF1
expression was first inhibited in miR-552 downregulated Huh-7 cells
(Fig. 5A; P<0.05). The
wound-healing and Transwell assays were then repeated to indicate
that WIF1 knockdown abrogated the inhibitory effects of migration,
invasion and EMT induced by miR-552 downregulation (Fig. 5B and C; P<0.05). Subsequently,
the same experiments were used to demonstrate that WIF1
overexpression decreased the migration, invasion and EMT in
miR-552-upregulated Hep3B cells (Fig.
5D-F; P<0.05).
GSK3β/β-catenin signaling is crucial to
the functions of miR-552 in HCC
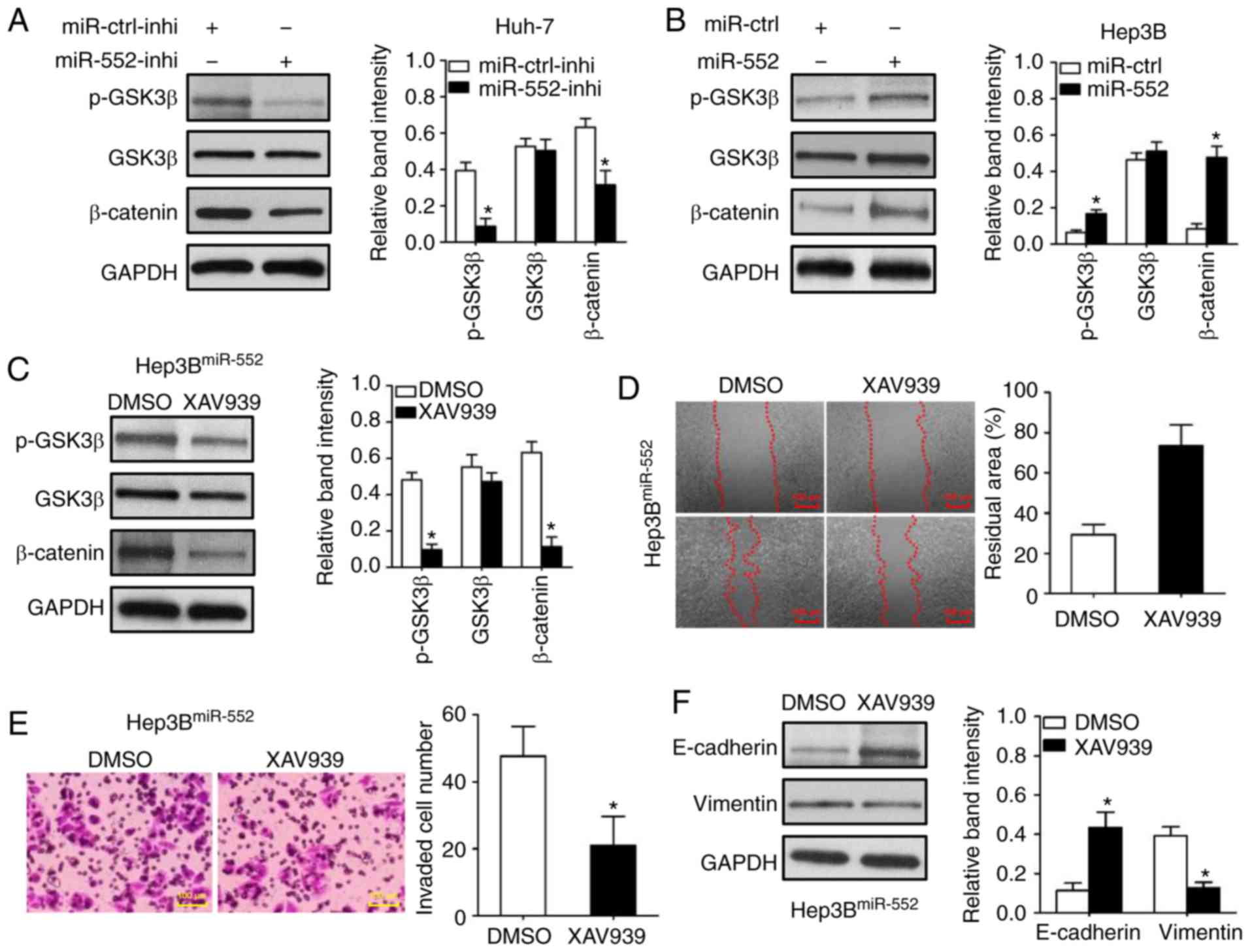
Western blot analysis was used to demonstrate that
miR-552 knockdown decreased GSK3β phosphorylation at Ser9 in Huh-7
cells (Fig. 6A; P<0.05). The
dephosphorylation of GSK3β inhibited β-catenin expression (Fig. 6A; P<0.05). Conversely, miR-552
overexpression increased GSK3β phosphorylation and β-catenin
expression in Hep3B cells (Fig.
6B; P<0.05). Additionally, the inhibition of the
GSK3β/β-catenin signaling pathway by XAV939 (Fig. 6C; P<0.05) abrogated the
oncogenic functions of miR-552 overexpression on Hep3B cell
migration and invasion (Fig. 6D and
E; P<0.05). XAV939 treatment also inhibited miR-552-induced
EMT in Hep3B cells (Fig. 6F;
P<0.05). Taken together, these data demonstrate that the
oncogenic functions of miR-552 in HCC cells were attributed to a
decreased activation of the GSK3β/β-catenin signaling pathway,
mediated by WIF1.
Discussion
The ectopic expressions of miRNAs in benign and
malignant diseases may deregulate cell adhesion and movement
functions. Anton et al (14) suggested that 2 adhesion associated
genes, junctional adhesion molecule 1 and fascin actin-bundling
protein 1, were inhibited following an overexpression of miR-143
and miR-145 in cervical cells. This abnormal regulation may destroy
the cervical epithelial barrier, which is an important protector of
preterm birth (15). A number of
types of cancer (16), including
HCC (17), also exhibited
increased migration and invasion abilities caused by aberrant miRNA
expression profiles, which are associated with cancer metastasis
and poor prognosis.
Among these miRNAs, miR-552 was identified as an
upregulated miRNA in colon cancer by principal component analysis
(18). Notably, compared with the
level of miR-552 in defective DNA mismatch repair (MMR) tumors,
miR-552 expression is upregulated in proficient DNA MMR tumors
(19). This indicates that
miR-552 has potential to regulate colon cancer cell proliferation
and invasion. In the present study, it was identified that miR-552
was increased in HCC tissues, particularly in tumors with portal
vein invasion, lymph node metastasis and other features of
aggressive behavior. The in vitro results of the present
study also indicated that miR-552 may serve a pro-invasive role in
HCC cells: The inhibition of miR-552 not only decreased migration
and invasion of Huh-7 cells but also impaired the EMT markers’
expression, whereas an overexpression of miR-552 increased the
migration, invasion and EMT of Hep3B cells.
The Wnt/β-catenin pathway involves a family of
proteins that is critical to a number of biological functions
including cell differentiation (20) and organogenesis (21). In HCC, the Wnt/β-catenin pathway
has been demonstrated to promote cell proliferation, invasion and
angiogenesis (22). The WIF1
gene, located on human chromosome 12q14, encodes a secreted protein
that inactivates the Wnt/β-catenin signaling pathway by binding to
Wnt proteins (23). In the
present study, qPCR, western blot analysis and dual-luciferase
reporter assay methods were used to confirm that WIF1 is a
downstream target of miR-552. The clinical analysis also revealed
an inverse correlation between miR-552 and WIF1 protein expression
in HCC tissues. As miR-552 binds to the 3′-UTR of WIF1 mRNA to form
the RNA-induced silencing complex (RISC), the miR-552/WIF1 RISC may
lead to the degradation of WIF1 mRNA and arrest the subsequent
translation process. Therefore, miR-552 may downregulate WIF1 mRNA
and protein expression. As a critical Wnt antagonist, WIF1 is
involved in the suppression of invasion in various cancer cell
lines (24). Similarly, the
present study identified that the altered expression of WIF1
abrogated the pro-invasive functions of miR-552 on HCC cells. As
WIF1 may abolish the binding of the Wnt ligand to the LRP-5/6
receptors (25), we hypothesized
whether miR-552 knockdown may cause the dephosphorylation of GSK3β
at Ser9. As expected, miR-552 knockdown decreased GSK3β
phosphorylation and β-catenin expression. XAV939 has been
previously confirmed as an inhibitor of β-catenin (26). In the present study, XAV939 was
used to treat Hep3B cells with miR-552 overexpression. The present
study demonstrated that XAV939 also inhibited the cell migration,
invasion and EMT processes induced by miR-552. As a transcription
factor, β-catenin may increase the expression of a number of
EMT-associated transcription factors (27), including zinc finger protein
SNAI1, zinc finger protein SNAI2 and twist-related protein 1. This
may explain why miR-552 exhibits pro-invasive functions in HCC.
In conclusion, miR-552 is highly associated with HCC
malignancies and has potential value to be a prognostic biomarker
for patients with HCC.
Funding
The present study was supported by a grant from the
National Natural Scientific Foundation of China (grant no.
81472247).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors’ contributions
CLi contributed to the design of the study and wrote
the manuscript. ZW, SC, JZ and KQ performed the study; and CLiu
contributed to the design of the study. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The use of clinical samples was approved by the
Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong
University. Written informed consent was obtained from all patients
enrolled in the present study.
Patient consent for publication
Written informed consent was obtained and signed by
each patient enrolled in the present study.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Fang LL, Wang XH, Sun BF, Zhang XD, Zhu
XH, Yu ZJ and Luo H: Expression, regulation and mechanism of action
of the miR-17-92 cluster in tumor cells (Review). Int J Mol Med.
40:1624–1630. 2017.PubMed/NCBI
|
|
2
|
Liu H, Lei C, He Q, Pan Z, Xiao D and Tao
Y: Nuclear functions of mammalian MicroRNAs in gene regulation,
immunity and cancer. Mol cancer. 17:642018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tang A, Hallouch O, Chernyak V, Kamaya A
and Sirlin CB: Epidemiology of hepatocellular carcinoma: target
population for surveillance and diagnosis. Abdom Radiol. 43:13–25.
2018. View Article : Google Scholar
|
|
4
|
Yegin EG, Oymaci E, Karatay E and Coker A:
Progress in surgical and nonsurgical approaches for hepatocellular
carcinoma treatment. Hepatobiliary Pancreat Dis Int. 15:234–256.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tian Q, Xiao Y, Wu Y, Liu Y, Song Z, Gao
W, Zhang J, Yang J, Zhang Y, Guo T, et al: MicroRNA-33b suppresses
the proliferation and metastasis of hepatocellular carcinoma cells
through the inhibition of Sal-like protein 4 expression. Int J Mol
Med. 38:1587–1595. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu WL, Wang WY, Yao WQ and Li GD:
Suppressive effects of microRNA-16 on the proliferation, invasion
and metastasis of hepatocellular carcinoma cells. Int J Mol Med.
36:1713–1719. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li C, Jiang Y, Miao R, Qu K, Zhang J and
Liu C: MicroRNA-1271 functions as a metastasis and
epithelial-mesenchymal transition inhibitor in human HCC by
targeting the PTP4A1/c-Src axis. Int J Oncol. 52:536–546.
2018.PubMed/NCBI
|
|
8
|
Xia ZS, Wang L, Yu T, Zhong W, Lian GD, Wu
D, Zhou HM and Chen GC: MiR-5000-3p and miR-5009 3P and miR-552:
potential microRNA biomarkers of side population cells in colon
cancer. Oncol Rep. 32:589–596. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Li H, Wang Y, Wang L, Yan X, Zhang
D, Ma X, Du Y, Liu X and Yang Y: MicroRNA-552 enhances metastatic
capacity of colorectal cancer cells by targeting a disintegrin and
metal-loprotease 28. Oncotarget. 7:70194–70210. 2016.PubMed/NCBI
|
|
10
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak and Schmittgen: Analysis of relative
gene expression data using real-time quantitative PCR and the
2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
12
|
Li C, Jiang Y, Miao R, Qu K, Zhang J and
Liu C: MicroRNA-1271 functions as a metastasis and
epithelial-mesenchymal transition inhibitor in human HCC by
targeting the PTP4A1/c-Src axis. Int J Oncol. 52:536–546.
2018.PubMed/NCBI
|
|
13
|
Duseja A: Staging of hepatocellular
carcinoma. J Clin Exp Hepatol. 4:S74–S79. 2014. View Article : Google Scholar
|
|
14
|
Anton L, DeVine A, Sierra LJ, Brown AG and
Elovitz MA: miR-143 and miR-145 disrupt the cervical epithelial
barrier through dysregulation of cell adhesion, apoptosis and
proliferation. Sci Rep. 7:30202017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Akgul Y, Word RA, Ensign LM, Yamaguchi Y,
Lydon J, Hanes J and Mahendroo M: Hyaluronan in cervical epithelia
protects against infection-mediated preterm birth. J Clin Invest.
124:5481–5489. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lou W, Liu J, Gao Y, Zhong G, Chen D, Shen
J, Bao C, Xu L, Pan J, Cheng J, et al: MicroRNAs in cancer
metastasis and angiogenesis. Oncotarget. 8:115787–115802. 2017.
View Article : Google Scholar
|
|
17
|
Zhang Y, Wei Y, Li X, Liang X, Wang L,
Song J, Zhang X, Zhang C, Niu J, Zhang P, et al: microRNA-874
suppresses tumor proliferation and metastasis in hepatocellular
carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis.
9:1302018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang XY, Wu MH, Liu F, Li Y, Li N, Li GY
and Shen SR: Differential miRNA expression and their target genes
between NGX6-positive and negative colon cancer cells. Mol Cell
Biochem. 345:283–290. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oberg AL, French AJ, Sarver AL,
Subramanian S, Morlan BW, Riska SM, Borralho PM, Cunningham JM,
Boardman LA, Wang L, et al: miRNA expression in colon polyps
provides evidence for a multihit model of colon cancer. PLoS One.
6:e204652011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui YM, Han XH, Lin YY, Lv WW and Wang YL:
TNF-alpha was involved in calcium hydroxide-promoted osteogenic
differentiation of human DPSCs through NF-kappaB/p38MAPK/Wnt
pathway. Die Pharmazie. 72:329–333. 2017.
|
|
21
|
Rankin SA, McCracken KW, Luedeke DM, Han
L, Wells JM, Shannon JM and Zorn AM: Timing is everything:
Reiterative Wnt, BMP and RA signaling regulate developmental
competence during endoderm organogenesis. Dev Biol. 434:121–132.
2018. View Article : Google Scholar
|
|
22
|
Chen J, Rajasekaran M and Hui KM: Atypical
regulators of Wnt/beta-catenin signaling as potential therapeutic
targets in hepatocellular carcinoma. Exp Biol Med. 242:1142–1149.
2017. View Article : Google Scholar
|
|
23
|
Zheng Y, Li X, Jiang Y, Xu Y, Song B, Zhou
Q, Liang X and Yang X: Promoter hypermethylation of Wnt inhibitory
factor-1 in patients with lung cancer: A systematic meta-analysis.
Medicine. 95:e54332016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo H, Zhou S, Tan L, Wu X, Wu Z and Ran
R: Clinicopathological significance of WIF1 hypermethylation in
NSCLC, a meta-analysis and literature review. Oncotarget.
8:2550–2557. 2017.
|
|
25
|
Schmid SC, Sathe A, Guerth F, Seitz AK,
Heck MM, Maurer T, Schwarzenböck SM, Krause BJ, Schulz WA, Stoehr
R, et al: Wntless promotes bladder cancer growth and acts
synergistically as a molecular target in combination with
cisplatin. Urol Oncol. 35:544.e1-544.e102017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo W, Shen F, Xiao W, Chen J and Pan F:
Wnt inhibitor XAV939 suppresses the viability of small cell lung
cancer NCI-H446 cells and induces apoptosis. Oncol Lett.
14:6585–6591. 2017.
|
|
27
|
Wu C, Zhuang Y, Jiang S, Liu S, Zhou J, Wu
J, Teng Y, Xia B, Wang R and Zou X: Interaction between
Wnt/beta-catenin pathway and microRNAs regulates
epithelial-mesenchymal transition in gastric cancer (Review). Int J
Oncol. 48:2236–2246. 2016. View Article : Google Scholar : PubMed/NCBI
|