Introduction
Glioblastoma is the most frequent primary malignant
brain tumor among adults. The median survival is generally <1
year from the time of diagnosis, and even in the most favorable
situations, the majority of patients succumb to the disease within
2 years (1-3). Standard therapy consists of surgical
resection if that is safely feasible, followed by radiotherapy.
However, the 5-year survival rate is <3% (4). One of the reasons for the dismal
prognosis is that current treatment strategies cannot eliminate
glioblastoma-initiating cells (GICs) (5-7). A
comprehensive understanding of the molecular basis of GICs may
contribute to the identification of novel therapeutic targets.
Yin Yang 1 (YY1) is an ubiquitously expressed zinc
finger transcription factor encoded by the 23 kb YY1 gene (8-12).
Comprised of 414 amino acids, YY1 carries out various cellular
functions, including transcriptional regulation, cell
proliferation, chromatin remodeling and apoptosis (12-16). YY1 regulates multiple targets,
including Erb-B2 receptor tyrosine kinase 2 (ERBB2), p53, caspases
and histone deacetylases (HDACs), which have been implicated in
cancer progression (15). YY1
expression has been shown to be increased in many types of cancer,
including metastatic breast cancer (17,18), colon cancer (19), gastric cancer (20) and prostate cancer (21). However, its roles have not yet
been fully elucidated as regards the formation of GICs.
MicroRNAs (miRNAs or miRs), which are
single-stranded long non-coding RNAs of 19-25 nucleotides in
length, play important roles in the regulation of drug resistance
and GICs (22,23). miR-186 has been demonstrated to
play a significant role as a tumor suppressor in many types of
cancer (24-26). For example, miR-186 is a novel
tumor suppressor miRNA that functions to inhibit tumorigenesis in
glioblastoma multiforme (GBM) both in vitro and in
vivo, by targeting both FGF2 and RelA (27); miR-186 may be a molecular target
of glioblastoma (27). However,
the role of miR-186 in GIC and drug resistance remains elusive. In
this study, we observed that miR-186 reversed cisplatin resistance
and inhibited the formation of the GIC phenotype by degrading YY1
in glioblastoma.
Materials and methods
Human glioblastoma cell lines
U87MG cells (glioblastoma of unknown origin) and
LN-229 glioblastoma cells were purchased from then Biochemistry and
Cell Biology Institute of Shanghai, Chinese Academy of Sciences
(Shanghai, China), within 3 months of the experiments. Of note, it
has been reported that the U87MG cell line has been misidentified
(28). The U87 cell line used has
been authenticated by STR profiling; thus, misidentification is not
likely to affect the outcomes of this study. To obtain
cisplatin-resistant glioblastoma U87MG cells (U87MG-CR cells), the
U87MG cells were treated with escalating concentrations of
cisplatin from 107 to 105 M as previously
reported (29). The established
U87MG-CR cells grew at a similar rate in the presence or absence of
105 M cisplatin for 3 days (data not shown). The half
maximal inhibitory concentration (IC50) of the U87MG-CR
cells increased by 12-fold, as compared with that of the U87MG
cells (data not shown). The cells were cultured in Dulbecco’s
modified Eagle’s medium (DMEM; Invitrogen, Shanghai, China)
supplemented with 10% fetal bovine serum (FBS; Invitrogen) and
antibiotics (100 mg/ml penicillin/100 U/ml streptomycin
(Invitrogen) in a 5% CO2 incubator at 37°C.
shYY1 plasmids and pre-miR-186 and
control miR
The shYY1 plasmids and scramble control were
purchased from Tiangen (Beijing, China). Pre-miR-186 and control
miR were purchased from Ambion, Inc. (Ambion, Austin, TX, USA).
Transfection experiment
Cell transfection was performed as previously
described (30). For the
transfection experiments, the cells were cultured in serum-free
medium without antibiotics at 60% confluence for 24 h, and then
transfected using FuGENE HD transfection reagent (Roche,
Indianapolis, IN, USA) according to the manufacturer’s
instructions. Following incubation for 6 h in a 5% CO2
incubator at 37°C, the medium was removed and replaced with normal
culture medium (serum-free medium without antibiotics) for 24 h.
Subsequently, western blot analysis, MTT assay, immunostaining
assay, PCR and immunofluorescence staining were performed as
described below.
Western blot analysis
This was performed as previously described (30,31). Total protein was prepared using
extraction buffer comprising NaCl/Pi containing 0.5%
Triton X-100, 1 mM EDTA, 1 mM phenylmethyl sulfonyl fluoride, and
complete protease inhibitors (Roche, Shanghai, China). The
concentration of each protein lysate was determined using a BCA™
protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA).
Equal amounts of total protein were subjected to 12% SDS/PAGE. The
samples were then transferred onto nitrocellulose membranes and
blocked for 60 min at room temperature in 5% skim milk powder (w/v)
in NaCl/Pi and protein was probed with antibodies
against human YY1 (ab109228; 1:500) mouse double minute 2
homolog (ab38618; 1:500), ATPase copper transporting beta
(ab124973, 1:500), integrinα6 (ab235905, 1:500), signal transducer
and activator of transcription 3 (ab68153, 1:500) or β-actin
(ab8227, 1:500) (all from Abcam, Cambridge, MA, USA) and then with
IRDyeTM-800 conjugated anti-rabbit secondary antibodies (1:10,000;
ab150077; Abcam) all for 30 min at room temperature. The specific
proteins were visualized using the Odyssey™ Infrared Imaging System
(Gene Company, Lincoln, NE, USA).
MTT assay
To monitor the resistance to cisplatin, the U87MG,
U87MG-CR and LN-229 cells were treated with 20 µM cisplatin
or DSMO for 24 h. MTT assay was performed as previously described
(32). Data were analyzed using
software origin 7.5 (OriginLab, Northampton, MA, USA) to fit the
sigmodial curve.
Sphere formation assay
The cells (103/ml) in serum-free
RPMI-1640/1 mM Na-pyruvate were seeded on 0.5% agar pre-coated
6-well plates. After 1 week, half the medium was exchanged every
3rd day. Single spheres were selected and counted by an inverted
microscope (TE2000-E2, Nikon Corporation, Tokyo, Japan).
Immunostaining assay for YY1 and CD133 in
glioblastoma spheres
Single cell suspensions of glioblastoma cells
transfected as indicated above were prepared and plated using ultra
low adherent wells of 6-well plate at 5,000 cells/well in sphere
formation medium (serum-free RPMI-1640/1 mM Na-pyruvate;
Invitrogen), as described above. Following 7 days of treatment, the
spheres were collected by centrifugation (10,00 × g, 10 min, 4°C),
washed with 1X PBS, and fixed with 3.7% parformaldehyde for
immunofluorescence staining. Anti-YY1 (ab109228; 1:500; Abcamand
anti-CD133 antibodies (ab19898, 1:500) were used for immunostaining
assay following the manufacturer’s instructions and as previously
described (33,34). The coverslips were counterstained
with 4′6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific)
for visualization of the nuclei. Microscopic analysis was performed
with a confocal laser-scanning microscope (Leica Microsystems,
Bensheim, Germany). The fluorescence intensities were measured in a
few viewing areas for 300 cells per coverslip and analyzed using
ImageJ 1.37v software (http://rsb.info.nih.gov/ij/index.html).
Real-time PCR for miRNA expression
Total RNA was isolated from the cells using the
mirVana miRNA Isolation kit (Ambion, Austin, TX, USA). The
detection of the mature form of miRNAs was performed using the
mirVana qRT-PCR miRNA Detection kit and qRT-PCR Primer Sets,
according to the manufacturer’s instructions (Ambion). For the
quantification PCR of miR-186, the forward primer was as follows:
5′-GCG GCG CAA AGA ATT CTC CT-3′, and the reverse primer was as
follows: 5′-GTG CAG GGT CCG AGG T-3′. The quantification of PCR
performed was performed using the ΔΔCq method (35). The U6 small nuclear RNA was used
as an internal control.
Immunofluorescence staining
This was performed as previously described (36). Following transfection, the cells
were fixed in 4% paraformaldehyde for 15 min, and then blocked with
goat serum blocking solution for 20 min at room temperature.
Subsequently, rabbit antibody against YY1 (ab109228; 1:500; Abcam)
were added, and the mixtures were incubated in a humid chamber
overnight. After washing 3 times with NaCl/Pi, the cells were
incubated with appropriate secondary antibodies (1:10,000;
ab150077; Abcam) for 30 min at 37°C. After washing with NaCl/Pi,
the samples were observed under a laser scanning confocal
microscope (Olympus, Tokyo, Japan). DAPI staining (blue) was used
to highlight the nuclei.
Reverse transcription-quantitative
polymerase chain reaction PCR (RT-qPCR) for mRNA expression
Total RNA was isolated from the cells using TRIzol
reagent (Invitrogen/Thermo Fisher Scientific). cDNA was synthesized
from 1 µg of total RNA in a 20 µl reverse
transcription (RT) system followed by PCR amplification in a 50
µl PCR system performed using an RT-PCR kit (Cat no. A3500,
Promega, Madison, WI, USA). The housekeeping gene,
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used as the
RNA loading control. The PCR primer sequences were as follows: YY1
forward, 5′-CAG AAG CAG GTG CAG ATC AAG-3′ and reverse, 5′-GAC CAC
ATG GTG ACC GAG AAC-3′; and GAPDH forward, 5′-ATT CAA CGG CAC AGT
CAA GG-3′ and reverse, 5′-GCA GAA GGG GCG GAG ATG A-3′. PCR was
conducted according to the manufacturer’s instructions: The thermal
cycle profile was as follows: Denaturation for 30 sec at 95 °C,
annealing for 45 sec at 52-58°C depending on the primers used, and
extension for 45 sec at 72°C. Each PCR reaction was performed for
28-32 cycles. The PCR products were analyzed by agarose gel
electrophoresis. Gels were photographed and densities of the bands
were determined with a computerized image analysis system (Alpha
Innotech, San Leandro, CA, USA). The area of each band was
calculated as the integrated density value (IDV). qPCR for YY1 was
performed using Power SYBR-Green PCR Master Mix (Applied
Biosystems, Carlsbad, CA, USA) according to the manufacturer’s
instructions. Quantification of PCR performed was performed using
the ΔΔCq method (35).
Methods of bioinformatics
The analysis of potential miRNA target sites was
carried out using the commonly used prediction algorithm, miRDB
(http://mirdb.org/).
Northern blot analysis
Northern blot analysis of miRNAs, was performed as
previously described (37).
Probes were labeled with [γ-32P] ATP complementary to
miR-186 and U6 snRNA.
Statistical analysis
Data are presented as the means ± SEM. The Student’s
t-test (two-tailed) was used for comparisons between 2 groups. A
value of P<0.05 was considered to indicate a statistically
significant difference.
Results
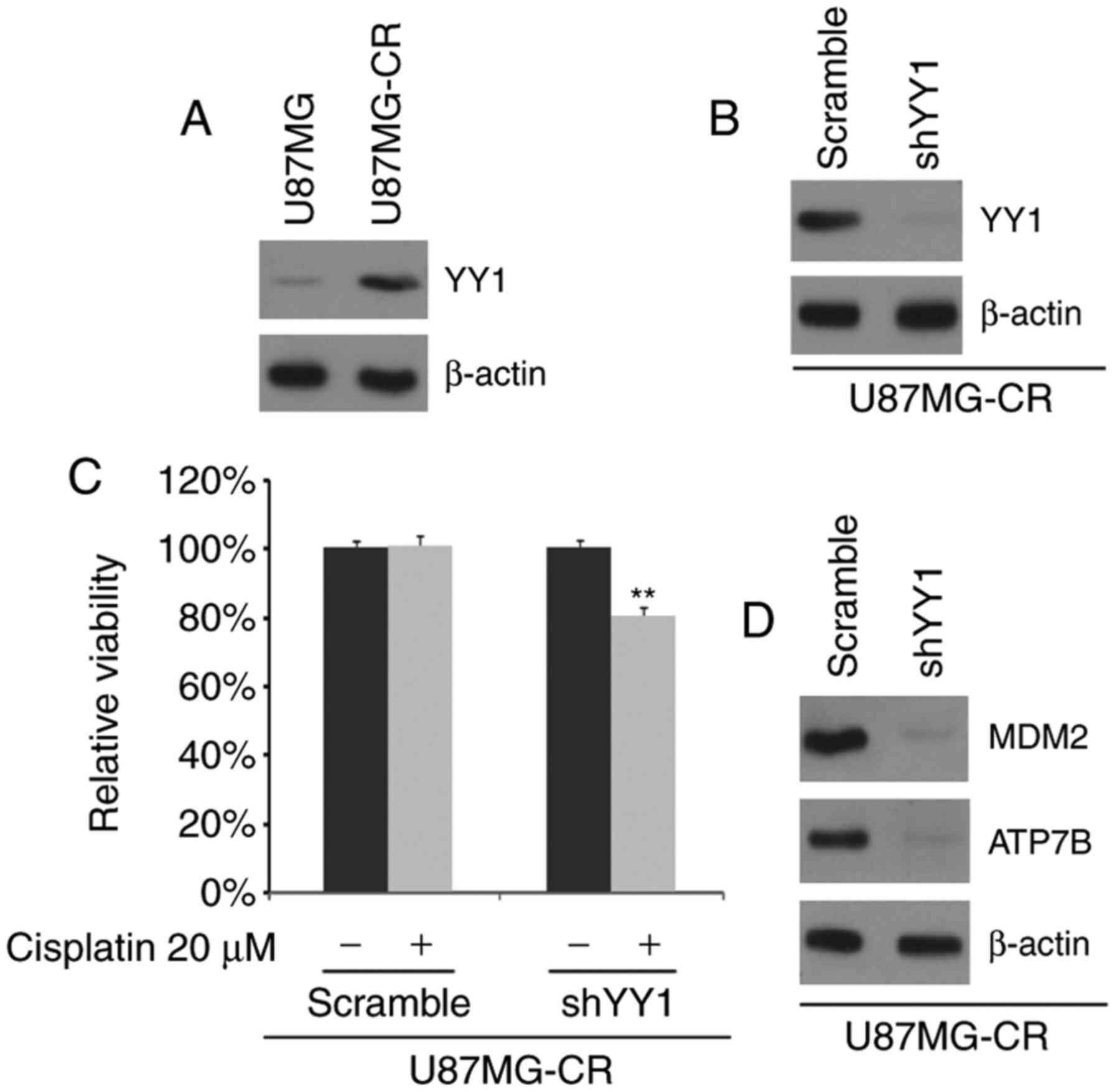
YY1 expression is increased in U87MG-CR
cells and the silencing of YY1 sensitizes the U87MG-CR to
cisplatin
In order to determine whether cisplatin resistance
is associated with YY1 expression, we examined the YY1 protein
concentrations in the U87MG and U87MG-CR cells. We observed that
YY1 protein expression was increased in the U87MG-CR cells
(Fig. 1A). To identify the role
of YY1, we examined whether transfection with shYY1 plasmid would
downregulate YY1 protein expression in the U87MG-CR cells. The
results revealed that YY1 protein expression was inhibited by
transfection with the shYY1 plasmid (Fig. 1B). To further determine whether
YY1 affects the sensitivity of glioblastoma cells to cisplatin, we
transfected the U87MG-CR cells with shYY1 plasmid or the scramble
control and then performed MTT assay. We found that the silencing
of YY1 transformed the U87MG-CR to cells to cisplatin-sensitive
cells (U87MG cells), as evidenced by the decreased viability of the
shYY1-transfected cells (Fig.
1C). We then examined the expression of MDM2 and ATP7B as MDM2
protein can confer the resistance of a human glioblastoma cell line
to cisplatin-induced apoptosis and ATP7B is associated with
cisplatin resistance (38,39).
In this study, we observed that the MDM2 and ATP7B protein
expression levels were decreased in the U87MG-CR cells following
transfection with shYY1 (Fig.
1D).
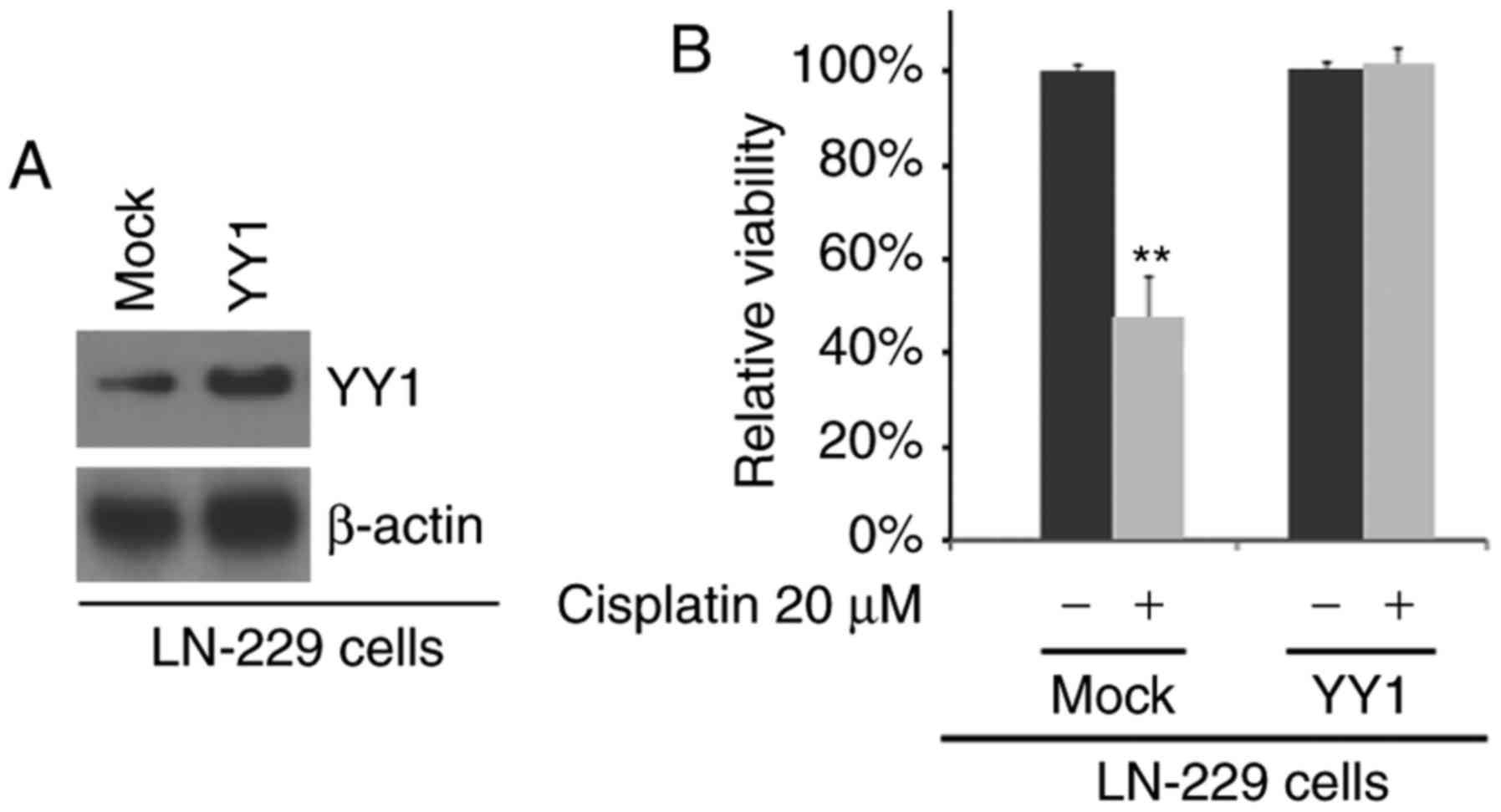
Overexpression of YY1 promotes the
resistance of LN-229 cells to cisplatin
To examine the effects of YY1, we examined whether
YY1 protein expression was increased by YY1-expressing plasmids in
LN-229 cells (cisplatin-sensitive cells). We observed that YY1
protein expression was increased following transfection with
YY1-expressing plasmids (Fig.
2A). To identify whether the responses to cisplatin can be
altered by YY1, we transfected the LN-229 cells with YY1-expressing
plasmids and we then performed MTT assay. We found that the
overexpression of YY1 promoted the resistance of LN-229 cells to
cisplatin, as no marked difference in cell viability was observed
between the cisplatin-treated or untreated LN-229-expressing cells
(Fig. 2B).
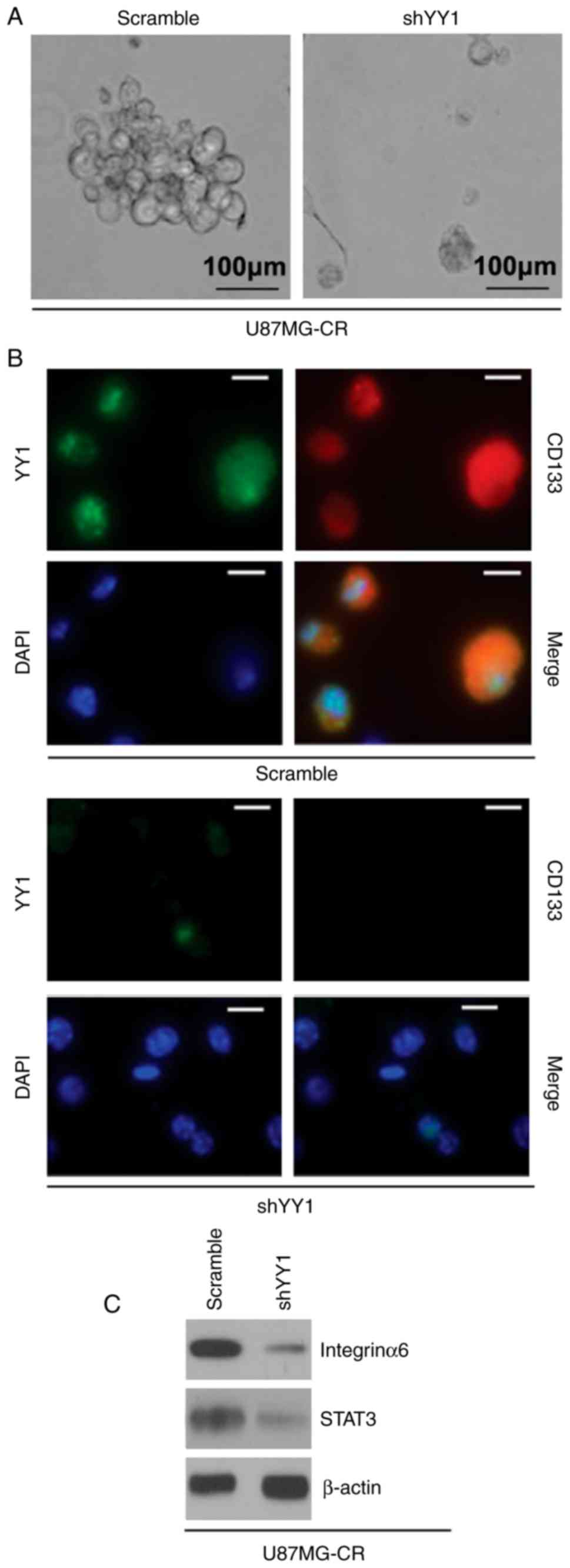
Silencing of YY1 inhibits the formation
of the GIC phenotype in U87MG-CR cells
To determine whether the silencing of YY1 affects
the GIC phenotype of the U87MG-CR cells, we performed a sphere
formation assay to assess the formation of GICs in the U87MG-CR
cells. We observed that the cells transfected with shYY1 formed
much smaller spheres after 14 days of culture as compared with the
control cells (Fig. 3A). As CD133
expression is associated with the GIC phenotype in glioblastoma
(40), in this study, we examined
whether YY1 regulates CD133 protein expression. We performed
immunostaining assay in the spheres isolated from the U87MG-CR
cells transfected with the shYY1 plasmid or the scramble control.
The results revealed that CD133 protein expression was decreased in
the spheres isolated from the U87MG-CR cells transfected with the
shYY1 plasmid (Fig. 3B). In
addition, as integrinα6 regulates GICs and targeting integrinα6 in
GICs inhibits self-renewal, proliferation and tumor formation
capacity (41); thus, in this
study, we also examined the expression of integrinα6. Moreover, as
STAT3 is required for the proliferation and maintenance of
multi-potency in glio-blastoma stem cells (42), we also examined its expression. We
found that the silencing of YY1 downregulated integrinα6 and STAT3
protein expression in the U87MG-CR cells (Fig. 3C).
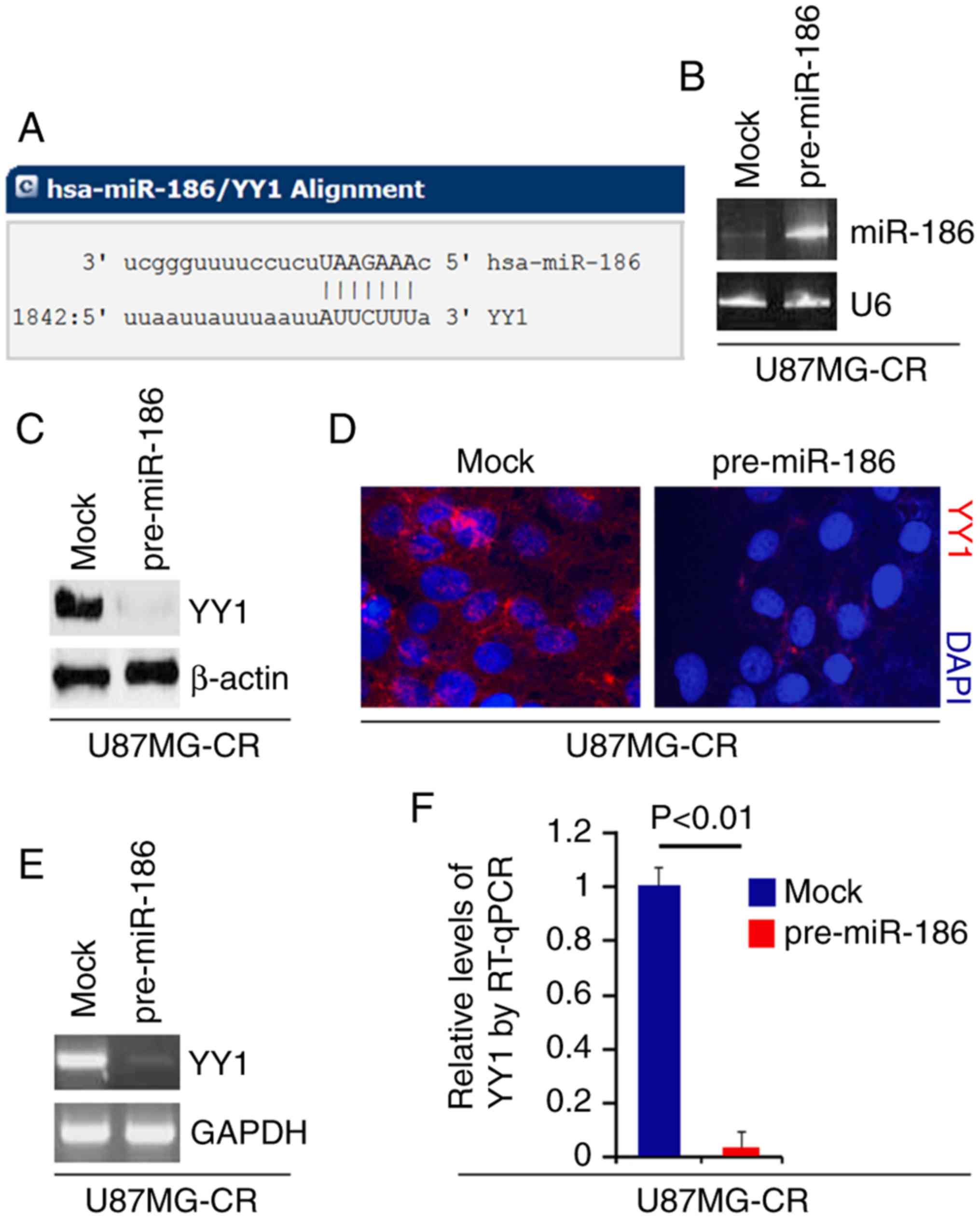
miR-186 inhibits YY1 protein expression
in U87MG-CR cells
To confirm whether YY1 is regulated by miRNAs, we
used a commonly used prediction algorithm, miRDB (http://mirdb.org/) to analyze the 3′UTR of YY1. A
total of 38 miRNAs were found by the algorithm. However, we were
interested in miR-186, as miR-186 is a tumor suppressor gene by
inhibiting oncogene expression (24,26,43). Moreover, miR-186 may sensitize
cancer cells to paclitaxel and cisplatin (25). The target sites on the 3′UTR of
YY1 are shown in Fig. 4A. In an
attempt to identify the role of miR-186 in regulating YY1
expression in glioblastoma, we transfected the U87MG-CR cells with
pre-miR-186 and control miR. Following transfection, miR-186
expression was detected by real-time PCR and the results revealed
that miR-186 expression was increased by transfection with
pre-miR-186 (Fig. 4B). We then
performed western blot analysis to detect YY1 protein expression in
the U87MG-CR cells transfected with pre-miR-186 or control miR. We
found that YY1 protein expression was inhibited by miR-186
(Fig. 4C). We then performed
immunofluorescence analyses of the U87MG-CR cells transfected with
pre-miR-186 anord control miR. We observed that YY1 protein
expression was inhibited in the cells transfected with pre-miR-186
(Fig. 4D). To examine whether
miR-186 degrades YY1 mRNA, we performed RT-qPCR and real-time PCR
and we found that the overexpression of miR-186 degraded YY1 mRNA
expresoin (Fig. 4E and F).
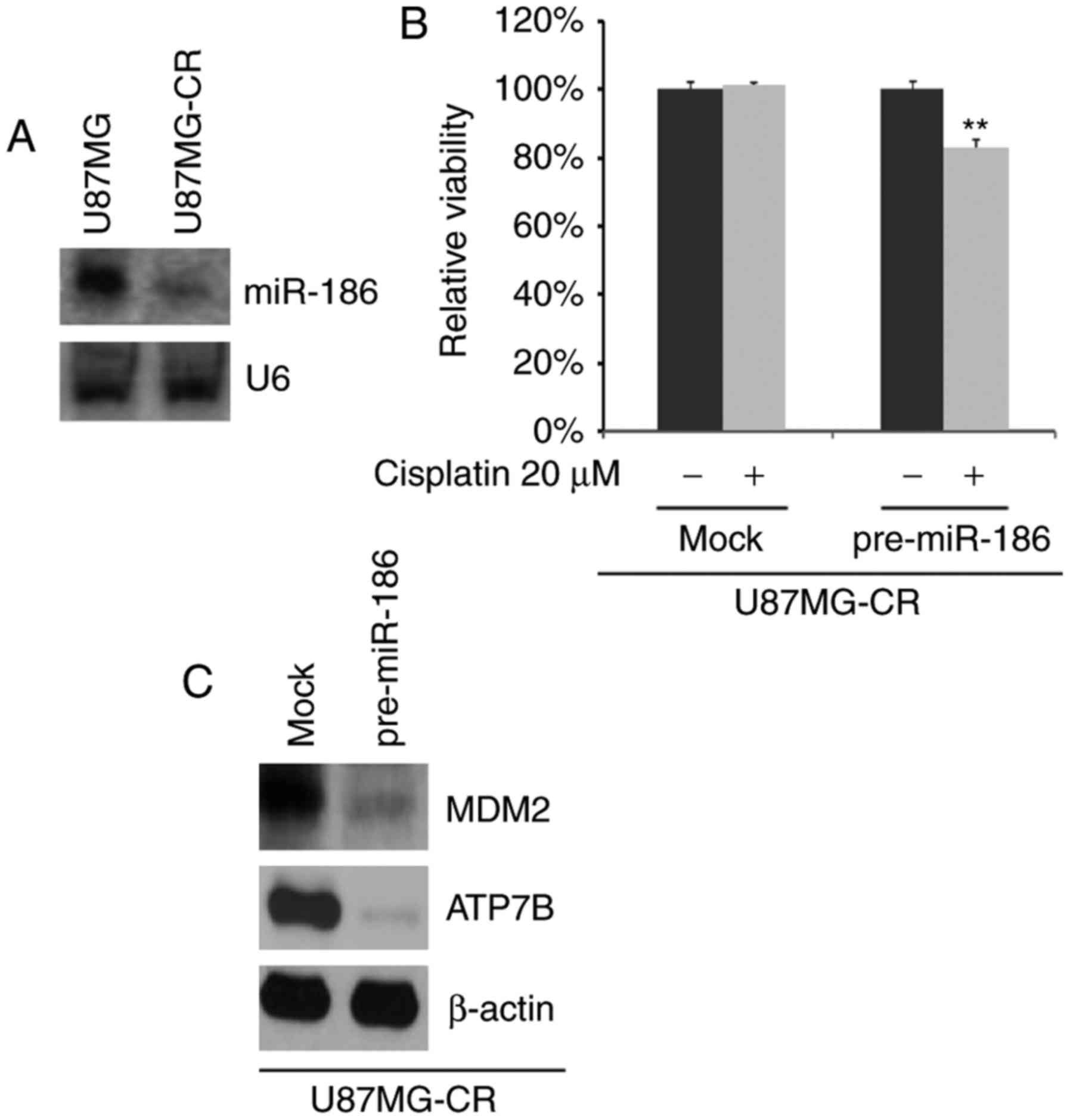
miR-186 expression is decreased in
U87MG-CR cells and its overexpression reverses cisplatin
resistance
To determine whether cisplatin resistance is
associated with miR-186 expression, we performed northern blot
analysis to detect miR-186 expression in U87MG cells and U87MG-CR
cells. We observed that miR-186 expression was markedly decreased
in the U87MG-CR cells (Fig. 5A).
To further identify whether miR-186 can affect the
resistance/sensitivity of U87MG-CR cells to cisplatin, we
transfected the U87MG-CR cells with pre-miR-186 or control miR. We
then performed MTT assay with the U87MG-CR cells treated as
indicated (Fig. 5B). We found
that the overexpression of miR-186 reversed cisplatin resistance,
evidenced by the decreased viability of the U87MG-CR cells treated
with cisplatin and transfected with pre-miR-186 (Fig. 5B). We also performed western blot
analysis to examine MDM2 and ATP7B protein expression in the
U87MG-CR cells transfected with pre-miR-186 or control miR. The
results revealed that MDM2 and ATP7B protein expression was
inhibited by miR-186 (Fig.
5C).
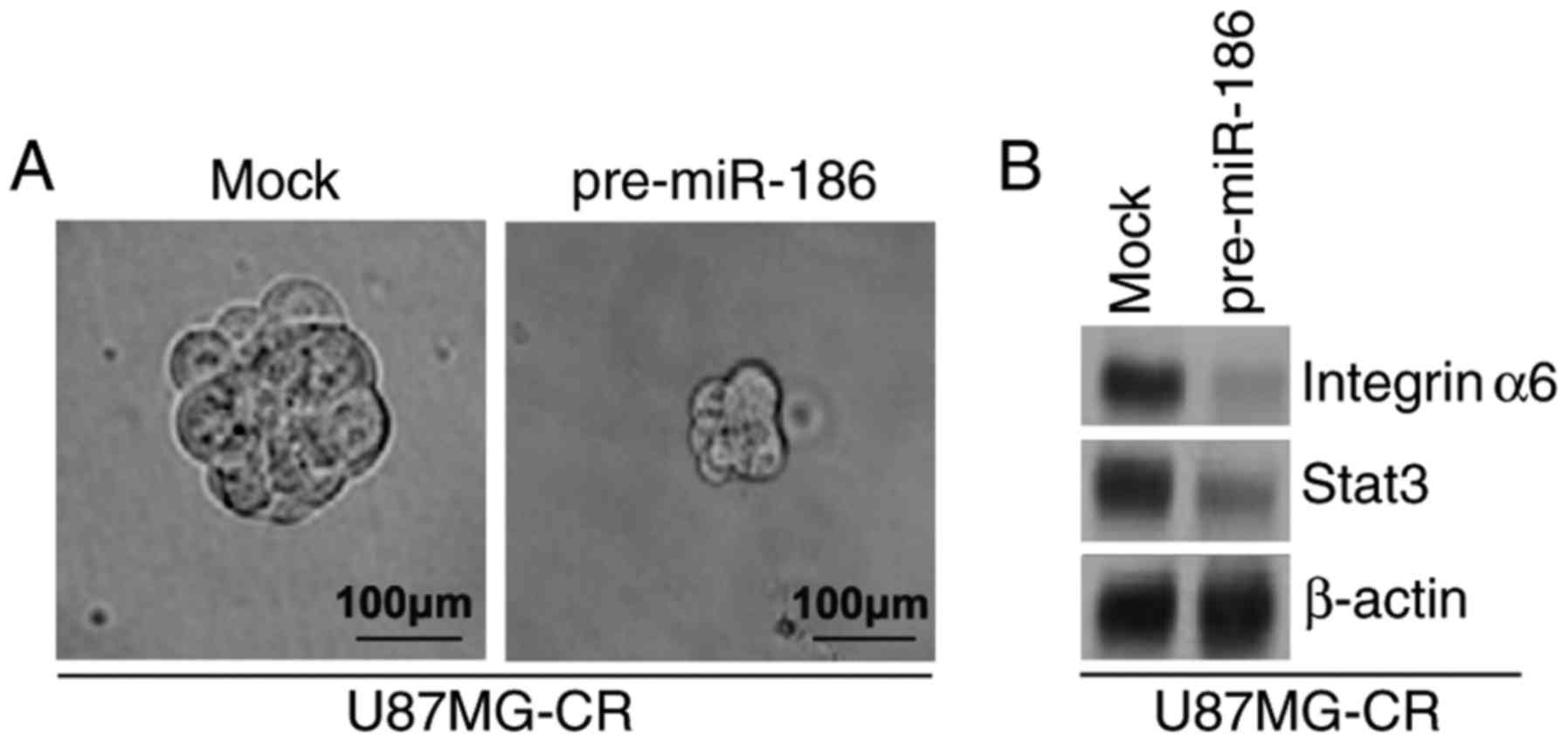
miR-186 inhibits the formation of the GIC
phenotype of U87MG-CR cells
To identify whether miR-186 can affect the GIC
phenotype of U87MG-CR cells, we performed a sphere formation assay
to assess the formation of GICs in the U87MG-CR cells. Sphere
formation assay revealed that the overexpression of miR-186
inhibited the formation of GICs in U87MG-CR cells (Fig. 6A). Subsequently, to determine
whether miR-186 regulates integrinα6 and STAT3 protein expression,
we performed western blot analysis of the U87MG-CR cells
transfected with pre-miR-186 or control miR. We observed that
integerinα6 and stat3 protein expression levels were inhibited by
miR-186 (Fig. 6B).
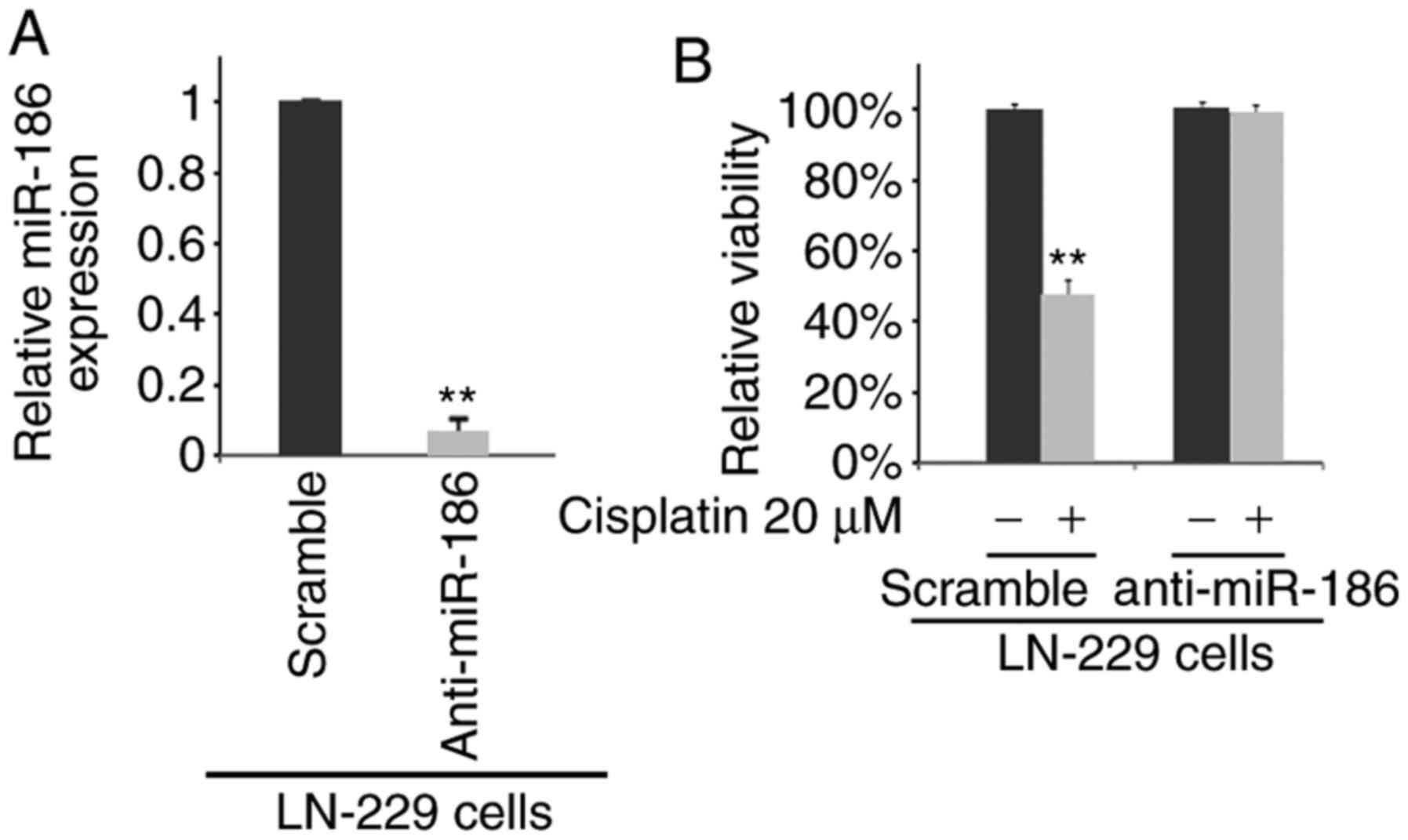
Silencing of miR-186 promotes the
resistance of LN-229 cells to cisplatin
To determine whether miR-186 affects the sensitivity
of the LN-229 cells to cisplatin, we transfected the LN-229 cells
with anti-miR-186. We then performed real-time PCR to detect
miR-186 expression in the LN-229 cells transfected with
anti-miR-186 and scramble (mock). We observed that miR-186
expression was evidently decreased in the LN-229 cells transfected
with anti-miR-186 (Fig. 7A). We
then performed MTT assay of the LN-229 cells treated as indicated
(Fig. 7B). The results revealed
that the overexpression of miR-186 promoted cisplatin resistance,
as the cells transfected with anti-miR-186 and treated with
cisplatin exhibited no marked difference in viability compared with
the anti-miR-186-transfected cells not treated with cisplatin
(Fig. 7B).
Discussion
Cisplatin is a neutral, square planar platinum (II)
complex containing two chloride ligands oriented in a cis
configuration. It has become one of the most effective
chemotherapeutic agents for the treatment of glioblastoma (44). However, intrinsic or acquired
resistance to cisplatin reduces its efficacy (45). The mechanisms of resistance
include miRNA deregulation and the formation of GICs (46-48). miR-186 has been demonstrated to
play a significant role as a tumor suppressor in many types of
cancer (24-26). Nevertheless, its biological
function in glioblastoma remains unknown. In the current study, we
found that miR-186 played an important role in the formation of
GICs and in the regulation of cisplatin resistance. These findings
provide novel insight into the potential roles of miR-186 in
promoting the formation of GICs and conferring chemoresistance in
glioblastoma. MDM2 protein can confer the resistance of a human
glioblastoma cell line to cisplatin (39). We demonstrated that the
overexpression of miR-186 inhibited MDM2 protein expression. The
ATP7B product, a protein of 1465 amino acids (ATP7B), is expressed
pre-dominantly in the liver, kidneys and placenta in humans
(49). ATP7B expression is
associated with cisplatin resistance (38). In this study, we found that ATP7B
expression was inhibited by miR-186 in U87MG-CR cells.
A number of studies have relied on the enrichment of
GICs based on the expression of the cell surface protein CD133
(prominin-1) (50,51), which has also been used as a
selection marker for neural stem cells (51). In this study, we demonstrated that
miR-186 inhibited CD133 expression. Moreover, integrinα6 is
co-expressed with conventional GIC markers (41); STAT3 is required for maintenance
of multipotency in GICs (42).
Herein, we observed that the over-expression of miR-186
significantly inhibited integrinα6 and STAT3 protein expression in
the U87MG-CR cells.
YY1 plays an important role in the EGFR-Src-p38
signaling cascade in glioblastoma. However, its roles and
regulatory mechanisms have not yet been fully elucidated. EGFR
signaling plays an important role in drug resistance for the
treatment of glioblastoma (52)
and EGFR inhibitor can enhance cisplatin sensitivity of human
glioma cells (52). We
demonstrated herein that YY1 expression was increased in
cisplatin-resistant U87MG cells and that the silencing of YY1
sensitized the U87MG-CR cells to cisplatin. In addition, we
observed that YY1 expression was regulated by miR-186 in U87MG-CR
cells.
Recently, the U-87 MG cell line from ATCC was
reported to be contaminated or misidentified (28). It has been proposed as a
glioblastoma cell line whose origin is unknown (28). However, the U-87 MG cell line is
still widely used for glioblastoma research (25). In the present study, the U87MG and
LN-229 cells were used. The results were same from the 2 cell
lines. Thus, the contamination or misidentification may not affect
the conclusions presented herein.
In conclusion, elucidating the mechanisms through
which miR-186 reverses cisplatin resistance and inhibits the
formation of the GIC phenotype by degrading YY1 in glioblastoma may
enhance our understanding of the molecular mechanisms of cisplatin
resistance in glioblastoma. As shown by our findings, the
restoration of miR-186 expression may represent a promising
therapeutic strategy with which to inhibit YY1-mediated cisplatin
resistance. However, the roles of miR-186 and YY1 require further
confirmation by in vivostudies.
Acknowledgments
Not applicable.
Funding
The present study was supported by Linyi People’s
Hospital.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
JL and FG conceived the study, collected the
experimental data and wrote a draft of the manuscript. JS
contributed to the experimental work and data analysis. All authors
edited and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Buckner JC: Factors influencing survival
in high-grade gliomas. Semin Oncol. 30:10–14. 2003. View Article : Google Scholar
|
|
2
|
Curran WJ Jr, Scott CB, Horton J, Nelson
JS, Weinstein AS, Fischbach AJ, Chang CH, Rotman M, Asbell SO,
Krisch RE, et al: Recursive partitioning analysis of prognostic
factors in three Radiation Therapy Oncology Group malignant glioma
trials. J Natl Cancer Inst. 85:704–710. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeAngelis LM: Brain tumors. N Eng J Med.
344:114–123. 2001. View Article : Google Scholar
|
|
4
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bao S, Wu Q, Sathornsumetee S, Hao Y, Li
Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD and Rich JN: Stem
cell-like glioma cells promote tumor angiogenesis through vascular
endothelial growth factor. Cancer Res. 66:7843–7848. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeppernick F, Ahmadi R, Campos B, Dictus
C, Helmke BM, Becker N, Lichter P, Unterberg A, Radlwimmer B and
Herold-Mende CC: Stem cell marker CD133 affects clinical outcome in
glioma patients. Clin Cancer Res. 14:123–129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi Y, Seto E, Chang LS and Shenk T:
Transcriptional repression by YY1, a human GLI-Krüippel-related
protein, and relief of repression by adenovirus E1A protein. Cell.
67:377–388. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park K and Atchison ML: Isolation of a
candidate repressor/activator, NF-E1 (YY-1, delta), that binds to
the immunoglobulin kappa 3′enhancer and the immunoglobulin
heavy-chain mu E1 site. Proc Natl Acad Sci USA. 88:9804–9808. 1991.
View Article : Google Scholar
|
|
10
|
Hariharan N, Kelley DE and Perry RP:
Delta, a transcription factor that binds to downstream elements in
several polymerase II promoters, is a functionally versatile zinc
finger protein. Proc Natl Acad Sci USA. 88:9799–9803. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Flanagan JR, Becker KG, Ennist DL, Gleason
SL, Driggers PH, Levi BZ, Appella E and Ozato K: Cloning of a
negative transcription factor that binds to the upstream conserved
region of Moloney murine leukemia virus. Mol Cell Biol. 12:38–44.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thomas MJ and Seto E: Unlocking the
mechanisms of transcription factor YY1: Are chromatin modifying
enzymes the key? Gene. 236:197–208. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi Y, Lee JS and Galvin KM: Everything
you have ever wanted to know about Yin Yang 1…. Biochim Biophys
Acta. 1332:F49–F66. 1997.PubMed/NCBI
|
|
14
|
Iuchi S and Kuldell N: Zinc Finger
Proteins: From Atomic Contact to Cellular Function. Springer
Science Business Media; New York, NY: 2007
|
|
15
|
Gordon S, Akopyan G, Garban H and Bonavida
B: Transcription factor YY1: Structure, function, and therapeutic
implications in cancer biology. Oncogene. 25:1125–1142. 2006.
View Article : Google Scholar
|
|
16
|
Wilkinson FH, Park K and Atchison ML:
Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc
Natl Acad Sci USA. 103:19296–19301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thomassen M, Tan Q and Kruse TA: Gene
expression meta-analysis identifies metastatic pathways and
transcription factors in breast cancer. BMC Cancer. 8:3942008.
View Article : Google Scholar
|
|
18
|
Wan M, Huang W, Kute TE, Miller LD, Zhang
Q, Hatcher H, Wang J, Stovall DB, Russell GB, Cao PD, et al: Yin
Yang 1 plays an essential role in breast cancer and negatively
regulates p27. Am J Pathol. 180:2120–2133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chinnappan D, Xiao D, Ratnasari A, Andry
C, King TC and Weber HC: Transcription factor YY1 expression in
human gastrointestinal cancer cells. Int J Oncol. 34:1417–1423.
2009.PubMed/NCBI
|
|
20
|
Kang W, Tong J, Chan A, Zhao J, Dong Y,
Wang S, Yang W, Sin FM, Ng SS, Yu J, et al: Yin Yang 1 contributes
to gastric carcinogenesis and its nuclear expression correlates
with shorter survival in patients with early stage gastric
adenocarcinoma. J Transl Med. 12:802014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Seligson D, Horvath S, Huerta-Yepez S,
Hanna S, Garban H, Roberts A, Shi T, Liu X, Chia D, Goodglick L and
Bonavida B: Expression of transcription factor Yin Yang 1 in
prostate cancer. Int J Oncol. 27:131–141. 2005.PubMed/NCBI
|
|
22
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gal H, Pandi G, Kanner AA, Ram Z,
Lithwick-Yanai G, Amariglio N, Rechavi G and Givol D: MIR-451 and
Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells.
Biochem Biophys Res Commun. 376:86–90. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ruan T, He X, Yu J and Hang Z:
MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo
signaling and tumorigenesis in hepatocellular carcinoma. Oncol
Lett. 11:2941–2945. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun KX, Jiao JW, Chen S, Liu BL and Zhao
Y: MicroRNA-186 induces sensitivity of ovarian cancer cells to
paclitaxel and cisplatin by targeting ABCB1. J Ovarian Res.
8:802015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang TJ, Wang YX, Yang DQ, Yao DM, Yang
L, Zhou JD, Deng ZQ, Wen XM, Guo H, Ma JC, et al: Down-regulation
of miR-186 correlates with poor survival in de novo acute myeloid
leukemia. Clin Lab. 62:113–120. 2015.
|
|
27
|
Wang F, Jiang H, Wang S and Chen B: Dual
functional MicroRNA-186-5p targets both FGF2 and RelA to suppress
tumorigenesis of glioblastoma multiforme. Cell Mol Neurobiol.
37:1433–1442. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M,
Zhang W, Chen W, Pan C, Liu Q, et al: miR-200b and miR-15b regulate
chemotherapy-induced epithelial-mesenchymal transition in human
tongue cancer cells by targeting BMI1. Oncogene. 31:432–445. 2012.
View Article : Google Scholar
|
|
30
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor α mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942. 2014.
View Article : Google Scholar
|
|
31
|
Xiang Y, Lu DL, Li JP, Yu CX, Zheng DL,
Huang X, Wang ZY, Hu P, Liao XH and Zhang TC: Myocardin inhibits
estrogen receptor alpha-mediated proliferation of human breast
cancer MCF-7 cells via regulating MicroRNA expression. IUBMB Life.
68:477–487. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kataoka J, Shiraha H, Horiguchi S,
Sawahara H, Uchida D, Nagahara T, Iwamuro M, Morimoto H, Takeuchi
Y, Kuwaki K, et al: Loss of Runt-related transcription factor 3
induces resistance to 5-fluorouracil and cisplatin in
hepatocellular carcinoma. Oncol Rep. 35:2576–2582. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ali S, Ahmad A, Banerjee S, Padhye S,
Dominiak K, Schaffert JM, Wang Z, Philip PA and Sarkar FH:
Gemcitabine sensitivity can be induced in pancreatic cancer cells
through modulation of miR-200 and miR-21 expression by curcumin or
its analogue CDF. Cancer Res. 70:3606–3617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kong D, Li Y, Wang Z, Banerjee S, Ahmad A,
Kim HR and Sarkar FH: miR-200 regulates PDGF-D-mediated
epithelial-mesenchymal transition, adhesion, and invasion of
prostate cancer cells. Stem Cells. 27:1712–1721. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
36
|
Jia Y, Chen J, Zhu H, Jia ZH and Cui MH:
Aberrantly elevated redox sensing factor Nrf2 promotes cancer stem
cell survival via enhanced transcriptional regulation of ABCG2 and
Bcl-2/Bmi-1 genes. Oncol Rep. 34:2296–2304. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu J, Ryan DG, Getsios S,
Oliveira-Fernandes M, Fatima A and Lavker RM: MicroRNA-184
antagonizes microRNA-205 to maintain SHIP2 levels in epithelia.
Proc Natl Acad Sci USA. 105:19300–19305. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Komatsu M, Sumizawa T, Mutoh M, Chen ZS,
Terada K, Furukawa T, Yang XL, Gao H, Miura N, Sugiyama T and
Akiyama S: Copper-transporting P-type adenosine triphosphatase
(ATP7B) is associated with cisplatin resistance. Cancer Res.
60:1312–1316. 2000.PubMed/NCBI
|
|
39
|
Kondo S, Barnett GH, Hara H, Morimura T
and Takeuchi J: MDM2 protein confers the resistance of a human
glioblastoma cell line to cisplatin-induced apoptosis. Oncogene.
10:2001–2006. 1995.PubMed/NCBI
|
|
40
|
Brescia P, Ortensi B, Fornasari L, Levi D,
Broggi G and Pelicci G: CD133 is essential for glioblastoma stem
cell maintenance. Stem Cells. 31:857–869. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lathia JD, Gallagher J, Heddleston JM,
Wang J, Eyler CE, Macswords J, Wu Q, Vasanji A, McLendon RE,
Hjelmeland AB and Rich JN: Integrin alpha 6 regulates glioblastoma
stem cells. Cell Stem Cell. 6:421–432. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sherry MM, Reeves A, Wu JK and Cochran BH:
STAT3 is required for proliferation and maintenance of multipotency
in glioblastoma stem cells. Stem Cells. 27:2383–2392. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He W, Feng J, Zhang Y, Wang Y, Zang W and
Zhao G: MicroRNA-186 inhibits cell proliferation and induces
apoptosis in human esophageal squamous cell carcinoma by targeting
SKP2. Lab Invest. 96:317–324. 2016. View Article : Google Scholar
|
|
44
|
Brandes AA, Basso U, Reni M, Vastola F,
Tosoni A, Cavallo G, Scopece L, Ferreri AJ, Panucci MG, Monfardini
S, et al: First-line chemotherapy with cisplatin plus fractionated
temozolomide in recurrent glioblastoma multiforme: A phase II study
of the Gruppo Italiano Cooperativo di Neuro-Oncologia. J Clin
Oncol. 22:1598–1604. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Timmer-Bosscha H, Mulder NH and de Vries
E: Modulation of cis-diamminedichloroplatinum(II) resistance: A
review. Br J Cancer. 66:227–238. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen W, Yang Y, Chen B, Lu P, Zhan L, Yu
Q, Cao K and Li Q: miR-136 targets E2F1 to reverse cisplatin
chemosensitivity in glioma cells. J Neurooncol. 120:43–53. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Q, Wang Z, Chu L, Li X, Kan P, Xin X,
Zhu Y and Yang P: The effects and molecular mechanisms of miR-106a
in multidrug resistance reversal in human glioma U87/DDP and U251/G
cell lines. PLoS One. 10:e01254732015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Eramo A, Ricci-Vitiani L, Zeuner A,
Pallini R, Lotti F, Sette G, Pilozzi E, Larocca LM, Peschle C and
De Maria R: Chemotherapy resistance of glioblastoma stem cells.
Cell Death Differ. 13:1238–1241. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Terada K, Schilsky ML, Miura N and
Sugiyama T: ATP7B (WND) protein. Int J Biochem Cell Biol.
30:1063–1067. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bidlingmaier S, Zhu X and Liu B: The
utility and limitations of glycosylated human CD133 epitopes in
defining cancer stem cells. J Mol Med (Berl). 86:1025–1032. 2008.
View Article : Google Scholar
|
|
51
|
Uchida N, Buck DW, He D, Reitsma MJ, Masek
M, Phan TV, Tsukamoto AS, Gage FH and Weissman IL: Direct isolation
of human central nervous system stem cells. Proc Natl Acad Sci USA.
97:14720–14725. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang Y, Xing X, Zhan H, Li Q, Fan Y, Zhan
L, Yu Q and Chen J: EGFR inhibitor enhances cisplatin sensitivity
of human glioma cells. J Huazhong Univ Sci Technol Med Sci.
31:773–778. 2011. View Article : Google Scholar : PubMed/NCBI
|