Introduction
Type 2 diabetes mellitus (T2DM) accounts for ~90% of
all DM cases (1), a global
epidemic posing a serious health burden to society (2). Insulin resistance is the primary
cause of T2DM, and has therefore become the primary target of T2DM
treatment (3,4). It is well known that the liver is
the primary glycogen storage site and serves a central role in
glucose homeostasis (5,6). Once glucose homeostasis is
disrupted, additional insulin is secreted to control the elevated
glucose levels, further promoting insulin resistance (7). However, the cellular and molecular
mechanisms responsible for hepatic insulin resistance are not yet
known.
Long noncoding RNAs (lncRNAs) are commonly defined
as non-protein-coding RNA transcripts measuring >200 nucleotides
(8). Previous studies have
indicated that lncRNAs are involved in the regulation of hepatic
glucose and lipid metabolism, with dysregulated lncRNA expression
potentially associated with T2DM pathogenesis (9,10).
One particular lncRNA, maternally expressed gene 3 (MEG3), is an
imprinted gene belonging to the imprinted delta like non-canonical
notch ligand 1 (DLK1)-MEG3 locus located on human chromosome
14q32.3 (11). In mice, MEG3 is
referred to as gene trap locus 2 and is located on distal
chromosome 12 (11). MEG3 has
been implicated as a putative tumour suppressor due to the loss of
MEG3 expression in a range of cancer types, and its ability to
inhibit tumour cell proliferation (11). Our previous study demonstrated
that MEG3 overexpression significantly enhanced hepatic insulin
resistance via increased forkhead box protein O1 (FoxO1) expression
(5), which is a key transcription
factor in regulating hepatic gluconeogenesis (12). However, the mechanisms by which
MEG3 increases FoxO1 expression remain unclear.
lncRNAs may serve as competing endogenous RNAs
(ceRNAs) that either segregate or competitively bind microRNAs
(miRNAs) to regulate target mRNA expression (13). miRNAs are a class of small RNA
molecules measuring 18-24 nucleotides that regulate gene expression
post-transcriptionally via mRNA destabilization or translational
repression (14). Fan et
al (15) revealed that
miR-214 is a direct target of MEG3. An additional study
demonstrated that miR-214 suppressed gluconeogenesis by targeting
activating transcription factor 4 (ATF4), a potential coactivator
of FoxO1 in the regulation of gluconeogenesis (16). Li et al (17) stated that ATF4 deficient
(ATF4−/−) mice exhibited decreased levels of fasting
blood glucose compared with the wild-type (ATF4+/+) mice
and that ATF4 served a role in high- carbohydrate diet-induced
insulin resistance.
Increased FoxO1 activity resulted in hyperglycaemia-
associated insulin resistance by promoting the transcription of two
key gluconeogenic enzymes, namely glucose-6-phosphatase catalytic
subunit (G6pc) and phosphoenolpyruvate carboxykinase (Pepck)
(18,19). G6pc and Pepck are rate-limiting
enzymes that are highly upregulated during fasting, but suppressed
in the fed state and by insulin (5). Based on these aforementioned
studies, we hypothesized that MEG3 serves as a ceRNA of miR-214 to
facilitate ATF4 expression, resulting in the promotion of FoxO1
expression and its downstream gluconeogenic enzymes G6pc and Pepck,
thereby increasing gluconeogenesis and promoting insulin
resistance. To address this, the present study evaluated the
expression of MEG3, miR-214 and ATF4 in ob/ob and high-fat diet
(HFD)-fed mice using reverse transcription quantitative polymerase
chain reaction (RT-qPCR) and western blot analysis.
Leptin-deficient ob/ob mice are overweight, develop insulin
resistance, and serve as a model for T2DM. Furthermore, their
interactions were examined using the luciferase reporter assay and
their effects on insulin resistance in T2DM were also investigated.
The results of the present study demonstrated that MEG3 promoted
hepatic insulin resistance by serving as a ceRNA of miR-214 to
facilitate ATF4 expression.
Materials and methods
Animals and treatment
Male C57BL/6 (3-5-week-old) leptin-deficient ob/ob
(T2DM model), and control mice (8 weeks) were obtained from Jackson
Laboratory (Bar Harbor, ME, USA). All animals were housed under
controlled temperatures (25±1°C) and humidity (50%), with 12 h
light-dark cycles and ad libitum access to food and water.
All animal experiments were approved by the Ethics Committees of
the First Affiliated Hospital of University of Science and
Technology of China (Hefei, China). All experimental procedures
were performed in strict accordance with the Institutional Animal
Care and Use Committees of Anhui Provincial Hospital and the First
Affiliated Hospital of University of Science and Technology of
China.
To induce insulin resistance, four-week-old male
C57BL/6 mice were fed an HFD (containing 45 kcal% fat; cat. no.,
D12451; Research Diets; New Brunswick, NJ, USA) or low-fat diet
(LFD; containing 10 kcal% fat) for 8 weeks. Next, liver tissues
were isolated and examined for the expression of MEG3, miR-214, and
ATF4 using RT-qPCR and western blot analysis. HFD-fed male C57BL/6
mice also received an 800 µl injection of the small
interfering (si)-MEG3 fragment (si-MEG), described subsequently, in
PBS (100 µg/ml) via the tail vein twice a week for 10 weeks.
At week 10, glucose tolerance tests (GTTs) and insulin tolerance
tests (ITTs) were performed. A summary of the experimental model is
presented in Fig. 1.
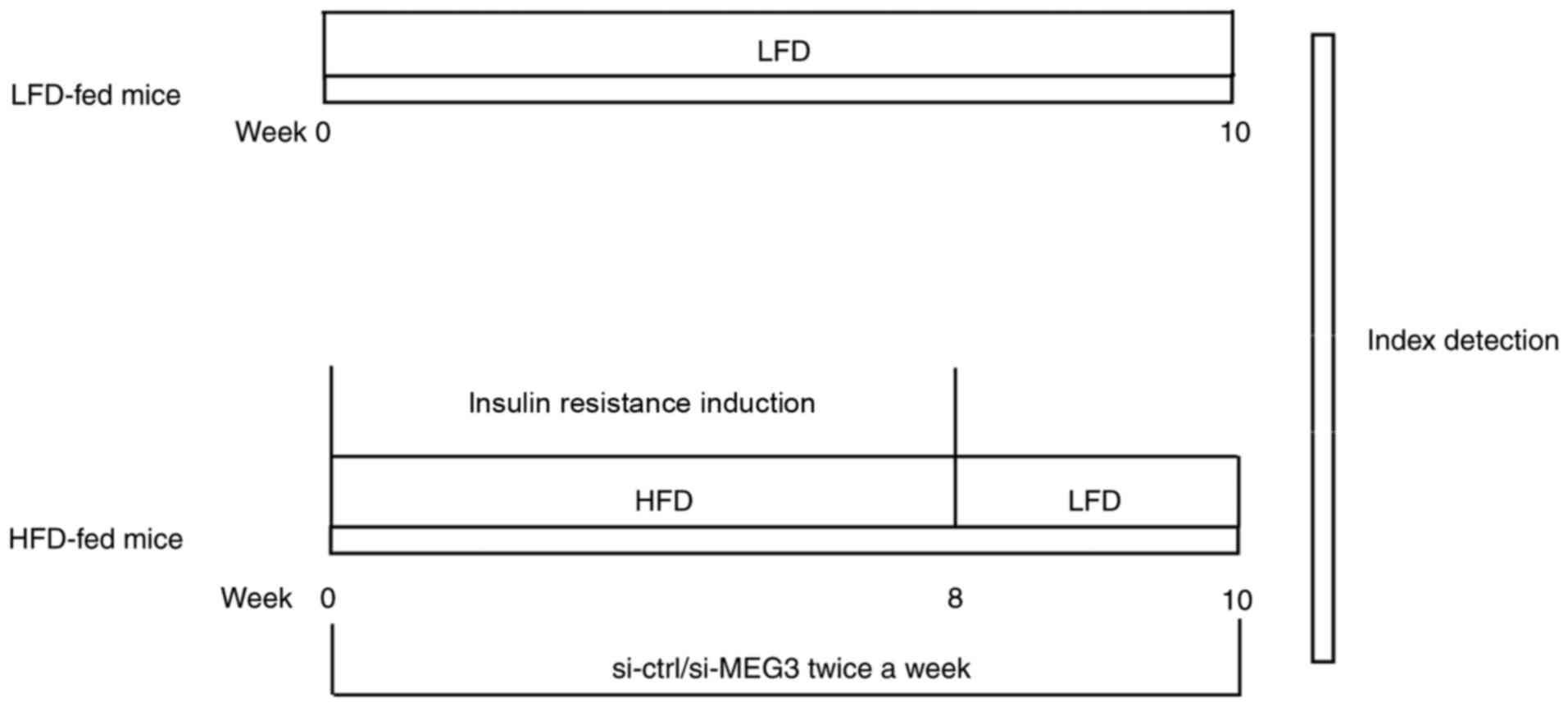 | Figure 1Summary of the experimental animal
model. Mice in the HFD group were fed HFD for 8 weeks from weeks
0-8 to induce insulin resistance. Mice in the HFD group also
received an injection of the si-MEG3 fragment via the tail vein
twice a week for 10 weeks from weeks 0-10. In parallel experiments,
mice in the LFD group were fed LFD for 10 weeks from weeks 0-10. At
week 10, GTTs and ITTs were performed. The relative expression of
MEG3, miR-214, and ATF4 in mouse liver tissues was evaluated by
reverse transcription-quantitative polymerase chain reaction. ATF4
expression in mouse liver tissues was measured by western blotting.
LFD, low-fat diet; HFD, high-fat diet; si, small interfering RNA;
ctrl, control; MEG3, maternally expressed gene 3; GTTs, glucose
tolerance tests; ITTs, insulin tolerance tests. |
GTTs and ITTs
For GTTs, mice received an intraperitoneal (IP)
injection of D-glucose (2 g/kg) following overnight fasting, as
described previously (20). For
ITTs, mice received an IP injection of 0.75 units/kg insulin, and
insulin levels were measured via a radioimmunoassay (RIA) using an
ultrasensitive rat insulin RIA kit (100% cross-reactivity with
mouse insulin; Linco; EMD Millipore, Billerica, MA, USA) following
4 h of fasting. Blood glucose levels were measured using a Bayer
Glucometer Elite monitor (Bayer AG, Leverkusen, Germany).
Plasmid construction and cell
transfection
To overexpress MEG3 or ATF4, the full-length MEG3 or
ATF4 cDNA fragments (20 ng; Shanghai GenePharma Co., Ltd.,
Shanghai, China) were cloned into the pcDNA 3.1 plasmid
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
generating pcDNA3.1-MEG3 or pcDNA3.1-ATF4 plasmids. An empty
pcDNA3.1 vector was used as the control. Primary hepatocytes were
transfected with these constructs using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), following the
manufacturer’s protocol. To knock down MEG3 expression, small
interfering RNA si-MEG3 (5′-AUU GGA GGU GAG GAA GGA AAG CAG C-3′)
was designed and synthesized by Shanghai GenePharma Co., Ltd. A
scramble siRNA (5′-UUC UCC GAA CGU GUC ACG UTT-3′) was used as the
negative control (si-Ctrl). Primary hepatocytes were transfected
with 50 nM siRNAs using Lipofectamine® 2000, according
to the manufacturer’s protocol.
The miRNA double-stranded mimics for miR-214 or
miR-214 inhibitors were purchased from Shanghai GenePharma Co.,
Ltd. When cells grew to 80-90% confluence, miR-214 mimics
(sequence: 5′-ACA GCA GGC ACA GAC AGG CAG U-3′) were transfected
into primary mouse hepatocytes using Lipofectamine™ RNAiMAX
Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
in 12-well plates at a dose of 0.5 ng/well.
Following transfection for 48 h, primary hepatocytes
were harvested for RT-qPCR analysis to examine the knockdown or
overexpression efficiency.
Isolation and culture of mouse primary
hepatocytes
Hepatocytes were isolated from male C57BL/6 mice
using the two-step collagenase perfusion method, as previously
described (21). Isolated
hepatocytes were cultured for 5 days in medium 199 (Gibco; Thermo
Fisher Scientific, Inc.), as previously described (22). Prior to all experiments,
hepatocytes were serum starved for 5 h. Cells were stimulated with
either 0.5 mM palmitate (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) or transfected with the DNA plasmids.
RNA extraction and RT-qPCR
RT-qPCR was used to detect the relative expression
of MEG3, miR-214, ATF4, FoxO1, G6pc and Pepck in mouse liver
tissues or primary mouse hepatocytes, as described in our previous
study (5). In brief, total RNA
was extracted from liver tissues or primary hepa-tocytes using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer’s protocol. Total RNA was then
reverse transcribed using the iScript kit (Bio-Rad Laboratories,
Inc., Hercules, CA, USA), and qPCR reactions were performed on the
CFX96™ real-time PCR detection system (Bio-Rad Laboratories, Inc.)
using Power SYBR-Green RT-qPCR reagents. PCR assays were performed
using a TaqMan Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The primers used were as follows: MEG3 forward
(F), 5′-CTG CCC ATC TAC ACC TCA CG-3′; reverse (R), 5′-CTC TCC GCC
GTC TGC GCT AGG GGC T-3′; miR-214 F, 5′-GCA CAG CAG GCA CAG ACA-3′;
miR-214 R, 5′-CAG AGC AGG GTC AGC GGT A-3′; ATF4 F, 5′-CCT GAA CAG
CGA AGT GTT GG-3′; ATF4 R, 5′-TGG AGA ACC CAT GAG GTT TCA A-3′;
FoxO1 F, 5′-AAG AGG CTC ACC CTG TCG C-3′; FoxO1 R, 5′-GCA TCC ACC
AAG AAC TTT TCC-3′; G6pc F, 5′-GTG CAG CTG AAC GTC TGT CTG TC-3′;
G6pc R, 5′-TCC GGA GGC TGG CAT TGT A-3′; Pepck F, 5′-CTT CTC TGC
CAA GGT CAT CC-3′; Pepck R, 5′-TTT TGG GGA TGG GCA C-3′; GAPDH F,
5′-ACC ACA GTC CAT GCC ATC AC-3′; GAPDH R, 5′-TCC ACC ACC CTG TTG
CTG TA-3′; 18S F, 5′-AGG GTT CGA TTC CGG AGA GG-3′; 18S R, 5′-CAA
CTT TAA TAT ACG CTA T TG G-3′. The thermocycling conditions used
were as follows: Initial denaturation at 95°C for 3 min; followed
by 40 cycles of denaturation at 95°C for 5 sec, annealing at 60°C
for 30 sec and extension at 72°C for 30 sec, and a final extension
of 72°C for 5 min. The relative expression levels of miR-214 and
MEG-3 were normalized to 18S, while ATF4, FoxO1, G6pc and Pepck
were normalized to GAPDH expression. The 2−ΔΔCq method
was used to detect mRNA expression levels (23).
Western blot analysis
Western blot analysis was used to detect ATF4,
FoxO1, G6pc and Pepck protein expression in mouse liver tissues or
primary mouse hepatocytes. Separated liver tissues or primary
hepatocytes were lysed in ice-cold radioimmunoprecipitation assay
lysis buffer (Beyotime Institute of Biotechnology, Haimen, China),
supplemented with protease inhibitors, for 60 min to extract total
liver or cell proteins. Protein concentrations were quantified
using a Bio-Rad protein assay with a Bio-Rad Protein assay kit
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Following
centrifugation at 12,000 x g for 5 min at 4°C, an equal amount (20
µg/lane) of protein from each lysate was separated by 10%
SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF)
membranes (Bio-Rad Laboratories, Inc.). The PVDF membranes were
then blocked with 5% fat-free milk for 1 h at room temperature and
sequentially incubated at 4°C overnight with the corresponding
primary antibodies against ATF4 (1:1,000; cat. no. ab23760; Abcam,
Cambridge, MA, USA), FoxO1 (1:1,000; cat. no. ab39670; Abcam),
Pepck (1:200; cat. no. sc-377027; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), and G6pc (1:1,000; cat. no. TA334651; OriGene
Technologies, Inc., Rockville, MD, USA). Subsequent to washing
three times with TBS + 0.1% Tween, the membranes were then
incubated at room temperature with appropriate horseradish
peroxidase-coupled secondary antibody (1:1,000; cat. no. NA931-1ML;
GE Healthcare, Chicago, IL, USA) for 1 h. Blots were developed
using an enhanced chemiluminescence kit (Pierce; Thermo Fisher
Scientific, Inc.) and band intensity was quantified with Quantity
One software (4.62 version; Bio-Rad Laboratories, Inc.). β-actin
served as a loading control.
Luciferase activity assay
The luciferase activity assay was performed as
previously described (13), with
minor alterations. Fragments of MEG3 and the 3′-untranslated region
(UTR) of ATF4, containing the predicted wild-type (Wt) or mutated
(Mut) binding sites of miR-214, were amplified from mouse genomic
DNA by PCR using DreamTaq DNA Polymerase (Thermo Fisher Scientific,
Inc.). The primers were as follows: ATF4-Wt 3′UTR construct primer
F, 5′-CCA GAA TTC GTA GTC AGG TGC TTT GTG-3′; ATF4-Wt 3′UTR
construct primer R, 5′-CCA CTC GAG AAC ATC ACT TTC ATG GA-3′;
ATF4-Mut primer F, 5′-AGT CTT GTG TTG CTG TGT TTG CTG TAA TAA ATT
ATT TTG TAG T-3′; ATF4-Mut primer R, 5′-ACT ACA AAA TAA TTT ATT ACA
GCA AAC ACA GCA ACA CAA GAC T-3′; MEG3 construct primer F, 5′-CCA
GAA TTC AGG ATG GCA AAG GAT GAA-3′; MEG3 construct primer R, 5′-CCA
CTC GAG TTT AGG GAG GTG TCG GTG-3′; MEG3 Mut primer F, 5′-CTT CTT
GAA AGG CCT GTC TAC ACT TGC TGT CTT CCT TCC TCA CCT CCA ATT TCC
TC-3′; and MEG3 Mut primer R, 5′-GAG GAA ATT GGA GGT GAG GAA GGA
AGA CAG CAA GTG TAG ACA GGC CTT TCA AGA AG-3′. The thermocycling
conditions used were as follows: Initial denaturation at 95°C for 2
min, followed by 40 cycles of denaturation at 95°C for 30 sec,
annealing at 60°C for 30 sec and extension at 72°C for 1 min, and a
final extension at 72°C for 5 min. The PCR products were then
cloned into a pMIR-REPORT lucif-erase reporter vector (Ambion;
Thermo Fisher Scientific, Inc.). The constructs were termed
MEG3-Wt, MEG3-Mut, ATF4-Wt and ATF4-Mut constructs. The
corresponding mutants were generated using a QuikChange Site
Directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara,
CA, USA). For the luciferase reporter assay, 293 cells were
co-transfected with 200 ng luciferase reporter vector constructs,
25 ng pRL-TK (expressing Renilla luciferase as the internal
control), and 20 µM miR-214 mimic or miR-negative control
(NC), using Lipofectamine® 2000. A Dual-Luciferase
Reporter assay kit (Promega Corporation, Madison, WI, USA) was
applied to analyse luciferase activity at 24 h
post-transfection.
Statistical analysis
All statistical analyses were performed using the
SPSS statistical software package standard version 16.0 (SPSS,
Inc., Chicago, IL, USA). The data are presented as the mean ±
standard deviation from three independent experiments. Unpaired
Student’s t-tests were used to analyse differences between two
groups. A one-way analysis of variance followed by Tukey’s post hoc
test was used to analyse differences among multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
MEG3, miR-214 and ATF4 expression in
HFD-fed and ob/ob mice
As demonstrated in Fig. 2A and B, HFD-fed mice exhibited
increased MEG3 expression and decreased miR-214 expression in liver
tissues compared with LFD-fed mice. Furthermore, ATF4 mRNA and
protein expression levels in the liver tissues were significantly
increased in HFD-fed compared to LFD-fed mice. The expression
patterns of MEG-3, miR-214 and ATF4 in the liver tissues of ob/ob
mice, a model of severe obesity, insulin resistance and T2DM caused
by leptin deficiency, was consistent with those in HFD-fed mice
(Fig. 2C and D). Together,
HFD-fed and ob/ob mice exhibited upregulated MEG3 and ATF4
expression, but downregulated miR-214 expression compared with the
LFD-fed mice and control mice, respectively.
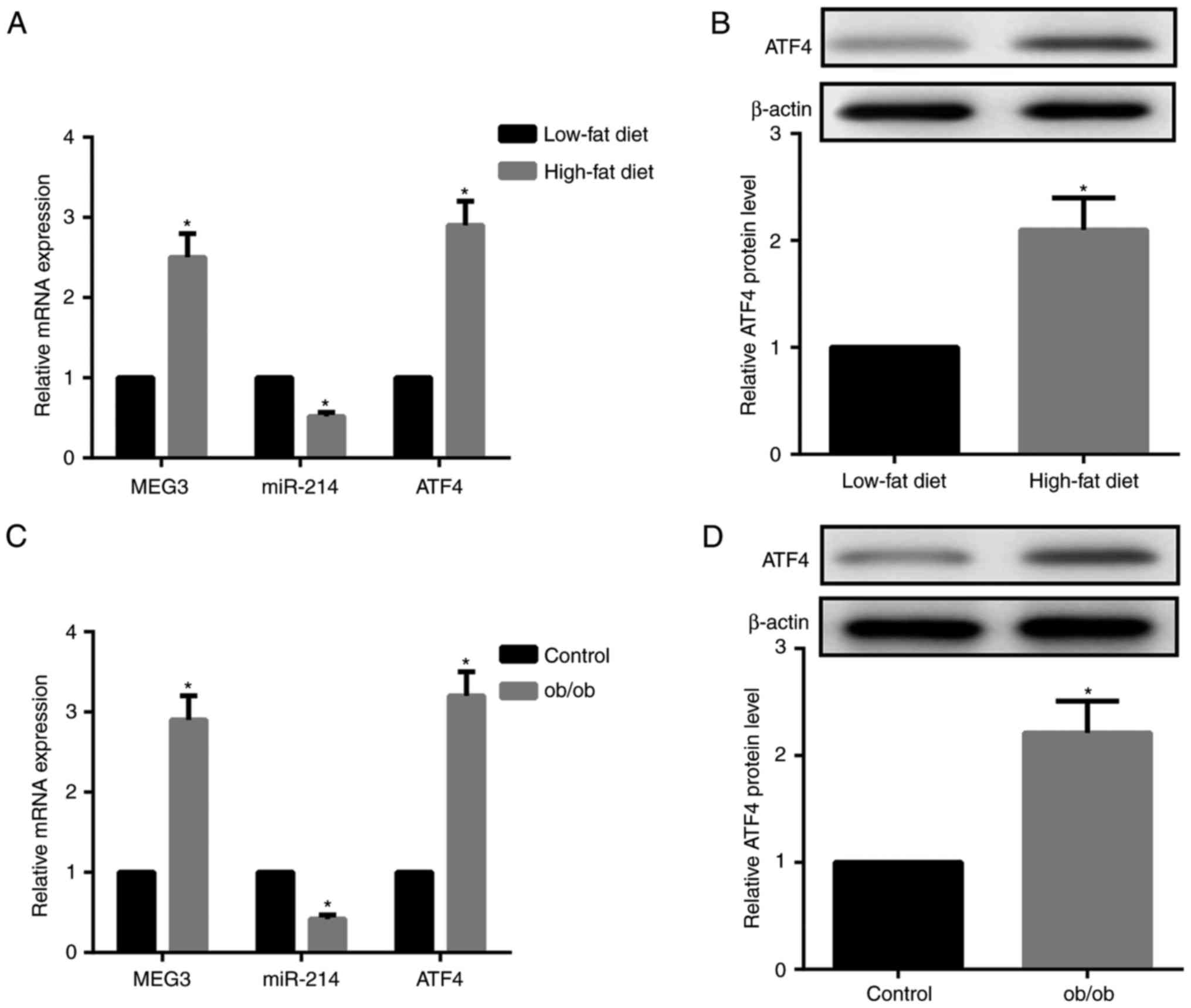 | Figure 2HFD-fed and ob/ob mice demonstrate
upregulated MEG3 and ATF4, downregulated miR-214. (A) Compared with
LFD-fed mice, RT-qPCR results indicated that MEG3 and ATF4 mRNA
expression were upregulated, while miR-214 expression was
downregulated, in liver tissues from HFD-fed mice. (B) Western blot
analysis revealed that ATF4 protein expression in the liver tissues
from HFD-fed mice was upregulated compared with LFD-fed mice. (C)
Compared with the control mice, RT-qPCR results demonstrated that
MEG3 and ATF4 mRNA expression was upregulated, while miR-214
expression was downregulated, in liver tissues from ob/ob mice. (D)
Western blot analysis revealed that ATF4 protein expression in
liver tissues from ob/ob mice was upregulated compared with control
mice. β-actin served as the loading control in western blots. N=10
for each group. *P<0.05 vs. the HFD or control
groups. LFD, low-fat diet; HFD, high-fat diet; miR, microRNA; MEG3,
maternally expressed gene 3; ATF4, activating transcription factor
4; RT-qPCR, reverse transcription quantitative polymerase chain
reaction. |
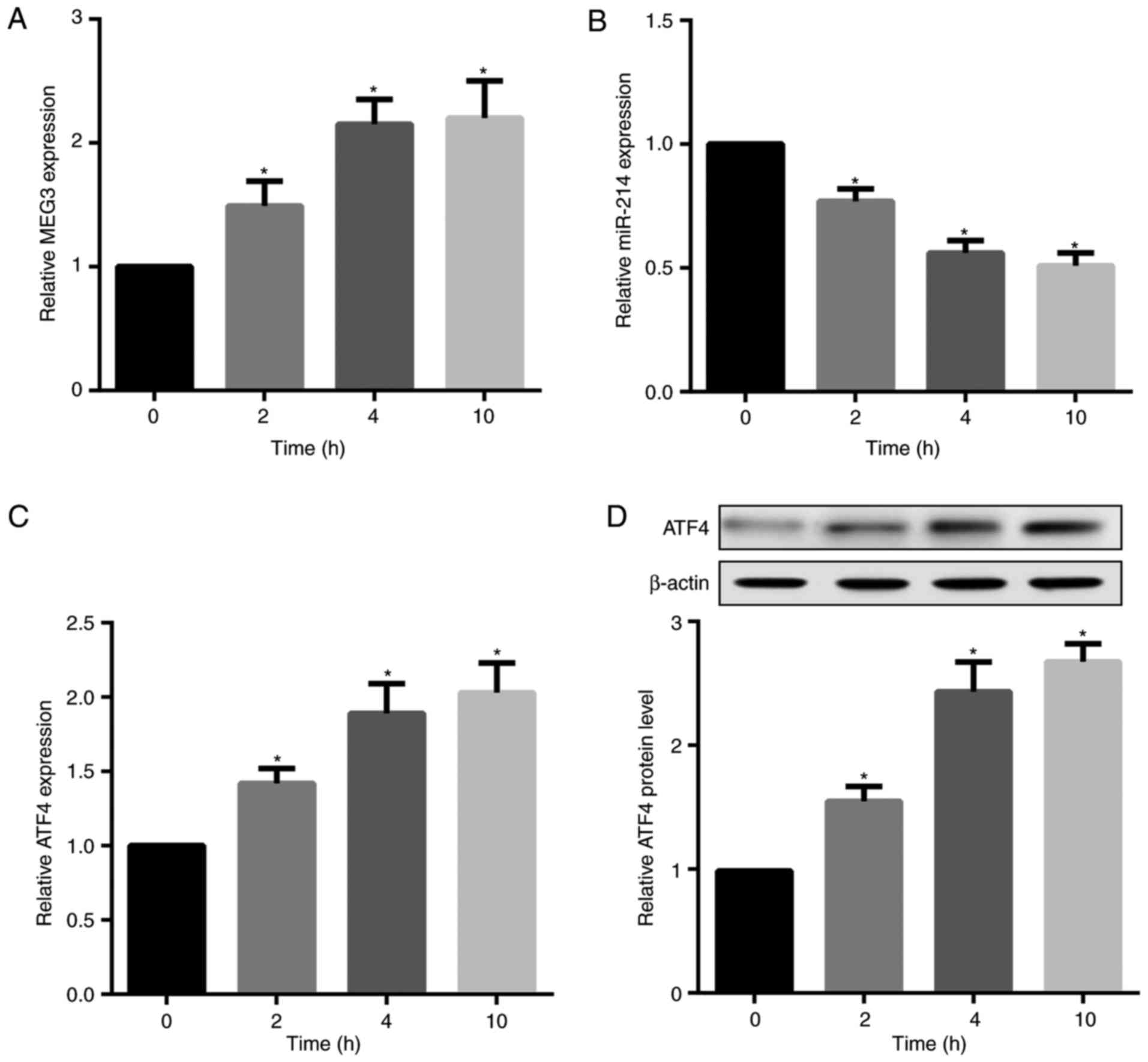
Palmitate time-dependently increases MEG3
and ATF4 but decreases miR-214 expression
Saturated fatty acid (SFA) has been indicated to
induce a proinflammatory response associated with obesity, T2DM,
insulin resistance and dyslipidaemia (24,25). Accordingly, the present study
investigated the in vitro effects of palmitate, a major SFA
in plasma, on MEG3, ATF4 and miR-214 expression in mouse primary
hepatocytes. The results revealed that palmitate treatment
time-dependently increased MEG3, but decreased miR-214 expression
in hepatocytes (Fig. 3A and B).
Furthermore, palmitate significantly increased ATF4 mRNA and
protein expression levels (Fig. 3C
and D).
MEG3 serves as a ceRNA of miR-214 to
facilitate ATF4 expression
To investigate the effects of MEG3 expression on
miR-214 and ATF4 expression, MEG3 was overexpressed or silenced by
transfecting hepatocytes with pcDNA3.1-MEG3 or si-MEG3,
respectively. The overexpression and knockdown efficiency of MEG3
was confirmed by RT-qPCR (Fig.
4A). MEG3 overexpression significantly decreased relative
miR-214 expression, but markedly increased ATF4 mRNA and protein
expression. By contrast, MEG3 knockdown exerted the opposite
effects on miR-214 and ATF4 expression (Fig. 4B-D). Next, the effects of miR-214
expression on MEG3 and ATF4 expression were assessed. The
overexpression and knockdown efficiency of miR-214 was also
confirmed by RT-qPCR (Fig. 4E).
Compared with their corresponding controls, hepatocytes transfected
with the miR-214 mimic demonstrated significantly decreased MEG3
and ATF4 expression, whereas miR-214 inhibitor-transfected
hepatocytes exhibited markedly increased MEG3 and ATF4 expression
(Fig. 4F–H).
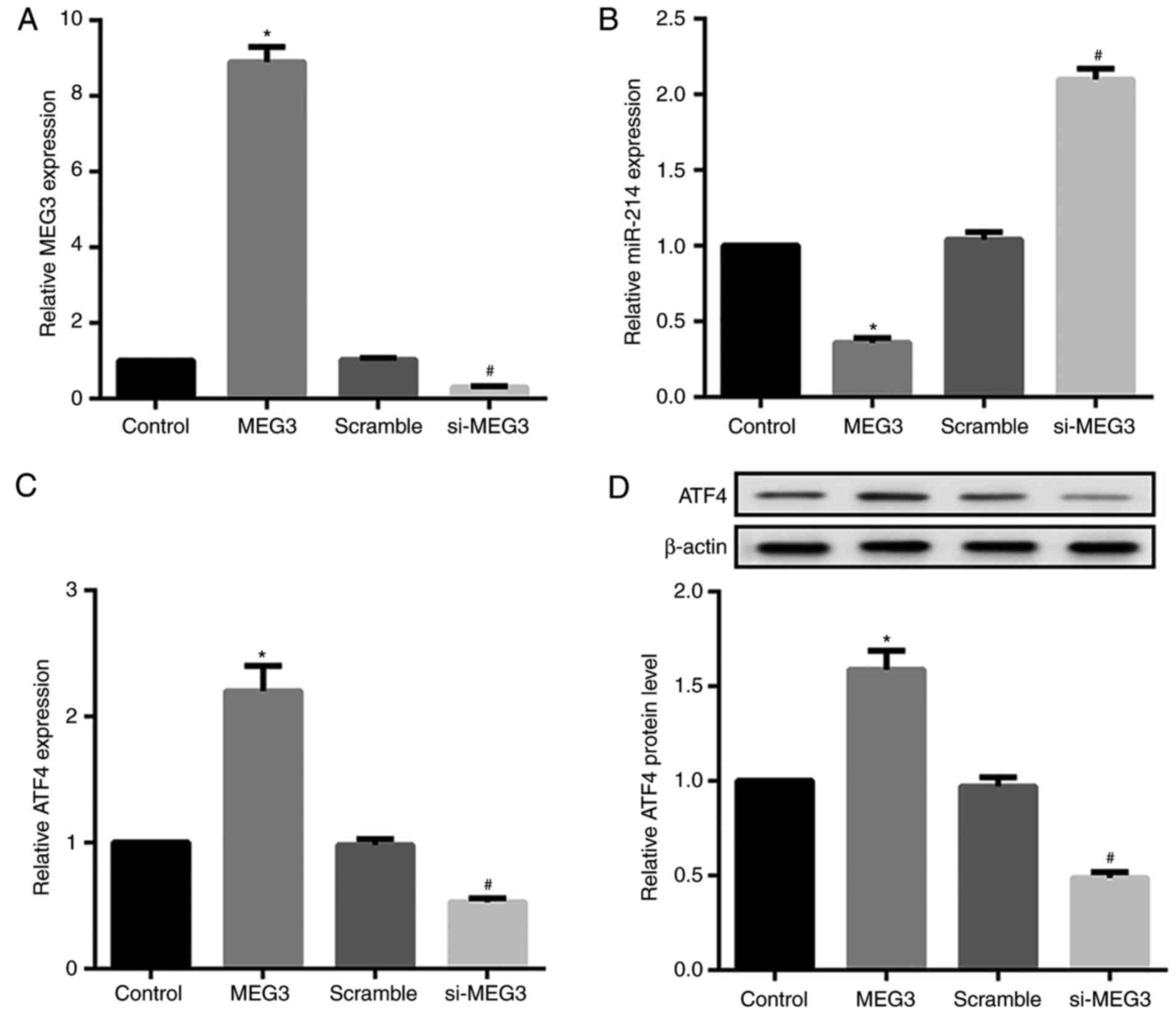 | Figure 4Effect of MEG3 and miR-214
overexpression and knockdown on ATF4 expression in primary
hepatocytes. Mouse primary hepatocytes were transfected with either
pcDNA3.1-MEG3, si-MEG3, and corresponding controls, or with miR-214
mimics/inhibitor and their controls. (A) The overexpression and
knockdown efficiency of MEG3 was confirmed by RT-qPCR. RT-qPCR and
western blot analysis results revealed that MEG3 overexpression
significantly decreased (B) relative miR-214 expression, but
markedly increased ATF4 (C) mRNA and (D) protein expression. MEG3
knockdown exerted the opposite effects. *P<0.05 vs.
NC or control groups. #P<0.05 vs. the scramble group.
MEG3, maternally expressed gene 3; ATF4, activating transcription
factor 4; si, small interfering RNA. Effect of MEG3 and miR-214
overexpression and knockdown on ATF4 expression in primary
hepatocytes. (E) RT-qPCR results indicated that the miR-214 mimic
increased miR-214 expression, whereas the miR-214 inhibitor
decreased miR-214 expression. RT-qPCR and western blot analysis
results revealed that the miR-214 mimic significantly decreased (F)
relative MEG3 expression, and ATF4 (G) mRNA and (H) protein
expression. The miR-214 inhibitor exerted the opposite effects.
β-actin served as the loading control in the western blot analyses.
*P<0.05 vs. NC or control groups.
#P<0.05 vs. the scramble group. miR, microRNA; NC,
negative control. |
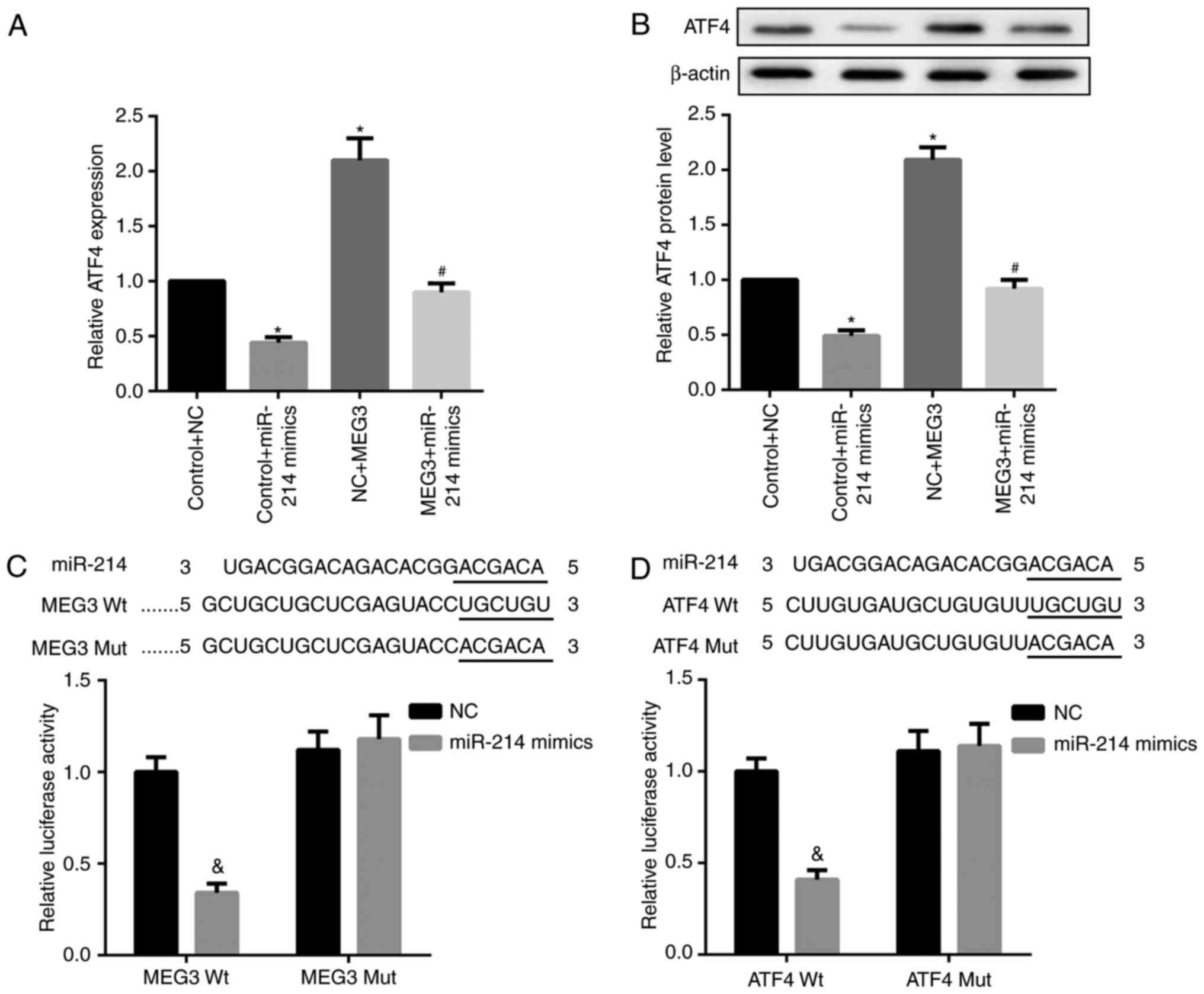
It is well-established that lncRNAs may function as
ceRNA by competitively binding miRNAs. Accordingly, whether MEG3
functioned similarly in regulating miR-214 was explored. Compared
with the corresponding controls, miR-214 mimics significantly
decreased ATF4 mRNA and protein expression, whereas MEG3
overexpression markedly increased ATF4 mRNA and protein expression
levels. Furthermore, the simultaneous overexpression of miR-214 and
MEG3 significantly decreased MEG3-mediated upregulation of ATF4
mRNA and protein expression (Fig. 5A
and B). In addition, the results of the luciferase reporter
assay indicated that the miR-214 mimic caused a marked decrease in
luciferase activity in the MEG3-Wt reporter group compared with the
miR-NC group. By contrast, the miR-214 mimic exhibited no obvious
effect on luciferase activity in the MEG3-Mut reporter group,
verifying the direct binding between MEG3 and miR-214 (Fig. 5C). In addition, decreased
luciferase activity in 293 cells co-transfected with WT ATF4 3′UTR
luciferase reporter plasmids and miR-214 mimics (Fig. 5D) was observed, suggesting the
3′UTR of ATF4 is directly targeted by miR-214. Collectively, these
results indicate that MEG3 serves as a ceRNA of miR-214 to
facilitate ATF4 expression.
miR-214 inhibition and ATF4
overexpression reverse the MEG3 knockdown-mediated decrease in
FoxO1, G6pc and Pepck expression
FoxO1 serves an important role in retinoid
metabolism within hepatic gluconeogenesis. FoxO1 is also a critical
regulator of hepatic glucose and lipid metabolism via its ability
to drive the transcription of two key gluconeogenic enzymes, G6pc
and Pepck (26). The results from
the present study revealed that palmitate significantly increased
FoxO1, Pepck and G6pc protein expression in primary hepatocytes
(Fig. 6A–D). Furthermore, MEG3
knockdown significantly suppressed the palmitate-mediated
upregulation of these 3 proteins (Fig. 6A–D), indicating an attenuation of
hepatocyte gluconeogenesis. By contrast, miR-214 inhibitor and ATF4
overexpression additionally induced the palmitate-mediated
upregulation of FoxO1, Pepck and G6pc (Fig. 6E–L). Similar results were observed
for FoxO1, Pepck, and G6pc mRNA expression (Fig. 7). These results indicated that
miR-214 inhibition and ATF4 overexpression significantly reversed
the MEG3 knockdown-mediated decrease in palmitate-induced FoxO1,
G6pc and Pepck protein expression.
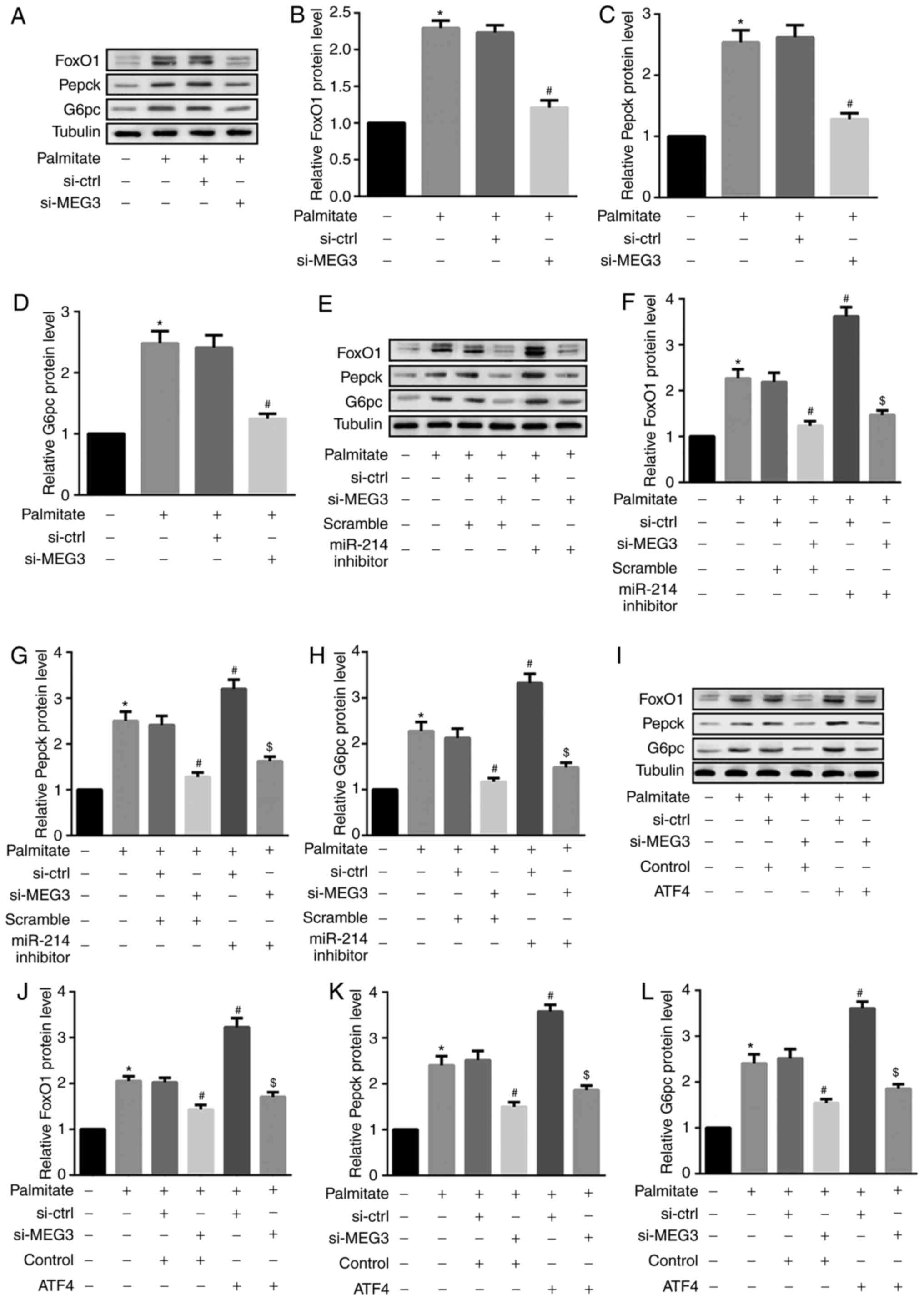 | Figure 6miR-214 inhibition and ATF4
overexpression reverse MEG3 knockdown-mediated decreases in FoxO1,
G6pc and Pepck protein expression. Cells were transfected with the
indicated plasmids, followed by treatment with 0.5 mM palmitate for
4 h. (A–D) Western blot analysis revealed that MEG3 knockdown
significantly suppressed the palmitate-mediated increase in (B)
FoxO1, (C) Pepck, and (D) G6pc protein expression. (E–H) By
contrast, the miR-214 inhibitor additionally induced the
palmitate-mediated increase in (F) FoxO1, (G) Pepck and (H) G6pc
protein expression. (I–L) ATF4 overexpression also induced the
palmitate-mediated increase in (J) FoxO1, (K) Pepck and (L) G6pc
protein expression. β-tubulin served as the loading control in the
western blot analyses. *P<0.05 vs. group 1;
#P<0.05 vs. group 3;$P<0.05 vs.
group 4. si, small interfering RNA; ctrl, control; MEG3, maternally
expressed gene 3; miR, microRNA; FoxO1, forkhead box protein O1;
Pepck, phosphoenolpyruvate carboxykinase; G6pc,
glucose-6-phosphatase catalytic subunit; ATF4, activating
transcription factor 4. |
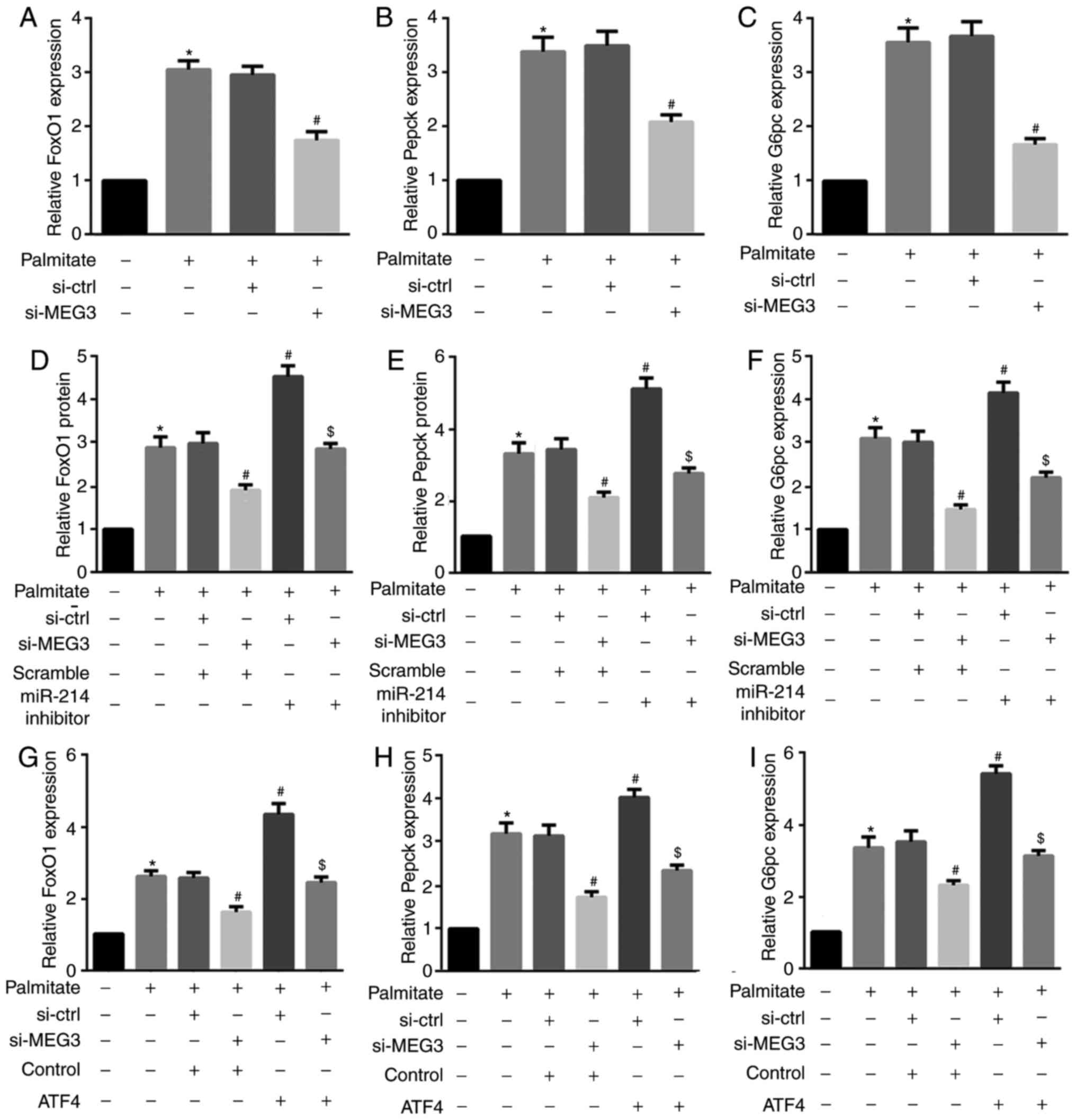 | Figure 7miR-214 inhibition and ATF4
overexpression reverse the MEG3 knockdown-mediated decrease in
FoxO1, G6pc and Pepck mRNA expression. Cells were transfected with
the indicated plasmids, followed by treatment with 0.5 mM palmitate
for 4 h. (A–C) RT-qPCR results revealed that MEG3 knockdown
significantly suppressed the palmitate-mediated increase in (A)
FoxO1, (B) Pepck, and (C) G6pc mRNA expression. (D–F) By contrast,
the miR-214 inhibitor induced the palmitate-mediated increase in
(D) FoxO1, (E) Pepck, and (F) G6pc mRNA expression. (G–I) ATF4
overexpression also induced the palmitate-mediated increase in (G)
FoxO1, (H) Pepck, and (I) G6pc mRNA expression.
*P<0.05 vs. group 1; #P<0.05 vs.
group 3; $P<0.05 vs. group 4. miR, microRNA;
ATF, activating transcription factor 4; MEG3, maternally expressed
gene 3; FoxO1, forkhead box protein O1; Pepck, phosphoenolpyruvate
carboxykinase; G6pc, glucose-6-phosphatase catalytic subunit; si,
small interfering RNA; ctrl, control; MEG3. |
MEG3 knockdown improves glucose and
insulin tolerance in HFD-fed mice
Next, HFD-fed male C57BL/6 mice were injected with
si-MEG3 fragments via the tail vein to examine the effects of MEG3
knockdown on glucose and insulin tolerance. As indicated in
Fig. 8A, HFD-fed mice
demonstrated significantly increased fasting blood glucose levels
compared with the control LFD-fed mice. However, the treatment of
HFD-fed mice with si-MEG3 substantially improved impaired glucose
tolerance. Additionally, HFD-fed mice treated with si-MEG3 also
improved impaired insulin tolerance (Fig. 8B).
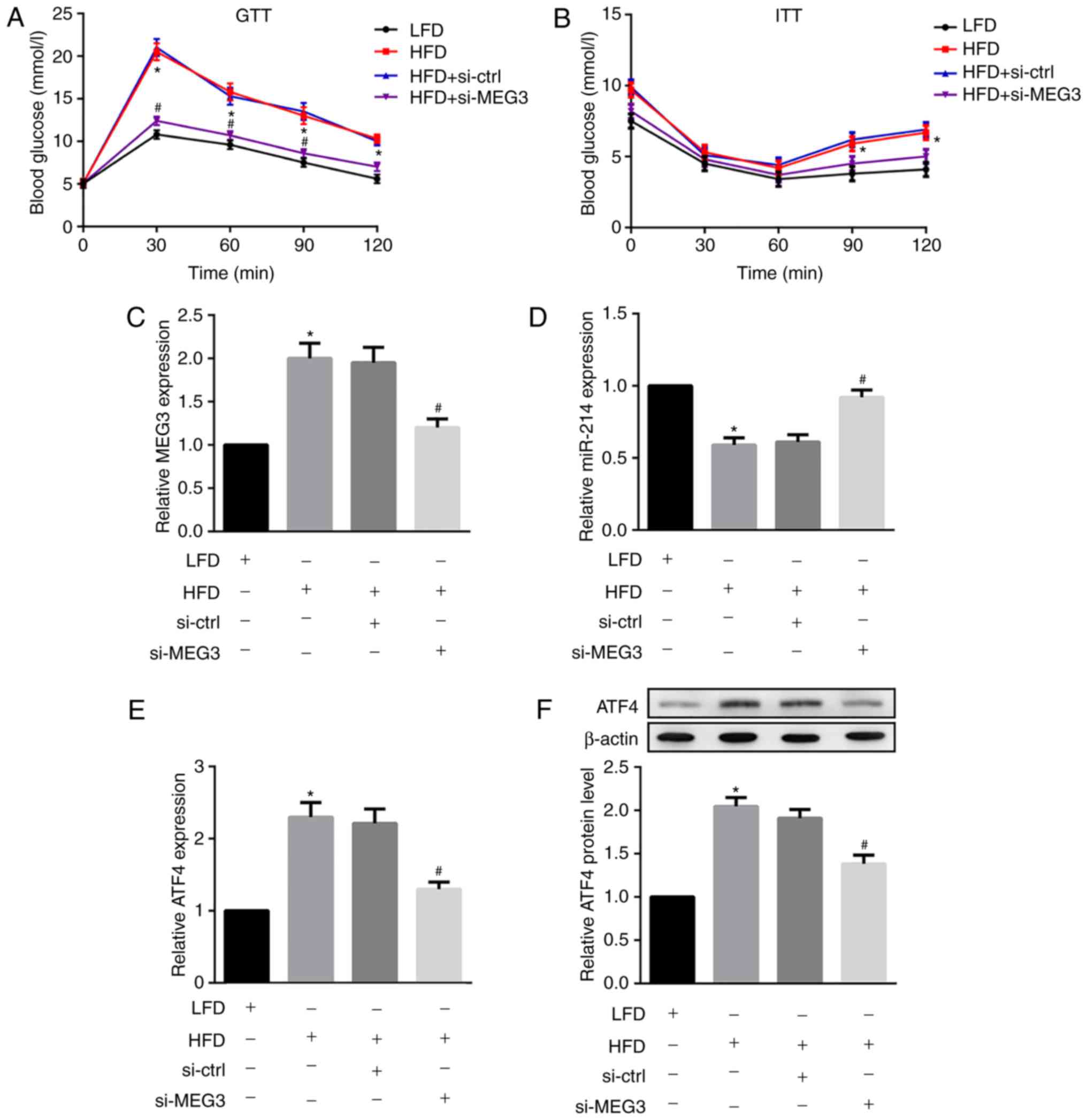 | Figure 8Effects of MEG3 knockdown on miR-214
and ATF4 expression, glucose tolerance and insulin tolerance in
HFD-fed mice. HFD-fed male C57BL/6 received a si-MEG3 fragment
injection via the tail vein twice a week for 10 weeks. At week 10,
following overnight fasting, (A) GTTs and (B) ITTs were performed
to analyse glucose and insulin tolerance, respectively. MEG3
knockdown improved glucose and insulin tolerance in HFD-fed mice.
Liver tissues were separated and the relative mRNA expression of
(C) MEG3, (D) miR-214 and (E) expression of ATF4 were examined by
reverse transcription polymerase chain reaction. (F) Protein
expression of ATF4 was examined by western blot analysis. MEG3
knockdown upregulated miR-214 but downregulated ATF4 expression in
HFD-fed mice. β-actin served as the loading control. N=10 for each
group.*P<0.05 vs. the LFD group;
#P<0.05 vs. the HFD + si-Ctrl group. MEG3,
maternally expressed gene 3; miR, microRNA; ATF, activating
transcription factor 4; HFD, high-fat diet; LFD, low-fat diet; si,
small interfering RNA; ctrl, control; GTTs, glucose tolerance
tests; ITTs, insulin tolerance tests. |
MEG3 knockdown upregulates miR-214 but
downregulates ATF4 expression in HFD-fed mice
Finally, the in vivo effects of MEG3
knockdown on miR-214 and ATF4 expression in liver tissues from
HFD-fed mice were investigated. MEG3 knockdown was demonstrated to
significantly down-regulate the HFD-induced expression of MEG3
(Fig. 8C) and ATF4 (Fig. 8E and F) in mouse liver tissues. By
contrast, MEG3 knockdown fragments significantly upregulated
HFD-suppressed miR-214 expression in mouse liver tissues (Fig. 8D). In agreement with in
vitro results, the in vivo results also indicated that
MEG3 knockdown upregulated miR-214 but downregulated ATF4
expression in HFD-fed mice.
Discussion
The aim of the present study was to evaluate the
expression of MEG3, miR-214 and ATF4 and their interaction and
effects on insulin resistance in T2DM. Leptin-deficient ob/ob mice
are overweight, develop insulin resistance, and serve as a model
for T2DM. Insulin resistance is a characteristic feature of T2DM.
The results of the present study provided evidence that MEG-3 and
ATF4 were upregulated, while miR-214 was downregulated, in the
livers of HFD-fed and ob/ob mice. Furthermore, in mouse primary
hepatocytes, palmitate time-dependently increased MEG3 and ATF4
expression, but decreased miR-214 expression. It was also
identified that MEG3 served as a ceRNA of miR-214 to facilitate
ATF4 expression. In addition, miR-214 inhibition and ATF4
overexpression reversed the MEG3 knockdown-mediated decrease in the
expression of FoxO1 and FoxO1-downstream targets Pepck and G6pc.
Finally, in HFD-fed mice, MEG3 knockdown substantially improved
impaired glucose and insulin tolerance, while downregulating
induced ATF4 expression and upregulating suppressed miR-214
expression.
MEG3, an imprinted lncRNA within the DLK1-MEG3 locus
on chromosome 14q32 in humans, is expressed in a number of normal
tissues (27). MEG3 has been
demonstrated to serve as a tumour suppressor in several types of
cancer, including glioma (28),
bladder cancer (29) and
colorectal cancer (30). However,
the functional role of MEG3 genes in hepatic insulin resistance in
T2DM is poorly understood. In our previous study, it was identified
that lncRNA MEG3 upregulation enhanced hepatic insulin resistance
via increased FoxO1 expression (5). In the present study, the specific
mechanism by which MEG3 increased FoxO1 expression was
explored.
Previous evidence has confirmed that lncRNAs may
serve as ceRNA to absorb miRNAs away from target genes, and thereby
alter the expression of miRNA target genes (31). Notably, MEG3 was recently
demonstrated to serve as a ceRNA of certain miRNAs during the
tumorigenesis of several types of cancer: For example, Qin et
al (28) stated that MEG3
functioned as a ceRNA of miR-19a that repressed phosphatase and
tensin homolog expression to inhibit glioma cell proliferation,
migration and invasion. Peng et al (32) demonstrated that MEG3 may
upregulate B-cell lymphoma-2 via its ceRNA activity on miR-181 to
regulate gastric carcinogenesis. Yan et al (33) identified that MEG3 served as a
ceRNA and competed with programmed cell death 4 mRNA to directly
bind miR-21 to regulate ischemic neuronal death. MEG3 also
positively regulated the expression of sex-determining region
Y-box7 by functioning as a ceRNA of miR-21-5p to regulate cisplatin
resistance in non-small cell lung cancer (13). The present study attempted to
investigate whether MEG3 may also function as a ceRNA to regulate
FoxO1 expression.
miR-214 was recently identified as a direct target
of MEG3 (15). Li et al
(16) revealed that the
expression of miR-214 was downregulated in the livers of fasting
and HFD-induced diabetic mice, which was consistent with the
results of the present study. That study also suggested that
miR-214 suppressed gluconeogenesis by targeting ATF4, which
regulates gluconeogenesis by affecting FoxO1 transcriptional
activity (16). ATF4 serves a
role in high-carbohydrate diet-induced insulin resistance (17). Notably, the present study provided
evidence of a novel MEG3/miR-214/ATF4 axis: MEG3 was identified to
serve as a ceRNA of miR-214 to facilitate ATF4 expression.
Abnormally elevated hepatic gluconeogenesis is
largely responsible for the overproduction of glucose in patients
with T2DM (16). Insulin
represses hepatic gluconeogenesis by suppressing the
transcriptional activity of FoxO1 via protein kinase B
phosphorylation-mediated protein degradation (34). Increased FoxO1 activity leads to
hyperglycaemia and increased cellular production of hepatic
glucose, which is associated with insulin resistance (18). FoxO1 is a key regulator of hepatic
gluconeogenesis that drives G6pc and Pepck mRNA transcription
(26). The results from the
present study from mouse primary hepatocytes indicated that MEG3
directly bound to miR-214 and suppressed its expression. ATF4 was a
direct target of miR-214 and MEG3 positively regulated ATF4
expression by inhibiting miR-214. In addition, miR-214 inhibition
and ATF4 overexpression reversed the MEG3 knockdown-mediated
decrease in the expression of FoxO1 and FoxO1-downstream targets
Pepck and G6pc. Furthermore, in HFD-fed mice, MEG3 knockdown led to
substantial improvements in impaired glucose and insulin tolerance,
while downregulating HFD-induced ATF4 expression and upregulating
HFD-suppressed miR-214 expression. Collectively, the results from
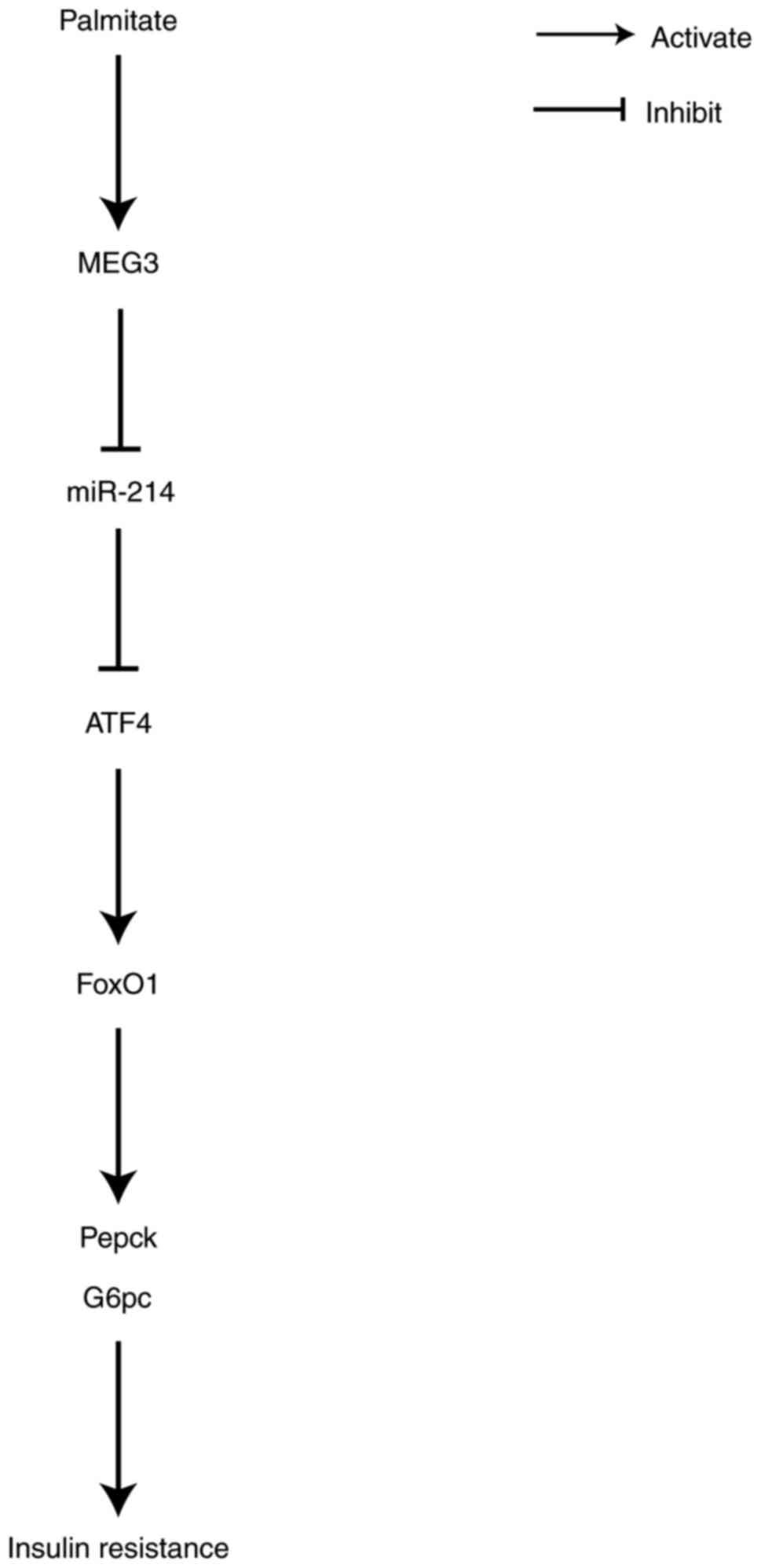
the present study indicated that MEG3 functioned as a ceRNA of
miR-214 to upregulate ATF4 expression, leading to the promotion of
FoxO1 expression and its downstream gluconeogenic enzymes, G6pc and
Pepck. This, in turn, increased gluconeogenesis and promoted
insulin resistance (Fig. 9).
In conclusion, the data from the present study
demonstrate that MEG3 promoted hepatic insulin resistance by
serving as a ceRNA of miR-214 to facilitate ATF4 expression. These
results provide insight into the molecular mechanism of MEG3
involvement in the development of T2DM.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
XZ was responsible for the literature review,
development of the intellectual content of the manuscript and data
analysis. XZ and HL performed the experiments. XZ prepared the
manuscript, and XZ and HL were involved in editing the manuscript.
YW and JZ were responsible for data acquisition. XZ and GY
performed the statistical analysis. WW was involved in developing
the concept and design of the study, and is guarantor of integrity
of the entire study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The animal experiments performed in the present
study were approved by the Animal Ethics Committee of Anhui
Provincial Hospital, The First Affiliated Hospital of University of
Science and Technology of China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stumvoll M, Goldstein BJ and van Haeften
TW: Type 2 diabetes: Principles of pathogenesis and therapy.
Lancet. 365:1333–1346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng Y, Ley SH and Hu FB: Global
aetiology and epidemiology of type 2 diabetes mellitus and its
complications. Nat Rev Endocrinol. 14:88–98. 2018. View Article : Google Scholar
|
|
3
|
Garabadu D and Krishnamurthy S: Metformin
attenuates hepatic insulin resistance in type-2 diabetic rats
through PI3K/Akt/GLUT-4 signalling independent to
bicuculline-sensitive GABAA receptor stimulation. Pharm
Biol. 55:722–728. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang Y, Thakran S, Bheemreddy R, Coppess
W, Walker RJ and Steinle JJ: Sodium salicylate reduced insulin
resistance in the retina of a type 2 diabetic rat model. PLoS One.
10:e01255052015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu X, Wu YB, Zhou J and Kang DM:
Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via
increasing FoxO1 expression. Biochem Biophys Res Commun.
469:319–325. 2016. View Article : Google Scholar
|
|
6
|
Burchell A: Glycogen storage diseases and
the liver. Baillieres Clin Gastroenterol. 12:337–354. 1998.
View Article : Google Scholar
|
|
7
|
Pan T, Guo JH, Ling L, Qian Y, Dong YH,
Yin HQ, Zhu HD and Teng GJ: Effects of Multi-electrode renal
denervation on insulin sensitivity and glucose metabolism in a
canine model of type 2 diabetes mellitus. J Vasc Interv Radiol.
29:731–738.e2. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li P, Ruan X, Yang L, Kiesewetter K, Zhao
Y, Luo H, Chen Y, Gucek M, Zhu J and Cao H: A liver-enriched long
non-coding RNA, lncLSTR, regulates systemic lipid metabolism in
mice. Cell Metab. 21:455–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morá I, Akerman İ, van de Bunt M, Xie R,
Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J,
Rodríguez-Seguí S, et al: Human β cell transcriptome analysis
uncovers lncRNAs that are tissue-specific, dynamically regulated,
and abnormally expressed in type 2 diabetes. Cell Metab.
16:435–448. 2012. View Article : Google Scholar
|
|
11
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oh KJ, Han HS, Kim MJ and Koo SH: CREB and
FoxO1: Two transcription factors for the regulation of hepatic
gluconeogenesis. BMB Rep. 46:567–574. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang P, Chen D, Ma H and Li Y: LncRNA MEG3
enhances cisplatin sensitivity in non-small cell lung cancer by
regulating miR-21-5p/SOX7 axis. Onco Targets Ther. 10:5137–5149.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meister G and Tuschl T: Mechanisms of gene
silencing by double-stranded RNA. Nature. 431:343–349. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan FY, Deng R, Yi H, Sun HP, Zeng Y, He
GC and Su Y: The inhibitory effect of MEG3/miR-214/AIFM2 axis on
the growth of T-cell lymphoblastic lymphoma. Int J Oncol.
51:316–326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li K, Zhang J, Yu J, Liu B, Guo Y, Deng J,
Chen S, Wang C and Guo F: MicroRNA-214 suppresses gluconeogenesis
by targeting activating transcriptional factor 4. J Biol Chem.
290:8185–8195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li H, Meng Q, Xiao F, Chen S, Du Y, Yu J,
Wang C and Guo F: ATF4 deficiency protects mice from
high-carbohydrate-diet-induced liver steatosis. Biochem J.
438:283–289. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Matsumoto M, Pocai A, Rossetti L, Depinho
RA and Accili D: Impaired regulation of hepatic glucose production
in mice lacking the forkhead transcription factor foxo1 in liver.
Cell Metabolism. 6:208–216. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang W, Yu J, Jia X, Xiong L, Li N and
Wen X: Zhenqing recipe improves glucose metabolism and insulin
sensitivity by repressing hepatic FOXO1 in type 2 diabetic rats. Am
J Chin Med. 40:721–733. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang J, Liu C, Qiao A, Cui Y, Zhang H,
Cui A, Zhang S, Yang Y, Xiao X, Chen Y, et al: MicroRNA-29a-c
decrease fasting blood glucose levels by negatively regulating
hepatic gluconeogenesis. J Hepatol. 58:535–542. 2013. View Article : Google Scholar
|
|
21
|
Morris EM, Meers GM, Booth FW, Fritsche
KL, Hardin CD, Thyfault JP and Ibdah JA: PGC-1α overexpression
results in increased hepatic fatty acid oxidation with reduced
triacylglycerol accumulation and secretion. Am J Physiol
Gastrointest Liver Physiol. 303:G979–G992. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Galbo T, Perry RJ, Nishimura E, Samuel VT,
Quistorff B and Shulman GI: PP2A inhibition results in hepatic
insulin resistance despite Akt2 activation. Aging (Albany NY).
5:770–781. 2013. View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Bunn RC, Cockrell GE, Ou Y, Thrailkill KM,
Lumpkin CK Jr and Fowlkes JL: Palmitate and insulin synergistically
induce IL-6 expression in human monocytes. Cardiovasc Diabetol.
9:732010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Volpe CM, Abreu LF, Gomes PS, Gonzaga RM,
Veloso CA and Nogueiramachado JA: The production of nitric oxide,
IL-6, and TNF-alpha in palmitate-stimulated PBMNCs is enhanced
through hyperglycemia in diabetes. Oxid Med Cell Longev.
2014.479587:2014.
|
|
26
|
Gross DN, Wan M and Birnbaum MJ: The role
of FOXO in the regulation of metabolism. Curr Diab Rep. 9:208–214.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miyoshi N, Wagatsuma H, Wakana S,
Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino
T and Ishino F: Identification of an imprinted gene, Meg3/Gtl2 and
its human homologue MEG3, first mapped on mouse distal chromosome
12 and human chromosome 14q. Genes Cells. 5:211–220. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qin N, Tong GF, Sun LW and Xu XL: Long
noncoding RNA MEG3 suppresses glioma cell proliferation, migration,
and invasion by acting as a competing endogenous RNA of miR-19a.
Oncol Res. 25:1471–1478. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ying L, Huang Y, Chen H, Wang Y, Xia L,
Chen Y, Liu Y and Qiu F: Downregulated MEG3 activates autophagy and
increases cell proliferation in bladder cancer. Mol Biosyst.
9:407–411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH
and Guo RH: Decreased expression of long noncoding RNA MEG3 affects
cell proliferation and predicts a poor prognosis in patients with
colorectal cancer. Tumour Biol. 36:4851–4859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu
YY, Wang SM, He FT and Yang SM: Long noncoding RNA BC032469, a
novel competing endogenous RNA, upregulates hTERT expression by
sponging miR-1207-5p and promotes proliferation in gastric cancer.
Oncogene. 35:3524–3534. 2016. View Article : Google Scholar
|
|
32
|
Peng W, Si S, Zhang Q, Zhao F, Wang F, Yu
J and Ma R: Long non-coding RNA MEG3 functions as a competing
endogenous RNA to regulate gastric cancer progression. J Exp Clin
Cancer Res. 34:792015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan H, Rao J, Yuan J, Gao L, Huang W, Zhao
L and Ren J: Long non-coding RNA MEG3 functions as a competing
endogenous RNA to regulate ischemic neuronal death by targeting
miR-21/PDCD4 signaling pathway. Cell Death Dis. 8:32112017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matsuzaki H, Daitoku H, Hatta M, Tanaka K
and Fukamizu A: Insulin-induced phosphorylation of FKHR (Foxo1)
targets to proteasomal degradation. Proc Natl Acad Sci USA.
100:11285–11290. 2003. View Article : Google Scholar : PubMed/NCBI
|