Introduction
Mesenchymal stem cells (MSCs) have been widely used
for cell-based treatment of various diseases due to their
immunomodulatory and anti-inflammatory properties (1). As a type of multipotent cell
originating from bone marrow, MSCs can differentiate into various
cell types including osteoblast, chondrocyte, myocyte and adipocyte
cells (2-5). Dental stem cells are MSC-like cells
isolated from dental tissues, and are classified into stem cells
from the apical papilla (SCAPs), stem cells from human exfoliated
deciduous teeth, periodontal ligament stem cells and dental pulp
stem cells according to the subsites of dental tissues from which
the cells are isolated (6-8).
Dental tissues including wisdom and primary teeth are a reservoir
of MSCs, contributing to the regeneration of dental tissues
(9). Thus, MSCs are considered as
a promising source in the context of regenerative medicine
(10-13). Among the MSC types, SCAPs are
present in the root apex of developing teeth, where they are
considered to be associated with root development (14). SCAPs can be readily isolated from
extracted wisdom teeth that are generally considered useless
(15). In response to
inflammatory stimuli, SCAPs have been identified to exhibit
increased osteogenic and angiogenesis potential due to enhanced
vitality and stemness (16).
Guanine nucleotide binding proteins (G proteins) are
a family of regulatory proteins responsible for molecular signal
transduction of extracellular signals to the intracellular
environment (17). G proteins are
typically composed of α, β and γ subunits. Gα subunits are
categorized into 4 major classes based on sequence similarity and
function of the α-subunit, termed Gαs, Gαi/o, Gαq/11 and Gα12/13
(18). The guanine nucleotide
binding protein 3 (GNAI3) belongs to the Gαi subunit class and is
located on chromosome 17q22-24 (19). Increasing evidence has
demonstrated that GNAI3 is involved in a variety of cellular
processes including proliferation, apoptosis and differentiation
(19-22). GNAI3 has also been reported to
regulate craniofacial growth and development, and homeotic
modifications in the human GNAI3 gene may lead to auriculo-condylar
syndrome (ACS), an uncommon craniofacial natal defect characterized
by mandible hypoplasia (23).
Mitogen-activated protein kinase (MAPK) has three
major subfamilies, extracellular-signal regulated kinases (ERKs),
c-Jun N-terminal kinases (JNKs) and p38 MAPKs. MAPK signaling
serves an essential role in regulating cell proliferation and
differentiation, and accumulating experimental data have
demonstrated that MAPK is associated with osteogenic
differentiation (24,25). Furthermore, bioinformatic analysis
has identified that MAPK signaling pathways may play an important
role in the SCAPs niche (26).
A previous study by our group demonstrated that GATA
binding protein 4 (GATA4) regulated root development via GNAI3
(27). However, to our knowledge
the role and mechanism of GNAI3 remain unknown. In the present
study, the role of GNAI3 in regulating the proliferation, cell
cycle progression, apoptosis, migration and odonto/osteogenic
differentiation of SCAPs was investigated using loss-of-function
assays. The molecular mechanism underlying the function of GNAI3
was also examined.
Materials and methods
Animals and histological analysis
A total of 20, 14 day-old male mice (C57BL/6J;
weight 5.2-8.7g) were provided by the Animal Center of Nanjing
Medical University (Nanjing, China) and maintained in a
temperature-controlled room (room temperature 24±1°C; relative
humidity 50±10%; 12-h light/dark cycle; free access to mouse chow
and water). The maxillary halves of the mice were isolated, fixed
in 4% buffered formalin for 24 h at 4°C, decalcified in 10% EDTA
(pH 7.4) for 7 days at room temperature, paraffin-embedded and
sectioned into 5-µm slices. Ethical approval for the animal
experimentation was provided by the Experimental Animal Care and
Use Committee of Nanjing Medical University (approval no.
2015-03-40).
The tissue sections were treated with 3%
H2O2 for 30 min followed by normal goat serum
(Wuhan Boster Biological Technology, Ltd., Wuhan, China) for 30 min
at 37°C, and subject to immunohistochemical staining for
histological investigation. The sections were incubated with
primary rabbit anti-mouse GNAI3 antibody (1:200 dilution; cat. no.
ab154024) and primary monoclonal mouse anti-mouse nestin (1:100
dilution; cat. no. ab11306; both from Abcam, Cambridge, MA, USA)
overnight at 4°C. Nestin was used as a biomarker of mature and
differentiated odontoblasts (28). Following three washes with 0.1
mol/l phosphate-buffered saline (PBS), the sections were incubated
with secondary horseradish peroxidase (HRP)-conjugated antibody
(KIT-5020; MaxVision Biosciences Inc., Fuzhou, China) for 30 min at
37°C. The immunoreaction was localized using a diaminobenzidine kit
(Beyotime Institute of Biotechnology, Haimen, China) according to
the manufacturer’s protocol and the sections were counterstained
with hematoxylin for 5 min at 37°C. Stained sections were observed
and analyzed by ImageJ v.1.45 software (National Institutes of
Health, Bethesda, NJ, USA) under a light microscope (DM4000; Leica
Microsystems GmbH, Wetzlar, Germany) at ×400 magnification.
Cell cultures and stem cell
characterization
Between January and December 2017, a total of 20
male healthy patients (14-18 years of age) underwent extraction
surgery at Stomatological Hospital of Jiangsu Province, Nanjing,
China, and 20 wisdom teeth (one tooth per patient) with open apices
were acquired. The use of human samples for the study was approved
by the Medical Ethics Committee of Stomatological Hospital of
Jiangsu Province (approval no. PJ2016-038-001). Consent was
provided by the patients or their next of kin for the collection of
the dental samples. Soft tissues of the root apex were collected
immediately and washed with PBS. The separated apex tissues were
minced with sterilized scissors and then digested with 3 mg/ml
collagenase type I (Sigma Aldrich Chemie, Taufkirchen, Germany) and
4 mg/ml dispase (Roche Applied Science, Penzberg, Germany) for 1 h
at 37°C, and then the single cell suspensions were grown in
α-minimum essential medium supplemented with 10% fetal bovine serum
(FBS), 2 mmol/l glutamine, 100 U/ml penicillin and 100 mg/ml
streptomycin (all from Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) in a humidified 5% CO2 incubator at
37°C. The culture medium was changed every 3 days. SCAPs at passage
2-5 were used in the subsequent experiments. To address the
potential roles of JNK and ERK signaling pathways, specific JNK
inhibitor (20 µM; SP600125), specific ERK inhibitor (20
µM; U0126; both from Beyotime Institute of Biotechnology)
and dimethyl sulfoxide (20 µM; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) as a control were respectively added to the
SCAPs, and the cells were incubated at 37°C for 5 min (29,30). As for the characterization of
cultured SCAPs, the cells were digested with trypsin (Roche Applied
Science), collected and incubated with fluorochrome-conjugated
rabbit anti-human antibodies: Cluster of differentiation
(CD)90-fluorescein isothiocyanate (FITC; 561969),
CD44-phycoerythrin (PE; 561858), CD45-allophycocyanin (560973) and
CD14-PE-cyanine 7 (560919; all at 0.5 mg/ml; BD Biosciences, San
Jose, CA, USA) for 1 h at 4°C in the dark to detect the SCAPs
phenotype (31,32). Following fixation with 4%
paraformaldehyde buffer for 30 min at 4°C, cells were washed twice
with PBS containing 2% FBS (Hyclone; GE Healthcare Life Science,
Logan, UT, USA) and stained cells were characterized and identified
by flow cytometry (BD FACSCalibur™; BD Biosciences) (33). Data were analyzed with Summit 5.1
software (Beckman Coulter, Inc., Brea, CA, USA).
Lentiviral transduction
Recombinant lentiviral LV-3 [pGLVH1/green
fluorescent protein (GFP)+Puro] vectors expressing GAIN3
short-hairpin RNAs (shGNAI3-GFP) were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). The short-hairpin sequence
was 5′-CCA GGG AAU AUC AGC UCA ATT-3′, and the sequence of 5′-TTC
TCC GAA CGT GTC ACG T-3′ was used as the negative control
(shCTRL-GFP). When SCAPs were at 60-80% confluence, lentiviral
particles were added to the medium with a multiplicity of infection
of 30. The virus-containing medium was replaced with fresh normal
medium after 24 h at 37°C. The transduced cells (GFP-positive) were
observed under a fluorescence microscope (DMI3000B; Leica
Microsystems GmbH) at ×100 magnification 72 h after transduction.
GNAI3 inhibition efficiency was confirmed by western blotting and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) assays.
Cell proliferation assay
The cell proliferation capacity of SCAPs following
different treatments was evaluated by Cell Counting Kit-8 assay
(CCK8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
according to the manufacturer’s protocol. At 72 h after
transduction of lentiviral vectors or exposure to inhibitors, the
cells were seeded at a density of 1×103 cells/well in
96-well plates. A total of 100 µl CCK8 solution was added on
days 0, 1, 2, 3, 4, 5 and 6 days of cell plating, and the plates
were incubated for 1 h at 37°C. Cell number was assessed based on
optical density (OD) measurements at 450 nm using a microplate
reader. To assess cell cycle arrest, SCAPs transduced with
lentiviral vectors or exposed to inhibitors were grown for 72 h,
and then collected, washed with PBS and fixed overnight with 70%
ethanol at 4°C. Subsequently, the cells were washed and treated
with RNase, then stained with 20 mg/ml propidium iodide (PI) for an
additional 1 h at room temperature. Cell cycle distribution
(populations in G1, G2, and S phases) was determined by flow
cytometry (BD FACSverse™; BD Biosciences) and analyzed using FlowJo
V7 software (Tree Star, Oregon, USA). For analysis of apoptosis,
SCAPs were digested with 4 mg/ml trypsin and washed with ice-cold
PBS at 72 h after transduction with shGNAI3-GFP or shCTRL-GFP. The
SCAPs were then resuspended in ice-cold 0.1 mol/l PBS and stained
with an Annexin V-FITC/PI kit (Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China) for 15 min in the dark at room temperature,
according to the manufacturer’s instructions. The apoptotic rates
of the SCAPs were determined by flow cytometry (BD FACSverse™) and
analyzed with FlowJo V7.
Cell migration assay
The cell migration capacity of SCAPs following
different treatments was evaluated by a scratch wound healing
assay. Cultured cells in 6-well plates at 80% confluence were
mechanically wounded with a 200 µl sterile pipette tip. Cell
debris was removed gently and the remaining cells were incubated
with normal media. After 0, 12 and 24 h of incubation, photographs
were taken under a light microscope (DMIL LED; Leica Microsystems
GmbH) at ×100. Cell migration rate was quantified by counting the
migrating cells.
Alkaline phosphatase (ALP) activity
assay
ALP activity was evaluated using a BCIP/NBT Alkaline
Phosphatase Color Development kit (Beyotime Institute of
Biotechnology) according to the manufacturer’s protocol. SCAPs
transduced with shGNAI3-GFP or shCTRL-GFP after 72 h were seeded at
a density of 1×105 cells/well in 6-well plates and
cultured in mineralization-inducing media containing α-minimum
essential medium (Gibco; Thermo Fisher Scientific, Inc.), 10% FBS
(Hyclone; GE Healthcare Life Science), 100 U/ml penicillin, 100
µg/ml streptomycin, 100 µM ascorbic acid, 2 mM
2-glycerophosphate and 10 nm dexamethasone for 5 days at 37°C.
Images were captured using a light microscope (DMIL LED) at ×100
magnification. The staining intensity was determined using ImageJ
v.1.45 software (National Institutes of Health, Bethesda, NJ,
USA).
Matrix mineralization assay
Alizarin red S (ARS) staining was performed to
assess extracellular matrix mineralization. SCAPs transduced with
shGNAI3-GFP or shCTRL-GFP after 72 h were seeded at a density of
1×105 cells/well in 6-well plates and cultured in
mineralization-inducing media for 2 weeks at 37°C, followed by
fixation with 75% ethanol for 30 min at room temperature. A 2% ARS
solution was used for detection of calcium deposits. Images were
captured using a light microscope (DMIL LED) at ×100 magnification.
The OD of ARS staining was measured using a microplate reader at
560 nm.
Western blot analysis
SCAPs following the indicated treatments were
harvested and lysed in radioimmunoprecipitation assay buffer
(Beyotime Institute of Biotechnology) containing 1 mM
phenylmethylsulfonyl fluoride for 30 min at 4°C. Cell debris was
eliminated by centrifugation at 240 × g for 15 min at 4°C and
protein concentration was determined by the bicinchoninic acid
method. A total of 30 µg protein per lane was loaded on 10%
sodium dodecyl sulfate-polyacrylamide gels for electrophoresis, and
then transferred to polyvinylidene fluoride membranes followed by
blocking in 5% nonfat milk in tris-buffered saline/Tween-20. The
membranes were incubated with primary antibodies overnight at 4°C,
then with secondary antibodies for 1 h at room temperature. The
protein bands were visualized with electrochemiluminescence
substrate solution (GE Healthcare Life Sciences, Little Chalfont,
UK), and images were captured using a Micro Chemiluminescence
system 4.2 (DNR Bio-Imaging Systems, Ltd., Jerusalem, Israel).
Semi-quantitative analysis were performed using ImageJ v.1.45
software.
Protein expression of dentin sialophosphoprotein
(DSPP), runt-related transcription factor 2 (RUNX2), osterix (OSX),
osteopontin (OPN), osteocalcin (OCN) and bone morphogenetic protein
4 (BMP4), as established osteogenic and odontogenic markers
(34-38), was determined. Activity of MAPK
and JNK signaling was also assessed based on phosphorylation
levels. The primary antibodies used were as follows: Rabbit
anti-GNAI3 (1:1,000 dilution; cat. no. ab154024; Abcam), mouse
anti-DSPP (1:1,000 dilution; cat. no. sc73632; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), mouse anti- RUNX2 (1:1000;
dilution; cat. no. ab76956), polyclonal rabbit anti-OSX (1:1,000
dilution; cat. no. ab22552), rabbit anti-OPN (1:1,000 dilution;
cat. no. ab63856), polyclonal rabbit anti-OCN (1:1,000 dilution;
cat. no. ab93876), rabbit anti-BMP4; 1:1,000 dilution; cat. no.
ab39973; all from Abcam), rabbit anti-GAPDH (1:1,000 dilution; cat.
no. AP0063; Bioworld Technology, Inc., St. Louis Park, MN, USA),
anti-phosphorylated (p)-JNK (dilution 1:1,000; cat. no. 4668),
anti-JNK (dilution 1:1,000; cat. no. 9252), anti-p-p38 (dilution
1:1,000; cat. no. 9211), anti-p38 (dilution 1:1,000; cat. no.
9212), anti-p-ERK (dilution 1:1,000; cat. no. 4370) and anti-ERK
(dilution 1:1,000; cat. no. 4694; all from Cell Signaling
Technology, Inc., Danvers, MA, USA). HRP-conjugated secondary
antibodies were used prior to detection (1:8,000 dilution; cat. no.
ZB-2301; Origene Technologies, Beijing, China) for 1 h at room
temperature. Band densities were quantified relative to that of
GADPH.
RT-qPCR
RNA expression of the osteogenic and odontogenic
gene markers dentin matrix acidic phosphoprotein 1 (DMP1), DSPP,
OCN, OPN, OSX and RUNX2 (39) was
determined. Total RNA was extracted from SCAPs using an RNApure
High-purity Total RNA Rapid Extraction kit (Invitrogen; Thermo
Fisher Scientific, Inc.) and then reverse transcribed into
complementary DNA using a PrimeScript RT reagent kit (Takara Bio,
Inc., Otsu, Japan) according to the manufacturer’s protocol. The
qPCR reactions were performed using a SYBR-Green RT-qPCR kit
(Vazyme, Piscataway, NJ, USA) in 96-wells plates according to the
manufacturer’s protocol on a 7300 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Each 20-µl qPCR
reaction system contained 10 µl SYBR-Green qPCR Master Mix,
2 µl cDNA and 0.4 µl of each primer (final primer
concentration of 0.2 µM). The amplifications were performed
under the following conditions: 95°C for 30 sec, followed by 40
cycles of 95°C for 10 sec, 60°C for 30 sec, 95°C for 15 sec, 60°C
for 1 min and 95°C for 15 sec. GAPDH was used as the internal
control. The primer sequences were as follows: GNAI3 forward,
5′-GGGAAG ACAAATGAAAGAGAA-3′ and reverse, 5′-CCAACAAAG
GCACTGAAC-3′; DMP1 forward, 5′-GGAAGAGGTGGT GAGTGAG-3′ and reverse,
5′-TTGAGTGGGAGAGTGTG TG-3′; DSPP forward, 5′-CCATTCCAGTTCCTCAAA-3′
and reverse, 5′-GCCTTCCTCTATCACCTTC-3′; OCN forward,
5′-CACACTCCTCGCCCTATT-3′ and reverse, 5′-GGTCTCTTCACTACCTCGCT-3′;
OPN forward, 5′-CTC CAATCGTCCCTACAGTCG-3′ and reverse, 5′-CCAAGC
TATCACCTCGGCC-3′; OSX forward, 5′-CTACCCATC TGACTTTGCTC-3′ and
reverse, 5′-CACTATTTCCCA CTGCCTT-3′; RUNX2 forward, 5′-AGTTCCCAAGCA
TTTCATC-3′ and reverse, 5′-GGCAGGTAGGTGTGG TAGT-3′; and GAPDH
forward, 5′-TGAACCATGAGAAGT ATGACAACA-3′ and reverse,
5′-TCTTCTGGGTGG CAGTG-3′. RT-qPCR for each sample was performed in
triplicate and repeated three times. The relative expression of
target mRNAs was determined using the 2−∆∆Cq method
(40).
Statistical analysis
Data are expressed as the mean ± standard deviation
and were analyzed using SPSS 13 software (SPSS, Inc., Chicago, IL
USA). The column graphs and line charts were generated using
GraphPad Prism 7 (GraphPad, Inc., La Jolla, CA, USA). Independent
two-sample t-tests were applied to compare the differences between
the experimental and control groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
GNAI3 is induced in tooth root
development in vivo and in SCAP mineralization in vitro
To determine the role of GNAI3 in tooth root
development (dentin mineralization), the expression pattern of
GNAI3 during root development and dentin mineralization was first
examined. As depicted in Fig. 1A,
GNAI3 was primarily expressed in Hertwig’s epithelial root sheath
(HERS) and the surrounding mesenchyme in mice, suggesting the
possible involvement of GNAI3 in tooth root development. In
addition, it was identified that GNAI3 expression was significantly
upregulated in human SCAPs as early as 3 days after mineralization
induction, with a peak at 7 days of induction (P<0.01; Fig. 1B and C). These results suggested
that GNAI3 may be involved in tooth root development in vivo
and in vitro.
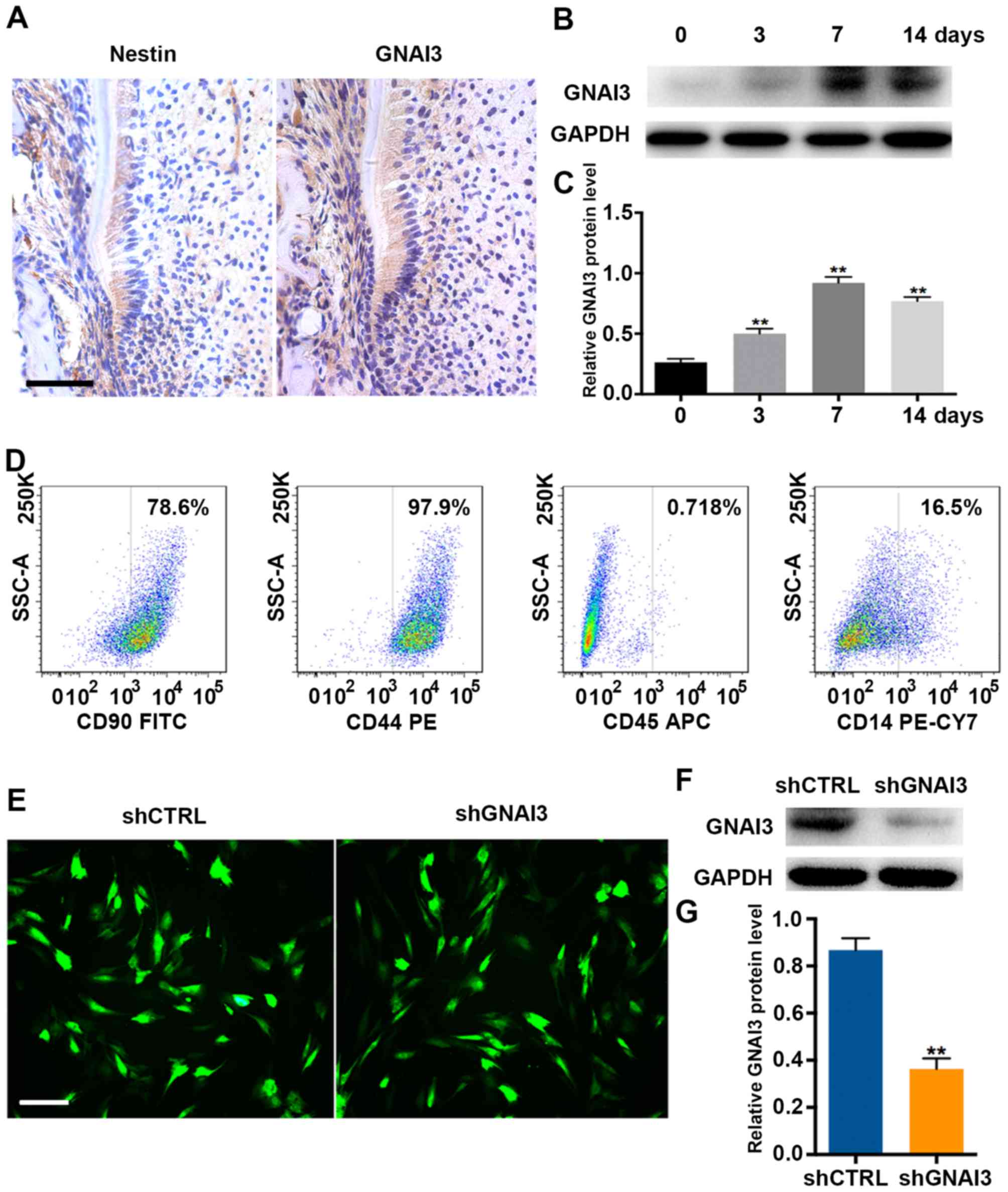 | Figure 1Upregulation of GNAI3 in mouse tooth
root development in vivo and in human SCAPs mineralization
in vitro. (A) Immunohistochemical staining for nestin (a
biomarker of differentiated odontoblasts) and GNAI3 in mouse tooth
roots on postnatal day 14. (B) Western blot analysis of GNAI3
expression in SCAPs following mineralization induction for the
times indicated. (C) Quantification of western blot results in (B)
by normalization to GAPDH. (D) Characterization of isolated human
SCAPs by flow cytometry. (E) Detection of transduction efficiency
of lentiviral particles expressing shRNA (multiplicity of infection
of 30) in human SCAPs. After 3 days of transduction, GFP
fluorescence was observed under a fluorescence microscope. (F)
Western blot analysis of GNAI3 expression in SCAPs transduced with
shGNAI3-GFP or shCTRL-GFP. (G) Quantification of western blot
results in (F) by normalization to GAPDH. Scale bars: (A) 100
µm; (E) 400 µm. **P<0.01 vs. (B) day 0
group or (G) shCTRL group. GNAI3, guanine and nucleotide binding
protein 3; SCAPs, stem cells of the apical papilla; sh[RNA], short
hairpin [RNA]; GFP, green fluorescent protein; CTRL, control; CD,
cluster of differentiation; FITC, fluorescein isothiocyanate; PE,
phycoerythrin; APC, allophycocyanin; CY7, cyanine 7. |
To further investigate whether GNAI3 plays a role in
mineralization induction of human SCAPs that were defined as
CD90+/CD44+/CD45-/CD14- (Fig.
1D), lentiviral vector expressing shGNAI3 (shGNAI3-GFP) was
used to knock down the expression of GNAI3. Lentiviral transduction
efficiency (~90%) is shown in Fig.
1E. Importantly, shGNAI3-GFP effectively inhibited the
expression of GNAI3 in SCAPs, compared with the control group
(P<0.01; Fig. 1F and G),
indicating that shGNAI3-GFP could be used for loss-of-function
assays to detect the function of GNAI3 in SCAPs.
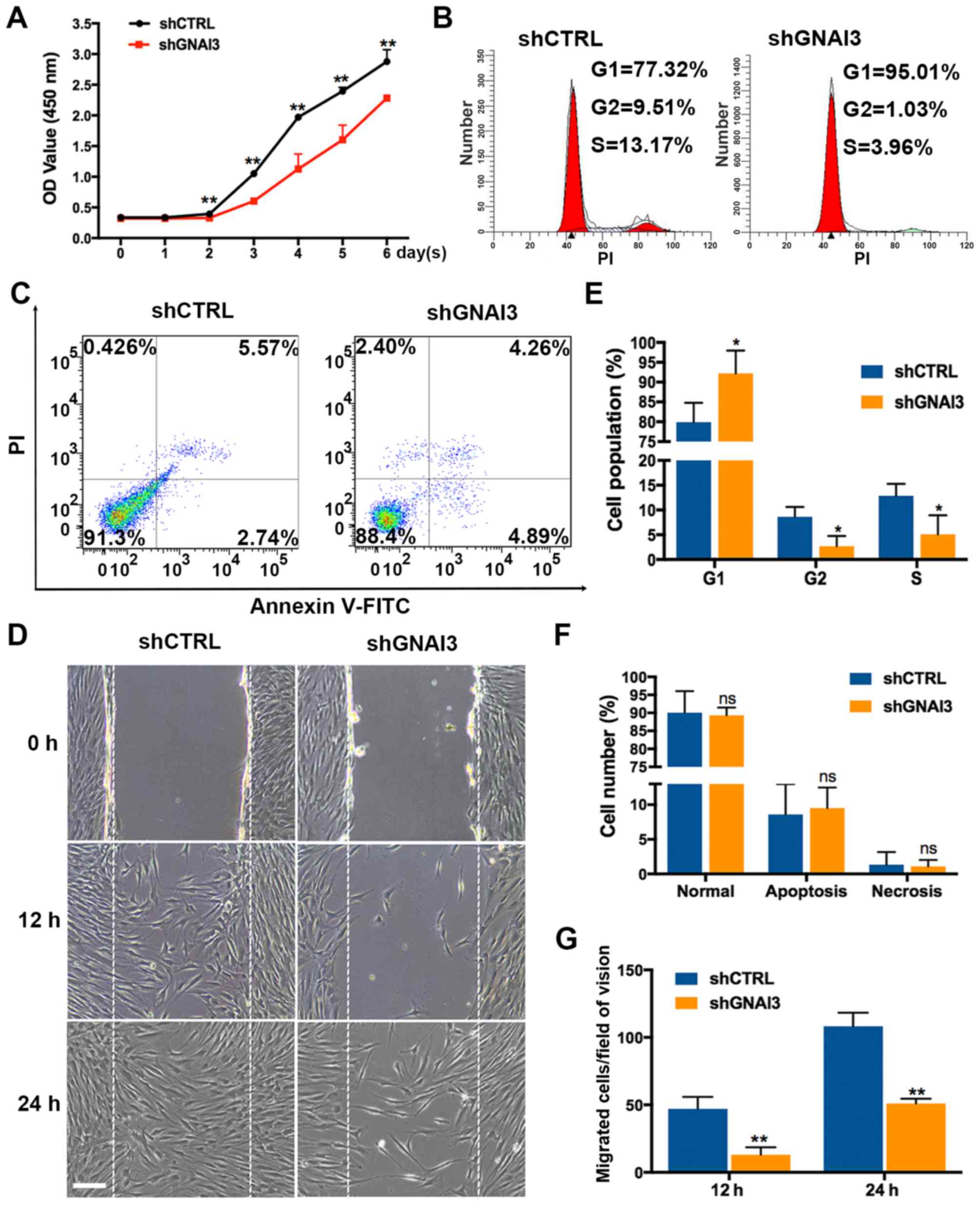
GNAI3 knockdown suppresses the
proliferation and migration of SCAPs
It was subsequently determined whether GNAI3
regulates the proliferation and migration of SCAPs, two important
processes in tooth root development. The results (Fig. 2) showed that GNAI3 knockdown by
shGNAI3-GFP inhibited SCAPs growth over 2-6 days after transduction
(P<0.01) in an apparent time-dependent manner (Fig. 2A), as well as cell cycle
progression, as evidenced by G1 phase arrest (increased G1-phase
cells) and decreased S and G2-phase populations of GNAI3-deficient
SCAPs (P<0.05; Fig. 2B and E).
However, GNAI3 knockdown had no apparent effect on the apoptosis or
necrosis of SCAPs (Fig. 2C and
F). In addition, GNAI3-deficient SCAPs exhibited markedly
reduced migration ability 12 and 24 h after wounding, compared with
control SCAPs (P<0.01; Fig. 2D and
G). Collectively, these data indicated that GNAI3 is required
for SCAPs proliferation and migration in vitro.
GNAI3 knockdown inhibits the
odonto/osteogenic differentiation of SCAPs
Since dentin mineralization is a key event in tooth
root development, the effects of GNAI3 knockdown on dentin
mineralization resulting from odonto/osteogenic differentiation of
SCAPs were investigated (Fig. 3).
As displayed in Fig. 3A and C,
GNAI3 knockdown significantly decreased ALP activity in SCAPs 5
days after mineralization induction (P<0.01). Consistently,
GNAI3 knockdown significantly reduced ARS staining intensity 2
weeks after the mineralization induction of SCAPs (P<0.01;
Fig. 3B and D). These results
demonstrated that GNAI3 deficiency may increase calcium loss in
SCAPs, suggesting an essential role of GNAI3 in the dentin
mineralization and/or odonto/osteogenic differentiation of
SCAPs.
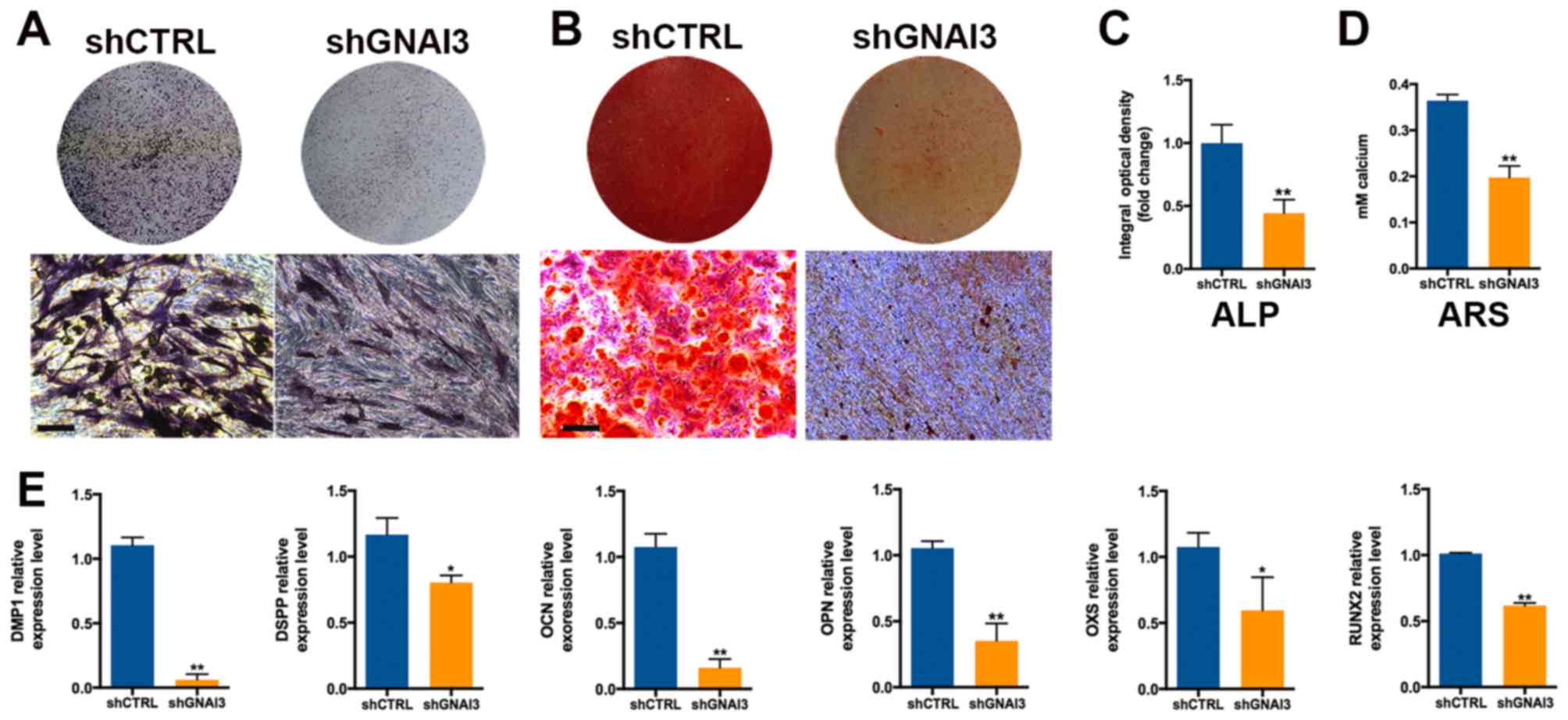 | Figure 3Effect of GNAI3 knockdown on the
odonto/osteogenic differentiation of SCAPs. (A) ALP staining assay
of SCAPs mineralization. (B) ARS staining assay for the
accumulation of calcium during SCAPs mineralization. (C)
Quantification of ALP staining in (A). (D) Quantification of ARS
staining in (B). (E) Quantitative polymerase chain reaction
detection of the mRNA expression of odonto/osteogenic markers as
indicated. Scale bar: 200 µm. *P<0.05 and
**P<0.01 vs. corresponding shCTRL group. GNAI3,
guanine and nucleotide binding protein 3; SCAPs, stem cells of the
apical papilla; sh[RNA], short hairpin [RNA]; CTRL, control; DMP1,
dentin matrix acidic phosphoprotein 1; DSPP, dentin
sialophosphoprotein; OCN, osteocalcin; OPN, osteopontin; OSX,
osterix; RUNX2, runt-related transcription factor 2. |
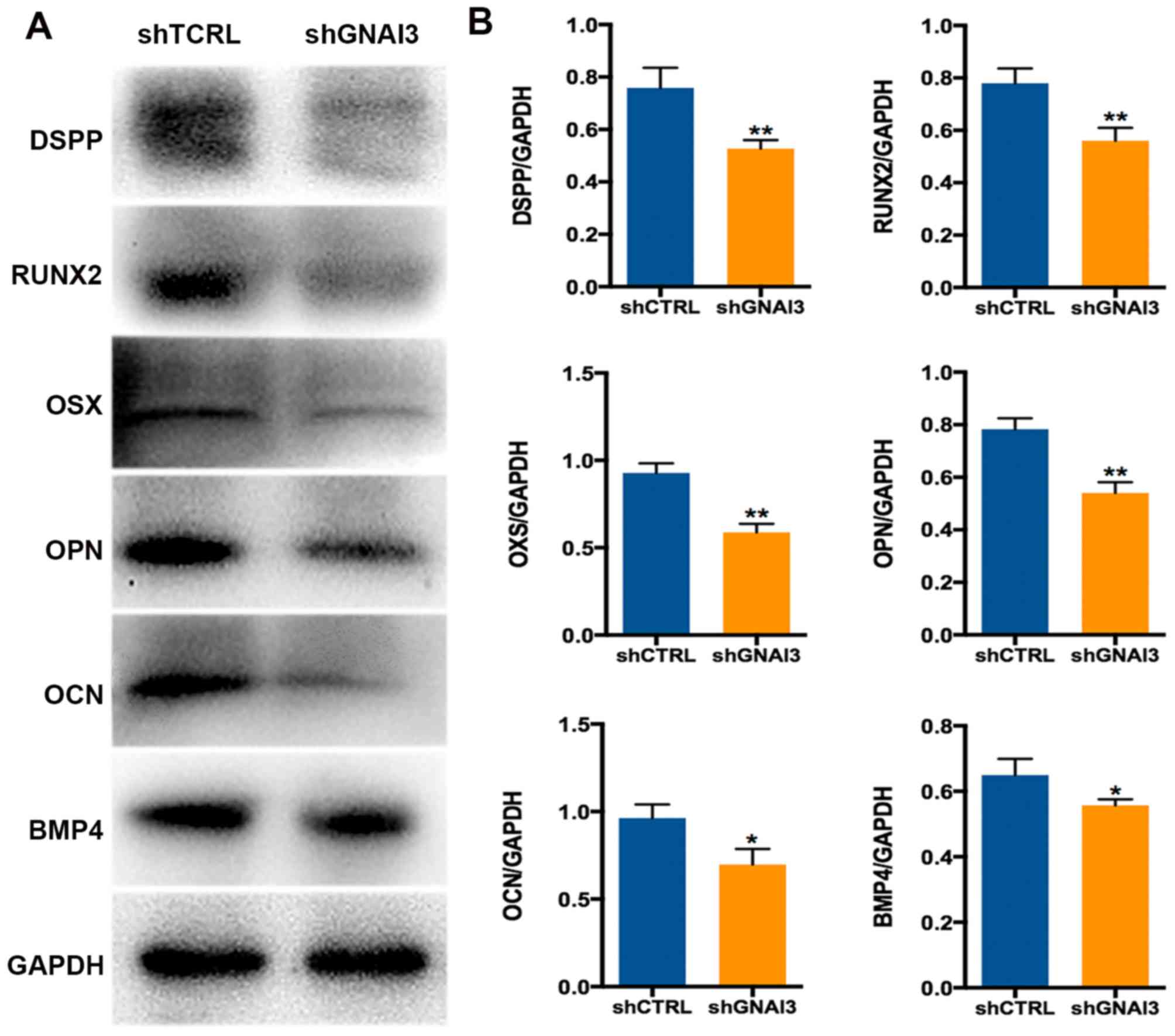
To further confirm the function of GNAI3 in the
odonto/osteogenic differentiation of SCAPs, the expression of
odonto/osteogenic differentiation and mineralization markers was
examined in GNAI3-deficient SCAPs. As shown in Fig. 3E, GNAI3-deficient SCAPs exhibited
significantly decreased mRNA expression of the markers DMP1
(P<0.01), DSPP (P<0.05), OCN (P<0.01), OPN (P<0.01),
OXS (P<0.05) and RUNX2 (P<0.01), compared with control SCAPs.
Similar results were observed at the level of protein expression
for the markers DSPP, RUNX2, OSX, OPN, OCN and BMP4 (all P<0.05
or <0.01; Fig. 4A and B). DMP1
protein expression was not detected as sufficient bands could not
be obtained despite multiple attempts with different antibodies.
Taken together, these data demonstrate that GNAI3 may be a critical
regulator of odonto/osteogenic differentiation in SCAPs, suggesting
a potential function in tooth root development.
GNAI3 knockdown inactivates JNK/ERK
signaling path- ways in the odonto/osteogenic differentiation of
SCAPs
Subsequently, the molecular mechanisms underlying
the function of GNAI3 in the odonto/osteogenic differentiation of
SCAPs were investigated. SCAPs transfected with shGNAI3 or shCTRL
lentivirus underwent osteogenic differentiation for 7 days before
extracting total protein. It was observed that GNAI3 knockdown
markedly downregulated the phosphorylation levels of JNK
(P<0.01) and ERK (P<0.05) 7 days after the mineralization
induction of SCAPs, while having no significant effect on p38
phosphorylation (Fig. 5A and B),
suggesting that JNK and ERK, but not p38, are the downstream
targets of GNAI3. Furthermore, JNK phosphorylation was found to be
induced in an apparent time-dependent manner between 0-7 days
during the odonto/osteogenic differentiation of SCAPs (Fig. 5C and D), suggesting that JNK
proteins may be actively involved in this differentiation process.
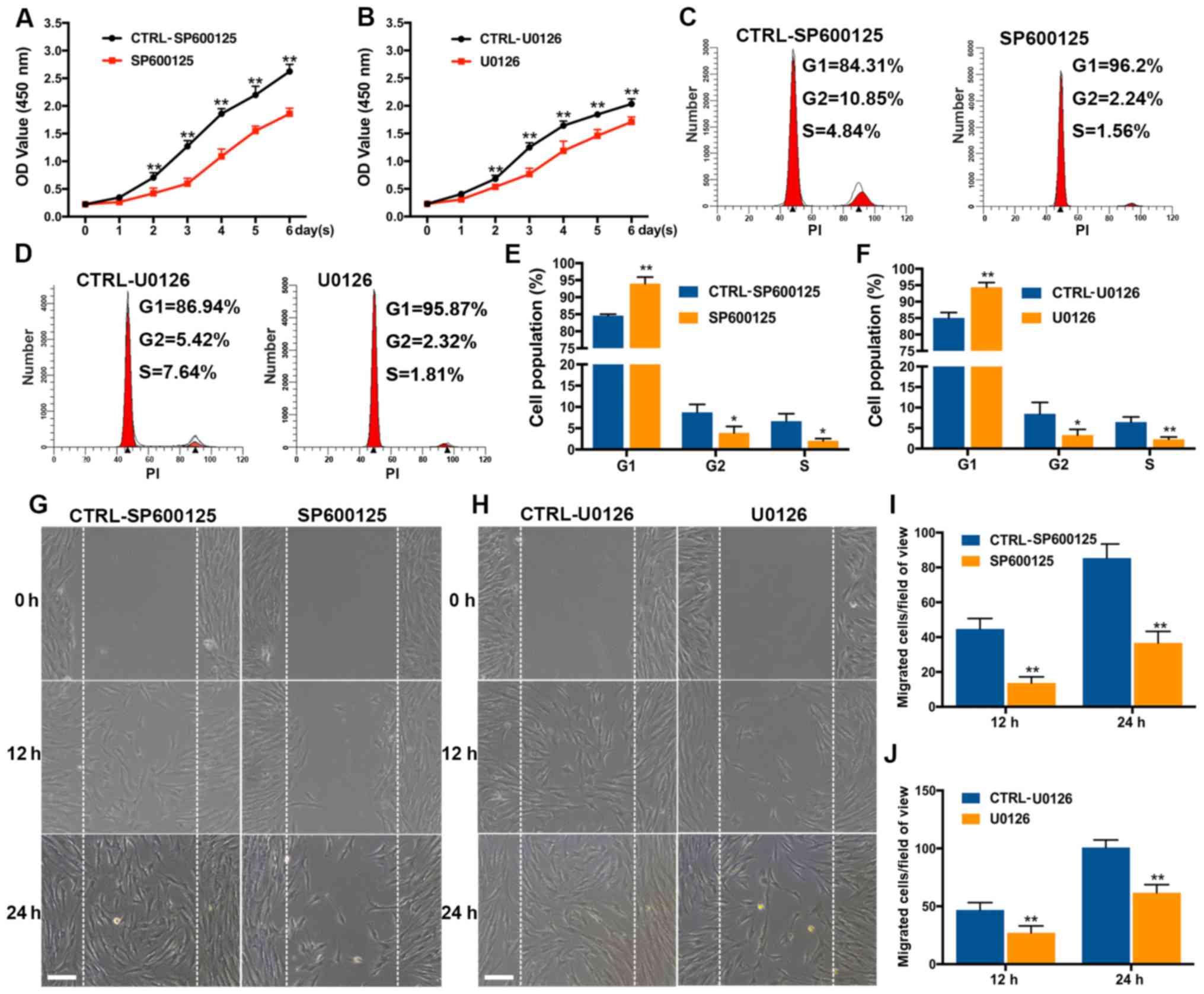
Additionally, it was found that JNK inhibitor SP600125 and ERK
inhibitor U0126 markedly suppressed the proliferation and migration
of the SCAPs. Specifically, results indicated that SP600125 and
U0126 significantly inhibited cell proliferation after 2-6 days in
an apparent time-dependent manner (P<0.01; Fig. 6A and B), as well as cell cycle
progression, as evidenced by G1 phase arrest, and S and G2-phase
cell reductions (all P<0.05 or <0.01; Fig. 6C-F). In addition, treatment with
SP600125 and U0126 caused inhibited migration ability at 12 and 24
h after exposure (P<0.01; Fig.
6G-J). The inhibitory effects of these specific JNK and ERK
inhibitors on SCAPs proliferation and migration were consistent
with the effects of GNAI3-deficiency in corresponding assays. Taken
together, these results suggested that GNAI3 promotes
odonto/osteogenic differentiation of SCAPs possibly via JNK and ERK
signaling pathways.
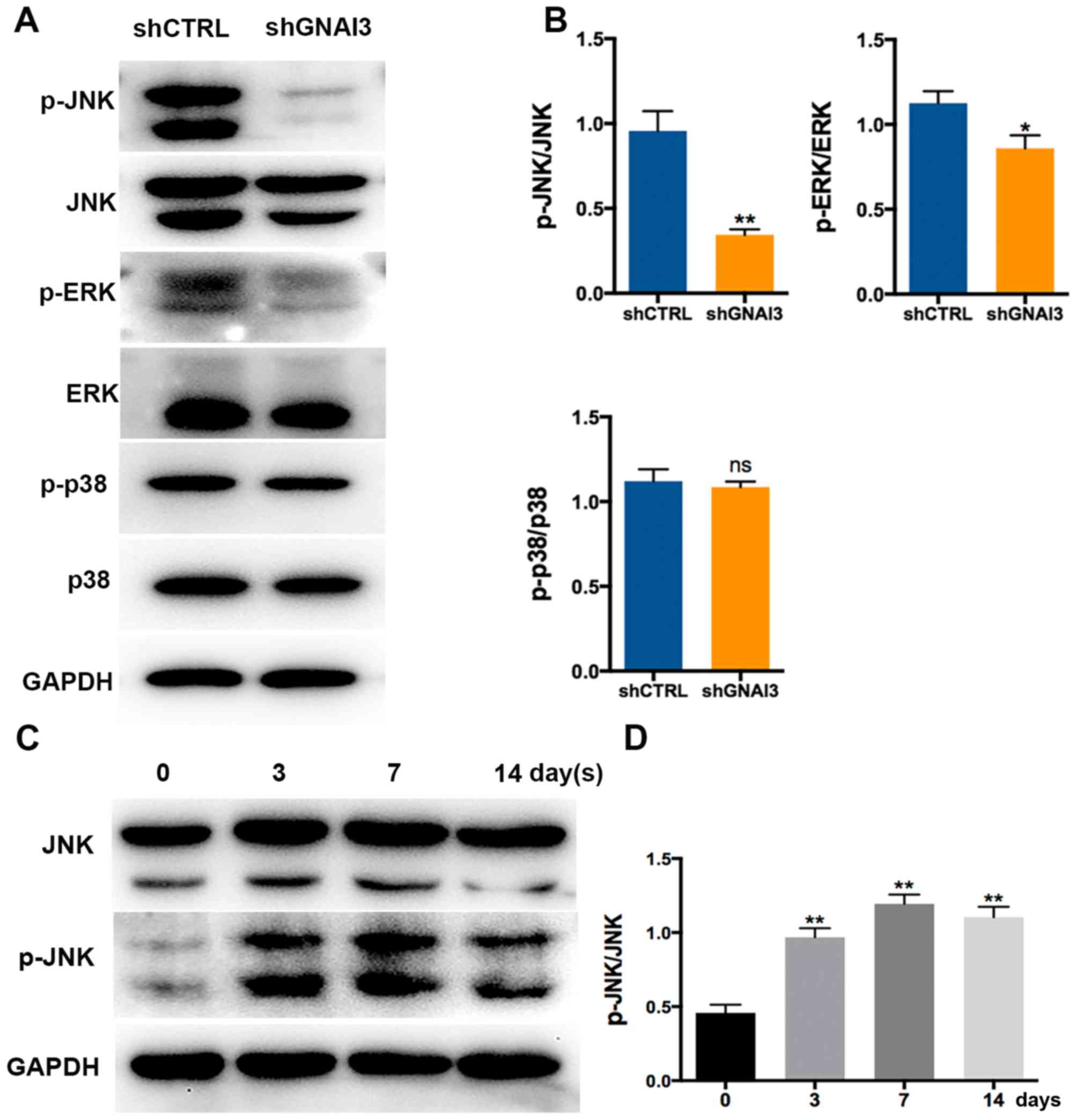 | Figure 5Effect of GNAI3 knockdown on JNK and
ERK activation. (A) Western blot analysis of the expression of
p-JNK, JNK, p-ERK, ERK, p-p38 and p38. (B) Quantification of
western blot results in (A) by normalization to GAPDH. (C)
Osteogenic differentiation of stem cells of the apical papilla was
induced for the times indicated. The protein levels of p-JNK and
JNK were determined by western blotting. (D) Quantification of
western blot results in (C). *P<0.05 and
**P<0.01 vs. corresponding shCTRL group. GNAI3,
guanine and nucleotide binding protein 3; sh[RNA], short hairpin
[RNA]; CTRL, control; JNK, c-Jun N-terminal kinase; ERK,
extracellular-signal regulated kinase; p-, phosphorylated; ns, not
significant. |
Discussion
Tooth root development is a complicated process
involving biomineralization, molecular signaling, the development
of dental tissue and tooth movement (41,42). Following crown formation, the
inner and outer epithelium of the enamel organ extends downwards
and forms a double-layered tip known as HERS. In response to the
occlusal movement of the teeth, HERS induces the formation and
mineralization of the root and determines the tooth morphology
(43). GNAI3 has been
demonstrated to play a role in regulating various cellular
processes including proliferation, cytokinesis, apoptosis,
migration and invasion (19,20,44). Causative variants in phospholipase
C beta 4, GNAI3 and endothelin 1 during early pharyngeal arch
patterning have been identified in ACS, which is characterized by
mandibular hypoplasia, suggesting that GNAI3 may be involved in
osteogenesis (45). Our previous
study demonstrated that GATA4 regulated tooth root development
through mediating the expression of GNAI3 (27), which prompted the current
investigation into the association of GNAI3 with tooth root
development. Since in mice, molar tooth crown and root formation
occurs on approximately postnatal day 11 (46), the expression of GNAI3 was
detected in the molar teeth of mice on postnatal day 14. It was
identified that GNAI3 was markedly upregulated in HERS and the
surrounding mesenchyme, which serve as two major contributors to
root formation, and this was consistent with our previous
observation (27), suggesting the
possible role of GNAI3 in tooth root development.
SCAPs are located around the open apex of premature
tooth root and have odonto/osteogenic potential (47-49). Therefore, we focused on the
expression and function of GNAI3 in the odonto/osteogenic
differentiation of SCAPs. Consistent with the in vivo data,
GNAI3 was significantly induced in SCAPs 7 days after
mineralization induction. Notably, knockdown of GNAI3 could inhibit
the proliferation, migration and odonto/osteogenic differentiation
of SCAPs, as key events involved in tooth root development. These
results indicated that GNAI3 is required for the mineralization
induction of SCAPs. Moreover, knockdown of GNAI3 also suppressed
the accumulation of calcium in SCAPs, as evidenced by the decreased
ALP activity and ARS staining intensity in GNAI3-deficient SCAPs,
suggesting a promotive role of GNAI3 in dentin mineralization.
DMP1, as an acidic non-collagenous protein expressed
in a variety of bone and tooth structures, plays a key role during
dentin and bone formation (39).
DSPP is expressed only in secretory odontoblasts, and regarded as a
tooth-specific marker contributing to the formation of dentin
(34). RUNX2 is a transcription
factor of OSX, and functions in the initial stage of osteogenesis,
whereas OPN, OCN and BMP4 have been identified as late stage
markers of osteogenesis (35-38). The current study indicated that
knockdown of GNAI3 suppressed the expression of these markers at
both transcriptional and translational levels, suggesting that
GNAI3 plays a universal role in regulating odonto/osteogenic
differentiation of SCAPs in vitro.
The MAPK signaling pathway is a major orchestrator
of complex networks, governing a number of cellular processes and
activities including epithelial-to-mesenchymal transition and cell
differentiation (24,25,50-55). Recently, it has been established
that MAPK signaling may be involved in odonto/osteogenic
differentiation of SCAPs (26).
However, it remains largely unknown whether GNAI3 functions via
MAPK signaling. In the current study, it was found that knockdown
of GNAI3 significantly inhibited the phosphorylation of JNK and
ERK, two main subfamilies of MAPKs, during SCAPs mineralization. We
further determined that specific JNK1/2/3 inhibitor (SP600125) and
specific ERK1/2 inhibitor (U0126) markedly inhibited the
proliferation and migration of the SCAPs, consistent with the
results that knockdown of GNAI3 inhibited the proliferation and
migration of the SCAPs. Thus, it appears that GNAI3 promotes
odonto/osteogenic differentiation of SCAPs and tooth root
development at least partially through activation of JNK and ERK
signaling pathways.
To further verify the role of GNAI3 in
odonto/osteogenic differentiation of SCAPs and tooth root
development, future studies by our group will perform
gain-of-function assays (overexpression of GNAI3) to investigate
whether GNAI3 itself is sufficient to induce the odonto/osteogenic
differentiation of SCAPs in vitro. Furthermore, GNAI3
transgenic mice or SCAPs-specific GNAI3 knockout mice should be
used for examination of the in vivo role of GNAI3 in tooth
root development, which is warranted in further studies.
In conclusion, the present study identified GNAI3 as
a novel regulator of odonto/osteogenic differentiation of SCAPs,
and provided insight for understanding the mechanisms underlying
the regeneration of dentin or other tissues.
Acknowledgments
Not applicable.
Funding
The current study was supported by the National
Natural Science Foundation of China (grant nos. 81700942 and
81771029) and the Priority Academic Program Development of Jiangsu
Higher Education Institutions (grant no. 2014-037).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors’ contributions
YZ, LW and JM conceived and designed the study. LY
performed the experiments and edited the manuscript. LM and MF
analyzed the data. YZ wrote the manuscript. YZ and LY revised the
manuscript for intellectual content. SG and DW provided guidance on
issues related to the experiments and manuscript writing. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The use of human samples for the study was approved
by the Medical Ethics Committee of Stomatological Hospital of
Jiangsu Province, Nanjing, China (approval no. PJ2016-038-001).
Informed consent was provided by all patients or their next of kin
for collection and use of the dental samples. Ethical approval for
the animal experimentation (approval no. 2015-03-40) was provided
by the Experimental Animal Care and Use Committee of Nanjing
Medical University, Nanjing, China.
Patient consent for publication
Written informed consent for publication of related
data on patient samples was obtained from all patients or their
parents prior to extraction surgery.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li Y, Wu Q, Wang Y, Li L, Bu H and Bao J:
Senescence of mesenchymal stem cells (Review). Int J Mol Med.
39:775–782. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garg P, Mazur MM, Buck AC, Wandtke ME, Liu
J and Ebraheim NA: Prospective review of mesenchymal stem cells
differentiation into osteoblasts. Orthop Surg. 9:13–19. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan AR and Hung CT: Concise review:
Mesenchymal stem cells for functional cartilage tissue engineering:
Taking cues from chondrocyte-based constructs. Stem Cells. Transl
Med. 6:1295–1303. 2017.
|
|
4
|
Liu GX, Zhu JC, Chen XY, Zhu AZ, Liu CC,
Lai Q and Chen ST: Inhibition of adipogenic differentiation of bone
marrow mesenchymal stem cells by erythropoietin via activating ERK
and P38 MAPK. Genet Mol Res. 14:6968–6977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng X, Sun B, Xue M, Xu P, Hu F and Xiao
Z: Comparative analysis of microRNA expression in human mesenchymal
stem cells from umbilical cord and cord blood. Genomics.
107:124–131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park YJ, Cha S and Park YS: Regenerative
applications using tooth derived stem cells in other than tooth
regeneration: A literature review. Stem Cells Int.
2016.9305986:2016.
|
|
7
|
Elahi KC, Klein G, Avci-Adali M, Sievert
KD, MacNeil S and Aicher WK: Human mesenchymal stromal cells from
different sources diverge in their expression of cell surface
proteins and display distinct differentiation patterns. Stem Cells
Int. 2016.5646384:2016.
|
|
8
|
Bakopoulou A and About I: Stem cells of
dental origin: Current research trends and key milestones towards
clinical application. Stem Cells Int. 2016.4209891:2016.
|
|
9
|
Ercal P, Pekozer GG and Kose GT: Dental
stem cells in bone tissue engineering: Current overview and
challenges. Adv Exp Med Biol: Mar. 2:2018Epub ahead of print.
|
|
10
|
Yadlapati M, Biguetti C, Cavalla F, Nieves
F, Bessey C, Bohluli P, Garlet GP, Letra A, Fakhouri WD and Silva
RM: Characterization of a vascular endothelial growth factor-loaded
bioresorbable delivery system for pulp regeneration. J Endod.
43:77–83. 2017. View Article : Google Scholar
|
|
11
|
Vanacker J, Viswanath A, De Berdt P,
Everard A, Cani PD, Bouzin C, Feron O, Diogenes A, Leprince JG and
des Rieux A: Hypoxia modulates the differentiation potential of
stem cells of the apical papilla. J Endod. 40:1410–1418. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zou XY, Yang HY, Yu Z, Tan XB, Yan X and
Huang GT: Establishment of transgene-free induced pluripotent stem
cells reprogrammed from human stem cells of apical papilla for
neural differentiation. Stem Cell Res Ther. 3:432012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prateeptongkum E, Klingelhoffer C and
Morsczeck C: The influence of the donor on dental apical papilla
stem cell properties. Tissue Cell. 47:382–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo L, Li J, Qiao X, Yu M, Tang W, Wang H,
Guo W and Tian W: Comparison of odontogenic differentiation of
human dental follicle cells and human dental papilla cells. PLoS
One. 8:e623322013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu G, Wang J, Lin X, Diao S, Cao Y, Dong
R, Wang L, Wang S and Fan Z: Demethylation of SFRP2 by histone
demethylase KDM2A regulated osteo-/dentinogenic differentiation of
stem cells of the apical papilla. Cell Prolif. 49:330–340. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chrepa V, Pitcher B, Henry MA and Diogenes
A: Survival of the apical papilla and its resident stem cells in a
case of advanced pulpal necrosis and apical periodontitis. J Endod.
43:561–567. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Downes GB and Gautam N: The G protein
subunit gene families. Genomics. 62:544–552. 1999. View Article : Google Scholar
|
|
18
|
Wang Y, Li Y and Shi G: The regulating
function of heterotrimeric G proteins in the immune system. Arch
Immunol Ther Exp (Warsz). 61:309–319. 2013. View Article : Google Scholar
|
|
19
|
Chen G, Li X, He G, Yu Z, Luo J, He J and
Huang Z: Low expression of GNAI3 predicts poor prognosis in
patients with HCC. Int J Clin Exp Med. 8:21482–21486. 2015.
|
|
20
|
Hwang IY, Park C, Luong T, Harrison KA,
Birnbaumer L and Kehrl JH: The loss of Gnai2 and Gnai3 in B cells
eliminates B lymphocyte compartments and leads to a hyper-IgM like
syndrome. PLoS One. 8:e725962013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Yao J, Huan L, Lian J, Bao C, Li
Y, Ge C, Li J, Yao M, Liang L, et al: GNAI3 inhibits tumor cell
migration and invasion and is post-transcriptionally regulated by
miR-222 in hepatocellular carcinoma. Cancer Lett. 356:978–984.
2015. View Article : Google Scholar
|
|
22
|
Kelly P, Moeller BJ, Juneja J, Booden MA,
Der CJ, Daaka Y, Dewhirst MW, Fields TA and Casey PJ: The G12
family of heterotrimeric G proteins promotes breast cancer invasion
and metastasis. Proc Natl Acad Sci USA. 103:8173–8178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rieder MJ, Green GE, Park SS, Stamper BD,
Gordon CT, Johnson JM, Cunniff CM, Smith JD, Emery SB, Lyonnet S,
et al: A human homeotic transformation resulting from mutations in
PLCB4 and GNAI3 causes auriculocondylar syndrome. Am J Hum Genet.
90:907–914. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu Y, Zhao Q, Liu Y, Zhang L, Li D, Zhu Z,
Gan X and Yu H: Vibration loading promotes osteogenic
differentiation of bone marrow-derived mesenchymal stem cells via
p38 MAPK signaling pathway. J Biomech. 71:67–75. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li S, Wang J, Han Y, Li X, Liu C, Lv Z,
Wang X, Tang X and Wang Z: Carbenoxolone inhibits mechanical
stress-induced osteogenic differentiation of mesenchymal stem cells
by regulating p38 MAPK phosphorylation. Exp Ther Med. 15:2798–2803.
2018.PubMed/NCBI
|
|
26
|
Diao S, Lin X, Wang L, Dong R, Du J, Yang
D and Fan Z: Analysis of gene expression profiles between apical
papilla tissues, stem cells from apical papilla and cell sheet to
identify the key modulators in MSCs niche. Cell Prolif.
50:e123372017. View Article : Google Scholar
|
|
27
|
Guo S, Zhang Y, Zhou T, Wang D, Weng Y,
Wang L and Ma J: Role of GATA binding protein 4 (GATA4) in the
regulation of tooth development via GNAI3. Sci Rep. 7:15342017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aurrekoetxea M, Irastorza I,
Garcia-Gallastegui P, Jimenez-Rojo L, Nakamura T, Yamada Y,
Ibarretxe G and Unda FJ: Wnt/beta-catenin regulates the activity of
epiprofin/Sp6, SHH, FGF, and BMP to coordinate the stages of
odontogenesis. Front Cell Dev Biol. 4:252016. View Article : Google Scholar
|
|
29
|
Inoue C, Sobue S, Aoyama Y, Mizutani N,
Kawamoto Y, Nishizawa Y, Ichihara M, Abe A, Hayakawa F, Suzuki M,
et al: BCL2 inhibitor ABT-199 and JNK inhibitor SP600125 exhibit
synergistic cytotoxicity against imatinib-resistant Ph+ ALL cells.
Biochem Biophys Rep. 15:69–75. 2018.PubMed/NCBI
|
|
30
|
Su X, Wang X, Zhang K, Yang S, Xue Q, Wang
P and Liu Q: ERK inhibitor U0126 enhanced SDT-induced cytotoxicity
of human leukemia U937 cells. Gen Physiol Biophys. 33:295–309.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saito MT, Silverio KG, Casati MZ, Sallum
EA and Nociti FH Jr: Tooth-derived stem cells: Update and
perspectives. World J Stem Cells. 7:399–407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang W, Zhang X, Ling J, Liu W, Zhang X,
Ma J and Zheng J: Proliferation and odontogenic differentiation of
BMP2 genet-ransfected stem cells from human tooth apical papilla:
An in vitro study. Int J Mol Med. 34:1004–1012. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bakopoulou A, Leyhausen G, Volk J, Koidis
P and Geurtsen W: Comparative characterization of
STRO-1(neg)/CD146(pos) and STRO-1(pos)/CD146(pos) apical papilla
stem cells enriched with flow cytometry. Arch Oral Biol.
58:1556–1568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iejima D, Sumita Y, Kagami H, Ando Y and
Ueda M: Odontoblast marker gene expression is enhanced by a
CC-chemokine family protein MIP-3alpha in human mesenchymal stem
cells. Arch Oral Biol. 52:924–931. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nakashima K, Zhou X, Kunkel G, Zhang Z,
Deng JM, Behringer RR and de Crombrugghe B: The novel zinc
finger-containing transcription factor osterix is required for
osteoblast differentiation and bone formation. Cell. 108:17–29.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang W, Yang S, Shao J and Li YP:
Signaling and transcriptional regulation in osteoblast commitment
and differentiation. Front Biosci. 12:3068–3092. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen D, Harris MA, Rossini G, Dunstan CR,
Dallas SL, Feng JQ, Mundy GR and Harris SE: Bone morphogenetic
protein 2 (BMP-2) enhances BMP-3, BMP-4, and bone cell
differentiation marker gene expression during the induction of
mineralized bone matrix formation in cultures of fetal rat
calvarial osteoblasts. Calcif Tissue Int. 60:283–290. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Twine NA, Chen L, Pang CN, Wilkins MR and
Kassem M: Identification of differentiation-stage specific markers
that define the ex vivo osteoblastic phenotype. Bone. 67:23–32.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hirst KL, Simmons D, Feng J, Aplin H,
Dixon MJ and MacDougall M: Elucidation of the sequence and the
genomic organization of the human dentin matrix acidic
phosphoprotein 1 (DMP1) gene: Exclusion of the locus from a
causative role in the pathogenesis of dentinogenesis imperfecta
type II. Genomics. 42:38–45. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
41
|
Thesleff I: Epithelial-mesenchymal
signalling regulating tooth morphogenesis. J Cell Sci.
116:1647–1648. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang Y, Chen N and Miao D:
Pyrroloquinoline quinone plays an important role in rescuing
Bmi-1(−/−) mice induced developmental disorders of teeth and
mandible-anti-oxidant effect of pyrroloquinoline quinone. Am J
Transl Res. 10:40–53. 2018.
|
|
43
|
Xiong J, Gronthos S and Bartold PM: Role
of the epithelial cell rests of Malassez in the development,
maintenance and regeneration of periodontal ligament tissues.
Periodontol 2000. 63:217–233. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rasheed SA, Teo CR, Beillard EJ, Voorhoeve
PM and Casey PJ: MicroRNA-182 and microRNA-200a control G-protein
subunit alpha-13 (GNA13) expression and cell invasion
synergistically in prostate cancer cells. J Biol Chem.
288:7986–7995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Romanelli Tavares VL, Gordon CT,
Zechi-Ceide RM, Kokitsu-Nakata NM, Voisin N, Tan TY, Heggie AA,
Vendramini-Pittoli S, Propst EJ, Papsin BC, et al: Novel variants
in GNAI3 associated with auriculocondylar syndrome strengthen a
common dominant negative effect. Eur J Hum Genet. 23:481–485. 2015.
View Article : Google Scholar :
|
|
46
|
Li J, Parada C and Chai Y: Cellular and
molecular mechanisms of tooth root development. Development.
144:374–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chueh LH and Huang GT: Immature teeth with
periradicular periodontitis or abscess undergoing apexogenesis: A
paradigm shift. J Endod. 32:1205–1213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sonoyama W, Liu Y, Fang D, Yamaza T, Seo
BM, Zhang C, Liu H, Gronthos S, Wang CY, Wang S and Shi S:
Mesenchymal stem cell-mediated functional tooth regeneration in
swine. PLoS One. 1:e792006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang GT, Gronthos S and Shi S:
Mesenchymal stem cells derived from dental tissues vs. those from
other sources: Their biology and role in regenerative medicine. J
Dent Res. 88:792–806. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cao Y, Xia DS, Qi SR, Du J, Ma P, Wang SL
and Fan ZP: Epiregulin can promote proliferation of stem cells from
the dental apical papilla via MEK/Erk and JNK signalling pathways.
Cell Prolif. 46:447–456. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liao X, Feng B, Zhang D, Liu P, Zhou X, Li
R and Ye L: The Sirt6 gene: Does it play a role in tooth
development. PLoS One. 12:e01742552017. View Article : Google Scholar
|
|
52
|
Morris MA, Laverick L, Wei W, Davis AM,
O’Neill S, Wood L, Wright J, Dawson CW and Young LS: The
EBV-encoded oncoprotein, LMP1, induces an epithelial-to-mesenchymal
transition (EMT) via Its CTAR1 domain through integrin-mediated
ERK-MAPK signalling. Cancers (Basel). 10:e1102018. View Article : Google Scholar
|
|
53
|
Yang S, Guo L, Su Y, Wen J, Du J, Li X,
Liu Y, Feng J, Xie Y, Bai Y, et al: Nitric oxide balances
osteoblast and adipocyte lineage differentiation via the JNK/MAPK
signaling pathway in periodontal ligament stem cells. Stem Cell Res
Ther. 9:1182018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu P, Wang J, Sun B and Xiao Z: Integrated
analysis of miRNA and mRNA expression data identifies multiple
miRNAs regulatory networks for the tumorigenesis of colorectal
cancer. Gene. 659:44–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen D, Wang H, Chen J, Li Z, Li S, Hu Z,
Huang S, Zhao Y and He X: MicroRNA-129-5p regulates glycolysis and
cell proliferation by targeting the glucose transporter SLC2A3 in
gastric cancer cells. Front Pharmacol. 9:5022018. View Article : Google Scholar : PubMed/NCBI
|