Introduction
Breast cancer is among the most common types of
cancer in female patients, with a high global mortality rate
(1). Over 1,000,000 women are
diagnosed each year, and ≥410,000 diagnosed patients succumb to
mortality, accounting for ~14.0% of cases of cancer-associated
mortality in women (2). In recent
years, the incidence of breast cancer has increased by 3% each year
in China, seriously threatening the health of women and causing a
burden on society (3). According
to gene expression profiling, breast cancer is categorized into
four major subtypes: Luminal A, luminal B, human epidermal growth
factor receptor 2 positive (Her2+) and basal-like
(4). Different subgroups are
associated with different clinical outcomes (1). However, triple negative-breast
cancer (TNBC), which is characterized by the lack of estrogen
receptor, progesterone receptor, and expression of Her-2, is not
effectively treated by endocrine therapy or Her-2-targeted therapy
(5). TNBC has been a popular
focus of investigation in clinical and scientific fields over the
last decade (6). Since 2014, the
correlation between the progression of TNBC and epigenetics has
been investigated thoroughly (7).
The identification of effective therapies for TNBC is urgent.
Long non-coding RNAs (lncRNAs), which are RNA
molecules >200 nt in length, are important in a large number of
cellular processes, including alterations of the cell- or
tissue-specific expression profile, and the promotion of cell
proliferation, metastasis and invasion (8). lncRNAs have been reported to be of
significance in the diagnosis and treatment of multiple types of
disease, specifically the tumorigenesis and progression of various
types of cancer, including osteo-sarcoma and TNBC (9,10).
The functions of lncRNAs include modulation of gene methylation,
activation of transcription, regulation of translation and other
processes in conjugation with mRNAs and microRNAs (miRNAs)
(11). Human ovarian
cancer-specific transcript 2 (HOST2) is an lncRNA of 2.9 kb, with
no identified open reading frame (12). The lncRNA HOST2 is expressed at
high levels in epithelial ovarian cancer, and the inhibition of
HOST2 has been demonstrated to reduce the proliferation, migration
and invasion of ovarian cancer cells (13).
Accordingly, the present study aimed to investigate
the effects of lncRNA HOST2 on TNBC cell proliferation. This may be
beneficial for understanding the developmental mechanisms of TNBC,
and to identify potential biomarkers for the diagnosis and therapy
of this disease.
Materials and methods
Cell lines and reagents
The MDA-MB-231 and MDA-MB-468 human TNBC cell lines,
and the MCF10A normal human mammary epithelial cell line were
purchased from American Type Culture Collection (Manassas, VA,
USA). The MCF10A cells were maintained in DMEM/Ham's F-12
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 100 ng/ml cholera toxin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany), 20 ng/ml epidermal growth factor
(Sigma-Aldrich, Merck KGaA), 0.01 mg/ml insulin (Sigma-Aldrich,
Merck KGaA), 500 ng/ml hydrocortisone (Sigma-Aldrich, Merck KGaA),
and 5% chelex-treated horse serum (Sigma-Aldrich, Merck KGaA). The
MDA-MB-468 and MDA-MB-231 cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) or L-15 (Gibco; Thermo
Fisher Scientific, Inc.) The culture medium was supplemented with
10% (v/v) fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml streptomycin (Gibco; Thermo Fisher Scientific,
Inc.), and 100 U/ml penicillin (Gibco; Thermo Fisher Scientific,
Inc.) in an incubator with 5% CO2 at 37°C.
Tissue collection
In total, 30 patients with primary TNBC, treated at
Wuhai People's Hospital, (Wuhai, China), participated in the
present study. The primary TNBC tumor tissues and adjacent normal
tissues were collected, washed, and frozen in liquid nitrogen
immediately following surgery. No patients had received other
treatments prior to surgery. Informed consent regarding the use of
these samples was obtained from each patient. The present study was
approved by the Ethics Committee of Wuhai People's Hospital.
Patient information is summarized in Table I.
 | Table IAssociation between the expression of
HOST2 and patient clinical and pathological characteristics. |
Table I
Association between the expression of
HOST2 and patient clinical and pathological characteristics.
| Clinicopathological
feature | Cases (n) | Expression of HOST2
| P-value |
|---|
| Low (n) | High (n) |
|---|
| Age (years) | | | | 0.699 |
| ≤45 | 10 | 4 | 6 | |
| >45 | 20 | 11 | 9 | |
| Tumor size
(cm) | | | | 0.710 |
| ≤2 | 12 | 7 | 5 | |
| >2 | 18 | 8 | 10 | |
| TNM stage | | | | 0.023a |
| I | 8 | 7 | 1 | |
| II | 10 | 5 | 5 | |
| III | 12 | 3 | 9 | |
| Ki-67 | | | | 0.139 |
| ≤20 | 17 | 11 | 6 | |
| >20 | 13 | 4 | 9 | |
Cell transfection
A total of 2×105 cells/well were seeded
in 6-well plates and incubated in an incubator at 37°C with 5%
CO2 for 24 h to a confluence of 50-60%. The cells were
transfected with control small interfering (si)RNA and HOST2 siRNA
(Invitrogen; Thermo Fisher Scientific, Inc.) using Lipofectamine
RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol, and incubated at room temperature for
20 min. The lnc-HOST2 siRNA sequences used were as follows: HOST2
siRNA, 5'-GAC TAA ACA AGG TCT TAA TTT-3' and HOST2 siRNA#, 5'-TGA
CTA AAC AAG GTC TTA ATT T-3'. The negative control siRNA sequence
used was 5'-TTC TCC GAA CGT GTC ACG TTT-3'. The transfected cells
were washed with PBS three times, followed by incubation with 1.5
ml fresh serum/antibiotic-free MEM for 48 h for the subsequent
experiments.
Western blot analysis
Proteins were extracted from the cell lysates using
RIPA buffer (Roche Diagnostics, Shanghai, China). The protein
concentration was determined using a BCA kit (Beyotime Institute of
Biotechnology, Shanghai, China). A total of 10 µg protein
per sample was separated by 10% polyacrylamide gel electrophoresis
Subsequently, the protein was transferred onto polyvinylidene
fluoride (PVDF) membranes (EMD Millipore). The PVDF membranes were
blocked with 5% bovine serum albumin (Sigma-Aldrich, Merck KGaA) at
room temperature for 1 h, and incubated with primary antibodies
against CDK6 (cat. no. 13331; 1:1,000) and GAPDH (cat. no. 5174;
1:5,000) (Cell Signaling Technologies, Inc., Danvers, MA, USA) at
4°C overnight. The following day, the PVDF membranes were washed
three times with TBS-Tween, incubated with a secondary antibody
(horseradish peroxidase-linked anti-rabbit IgG; Cell Signaling
Technologies, Inc., cat. no. 7074, 1:10,000) at room temperature
for 1 h, and washed three times again with TBST. Finally, the
proteins were visualized using chemiluminescence reagent and images
were captured with the Bio-Rad Gel Dol EZ system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the tissues or cells
with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA
was synthesized using a reverse transcription kit (Promega
Corporation, Madison, WI, USA).
To evaluate the expression of let-7b, the PCR
reaction contained 5 µl 2X SYBR-Green Taq reaction liquid,
0.2 µl Rox II, 0.2 µl sense primer, 0.2 µl
antisense primer, 1 µl cDNA and 3.4 µl
ddH2O. The thermocycling conditions were as follows:
94°C for 10 sec, 94°C for 5 sec, and 60°C for 34 sec (40 cycles).
The primers were designed using Primer 5.0 software (Premier
Biosoft International, Palo Alto, CA, USA) according to the gene
sequences in the Genebank database (www.ncbi.nlm.nih.gov/), the sequences were as follows:
let-7b, forward 5'-TGA GGT AGT AGG TTG TGT GGT T-3' and miR
universal reverse 5'-GTG CAG GGT CCG AGG T-3'; U6, forward 5'-CTC
GCT TCG GCA GCA CA-3' and reverse 5'-AAC GCT TCA CGA ATT TGC GT-3'.
U6 was used as an internal control.
For the evaluation of HOST2 and CDK6, the PCR
reaction consisted of 5 µl 2X super Real, 0.3 µl
forward primer, 0.3 µl reverse primer, 2 µl reverse
transcription products, 1 µl 10X ROX and 1.4 µl water
without RNase. The thermocycling conditions were as follows: 95°C
for 15 min, followed by 95°C for 10 sec, annealing for 30 sec and
extension for 30 sec (40 cycles). The primers were designed by
Primer 5.0 software, according to the gene sequences in the
Genebank database, and the sequences are as follows: HOST2 forward,
5'-CTC AAA TCA ATC ACG ACC CT-3' and reverse, 5'-AAT GTA GCA GGA
CGA GCC-3'; CDK6 forward, 5'-TGGA GAC CTT CGA GC A CC-3' and
reverse 5'-CAC TCC AGG CTC TGG AAC TT-3'; and GAPDH forward, 5'-GGT
GAA GGT CGG AGT CA A CG-3' and reverse, 5'-CAA AGT TGT CAT GGA TGH
ACC-3'. GAPDH was used as an internal control. The gene expression
levels were quantified using the 2−ΔΔCq method (14).
Flow cytometry with PI staining
At 48 h post-transfection, the cells were washed
three times with ice-cold PBS and centrifuged at 4°C for 10 mins
(1,000 × g). The cells were re-suspended and adjusted to a density
of 1×105 cells/ml. The cells were fixed with 1 ml
ice-cold 75% ethanol overnight at 4°C. The following day, the cells
were washed three times with PBS, followed by aspiration of the
supernatant. The cells were then treated with 100 µl RNaseA
in a water bath at 37°C for 30 min in the dark, followed by
staining with 400 µl PI and incubation at 4°C for 30 min in
the dark. The phases of the cell cycle were evaluated by flow
cytometry (Coulter Corporation Co., Ltd., California, USA) using
red fluorescence at 488 nm.
Cell Counting kit-8 (CCK-8) assay
When the transfected cells reached 60% confluence,
they were washed by PBS three times and digested into a single-cell
suspension using 0.25% trypsin. The cells were then seeded in a
96-well plate (200 µl/well) at a density of 6×103
cells/well, and cultured for 48 h at 37°C. Following this, 10
µl CCK-8 solution was added to each well, and the cells were
cultured for another 2 h at 37°C. The optical density (OD) in each
well was evaluated at a wavelength of 450 nm.
Overexpression of let-7b
miRNA (miR)-NC mimics and let-7b mimics were
synthesized by Guangzhou Ribobio Co., Ltd. (Guangzhou, China). To
induce let-7b overexpression, the let-7b mimics were mixed with
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in
serum-free medium for 15 mins, and then added to the cells.
Bioinformatics analysis and dual
luciferase activity assay
The bioinformatics analysis of the potential binding
site between let-7b and CDK6 3'UTR was performed on miRDB
(http://mirdb.org/). Oligonucleotides containing the
CDK6 cDNA fragment, including let-7b binding sites, were amplified
from the MDA-MB-231 cells and cloned into pmirGLO plasmids (Promega
Corporation). Mutant CDK6 (pmirGLO-CDK6-MUT) served as a negative
control and was generated by site-directed mutagenesis PCR using
platinum pfx DNA polymerase, according to the manufacturer's
protocol. The luciferase reporter plasmid and let-7b mimics or
miR-NC mimics were co-transfected into MDA-MB-231 cells using
Lipofectamine 2000. At 48 h post-transfection, the relative
luciferase activity was examined using a Dual-Luciferase Reporter
Assay system (Promega Corporation) in a luminometer. For the
analysis of let-7b and HOST2 binding, 1,500-2,000 nt of the HOST2
sequence was amplified from MCF10A cDNA and annealed into the pGL3
plasmid (Promega Corporation). Mutations were introduced via
site-directed mutagenesis PCR using platinum pfx DNA polymerase to
construct pGL3-HOST2 mutant (Mut). In the dual luciferase reporter
assay, the MAD-MB-231 and MDA-MB-468 cells were co-transfected with
pGL3-HOST2 wild-type (WT) or pGL3-HOST2 Mut in combination with
let-7b mimics or miR-NC mimics using Lipofectamine 2000. After 48
h, the relative luciferase activity was examined using a
Dual-Luciferase Reporter Assay system (Promega Corporation) in a
luminometer.
Statistical analysis
SPSS 21.0 (IBM SPSS, Armonk, NY, USA) was used for
statistical analysis. The data are presented as the mean ± standard
deviation. Comparisons of two groups were made using Student's
t-test. One-way analysis of variance followed by NewmanKeuls post
hoc analysis was used to compare the means of multiple groups. The
association between HOST2 levels and patient clinical and
pathological characteristics were analysed using Chi-square test.
P<0.05 was considered to indicate a statistically significant
difference. The experiments were repeated three times.
Results
Overexpression of HOST2 in TNBC tumor
tissues and cell lines
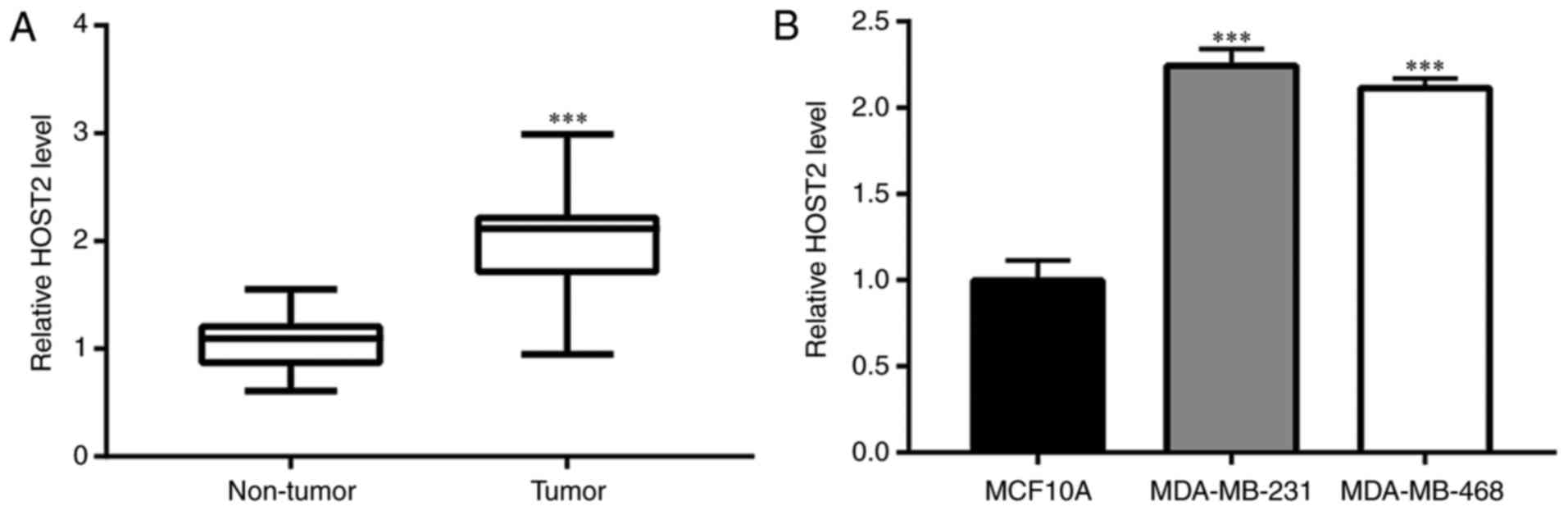
To evaluate the role of HOST2 in TNBC, RT-qPCR
analysis was performed to compare the expression of HOST2 between
TNBC tumor tissues and adjacent normal tissues. Compared with the
adjacent normal tissues, the expression of HOST2 was significantly
increased in the TNBC tumor tissues (Fig. 1A). The expression of HOST2 was
also detected in MCF10A normal breast epithelial cells and in the
MDA-MB-231 and MDA-MB-468 TNBC cell lines. Increased expression of
HOST2 was observed in the TNBC cells, compared with the MCF10A
cells (Fig. 1B). The potential
clinicopathological implications of the expression of HOST2 were
also investigated in 30 patients with TNBC. The patients were
divided into low and high HOST2 expression groups. Analysis of the
association between the expression of HOST2 and patient
clinicopathological parameters revealed that a high expression of
HOST2 was closely associated with TNBC tumor-node-metastasis stage.
However, the expression of HOST2 was not associated with age, tumor
size or the expression of Ki-67 (Table I).
Overall, the above results suggested the oncogenic
potential of HOST2, and that it is involved in the progression of
TNBC.
Silencing of HOST2 inhibits cell
proliferation and cell cycle arrest in TNBC cells
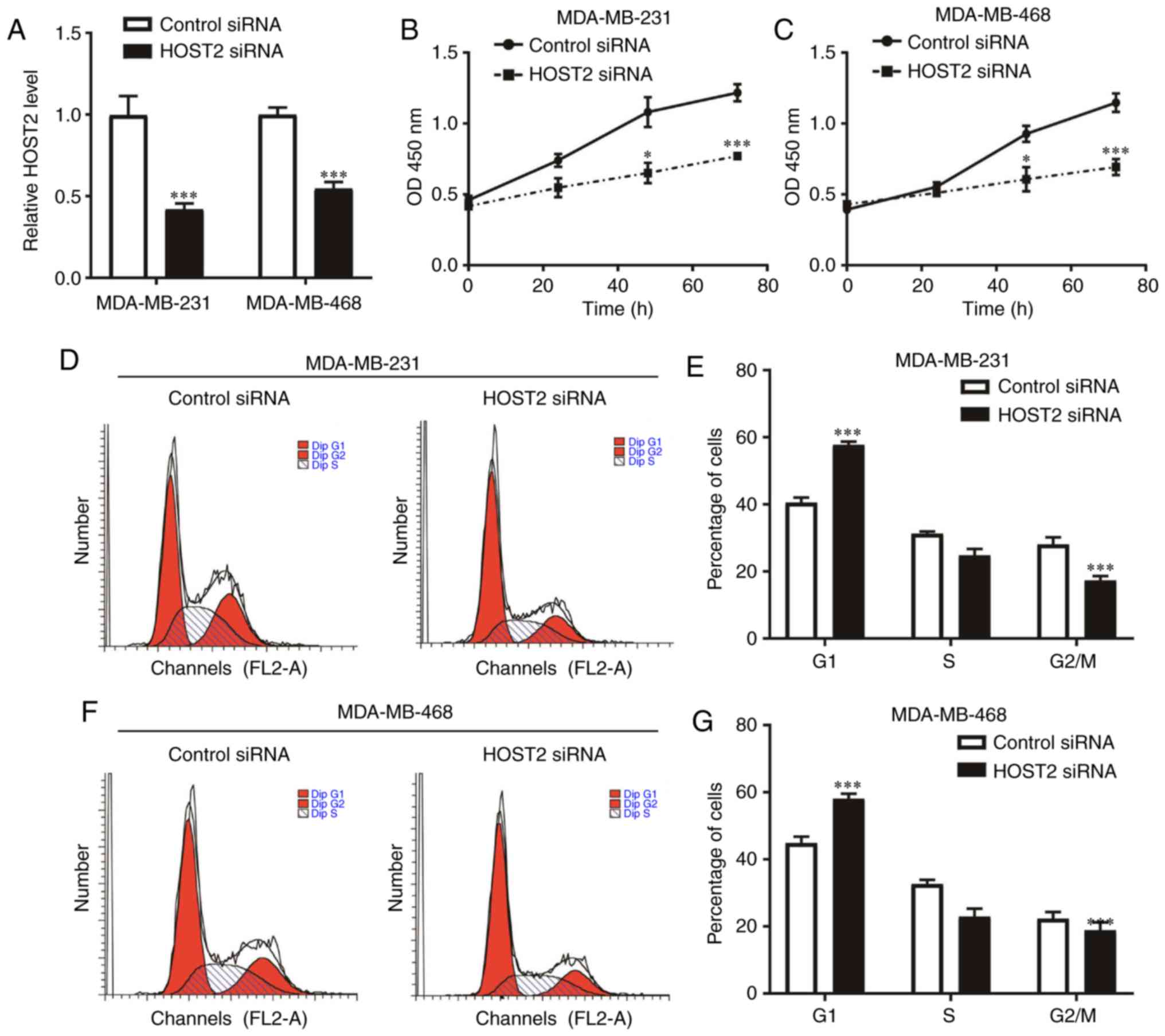
The silencing of HOST2 was induced to assess the
function of HOST2 in TNBC cells. HOST2 siRNA transfection decreased
the expression of HOST2 in the MDA-MB-231 and MDA-MB-468 cells
(Fig. 2A). The CCK-8 assay
indicated that HOST2 silencing led to a significant decrease in the
proliferative rate of the MDA-MB-231 and MDA-MB-468 cells (Fig. 2B and C). Cell cycle arrest was a
major reason for cell proliferation inhibition. Using flow
cytometric analysis, it was found that HOST2 silencing increased
the proportion of MDA-MB-231 and MDA-MB-468 cells in the G1 phase
(Fig. 2D-G), but did not affect
the apoptotic cell rate of the MDA-MB-231 and MDA-MB-468 cells
(data not shown). Therefore, HOST2 may promote TNBC progression
through regulation of the G1-S checkpoint.
Silencing of HOST2 represses the
expression of CDK6 in TNBC cells
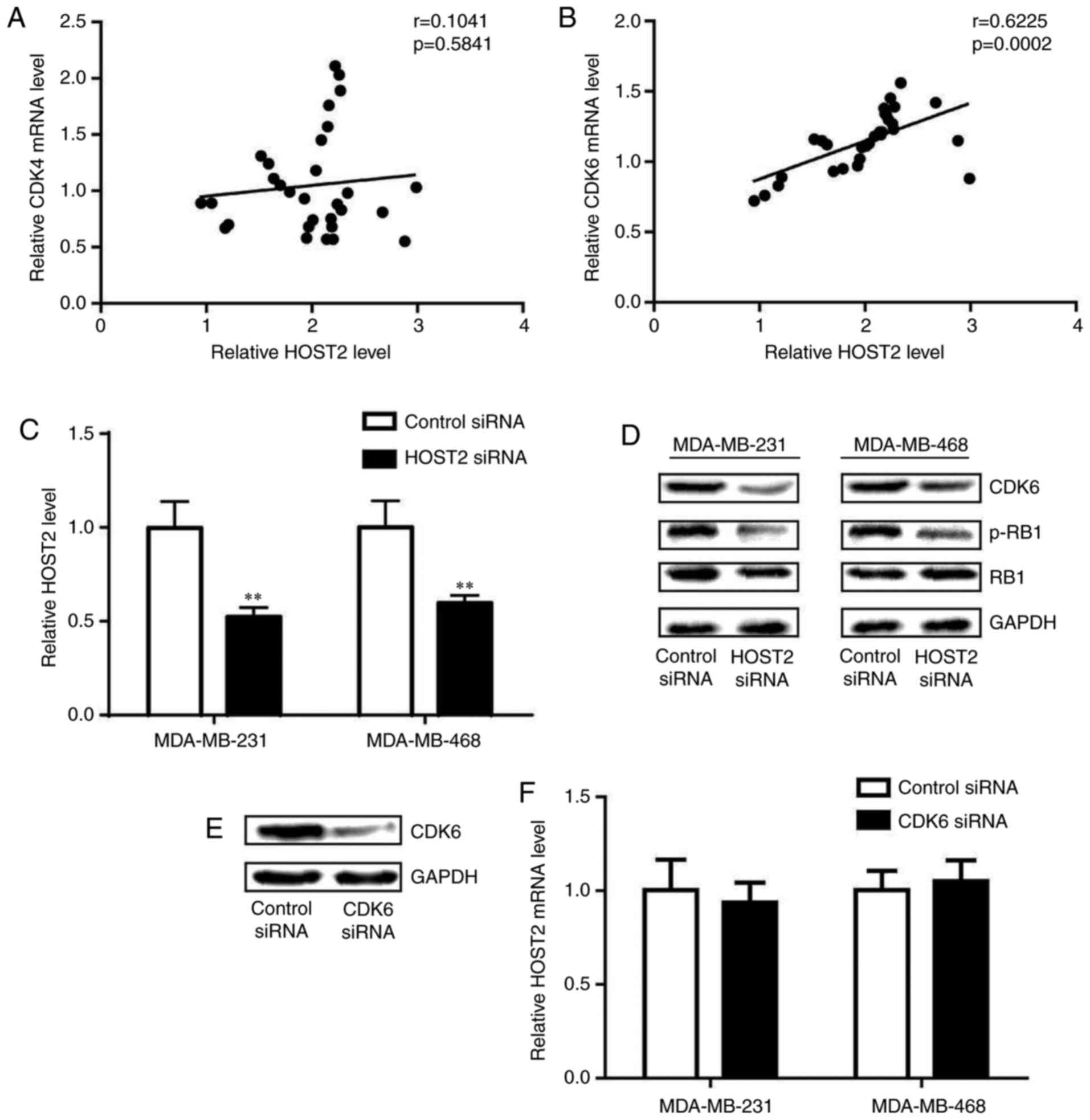
The cyclin D-CDK4/6 complex has been demonstrated to
be pivotal in controlling the G1-S checkpoint (15). The analysis of gene expression
levels in TNBC tumor tissues revealed that the expression of CDK6
was positively correlated with that of HOST2, whereas the
expression of CDK4 was not (Fig. 3A
and B). Consistently, silencing of the expression of HOST2
decreased the expression of CDK6 in the MDA-MB-231 and MDA-MB-468
cells (Fig. 3C). During the G1-S
phase, the phosphorylation of RB1 by CDK6 permits the transcription
of S phase genes, and is required for DNA replication (16). Western blot analysis revealed that
the protein expression level of CDK6 and the phosphorylation of RB1
were reduced following HOST2 silencing in the MDA-MB-231 and
MDA-MB-468 cells (Fig. 3D). To
determine whether CDK6 regulated HOST2, CDK6 siRNA was transfected
into TNBC cells. The transfection of CDK6 siRNA decreased the
expression of CDK6 (Fig. 3E).
CDK6 silencing did not alter the level of HOST2 in the MDA-MB-231
or MDA-MB-468 cells (Fig.
3F).
Collectively, the above data suggested that CDK6, a
key regulatory kinase in the cell cycle, was negatively regulated
by HOST2 in TNBC cells.
HOST2 regulates CDK6 via the repression
of let-7b
lncRNAs can function as sponge RNAs to regulate gene
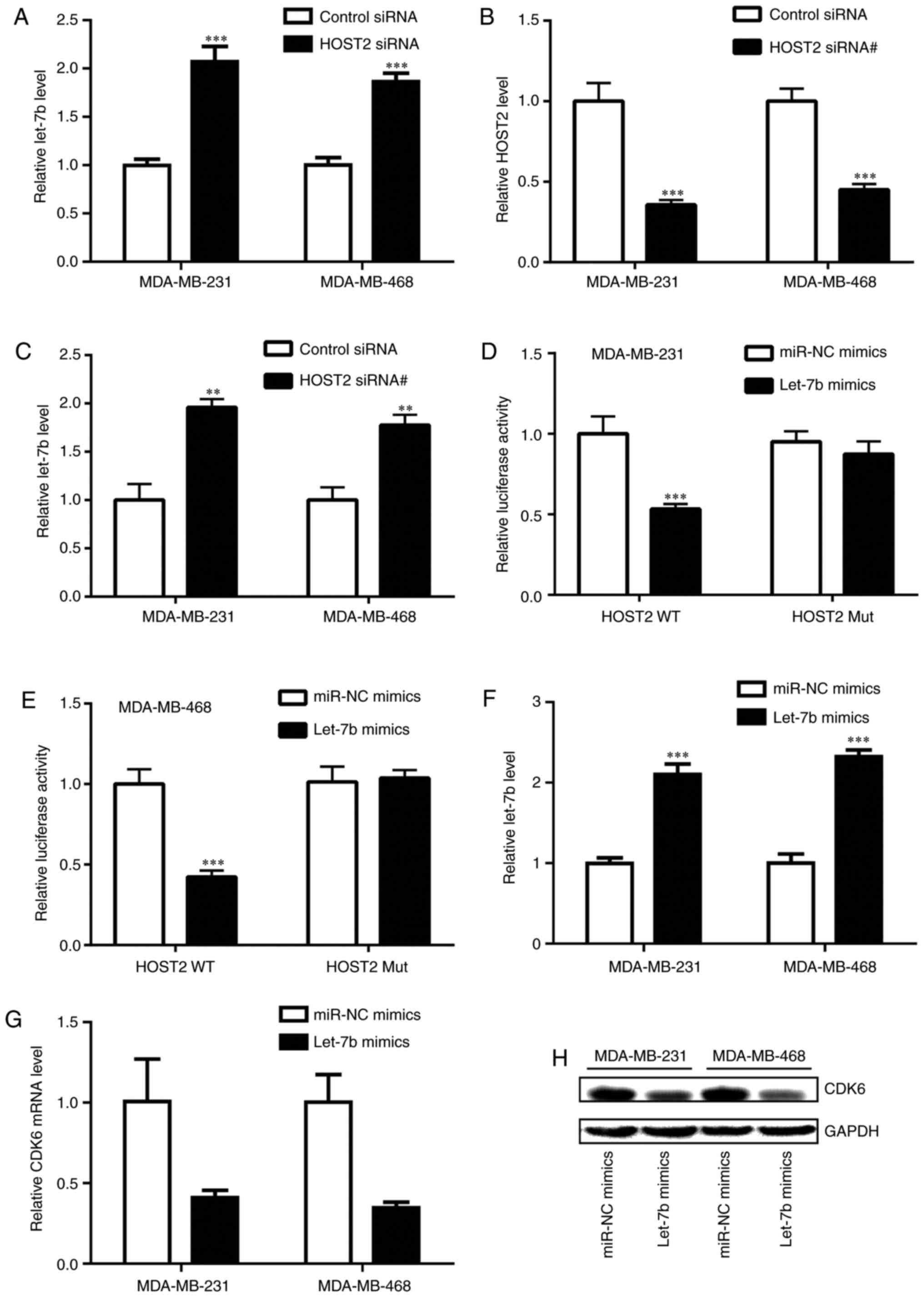
expression (17). A previous
study revealed that HOST2 repressed the expression of let-7b in
breast cancer (18). To determine
whether HOST2 controlled CDK6 levels through the sponging of
let-7b, the expression of HOST2 was silenced in the TNBC cells. The
knockdown of HOST2 increased the expression levels of let-7b in the
MDA-MB-231 and MDA-MB-468 cells (Fig.
4A). To further validate the regulation of let-7b by HOST2,
another independent HOST2 siRNA (HOST2 siRNA#) was used to knock
down HOST2 in the TNBC cells. Consistent with the HOST siRNA,
transfection with HOST2 siRNA# also increased the expression of
let-7b (Fig. 4B and C). In
addition, the transfection of let-7b mimics reduced the luciferase
activity of MDA-MB-231 cells transfected with pGL3-HOST2 WT
containing a putative binding site for let-7b (Fig. 4D). Similarly, in the MDA-MB-468
cells, the overexpression of let-7b repressed luciferase activity
in the cells transfected with pGL3-HOST2 WT (Fig. 4E), indicating that HOST2 directly
repressed the expression of let-7b. In addition, the enhanced
expression of let-7b caused by transfection of let-7b mimics
(Fig. 4F) reduced the expression
of CDK6 at the mRNA and protein levels in the MDA-MB-231 and
MDA-MB-468 cells (Fig. 4G and H).
These findings suggested that HOST2 controlled the expression of
CDK6 through sponging of let-7b in the TNBC cells.
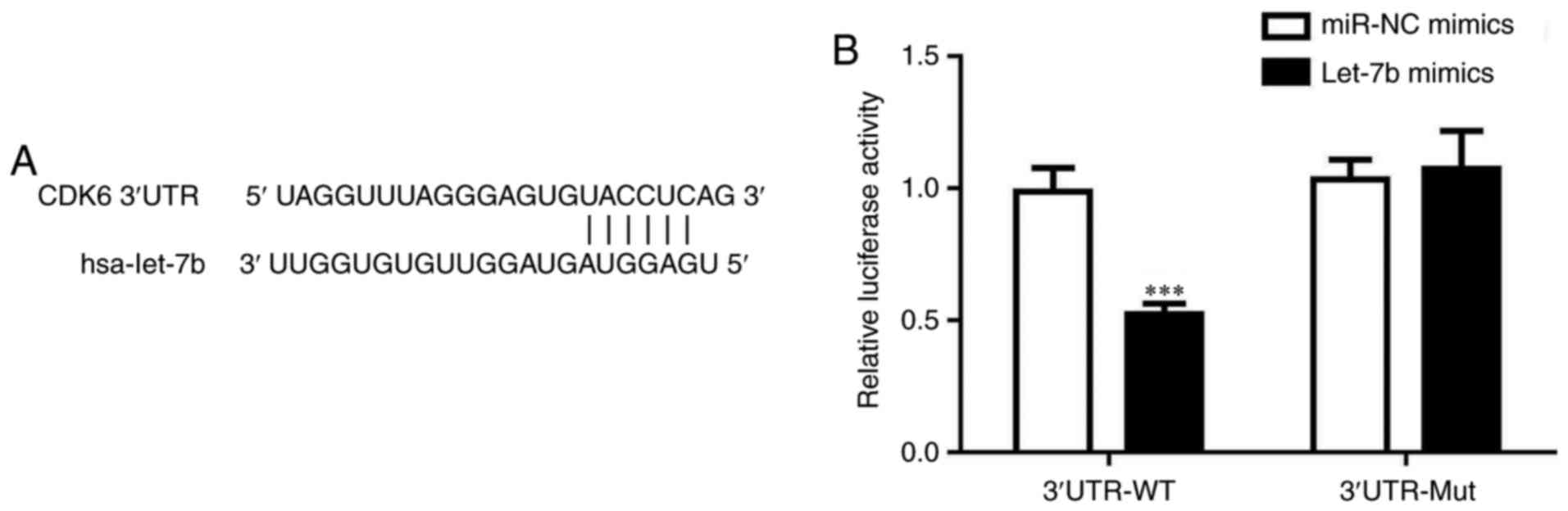
Let-7b directly binds to the 3'UTR of
CDK6 mRNA
Whether let-7b directly regulated the level of CDK6
was investigated. The prediction of let-7b binding sites indicated
that let-7b may directly bind to the 3'UTR of CDK6 mRNA (Fig. 5A). A dual luciferase assay was
used, which demonstrated that the over-expression of let-7b
decreased the relative luciferase activity of cells transfected
with CDK6 3'UTR-WT, but not 3'UTR-Mut (Fig. 5B). These results confirmed that
let-7b targeted the 3'UTR of CDK6 mRNA.
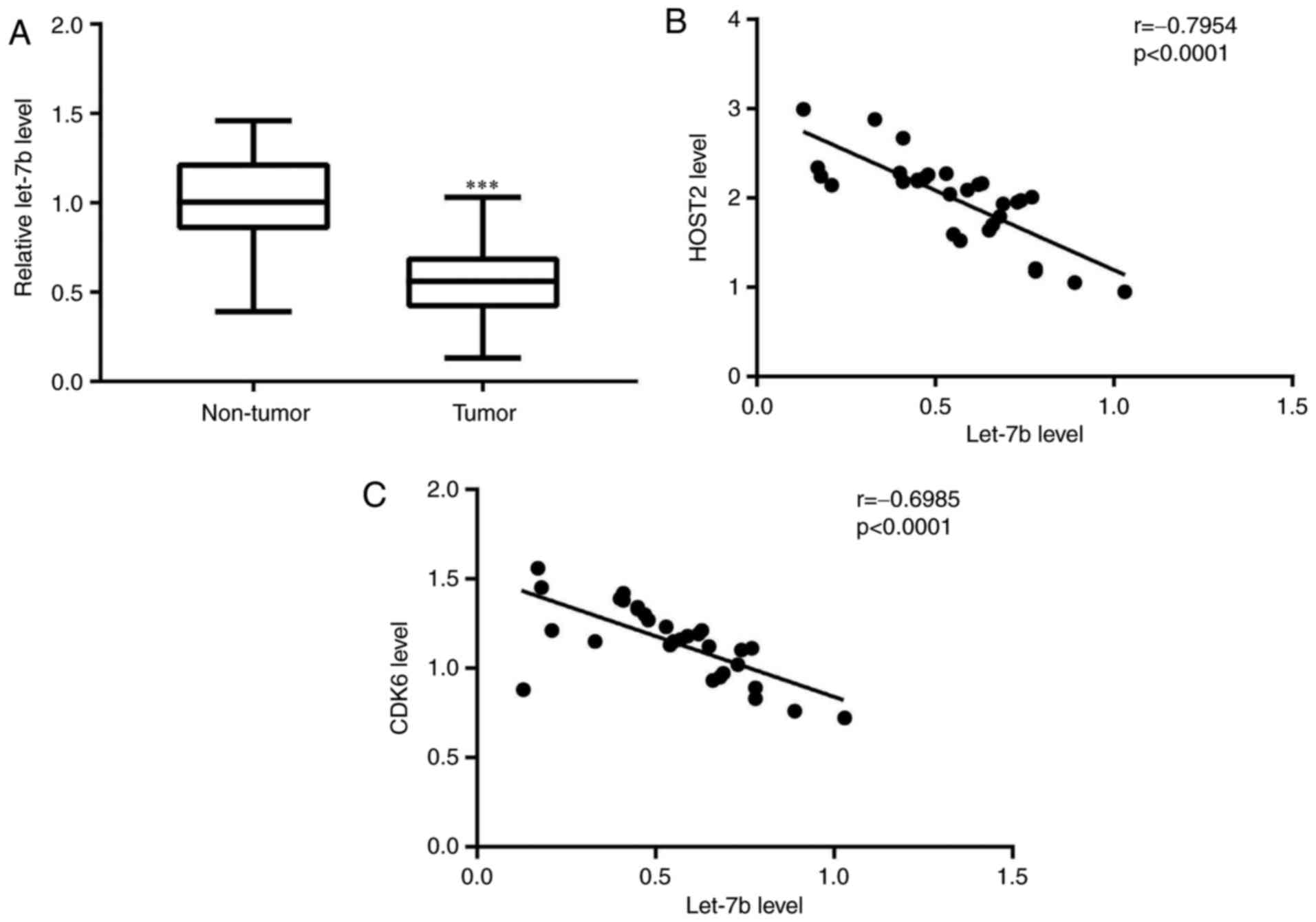
Expression of let-7b is negatively
associated with expression levels of HOST2 and CDK6 in TNBC tumor
tissues
To further evaluate the clinical significance of
HOST2/let7b/CDK6 in TNBC, RT-qPCR analysis was performed to detect
the expression of let-7b, and Pearson's correlation analysis was
used to analyze the results. Compared with matched normal tissues,
decreased expression of let-7b was observed in TNBC tumor tissues
(Fig. 6A). Additionally, a
significant negative correlation was identified between the
expression of let-7b and HOST2, and the expression of let-7b and
CDK6 in TNBC tumor tissues (Fig. 6B
and C).
Discussion
TNBC is characterized by an aggressive phenotype and
a high rate of relapse (19). In
the absence of well-defined molecular targets, chemotherapy is the
first line of treatment for TNBC, but this often results in
chemoresistance, and the mortality rate remains high (20). Several reports have indicated that
lncRNAs are important regulators of TNBC progression (21-23). In the present study, lncRNA-HOST2
was identified as an oncogene in TNBC, and may provide a novel
therapeutic target for TNBC.
HOST2 was first defined in human ovarian cancer and
functions as an oncogene through binding to let-7b (24,25). HOST2 has been demonstrated to
regulate cell proliferation, migration, invasion and cell apoptosis
in human osteosarcoma and hepatocellular carcinoma cells (12,26). A previous study showed that HOST2
was overexpressed in breast cancer and that the silencing of HOST2
inhibited the migration and invasion of MCF7 cells, a luminal
breast cancer cell line (18).
Through comparison of the expression of HOST in TNBC tumor tissues
and matched normal tissues, a marked increase in HOST2 was revealed
in TNBC tumor tissues. In addition, the expression of HOST2 was
significantly increased in TNBC cell lines compared with normal
breast epithelial cells. The silencing of HOST2 decreased the rate
of cell proliferation of the two TNBC cell lines. Flow cytometric
analysis indicated that HOST2 silencing led to an enrichment of
cells in the G0/G1 phase, but did not
significantly elevate apoptotic cell rate, suggesting that HOST2
may promote TNBC proliferation via accelerating the cell cycle.
The above results of the cell cycle assay showed
that HOST2 regulated the G1/S phase in cells. Through a
literature review, it was noted that CDK6 is a major
G0/G1 checkpoint regulator and frequently
deregulated in cancer cells (27); the expression of CDK6 has been
associated with poor survival outcomes and is considered to be a
promising therapeutic target for TNBC (28). The results suggested that CDK6 was
a key regulator in the G1/S phase and is pivotal in the
progression of TNBC. Other lncRNAs, including gadd7, GAS5 and MYU,
have been reported to regulate the expression of CDK6 in different
cellular contexts (29-31). In the present study, following the
silencing of HOST2, the expression of G1/S cell cycle
regulator CDK6, was detected. It was found that CDK6 was
downregulated following HOST silencing, and a positive correlation
between the expression of HOST2 and CDK6 was identified in the TNBC
tumour tissues. In addition, the silencing of HOST2 decreased the
expression of CDK6 in TNBC cell lines, indicating that HOST2 may
enhance the expression of CDK6. The regulatory association between
HOST2 and CDK6 may explain the cell cycle redistribution following
HOST2 knockdown. A previous study showed that several lncRNAs
function as potent miRNA sponges to regulate gene expression
(32). It has been suggested that
abundant lncRNAs can sequester miRNAs away from their targeted
mRNAs (33). A previous study
showed that let-7b was a target of HOST2 (25). The present study further confirmed
that HOST2 sponged let-7b to downregulate its expression in
MDA-MB-231 and MDA-MB-468. In addition, bioinfor-matics analysis
indicated complementary sequences between let-7b and the 3'UTR of
CDK6 mRNA. A dual luciferase assay confirmed let-7b as a direct
regulator of CDK6, and the results of western blot and RT-qPCR
analyses confirmed that let-7b suppressed the expression of CDK6.
Of note, the expression of let-7b was downregulated in TNBC tumour
tissues, and its expression was negatively correlated with that of
CDK6 and HOST2.
In conclusion, the present study suggested that an
HOST2/let-7b/CDK6 expression axis is involved in the promotion TNBC
proliferation. Mechanistically, downregulation of the expression of
HOST2 resulted in increased expression of let-7b, a tumor
suppressor, and decreased expression of CDK6, resulting in the
alteration of cell cycle distribution. Therefore, HOST2 may be a
candidate therapeutic target for the treatment of patients with
TNBC in the near future.
Funding
No funding was received.
Availability of data and materials
Details of data and materials are available from the
correspondence author on request.
Authors' contributions
YucZ was responsible for study design. YueZ, HZ and
YuZ performed most of experiments. HK, WH helped with collection of
clinical samples. YucZ supervised the study and prepared the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Wuhai People's Hospital. Informed consent regarding
the use of tissue samples was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors confirm that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Schettini F, Buono G, Cardalesi C,
Desideri I, De Placido S and Del Mastro L: Hormone receptor/human
epidermal growth factor receptor 2-positive breast cancer: Where we
are now and where we are going. Cancer Treat Rev. 46:20–26. 2016.
View Article : Google Scholar
|
|
2
|
Zheng S, Bai JQ, Li J, Fan JH, Pang Y,
Song QK, Huang R, Yang HJ, Xu F, Lu N and Qiao YL: The pathologic
characteristics of breast cancer in China and its shift during
1999-2008: a national-wide multicenter cross-sectional image over
10 years. Int J Cancer. 131:2622–2631. 2012. View Article : Google Scholar
|
|
3
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss P: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Comprehensive molecular portraits of human
breast tumours. Nature. 490:61–70. 2012. View Article : Google Scholar
|
|
5
|
Agrawal LS and Mayer IA: Platinum agents
in the treatment of early-stage triple-negative breast cancer: Is
it time to change practice? Clin Adv Hematol Oncol. 12:654–658.
2014.
|
|
6
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar
|
|
7
|
Pinto R, De Summa S, Pilato B and Tommasi
S: DNA methylation and miRNAs regulation in hereditary breast
cancer: Epigenetic changes, players in transcriptional and
post-transcriptional regulation in hereditary breast cancer. Curr
Mol Med. 14:45–57. 2014. View Article : Google Scholar
|
|
8
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li JP, Liu LH, Li J, Chen Y, Jiang XW,
Ouyang YR, Liu YQ, Zhong H, Li H and Xiao T: Microarray expression
profile of long noncoding RNAs in human osteosarcoma. Biochem
Biophys Res Commun. 433:200–206. 2013. View Article : Google Scholar
|
|
10
|
Lv M, Xu P, Wu Y, Huang L, Li W, Lv S, Wu
X, Zeng X, Shen R, Jia X, et al: LncRNAs as new biomarkers to
differentiate triple negative breast cancer from non-triple
negative breast cancer. Oncotarget. 7:13047–13059. 2016. View Article : Google Scholar
|
|
11
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu RT, Cao JL, Yan CQ, Wang Y, An CJ and
Lv HT: Effects of LncRNA-HOST2 on cell proliferation, migration,
invasion and apoptosis of human hepatocellular carcinoma cell line
SMMC-7721. Biosci Rep. 37:2017. View Article : Google Scholar
|
|
13
|
Zhong Y, Gao D, He S, Shuai C and Peng S:
Dysregulated expression of long noncoding RNAs in ovarian cancer.
Int J Gynecol Cancer. 26:1564–1570. 2016. View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
15
|
Malumbres M and Barbacid M: Mammalian
cyclin-dependent kinases. Trends Biochem Sci. 30:630–641. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lim S and Kaldis P: Cdks, cyclins and
CKIs: roles beyond cell cycle regulation. Development.
140:3079–3093. 2013. View Article : Google Scholar
|
|
17
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar
|
|
18
|
Lu PW, Li L, Wang F and Gu YT: Effects of
long non-coding RNA HOST2 on cell migration and invasion by
regulating microRNA let-7b in breast cancer. J Cell Biochem.
119:4570–4580. 2017. View Article : Google Scholar
|
|
19
|
Palma G, Frasci G, Chirico A, Esposito E,
Siani C, Saturnino C, Arra C, Ciliberto G, Giordano A and D'Aiuto
M: Triple negative breast cancer: Looking for the missing link
between biology and treatments. Oncotarget. 6:26560–26574. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oakman C, Viale G and Di Leo A: Management
of triple negative breast cancer. Breast. 19:312–321. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang
Z, Yuan J, Shan W, Li C, Hu X, et al: Long noncoding RNA LINP1
regulates repair of DNA double-strand breaks in triple-negative
breast cancer. Nat Struct Mol Biol. 23:522–530. 2016. View Article : Google Scholar :
|
|
22
|
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L,
Wang C, Hawke DH, Wang S, Zhang Y, et al: The LINK-A lncRNA
activates normoxic HIF1alpha signalling in triple-negative breast
cancer. Nat Cell Biol. 18:213–224. 2016. View Article : Google Scholar
|
|
23
|
Pickard MR and Williams GT: Regulation of
apoptosis by long non-coding RNA GAS5 in breast cancer cells:
implications for chemotherapy. Breast Cancer Res Treat.
145:359–370. 2014. View Article : Google Scholar
|
|
24
|
Rangel LB, Sherman-Baust CA, Wernyj RP,
Schwartz DR, Cho KR and Morin PJ: Characterization of novel human
ovarian cancer-specific transcripts (HOSTs) identified by serial
analysis of gene expression. Oncogene. 22:7225–7232. 2003.
View Article : Google Scholar
|
|
25
|
Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao
T, Liu Y, Ou J, Wang D, Yao L, et al: LncRNA-HOST2 regulates cell
biological behaviors in epithelial ovarian cancer through a
mechanism involving microRNA let-7b. Hum Mol Genet. 24:841–852.
2015. View Article : Google Scholar
|
|
26
|
Wang W, Li X, Meng FB, Wang ZX, Zhao RT
and Yang CY: Effects of the long non-coding RNA HOST2 on the
proliferation, migration, invasion and apoptosis of human
osteosarcoma cells. Cell Physiol Biochem. 43:320–330. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tadesse S, Yu M, Kumarasiri M, Le BT and
Wang S: Targeting CDK6 in cancer: State of the art and new
insights. Cell Cycle. 14:3220–3230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsu YH, Yao J, Chan LC, Wu TJ, Hsu JL,
Fang YF, Wei Y, Wu Y, Huang WC, Liu CL, et al: Definition of
PKC-alpha, CDK6, and MET as therapeutic targets in triple-negative
breast cancer. Cancer Res. 74:4822–4835. 2014. View Article : Google Scholar
|
|
29
|
Liu X, Li D, Zhang W, Guo M and Zhan Q:
Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6
mRNA decay. EMBO J. 31:4415–4427. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Z, Wang W, Jiang J, Bao E, Xu D, Zeng
Y, Tao L and Qiu J: Downregulation of GAS5 promotes bladder cancer
cell proliferation, partly by regulating CDK6. PLoS One.
8:e739912013. View Article : Google Scholar
|
|
31
|
Kawasaki Y, Komiya M, Matsumura K, Negishi
L, Suda S, Okuno M, Yokota N, Osada T, Nagashima T, Hiyoshi M, et
al: MYU, a target lncRNA for Wnt/c-Myc signaling, mediates
induction of CDK6 to promote cell cycle progression. Cell Rep.
16:2554–2564. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar
|
|
33
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar
|