Introduction
The complex process of embryogenesis involves tight
regulation of gene expression, which is controlled by different
genetic and epigenetic modifications (1,2).
Post-translational modifications of histones, including
acetylation, methylation, ubiquitination and phosphorylation, are
important epigenetic modifications regulating gene expression
(3).
Methylation of lysine residues of histone molecules
is an important part of epigenetics and is involved in the control
of gene expression (4). The
methylation state of a histone around a gene determines whether it
is transcriptionally active or not; histone methylation is strictly
controlled by histone lysine methyltransferases and demethylases
(KDMs) that are responsible for transfer and removal of a methyl
group (5). Methylation of histone
H3 lysine 9 (H3K9) is a distinctive feature of transcriptionally
repressed genes in eukaryotic chromatin (4,6).
H3K9, which can be mono-, di-and tri-methylated (me1, me2 and me3,
respectively), is well characterized for its roles in embryonic
development (7). H3K9me3 is
present in heterochromatin compartments (4) while H3K9me2 is present in
euchromatin regions of eukaryotic chromatin (1). The meth-ylation status of H3K9 is
maintained by two groups of KDMs containing Jumonji-C (JmjC)
domains (8). Demethylases of the
Kdm4 family catalyze H3K9me2/3 demethylation, while demethylation
of H3K9me1/2 is catalyzed by Kdm3a (9-11).
Lysine demethylase 3a (Kdm3a), also known as jumonji
domain containing 1A, JMJD1A, KIAA0742 and testis-specific gene A,
is a JmjC-domain containing demethylase (8). In addition to the JmjC domain, Kdm3a
contains a zinc finger domain and LXXLL motif; these domains/motifs
are specific for steroid hormone receptor interactions (11,12). Kdm3a catalyzes demethylation of
H3K9me1/2 and serves a role in the activation of gene transcription
(8). The catalytic activity of
Kdm3a, similar to other JmjC domain-containing proteins, involves a
hydroxylation reaction requiring ferrous ion and α-ketoglutarate as
cofactors (11,13).
Kdm3a has been implicated in the regulation of gene
expression during numerous biological functions, including
spermatogenesis, metabolism, sex determination, androgen
receptor-mediated transcription and cell differentiation (14-17). Aberrant expression of kdm3a
is associated with several malignancies, including colorectal
cancer, breast cancer, hepatocellular carcinoma and hypoxia
(18-21).
Multiple previous studies indicated the involvement
of H3K9me2 demethylation in neural development (22-26). In human embryonic stem cells,
Kdm3c was reported to serve roles in the repression of neural
differentiation and was necessary for the maintenance of miR-302
expression (22). miR-302 is an
epigenetic regulator of pluripotency and neural differentiation
(27). Furthermore, depletion of
lysine demethylase 1a splice variant catalyzing demethylation of
H3K9me1/2, increases the level of H3K9me2 at the target promoters
associated with the impairment of neuronal differentiation
(23). In addition, lysine
demethylases 7a and 7b which demethylate H3K9me1/2 are also
involved in the regulation of neural development (24,25).
Xenopus laevis is a model used to investigate
the mechanisms underlying neural development and differentiation
(28,29). The present study investigated the
developmental functions of Kdm3a and its effects on craniofacial
and neural development during Xenopus embryogenesis.
Whole-mount in situ hybridization (WISH) analysis revealed
the expression of kdm3a in the anterior regions of
Xenopus embryos, including the retina, central nervous
system (CNS) and branchial arches. Morpholino antisense
oligonucleotide (MO)-mediated knockdown of kdm3a resulted in
defective craniofacial development and neural deformities.
Furthermore, the results of WISH using neural crest-specific probes
indicated that Kdm3a regulated neural crest migration as
kdm3a morphant embryos exhibited decreased migration of
neural crest cells compared with control MO-injected embryos. In
addition, kdm3a depletion altered the expression of neural
markers and downregulated the expression of genes specific for
mesoderm development, cell adhesion and metabolism. The results of
the present study suggest that Kdm3a may serve important roles
during Xenopus embryogen-esis by regulating the expression
of neural-specific genes.
Materials and methods
Xenopus husbandry and in vitro
fertilization
Xenopus laevis (age, ~3 years old; 20 females
and 10 males) were obtained from the Korean Xenopus Resource
Center for Research and housed with a 12-h light/dark cycle at 18°C
in containers built specifically according to the requirements for
maintenance of laboratory organisms by the Institutional Review
Board of Ulsan National Institute of Science and Technology
(ethical approval no. UNISTACUC-16-14). To induce ovulation,
Xenopus females were injected with 1,000 IU/animal of human
chorionic gonadotropin (Dae Sung Microbiological Labs., Co., Ltd.,
Seoul, Korea) into the dorsal lymph sac in the evening. The
following day, the abdomens of female frogs were gently squeezed
and eggs were placed in 60-mm petri dishes containing 1X Modified
Barth's Saline (MBS; 88 mM NaCl, 5 mM Hepes, 2.5 mM
NaHCO3, 1 mM KCl, 1 mM MgSO4, and 0.7 mM
CaCl2, pH 7.8). After washing three times with 0.1X MBS,
eggs were fertilized using a suspension solution of sperm obtained
from isolated testes of sacrificed males. Following successful
fertilization, the embryos were swirled in a 2% L-cysteine solution
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) to remove the jelly
coat and washed five times with 0.5X MBS. Live and healthy embryos
were transferred to 0.5X MBS containing 2% Ficoll® PM
400 (GE Healthcare Life Sciences, Little Chalfont, UK) while
unfertilized eggs and non-viable embryos were removed by
observation under a light microscope (magnification, ×1).
Plasmids, mRNAs, MOs and microinjections
of Xenopus embryos
A full-length kdm3a cDNA clone (GeneBank ID:
NM_001095502) was obtained from the American Type Culture
Collection (Manassas, VA, USA). The amplification of Flag-tagged
kdm3a was performed by polymerase chain reaction (PCR), as
previously described (30). The
amplified fragment was sub-cloned into a pCS107 vector (Laboratory
of Protein Dynamics and Signaling, National Cancer Institute,
Frederick, MD, USA). Tagged kdm3a was subsequently
linearized with PvuII restriction endonucleases. SP6
mMes-sage mMachine kit (Ambion; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was used to synthesize capped mRNAs for
microinjections. The kdm3a MOs and control MOs were
synthesized by Gene Tools, LLC (Philomath, OR, USA; 5′-GTT CTC TTG
CTG AGT GAG CAC CAT A-3′; Control MO, 5′-CCT CTT ACC TCA GTT ACA
ATT TAT A-3′). Both blastomeres of two-cell staged embryos were
microinjected with MOs (30 ng/blastomere) and/or mRNAs (1 ng) and
incubated until the required stages (16, 22 and 32). For rescue
experiments, a MO-resistant mRNA (kdm3a*) with seven point
mutations in wobble codons followed by an ATG start codon (5′-ATG
GTA CTG ACG CAA CAG GAA AAT-3′) was synthesized. For
tissue-specific expression of neural crest-specific markers,
kdm3a MO was microinjected along with β-galactosidase
(Laboratory of Protein Dynamics and Signaling, National Cancer
Institute; 300 pg) into one of the two blastomeres of two-cell
stage embryos.
Whole-mount in situ hybridization (WISH)
and β-galactosidase staining
Xenopus embryos were collected at
developmental stage 16, 22 and 32 and prefixed in MEMFA (4%
parafor-maldehyde, 0.1 M MOPS pH 7.4, 1 mM MgSO4 and 2
mM EGTA) for 2 h at room temperature. WISH was performed on fixed
embryos by hybridization with neural crest-specific probes,
including Twist-family bHLH transcription factor 1 (twist),
snail family zinc finger 2 (snail2), transcription factor
AP-2α (tfap2a), early growth response 2 (egr2)
forkhead box d3 (foxd3), myc proto-oncogene, bHLH
transcription factor (c-myc), msh homeobox 1 (msx1)
and paired box 3 (pax3), as previously described (31). To distinguish the MO-injected side
of the embryos, β-galactosidase staining was performed, as
previously described (31).
Quantitative (q)PCR
Total RNA extraction was performed by lysing the
embryos in Isol-RNA lysis reagent (5 Prime GmbH, Hamburg, Germany)
and cDNA was prepared using the first strand cDNA synthesis kit
(Takara Bio, Inc., Otsu, Japan) at 65°C for 5 min followed by 42°C
for 1 h and 95°C for 5 min. qPCR reaction was performed using
specific primers (Table I) and
SYBR Premix Ex Taq, according to the manufacturer's protocol
(Takara Bio, Inc.). Thermocycler was adjusted at 95°C for 30 sec to
ensure denaturation, annealing temperature was set at 55°C for 30
sec followed by extension at 72°C for 1 min (30 cycles). The
analysis was performed using StepOnePlus™ Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative
expression levels of target genes were analyzed using the
2-ΔΔCq method (32).
All data are representative of at least three experiments. Primers
were designed using Primer3 software (33) and ornithine decarboxylase was used
as an internal control.
 | Table IPrimers used for qPCR. |
Table I
Primers used for qPCR.
| Gene | Primer sequence
(5′→3′)
|
|---|
| Forward | Reverse |
|---|
| odc | CAG CTA GCT GTG GTG
TGG | CAA CAT GGA AAC TCA
CAC C |
| kdm3a | TCC TAC ACC TGC CTG
CTC T | CAA GCT GGG CTG CAT
TGG A |
| ag1 | AAG GCC TGA AGA CCC
TGG A | TGC GAG AGT CAG GGA
TGG A |
| hesx1 | AGC TTT CAC TAG GAG
CCA GA | AGG TCC AAG GCT CTA
TCA |
| zic3 | CAA CAG TGA GGA ACC
TTC CA | GGG CTT TGT TAG TCT
GTA GC |
| six3 | AAC ACG AGT CCA TCC
TGC G | CCT GCT GGG GTT GGG
GTA G |
| otx2 | GGA TGG ATT TGT TAC
ATC CGT C | CAC TCT CCG AGC TCA
CTT CCC |
| en2 | ATG AGC AGA ATA ACA
GGG AAG TGG A | CCT CGG GGA CAT TGA
CTC GGT GGT G |
| n-cam | GCG GGT ACC TTC TAA
TAG TCA C | GGC TTG GCT GTG GTT
CTG AAG G |
| foxg1 | AGC AGC GAC GAT GTT
TCA | GGC CAT TGG ACG TGG
AGA A |
Animal cap assay
Xenopus embryos were microinjected with
kdm3a MO and/or 2 ng of dominant negative bone morphogenetic
protein 4 receptor (dnbr) mRNA (Laboratory of Protein
Dynamics and Signaling, National Cancer Institute) which induced
neural tissue formation from the prospective ectoderm. The animal
caps were excised using forceps following removal of the chorion
membrane from the vegetal side of late blastula (stage 8.5-9
embryos). Excised animal caps were cultured at 18°C in 0.5X MBS
containing 2% Ficoll PM 400 until developmental stage 16 of
Xenopus embryogenesis (34).
Alcian blue staining
For alcian blue staining, Xenopus embryos
were collected at the late tadpole stage (stage 43) and fixed in
Bouin's solution for 2 h at room temperature. Following fixation,
embryos were washed in 70% ethanol + 0.1% NH4OH and
stained with 0.05% Alcian Blue 8GX (Sigma-Aldrich; Merck KGaA) in
5% acetic acid for >2 h at room temperature. The embryos were
subsequently washed in 5% acetic acid for 2 h at room temperature
and cleared in 100% methanol. Following clearing, the embryos were
kept in benzyl benzoate and benzyl alcohol at a ratio of 2:1,
respectively.
Western blot analysis
The kdm3a MO-injected embryos were treated
with the lysis buffer (137 mM NaCl, 20 mM Tris-HCl pH 8.0, 1%
Nonidet-P40 and 10% glycerol) and 1 mM phenyl-methylsulfonyl
fluoride (Amresco, LLC, Solon, OH, USA), 5 mM sodium orthovanadate
(Sigma-Aldrich; Merck KGaA) and 1X protease inhibitor mix (Roche
Diagnostics, Basel, Switzerland), followed by heating of lysates at
95°C in loading buffer for 5 min. Bradford assay was used for
protein determination; lysates (40 µg/lane) were analyzed using
SDS-PAGE (12% gel) followed by transfer to nitrocellulose
membranes. The membranes were blocked with skim milk (5%) for 1 h
at room temperature and proteins were detected with anti-Flag tag
(cat. no. G188; Applied Biological Materials, Inc., Richmond, BC,
Canada) for 1 h at room temperature. Subsequently, the membranes
were incubated with goat anti-mouse (cat. no. sc2005; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and goat anti-rabbit (cat.no.
sc2004; Santa Cruz Biotechnology, Inc.) horseradish
peroxidase-conjugated antibodies by incubating for 1 h at room
temperature. Primary and secondary antibodies were used at a
dilution of 1:1,000. The immunoreactive bands were detected using
an enhanced chemiluminescence kit (HyGLO™ kit; Denville Scientific,
Inc., South Plainfield, NJ, USA). Histone H3 (cat. no. ab1791;
Abcam, Cambridge, UK) antibody is used as a loading control.
RNA sequencing
Total RNA was harvested from each sample (3
embryos/sample) at tadpole stage (stage 32) and RNA sequencing
library was constructed according to manufacturer's protocol
(TruSeq RNA Sample Prep kit v.2; cat. nos. RS-122-2001 and
RS-122-2002; Illumina, Inc., San Diego, CA, USA) using polyA
enrichment. For estimation of mRNA abundance, the reads were mapped
to the Xenopus laevis cDNA sequences from the genome project
consortium (35) using BWA
software (v.0.7.15) (36), and
the significant differential expression of genes was estimated
using edgeR software (v.3.3.1). Genes exhibiting >4-fold change
and false discovery rate <0.01 were considered significantly
differentially expressed. To determine biological processes in
which the differentially expressed genes were enriched, Fisher's
test provided by the PANTHER database (release 20171205) (37) was used with human orthologous
genes based on the best-hit results using BLASTP search (38). RNA sequencing raw data are
available at the National Center for Biotechnology Information Gene
Expression Omnibus database (accession no. GSE117754).
Statistical analysis
ImageJ software (v.1.45; National Institutes of
Health, Bethesda, MD, USA) was used to analyze the data from the
WISH and PCR analyses, and the results are presented as the mean ±
standard error of the mean. Results obtained from three independent
experiments. The level of significance was calculated using an
unpaired t-test or one-way analysis of variance followed by Tukey's
post hoc test using GraphPad Prism software (v.7; GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
kdm3a is expressed at the anterior
regions during Xenopus embryogenesis
To study the function of Kdm3a during Xenopus
embryonic development, the present study investigated the
expression pattern of kdm3a in early Xenopus embryos.
qPCR analysis indicated that kdm3a was expressed at early
stages of embryogenesis, ranging from the egg cell to the tadpole
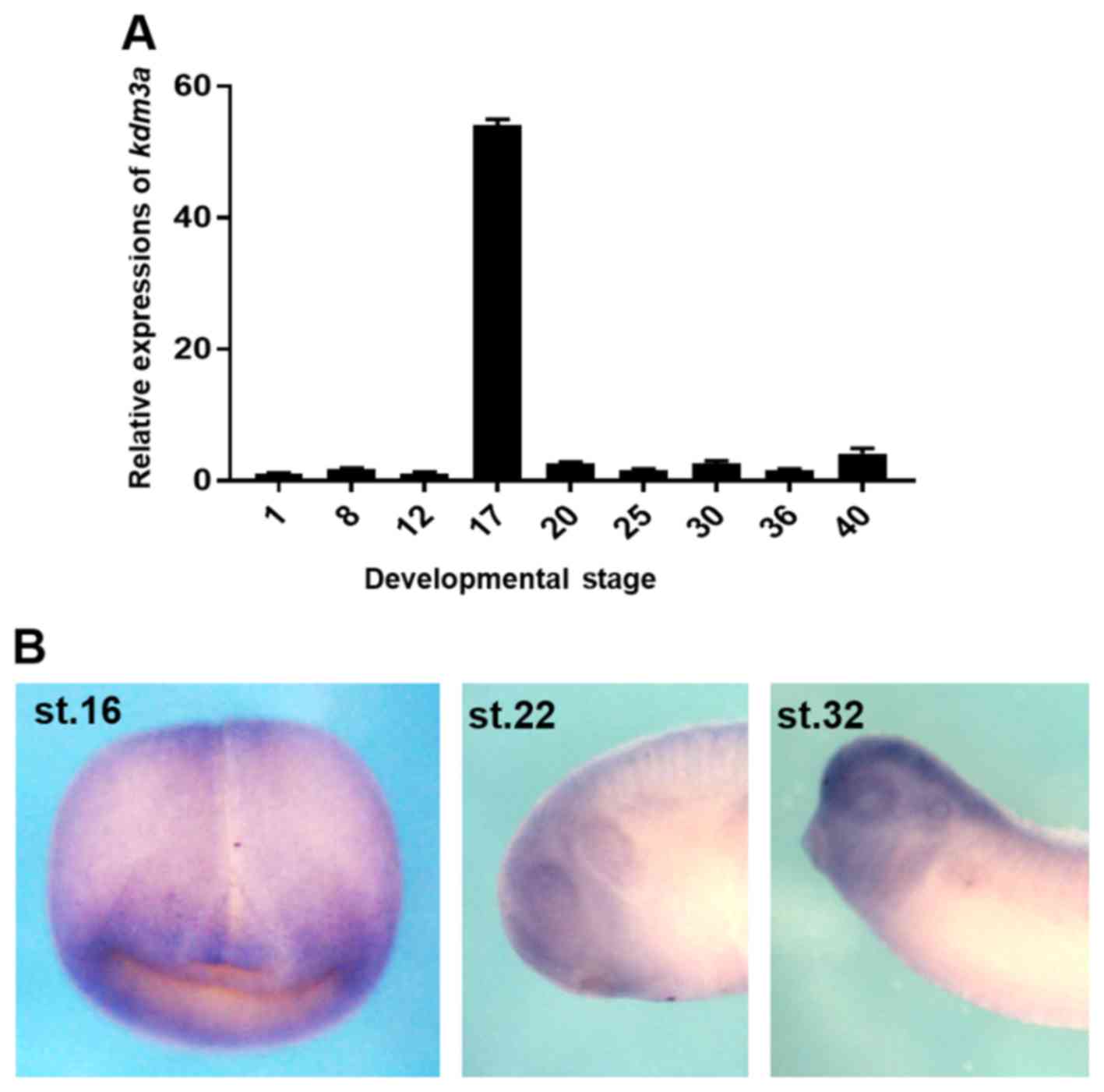
stage of development (Fig. 1A).
However, markedly increased level of kdm3a expression was
observed at the neurula stage (stage 17; Fig. 1A). The spatial expression of
kdm3a was analyzed using WISH, and kdm3a mRNA
localized to the CNS during the neurula stage of embryogenesis
(stage 16; Fig. 1B). Furthermore,
kdm3a was expressed at the anterior region, including the
eye and the CNS at stage 22 (early tailbud stage), and was observed
in the retina, otic vesicle and branchial arches at stage 32 (late
tailbud stage) of Xenopus embryogenesis (Fig. 1B). The results of qPCR and WISH
analysis indicate that kdm3a may serve roles during
Xenopus embryogenesis and regulate neurogenesis during
embryonic development.
Kdm3a knockdown arrests craniofacial
formation during Xenopus embryogenesis
To investigate the physiological functions of Kdm3a
during Xenopus embryogenesis, loss-of-function experiments
were performed using kdm3a MO to inhibit kdm3a
translation. Embryos at the two-cell stage were injected with
kdm3a MO to repress kdm3a expression.
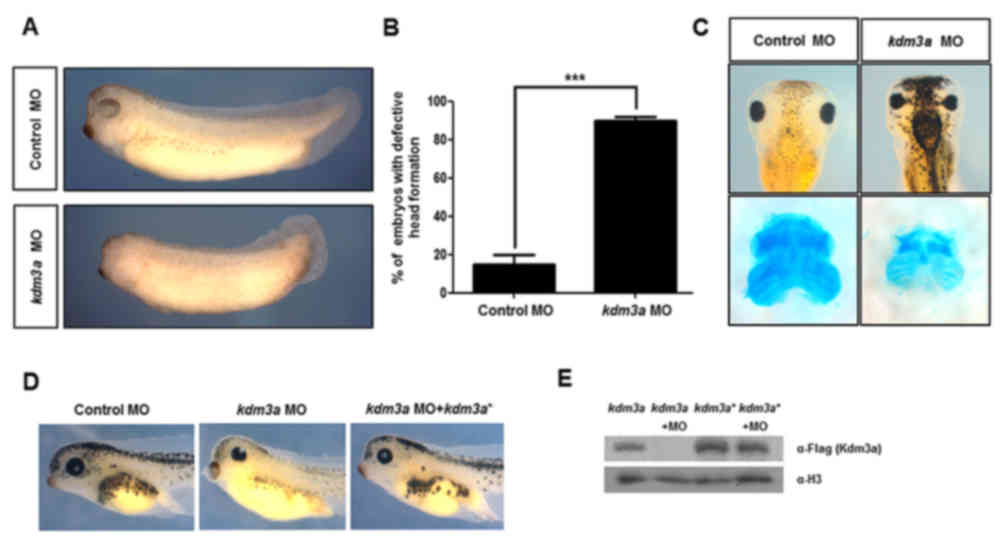
Embryos injected with the kdm3a MO exhibited
malformed phenotypes, including defective head formation and
markedly smaller eyes compared with the control MO-injected embryos
(Fig. 2A). Anterior defects were
identified in >90% of kdm3a morphants (Fig. 2B). Furthermore, abnormal
pigmentation was observed among kdm3a morphants (Fig. 2C). Alcian blue staining was
performed to visualize the effects of kdm3a MOs on
craniofacial development. kdm3a morphants exhibited markedly
smaller cartilage compared with cartilage formation in control
embryos (Fig. 2C).
To confirm whether the malformed phenotypes were
specifically induced by the depletion of kdm3a, rescue
experiments were performed by microinjecting kdm3a* into
two-cell stage embryos. Injection of kdm3a* recovered the
malformed phenotypes observed in kdm3a morphants (Fig. 2D). This validation experiment
confirmed that the defects observed in kdm3a morphants were
caused by kdm3a knockdown during Xenopus
embryogenesis. Furthermore, western blot analysis was used to
examine protein expression of kdm3a. No kdm3a
expression was observed in the group treated with MO, confirming
the efficiency of knockdown (Fig.
2E). In addition, protein expression in embryos injected with
kdm3a* or kdm3a* + MO, verified that MO did not bind
to kdm3a* (Fig. 2E). These
results suggested that the malformed phenotypes including defective
craniofacial formation were caused by the loss of kdm3a,
validating the role of this gene during Xenopus embryonic
development.
Depletion of kdm3a affects neural crest
migration during Xenopus embryonic development
The cells of neural crest emanate from neural tube
immediately after its closure, and subsequently, migrate to
specific regions of the embryo where they differentiate to the
peripheral nervous system, facial skeleton cells and pigment cells
(39,40). As described above, kdm3a
morphants exhibited defects in craniofacial formation, abnormal
head development and dense pigmentation (Fig. 2C), and kdm3a expression was
observed at the neural plate during the neural stage of development
(Fig. 1B). The present study
further investigated whether kdm3a may affect neural crest
migration.
To examine the role of Kdm3a in neural crest
migration, kdm3a MO and β-galactosidase mRNA were
injected into one blastomere of two-cell stage embryos. The
injected sides of the embryos were visualized by
β-galactosidase assays and WISH analysis was performed using
neural crest marker genes. twist, snail2,
tfap2a, egr2 and foxd3 are widely used neural
crest marker genes (41,42). WISH expression patterns of
twist (stage 32; late tailbud), and snail2,
foxd3 and tfap2a (all at stage 20; neurula) indicated
that injection with kdm3a MO significantly reduced neural
crest migration on the injected side of the embryos (Fig. 3). In addition, kdm3a
MO-injected sides of the embryos exhibited abnormal expression
patterns of egr2-a, a hindbrain-specific marker; however,
normal development of rhombomere 3 and 5, and neural crest was
observed in the uninjected sides of the embryos (Fig. 3). These genes serve important
roles during development of progenitor cells of neural crest into
bona fide neural crest cells (43). Furthermore, twist is
involved in neural crest cell migration as a repressor of
transcription (44). Injected
embryos exhibited abnormal expression patterns of twist,
snail2 and foxd3, compared with the uninjected
control (Fig. 3B).
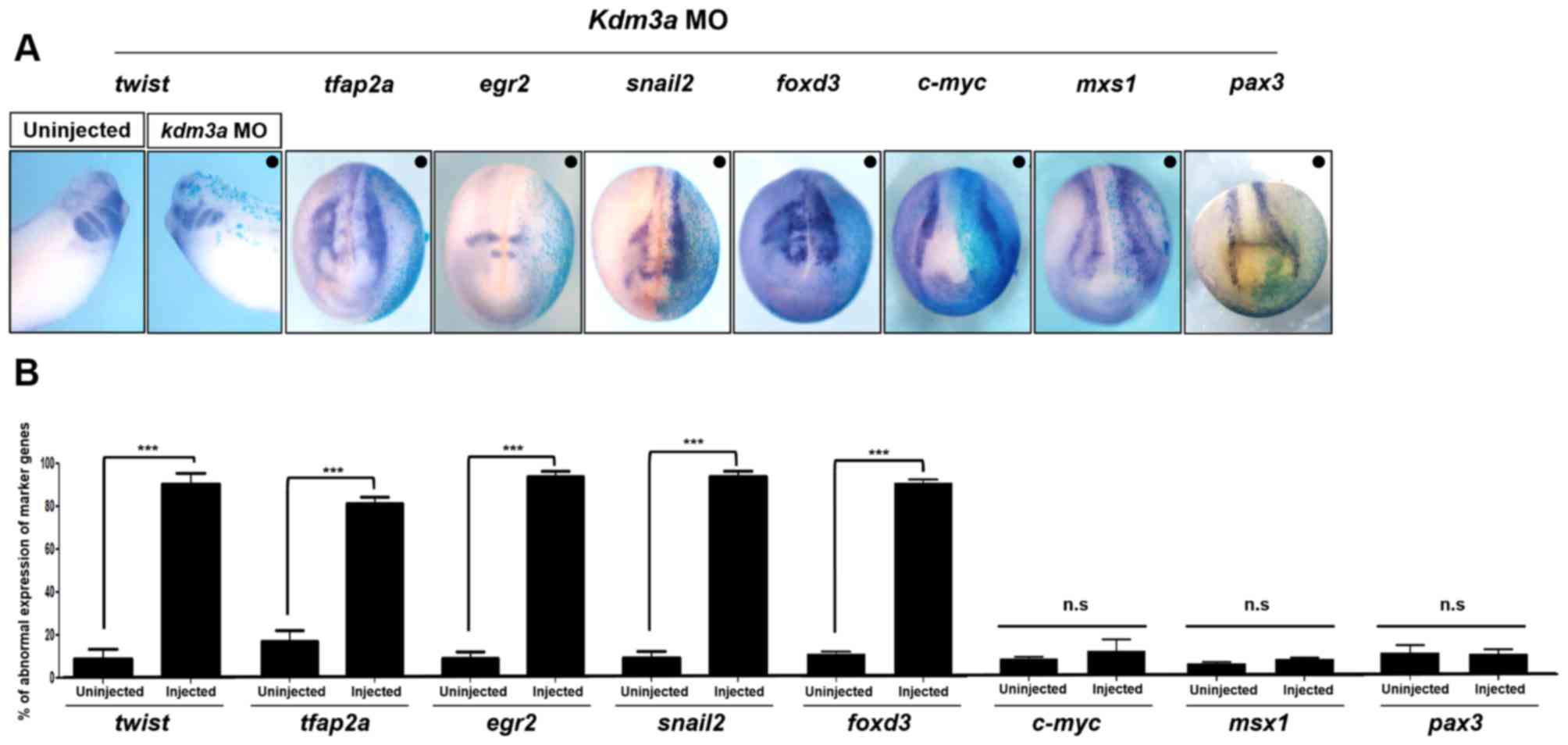 | Figure 3Kdm3a is required for neural crest
migration by regulating the expression of neural crest specifiers.
(A) kdm3a MO and β-galactosidase mRNA were
co-injected into one blastomere of two-cell stage embryos. Embryos
were harvested at stage 16, 20 (neurula), and stage 32 (late
tailbud). kdm3a depletion led to defective neural crest
migration as indicated by the expression patterns of twist
(stage 32; late tailbud), tfap2a, egr2,
snail2, foxd3 (stage 20; neurula stage),
c-myc, msx1 and pax3 (stage 16; neurula
stage). Injected sides of the embryos exhibited reduced neural
crest cell migration compared with the uninjected sides of the
embryos. The black dot (•) represents the injected side. Embryos
were analyzed under a light microscope (magnification, x2.5). (B)
Expression patterns of neural crest specifiers, including
twist, tfap2a, egr2, snail2,
foxd3, c-myc, msx1 and pax3 induced by
kdm3a knockdown. Data are presented as the mean ± standard
error of the mean. ***P<0.001, as indicated. n.s, not
significant; twist, twist-family bHLH transcription factor
1; snail2, snail family zinc finger 2; tfap2a,
transcription factor AP-2α; egr2, early growth response 2;
foxd3, forkhead box d3; c-myc, myc proto-oncogene
bHLH transcription factor; msx1, msh homeobox 1;
pax3, paired box 3; kdm3a, lysine demethylase 3a; MO,
morpholino antisense oligonucleotide. |
It has been previously demonstrated that Kdm3a may
serve important roles in primary neuron formation; however, it was
not required for neural induction (45). To verify these previous results,
the present study observed the expression patterns of early markers
of neural crest progenitor cells, including c-myc (46), msx1 and pax3
(47). Expression of c-myc
was not affected by kdm3a knockdown (Fig. 3). Expression patterns of
msx1 and pax3-neural crest induction markers
(47,48) remained unaltered on the
kdm3a MO-injected side (Fig.
3). Therefore, WISH analysis results together with the
β-galactosidase assay demonstrated that depletion of
kdm3a may affect the migration of neural crest cells by
disturbing the expression of neural crest specifiers. Since facial
cartilage is formed by the cranial neural crest and trunk neural
crest cells differentiate into pigment cells (49,50); therefore, it may be hypothesized
that the reduced size of cartilage and abnormal pigmentation in
kdm3a morphants resulted from abnormal neural crest
migration.
kdm3a knockdown alters the expression of
neural-specific genes
Kdm3a serves essential roles in primary neuron
formation and its depletion downregulates the expression of
neural-differentiation associated genes (45). Therefore, the present study aimed
to investigate the effect of Kdm3a on neural-specific (anterior,
posterior and pan) genes. For this purpose, kdm3a MO and
dnbr mRNA were co-injected into the animal pole region of
two-cell stage embryos. Animal caps were excised from the
microinjected embryos at blastula stage (stage 8.5-9) and cultured
until neurula stage (stage 16). Xenopus animal caps,
equivalent to embryonic stem cells, develop into the ectoderm,
however, these cells are pluripotent and can differentiate into
neural, mesodermal and endo-dermal tissues depending on the level
and type of specific inducers (34,51). dnbr induces the formation
of neural tissue from animal cap explants by inhibiting bone
morphogenetic protein (BMP) signaling during early development of
Xenopus and increasing expression of neurogenesis-specific
genes (34).
Gene expression levels in animal caps dissected from
kdm3a MO and dnbr mRNA co-injected embryos were
analyzed by qPCR to determine the neural marker gene expression.
Depletion of kdm3a led to the inhibition of expression of
cement gland marker, anterior gradient 1 (ag1) (52), anterior markers, orthodenticle
homeobox 2 (otx2), HESX hmeobox1 (hesx1) and forkhead
box G1 (foxg1) (53),
pan-neural marker, neural cell adhesion molecule 1 (n-cam)
(48), and retina marker, SIX
homeobox 3 (six3) (54),
consistent with the kdm3a morphant phenotype (Fig. 4). By contrast, the expression
levels of neural crest marker, ZIC family member 3 and posterior
neural marker, engrailed homeobox 2 (55), were not suppressed by the
knockdown of kdm3a (Fig.
4). Our qPCR data for neural marker gene expression are
consistent with the previously published data (45). Furthermore, the present study
indicated that kdm3a depletion inhibited the expression of
six3. It may be hypothesized that kdm3a serves a role
in the expression of retina specific markers at the early stage of
embryonic development.
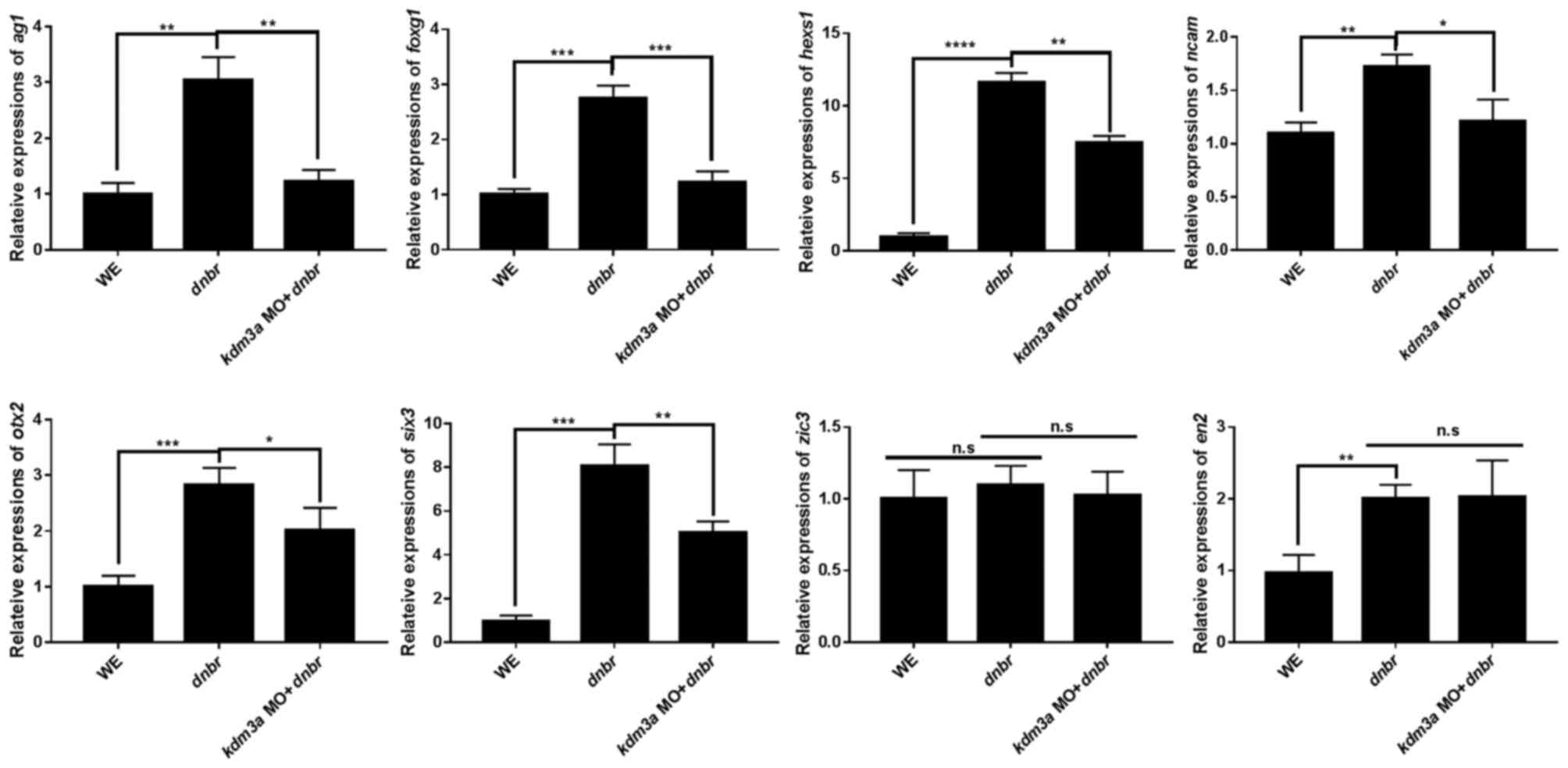 | Figure 4Kdm3a affects the expression of
neural specific markers. Embryos were co-injected with kdm3a
MO and dnbr mRNA into the animal pole region of two-cell
stage embryos. Animal caps were dissected from injected embryos at
stage 8.5-9 (blastula) and incubated in 0.5X MBS. Animal caps were
collected at stage 16 (neurula) and processed for reverse
transcription-quantitative polymerase chain reaction using standard
methods to detect the expression levels of hesx1,
ag1, foxg1, otx2, n-cam, zic3,
six3 and en2. odc was used as the loading
control. *P<0.05, **P<0.01 and
***P<0.001, as indicated. n.s, not significant;
kdm3a, lysine demethylase 3a; MO, morpholino antisense
oligonucleotide; dnbr, dominant negative bone morphogenetic
protein 4 receptor; WE, whole embryos; ag1, anterior
gradient 1; odc, ornithine decarboxylase; hesx1, HESX
hmeobox1; zic3, ZIC family member 3; six3, SIX
homeobox 3; otx2, orthodenticle homeobox 2; en2,
engrailed homeobox 2; n-cam, neural cell adhesion molecule
1; foxg1, forkhead box G1. |
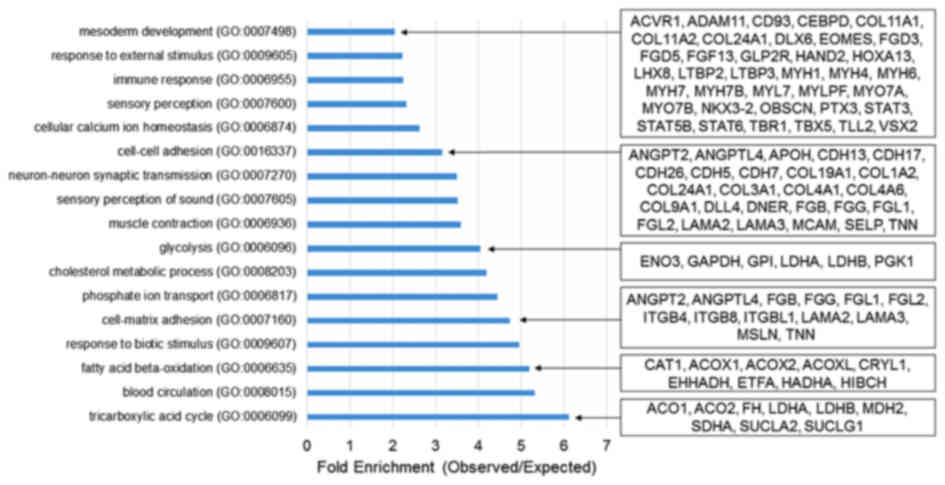
The present study further conducted a
transcriptomics analysis to elucidate the role of Kdm3a during
Xenopus embryogenesis. Transcriptomics analysis demonstrated
that kdm3a is important for metabolism, cell-cell adhesion
and mesoderm development during Xenopus embryogen-esis
(Fig. 5). Knockdown of
kdm3a led to downregulation of a number of genes involved in
mesoderm patterning and development, including fibroblast growth
factor 13 (fgf13), signal transducer and activator of
transcription 3 gene 1 (stat3) and T-box 5 (tbx5)
(56). Furthermore, the depletion
of kdm3a induced the downregulation of multiple genes
involved in mesoderm induction and patterning (Fig. 5). Knockdown of kdm3a
suppressed the expression of genes involved in cell-cell adhesion,
including angiopoietin 2 (angpt2), cadherin 5 (cdh5)
and fibrinogen β chain (fgb) (Fig. 5).
qPCR analysis using animal caps and the
transcriptomics data indicated that kdm3a depletion altered
the expression of various genes involved in fundamental
developmental processes. Therefore, Kdm3a may serve an important
role in normal embryonic development of Xenopus laevis.
Discussion
Epigenetic regulator Kdm3a catalyzes demethylation
of H3K9me1/2, and is involved in the maintenance of the histone
code (15). It has been reported
that this demethylase is involved in numerous biological and
pathological processes, including germ cell development, sex
determination, metabolism, stem cell differentiation, stem cell
self-renewal and cancer epigenetics (2,57).
In the present study, the expression pattern and physiological
functions of Kdm3a were examined during embryonic development of
Xenopus.
Appropriately regulated gene expression is
necessary in the process of embryonic growth and development.
Maternal and zygotic kdm3a was expressed during early stages
of Xenopus embryonic development, until the tadpole stage of
embryogen-esis. The kdm3a expression pattern observed in the
present study is consistent with a previously conducted study
(35). The qPCR analysis
demonstrated that expression levels of kdm3a varied across
developmental stages and were highest during the neurula stage
(stage 17) of embryogenesis. kdm3a was expressed in the CNS
during the neurula stage of embryonic development. The expression
of kdm3a was observed in the anterior region, including the
eye and the CNS at stage 22 (early tailbud), retina, otic vesicle,
and branchial arches at stage 32 (late tailbud) of Xenopus
embryogenesis. Microinjection of kdm3a MO into the two-cell
stage Xenopus embryos induced phenotypic abnormalities,
including reduced head size and smaller eyes, compared with control
MO-injected embryos. Craniofacial development of kdm3a
morphants was also affected, exhibiting markedly reduced cartilage
and abnormal pigmentation, compared with the control embryos.
Rescue experiments confirmed the role of Kdm3a during
Xenopus embryogenesis. All malformed phenotypes of
kdm3a morphants were rescued following co-injection of
kdm3a MO and kdm3a*. These results indicate that
Kdm3a may serve important roles during neural crest migration of
Xenopus embryos.
Jmjc domain-containing demethylases are involved in
neural development. Kdm7 is necessary for brain formation through
its interaction with a BMP antagonist, the follistatin gene locus
(26). Furthermore, Kdm6b is
specific for demethylation of H3K27me3 and is involved in neural
development (58). Based on the
previous results regarding Kdm3a and its expression at the neurula
stage (45), the present study
investigated the physiological effect of Kdm3a during neural
development. Injection of kdm3a MO altered the expression of
neural crest specifiers. The WISH analysis indicated markedly
altered expression of neural crest specifiers as a result of
kdm3a knockdown. These results indicated that kdm3a
depletion may be associated with impaired neural crest migration
and may be required for facial cartilage formation.
Kdm3a serves a role in primary neuron formation and
is known to regulate the expression of neuronal
differentiation-associated genes; however the depletion of
kdm3a exhibits no effect on the expression of neural stem
progenitor markers pax6 and sox3 (45). The results of the present study
supported these previously published data and indicated that Kdm3a
may not be involved in the induction of neural crest cells, as
demonstrated by the expression levels of early markers of neural
progenitor cells, including c-myc, msx1 and
pax3.
The present study analyzed the effect of Kdm3a on
neural marker gene expression in animal caps of
dnbr-injected embryos. Loss-of-function of kdm3a
resulted in reduced expression of anterior neural markers
hesx1, otx2 and foxg1, and pan-neural marker
n-cam, suggesting that kdm3a may serve a role in the
neural development during Xenopus embryogenesis.
Transcriptomics data analysis of kdm3a morphants indicated
that kdm3a may be required for mesoderm formation. The
mesoderm is one of the earliest germinal layers and is essential
for normal development of the skeletal system, muscular system and
a major part of the neural system (59). Knockdown of kdm3a
downregulated the expression of fgf13, stat3 and
tbx5, and other genes associated with mesoderm development.
The expression levels of angpt2, cdh5, fgb and
other genes involved in cell-cell adhesion were also suppressed in
kdm3a morphants.
In conclusion, the present study indicated that
Kdm3a, an epigenetic modifier associated with histone code
maintenance, is physiologically relevant as a regulator of
neurogenesis during Xenopus embryonic development.
Additional investigation is required to determine the interaction
between this epigenetic regulator and developmental pathways, which
may provide novel methods for the treatment of developmental
disorders.
Funding
This study was supported by grants from the
National Research Foundation of Korea (grant no.
NRF-2015R1A2A1A10053265) and the Ministry of Science, ICT and
Future Planning, the Republic of Korea (grant no.
2015R1A4A1042271).
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request. RNA sequencing raw data are available at the
National Center for Biotechnology Information Gene Expression
Omnibus database (accession no. GSE117754).
Authors' contributions
HKL, TI, CK and YK performed the experiments. HKL,
TI, CK, JWP, OSK, BSK, DSL and HSL performed the data analysis and
wrote the manuscript. TK, TJP and HSL designed the study,
interpreted the results and critically analyzed the manuscript.
Ethics approval and consent to
participate
Experiments were conducted according to the
guidelines of the Animal Care and Use Committee consistent with
international laws and policies (National Institute of Health Guide
for the Care and Use of Laboratory Animals; NIH publication no.
85-23, 1985). The Institutional Review Board of Ulsan National
Institute of Science and Technology in Korea approved the
experimental use of amphibians (approval no. UNISTACUC-16-14). All
members of the research group were trained for the appropriate care
and use of experimental organisms. There were no unexpected cases
of mortality of adult Xenopus during the present study.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Meissner A: Epigenetic modifications in
pluripotent and differentiated cells. Nat Biotechnol. 28:1079–1088.
2010. View Article : Google Scholar
|
|
2
|
Mohn F and Schübeler D: Genetics and
epigenetics: Stability and plasticity during cellular
differentiation. Trends Genet. 25:129–136. 2009. View Article : Google Scholar
|
|
3
|
Mattout A and Meshorer E: Chromatin
plasticity and genome organization in pluripotent embryonic stem
cells. Curr Opin Cell Biol. 22:334–341. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jenuwein T: The epigenetic magic of
histone lysine methylation. FEBS J. 273:3121–3135. 2006. View Article : Google Scholar
|
|
5
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012. View Article : Google Scholar
|
|
6
|
Bernstein BE, Mikkelsen TS, Xie X, Kamal
M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al:
A bivalent chromatin structure marks key developmental genes in
embryonic stem cells. Cell. 125:315–326. 2006. View Article : Google Scholar
|
|
7
|
Tachibana M, Sugimoto K, Nozaki M, Ueda J,
Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al: G9a
histone meth-yltransferase plays a dominant role in euchromatic
histone H3 lysine 9 methylation and is essential for early
embryogenesis. Genes Dev. 16:1779–1791. 2002. View Article : Google Scholar
|
|
8
|
Klose RJ, Kallin EM and Zhang Y:
JmjC-domain-containing proteins and histone demethylation. Nat Rev
Genet. 7:715–727. 2006. View Article : Google Scholar
|
|
9
|
Fodor BD, Kubicek S, Yonezawa M,
O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K,
Schotta G and Jenuwein T: Jmjd2b antagonizes H3K9 trimethylation at
peri-centric heterochromatin in mammalian cells. Genes Dev.
20:1557–1562. 2006. View Article : Google Scholar :
|
|
10
|
Wissmann M, Yin N, Müller JM, Greschik H,
Fodor BD, Jenuwein T, Vogler C, Schneider R, Günther T, Buettner R,
et al: Cooperative demethylation by JMJD2C and LSD1 promotes
androgen receptor-dependent gene expression. Nat Cell Biol.
9:347–353. 2007. View Article : Google Scholar
|
|
11
|
Yamane K, Toumazou C, Tsukada Y,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: JHDM2A, a
JmjC-containing H3K9 demethylase, facilitates transcription
activation by androgen receptor. Cell. 125:483–495. 2006.
View Article : Google Scholar
|
|
12
|
Knebel J, De Haro L and Janknecht R:
Repression of transcription by TSGA/Jmjd1a, a novel interaction
partner of the ETS protein ER71. J Cell Biochem. 99:319–329. 2006.
View Article : Google Scholar
|
|
13
|
Cai C, Yuan X and Balk SP: Androgen
receptor epigenetics. Transl Androl Urol. 2:148–157. 2013.
|
|
14
|
Inagaki T, Tachibana M, Magoori K, Kudo H,
Tanaka T, Okamura M, Naito M, Kodama T, Shinkai Y and Sakai J:
Obesity and metabolic syndrome in histone demethylase JHDM2a-
deficient mice. Genes Cells. 14:991–1001. 2009. View Article : Google Scholar
|
|
15
|
Kuroki S, Matoba S, Akiyoshi M, Matsumura
Y, Miyachi H, Mise N, Abe K, Ogura A, Wilhelm D, Koopman P, et al:
Epigenetic regulation of mouse sex determination by the histone
demethylase Jmjd1a. Science. 341:1106–1109. 2013. View Article : Google Scholar
|
|
16
|
Okada Y, Scott G, Ray MK, Mishina Y and
Zhang Y: Histone demethylase JHDM2A is critical for Tnp1 and Prm1
transcription and spermatogenesis. Nature. 450:119–123. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tateishi K, Okada Y, Kallin EM and Zhang
Y: Role of Jhdm2a in regulating metabolic gene expression and
obesity resistance. Nature. 458:757–761. 2009. View Article : Google Scholar
|
|
18
|
Beyer S, Kristensen MM, Jensen KS,
Johansen JV and Staller P: The histone demethylases JMJD1A and
JMJD2B are transcriptional targets of hypoxia-inducible factor HIF.
J Biol Chem. 283:36542–36552. 2008. View Article : Google Scholar
|
|
19
|
Ueda J, Ho JC, Lee KL, Kitajima S, Yang H,
Sun W, Fukuhara N, Zaiden N, Chan SL, Tachibana M, et al: The
hypoxia-inducible epigenetic regulators Jmjd1a and G9a provide a
mechanistic link between angiogenesis and tumor growth. Mol Cell
Biol. 34:3702–3720. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wade MA, Jones D, Wilson L, Stockley J,
Coffey K, Robson CN and Gaughan L: The histone demethylase enzyme
KDM3A is a key estrogen receptor regulator in breast cancer.
Nucleic Acids Res. 43:196–207. 2015. View Article : Google Scholar
|
|
21
|
Yamada D, Kobayashi S, Yamamoto H,
Tomimaru Y, Noda T, Uemura M, Wada H, Marubashi S, Eguchi H,
Tanemura M, et al: Role of the hypoxia-related gene, JMJD1A, in
hepatocellular carcinoma: Clinical impact on recurrence after
hepatic resection. Ann Surg Oncol. 19(Suppl 3): S355–S364. 2012.
View Article : Google Scholar
|
|
22
|
Wang J, Park JW, Drissi H, Wang X and Xu
RH: Epigenetic regulation of miR-302 by JMJD1C inhibits neural
differentiation of human embryonic stem cells. J Biol Chem.
289:2384–2395. 2014. View Article : Google Scholar :
|
|
23
|
Laurent B, Ruitu L, Murn J, Hempel K,
Ferrao R, Xiang Y, Liu S, Garcia BA, Wu H, Wu F, et al: A specific
LSD1/KDM1A isoform regulates neuronal differentiation through H3K9
demethylation. Mol Cell. 57:957–970. 2015. View Article : Google Scholar
|
|
24
|
Huang C, Chen J, Zhang T, Zhu Q, Xiang Y,
Chen CD and Jing N: The dual histone demethylase KDM7A promotes
neural induction in early chick embryos. Dev Dyn. 239:3350–3357.
2010. View Article : Google Scholar
|
|
25
|
Huang C, Xiang Y, Wang Y, Li X, Xu L, Zhu
Z, Zhang T, Zhu Q, Zhang K, Jing N, et al: Dual-specificity histone
demethylase KIAA1718 (KDM7A) regulates neural differentiation
through FGF4. Cell Res. 20:154–165. 2010. View Article : Google Scholar
|
|
26
|
Tsukada Y, Ishitani T and Nakayama KI:
KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and
functions in brain development. Genes Dev. 24:432–437. 2010.
View Article : Google Scholar
|
|
27
|
Lipchina I, Studer L and Betel D: The
expanding role of miR-302-367 in pluripotency and reprogramming.
Cell Cycle. 11:1517–1523. 2012. View Article : Google Scholar
|
|
28
|
Hemmati-Brivanlou A, Kelly OG and Melton
DA: Follistatin, an antagonist of activin, is expressed in the
Spemann organizer and displays direct neuralizing activity. Cell.
77:283–295. 1994. View Article : Google Scholar
|
|
29
|
Sasai Y, Lu B, Steinbeisser H, Geissert D,
Gont LK and De Robertis EM: Xenopus chordin: A novel dorsalizing
factor activated by organizer-specific homeobox genes. Cell.
79:779–790. 1994. View Article : Google Scholar
|
|
30
|
Lorenz TC: Polymerase chain reaction:
Basic protocol plus troubleshooting and optimization strategies. J
Vis Exp. 63:e39982012.
|
|
31
|
Jones CM and Smith JC: Wholemount in situ
hybridization to Xenopus embryos. Methods Mol Biol. 461:697–702.
2008. View Article : Google Scholar
|
|
32
|
Yuan JS, Reed A, Chen F and Stewart CN Jr:
Statistical analysis of real-time PCR data. BMC Bioinformatics.
7:852006. View Article : Google Scholar
|
|
33
|
Rozen S and Skaletsky H: Primer3 on the
WWW for general users and for biologist programmers. Methods Mol
Biol. 132:365–386. 2000.
|
|
34
|
Yoon J, Kim JH, Lee OJ, Yu SB, Kim JI, Kim
SC, Park JB, Lee JY and Kim J: xCITED2 Induces Neural Genes in
Animal Cap Explants of Xenopus Embryos. Exp Neurobiol. 20:123–129.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Session AM, Uno Y, Kwon T, Chapman JA,
Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et
al: Genome evolution in the allotetraploid frog Xenopus laevis.
Nature. 538:336–343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar
|
|
37
|
Mi H, Huang X, Muruganujan A, Tang H,
Mills C, Kang D and Thomas PD: PANTHER version 11: Expanded
annotation data from Gene Ontology and Reactome pathways, and data
analysis tool enhancements. Nucleic Acids Res. 45(D1): D183–D189.
2017. View Article : Google Scholar
|
|
38
|
Altschul SF, Gish W, Miller W, Myers EW
and Lipman DJ: Basic local alignment search tool. J Mol Biol.
215:403–410. 1990. View Article : Google Scholar
|
|
39
|
Betancur P, Bronner-Fraser M and
Sauka-Spengler T: Assembling neural crest regulatory circuits into
a gene regulatory network. Annu Rev Cell Dev Biol. 26:581–603.
2010. View Article : Google Scholar
|
|
40
|
Sauka-Spengler T and Bronner-Fraser M: A
gene regulatory network orchestrates neural crest formation. Nat
Rev Mol Cell Biol. 9:557–568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ishii M, Merrill AE, Chan YS, Gitelman I,
Rice DP, Sucov HM and Maxson RE Jr: Msx2 and Twist cooperatively
control the development of the neural crest-derived skeletogenic
mesenchyme of the murine skull vault. Development. 130:6131–6142.
2003. View Article : Google Scholar
|
|
42
|
Huang C, Kratzer M-C, Wedlich D and Kashef
J: E-cadherin is required for cranial neural crest migration in
Xenopus laevis. Dev Biol. 411:159–171. 2016. View Article : Google Scholar
|
|
43
|
Bronner-Fraser M: Neural crest cell
formation and migration in the developing embryo. FASEB J.
8:699–706. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen ZF and Behringer RR: twist is
required in head mesenchyme for cranial neural tube morphogenesis.
Genes Dev. 9:686–699. 1995. View Article : Google Scholar
|
|
45
|
Lin H, Zhu X, Chen G, Song L, Gao L, Khand
AA, Chen Y, Lin G and Tao Q: KDM3A-mediated demethylation of
histone H3 lysine 9 facilitates the chromatin binding of Neurog2
during neurogenesis. Development. 144:3674–3685. 2017. View Article : Google Scholar
|
|
46
|
Bellmeyer A, Krase J, Lindgren J and
LaBonne C: The protoon-cogene c-myc is an essential regulator of
neural crest formation in xenopus. Dev Cell. 4:827–839. 2003.
View Article : Google Scholar
|
|
47
|
Monsoro-Burq A-H, Wang E and Harland R:
Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during
Xenopus neural crest induction. Dev Cell. 8:167–178. 2005.
View Article : Google Scholar
|
|
48
|
Ishimura A, Maeda R, Takeda M, Kikkawa M,
Daar IO and Maéno M: Involvement of BMP-4/msx-1 and FGF pathways in
neural induction in the Xenopus embryo. Dev Growth Differ.
42:307–316. 2000. View Article : Google Scholar
|
|
49
|
Basch ML, Bronner-Fraser M and
García-Castro MI: Specification of the neural crest occurs during
gastrulation and requires Pax7. Nature. 441:218–222. 2006.
View Article : Google Scholar
|
|
50
|
Mayor R, Young R and Vargas A: Development
of neural crest in Xenopus. Curr Top Dev Biol. 43:85–113. 1999.
View Article : Google Scholar
|
|
51
|
Dyson S and Gurdon JB: Activin signalling
has a necessary function in Xenopus early development. Curr Biol.
7:81–84. 1997. View Article : Google Scholar
|
|
52
|
Aberger F, Weidinger G, Grunz H and
Richter K: Anterior specification of embryonic ectoderm: The role
of the Xenopus cement gland-specific gene XAG-2. Mech Dev.
72:115–130. 1998. View Article : Google Scholar
|
|
53
|
Tereshina MB, Zaraisky AG and Novoselov
VV: Ras-dva, a member of novel family of small GTPases, is required
for the anterior ectoderm patterning in the Xenopus laevis embryo.
Development. 133:485–494. 2006. View Article : Google Scholar
|
|
54
|
Nakata K, Nagai T, Aruga J and Mikoshiba
K: Xenopus Zic3, a primary regulator both in neural and neural
crest development. Proc Natl Acad Sci USA. 94:11980–11985. 1997.
View Article : Google Scholar
|
|
55
|
Doniach T and Musci TJ: Induction of
anteroposterior neural pattern in Xenopus: Evidence for a
quantitative mechanism. Mech Dev. 53:403–413. 1995. View Article : Google Scholar
|
|
56
|
Slack JM, Darlington BG, Gillespie LL,
Godsave SF, Isaacs HV and Paterno GD: Mesoderm induction by
fibroblast growth factor in early Xenopus development. Philos Trans
R Soc Lond B Biol Sci. 327:75–84. 1990. View Article : Google Scholar
|
|
57
|
Tsukada Y, Fang J, Erdjument-Bromage H,
Warren ME, Borchers CH, Tempst P and Zhang Y: Histone demethylation
by a family of JmjC domain-containing proteins. Nature.
439:811–816. 2006. View Article : Google Scholar
|
|
58
|
Shpargel KB, Starmer J, Yee D, Pohlers M
and Magnuson T: KDM6 demethylase independent loss of histone H3
lysine 27 trimethylation during early embryonic development. PLoS
Genet. 10:e10045072014. View Article : Google Scholar :
|
|
59
|
Kinoshita T, Haruta Y, Sakamoto C and
Imaoka S: Antagonistic role of XESR1 and XESR5 in mesoderm
formation in Xenopus laevis. Int J Dev Biol. 55:25–31. 2011.
View Article : Google Scholar
|