Introduction
Immunoglobin (Ig)E-associated inhalational allergies
are the most common allergy diseases, affecting >25% of the
world's population. Allergy symptoms include asthma, rhinitis,
eczema and even life-threatening systemic allergic reaction.
Allergies to furred animals, especially dogs and cats, are key
factors in the development of rhinitis and asthma (1-3).
Dogs are considered family pets worldwide. However, even if there
is no dog in a person's house, canine allergens can also be found
in schools and other public places as they can be carried on
clothes, making them almost impossible to avoid. Canine allergens
can cause symptoms ranging from rhinoconjunctivitis to severe
asthma attacks (4,5). In one large study conducted in a
European population, 27% of people with suspected allergic disease
had a positive skin prick test when exposed to canine extract
(6). In a population-based
cross-sectional study in Germany, up to 9% of children and
adolescents were canine dandruff-susceptible, consistent with data
obtained from the Swedish birth cohort BAMSE (7,8).
To date, seven dog allergens have been identified
(www.allergen.org). Among these allergens, Can f 1
is a lipocalin, which is secreted from canine sebaceous glands and
is found in dog hair and dander as well as saliva; it is considered
a major canine allergen (9).
Additionally, other known allergens include albumin (Can f 3)
(10), prostatic kallikrein (Can
f 5) (11) and lipocalin [Can f 2
(12), Can f 4 (13) and Can f 6 (14); these show various molecular
weights ranging from 16 to 69 kDa. Can f 7 was initially identified
from canine allergen testing extracts by sequencing a distinct
protein from Coomassie-stained 2 dimension (2D)-PAGE gels. It
belongs to the Niemann pick type C2 protein family and reportedly
shows 16.9% sensitivity in dog-sensitized subjects (15). With the rapid increase in the
number of pet dogs, several typical allergic diseases caused by dog
allergens, such as asthma, atopic dermatitis and eczema, are
becoming increasingly frequent in China (16). Li et al (17) discovered that, among 6,304
patients aged 5-65 years in China, the prevalence of positive skin
prick responses was 14.0% for dog extract. In addition, given the
population's frequent contact with dogs, the incidence of allergic
diseases in children is increasing. Among 153 recruited Chinese
children with infant food protein allergies, 114 (74.51%)
manifested skin symptoms such as atopic dermatitis or urticaria,
and 28 (18.30%) were presented with early respiratory symptoms
(18). However, except for Can f
6 (3), there have been few
studies regarding specific dog allergens in China, particularly for
Can f 7, although these may seriously impact children's health.
Identification of an allergen's 3D structure can be
used to understand the body's immune response to allergenic
proteins, including cross-reactivity between allergens (19). Furthermore, predicting the
secondary structure of a protein from its amino acid sequence is an
important step in predicting its 3D structure (20). B cell epitopes show fundamental
differences depending on the location of the allergen molecule and
the subsequent initiation of the immune response. B cell epitopes
can be linear or conformational, and they are usually located on
the surface of allergens for easy binding to antibodies. The
primary purpose of epitope prediction is to design and propose
hypoallergenic molecules that can replace crude allergen extracts.
Therefore, identification of B cell epitopes in Can f 7 may aid the
design of novel therapeutic modalities and diagnostic tests for
canine allergies (21,22). Overall, these results could
provide critical information regarding the distribution and
characteristics of canine allergies and provide a foundation for
improving the diagnosis and treatment of canine allergies in
Chinese children.
Patients and methods
Patients and samples
Twenty pediatric patients with positive skin prick
test results (allergens were supplied by ALK-Abelló, Inc.,
Hørsholm, Denmark) and positive serum IgE test results to dog
extracts [via ImmunoCAP assay (Pharmacia Diagnostics AB, Uppsala,
Sweden) as described previously (23)], as well as six healthy children,
were recruited for the present study. Clinical information for
these patients, including specific IgE levels, is shown in Table I. Serum samples (4 ml) were
collected from peripheral venous blood from each patient and
healthy participant after centrifugation at 2,000 × g for 10 min at
4°C. The present study was approved by the Ethics Committee of the
First Affiliated Hospital of Nanjing Medical University
(2015-SRFA-055). According to the Declaration of Helsinki, all
participants written consent was obtained from their parents or
legal guardians for the use of their blood samples prior to
admission.
 | Table IInformation of participants. |
Table I
Information of participants.
| No. | Sex | Age | ImmunoCAP
(KU/L) | Associated
disease |
|---|
| 1 | Female | 1 | 0.96 | Asthma |
| 2 | Female | 1 | 59.6 | Asthma |
| 3 | Male | 2 | 2.16 | Pollinosis |
| 4 | Female | 3 | 91.7 | Asthma,
rhinitis |
| 5 | Female | 4 | 5.61 | Rhinitis |
| 6 | Female | 5 | 13.8 | Asthma,
rhinitis |
| 7 | Female | 6 | 21.1 | Rhinitis |
| 8 | Male | 6 | 56.1 | Asthma,
rhinitis |
| 9 | Male | 7 | 31.3 | Rhinitis |
| 10 | Male | 7 | 18.4 | Asthma |
| 11 | Female | 7 | 9.85 | Asthma,
rhinitis |
| 12 | Female | 8 | 24.5 | Cough |
| 13 | Female | 8 | >100 | Asthma |
| 14 | Male | 9 | 17.3 | Rhinitis |
| 15 | Male | 9 | 63.6 | Asthma |
| 16 | Female | 10 | 77.4 | Asthma |
| 17 | Female | 10 | 20.2 | Dermatitis |
| 18 | Female | 11 | 13.9 | Asthma,
rhinitis |
| 19 | Female | 12 | 8.57 | Asthma,
rhinitis |
| 20 | Female | 12 | 36.6 | Rhinitis |
| N1 | Female | 9 | 0.37 | None |
| N2 | Male | 11 | 0.57 | None |
| N3 | Male | 11 | 0.48 | None |
| N4 | Male | 14 | 0.22 | None |
| N5 | Female | 16 | 0.32 | None |
| N6 | Male | 16 | 0.44 | None |
Expression and purification of Can f 7 in
Escherichia coli (E. coli)
Can f 7 has 450 base pairs and encodes 149 amino
acids (GenBank accession no. 945178). The Can f 7 gene was
synthesized (GenScript, Nanjing, China) using the GenScript rare
codon analysis tool (www.genscript.com/tools/rare-codon-analysis/),
optimized on the basis of constant amino acid sequences, subcloned
into the pET-28a vector via NcoI and XhoI sites and
verified by DNA sequencing (24).
The pET28a-Can f 7 plasmid was transformed into BL21
(DE3) pLysS E. coli cells. Positive colonies were selected
and incubated overnight at 37°C. All broths were inoculated into
200 ml of LB-Kanamycin broth and cultured with shaking at 37°C
until the optical density value at 600 nm reached 0.6-0.8.
Isopropyl-β-D-thiogalactopyranoside (Takara Bio, Inc., Otsu, Japan)
was added to a final concentration of 0.5 mM in the broth and
incubated for a further 16 h. Bacterial cells were pelleted by
centrifugation at 2,000 × g for 30 min at 4°C, and the pellet was
collected. The pellet was lysed in lysis buffer (50 mM
NaH2PO4, 300 mM NaCl, pH 8.0) by sonication
with 50 short bursts of 10 sec at 100 W and a 10 sec cooling period
between each burst. The lysate was centrifuged at 13,523 × g for 30
min at 4°C. As recombinant Can f 7 is mainly found in inclusion
bodies, inclusion bodies were harvested by centrifugation at 13,523
× g for 20 min at 4°C. After solubilizing the inclusion bodies with
8 M urea, the supernatant was applied to High Affinity Ni-NTA Resin
(GenScript, Nanjing, China), then the resin was washed with a
buffer containing 20 mM Tris-HCl, 100 mM
NaH2PO4, 10 mM imidazole, and 8 M urea, and
eluted with a buffer containing 100 mM
NaH2PO4, 500 mM imidazole, and 8 M urea, pH
8.0 (25). Eluted fractions were
collected and reduced overnight by adding a final concentration of
50 mM dithiothreitol. Refolding was performed via 1:50 dilution of
the denatured protein in a redox refolding buffer (0.05 mg/ml final
concentration) consisting of 0.1 M Tris, pH 8.5, 0.5 Ml-arginine, 1
mM EDTA, 5 mM reduced glutathione (GSH) and 1 mM oxidized
glutathione (GSSG) for 48 h at 15°C. The refolded rCan f 7 was
dialyzed against PBS (pH 7.4), and the supernatant was concentrated
using Amicon® Ultra 15 ml Centrifugal Filters (MWCO 3
kDa; Merck KGaA, Darmstadt, Germany), the final concentration was
0.3 mg/ml.
Circular dichroism (CD) analysis of rCan
f 7 expressed in E. coli
CD analysis of rCan f 7 was carried out using a
Chirascan CD spectrometer (Applied Photophysics Ltd., Surrey, UK).
To record far ultra violet CD (200-250 nm) spectra, 0.1 mg/ml
refolded Can f 7 in PBS was analyzed in a 10-mm path-length quartz
cuvette at a 1 nm bandwidth and 0.5 sec per point. The final
spectra were averaged from three consecutive scans and
baseline-corrected by subtracting a control PBS spectrum. The
results were expressed as the mean residue ellipticity in
degxcm2 × dmol−1 and analyzed using the K2D3
program (cbdm-01.zdv.uni-mainz.de/~andrade/k2d3/) (26).
IgE binding activity of Can f 7
identified by western blot analysis
Western blot analysis was used to detect the
IgE-binding activity of Can f 7. Recombinant Can f 7 (2 µg)
was separated by 13% SDS-PAGE and then transferred to a
polyvinylidene difluoride (PVDF) membrane (Merck KGaA, Darmstadt,
Germany). The PVDF membrane was then blocked with 5% skim milk for
2 h at room temperature and subsequently incubated with sera from
20 children with dog allergies [diluted 1:20 in Tris-buffered
saline Tween-20 (TBST)] as the primary antibody and incubated
overnight at 4°C. After washing with TBST, the membrane was
incubated with horseradish peroxidase (HRP)-conjugated goat
anti-human IgE monoclonal antibody (cat. no. 074-1004; KPL, Inc.,
Gaithersburg, MD, USA) diluted 1:5,000 in TBST for 1 h at room
temperature. Positive protein bands were visualized by incubating
the membrane with Immobilon™ Western HRP Substrate Luminol Reagent
(Merck KGaA). Six healthy children's sera were used as negative
controls and PBST were used as a blank control.
Immunoreactivity of human sera with
recombinant Can f 7 by ELISA
A 96-well microplate (Corning Inc., NY, USA) was
coated with 100 µl 10 µg/ml recombinant Can f 7 in
PBS and incubated at 4°C overnight. The wells of the plate were
then blocked with bovine serum albumin (BSA; Biosharp, Hefei,
China; 1 mg/ml; 200 µl/well) for 1 h at 25°C and washed once
with PBS-0.05% Tween-20 (PBST). Subsequently, 100 µl of the
20 Can f 7-allergic children's sera (1:10 dilution in PBST with
0.1% BSA) was incubated for 2 h at 25°C. After incubation, the
wells were washed three times with PBST and incubated with
HRP-conjugated goat anti-human IgE monoclonal antibody (1:2,500)
for 1 h at 25°C. Subsequently, the wells of the plate were washed
and incubated with 100 µl Tetramethylbenzidine (TMB)
substrate solution (Nanjing KeyGEN Biotech Co., Ltd., Nanjing,
China). After incubating for 30 min at room temperature, the
reaction was stopped by adding 50 µl 2 M
H2SO4 in the wells. The absorbance value was
then read at 450 nm on a Eon microplate spectrophotometer (BioTek
Instruments Inc., Winooski, VT, USA; www.biotek.com/). Sera from six healthy subjects were
used as control normal human serum (NHS). Absorbance values were
regarded as positive if they have statistical significance compared
to the mean value of the healthy controls.
Basophil activation analysis
Allergens induce protein expression levels of the
Cluster of differentiation (CD)63 and the C-C motif chemokine
receptor (CCR)3 on basophils which is considered an indicator of
basophil activation (27). The
ability of rCan f 7 to activate basophils in a modified basophil
activation test, was examined (28-31). Briefly, a total of
4.3×107 peripheral blood mononuclear cells were obtained
from 20 ml of blood collected from healthy individuals using a
Ficoll-Paque density gradient (GE Healthcare, Uppsala, Sweden)
according to the manufacturer's protocol. Then, the cells were
incubated with 10 ml of lactic acid buffer (containing 1.3 M NaCl,
0.005 M KCl and 0.01 M lactic acid, pH 3.9) for 2 min on ice. After
neutralization with 12% Tris (pH 10.9), nonspecific IgE on
basophils was removed. The cells were washed and aliquots of a cell
suspension were passively sensitized by incubation in RPMI-1640
medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% patient serum for 2 h at 37°C. The six serum groups
used in this test consisted of six patients' sera (No. 2, No. 4,
No. 6, No. 13, No. 17 and No. 20 in Table I) and six normal sera. The cells
were then incubated with Can f 7 (1.0 µg/ml) for 15 min at
37°C. HRP-conjugated goat anti-human IgE antibody (1.0
µg/ml) was used as a positive control. After incubation, the
cells were washed and resuspended in 100 µl stain buffer,
then 20 µl of anti-human CD63-FITC antibody (cat. no.
MA1-19602; Invitrogen, Camarillo, CA, USA) and 5 µl of
CCR3-PE-labeled antibody (cat. no. A18365; eBioscience, Inc., San
Diego, CA, USA) were added to the cells for 15 min at 37°C in the
dark. Surface-labeled flow cytometric analysis was performed on a
FACSAria flow cytometer at 488 nm and analyzed with FACSDiva
software v.6.1.3 (both BD Biosciences, Franklin Lakes, NJ, USA)
(32).
Structure modeling and B cell epitopes
prediction
A structural model of Can f 7 protein was generated
by aligning patterns in SWISS-MODEL (swissmodel.expasy.org/interactive) (33). Briefly, the amino acid sequences
of Can f 7 were used to search the homologous template. A template
for Can f 7 was selected from the SWISS-MODEL server (34). TMFMM 2.0 software (http://www.cbs.dtu.dk/services/TMHMM/)
can predict transmembrane helices of Can f 7 (35). The secondary structure of Can f 7
(α-helix, β-sheet, and random coil) was predicted using the PyMOL
molecular graphics system (pymol.org/2/)
(36).
Using the DNAStar (www.dnastar.com/) protean system, the BepiPred 2.0
server (www.cbs.dtu.dk/services/BepiPred/) and the BPAP system
(imed.med.ucm.es/Tools/antigenic.pl) predicted B cell epitopes of
Can f 7 (37,38). The final consensus epitope results
were obtained by combining the results of these three tools with
previous methods (39-41). In the DNAStar Protean system, four
properties (hydrophilicity, flexibility, accessibility and
antigenicity) of the amino acid sequence were chosen as parameters
for epitope prediction. Peptide regions with good hydrophilicity,
high sensitivity, surface accessibility, and high antigen index
were selected as candidate epitopes for further study. The BPAP
system and the BepiPred 2.0 server require only amino acid
sequences, which provide more direct results, together with the
physicochemical properties of amino acids, such as hydrophilicity,
flexibility, accessibility, corners and exposed surfaces. Five
predicted B cell epitopes were synthesized by GenScript with a
purity >90% and named as P1, P2, P3, P4 and P5 respectively.
Verification of B cell epitopes by
ELISA
The IgE binding of the predicted B cell peptides was
detected using ELISA (42). A
96-well microplate was coated with each epitope in PBS at 2
µg/100 µl/well (0.02 µg/µl). The plate
was then blocked with 1 mg/ml BSA (200 µl/well) for 1 h at
25°C and washed once with PBST. Subsequently, 100 µl of four
Can f 7 allergic children's sera (No. 2, No. 4, No. 6 and No. 13 in
Table I; diluted 1:10 in PBST
with 0.1% BSA) was incubated for 2 h at 25°C. After incubation, the
plates were washed three times with PBST. The plates were incubated
with a 1:2,500-diluted HRP-conjugated goat anti-human IgE
monoclonal antibody for 1 h at 25°C and washed with PBST three
times. Subsequently, 100 µl TMB color liquid was added to
each well. After staining for 30 min at room temperature, the
reaction was stopped by the addition of 50 µl of 2 M
H2SO4, and the serum of four healthy
participants without a history of allergic symptoms was used as
control, NHS. After incubation, plates were processed as described
above, and absorbances were read at 450 nm.
ELISA inhibition of B cell epitopes
The five B cell epitopes, BSA and the rCan f 7 were
tested via inhibition ELISAs of Can f 7 with the sera of two Can f
7-allergic children (No. 4 and No. 13 in Table I). In this experiment, 96-well
microplates were coated with 2 µg recombinant Can f 7 (100
µl/well) in PBS overnight 4°C. The plate was washed three
times with PBST. The plate was then blocked with BSA (1 mg/ml; 200
µl/well) for 1 h at 25°C and washed once with PBST. The
plates were then incubated with 100 µl of 2 allergic
children's sera (1:10 diluted in PBST, which were previously
preincubated with synthesized epitopes for 1 h at 37°C with 8
gradient concentrations (0, 10−4, 10−3,
10−2, 10−1, 1, 10, and 102
µg/ml) at 25°C for 2 h. After incubation, the plates were
washed three times with PBST. The plates were incubated with a
1:2,500-diluted HRP-labeled anti-human IgE monoclonal antibody for
1 h at 25°C and washed with PBST three times. Subsequently, 100
µl TMB color liquid was added to each well. After staining
for 30 min at room temperature, the reaction was stopped by the
addition of 50 µl of 2 M H2SO4. After
incubation, plates were processed as described above, and the
absorbance was read at 450 nm (43).
Statistical analysis
Statistical significance between mean values of
three experiments was analyzed by Dunnett's t-test following
one-way analysis of variance or the Student's t-test utilizing the
SPSS 13.0 version (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression and purification of Can f 7 in
E. coli
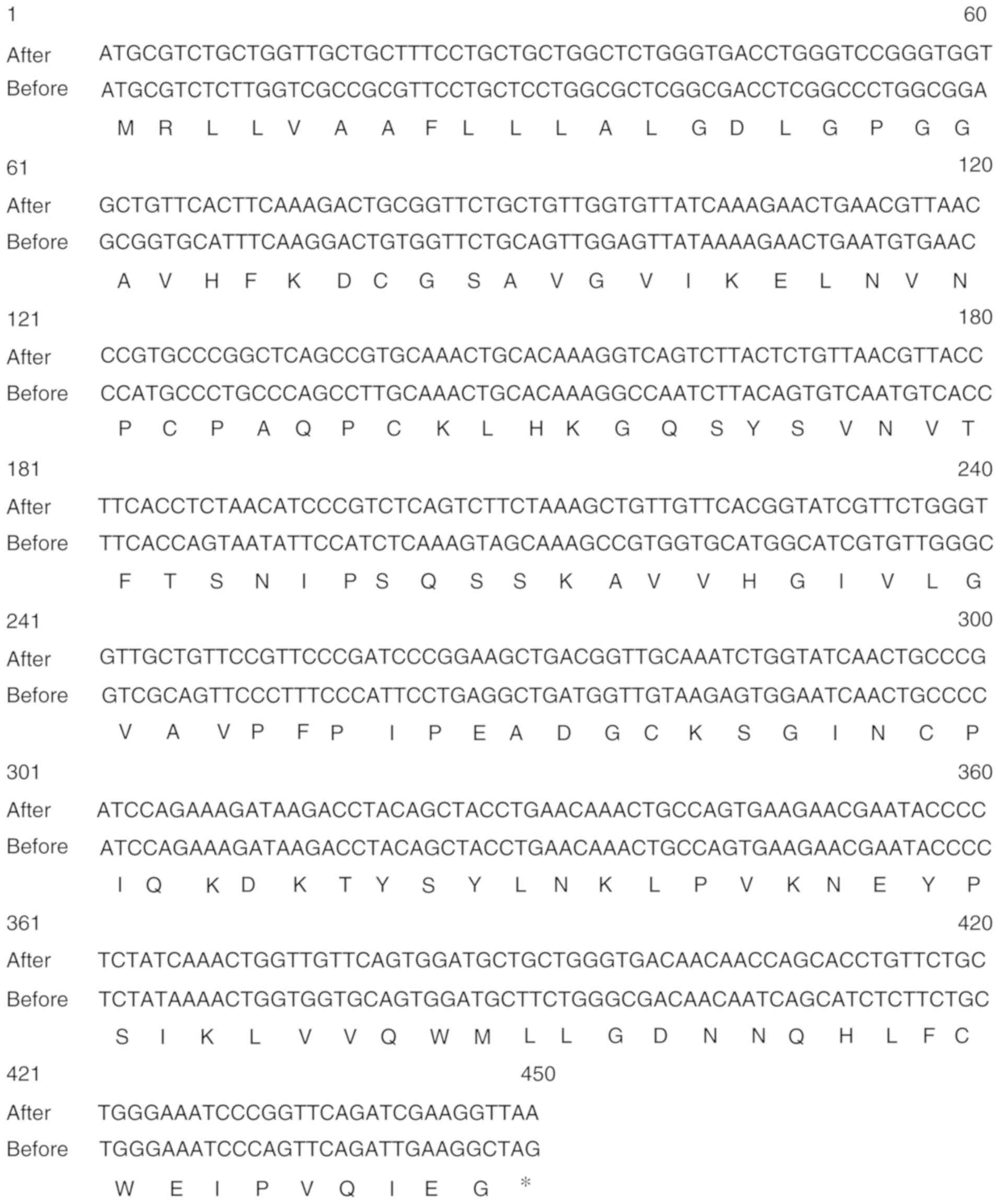
The optimized codon sequence of Can f 7 used for
expression in E. coli is shown in Fig. 1. The codon-optimized Can f 7 gene
was then subcloned into the pET-28a (+) vector and transformed into
BL21 (DE3) pLysS E. coli cells. Fig. 1 shows a comparison of the
sequences prior and following optimization. By SDS-PAGE, Can f 7
was shown to be primarily expressed in inclusion bodies (Fig. 2A). Recombinant Can f 7 was then
purified on a Ni-NTA resin under denaturing conditions. Purified
Can f 7 is shown as a single band in the subsequent SDS-PAGE
analysis. The apparent molecular weight of purified recombinant Can
f 7 was ~14 kDa (Fig. 2B). The
denatured proteins were refolded in a glutathione redox system. The
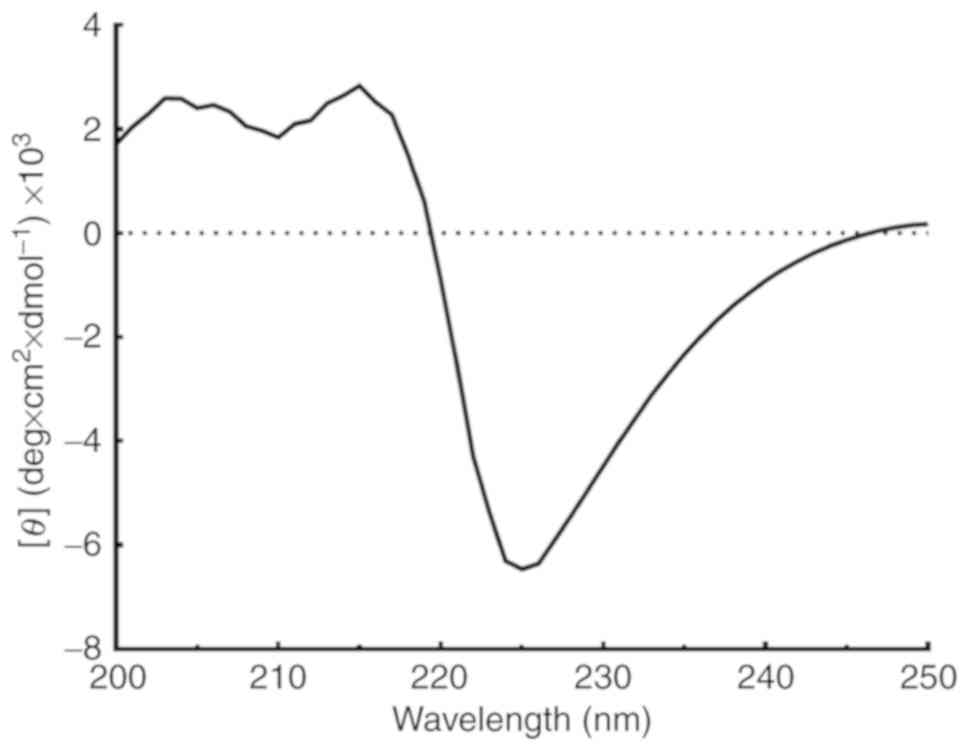
far UV spectra of refolded Can f 7 showed a trend towards β-sheet
structures (Fig. 3). Estimation
of the secondary structure using the K2D3 program resulted in a
determination of ~1.43% α-helices and 44.13% β-sheets.
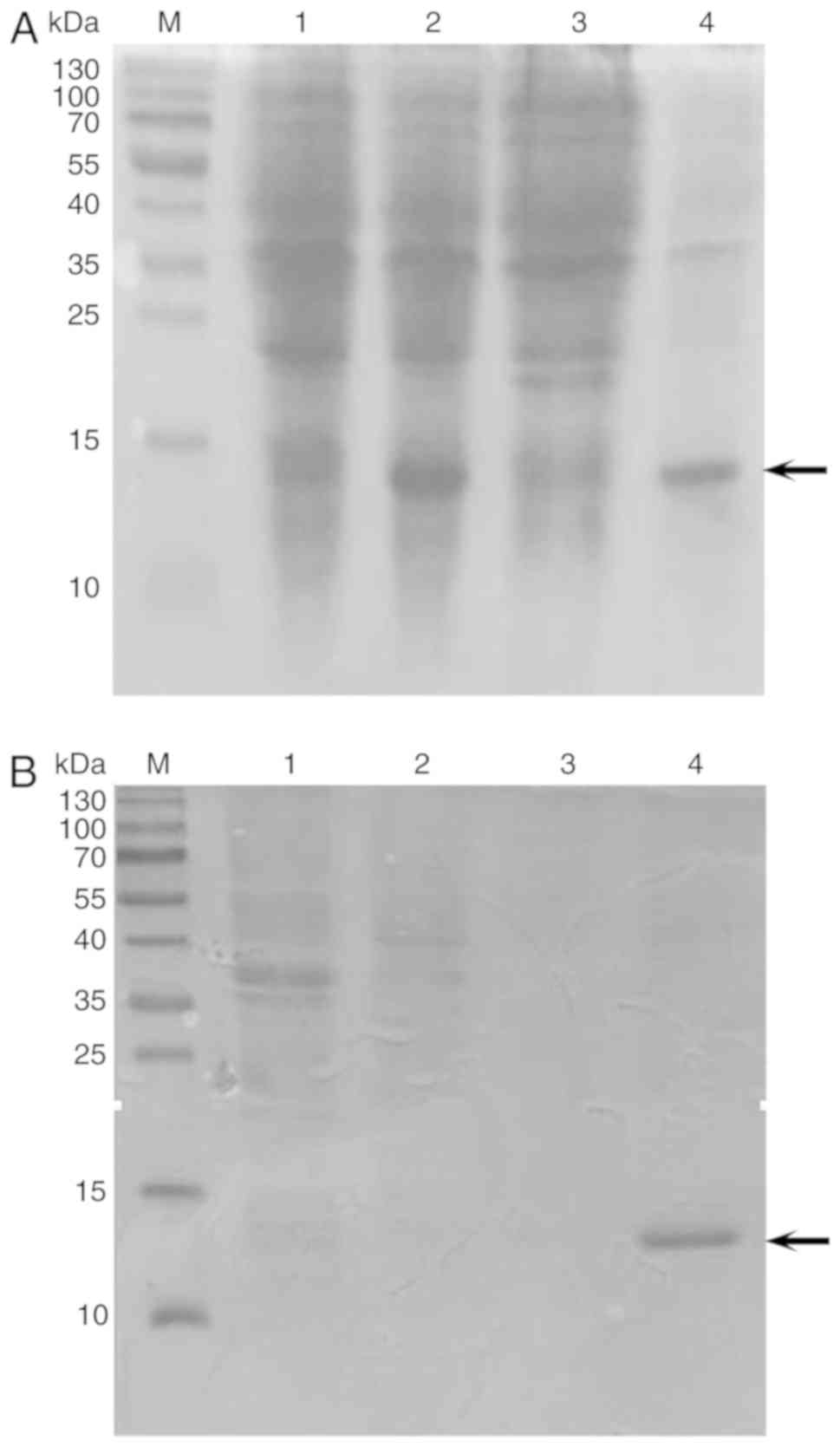 | Figure 2Expression and purification of Can f
7 in E. coli. (A) Lane M, protein molecular weight standard;
Lane 1, noninduced recombinant Can f 7 whole cell lysate; Lane 2,
isopropyl-β-D-thiogalactopyranoside-ind uced recombinant Can f 7
whole cell lysate; Lane 3, supernatant fraction after
ultrasonication; Lane 4, precipitated fraction (inclusion bodies)
after ultrasonication. (B) SDS-PAGE after affinity chromatography
of Can f 7. Lane M, protein molecular weight standard; Lane 1,
unbound fractions of the supernatant of Can f 7; Lane 2, first
elution fraction using 500 mM imidazole; Lane 3, second elution
fraction using 500 mM imidazole; Lane 4, third elution fraction
using 500 mM imidazole. The arrow indicates Can f7 and its the
purified version. Can f7, Canis familiaris. |
Immunoreactivity of Can f 7 to IgE by
western blot and ELISA
To determine the ability of Can f 7 to bind specific
IgE in sera from dog-allergic children, western blot analysis and
ELISA were performed. A total of 20 pediatric patients with dog
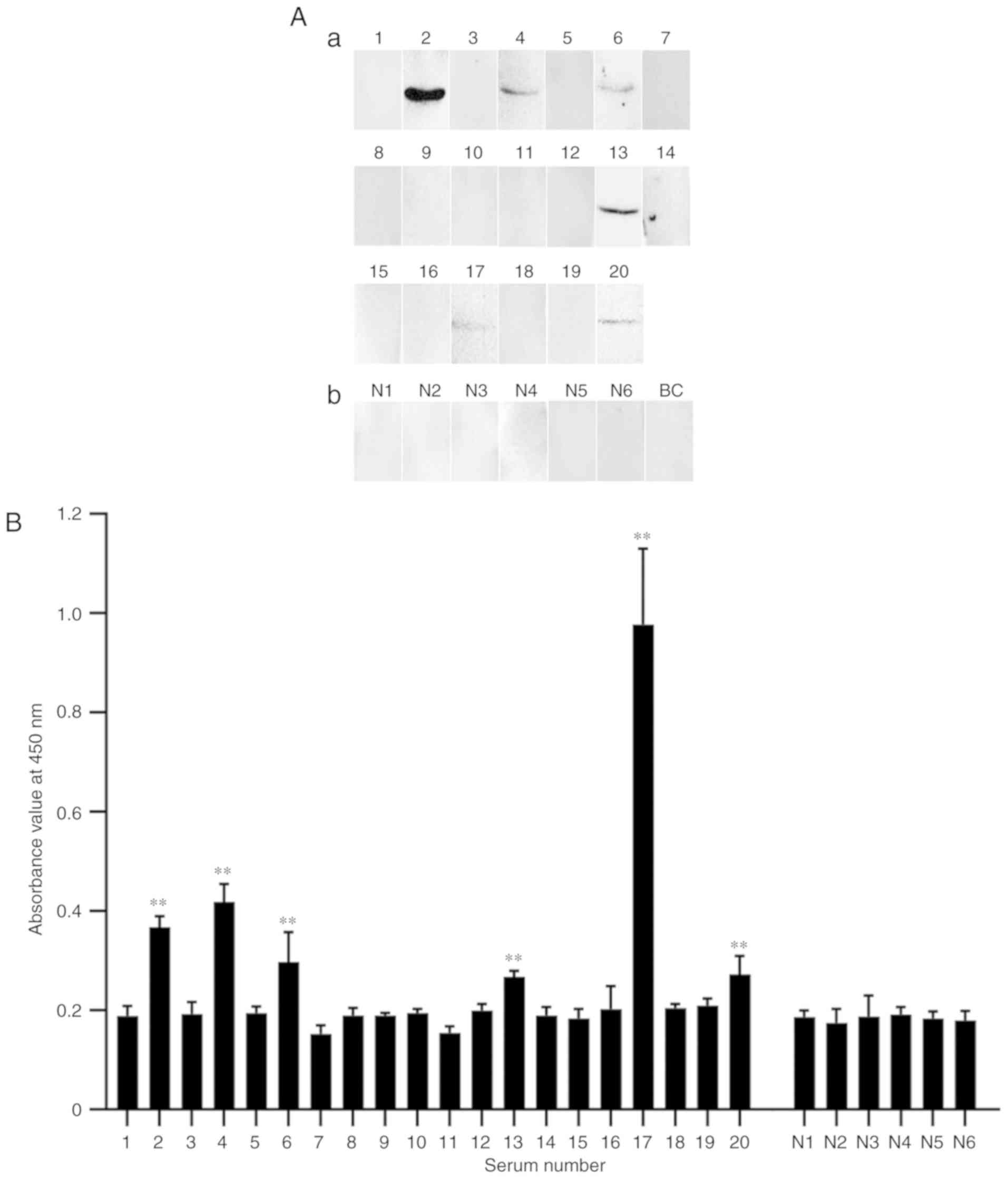
allergies were included. As shown in Fig. 4, six of 20 dog-allergic sera from
pediatric patients showed positive IgE binding to Can f 7 (numbers
of the positive children are No. 2, 4, 6, 13, 17 and 20 in Table I), whereas healthy control sera
failed to do so.
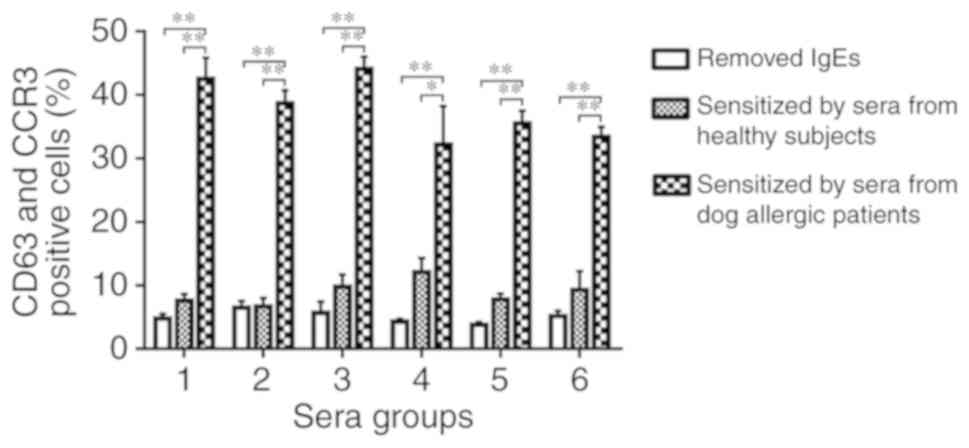
Basophil activation analysis
Can f 7 induced an ~4.0-fold increase in CD63 and
CCR3 expression in basophils sensitized by serum from dog-allergic
children compared with healthy controls (Fig. 5).
Structure modeling and B cell epitope
prediction
Looking for a protein with a similar amino acid
sequences to Can f 7 in SWISS-MODEL, an epididymal secretory
protein E1 (PDB ID: 2hka.1.A; www.rcsb.org/)
demonstrated the highest sequence identity with Can f 7 (76.15%).
Therefore, the template 2hka.1. A was selected as a template for
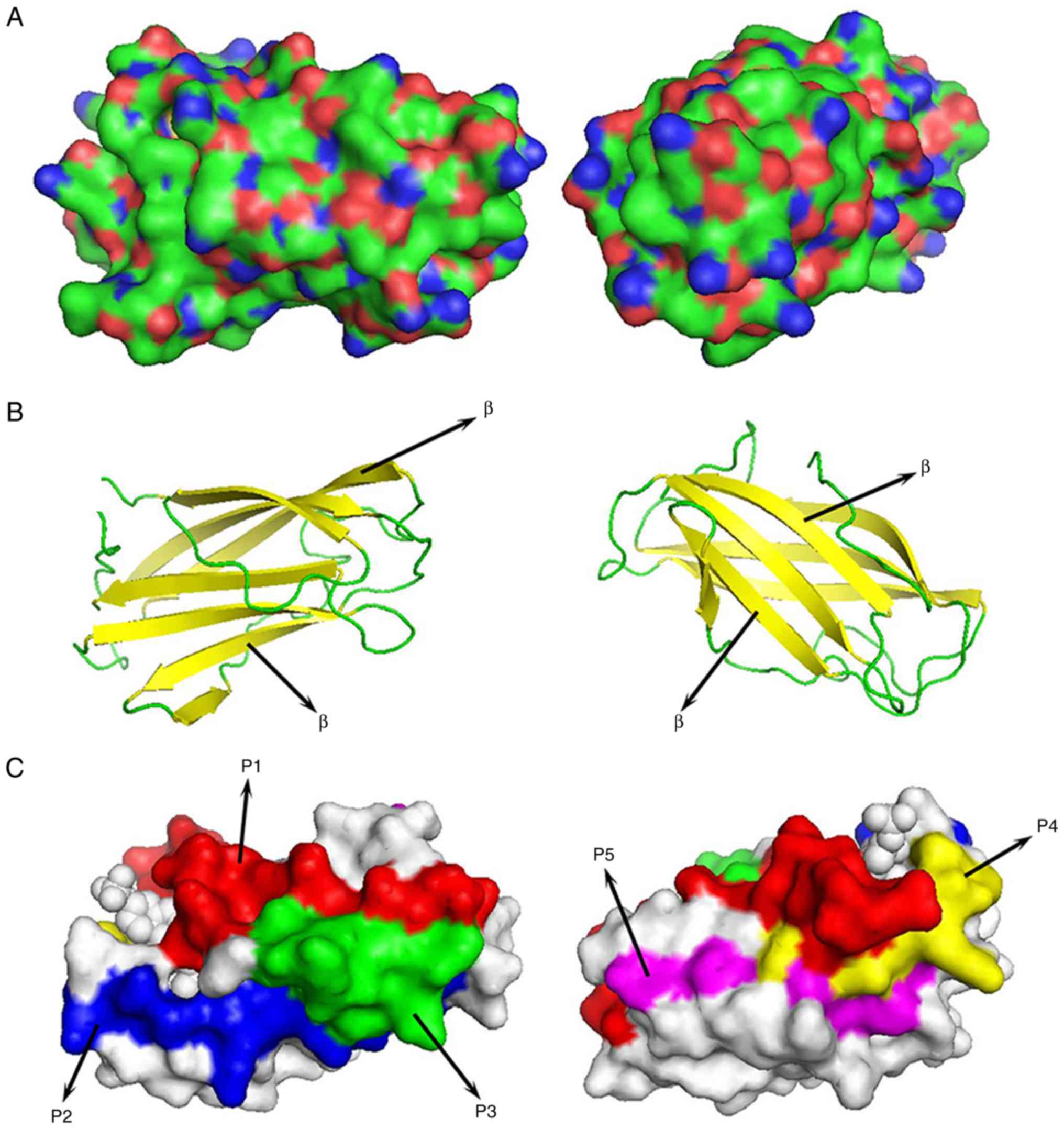
homology modeling. Fig. 6A shows
the overall 3D structure of Can f 7 determined via homology
modeling.
Can f 7 was predicted to contain no transmembrane
helix by TMHMM 2.0. Secondary structure prediction showed that Can
f 7 contains 6 β-sheets and no α-helices (Fig. 6B).
Based on four main properties (hydrophilicity,
flexibility, reachability and antigenicity), the final predicted
B-cell epit-opes in Can f 7 allergen by DNAstar were amino acid
regions 64-87, 89-106 and 117-127. In contrast, the BPAP system
predicted regions 18-29, 65-87, 89-101, 106-117, 120-128 and
136-145 as B cell epitopes in Can f 7. The BepiPred 2.0 server
predicted regions 34-47, 88-103, 105-118, and 136-148 as B cell
epitopes in Can f 7. The final potential B cell epitopes of Can f 7
were determined based on the results from all three tools. The
final results of the three immunological informatics tools
ultimately predicted five B cell epitopes in Can f 7: P1:
IPSQSSKAVVHGIVLGVAVPFPI; P2: EADGCKSGINCPI; P3: TYSYLNKLPVKN; P4:
PSIKLVVQW; and P5: QHLFCWEIPV (Table
II; Fig. 6C).
 | Table IILocation of the B cell epitopes of
the Can f 7 in Canis familaris predicted by
immunoinformatics tools. |
Table II
Location of the B cell epitopes of
the Can f 7 in Canis familaris predicted by
immunoinformatics tools.
| Peptide | Position | Sequence |
|---|
| P1 | 65-87 |
IPSQSSKAVVHGIVLGVAVPFPI |
| P2 | 89-101 | EADGCKSGINCPI |
| P3 | 106-117 | TYSYLNKLPVKN |
| P4 | 120-128 | PSIKLVVQW |
| P5 | 136-145 | QHLFCWEIPV |
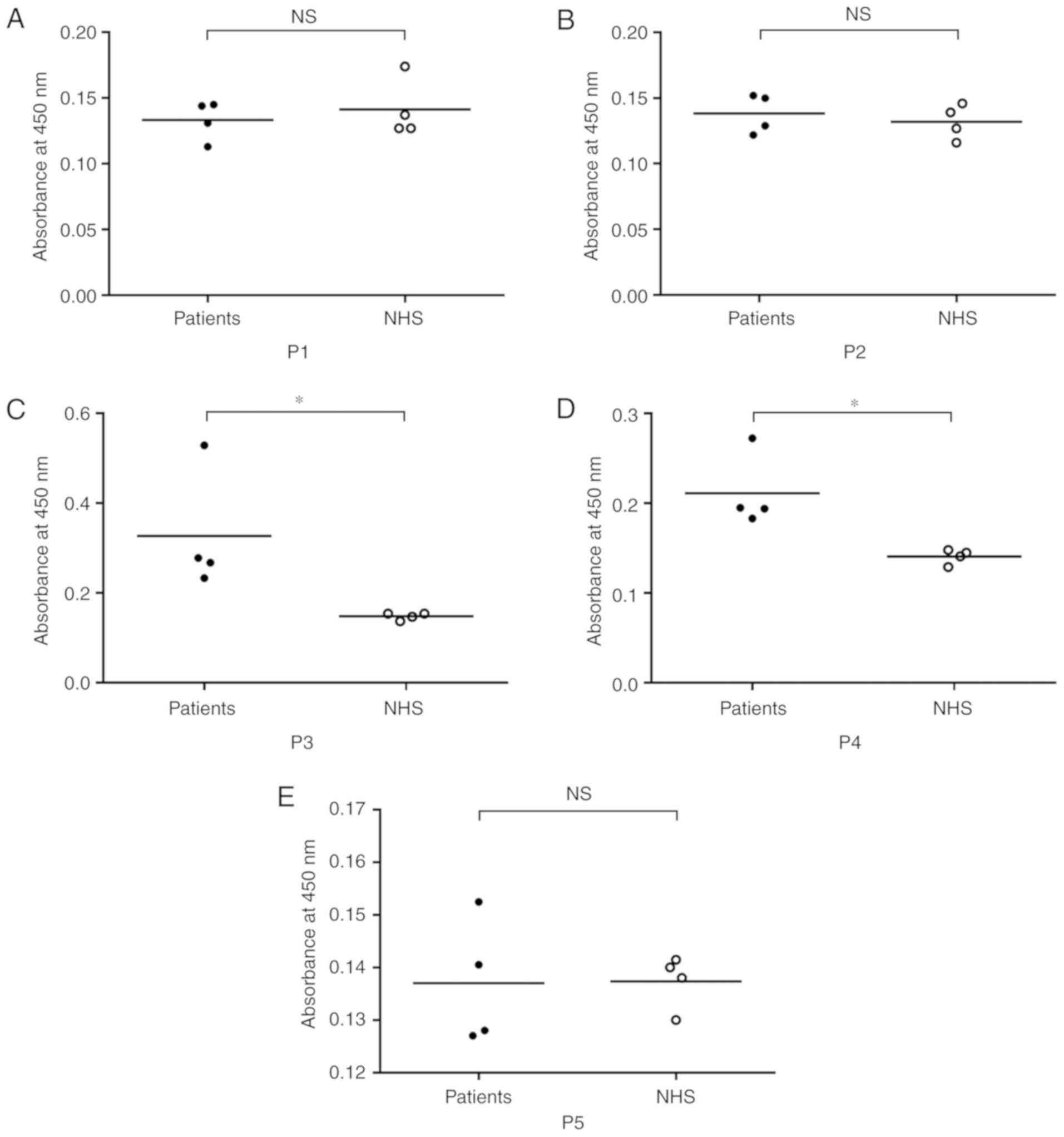
B cell epitope verification
The specific IgE binding of predicted B cell
peptides was detected by ELISA using sera from children with dog
allergies (patient No. 2, No. 4, No. 6 and No. 13 in Table I). The results showed that the
mean absorbance values in P3 and P4 groups were approximately twice
that of the control NHS. However, there was no prominent increase
in the absorbance values in P1, P2 and P5 groups. Therefore, among
the five predicted B cell peptides, P3 (TYSYLNKLPVKN) and P4
(PSIKLVVQW) reacted with four dog-allergic children's sera and
showed significantly different IgE binding affinity compared with
NHS (P<0.05; Fig. 7).
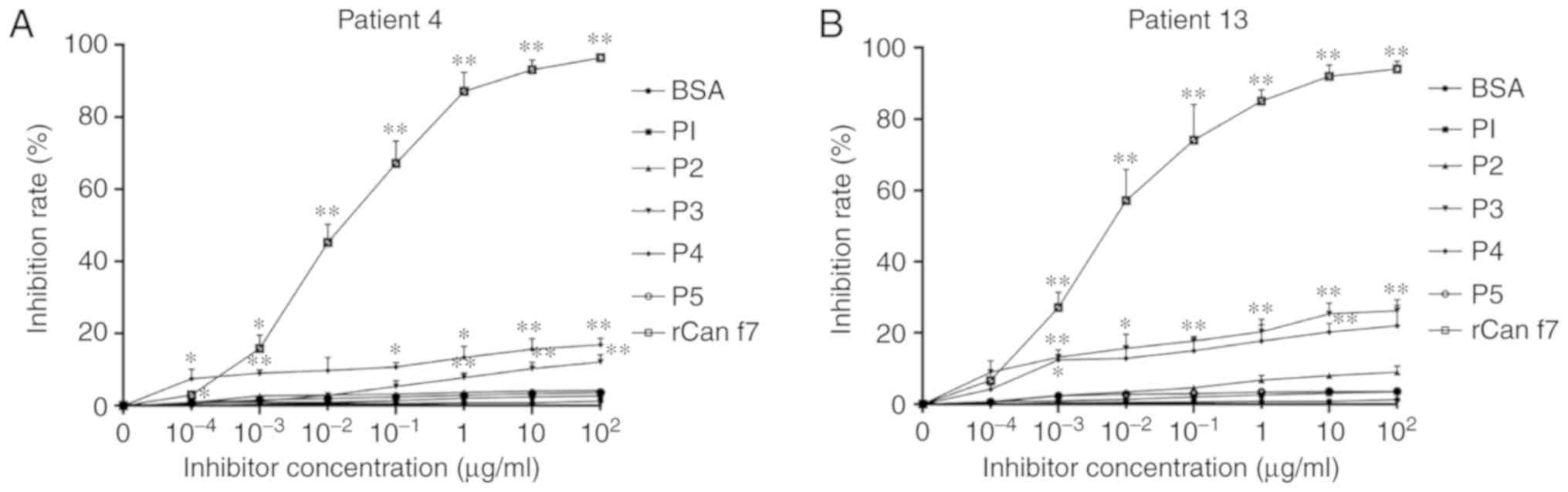
Inhibition ELISA experiments
Inhibition ELISA experiments were performed with the
five B cell epitopes, BSA and rCan f 7. P3 and P4 could inhibit the
binding of specific IgE antibodies to the recombinant fusion
protein Can f 7. In Patient 4, the maximum inhibition rates of P3
and P4 were 13% and 17% at a concentration of 100 µg/ml,
respectively, whereas 96% inhibition could be achieved by rCan f 7
(the inhibition rates of P1, P2, P5 and BSA were <5%). In
Patient 13, the maximal inhibition of P3 and P4 were 22 and 26%,
respectively. Similarly, the maximal inhibition of rCan f 7 was
93%, and the others were all <10% (Fig. 8).
Discussion
To better understand the prevalence and mechanism of
Can f 7-associated dog allergy in children in China, recombinant
Can f 7 was isolated from E. coli. Can f 7 has three
potential glycosylation sites and can reportedly be glycosylated in
yeast expression systems (15).
However, it has also been reported that glycosylation of Can f 7
does not affect IgE binding to Can f 7 (15). The present study primarily sought
to examine the sensitization rate of Can f 7 in Chinese children
with allergies to dogs; therefore, glycosylation of the protein was
not taken into consideration, which led to the use of the more
convenient E. coli expression system.
In the present study, Can f 7 was expressed in
inclusion bodies, while the same protein was reported to be
expressed in a soluble form in E. coli (15). This may be due to different
factors between the two studies, including the cloning vectors, the
optimized codons, the host cells, and the expression conditions,
among others. The inclusion bodies were purified on a Ni-NTA column
under denaturing conditions. To allow correct folding and disulfide
bond formation, a GSH/GSSG redox system was used (44). This system has been widely used
for refolding proteins with disulfide bonds (45-47). CD analysis of the refolded Can f 7
found that the secondary structure of this protein was estimated to
be ~1.43% α-helix and 44.13% β-sheets, which was consistent with
the predicted result that Can f 7 contains six β-sheets.
Can f 7 showed immunological activity, binding with
IgE in dog-allergic children's sera with a sensitization rate of
30% (6/20) in Chinese children. In Fig. 4, not all patients' sera show high
reactivity to rCan f 7 since the children were all reacted with dog
extracts and then they would be diagnosed with dog allergies.
However, dog extracts are complex substances which contain not only
Can f 7 but also other allergens, such as Can f 1-6 and even
unreported allergens. Thus, not all children were allergic to rCan
f 7. This is a preliminary assessment of the IgE-binding activity
of dog-allergic children in China and should be confirmed in
studies with larger populations in the future. In addition,
basophil activation testing, which is well-established as a
functional assay for allergenicity was performed. In this test, the
expression of CD63 and CCR3 on basophil surfaces is considered an
indicator of basophil activation; it was found that Can f 7 could
induce up to a 4.0-fold increase in their expression. Therefore, it
was confirmed that Can f 7 is an active canine allergen. Compared
with a previous study of Can f 7, a major innovation of this
experiment is that allergic reactions in Chinese children was
analyzed, identifying a higher sensitization rate (30%) compared
with that (16.9%) in a previous report (15). These findings indicate that the
sensitization rate of Can f 7 is higher in Chinese children than
adults and show that Can f 7 is also an important allergen in
Chinese children. Taking into account the increase in the number of
pet dogs in China, the present analysis of dog allergens is
significant.
α-Helices and β-sheets are the two major secondary
structures of proteins whose structure is maintained by hydrogen
bonding, making it impossible for the epitope sequence to be
located therein (3). In contrast,
random coils always contain epitope sequences as they are located
in areas of surface exposure (48). The secondary structure of Can f 7
is predicted to contain six β-sheets. In addition to a sequence
consisting of β-sheets, many other residues consist of random coils
and may also be associated with certain immunological features of
Can f 7. Greater analysis of the structure and function of Can f 7
will likely contribute to improved peptide-based vaccine design for
dog allergies.
In this study, B-cell epitopes of Can f 7 were
analyzed and projected them on the 3D structure of Can f 7. The
synthesis of allergen-specific IgE is a key step in the development
of allergic symptoms (3,49). IgE production requires B cells to
be in close contact with allergen-specific helper T cells for
translational recombination (50). Therefore, identification of
relevant epitopes is important to understand the development of
allergy symptoms. Computer prediction has become a useful method of
identifying epitopes from immune-related proteins (51). Hydrophilicity, antigenicity,
segmental motility, flexibility and accessibility have all been
used to predict linear B cell epitopes on protein sequences based
on the propensity values of amino acid properties in many
algorithms (43). Actually,
conformational epitopes are likely the most significant ones since
the results between Fig. 4A and B
are not fully consistent. For instance, in Fig. 4A, serum sIgE from patient 17 shows
the lowest reactivity to rCan f 7, but in Fig. 4B it shows the highest reactivity.
This may due to the conformational epitopes of rCan f 7 were fully
destroyed during the sample preparation step in WB and the sIgE in
the serum from patient 17 was mainly target to the conformational
epitopes of rCan f 7. Although the conformational epitopes in
allergen play an important role in allergy, their prediction
algorithms are still far less than satisfactory (52). Thus, in the present study, the
focus was on the prediction of five linear peptides (P1-P5) as
potential B cell epitopes of Can f 7. In addition, it was attempted
to confirm the predicted allergenicity of these B cell epitopes by
ELISA. The present results identified that only P3 and P4 showed a
positive association with sera from children with dog allergies.
Moreover, these B cell epitopes are composed of consecutive amino
acids, meaning that they belong to sequential B cell epitopes.
However, although the two predicted epitopes could be detected by
IgE raised against dog allergens, the positive signals in ELISA
were relatively weak compared with that of the negative control.
This is most likely an artifact of the testing methodology, as a
single epitope coated on an ELISA plate has limited ability to bind
with Can f 7-specific IgE, which may have led to a weaker ELISA
value. The inhibition ELSIA demonstrated that both P3 and P4 could
inhibit the binding of specific IgE antibodies to rCan f 7. In this
experiment, the sera from patient 4 and 13 were selected as they
had significant reactions to P3 and P4. Moreover, the inhibition
rate calculated from the low OD value would lead to large
deviation. Therefore, other patient's sera were not included in
this experiment. Finally, the present results suggest that
sequential epitopes may serve an important role in Can f 7-related
dog allergies.
In summary, Can f 7 recombinant protein was
expressed and purified, and its sensitization rate was determined
by Western blot, ELISA, and basophil activation analysis. The
homology of the Can f 7 protein was modeled, and two B cell
epitopes were predicted and confirmed by ELISA. Currently, there is
no commercially available method to detect sensitization to Can f
7. The present study provides a potential epitope prediction
strategy that may be used for the future diagnosis and treatment of
canine allergy in children.
Funding
The present study was sponsored by grants from the
National Natural Science Foundation of China (grant nos. 81571568,
81771725 and 81871265), the CAMS Innovation Fund for Medical
Sciences (CIFMS; no. 2016-I2M-1003), the Innovation team of the
Jiangsu provincial Commission of Health and Family Planning (grant
no. CXTDA2017049), and the Priority Academic Program Development of
Jiangsu Higher Education Institutions.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JLS and JFW contributed to the conception and design
of the study. RQW, YJW and ZQX performed all experiments and
verified the analytical data. YJZ, MDC and WZ contributed to the
statistical analysis and helped to interpret the results. YJW, ZQX
and WZ wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Nanjing Medical
University (2015-SRFA-055).
Patient consent for publication
All participants written consent was obtained from
their parents or legal guardians for the use of their blood samples
prior to admission.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Konradsen JR, Fujisawa T, van Hage M,
Hedlin G, Hilger C, Kleine-Tebbe J, Matsui EC, Roberts G, Rönmark E
and Platts-Mills TA: Allergy to furry animals: New insights,
diagnostic approaches, and challenges. J Allergy Clin Immunol.
135:616–625. 2015. View Article : Google Scholar
|
|
2
|
Nilsson OB, Hage MV and Grönlund H:
Mammalian-derived respiratory allergens-Implications for diagnosis
and therapy of individuals allergic to furry animals. Methods.
66:86–95. 2014. View Article : Google Scholar
|
|
3
|
Wang YJ, Li L, Song WJ, Zhou YJ, Cao MD,
Zuo XR and Wei JF: Canis familiaris allergen Can f 6: Expression,
purification and analysis of B-cell epitopes in Chinese dog
allergic children. Oncotarget. 8:90796–90807. 2017.PubMed/NCBI
|
|
4
|
Polovic N, Wadén K, Binnmyr J, Hamsten C,
Grönneberg R, Palmberg C, Milcic-Matic N, Bergman T, Grönlund H and
van Hage M: Dog saliva-an important source of dog allergens.
Allergy. 68:585–592. 2013. View Article : Google Scholar :
|
|
5
|
Vredegoor DW, Willemse T, Chapman MD,
Heederik DJJ and Krop EJM: Can f 1 levels in hair and homes of
different dog breeds: Lack of evidence to describe any dog breed as
hypoallergenic. J Allergy Clin Immunol. 130:904–909.e7. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Burbach GJ, Heinzerling LM, Edenharter G,
Bachert C, Bindslev-Jensen C, Bonini S, Bousquet J,
Bousquet-Rouanet L, Bousquet PJ, Bresciani M, et al: GA(2)LEN skin
test study II: Clinical relevance of inhalant allergen
sensitizations in Europe. Allergy. 64:1507–1515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmitz R, Ellert U, Dahm S and Thamm M:
Patterns of sensitization to inhalant and food allergens-findings
from the german health interview and examination survey for
children and adolescents. Int Arch Allergy Immunol. 162:263–270.
2013. View Article : Google Scholar
|
|
8
|
Asarnoj A, Ostblom E, Kull I, Lilja G,
Pershagen G, Hedlin G, van Hage M and Wickman M: Sensitization to
inhalant allergens between 4 and 8 years of age is a dynamic
process: results from the BAMSE birth cohort. Clin Exp Allergy.
38:1507–1513. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schou C, Svendsen UG and Løwenstein H:
Purification and characterization of the major dog allergen, Can f
1. Clin Exp Allergy. 21:321–318. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Basagaña M, Luengo O, Labrador M, Garriga
T, Mattsson L, Lidholm J and Cardona V: Component-resolved
diagnosis of dog allergy. J Investig Allergol Clin Immunol.
27:185–187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mattsson L, Lundgren T, Everberg H,
Larsson H and Lidholm J: Prostatic kallikrein: A new major dog
allergen. J Allergy Clin Immunol. 123:362–368.e363. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kamata Y, Miyanomae A, Nakayama E,
Miyanomae T, Tajima T, Nishimura K, Tada T and Hoshi H:
Characterization of dog allergens Can f 1 and Can f 2. 2. A
comparison of Can f 1 with Can f 2 regarding their biochemical and
immunological properties. Int Arch Allergy Immunol. 142:301–308.
2007. View Article : Google Scholar
|
|
13
|
Mattsson L, Lundgren T, Olsson P, Sundberg
M and Lidholm J: Molecular and immunological characterization of
Can f 4: A dog dander allergen cross-reactive with a 23 kDa
odorant-binding protein in cow dander. Clin Exp Allergy.
40:1276–1287. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nilsson OB, Binnmyr J, Zoltowska A, Saarne
T, Hage MV and Grönlund H: Characterization of the dog lipocalin
allergen Can f 6: The role in cross-reactivity with cat and horse.
Allergy. 67:751–757. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khurana T, Newmanlindsay S, Young PR and
Slater JE: The NPC2 protein: A novel dog allergen. Ann Allergy
Asthma Immunol. 116:440–446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo S, Sun Y, Hou J, Kong X, Wang P, Zhang
Q and Sundell J: Pet keeping in childhood and asthma and allergy
among children in Tianjin area, China. PLos One. 13:e01972742018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li J, Sun B, Huang Y, Lin X, Zhao D, Tan
G, Wu J, Zhao H, Cao L and Zhong N; China Alliance of Research on
Respiratory Allergic Disease: A multicentre study assessing the
prevalence of sensitizations in patients with asthma and/or
rhinitis in China. Allergy. 64:1083–1092. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao Q, Ren YX, Liu YG, Ma L, Gu XH, Zhang
WX, Liu L, Zhai XJ, Xiang L and Shen KL: Allergy march of Chinese
children with infantile allergic symptoms: A prospective
multi-center study. World J Pediatr. 13:1–6. 2017. View Article : Google Scholar
|
|
19
|
Negi SS and Braun W: Cross-React: A new
structural bioinformatics method for predicting allergen
cross-reactivity. Bioinformatics. 33:1014–1020. 2017.PubMed/NCBI
|
|
20
|
Feng Y, Lin H and Luo L: Prediction of
protein secondary structure using feature selection and analysis
approach. Acta Biotheor. 62:1–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fanuel S, Tabesh S, Sadroddiny E and
Kardar GA: Analysis of predicted B and T-cell epitopes in Der p 23,
allergen from Dermatophagoides pteronyssinus. Bioinformation.
13:307–312. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Curin M, Weber M, Hofer G, Apostolovic D,
Keller W, Reininger R, Swoboda I, Spitzauer S, Focke-Tejkl M, van
Hage M and Valenta R: Clustering of conformational IgE epitopes on
the major dog allergen Can f 1. Sci Rep. 7:121352017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Griffiths RLM, El-Shanawany T, Jolles SRA,
Selwood C, Heaps AG, Carne EM and Williams PE: Comparison of the
performance of skin prick, immunocap, and isac tests in the
diagnosis of patients with allergy. Int Arch Allergy Immunol.
172:215–223. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xue F, Guo H, Hu Y, Liu R, Huang L, Lv H,
Liu C, Yang M and Ma L: Expression of codon-optimized plant
glycosyltransferase UGT72B14 in escherichia coli enhances
salidroside production. Biomed Res Int. 2016:98459272016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ni WW, Huang W, Wu DQ, Zhou YJ, Ji CM, Cao
MD, Guo M, Sun JL and Wei JF: Expression and purification of a
major allergen, Pla a 1, from Platanus acerifolia pollen and the
preparation of its monoclonal antibody. Mol Med Rep. 16:2887–2892.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Louis-Jeune C, Andrade-Navarro MA and
Perez-Iratxeta C: Prediction of protein secondary structure from
circular dichroism using theoretically derived spectra. Proteins.
80:374–381. 2012. View Article : Google Scholar
|
|
27
|
Sanz ML, Gamboa PM, Antépara I, Uasuf C,
Vila L, Garcia-Avilés C, Chazot M and De Weck AL: Flow cytometric
basophil activation test by detection of CD63 expression in
patients with immediate-type reactions to betalactam antibiotics.
Clin Exp Allergy. 32:277–286. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wallowitz ML, Chen RJ, Tzen JT and Teuber
SS: Using human basophil donors to assess the clinical relevance of
sesame 11S Globulin, Ses i 6. J Allergy Clin Immunol. 117:S50.
2006. View Article : Google Scholar
|
|
29
|
Wallowitz ML, Chen RJ, Tzen JT and Teuber
SS: Ses i 6, the sesame 11S globulin, can activate basophils and
shows cross-reactivity with walnut in vitro. Clin Exp Allergy.
37:929–938. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
An S, Ma D, Wei JF, Yang X, Yang HW, Yang
H, Xu X, He S and Lai R: A novel allergen Tab y 1 with inhibitory
activity of platelet aggregation from salivary glands of
horseflies. Allergy. 66:1420–1427. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei JF, Yang H, Li D, Gao P and He S:
Preparation and identification of Per a 5 as a novel American
cockroach allergen. Mediators Inflamm. 2014:5914682014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garcia OP and Marin A: Usefulness of the
basophil activation test (BAT) in the diagnosis of life-threatening
drug anaphylaxis. Allergy. 65:1204. 2010.
|
|
33
|
Arnold K, Bordoli L, Kopp J and Schwede T:
The SWISS-MODEL workspace: A web-based environment for protein
structure homology modelling. Bioinformatics. 22:195–201. 2006.
View Article : Google Scholar
|
|
34
|
Kiefer F, Arnold K, Künzli M, Bordoli L
and Schwede T: The SWISS-MODEL Repository and associated resources.
Nucleic Acids Res. 37:387–392. 2009. View Article : Google Scholar
|
|
35
|
Krogh A, Larsson B, Von HG and Sonnhammer
EL: Predicting transmembrane protein topology with a hidden Markov
model: Application to complete genomes. J Mol Biol. 305:567–580.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bramucci E, Paiardini A, Bossa F and
Pascarella S; PyMod: Sequence similarity searches, multiple
sequence-structure alignments, and homology modeling within PyMOL.
Bmc Bioinformatics. 13(Suppl 4): pp. S22012, View Article : Google Scholar :
|
|
37
|
Ni WW, Wang LB, Zhou YJ, Cao MD, Huang W,
Guo M, Ji CM, Sun JL and Wei JF: Expression, purification and
epitope analysis of Pla a 3 allergen from Platanus acerifolia
pollen. Mol Med Rep. 16:2851–2255. 2018. View Article : Google Scholar
|
|
38
|
Chen H, Yang HW, Wei JF and Tao AL: In
silico prediction of the T-cell and IgE-binding epitopes of Per a 6
and Bla g 6 allergens in cockroaches. Mol Med Rep. 10:2130–3136.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li X, Yang HW, Chen H, Wu J, Liu Y and Wei
JF: In Silico prediction of T and B cell epitopes of Der f 25 in
Dermatophagoides farinae. Int J Genomics. 2014:4839052014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang H, Chen H, Jin M, Xie H, He S and Wei
JF: Molecular cloning, expression, IgE binding activities andin
silicoepitope prediction of Per a 9 allergens of the American
cockroach. Int J Mol Med. 38:1795–1805. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tong X, Guo M, Jin M, Chen H, Li Y and Wei
JF: In silico epitope prediction, expression and functional
analysis of Per a 10 allergen from the American cockroach. Int J
Mol Med. 38:1806–1814. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bublin M, Kostadinova M, Radauer C, Hafner
C, Szépfalusi Z, Varga EM, Maleki SJ, Hoffmann-Sommergruber K and
Breiteneder H: IgE cross-reactivity between the major peanut
allergen Ara h 2 and the non-homologous allergens Ara h 1 and Ara h
3. J Allergy Clin Immunol. 3:118–124. 2013. View Article : Google Scholar
|
|
43
|
Govindaraj D, Sharma S, Gaur SN, Lavasa S,
Prasad N and Arora N: Immunogenic peptides: B & T Cell Epitopes
of Per a 10 Allergen of Periplaneta americana. Mol Immunol.
80:24–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rudolph R and Lilie H: In vitro folding of
inclusion body proteins. FASEB J. 10:49–56. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rattenholl A, Ruoppolo M, Flagiello A,
Monti M, Vinci F, Marino G, Lilie H, Schwarz E and Rudolph R:
Pro-sequence assisted folding and disulfide bond formation of human
nerve growth factor. J Mol Biol. 305:523–533. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rattenholl A, Lilie H, Grossmann A, Stern
A, Schwarz E and Rudolph R: The pro-sequence facilitates folding of
human nerve growth factor from Escherichia coli inclusion bodies.
Eur J Biochem. 268:3296–3303. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Clewes O, Fahey MS, Tyler SJ, Watson JJ,
Seok H, Catania C, Cho K, Dawbarn D and Allen SJ: Human ProNGF:
Biological effects and binding profiles at TrkA, P75NTR and
sortilin. J Neurochem. 107:1124–1135. 2008.PubMed/NCBI
|
|
48
|
Wang DW, Ni WW, Zhou YJ, Huang W, Cao MD,
Meng L and Wei JF: Expression, purification and epitope analysis of
Pla a 2 allergen from Platanus acerifolia pollen. Mol Med Rep.
17:394–399. 2018.
|
|
49
|
Galli SJ and Mindy T: IgE and mast cells
in allergic disease. Nat Med. 18:693–704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pomes A and Xe A: Relevant B cell epitopes
in allergic disease. Int Arch Allergy Immunol. 152:1–11. 2010.
View Article : Google Scholar :
|
|
51
|
Li GF, Wang Y, Zhang ZS, Wang XJ, Ji MJ,
Zhu X, Liu F, Cai XP, Wu HW and Wu GL: Identification of
immunodominant Th1-type T cell epitopes from Schistosoma japonicum
28 kDa glutathione-S-transferase, a vaccine candidate. Acta Biochim
Biophys Sin (Shanghai). 37:751–758. 2005. View Article : Google Scholar
|
|
52
|
Yao B, Zheng D, Liang S and Zhang C:
Conformational B-cell epitope prediction on antigen protein
structures: A review of current algorithms and comparison with
common binding site prediction methods. PLoS One. 8:e622492013.
View Article : Google Scholar : PubMed/NCBI
|