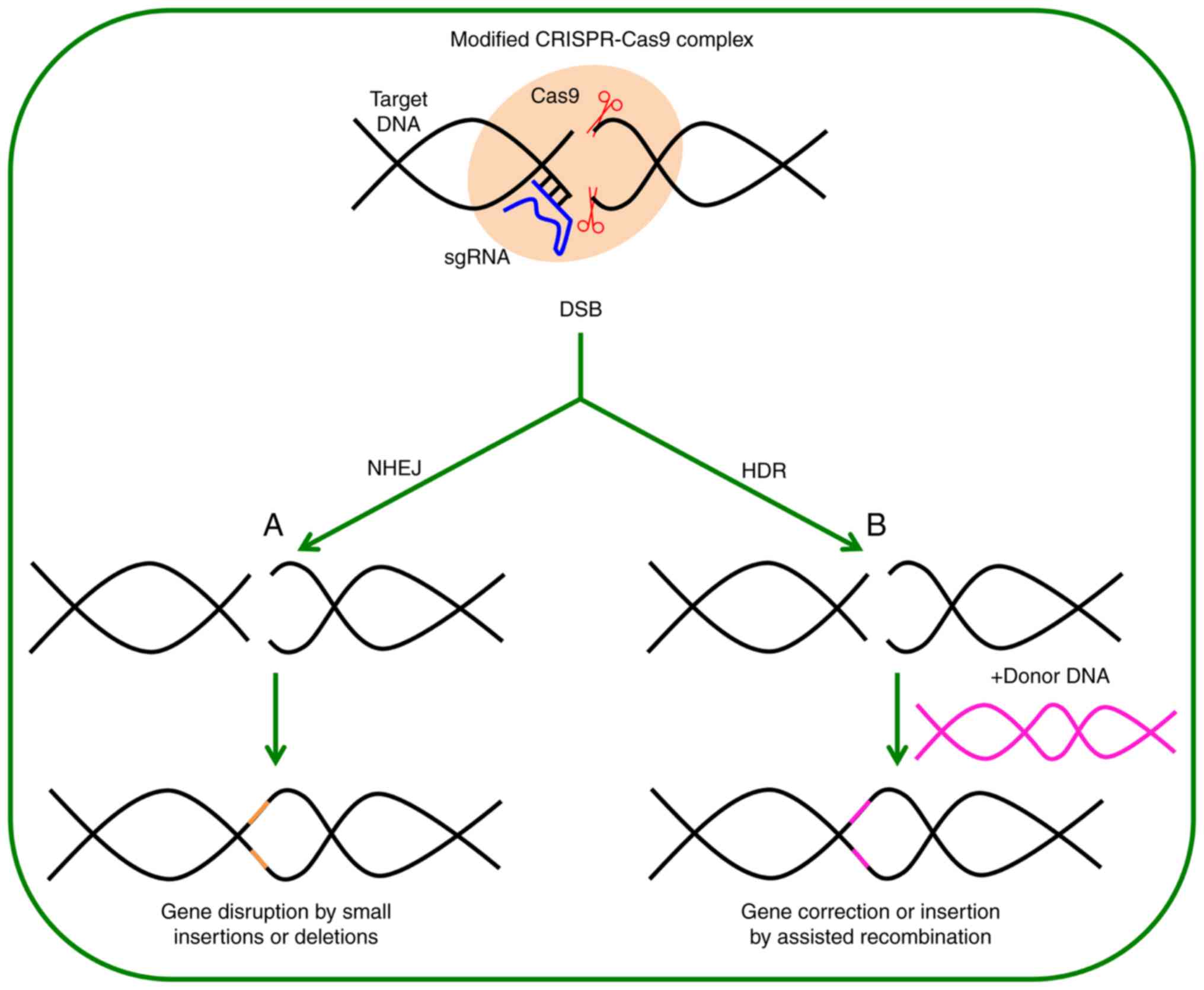
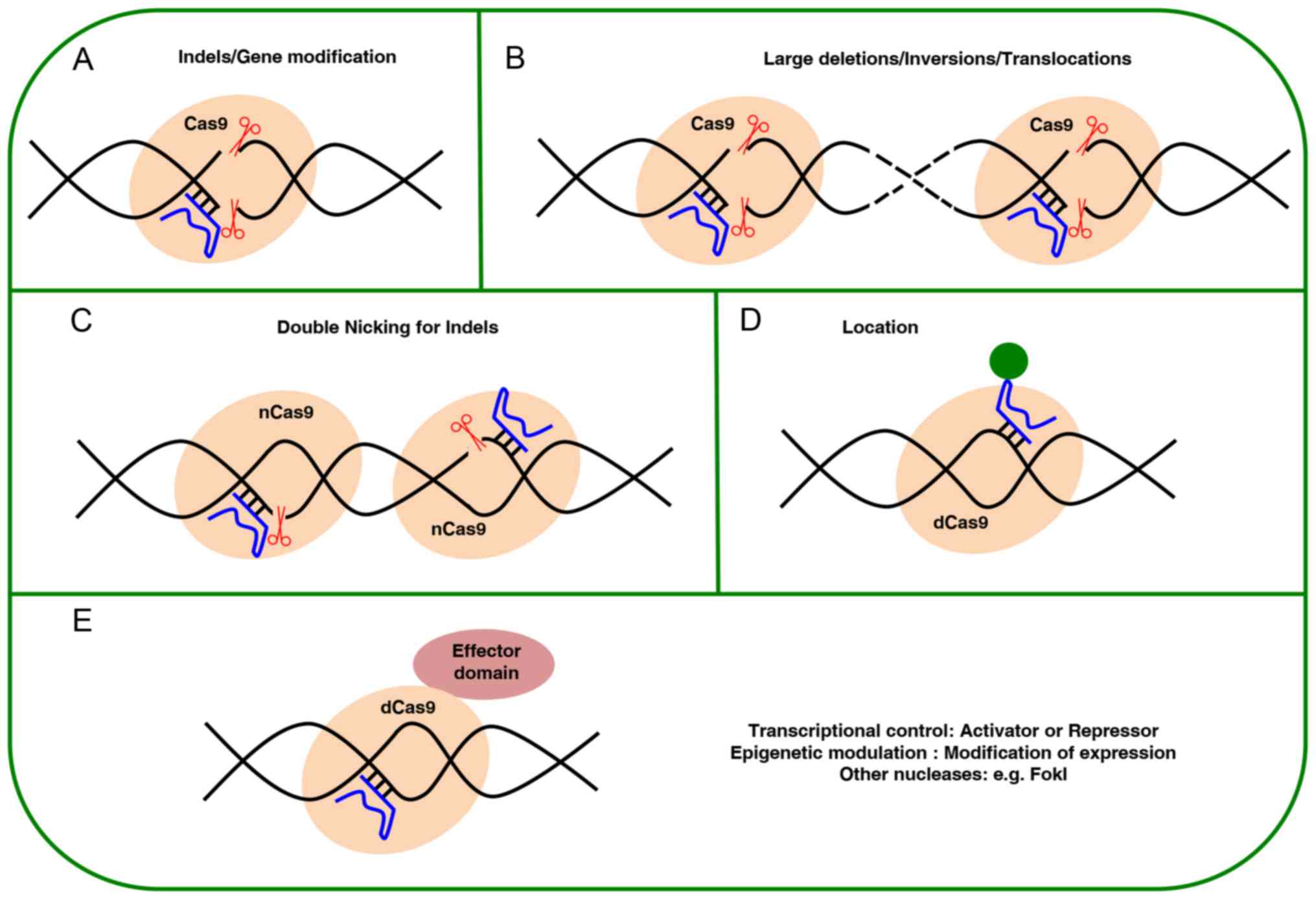
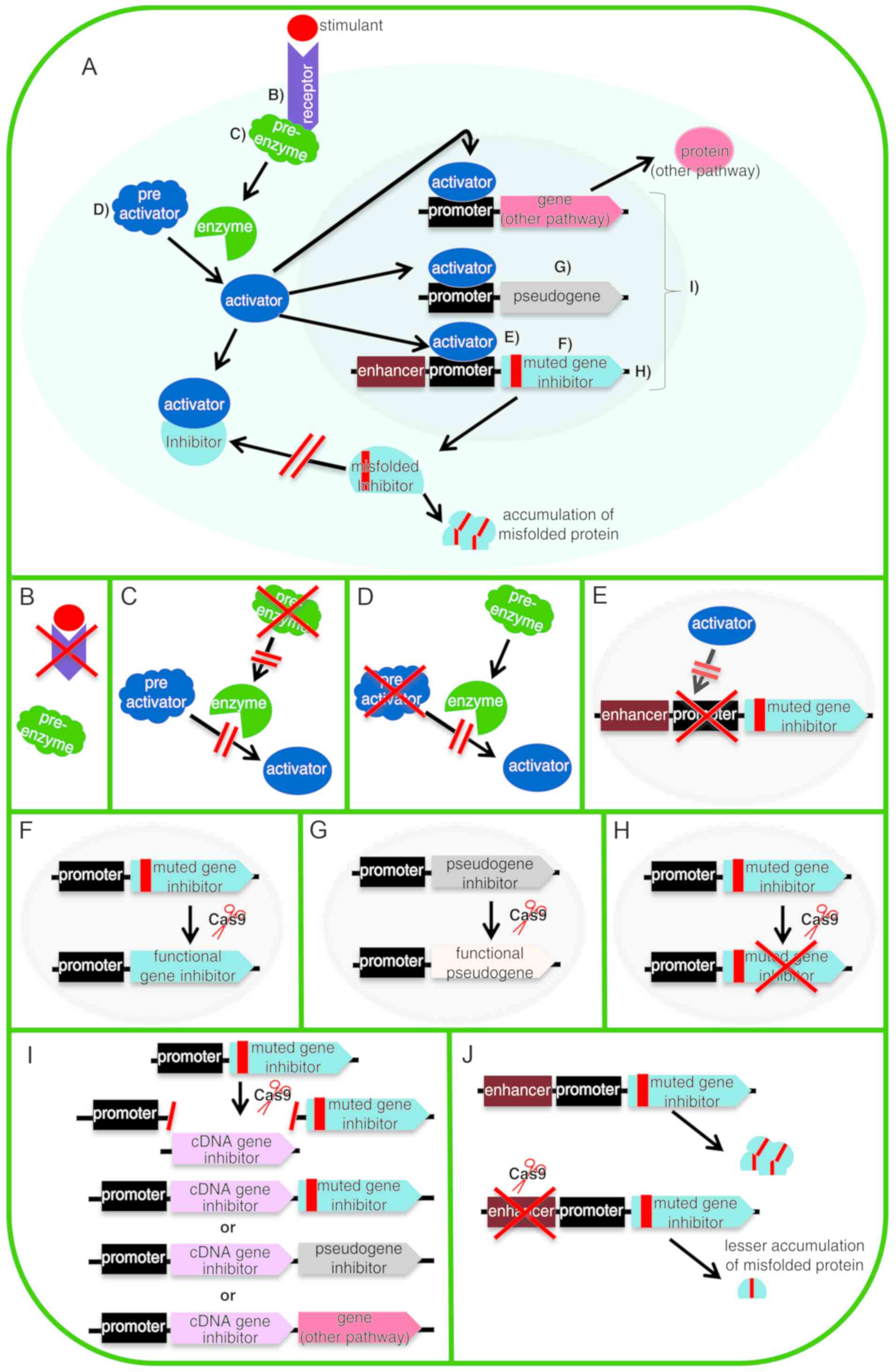
|
1
|
Im W, Moon J and Kim M: Applications of
CRISPR/Cas9 for gene editing in hereditary movement disorders. J
Mov Disord. 9:136–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gaj T, Gersbach CA and Barbas CF III: ZFN,
TALEN, and CRISPR/Cas-based methods for genome engineering. Trends
Biotechnol. 31:397–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sander JD and Joung JK: CRISPR-Cas systems
for editing, regulating and targeting genomes. Nat Biotechnol.
32:347–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cox DBT, Platt RJ and Zhang F: Therapeutic
genome editing: Prospects and challenges. Nat Med. 21:121–131.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Epinat JC, Arnould S, Chames P, Rochaix P,
Desfontaines D, Puzin C, Patin A, Zanghellini A, Pâques F and
Lacroix E: A novel engineered meganuclease induces homologous
recombination in yeast and mammalian cells. Nucleic Acids Res.
31:2952–2962. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Urnov FD, Rebar EJ, Holmes MC, Zhang HS
and Gregory PD: Genome editing with engineered zinc finger
nucleases. Nat Rev Genet. 11:636–646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miller JC, Tan S, Qiao G, Barlow KA, Wang
J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al: A TALE
nuclease architecture for efficient genome editing. Nat Biotechnol.
29:143–148. 2011. View Article : Google Scholar
|
|
8
|
Jinek M, Chylinski K, Fonfara I, Hauer M,
Doudna JA and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Horvath P and Barrangou R: CRISPR/Cas, the
immune system of bacteria and archaea. Science. 327:167–170. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sorek R, Kunin V and Hugenholtz P:
CRISPR-a widespread system that provides acquired resistance
against phages in bacteria and archaea. Nat Rev Microbiol.
6:181–186. 2008. View Article : Google Scholar
|
|
11
|
Singh V, Braddick D and Dhar PK: Exploring
the potential of genome editing CRISPR-Cas9 technology. Gene.
599:1–18. 2017. View Article : Google Scholar
|
|
12
|
Makarova KS, Wolf YI, Alkhnbashi OS, Costa
F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E,
Haft DH, et al: An updated evolutionary classification of
CRISPR-Cas systems. Nat Rev Microbiol. 13:722–736. 2015. View Article : Google Scholar
|
|
13
|
Mojica FJ, Ferrer C, Juez G and
Rodríguez-Valera F: Long stretches of short tandem repeats are
present in the largest replicons of the Archaea Haloferax
mediterranei and Haloferax volcanii and could be involved in
replicon partitioning. Mol Microbiol. 17:85–93. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Riehle MM, Bennett AF and Long AD: Genetic
architecture of thermal adaptation in Escherichia coli. Proc Natl
Acad Sci USA. 98:525–530. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
DeBoy RT, Mongodin EF, Emerson JB and
Nelson KE: Chromosome evolution in the Thermotogales: Large-scale
inversions and strain diversification of CRISPR sequences. J
Bacteriol. 188:2364–2374. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Makarova KS, Aravind L, Grishin NV,
Rogozin IB and Koonin EV: A DNA repair system specific for
thermophilic Archaea and bacteria predicted by genomic context
analysis. Nucleic Acids Res. 30:482–496. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ishino Y, Shinagawa H, Makino K, Amemura M
and Nakata A: Nucleotide sequence of the iap gene, responsible for
alkaline phosphatase isozyme conversion in Escherichia coli, and
identification of the gene product. J Bacteriol. 169:5429–5433.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mojica FJ, Díez-Villaseñor C, Soria E and
Juez G: Biological significance of a family of regularly spaced
repeats in the genomes of Archaea, Bacteria and mitochondria. Mol
Microbiol. 36:244–246. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jansen R, Embden JD, Gaastra W and Schouls
LM: Identification of genes that are associated with DNA repeats in
prokaryotes. Mol Microbiol. 43:1565–1575. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mojica FJ, Díez-Villaseñor C,
García-Martínez J and Soria E: Intervening sequences of regularly
spaced prokaryotic repeats derive from foreign genetic elements. J
Mol Evol. 60:174–182. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bolotin A, Quinquis B, Sorokin A and
Ehrlich SD: Clustered regularly interspaced short palindrome
repeats (CRISPRs) have spacers of extrachromosomal origin.
Microbiology. 151:2551–2561. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pourcel C, Salvignol G and Vergnaud G:
CRISPR elements in Yersinia pestis acquire new repeats by
preferential uptake of bacteriophage DNA, and provide additional
tools for evolutionary studies. Microbiology. 151:653–663. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barrangou R, Fremaux C, Deveau H, Richards
M, Boyaval P, Moineau S, Romero DA and Horvath P: CRISPR provides
acquired resistance against viruses in prokaryotes. Science.
315:1709–1712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brouns SJJ, Jore MM, Lundgren M, Westra
ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV
and van der Oost J: Small CRISPR RNAs guide antiviral defense in
prokaryotes. Science. 321:960–964. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garneau JE, Dupuis MÈ, Villion M, Romero
DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH and
Moineau S: The CRISPR/Cas bacterial immune system cleaves
bacteriophage and plasmid DNA. Nature. 468:67–71. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deltcheva E, Chylinski K, Sharma CM,
Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J and Charpentier
E: CRISPR RNA maturation by trans-encoded small RNA and host factor
RNase III. Nature. 471:602–607. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sapranauskas R, Gasiunas G, Fremaux C,
Barrangou R, Horvath P and Siksnys V: The Streptococcus
thermophilus CRISPR/Cas system provides immunity in Escherichia
coli. Nucleic Acids Res. 39:9275–9282. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mali P, Yang L, Esvelt KM, Aach J, Guell
M, DiCarlo JE, Norville JE and Church GM: RNA-guided human genome
engineering via Cas9. Science. 339:823–826. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cong L, Ran FA, Cox D, Lin S, Barretto R,
Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA and Zhang F:
Multiplex genome engineering using CRISPR/Cas systems. Science.
339:819–823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cornu TI, Mussolino C and Cathomen T:
Refining strategies to translate genome editing to the clinic. Nat
Med. 23:415–423. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cyranoski D: CRISPR gene-editing tested in
a person for the first time. Nature. 539:479. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cyranoski D: Chinese scientists to pioneer
first human CRISPR trial. Nature. 535:476–477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shub DA, Goodrich-Blair H and Eddy SR:
Amino-acid-sequence motif of group I intron endonucleases is
conserved in open reading frames of group II introns. Trends
Biochem Sci. 19:402–404. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Al-Attar S, Westra ER, van der Oost J and
Brouns SJ: Clustered regularly interspaced short palindromic
repeats (CRISPRs): The hallmark of an ingenious antiviral defense
mechanism in prokaryotes. Biol Chem. 392:277–289. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wright AV, Nuñez JK and Doudna JA: Biology
and applications of CRISPR systems: Harnessing nature’s toolbox for
genome engineering. Cell. 164:29–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang F, Wen Y and Guo X: CRISPR/Cas9 for
genome editing: Progress implications and challenges. Hum Mol
Genet. 23:R40–R46. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Canver MC, Bauer DE and Orkin SH:
Functional interrogation of non-coding DNA through CRISPR genome
editing. Methods. 121–122. 118–129. 2017.
|
|
38
|
Karvelis T, Gasiunas G, Miksys A,
Barrangou R, Horvath P and Siksnys V: crRNA and tracrRNA guide
Cas9-mediated DNA interference in Streptococcus thermophilus. RNA
Biol. 10:841–851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gasiunas G, Barrangou R, Horvath P and
Siksnys V: Cas9-crRNA ribonucleoprotein complex mediates specific
DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci
USA. 109:E2579–E2586. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Friedland AE, Baral R, Singhal P, Loveluck
K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, et
al: Characterization of Staphylococcus aureus Cas9: A smaller Cas9
for all-in-one adeno-associated virus delivery and paired nickase
applications. Genome Biol. 16:2572015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ran FA, Cong L, Yan WX, Scott DA,
Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et
al: In vivo genome editing using Staphylococcus aureus Cas9.
Nature. 520:186–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hou Z, Zhang Y, Propson NE, Howden SE, Chu
LF, Sontheimer EJ and Thomson JA: Efficient genome engineering in
human pluripotent stem cells using Cas9 from Neisseria
meningitidis. Proc Natl Acad Sci USA. 110:15644–15649. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen F, Ding X, Feng Y, Seebeck T, Jiang Y
and Davis GD: Targeted activation of diverse CRISPR-Cas systems for
mammalian genome editing via proximal CRISPR targeting. Nat Commun.
8:149582017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Price AA, Sampson TR, Ratner HK, Grakoui A
and Weiss DS: Cas9-mediated targeting of viral RNA in eukaryotic
cells. Proc Natl Acad Sci USA. 112:6164–6169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Murovec J, Pirc Ž and Yang B: New variants
of CRISPR RNA-guided genome editing enzymes. Plant Biotechnol J.
15:917–926. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Oliveros JC, Franch M, Tabas-Madrid D,
San-León D, Montoliu L, Cubas P and Pazos F:
Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas
experiments for ENSEMBL genomes. Nucleic Acids Res. 44:W267–W271.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kaur K, Gupta AK, Rajput A and Kumar M:
ge-CRISPR-an integrated pipeline for the prediction and analysis of
sgRNAs genome editing efficiency for CRISPR/Cas system. Sci Rep.
6:308702016. View Article : Google Scholar
|
|
48
|
Naito Y, Hino K, Bono H and Ui-Tei K:
CRISPRdirect: Software for designing CRISPR/Cas guide RNA with
reduced off-target sites. Bioinformatics. 31:1120–1123. 2015.
View Article : Google Scholar :
|
|
49
|
Heigwer F, Kerr G and Boutros M: E-CRISP:
Fast CRISPR target site identification. Nat Methods. 11:122–123.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wong N, Liu W and Wang X: WU-CRISPR:
Characteristics of functional guide RNAs for the CRISPR/Cas9
system. Genome Biol. 16:2182015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Doench JG, Hartenian E, Graham DB, Tothova
Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ and Root DE:
Rational design of highly active sgRNAs for CRISPR-Cas9-mediated
gene inactivation. Nat Biotechnol. 32:1262–1267. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chari R, Mali P, Moosburner M and Church
GM: Unraveling CRISPR-Cas9 genome engineering parameters via a
library-on-library approach. Nat Methods. 12:823–826. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chari R, Yeo NC, Chavez A and Church GM:
sgRNA scorer 2.0: A species-independent model to predict
CRISPR/Cas9 activity. ACS Synth Biol. 6:902–904. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Moreno-Mateos MA, Vejnar CE, Beaudoin JD,
Fernandez JP, Mis EK, Khokha MK and Giraldez AJ: CRISPRscan:
Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in
vivo. Nat Methods. 12:982–988. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu H, Wei Z, Dominguez A, Li Y, Wang X
and Qi LS: CRISPR-ERA: A comprehensive design tool for
CRISPR-mediated gene editing, repression and activation.
Bioinformatics. 31:3676–3678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Stemmer M, Thumberger T, Del Sol, Keyer M,
Wittbrodt J and Mateo JL: CCTop: An intuitive, flexible and
reliable CRISPR/Cas9 target prediction tool. PLoS One.
10:e01246332015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Haeussler M, Schönig K, Eckert H,
Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava
A, Teboul L, Kent J, et al: Evaluation of off-target and on-target
scoring algorithms and integration into the guide RNA selection
tool CRISPOR. Genome Biol. 17:1482016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Montague TG, Cruz JM, Gagnon JA, Church GM
and Valen E: CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome
editing. Nucleic Acids Res. 42:W401–W407. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Prykhozhij SV, Rajan V, Gaston D and
Berman JN: CRISPR multitargeter: A web tool to find common and
unique CRISPR single guide RNA targets in a set of similar
sequences. PLoS One. 10:e01193722015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
O’Brien A and Bailey TL: GT-Scan:
Identifying unique genomic targets. Bioinformatics. 30:2673–2675.
2014. View Article : Google Scholar :
|
|
61
|
Hsu PD, Scott DA, Weinstein JA, Ran FA,
Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al: DNA
targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol.
31:827–832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Park J, Bae S and Kim JS: Cas-Designer: A
web-based tool for choice of CRISPR-Cas9 target sites.
Bioinformatics. 31:4014–4016. 2015.PubMed/NCBI
|
|
63
|
Bae S, Park J and Kim JS: Cas-OFFinder: A
fast and versatile algorithm that searches for potential off-target
sites of Cas9 RNA-guided endonucleases. Bioinformatics.
30:1473–1475. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cradick TJ, Qiu P, Lee CM, Fine EJ and Bao
G: COSMID: A web-based tool for identifying and validating
CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 3:e2142014.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hough SH, Kancleris K, Brody L,
Humphryes-Kirilov N, Wolanski J, Dunaway K, Ajetunmobi A and
Dillard V: Guide Picker is a comprehensive design tool for
visualizing and selecting guides for CRISPR experiments. BMC
Bioinformatics. 18:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Güell M, Yang L and Church GM: Genome
editing assessment using CRISPR genome analyzer (CRISPR-GA).
Bioinformatics. 30:2968–2970. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cao J, Wu L, Zhang SM, Lu M, Cheung WK,
Cai W, Gale M, Xu Q and Yan Q: An easy and efficient inducible
CRISPR/Cas9 platform with improved specificity for multiple gene
targeting. Nucleic Acids Res. 44:e1492016.PubMed/NCBI
|
|
68
|
Ran FA, Hsu PD, Lin CY, Gootenberg JS,
Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y and
Zhang F: Double nicking by RNA-guided CRISPR Cas9 for enhanced
genome editing specificity. Cell. 154:1380–1389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Sato T, Sakuma T, Yokonishi T, Katagiri K,
Kamimura S, Ogonuki N, Ogura A, Yamamoto T and Ogawa T: Genome
editing in mouse spermatogonial stem cell lines Using TALEN and
double-nicking CRISPR/Cas9. Stem Cell Reports. 5:75–82. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sakuma T, Masaki K, Abe-Chayama H, Mochida
K, Yamamoto T and Chayama K: Highly multiplexed
CRISPR-Cas9-nuclease and Cas9-nickase vectors for inactivation of
hepatitis B virus. Genes Cells. 21:1253–1262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Slaymaker IM, Gao L, Zetsche B, Scott DA,
Yan WX and Zhang F: Rationally engineered Cas9 nucleases with
improved specificity. Science. 351:84–88. 2016. View Article : Google Scholar :
|
|
72
|
Tsai SQ, Wyvekens N, Khayter C, Foden JA,
Thapar V, Reyon D, Goodwin MJ, Aryee MJ and Joung JK: Dimeric
CRISPR RNA-guided FokI nucleases for highly specific genome
editing. Nat Biotechnol. 32:569–576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang H, Yan Z, Li M, Peabody M and He TC:
CRISPR clear? Dimeric Cas9-Fok1 nucleases improve genome-editing
specificity. Genes Dis. 1:6–7. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Guilinger JP, Thompson DB and Liu DR:
Fusion of catalytically inactive Cas9 to FokI nuclease improves the
specificity of genome modification. Nat Biotechnol. 32:577–582.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wright AV, Sternberg SH, Taylor DW, Staahl
BT, Bardales JA, Kornfeld JE and Doudna JA: Rational design of a
split-Cas9 enzyme complex. Proc Natl Acad Sci USA. 112:2984–2989.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Chew WL, Tabebordbar M, Cheng JK, Mali P,
Wu EY, Ng AH, Zhu K, Wagers AJ and Church GM: A multifunctional
AAV-CRISPR-Cas9 and its host response. Nat Methods. 13:868–874.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Komor AC, Kim YB, Packer MS, Zuris JA and
Liu DR: Programmable editing of a target base in genomic DNA
without double-stranded DNA cleavage. Nature. 533:420–424. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Murugan K, Babu K, Sundaresan R, Rajan R
and Sashital DG: The revolution continues: Newly discovered systems
expand the CRISPR-Cas toolkit. Mol Cell. 68:15–25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Shmakov S, Abudayyeh OO, Makarova KS, Wolf
YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S,
Severinov K, et al: Discovery and functional characterization of
diverse class 2 CRISPR-Cas systems. Mol Cell. 60:385–397. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Abudayyeh OO, Gootenberg JS, Konermann S,
Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E,
Minakhin L, et al: C2c2 is a single-component programmable
RNA-guided RNA-targeting CRISPR effector. Science. 353:aaf55732016.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Abudayyeh OO, Gootenberg JS,
Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT,
Kellner MJ, Regev A, et al: RNA targeting with CRISPR-Cas13.
Nature. 550:280–284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Cox DBT, Gootenberg JS, Abudayyeh OO,
Franklin B, Kellner MJ, Joung J and Zhang F: RNA editing with
CRISPR-Cas13. Science. 358:1019–1027. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Balboa D, Weltner J, Eurola S, Trokovic R,
Wartiovaara K and Otonkoski T: Conditionally stabilized dCas9
activator for controlling gene expression in human cell
reprogramming and differentiation. Stem Cell Reports. 5:448–459.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Konermann S, Brigham MD, Trevino AE, Joung
J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS,
Nishimasu H, et al: Genome-scale transcriptional activation by an
engineered CRISPR-Cas9 complex. Nature. 517:583–588. 2015.
View Article : Google Scholar :
|
|
85
|
Piatek A, Ali Z, Baazim H, Li L, Abulfaraj
A, Al-Shareef S, Aouida M and Mahfouz MM: RNA-guided
transcriptional regulation in planta via synthetic dCas9-based
transcription factors. Plant Biotechnol J. 13:578–589. 2015.
View Article : Google Scholar
|
|
86
|
Fu Y, Rocha PP, Luo VM, Raviram R, Deng Y,
Mazzoni EO and Skok JA: CRISPR-dCas9 and sgRNA scaffolds enable
dual-colour live imaging of satellite sequences and repeat-enriched
individual loci. Nat Commun. 7:117072016. View Article : Google Scholar :
|
|
87
|
Qin P, Parlak M, Kuscu C, Bandaria J, Mir
M, Szlachta K, Singh R, Darzacq X, Yildiz A and Adli M: Live cell
imaging of low- and non-repetitive chromosome loci using
CRISPR-Cas9. Nat Commun. 8:147252017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Pankowicz FP, Barzi M, Legras X, Hubert L,
Mi T, Tomolonis JA, Ravishankar M, Sun Q, Yang D, Borowiak M, et
al: Reprogramming metabolic pathways in vivo with CRISPR/Cas9
genome editing to treat hereditary tyrosinaemia. Nat Commun.
7:126422016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kang H, Minder P, Park MA, Mesquitta WT,
Torbett BE and Slukvin II: CCR5 disruption in induced pluripotent
stem cells using CRISPR/Cas9 provides selective resistance of
immune cells to CCR5-tropic HIV-1 virus. Mol Ther Nucleic Acids.
4:e2682015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Brunger JM, Zutshi A, Willard VP, Gersbach
CA and Guilak F: Genome engineering of stem cells for autonomously
regulated, closed-loop delivery of biologic drugs. Stem Cell
Reports. 8:1202–1213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY,
Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, et al: The CRISPR/Cas9
system facilitates clearance of the intrahepatic HBV templates in
vivo. Mol Ther Nucleic Acids. 3:e1862014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhen S, Hua L, Takahashi Y, Narita S, Liu
YH and Li Y: In vitro and in vivo growth suppression of human
papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9.
Biochem Biophys Res Commun. 450:1422–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhen S, Lu JJ, Wang LJ, Sun XM, Zhang JQ,
Li X, Luo WJ and Zhao L: In vitro and in vivo synergistic
therapeutic effect of cisplatin with human papillomavirus16 E6/E7
CRISPR/Cas9 on cervical cancer cell line. Transl Oncol. 9:498–504.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Das D, Smith N, Wang X and Morgan IM: The
deacetylase SIRT1 regulates the replication properties of human
papilloma-virus 16 E1 and E2. J Virol. 91:e00102-e001172017.
View Article : Google Scholar
|
|
95
|
Merling R, Kuhns D, Sweeney C, Wu X,
Burkett S, Chu J, Lee J, Koontz S, Di Pasquale G, Afione S, et al:
Gene-edited pseudogene resurrection corrects p47 phox-deficient
chronic granulomatous disease. Blood Adv. 1:270–278. 2016.
View Article : Google Scholar
|
|
96
|
Paquet D, Kwart D, Chen A, Sproul A, Jacob
S, Teo S, Olsen KM, Gregg A, Noggle S and Tessier-Lavigne M:
Efficient introduction of specific homozygous and heterozygous
mutations using CRISPR/Cas9. Nature. 533:125–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Schwank G, Koo BK, Sasselli V, Dekkers JF,
Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK,
et al: Functional repair of CFTR by CRISPR/Cas9 in intestinal stem
cell organoids of cystic fibrosis patients. Cell Stem Cell.
13:653–658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yin H, Xue W, Chen S, Bogorad RL,
Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T and
Anderson DG: Genome editing with Cas9 in adult mice corrects a
disease mutation and phenotype. Nat Biotechnol. 32:551–553. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Gui H, Schriemer D, Cheng WW, Chauhan RK,
Antiňolo G, Berrios C, Bleda M, Brooks AS, Brouwer RW, Burns AJ, et
al: Whole exome sequencing coupled with unbiased functional
analysis reveals new Hirschsprung disease genes. Genome Biol.
18:482017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Halim D, Wilson MP, Oliver D, Brosens E,
Verheij JB, Han Y, Nanda V, Lyu Q, Doukas M, Stoop H, et al: Loss
of LMOD1 impairs smooth muscle cytocontractility and causes
megacystis microcolon intestinal hypoperistalsis syndrome in humans
and mice. Proc Natl Acad Sci USA. 114:E2739–E2747. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Dever DP, Bak RO, Reinisch A, Camarena J,
Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB,
Mantri S, et al: CRISPR/Cas9 β-globin gene targeting in human
haematopoietic stem cells. Nature. 539:384–389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Ou Z, Niu X, He W, Chen Y, Song B, Xian Y,
Fan D, Tang D and Sun X: The combination of CRISPR/Cas9 and iPSC
technologies in the gene therapy of human β-thalassemia in mice.
Sci Re. 6:324632016.
|
|
103
|
Xie F, Ye L, Chang JC, Beyer AI, Wang J,
Muench MO and Kan YW: Seamless gene correction of β-thalassemia
mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac.
Genome Res. 24:1526–1533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Xu P, Tong Y, Liu XZ, Wang TT, Cheng L,
Wang BY, Lv X, Huang Y and Liu DP: Both TALENs and CRISPR/Cas9
directly target the HBB IVS2 654 (C > T) mutation in
β-thalassemia-derived iPSCs. Sci Rep. 5:120652015. View Article : Google Scholar
|
|
105
|
Traxler EA, Yao Y, Wang YD, Woodard KJ,
Kurita R, Nakamura Y, Hughes JR, Hardison RC, Blobel GA, Li C and
Weiss MJ: A genome-editing strategy to treat β-hemoglobinopathies
that recapitulates a mutation associated with a benign genetic
condition. Nat Med. 22:987–990. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Canver MC, Smith EC, Sher F, Pinello L,
Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP,
et al: BCL11A enhancer dissection by Cas9-mediated in situ
saturating mutagenesis. Nature. 527:192–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zhang N, Zhi H, Curtis BR, Rao S, Jobaliya
C, Poncz M, French DL and Newman PJ: CRISPR/Cas9-mediated
conversion of human platelet alloantigen allotypes. Blood.
127:675–680. 2016. View Article : Google Scholar :
|
|
108
|
Osborn MJ, Gabriel R, Webber BR, DeFeo AP,
McElroy AN, Jarjour J, Starker CG, Wagner JE, Joung JK, Voytas DF,
et al: Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum
Gene Ther. 26:114–126. 2015. View Article : Google Scholar :
|
|
109
|
Park CY, Kim DH, Son JS, Sung JJ, Lee J,
Bae S, Kim JH, Kim DW and Kim JS: Functional correction of large
factor VIII gene chromosomal inversions in hemophilia a
patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell.
17:213–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhang H and McCarty N: CRISPR-Cas9
technology and its application in haematological disorders. Br J
Haematol. 175:208–225. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Chen J, Jiang N, Wang T, Xie G, Zhang Z,
Li H, Yuan J, Sun Z and Chen J: DNA shuffling of uricase gene leads
to a more ‘human like’ chimeric uricase with increased uricolytic
activity. Int J Biol Macromol. 82:522–529. 2016. View Article : Google Scholar
|
|
112
|
Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L,
Wang L, Zeng L, Shao Y, Chen Y, et al: CRISPR/Cas9-mediated somatic
correction of a novel coagulator factor IX gene mutation
ameliorates hemophilia in mouse. EMBO Mol Med. 8:477–488. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Hai T, Teng F, Guo R, Li W and Zhou Q:
One-step generation of knockout pigs by zygote injection of
CRISPR/Cas system. Cell Res. 24:372–375. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Puschnik AS, Majzoub K, Ooi YS and Carette
JE: A CRISPR toolbox to study virus-host interactions. Nat Rev
Microbiol. 15:351–364. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Kennedy EM, Kornepati AVR, Goldstein M,
Bogerd HP, Poling BC, Whisnant AW, Kastan MB and Cullen BR:
Inactivation of the human papillomavirus E6 or E7 gene in cervical
carcinoma cells by using a bacterial CRISPR/Cas RNA-guided
endonuclease. J Virol. 88:11965–11972. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Wang W, Ye C, Liu J, Zhang D, Kimata JT
and Zhou P: CCR5 gene disruption via lentiviral vectors expressing
Cas9 and single guided RNA renders cells resistant to HIV-1
infection. PLoS One. 9:e1159872014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan
DY, Dong LH, Song HF and Gao X: Harnessing the clustered regularly
inter-spaced short palindromic repeat (CRISPR)/CRISPR-associated
Cas9 system to disrupt the hepatitis B virus. Gene Ther.
22:404–412. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Ramanan V, Shlomai A, Cox DBT, Schwartz
RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM and Bhatia
SN: CRISPR/Cas9 cleavage of viral DNA efficiently suppresses
hepatitis B virus. Sci Rep. 5:108332015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Karimova M, Beschorner N, Dammermann W,
Chemnitz J, Indenbirken D, Bockmann JH, Grundhoff A, Lüth S,
Buchholz F, Schulze zur Wiesch J and Hauber J: CRISPR/Cas9
nickase-mediated disruption of hepatitis B virus open reading frame
S and X. Sci Rep. 5:137342015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Qi Xu Y, Luo Y, Yang J, Xie J, Deng Q, Su
C, Wei N, Shi W, Xu DF, et al: Hepatitis B virus X protein
stimulates proliferation, wound closure and inhibits apoptosis of
HuH-7 cells via CDC42. Int J Mol Sci. 18:E5862017. View Article : Google Scholar
|
|
121
|
Ren Q, Li C, Yuan P, Cai C, Zhang L, Luo
GG and Wei W: A dual-reporter system for real-time monitoring and
high-throughput CRISPR/Cas9 library screening of the hepatitis C
virus. Sci Rep. 5:88652015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yuen KS, Chan CP, Wong N-HM, Ho CH, Ho TH,
Lei T, Deng W, Tsao SW, Chen H, Kok KH and Jin DY:
CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human
cells. J Gen Virol. 96:626–636. 2015. View Article : Google Scholar
|
|
123
|
Wang J and Quake SR: RNA-guided
endonuclease provides a therapeutic strategy to cure latent
herpesviridae infection. Proc Natl Acad Sci USA. 111:13157–13162.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kistler KE, Vosshall LB and Matthews BJ:
Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti.
Cell Rep. 11:51–60. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhang C, Xiao B, Jiang Y, Zhao Y, Li Z,
Gao H, Ling Y, Wei J, Li S, Lu M, et al: Efficient editing of
malaria parasite genome using the CRISPR/Cas9 system. MBio.
5:e01414-142014. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Wagner JC, Platt RJ, Goldfless SJ, Zhang F
and Niles JC: Efficient CRISPR/Cas9-mediated genome editing in P.
falciparum. Nat Methods. 11:915–918. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Ghorbal M, Gorman M, Macpherson CR,
Martins RM, Scherf A and Lopez-Rubio JJ: Genome editing in the
human malaria parasite Plasmodium falciparum using the crisPr-cas9
system. Nat Biotechnol. 32:819–821. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Burt A: Site-specific selfish genes as
tools for the control and genetic engineering of natural
populations. Proc Biol Sci. 270:921–928. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Webber BL, Raghu S and Edwards OR:
Opinion: Is CRISPR-based gene drive a biocontrol silver bullet or
global conservation threat? Proc Natl Acad Sci USA.
112:10565–10567. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Hammond A, Galizi R, Kyrou K, Simoni A,
Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S,
et al: A CRISPR-Cas9 gene drive system targeting female
reproduction in the malaria mosquito vector Anopheles gambiae. Nat
Biotechnol. 34:78–83. 2016. View Article : Google Scholar :
|
|
131
|
Sánchez-Rivera FJ and Jacks T:
Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev
Cancer. 15:387–395. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Kawamura N, Nimura K, Nagano H, Yamaguchi
S, Nonomura N and Kaneda Y: CRISPR/Cas9-mediated gene knockout of
NANOG and NANOGP8 decreases the malignant potential of prostate
cancer cells. Oncotarget. 6:22361–22374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
García-Tuñón I, Hernández-Sánchez M,
Ordoñez JL, Alonso-Pérez V, Álamo-Quijada M, Benito R, Guerrero C,
Hernández-Rivas JM and Sánchez-Martín M: The CRISPR/Cas9 system
efficiently reverts the tumorigenic ability of BCR/ABL in vitro and
in a xenograft model of chronic myeloid leukemia. Oncotarget.
8:26027–26040. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Zuckermann M, Hovestadt V, Knobbe-Thomsen
CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida
B, Moriarity B, et al: Somatic CRISPR/Cas9-mediated tumour
suppressor disruption enables versatile brain tumour modelling. Nat
Commun. 6:73912015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Maddalo D, Manchado E, Concepcion CP,
Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N,
de Stanchina E, et al: In vivo engineering of oncogenic chromosomal
rearrangements with the CRISPR/Cas9 system. Nature. 516:423–427.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Pathak S, McDermott MF and Savic S:
Autoinflammatory diseases: Update on classification diagnosis and
management. J Clin Pathol. 70:1–8. 2017. View Article : Google Scholar
|
|
137
|
Sá DC: Inflammasomes and dermatology. An
Bras Dermatol. 91:566–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Kim K, Bang SY, Lee HS and Bae SC: Update
on the genetic architecture of rheumatoid arthritis. Nat Rev
Rheumatol. 13:13–24. 2017. View Article : Google Scholar
|
|
139
|
Yang M, Zhang L, Stevens J and Gibson G:
CRISPR/Cas9 mediated generation of stable chondrocyte cell lines
with targeted gene knockouts; analysis of an aggrecan knockout cell
line. Bone. 69:118–125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Tang S, Chen T, Yu Z, Zhu X, Yang M, Xie
B, Li N, Cao X and Wang J: RasGRP3 limits toll-like
receptor-triggered inflammatory response in macrophages by
activating Rap1 small GTPase. Nat Commun. 5:46572014. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Jing W, Zhang X, Sun W, Hou X, Yao Z and
Zhu Y: CRISPR/CAS9-mediated genome editing of miRNA-155 inhibits
proinflammatory cytokine production by RAW264.7 cells. Biomed Res
Int. 2015.326042:2015.
|
|
142
|
Ott de Bruin LM, Volpi S and Musunuru K:
Novel genome-editing tools to model and correct primary
immunodeficiencies. Front Immunol. 6:2502015. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Cowan MJ, Neven B, Cavazanna-Calvo M,
Fischer A and Puck J: Hematopoietic stem cell transplantation for
severe combined immunodeficiency diseases. Biol Blood Marrow
Transplant. 14(1 Suppl 1): S73–S75. 2008. View Article : Google Scholar
|
|
144
|
Deakin CT, Alexander IE and Kerridge I:
Accepting risk in clinical research: Is the gene therapy field
becoming too risk-averse. Mol Ther. 17:1842–1848. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Kohn DB and Kuo CY: New frontiers in the
therapy of primary immunodeficiency: From gene addition to gene
editing. J Allergy Clin Immunol. 139:726–732. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Goodman MA, Moradi Manesh D, Malik P and
Rothenberg ME: CRISPR/Cas9 in allergic and immunologic diseases.
Expert Rev Clin Immunol. 13:5–9. 2017. View Article : Google Scholar
|
|
147
|
Wang M, Glass ZA and Xu Q: Non-viral
delivery of genome-editing nucleases for gene therapy. Gene Ther.
24:144–150. 2017. View Article : Google Scholar
|
|
148
|
Chang CW, Lai YS, Westin E,
Khodadadi-Jamayran A, Pawlik KM, Lamb LS Jr, Goldman FD and Townes
TM: Modeling human severe combined immunodeficiency and correction
by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 12:1668–1677.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Flynn R, Grundmann A, Renz P, Hänseler W,
James WS, Cowley SA and Moore MD: CRISPR-mediated genotypic and
phenotypic correction of a chronic granulomatous disease mutation
in human iPS cells. Exp Hematol. 43:838–848. e32015. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Wrona D, Siler U and Reichenbach J:
CRISPR/Cas9-generated p47(phox)-deficient cell line for chronic
granulomatous disease gene therapy vector development. Sci Rep.
7:441872017. View Article : Google Scholar
|
|
151
|
Yan Q, Zhang Q, Yang H, Zou Q, Tang C, Fan
N and Lai L: Generation of multi-gene knockout rabbits using the
Cas9/gRNA system. Cell Regen (Lond). 3:122014.
|
|
152
|
Fan Z, Li W, Lee SR, Meng Q, Shi B, Bunch
TD, White KL, Kong IK and Wang Z: Efficient gene targeting in
golden Syrian hamsters by the CRISPR/Cas9 system. PLoS One.
9:e1097552014. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Chen F, Wang Y, Yuan Y, Zhang W, Ren Z,
Jin Y, Liu X, Xiong Q, Chen Q, Zhang M, et al: Generation of B
cell-deficient pigs by highly efficient CRISPR/Cas9-mediated gene
targeting. J Genet Genomics. 42:437–444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Chu HW, Rios C, Huang C,
Wesolowska-Andersen A, Burchard EG, O’Connor BP, Fingerlin TE,
Nichols D, Reynolds SD and Seibold MA: CRISPR-Cas9-mediated gene
knockout in primary human airway epithelial cells reveals a
proinflammatory role for MUC18. Gene Ther. 22:822–829. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Kuo CY, Hoban MD, Joglekar AV and Kohn DB:
Site specific gene correction of defects in CD40 ligand using the
Crispr/Cas9 genome editing platform. J Allergy Clin Immunol.
135:AB172015. View Article : Google Scholar
|
|
156
|
Cheong TC, Compagno M and Chiarle R:
Editing of mouse and human immunoglobulin genes by CRISPR-Cas9
system. Nat Commun. 7:109342016. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Rodriguez-Sanchez IP, Guindon J, Ruiz M,
Tejero ME, Hubbard G, Martinez-de-Villarreal LE, Barrera-Saldaña
HA, Dick EJ Jr, Comuzzie AG and Schlabritz-Loutsevitch NE: The
endocannabinoid system in the baboon (Papio spp.) as a complex
framework for developmental pharmacology. Neurotoxicol Teratol.
58:23–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang
L, Kang Y, Zhao X, Si W, Li W, et al: Generation of gene-modified
cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell
embryos. Cell. 156:836–843. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Kang Y, Zheng B, Shen B, Chen Y, Wang L,
Wang J, Niu Y, Cui Y, Zhou J, Wang H, et al: CRISPR/Cas9-mediated
Dax1 knockout in the monkey recapitulates human AHC-HH. Hum Mol
Genet. 24:7255–7264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Kim S, Huang LW, Snow KJ, Ablamunits V,
Hasham MG, Young TH, Paulk AC, Richardson JE, Affourtit JP,
Shalom-Barak T, et al: A mouse model of conditional lipodystrophy.
Proc Natl Acad Sci USA. 104:16627–16632. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Croasdell A, Duffney PF, Kim N, Lacy SH,
Sime PJ and Phipps RP: PPARγ and the innate immune system mediate
the resolution of inflammation. PPAR Res. 2015.549691:2015.
|
|
162
|
Huang J, Guo X, Fan N, Song J, Zhao B,
Ouyang Z, Liu Z, Zhao Y, Yan Q, Yi X, et al: RAG1/2 knockout pigs
with severe combined immunodeficiency. J Immunol. 193:1496–1503.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Chen Y, Xiong M, Dong Y, Haberman A, Cao
J, Liu H, Zhou W and Zhang SC: Chemical control of grafted human
PSC-derived neurons in a mouse model of Parkinson’s disease. Cell
Stem Cell. 18:817–826. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Zhou X, Xin J, Fan N, Zou Q, Huang J,
Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, et al: Generation of
CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear
transfer. Cell Mol Life Sci. 72:1175–1184. 2015. View Article : Google Scholar
|
|
165
|
Wang L, Yi F, Fu L, Yang J, Wang S, Wang
Z, Suzuki K, Sun L, Xu X, Yu Y, et al: CRISPR/Cas9-mediated
targeted gene correction in amyotrophic lateral sclerosis patient
iPSCs. Protein Cell. 8:365–378. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Bhinge A, Namboori SC, Zhang X, VanDongen
AMJ and Stanton LW: Genetic correction of SOD1 mutant iPSCs reveals
ERK and JNK activated AP1 as a driver of neurodegeneration in
amyotrophic lateral sclerosis. Stem Cell Reports. 8:856–869. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Merienne N, Vachey G, de Longprez L,
Meunier C, Zimmer V, Perriard G, Canales M, Mathias A, Herrgott L,
Beltraminelli T, et al: The self-inactivating KamiCas9 system for
the editing of CNS disease genes. Cell Rep. 20:2980–2991. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Page SC, Hamersky GR, Gallo RA, Rannals
MD, Calcaterra NE, Campbell MN, Mayfield B, Briley A, Phan BN,
Jaffe AE and Maher BJ: The schizophrenia- and autism-associated
gene, transcription factor 4 regulates the columnar distribution of
layer 2/3 prefrontal pyramidal neurons in an activity-dependent
manner. Mol Psychiatry. 23:304–315. 2018. View Article : Google Scholar
|
|
169
|
Long C, Amoasii L, Mireault AA, McAnally
JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby
R and Olson EN: Postnatal genome editing partially restores
dystrophin expression in a mouse model of muscular dystrophy.
Science. 351:400–403. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Long C, McAnally JR, Shelton JM, Mireault
AA, Bassel-Duby R and Olson EN: Prevention of muscular dystrophy in
mice by CRISPR/Cas9-mediated editing of germline DNA. Science.
345:1184–1188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Nelson CE, Hakim CH, Ousterout DG, Thakore
PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan
WX, et al: In vivo genome editing improves muscle function in a
mouse model of Duchenne muscular dystrophy. Science. 351:403–407.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Tabebordbar M, Zhu K, Cheng JKW, Chew WL,
Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al: In
vivo gene editing in dystrophic mouse muscle and muscle stem cells.
Science. 351:407–411. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Ackermann AM, Zhang J, Heller A, Briker A
and Kaestner KH: High-fidelity Glucagon-CreER mouse line generated
by CRISPR-Cas9 assisted gene targeting. Mol Metab. 6:236–244. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Jarrett KE, Lee CM, Yeh YH, Hsu RH, Gupta
R, Zhang M, Rodriguez PJ, Lee CS, Gillard BK, Bissig KD, et al:
Somatic genome editing with CRISPR/Cas9 generates and corrects a
metabolic disease. Sci Rep. 7:446242017. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Mookherjee Yu W, Chaitankar S, Hiriyanna
V, Kim S, Brooks JW, Ataeijannati M, Sun Y, Dong X, Li LT, et al:
Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal
degeneration in mice. Nat Commun. 8:147162017. View Article : Google Scholar : PubMed/NCBI
|