1. Introduction
Gene regulation via DNA and protein modifications
has been intensively studied. However, the knowledge on RNA
modifications remains insufficient (1). Among all mRNA modifications,
N6-methyladenosine (m6A) modification is the
most common type in eukaryotic RNA and occurs in three to five
sites per transcript on average (2-4).
m6A was discovered in the 1970s in a wide
range of cellular mRNAs (4-6).
Methylation occurs at the sixth position of nitrogen atoms of
adenosine at the post-transcriptional level with
S-adenosylmethionine serving as the methyl donor for m6A
formation, which is termed m6A modification (7-9).
The reversible activity of the m6A
modification is regulated by the combined action of methylase and
demethylase (10). The
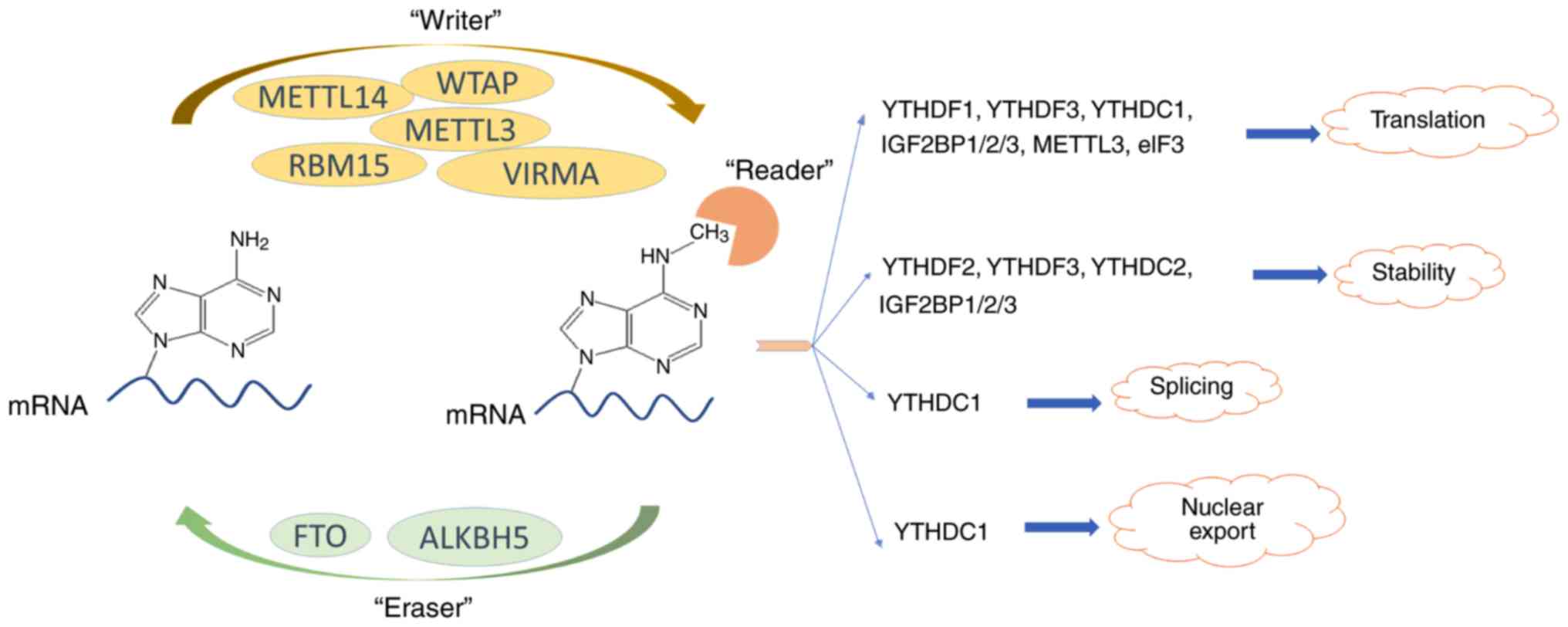
m6A methylase complex consists of at least five 'writer'
proteins (11-14), among which methyltransferase like
3 (METTL3) protein serves a central role. METTL14 protein supports
METTL3 protein structurally (15), WT1-associated protein (WTAP)
regulates the recruitment of the m6A methyltransferase
complex to mRNA targets (12) and
the RNA-binding motif protein 15 serves a role in helping this
complex move towards the appropriate m6A sites (16). Vir like m6A
methyltransferase associated (VIRMA) is also a component of this
m6A methylase complex; however, its molecular function
remains largely unknown (13).
METTL16 is a newly discovered m6A methyltransferase that
primarily methylates m6A sites in the 3′-untranslated
region (3′-UTR) of RNA. When METTL16 is knocked down, the level of
m6A in the cell decreases by ~20% (17). Two reported demethylases that
reverse m6A modification are FTO (18) and alkB homolog 5 (ALKBH5)
(19). It has been confirmed that
m6A levels increased following the knockdown of FTO and
ALKBH5 expression (18,19). The identified 'readers' of
m6A are YT521-B homology (YTH) domain-containing
proteins, including YTHDF1-3, YTHDC1 and YTHDC2, which participate
in the translation (20),
stabilization (21), splicing
(22) or nuclear export (23) of mRNA. The affinity of 'readers'
has been reported to be higher compared with unmethylated mRNA for
m6A-methylated mRNA (24).
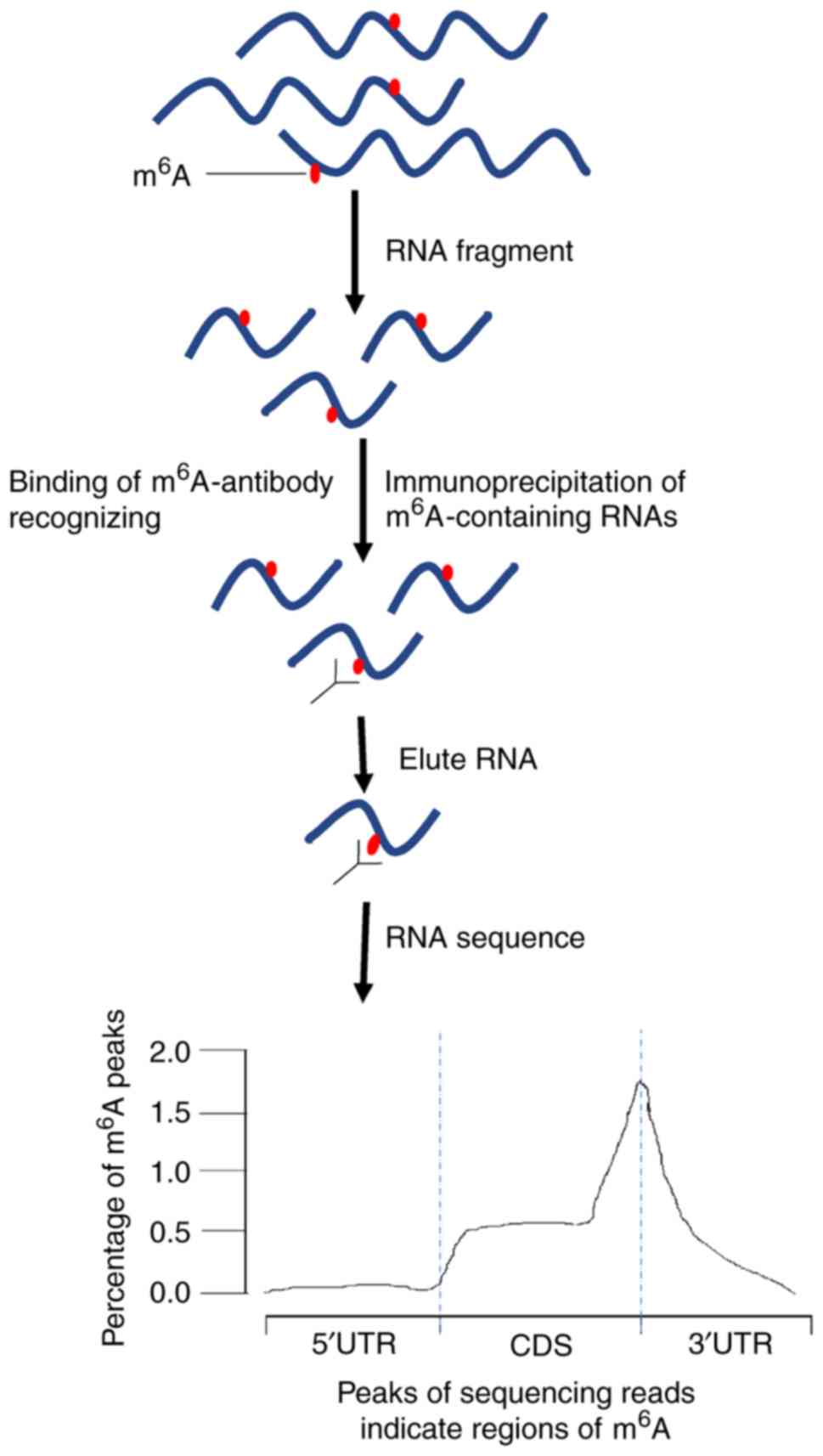
m6A bases cannot be detected directly by
sequencing because the m6A modification does not change
the base pairing properties and cannot be distinguished from
regular bases by reverse transcription (25,26). Previously, new methods have been
developed to identify m6A modification in cells, which
are based on immunoprecipitation or selective RNA chemistry to
isolate modified RNA fragments and combined with high-throughput
sequencing (3,25,27). Dot blot technology is frequently
used to observe changes in m6A. However, dot blots
cannot determine the quantitation and precise location of
m6A (28). RNA
photo-crosslinkers, quantitative proteomics and electrochemical
immunosensor methods may be applied to detect the presence of
m6A in cells; however, they cannot precisely determine
the m6A modification sites (29,30). The newly developed methylated RNA
immunoprecipitation sequencing (MeRIP-seq) method combines
m6A antibody immunoprecipitation and deep sequencing to
identify the m6A residues in 100-200 nucleotide RNA
segments (3,25). However, this approach is
complicated as m6A often appears in clusters since
multiple different m6A-containing fragments generate
overlapping reads, which can result in large peaks spanning several
m6A residues (2).
Thus, the summit of these peaks may not accurately reflect the
positions of m6A residues. By contrast, when using the
m6A individual-nucleotide-resolution cross-linking and
immunoprecipitation (miCLIP) technique, identification of
m6A residues is not influenced by the peak shapes
(31). Furthermore,
identification of m6A residues by miCLIP is not
restricted to a specified subset of DRACH (D=A, G) motifs (2,3,13).
Therefore, miCLIP can properly identify m6A residues
(31).
High-performance liquid chromatography (HPLC) or
mass spectrometry are applied in some of the abovementioned methods
to detect specific modified RNA or RNA bases (3,29).
Isolated RNA is fragmented into nucleosides prior to analysis,
which may change the original RNA structure. Dot blot technology,
which does not require fragmentation, is used as an alternative
method to detect specific modifications. However, dot blot
technology is limited in that RNA samples cannot be separated by
the size, and hence, it is impossible to distinguish the targeted
specific RNA in the RNA samples (32,33). Notably, HPLC can be used alone to
measure m6A modification rate (18).
The abovementioned methods and specific references
are provided in Table I (18,26,27,29-51). In the present review, the most
recent information on the currently established m6A
detection methods is presented and discussed.
 | Table IMethods for detecting m6A
residues. |
Table I
Methods for detecting m6A
residues.
| Method | (Refs.) |
|---|
|
Semi-quantitative | |
| Dot blot
technology | (32,33) |
| Methyl sensitivity
of MazF RNA endonucleases | (34) |
| Immune-northern
blot | (35,36) |
| Quantitative | |
| RNA
photo-crosslinkers and quantitative proteomics | (29) |
| Electrochemical
immunosensor method | (30) |
| Support vector
machine-based method | (37) |
| Detection of
precise locations | |
| HRM | (38) |
| LAIC | (27,39) |
| SCARLET | (39-41) |
| MeRIP-seq | (42-46) |
| MiClip | (31,39,47) |
| DNA polymerase for
direct m6A sequencing | (26,48) |
| HPLC | (18,49-51) |
2. Functions of m6A
modifications
m6A is the most common type of RNA
residue modification in eukaryotes (2-4).
However, its specific biological roles remain largely unknown.
Studies have indicated that the dynamic regulation of
m6A has a significant impact on the control of gene
expression (20,21). m6A-seq has revealed
that m6A predominantly exists on exons and the 3′-UTR of
mRNA (2). METTL3, FTO and ALKBH5
have been identified to serve significant roles in biological
regulation, including development, metabolism and fertility
(18,19).
FTO, an m6A demethylase, is recognized as
a major obesity factor. It belongs to the ALKB enzyme family, which
oxidatively demethylates m6A on mRNA (32,52). ALKBH5 is another mammalian
m6A demethylase, and is associated with certain
mRNA-processing factors in nuclear speckles, which may influence
RNA export and metabolism (19).
ALKBH5-deficient mice have differential expression of genes
involved in the p53 signaling pathway and spermatogenesis (19), suggesting a global role for
m6A in human health. METTL3 is expressed in all human
tissues and is highly expressed in the testis (10,11). It has been identified that METTL3
can participate in tumor growth and progression by regulating the
cell cycle of cancer cells (53,54). WTAP was primarily recognized as a
protein associated with Wilm's tumor and has been indicated to
exhibit a significant role in cell cycle progression (12). WTAP also participates in RNA
splicing and results in embryonic defects (12). Cooperation between the RNA
m6A methyltransferase complex and the demethylases
establishes a reversible regulation of RNA m6A
modifications. Taken together, RNA m6A modifications
exhibit significant roles in the molecular mechanisms of gene
biology, as illustrated in Fig.
1.
3. Methods used to analyze m6A
modifications
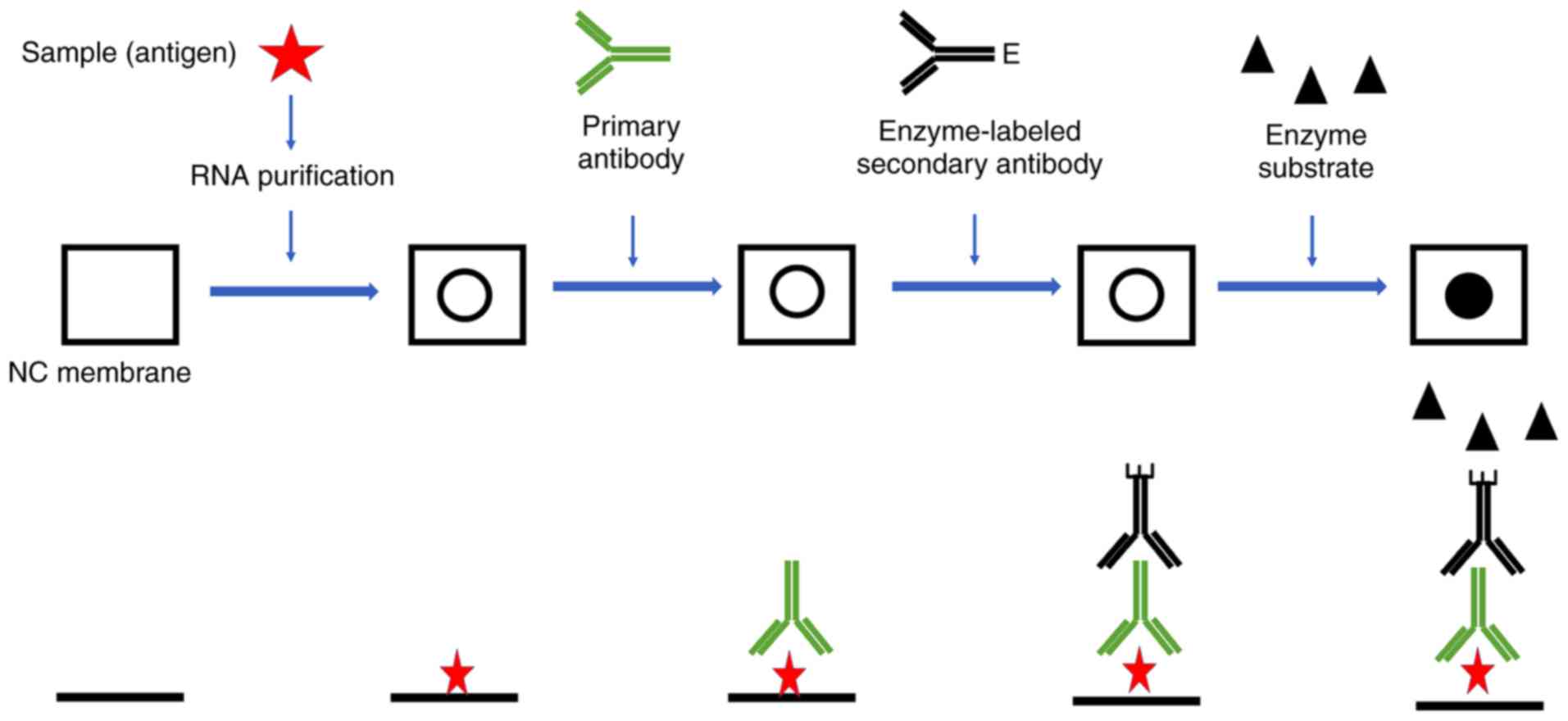
Dot blot
Dot blot (or slot blot) technology is used in
molecular biology as a semiquantitative or quantitative detection
method for DNA, RNA and protein samples, and is predominantly used
semiquantitatively in m6A analysis (32,33,55-57). Compared with western blotting,
northern blotting or Southern blotting methods, dot blot technology
has a simpler operation with a similar working principle. The
biomolecules are not subjected to electrophoretic separation prior
to detection with the dot blot method. Instead, samples containing
a mixture of RNAs are directly applied to a membrane through an
apparatus with circular templates that form a dot when the sample
is applied. After applying a vacuum to embed RNAs and dry the
membrane, biomolecules are detected with antibodies. When detecting
RNA, the DNA must be removed prior to loading to eliminate its
influence on m6A (32,33). A schematic diagram of the
determination of m6A modification residues by dot blot
is presented in Fig. 2.
Dot blot technology markedly saves time, since it
does not require chromatography, gel electrophoresis or complex gel
blocking procedures (32,33). However, regarding m6A
detection, dot blot technology is only able to verify the presence
of m6A or compare the amounts of m6A between
different groups (32,33). Li et al (32) improved the traditional dot blot
method to measure the global m6A abundance in the
transcriptomes of four acute myeloid leukemia cell lines. However,
this method is still not able to quantitate or precisely determine
the location of m6A.
Methyl-sensitive MazF RNA
endonucleases
The regulatory enzymes of m6A have been
reported to contribute to tumorigenesis (58-60). While the significance of
m6A has been confirmed, no convenient approach has been
developed to analyze m6A methyltransferase and
demethylase activities or to monitor the inhibitors of these
activities.
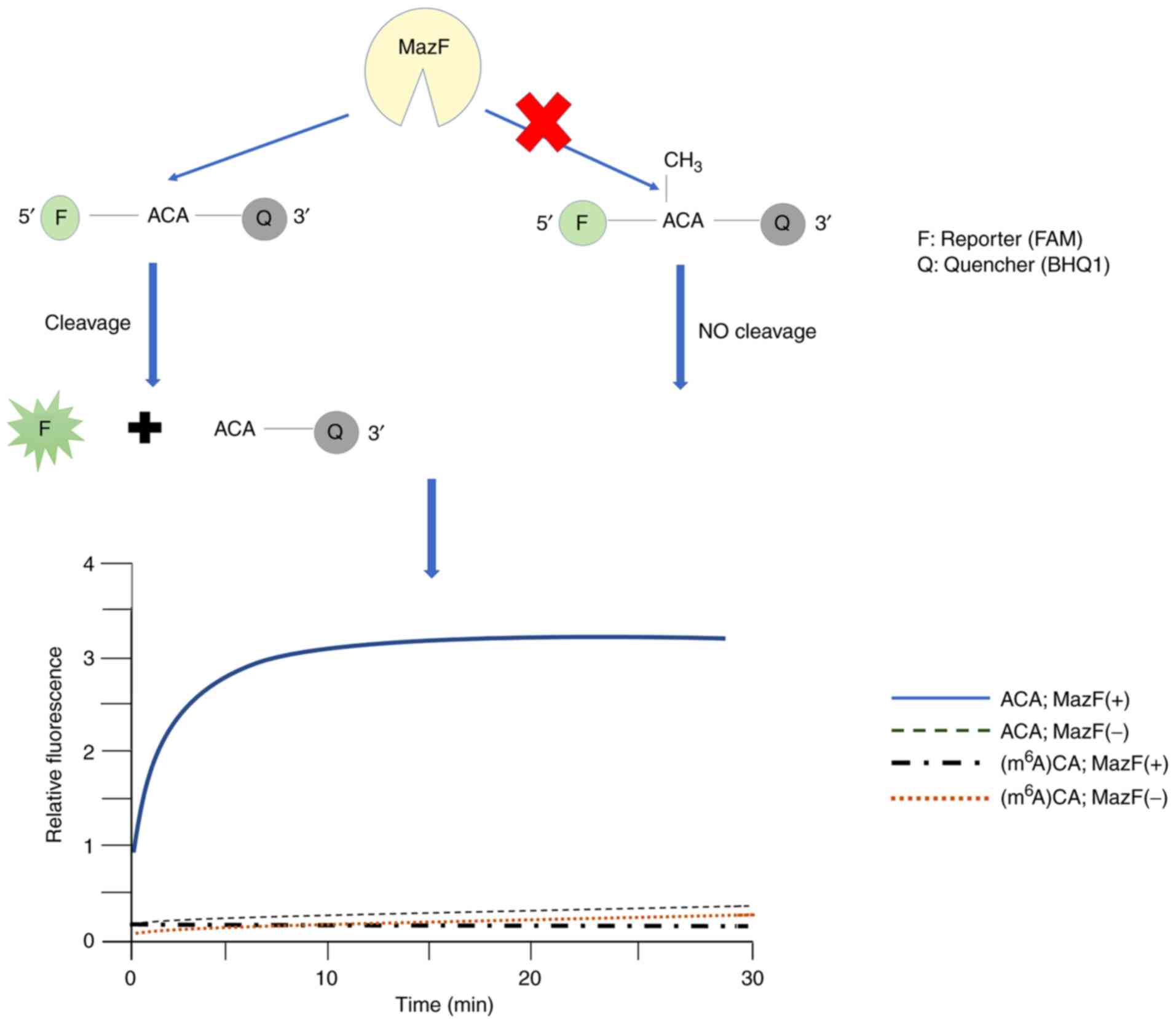
The Escherichia coli toxin MazF exhibits
endoribonuclease activity specific against ACA sequences and is
susceptible to m6A. MazF is the first enzyme discovered
with the ability to specifically cleave RNA containing
m6A, and is used to study activities of m6A
demethylase and methyltransferase (34). Furthermore, MazF has an
application for monitoring the inhibitors of m6A
methyltransferase and demethylase. RNA cleavage by MazF may be
detected by polyacrylamide gel electrophoresis and the
fluorescence-resonance energy transfer-based plate assay (61). A schematic diagram of the
determination of m6A modification residues by
methyl-sensitive MazF RNA endonucleases is presented in Fig. 3.
At the current stage of development, the MazF
cleavage methods are restricted regarding the evaluation of
m6A, as MazF is only able to cleave the 5′-ACA-3′ site
in single-stranded RNA, frequently occurring in endogenous RNAs.
However, MazF is not able to cleave the 5′-ACA-3′ site in
double-stranded RNA (61), and
thus, it may not accurately determine the presence of
m6A in structured RNA.
Immuno-northern blot
The immuno-northern blot combines a northern
blotting experimental program and m6A-binding antibody,
which is different from conventional northern blot techniques that
use DNA probes. Immuno-northern blot does not require RNA
fragmentation prior to analysis and the RNAs are separated based on
their molecular weights (35).
Thus, the detection of m6A modifications by
immuno-northern blot is applied in various types of RNA (35).
In brief, RNAs are separated in a denaturing
acryl-amide gel or an agarose gel and then transferred onto nylon
membranes. The RNA strands on the membranes are exposed to
ultraviolet (UV) irradiation for cross-linking, followed with
incubation with primary antibodies against m6A
modifications, corresponding secondary antibody and
chemiluminescent detection (35).
4. Quantification of m6A
modifications
RNA photo-crosslinkers and quantitative
proteomics
The regulatory role of mRNA predominantly depends on
the interaction between mRNA and RNA-binding proteins to regulate
RNA splicing, stability, localization and translation (62). Photo-crosslinking technologies are
diffusely applied to stabilize direct protein-RNA interactions
(63). These technologies depend
on the tendency for UV-induced photochemistry of nucleobases, which
are natural or derivatives containing sulfur or halogen
substituents. Photo-affinity labels, including diazirine (64) or benzophenone (65), are not widely used in the analysis
of protein-RNA interactions; however, they may be activated by
longer wavelengths and provide more efficient crosslinking.
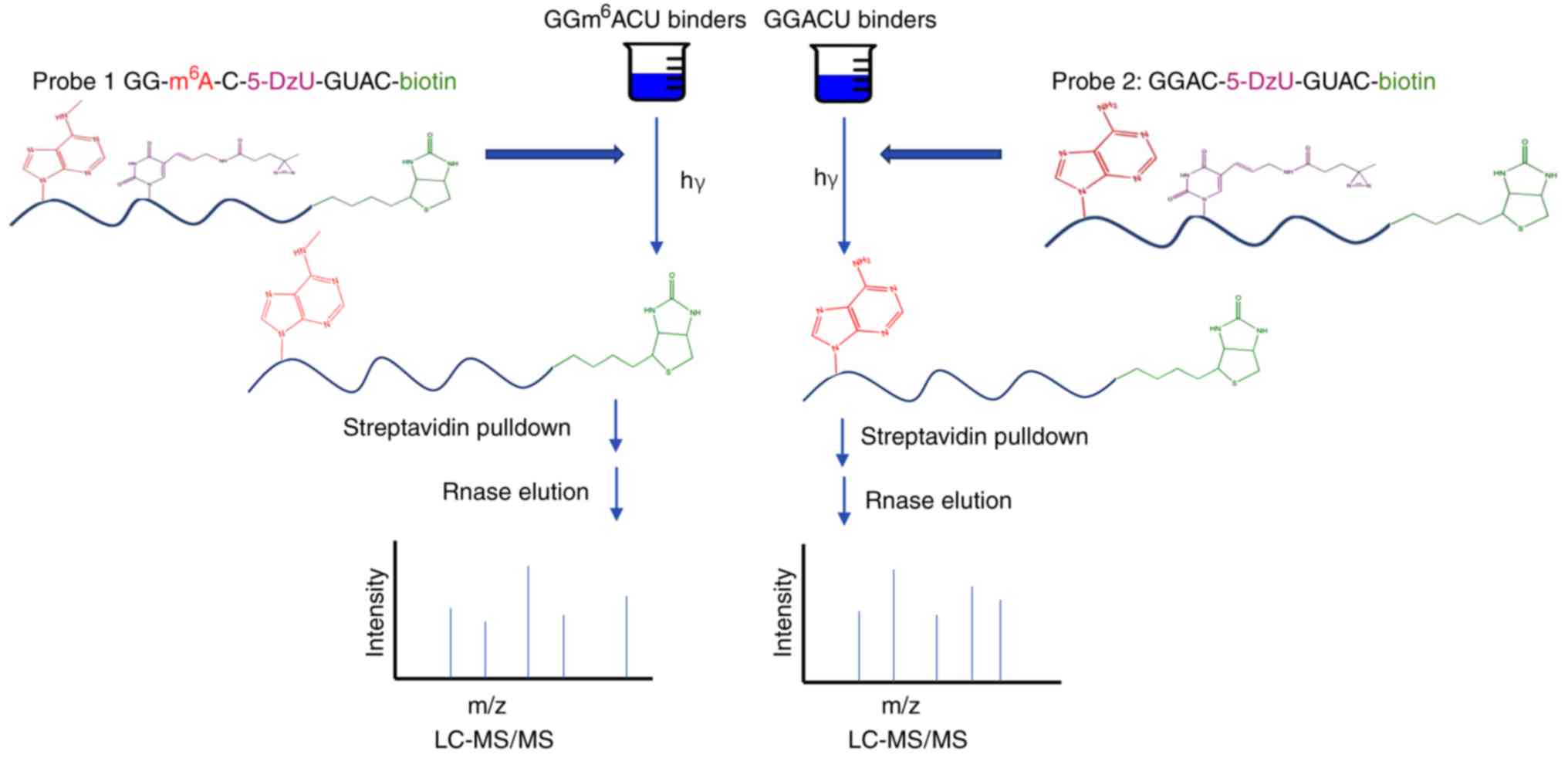
Arguello et al (29) developed a chemical proteomics
approach based on photo-crosslinking of the RNA base and diazirine,
which was highly efficient in quantitatively analyzing protein-RNA
interactions regulated by m6A modification. By using
this method, novel m6A 'readers' have been discovered.
To isolate m6A readers with photosynthetic and
quantitative proteomics, RNA probes are required to contain the
following: i) m6A molecules; ii) a photo-crosslinker
that is efficient and does not influence the protein-RNA
interactions; iii) streptavidin as an affinity handle for protein
enrichment. A probe was prepared that contains the sequence
GGm6ACU, the common recognition pattern of the
m6A site in mammalian cells. This sequence is
indispensable for binding the YTH-domain proteins (66,67). The probe was validated by known
m6A RNA 'readers', including YTHDF1 (20), YTHDF2 (21), YTHDF3 (3,22,68) and YTHDC1 (22). A schematic diagram of the
detection of m6A modification residues by the RNA
photo-crosslinkers and quantitative proteomics technologies is
presented in Fig. 4. We
hypothesize that the requirement of the synthesis of the sensor may
be a limitation regarding these techniques.
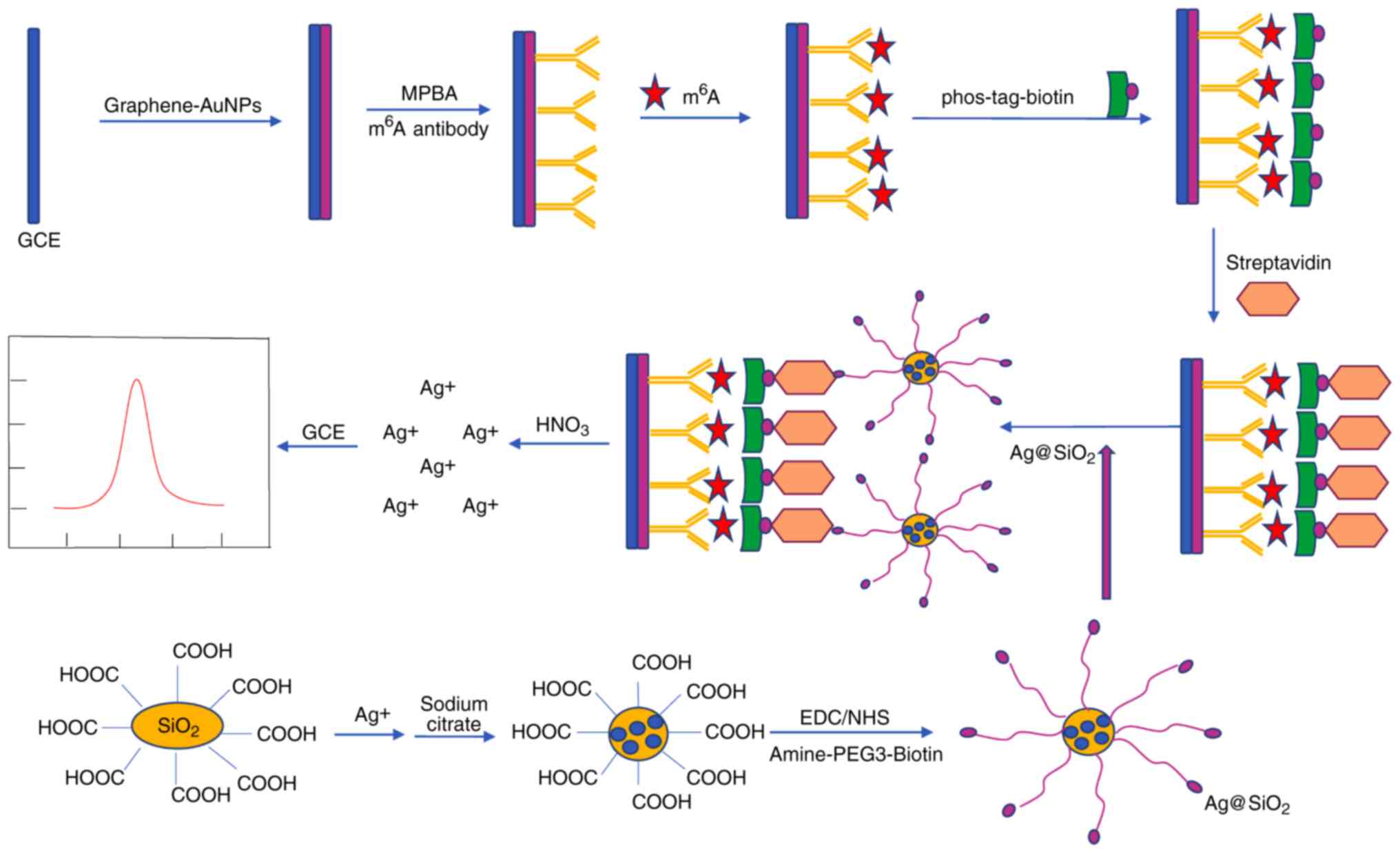
Electrochemical immunosensor method
The majority of the abovementioned analytic methods
are difficult to perform and expensive. The electrochemical
immunosensor method was developed to provide convenience and high
sensitivity (30). An
anti-m6A antibody has been used to detect m6A
by targeting m6A-5′-triphosphate (m6ATP). The
detection and capture of m6A relies on silver
nanoparticles and SiO2 (Ag@SiO2) nano-spheres
with amine-polyethylene glycol 3-biotin. Ag@SiO2
nanospheres were prepared to amplify signals. Phos-tag-biotin was
prepared to link m6ATP and Ag@SiO2 through
two types of specific interaction between phosphate group of
m6ATP and phos-tag, biotin and streptavidin,
respectively. Experiments for evaluating this strategy indicated
that the immuno-sensor has acceptable reproducibility and
specificity with a wide linear range and a low detection limit. A
schematic diagram for the determination of m6A
modification residues by the electrochemical immunosensor method is
presented in Fig. 5.
The efficacy of the detection of the m6A
content using the electrochemical immunosensor method had been
verified in human cell lines (30). It also provides a technological
basis for the detection of RNAs and DNAs with the advantages of
convenience, low cost, and high specificity and sensitivity.
Vector method to detect m6A
sites
High-throughput next-generation sequencing-based
technology for identifying m6A sites based on where
adenosine is methylated has not been applied in most species. In
recent years, a vector machine method was developed to identify
m6A sites in Arabidopsis thaliana (37). When combining anti-m6A
antibodies and high-throughput sequencing, Luo et al
(69) obtained thousands of
m6A peaks for A. thaliana, including 'common'
m6A peaks. Since the RRACH motif, where R resembles
purine, A stands for m6A and H resembles a non-guanine
base (69), was identified in
most of the m6A peaks, Chen et al (37) collected segments containing RRACH
at the center of the 'common' m6A peaks and proposed a
model that may accurately identify specific m6A sites
with high accuracy.
If the model is adapted to other plant species other
than A. thaliana, this vector machine-based method may be
used for the detection of m6A and other
post-transcriptional modifications in these other plants as
well.
5. Methods to determine m6A
residue locations
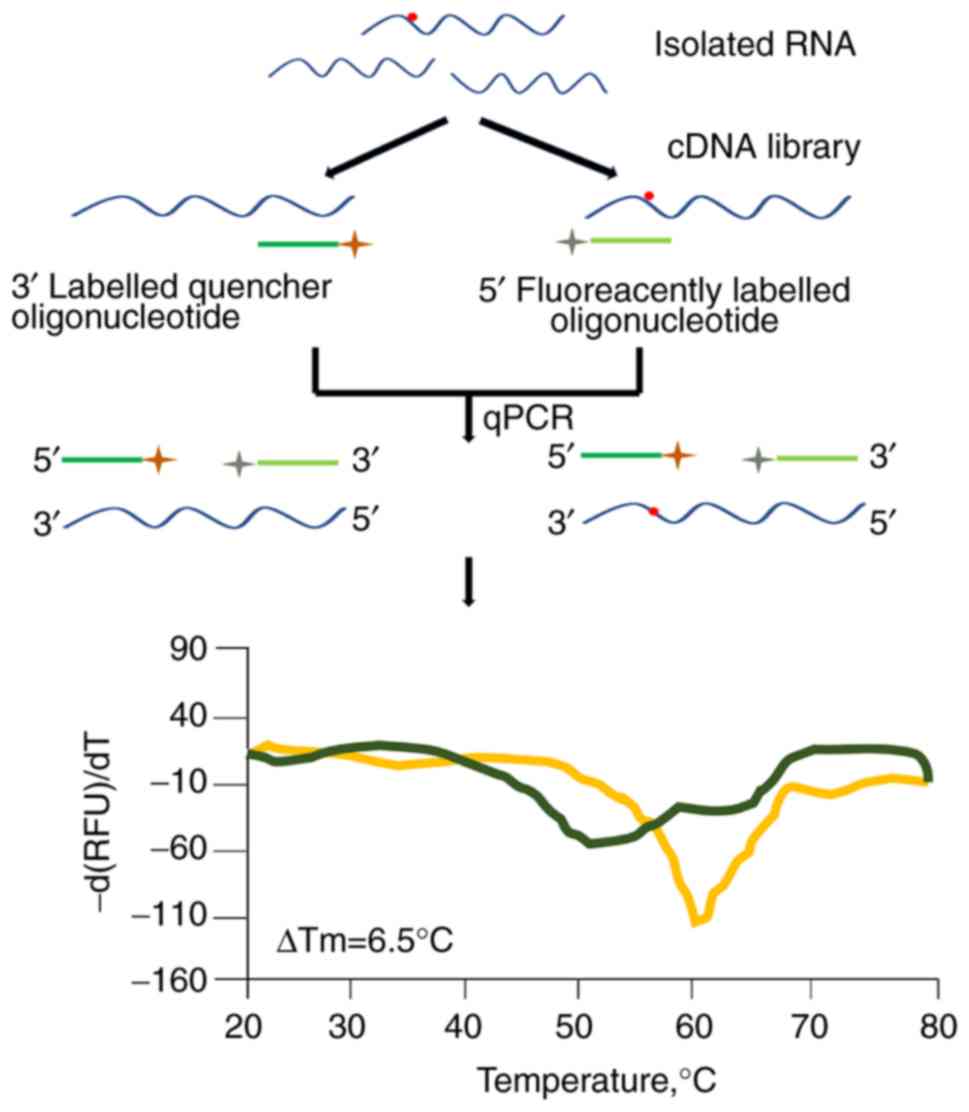
HRM
HRM analysis is a simple method to detect
m6A modification residues at a specific location in RNAs
(38). HRM may be applied to
high-throughput measurement. The resulting HRM curves of the
samples of RNA mixtures change steadily from 100% of methylated RNA
to 100% of unmethylated RNA (38). As presented in Fig. 6, the detection of m6A
modification residues by the HRM method relies on the modified
nucleoside position at a particular site of RNA and is followed by
rapid screening for conditions or genes necessary for analysis of
that modification (38).
According to the specificity of the oligonucleotide
probe hybridization, bulk cellular RNA, as opposed to purified
specific RNA, has been designed for detecting m6A at a
pre-defined position (38). In
addition, partial non-ribosomal target enrichments may be easily
accomplished using commercially available kits.
A possible application for this method would be to
screen knockout/knockdown strain libraries to identify genes
contributing to the formation of a specific m6A
nucleoside. Another possible application is to detect the presence
of a particular m6A nucleoside under different growth or
environmental conditions (2,3).
HRM analysis may help to elucidate the dynamic events that result
in the modification of certain RNAs.
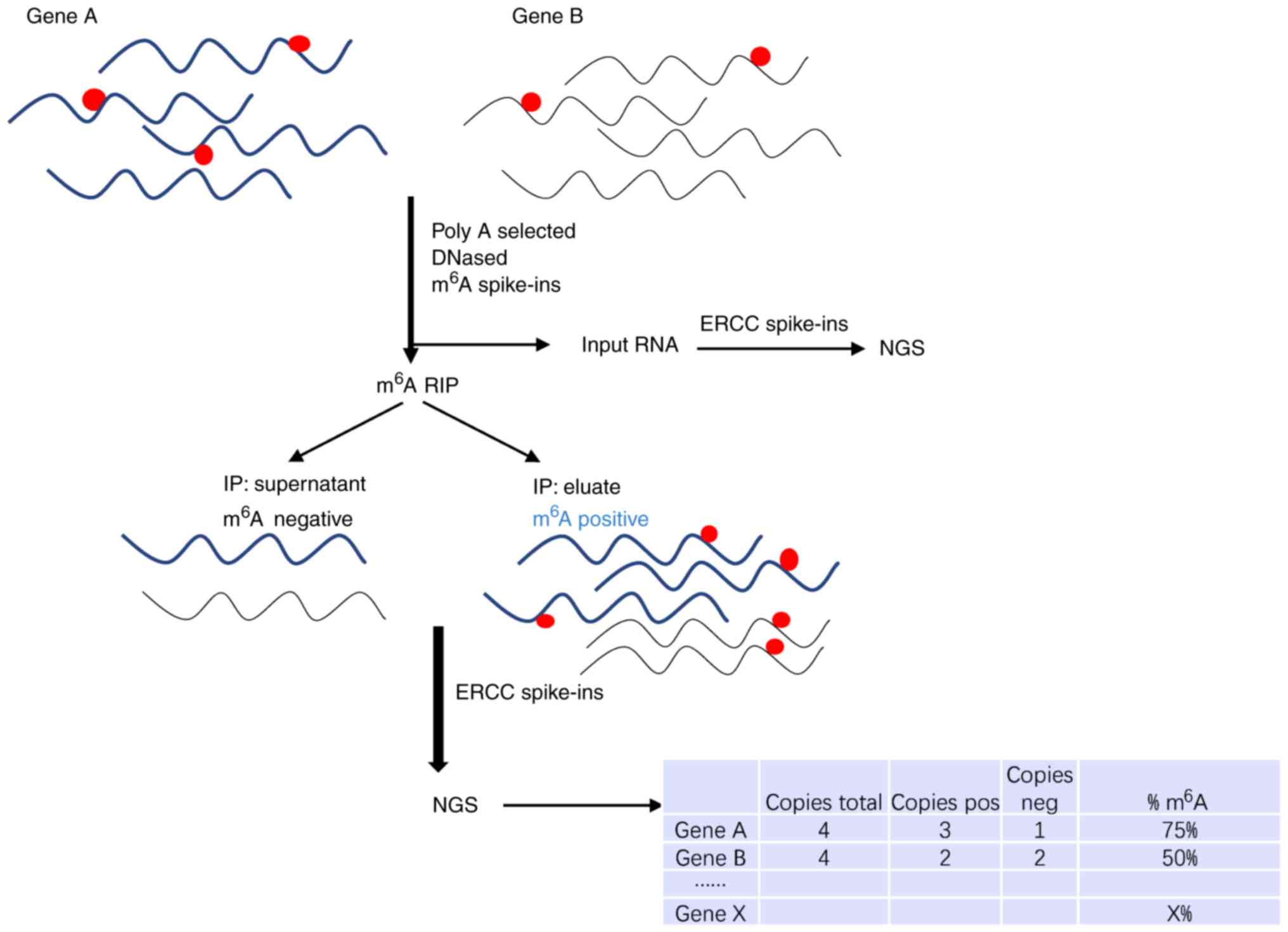
m6A level and
isoform-characterization sequencing (m6A-LAIC-seq)
For thorough investigation of the m6A
epitranscriptome, Molinie et al (27) invented the m6A-LAIC-seq
technique. Combined with RNA IP whole-transcriptome sequencing,
m6A-LAIC-seq may quantify m6A contents, with
spike-in RNAs as an internal standard. A schematic diagram for the
determination of m6A modification residues by
m6A-LAIC-seq is presented in Fig. 7. The results demonstrate a
quantitative road map to which genes are the most or the least
likely to be influenced by m6A-dependent regulatory
networks. This method may determine the m6A levels in
each gene but cannot stoichiometrically analyze the methylation of
a single modified nucleotide. m6A-LAIC-seq complements
m6A-seq identification of methylation sites and helps to
expand the understanding of the biology of m6A.
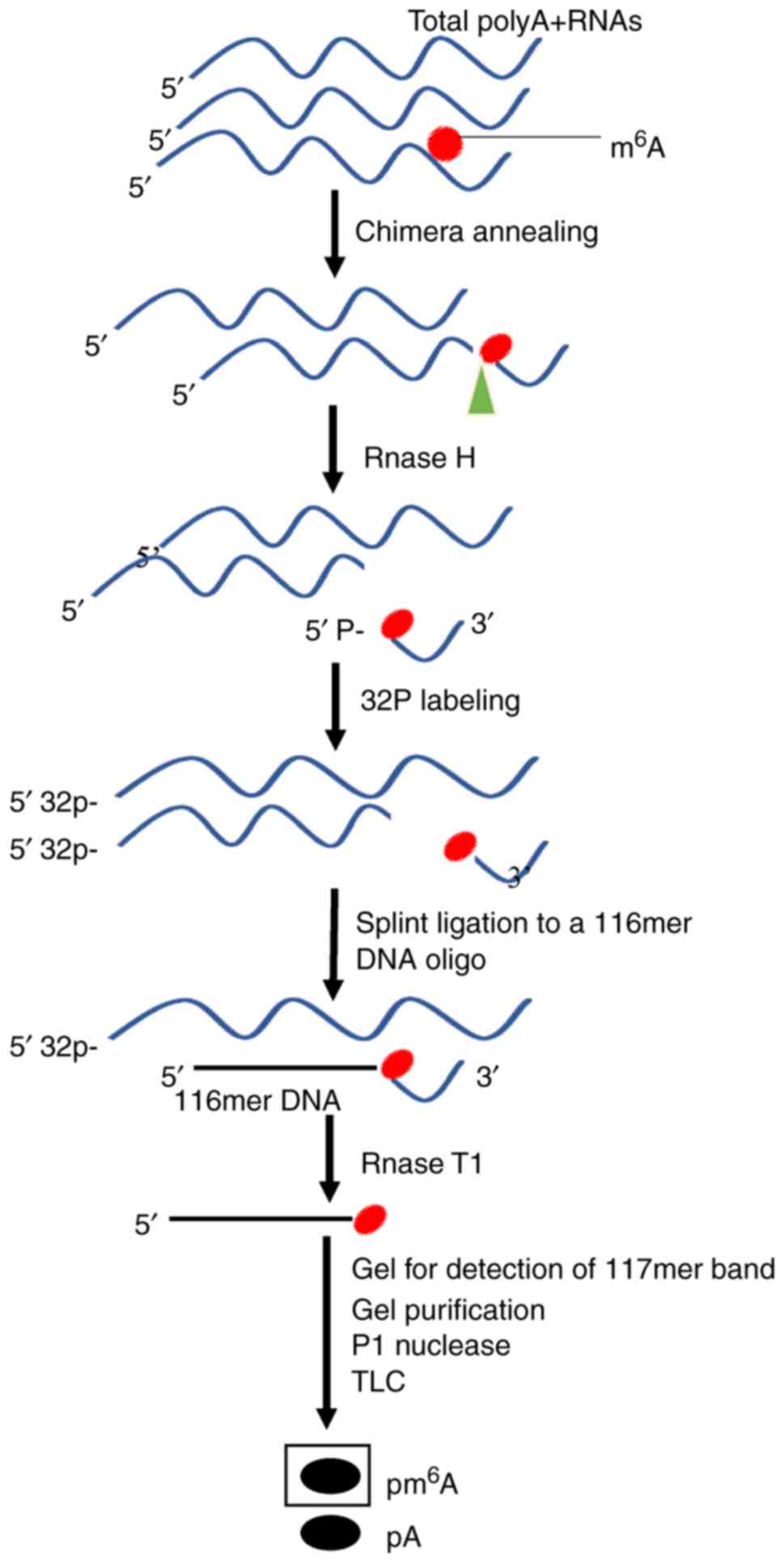
Site-specific cleavage and radioactive
labeling followed by ligation-assisted extraction and thin-layer
chromatography (SCARLET)
To elucidate the dynamic biological functions of
m6A, Liu et al (40) provided the SCARLET method that
directly measures the precise location and the status of
m6A modification at any candidate site of mRNA/lncRNA at
a single nucleotide resolution. In addition to m6A,
SCARLET may be used to observe other RNA modifications, including
5-methylcytosine, pseudouridine and 2′-O-methyl ribonucleosides.
SCARLET is available to study the biological functions of RNA
modifications with general experimental equipment and materials. A
schematic diagram for the determination of m6A
modification residues by SCARLET is presented in Fig. 8.
The feasibility of using the SCARLET method has
been confirmed in HeLa cell RNA samples, which produced similar
results compared with previously reported m6A sites in
Hela samples. Using this method, a minimally modified
m6A site that was not precisely determined in a
preceding study was also identified, demonstrating that SCARLET is
able to easily resolve the ambiguity of modification sites
(70).
MeRIP-Seq
Although RNA methylation has been identified and
verified in the 1970s, the relevant modification mechanism,
regulatory means and biological significance have not been
clarified due to technical limitations. The recent emergence of
MeRIP-Seq technology makes it possible to study m6A
methylation at the transcriptome level by high-throughput
sequencing (2,3).
MeRIP-Seq is a combination of ChIP-Seq and RNA-Seq
that is able to elucidate global mRNA m6A sites in
mammalian cells. MeRIP-seq is a novel type of IP-seq technology, in
which the known ChIP-seq, photoactivatable ribonucleo-side-enhanced
crosslinking and IP (a more mature sequence), is applied. MeRIP-seq
has been successfully used to detect whole-genome m6A
modifications (42-46). The major principle of this
technique is as follows: The anti-m6A antibody is
incubated with randomly interrupted RNA fragments using a co-IP
method resulting in an m6A modification fragment that is
precipitated and sequenced. Concurrently, a control sample is run
that eliminates the background during antibody capture (2).
Successful preparations of each library should be
evaluated prior to massive parallel sequencing. To identify and
localize the m6A sites at a transcriptome-wide level by
m6A IP, fragmentation of poly(A)+-selected
RNAs (input) is required prior to IP with anti-m6A
antibodies. The input and m6A IP RNAs are then
separately processed for next-generation sequencing (2). Zhang et al (48) first proposed a Bayesian
statistical model, BaySeqPeak, to analyze MeRIP-Seq data to help
discover methylation site signals in the transcriptome. A reference
transcriptome was prepared, which contains a single, non-intron
splice variant of each gene. The resultant reads were then
specifically compared with the reference to identify m6A
sites with a low false-detection rate (3). A schematic diagram for the
determination of m6A modification residues by MeRIP-Seq
is presented in Fig. 9.
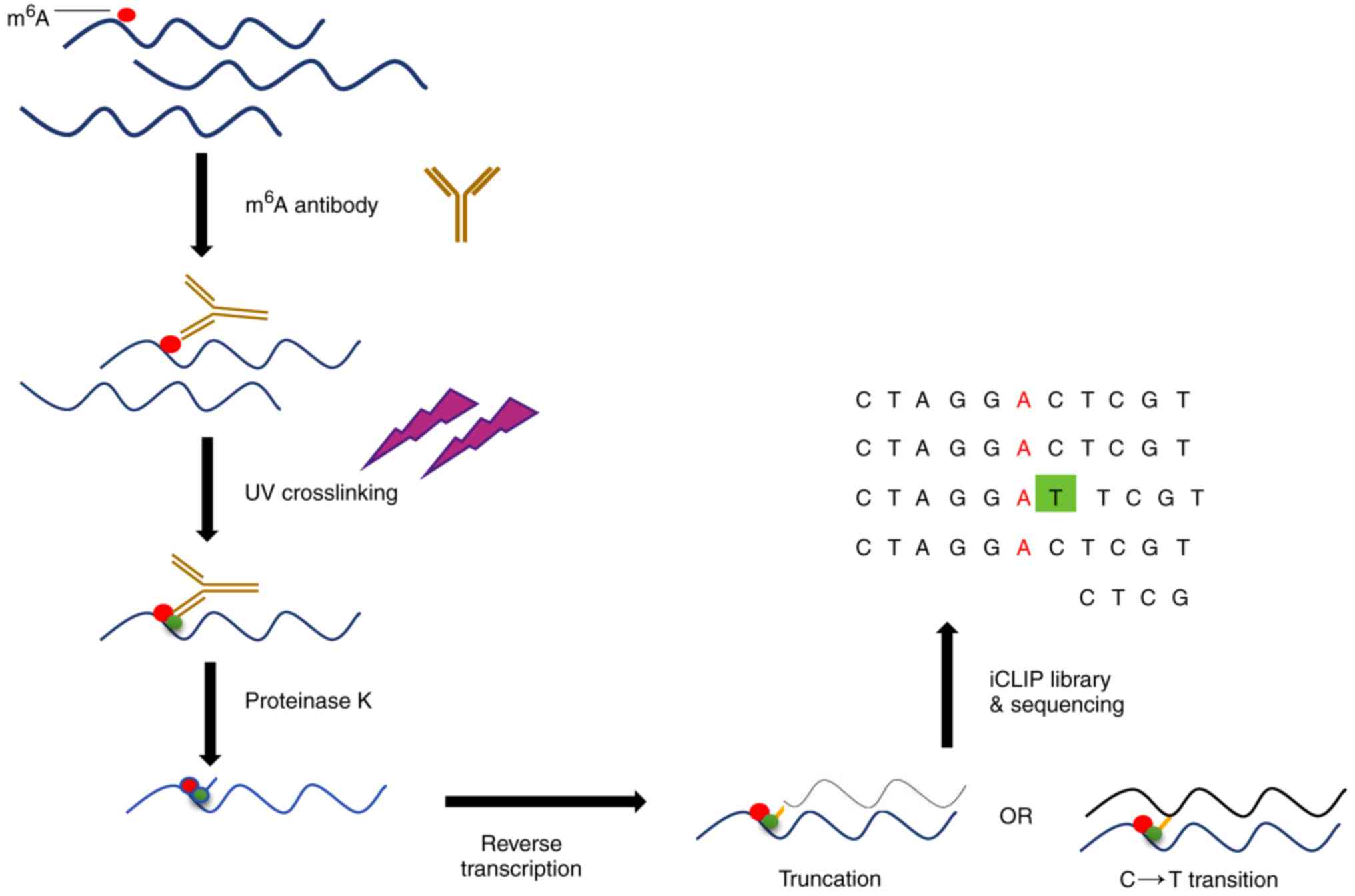
MiClip
It has been reported that m6A residues
may be located by producing unique signature mutations with
anti-m6A antibodies and UV crosslinking techniques
(31). m6A residues
were mapped with two antibodies, one of which translates C to T to
detect single and clustered m6A residues, and the other
antibody that produces truncations is used to determine the
position of m6A sites and detect m6A residues
concurrently. A schematic diagram for the determination of
m6A modification residues by miClip is provided in
Fig. 10.
Identification of m6A residues by direct
detection is superior to that by bioinformatics predictions from
MeRIP-Seq peaks (31). The
reliability of bioinformatics prediction depends on the
characteristics of the m6A peak. MeRIP-Seq can
accurately predict m6A residues only with a single clear
peak of a single m6A residue and is limited to the
centrally-located DRACH motif (31). m6A residues usually
cluster in mRNAs, resulting in multiple MeRIP-Seq peaks (2). Identification of m6A
residues using miCLIP technology is not influenced by peak shapes
and not confined to the centrally-located DRACH motif. Therefore,
miCLIP can correctly identify m6A residues.
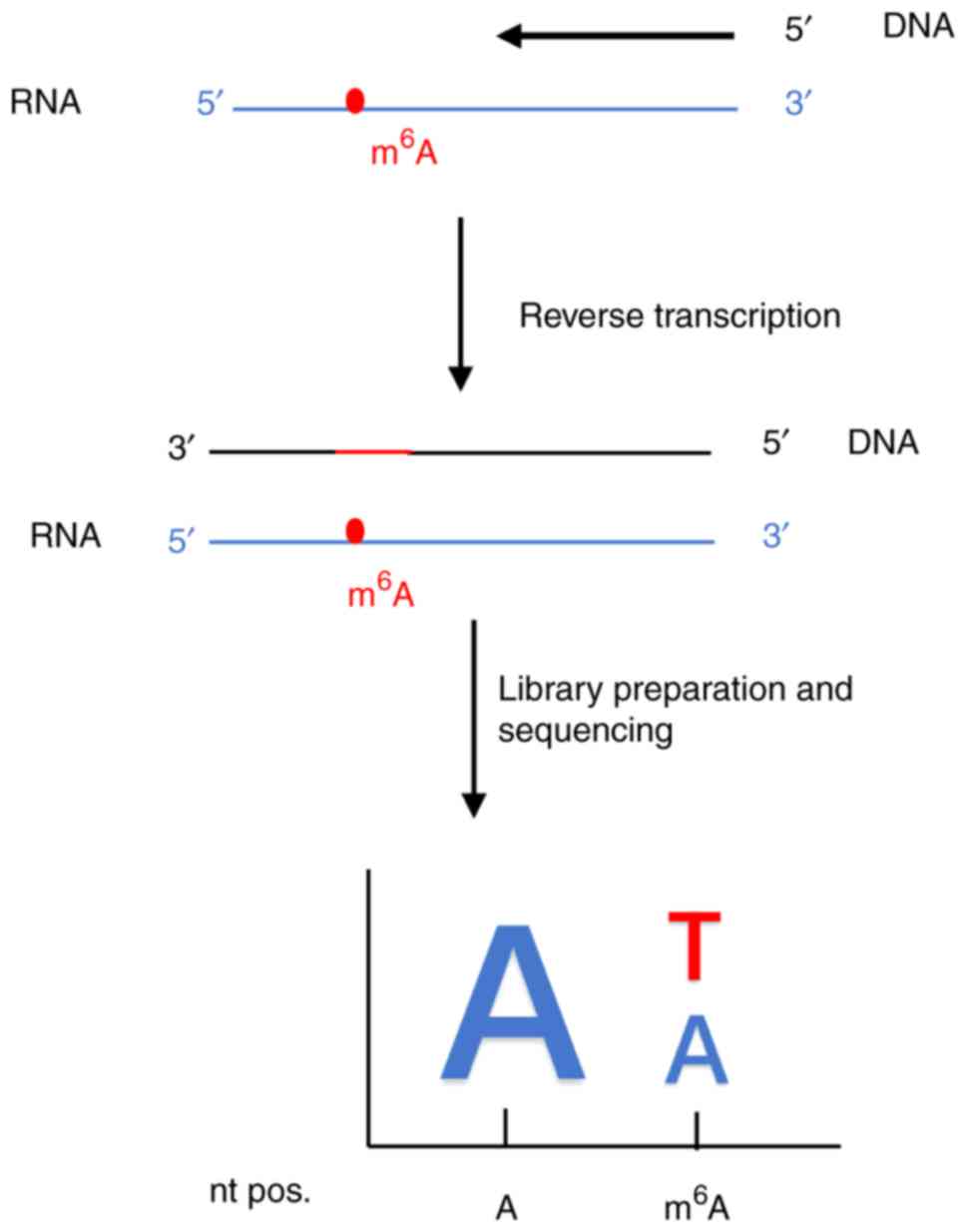
A DNA polymerase for direct
m6A sequencing
At present, RNA samples are prepared by
antibody-based enrichment of m6A residues prior to
sequencing, as m6A modifications are usually lost after
reverse transcription (25,26). The indirect detection may lead to
a higher error rate, pushing the generation of novel DNA polymerase
to sequence m6A directly (26). In this light, Aschenbrenner et
al (26) developed a
screening method to develop a reverse transcription-active KlenTaq
DNA polymerase variant for labeling N6-methylation
residues. A schematic diagram for the determination of
m6A modification residues by the DNA polymerase for
direct N6-methyladenosine sequencing is presented in
Fig. 11.
HPLC
The development of HPLC occurred only 30 years ago;
however, the development of this separation analysis technology is
very rapid, and is now widely used, including in the detection of
m6A modification (50). HPLC was firstly used to detect
m6A modification in nine DNAs (50). Subsequently, Rana and Tuck
(49) applied HPLC in the
detection of m6A modification in a T7 RNA transcript
coding for mouse dihydrofolate reductase by separating
m6A from adenine, cytosine, uracil and so on. In a study
by Jia et al (18), HPLC
helped observe the changes of the m6A ratio in mRNAs
following FTO treatment to better understand the function of FTO.
The development of this in vitro methylation assay opened
the door for studies investigating m6A levels in
specific mRNAs or learn the biological significance of
m6A modification.
6. Discussion
Detection of RNA modifications and study of their
functions are emerging fields of research. The potential role of
m6A modifications in regulating molecular and
physiological processes in several organisms, particularly in RNA
stability, splicing, transport, localization and translation, is
valued (19,20,52,21,70-73). With the discovery of 'reader'
proteins, the downstream molecular mechanisms of m6A
modifications are gradually being clarified (21). There is strong evidence that
m6A methylation is associated with RNA splicing and that
'readers' of m6A reduce the stability of RNA transcripts
(21). After removal of
m6A, a second class of proteins may bind to the RNAs,
which may be affected by changes of the RNA secondary structure
from m6A additions (74). Thus, the physiological effects of
m6A modifications should be observed at multiple levels,
including the tissue level, the pathway level, the cellular level
and the molecular level (75). An
increasing number of studies have suggested a link between
m6A RNA modifications with cancer and other similar
disease-associated processes (32,53,54,73,74-76). Detection of m6A
modification in vitro can help identify the precise
regulatory forms and synergistic roles of m6A
modifications in cancer and other diseases (32,53,54,74-76). Certain m6A methylases,
m6A demethylases or downstream genes have become
prognostic factors for different cancer types (77-79). Certain abovementioned methods,
including MeRIP-seq, could help identify the downstream genes and
mutation sites (54,80). Therefore, detection of them in
vitro may help diagnose and predict the final progress. The
present review provides an overview of these methods to drive the
elucidation of the biological roles of m6A and encourage
their further development.
Funding
This project was supported by grants from the
National Natural Science Foundation of China (grant nos. 81571568
and 81871265), the CAMS Innovation Fund for Medical Sciences (grant
no. CIFMS:2016-I2M-1003), the Innovation Team of Jiangsu Provincial
Commission of Health and Family Planning (grant no.
CXTDA2017049).
Availability of data and materials
Not applicable.
Authors' contributions
JWe and HY designed the study and revised the
article. WZ and JWa reviewed the literature and wrote the article.
ZX, MC, QH, CP and MG reviewed the literature and revised the
article. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of
interests.
Acknowledgments
Not applicable.
References
|
1
|
He C: Grand challenge commentary: RNA
epigenetics. Nat Chem Biol. 6:863–865. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meye KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar
|
|
3
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Adams JM and Cory S: Modified nucleosides
and bizarre 5′-termini in mouse myeloma mRNA. Nature. 255:28–33.
1975. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei CM, Gershowitz A and Moss B:
Methylated nucleotides block 5′ terminus of HeLa cell messenger
RNA. Cell. 4:379–386. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Narayan P and Rottman FM: Methylation of
mRNA. Adv Enzymol Relat Areas Mol Biol. 65:255–285. 1992.PubMed/NCBI
|
|
8
|
Dubin DT and Taylor RH: The methylation
state of poly A-containing messenger RNA from cultured hamster
cells. Nucleic Acids Res. 2:1653–1668. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Haugland RA and Cline MG:
Post-transcriptional modifications of oat coleoptile ribonucleic
acids. 5′-Terminal capping and methylation of internal nucleosides
in poly(A)-rich RNA. Eur J Biochem. 104:271–277. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Niu Y, Zhao X, Wu YS, Li MM, Wang XJ and
Yang YG: N6-methyl-adenosine (m6A) in RNA: An old modification with
a novel epigenetic function. Genomics Proteomics Bioinformatics.
11:8–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bokar JA, Shambaugh ME, Polayes D, Matera
AG and Rottman FM: Purification and cDNA cloning of the
AdoMet-binding subunit of the human mRNA
(N6-adenosine)-methyltransferase. RNA. 3:1233–1247. 1997.PubMed/NCBI
|
|
12
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ear J and Lin S: RNA methylation regulates
hematopoietic stem and progenitor cell development. J Genet
Genomics. 44:473–474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m(6)A RNA methylation promotes
XIST-mediated transcriptional repression. Nature. 537:369–373.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pendleton KE, Chen B, Liu K, Hunter OV,
Xie Y, Tu BP and Conrad NK: The U6 snRNA m6A
Methyltransferase METTL16 Regulates SAM Synthetase Intron
Retention. Cell. 169:824–835.e814. 2017. View Article : Google Scholar
|
|
18
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Bio. 7:885–887. 2011. View Article : Google Scholar
|
|
19
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol cell. 49:18–29. 2013. View Article : Google Scholar :
|
|
20
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar
|
|
22
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roundtree IA, Luo GZ, Zhang Z, Wang X,
Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al: YTHDC1
mediates nuclear export of N6-methyladenosine methylated
mRNAs. Elife. 6:e313112017. View Article : Google Scholar
|
|
24
|
Theler D, Dominguez C, Blatter M, Boudet J
and Allain FH: Solution structure of the YTH domain in complex with
N6-methyladenosine RNA: A reader of methylated RNA. Nucleic Acids
Res. 42:13911–13919. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saletore Y, Meyer K, Korlach J, Vilfan ID,
Jaffrey S and Mason CE: The birth of the Epitranscriptome:
Deciphering the function of RNA modifications. Genome Biol.
13:1752012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aschenbrenner J, Werner S, Marchand V,
Adam M, Motorin Y, Helm M and Marx A: Engineering of a DNA
polymerase for direct m6A sequencing. Angew Chem Int Ed Engl.
57:417–421. 2018. View Article : Google Scholar :
|
|
27
|
Molinie B, Wang J, Lim KS, Hillebrand R,
Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon
P, et al: m6A level and isoform characterization sequencing
(m6A-LAICseq) reveals the census and complexity of the m6A
epitranscriptome. Nat Methods. 13:692–698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nagarajan A, Janostiak R and Wajapeyee N:
Dot blot analysis for measuring global
N6-methyladenosine modification of RNA. Methods Mol
Biol. 1870:263–271. 2019. View Article : Google Scholar
|
|
29
|
Arguello AE, DeLiberto AN and Kleiner RE:
RNA chemical proteomics reveals the N6-methyladenosine
(m6A)-regulated protein-RNA interactome. J Am Chem Soc.
139:17249–17252. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yin H, Wang H, Jiang W, Zhou Y and Ai S:
Electrochemical immunosensor for N6-methyladenosine detection in
human cell lines based on biotin-streptavidin system and
silver-SiO2 signal amplification. Biosens Bioelectron.
90:494–500. 2017. View Article : Google Scholar
|
|
31
|
Linder B, Grozhik AV, Olarerin-George AO,
Meydan C, Mason CE and Jaffrey SR: Single-nucleotide-resolution
mapping of m6A and m6Am throughout the transcriptome. Nat Methods.
12:767–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Weng Li Z, Su H, Weng R, Zuo X, Li Z,
Huang C, Nachtergaele H, Dong S, Hu LC, et al: FTO plays an
oncogenic role in acute myeloid leukemia as a
N6-methyladenosine RNA demethylase. Cancer Cell.
31:127–141. 2017. View Article : Google Scholar
|
|
33
|
Wang Y, Li Y, Yue M, Wang J, Kumar S,
Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G and Zhao
JC: N6-methyladenosine RNA modification regulates
embryonic neural stem cell self-renewal through histone
modifications. Nat Neurosci. 21:195–206. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Imanishi M, Tsuji S, Suda A and Futaki S:
Detection of N6-methyladenosine based on the
methyl-sensitivity of MazF RNA endonuclease. Chem Commun (Camb).
53:12930–12933. 2013. View Article : Google Scholar
|
|
35
|
Mishima E, Jinno D, Akiyama Y, Itoh K,
Nankumo S, Shima H, Kikuchi K, Takeuchi Y, Elkordy A, Suzuki T, et
al: Immuno-Northern blotting: Detection of RNA modifications by
using antibodies against modified nucleosides. PLoS One.
10:e01437562015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mishima E and Abe T: Immuno-northern
blotting: Detection of modified RNA using gel separation and
antibodies to modified nucleosides. Methods Mol Biol. 1870:179–187.
2019. View Article : Google Scholar
|
|
37
|
Chen W, Feng P, Ding H and Lin H:
Identifying N6-methyladenosine sites in the Arabidopsis
thaliana transcrip-tome. Mol Genet Genomics. 291:2225–2229. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Golovina AY, Dzama MM, Petriukov KS,
Zatsepin TS, Sergiev PV, Bogdanov AA and Dontsova OA: Method for
site-specific detection of m6A nucleoside presence in RNA based on
high-resolution melting (HRM) analysis. Nucleic Acids Res. 42:e27.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lopez CM, Lloyd AJ, Leonard K and
Wilkinson MJ: Differential effect of three base modifications on
DNA thermostability revealed by high resolution melting. Anal Chem.
84:7336–7342. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu N, Parisien M, Dai Q, Zheng G, He C
and Pan T: Probing N6-methyladenosine RNA modification status at
single nucleotide resolution in mRNA and long noncoding RNA. RNA.
19:1848–1856. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jacob R, Zander S and Gutschner T: The
dark side of the epitranscriptome: Chemical modifications in long
non-coding RNAs. Int J Mol Sci. 18:E23872017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li X, Zhu P, Ma S, Song J, Bai J, Sun F
and Yi C: Chemical pulldown reveals dynamic pseudouridylation of
the mammalian transcriptome. Nat Chem Biol. 11:592–597. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Antanaviciute A, Baquero-Perez B, Watson
CM, Harrison SM, Lascelles C, Crinnion L, Markham AF, Bonthron DT,
Whitehouse A and Carr IM: M6aViewer: Software for the detection,
analysis, and visualization of N6-methyladenosine peaks
from m6A-seq/ME-RIP sequencing data. RNA. 23:1493–1501.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cui X, Meng J, Zhang S, Chen Y and Huang
Y: A novel algorithm for calling mRNA m6A peaks by modeling
biological variances in MeRIP-seq data. Bioinformatics.
32:i378-i3852016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Meng J, Lu Z, Liu H, Zhang L, Zhang S,
Chen Y, Rao MK and Huang Y: A protocol for RNA methylation
differential analysis with MeRIP-Seq data and exomePeak
R/Bioconductor package. Methods. 69:274–281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu H, Wang H, Wei Z, Zhang S, Hua G,
Zhang SW, Zhang L, Gao SJ, Meng J, Chen X and Huang Y: MeT-DB V2.0:
Elucidating context-specific functions of N6-methyl-adenosine
methyltran-scriptome. Nucleic Acids Res. 46:D281–D287. 2017.
View Article : Google Scholar
|
|
47
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang M, Li Q and Xie Y: A Bayesian
hierarchical model for analyzing methylated RNA immunoprecipitation
sequencing data. Quant Biol. 6:275–286. 2018. View Article : Google Scholar
|
|
49
|
Rana AP and Tuck MT: Analysis and in vitro
localization of internal methylated adenine residues in
dihydrofolate reductase mRNA. Nucleic Acids Res. 18:4803–4808.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ehrlich M, Gama-Sosa MA, Carreira LH,
Ljungdahl LG, Kuo KC and Gehrke CW: DNA methylation in thermophilic
bacteria: N4-methylcytosine, 5-methylcytosine, and
N6-methyladenine. Nucleic Acids Res. 13:1399–1412. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Clancy MJ, Shambaugh ME, Timpte CS and
Bokar JA: Induction of sporulation in Saccharomyces cerevisiae
leads to the formation of N6-methyladenosine in mRNA: A potential
mechanism for the activity of the IME4 gene. Nucleic Acids Res.
30:4509–4518. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao X, Yang Y, Sun BF, Shi Y, Yang X,
Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al: FTO-dependent
demethylation of N6-methyladenosine regulates mRNA splicing and is
required for adipogenesis. Cell Res. 24:1403–1419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Barbieri I, Tzelepis K, Pandolfini L, Shi
J, Millán-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister
AJ, Han N, et al: Promoter-bound METTL3 maintains myeloid leukaemia
by m6A-dependent translation control. Nature.
552:126–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tang Li X, Huang J, Wang W, Li F, Qin P,
Qin C, Zou Z, Wei Q, Hua JL, et al: The M6A methyltransferase
METTL3: Acting as a tumor suppressor in renal cell carcinoma.
Oncotarget. 8:96103–96116. 2017.PubMed/NCBI
|
|
55
|
Miao Z, Xin N, Wei B, Hua X, Zhang G, Leng
C, Zhao C, Wu D, Li J, Ge W, et al: 5-hydroxymethylcytosine is
detected in RNA from mouse brain tissues. Brain Res. 1642:546–552.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rona G, Scheer I, Nagy K, Pálinkás HL,
Tihanyi G, Borsos M, Békési A and Vértessy BG: Detection of uracil
within DNA using a sensitive labeling method for in vitro and
cellular applications. Nucleic Acids Res. 44:e282016. View Article : Google Scholar :
|
|
57
|
Wehr NB and Levine RL: Quantitation of
protein carbonylation by dot blot. Anal Biochem. 423:241–245. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jaffrey SR and Kharas MG: Emerging links
between m6A and misregulated mRNA methylation in cancer. Genome
Med. 9:22017. View Article : Google Scholar :
|
|
59
|
Kwok CT, Marshall AD, Rasko JE and Wong
JJ: Genetic alterations of m6A regulators predict poorer
survival in acute myeloid leukemia. J Hematol Oncol. 10:392017.
View Article : Google Scholar
|
|
60
|
Zhang C, Zhi WI, Lu H, Samanta D, Chen I,
Gabrielson E and Semenza GL: Hypoxia-inducible factors regulate
pluripotency factor expression by ZNF217- and ALKBH5-mediated
modulation of RNA methylation in breast cancer cells. Oncotarget.
7:64527–64542. 2016.PubMed/NCBI
|
|
61
|
Inouye M: The discovery of mRNA
interferases: Implication in bacterial physiology and application
to biotechnology. J Cell Physiol. 209:670–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gerstberger S, Hafner M and Tuschl T: A
census of human RNA-binding proteins. Nat Rev Genet. 15:829–845.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Licatalosi DD, Mele A, Fak JJ, Ule J,
Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et
al: HITS-CLIP yields genome-wide insights into brain alternative
RNA processing. Nature. 456:464–469. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Dubinsky L, Krom BP and Meijler MM:
Diazirine based photoaffinity labeling. Bioorg Med Chem.
20:554–570. 2012. View Article : Google Scholar
|
|
65
|
Kauer JC, Erickson-Viitanen S, Wolfe HR Jr
and DeGrado WF: p-benzoyl-L-phenylalanine, a new photoreactive
amino acid. Photolabeling of calmodulin with a synthetic
calmodulin-binding peptide. J Biol Chem. 261:10695–10700.
1986.PubMed/NCBI
|
|
66
|
Zhu T, Roundtree IA, Wang P, Wang X, Wang
L, Sun C, Tian Y, Li J, He C and Xu Y: Crystal structure of the YTH
domain of YTHDF2 reveals mechanism for recognition of
N6-methyladenosine. Cell Res. 24:1493–1496. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xu C, Wang X, Liu K, Roundtree IA, Tempel
W, Li Y, Lu Z, He C and Min J: Structural basis for selective
binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol.
10:927–929. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N6-methyladenosine-modified RNA. Cell Res. 27:315–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Luo GZ, MacQueen A, Zheng G, Duan H, Dore
LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J and He C: Unique
features of the m6A methylome i. Arabidopsis thaliana Nat Commun.
5:56302014. View Article : Google Scholar
|
|
70
|
Piekna-Przybylska D, Decatur WA and
Fournier MJ: The 3D rRNA modification maps database: With
interactive tools for ribosome analysis. Nucleic Acids Res.
36:D178–D183. 2008. View Article : Google Scholar :
|
|
71
|
Wang Y, Li Y, Toth JI, Petroski MD, Zhang
Z and Zhao JC: N6-methyladenosine modification destabilizes
developmental regulators in embryonic stem cells. Nat Cell Biol.
16:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)a promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)a methyltransferase Mettl3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang
Z, Liu Y, Zhang X, Zhang W and Ye L: HBXIP-elevated
methyltransferase METTL3 promotes the progression of breast cancer
via inhibiting tumor suppressor let-7g. Cancer Lett. 415:11–19.
2018. View Article : Google Scholar
|
|
76
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation
regulates the self-renewal and tumorigenesis of glioblastoma stem
cells. Cell Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Y, Zheng D, Wang F, Xu Y, Yu H and
Zhang H: Expression of demethylase genes, fto and alkbh1, is
associated with prognosis of gastric cancer. Dig Dis Sci. 2019.
View Article : Google Scholar
|
|
78
|
Wang X, Li Z, Kong B, Song C, Cong J, Hou
J and Wang S: Reduced m6A mRNA methylation is correlated
with the progression of human cervical cancer. Oncotarget.
8:98918–98930. 2017.PubMed/NCBI
|
|
79
|
Zhou J, Wang J, Hong B, Ma K, Xie H, Li L,
Zhang K, Zhou B, Cai L and Gong K: Gene signatures and prognostic
values of m6A regulators in clear cell renal cell carcinoma-a
retrospective study using TCGA database. Aging (Albany NY).
11:1633–1647. 2019. View Article : Google Scholar
|
|
80
|
Chen M, Wei L, Law CT, Tsang FH, Shen J,
Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, et al: RNA
N6-methyladenosine methyltransferase-like 3 promotes liver cancer
progression through YTHDF2-dependent posttranscriptional silencing
of SOCS2. Hepatology. 67:2254–2270. 2018. View Article : Google Scholar
|