Introduction
There is increasing evidence that nociceptive
signals trigger the neuronal excitation of the spinal cord. For
instance, mechanical and cooling stimuli induced by spinal nerve
ligation results in the alteration of spinal 5-hydroxytryptophan
(HT) receptors (1), which are
involved in descending pain facilitation and inhibition (2-4).
There is unequivocal agreement on the important role of
cardiac-spinal cord reflexes in physiological and
pathophysiological conditions (5,6).
Cardiac ischemia induces the release of some chemical mediators
including bradykinin, protons and reactive oxygen species in the
epicardium (7-10), which activate cardiogenic
sympathetic afferents projecting to the upper thoracic spinal cord
(11-14), resulting in the release of
substance P and calcitonin gene-related peptide into the spinal
cord (15-20). Studies on the mechanism of
heart-spinal cord neural crosstalk have given attractive prospects
in recent years (21-24). Accumulating evidence has shown
that the thoracic spinal cord exerts an important role during
myocardial ischemia-reperfusion injury. A case report demonstrated
that spinal cord stimulation effectively alleviated chest pain
caused by myocardial ischemia, which revealed that the spinal cord
mechanism may be involved in cardioprotection (25). Moreover, a study by Southerland
et al (26) found that
isch-emia-reperfusion injury can be alleviated through adrenergic
neurons, resulting in myocardial protection by prior application of
spinal cord stimulation. Jiang et al and Lu et al
(27-29) found that pretreatment with
intrathecal opioids attenuated myocardial ischemia-reperfusion
injury, which may be associated with nitric oxide synthase
activation. Myocardial reperfusion injury can be attenuated by
ischemic preconditioning (IPC) (30). Using functional MRI, Huang et
al (31) revealed that the
nociceptive-related neuronal activity of the spinal dorsal horn was
decreased in the IPC group. Therefore, demonstrating the mechanisms
between heart and spinal cord has become a focal point that
deserves further study. However, there are still many challenges
remaining for systemic clarification of the spinal mechanisms after
myocardial ischemia-reperfusion injury, as a number of underlying
details still remain poorly understood.
Rapid advancements in high-throughput technologies
and computational frameworks offer an excellent opportunity to
quantify spinal nociception using neuronal activation induced by
noxious stimuli. The author's previous study showed that
transcriptomics and metabolomics enable the examination of spinal
biological systems in unprecedented detail (32-36). More recently, different patterns
were revealed in the metabolic and transcriptional levels of the
thoracic spinal cord under myocardial ischemia-reperfusion injury
(37-39). Variations in metabolomics and
transcriptomics are closely related to proteomics. This study was
designed to further explore the differentially expressed proteins
in the thoracic spinal cord after myocardial ischemia-reperfusion
injury. Up to now, proteomics has been shown to be able to robustly
detect various proteins with diverse biological functions in the
brain and spinal cord (40,41), offering new clues for central
molecular mechanisms with greater spatial and temporal coverage. In
this study, the spinal cord proteomes were systematically analyzed
after myocardial ischemia-reperfusion (IR) injury, attempting to
identify the proteins involved in the processes.
Materials and methods
Animals
A total of 30 adult male Sprague-Dawley rats
(250-300 g) were provided by the Experimental Animal Center of
Tongji Medical College, Huazhong University of Science and
Technology. All surgical and experimental procedures were performed
according to the guidelines of the Huazhong University of Science
and Technology Guide for the Care and Use of Laboratory Animals
(TJ-A20150804). The rats were maintained and habituated under
controlled conditions (12-h light-dark cycles, 22°C±0.5°C, relative
humidity, 40-60%, with free access to food and drinking).
Myocardial IR injury
Rats were randomly divided into sham and model
groups (n=9 in each group). To induce myocardial IR injury, a
previously reported procedure was followed (37,42). rats were anesthetized with
pentobarbital sodium (30 mg/kg, intraperitoneal). Before
intratracheal intubation, rats were maintained with 2% isoflurane
in 100% oxygen in an anesthetic chamber until losing righting
reflex. After intubation, continuous 2% isoflurane in 100% oxygen
were given and a small animal ventilator (tidal volume at 2.5
ml/100 g and a respiration rate 80/min) was used to control the
respiration of the animal during the surgical procedure. The chest
was opened via the third intercostal space, then the left anterior
descending artery (LAD) was ligated with 6-0 silk suture via a
silicon tube. A paleness in the appearance in the ischemic
myocardium was one proof of a successful LAD ligation. After 30-min
ischemia, the ligation was released and the silicon tube was
removed, then the reperfusion for 2 h was initiated. Sham rats were
operated as the model group but without LAD ligation. The serum
cardiac troponin I (cTnI) concentration was used to detect
myocardial injury and values are presented as the mean ± standard
error of the mean (SEM; n=4 rats per group, Mann-Whitney U
test).
Extraction and digestion of spinal cord
proteins
According to Hickman's method (43), rats were decapitated before tissue
harvest, which was considered a common physical method for
euthanasia. Normally, pulse oxygen saturation, respiration rate or
heartbeats were used to verify death, prior to tissue collections.
Spinal cord segments between T1 and T4 were obtained from the
operated spinal cord area of the IR rats and sham rats. Tissues
were washed in PBS, directly frozen in liquid nitrogen and stored
at −80°C until further use.
For proteomics experiments, tissues from each rat
were homogenized individually with a douncer in RIPA buffer
containing various protease inhibitors (25 mM Tris-HCl pH 7.6, 150
mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM NaF, 1 mM
Na3VO4, with an added protease inhibitor
cocktail mixture, COMPLETE; Roche Applied Science). The homogenates
were held at 4°C for 30 min and then centrifuged at 12,000 × g for
30 min at 4°C. The supernatants were collected for the following
in-solution digestion.
The in-solution digestion was performed as described
previously with minor modifications (40). Briefly, proteins were precipitated
and re-suspended with 8 M urea/4 mM CaCl2/0.2 M
Tris-HCl, pH 8.0, and then reduced with 10 mM DTT at 37°C for 30
min and alkylated with 40 mM iodoacet-amide in the dark for 30 min.
The resultant proteins were digested with trypsin at a ratio of
1:50 (trypsin/protein w/w) at 37°C for overnight and then desalted
using a SepPak C18 cartridge (Waters Corporation) and dried with a
SpeedVac.
Stable isotope dimethyl labeling and
strong cation exchange (SCX) fractionation
Desalted peptides were re-suspended in 0.1 M sodium
acetate, pH 6.0. Next, 4% formaldehyde (CH2O, which
serves as 'light labeled'), 4% deuterated formaldehyde
(CD2O, which serves as 'Heavy labeled') were added to
the peptides extracted from IR rats and sham rats, respectively.
After mixing, 0.6 M sodium cyanoborohydride (NaBH3CN)
was added and the mixtures were incubated at room temperature
(20±2°C) for 1 h. The samples were then quenched by adding 1%
ammonium hydroxide, followed by the addition of 5% formic acid.
After labeling, the peptides were mixed at a ratio of 1:1 and
desalted prior to separation via SCX chromatography as describe
previously (44).
Liquid chromatography-mass spectrometry
(LC-MS)/MS and data processing
All ESI-based LC-MS/MS experiments were performed on
a TripleTOF 5600+ System coupled with an Ultra 1D Plus nano-liquid
chromatography device (SCIEX) as previous reported (45). Dried peptides were dissolved in
0.1% formic acid, 2% acetonitrile and 98% H2O.
Subsequently, samples were loaded onto a C18 trap column (5 µm;
5x0.3 mm; Agilent Technologies, Inc.) at a flow rate of 5 µl/min
and eluted from the trap column over the C18 analytic column (75 µm
x150 mm; 3 µm particle size, 100 Å pore size; Eksigent
Technologies) at a flow rate of 300 nl/min using a 100 min
gradient. The mobile phase consisted of two components: Component A
comprised 3% DMSO and 97% H2O with 0.1% formic acid;
acomponent B contained 3% DMSO and 97% acetonitrile with 0.1%
formic acid. Data was acquired using a spray voltage of 2.3 kV,
curtain gas of 20 psi and an interface heater temperature of 150°C.
The information dependent acquisition mode was used to acquire mass
spectrometric data. Each scan cycle consisted of one full-scan mass
spectrum (with m/z ranging from 350-1,500; ion accumulation time,
250 msec) followed by 40 MS/MS events (m/z ranging from 100-1,500;
ion accumulation time, 50 msec). The threshold for MS/MS
acquisition activation was set to 120 cps for +2-+5 precursors.
Former target ion exclusion was set for 18 sec. The generated raw
MS spectra were analyzed with ProteinPilot 4.5 software (SCIEX)
using the Paragon algorithm. The uniprot database [Rattus
norvegicus (Rat), UP000002494] was used. The false discovery
rates of the peptide-spectra matches determined by a decoy database
search were set to 1.0% for all experimental datasets. Proteins
were considered to be successfully identified when at least two
correct assigned peptides (95% confidence) were obtained.
Reverse transcription-quantitative
PCR
Total RNA from T1-4 segments of the spinal cord
tissue was extracted using TRIzol reagent (Invitrogen, Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA samples were quantified using a spectrophotometer
(BioPhotometer; Eppendorf) and then synthesized to cDNA via reverse
transcription using the PrimeScript™ RT reagent kit (Takara Bio,
Inc.). The temperature protocol of reverse transcription was as
follows: 15 min at 37°C, 5 sec at 85 and 4°C. cDNA was quantified
by quantitative PCR using SYBR-Green Master Mix (Takara Bio, Inc.).
The thermocycling conditions for PCR were as follows: 30 sec at
95°C, followed by 40 cycles of 15 sec at 95°C, 15 sec at 60°C and
45 sec at 72°C. Relative gene expression was determined by
normalization to GAPDH using comparative (2-ΔΔCq) method
(46). The specific forward and
reverse primer sequences (Table
I) were designed. The experiments were performed in triplicate.
Data are expressed as the mean ± SEM, Mann-Whitney U test compared
with the sham group.
 | Table IPrimer sequences utilized for reverse
transcription-quantitative PCR. |
Table I
Primer sequences utilized for reverse
transcription-quantitative PCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| SOS1 |
GCTCGCCATTACATCTCCAACCTC |
CTTCCTGTGTCAGTGGTGGTGATG |
| Rptor |
TGTCCTGGTCTTCCTGCCTGTG |
AGCGTAGTCTCTGAACCTGGTGAG |
| Washc2c |
TGCTGTCTAACACCCAGTTCATT |
CGCTCATTGGCATCTTCCTCT |
| Fry |
CCACCCTTCTATCGGTTCACA |
GGGCTCGGATCACGTTGCT |
| Decorin |
ACAACCATGAAGGCAACTCTCGTC |
GAACACTGCACCACTCGGAGATG |
| Coro2b |
GTGTCTGCTTGCGAGGTCTTCC |
GCTCTGTGCCTGGCGTCATG |
| Gphn |
AGTAAAAGATGGCTATGCTGTTCG |
CCCGCATCACTTGTCCG |
| Aspn |
AACCAAAGAGCCAGTGAACCC |
AAGGTCAACCATTCGAGTATCAA |
| Lamc1 |
CTCCATGAAGCAACAGATTACCC |
ACTGCCCTCCATACCCCAC |
Western blotting
Total protein was extracted from T1-4 spinal cord
tissues with ice-cold RIPA lysis buffer (Wuhan Boster Biological
Technology, Ltd.). Protein concentrations were determined using the
bicinchoninic Protein Assay kit (Wuhan Boster Biological
Technology, Ltd.). A total of 30 µg protein samples were separated
by SDS-PAGE (5% stacking gel and 10% separating gel) and
transferred onto PVDF membranes (EMD Millipore). Then the membrane
was blocked in 5% skimmed milk in 0.1% TBST for 1.5 h at room
temperature and then incubated overnight at 4°C with the following
primary antibodies: SOS1 (1:500; cat. no. A3272; ABclonal Biotech
Co., Ltd.), Coro2b (1:1,000; A16670; ABclonal Biotech Co., Ltd.),
Gephyrin (Gphn; 1:500; cat. no. A8572; ABclonal Biotech Co., Ltd.),
Rptor (1:200; cat. no. 20984-1-AP; ProteinTech Group, Inc.) and
Decorin (1:1,000; cat. no. 14667-1-AP; ProteinTech Group, Inc.).
After incubation with horseradish peroxidase-conjugated goat
anti-rabbit secondary antibodies (cat. no. A5014; ABclonal Biotech
Co., Ltd.) at 1:5,000 concentration for 1.5 h at room temperature.
This was followed by protein detection using the ECL Western
Blotting Substrate (Thermo Fisher Scientific, Inc.). The mean
intensities of selected areas and the areas of these images were
calculated using ImageJ software (v.1.8.0_112; National Institutes
of Health) and normalized to values of GADPH (1:5,000;
ProteinTech). Values were expressed as means ± SEM, Mann-Whitney
test compared to the sham group.
Myocardial tissue staining
Each myocardial tissue sample was cut transversely.
Hematoxylin and eosin (H&E) staining was used to observe
myocardial pathology. After deparaf-finization, 4 µm thick sections
were immersed in hematoxylin (cat. no. H9627; Sigma-Aldrich; Merck
KGaA) for 5-7 min at room temperature, differentiated in 1% acid
alcohol for 2-5 sec and stained with 0.5% eosin (cat. no. 71014544;
Sinopharm Chemical Reagent Co., Ltd.) for 2 min at room
temperature. After rinsing with distilled water for 30 sec,
sections were dehydrated with graded alcohol and cleared in xylene.
Infarct size was assessed by TTC staining 2 h after reperfusion.
After surgery, the hearts were removed and frozen for 20 min at
-20°C, then transversally cut into sections with thickness of ~1 to
2 mm. Tissue sections were incubated in 2% TTC for 10 min in dark
conditions at 37°C and then fixed in 10% formaldehyde overnight at
4°C. The infarct area was white, while the normal tissues were
red.
Bioinformatics
Statistical analysis of the MS results derived from
all the experimental groups was performed using Microsoft Office
Excel. Pearson correlation analyses was performed to assess the
correlation among experimental replicates. Biological Networks Gene
Ontology (BiNGO) 3.03 (http://apps.cytoscape.org/apps/bingo) was used to
calculate the gene ontology term enrichment of differentia
proteins. The analyses were conducted using the default BiNGO
Rattus norvegicus database. For protein interaction network
analysis, the Uniprot functional annotations (http://www.uniprot.org/uniprot) were used to
classify the proteins into several clusters. Proteins matched in
any cluster of pathways were extracted and submitted to the Search
Tool for the Retrieval of Interacting Genes/Proteins (STRING 9.0;
https://string-db.org/) to qualify the physical
and functional interactions. The proteins and their interactions
were then uploaded to Cytoscape (www.cytoscape.org/) for data visualization.
Results
Validation of IR-induced myocardial
injury in rats
The histopathological and functional validation
analysis was performed to ensure that the IR rats used for the
experiments showed acute myocardial failure. It was observed that
30 min of ischemia resulted in a pale appearance of the ischemia
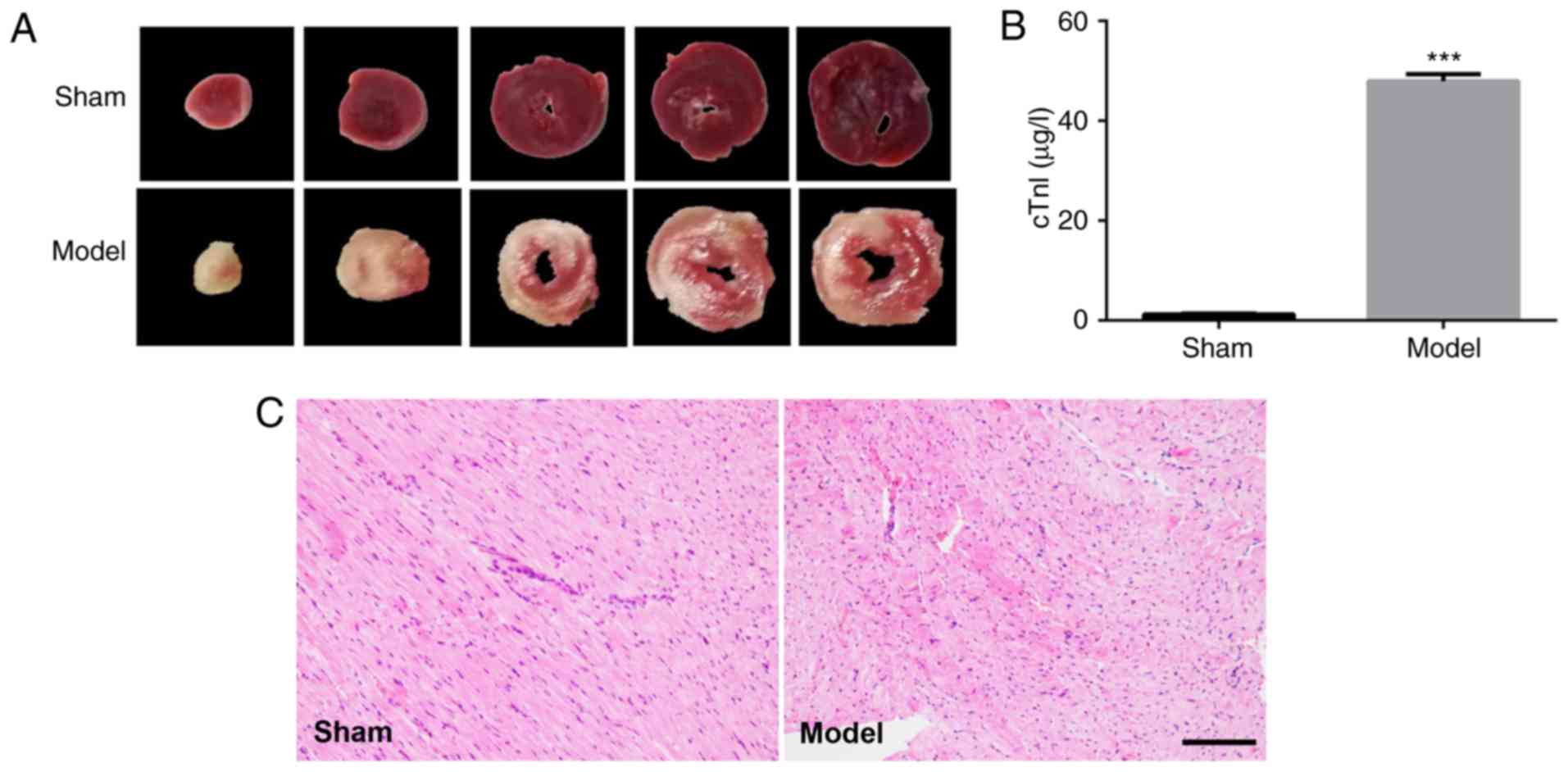
myocardium (Fig. 1A). Serum
cardiac troponin cTnI increased significantly (1.211±0.1815 µg/l in
sham group, n=4; 47.94±1.368 µg/l, n=4; P<0.001; Fig. 1B). Also, the IR rats showed
damaged myocardial cells in the H&E stained heart sections
(Fig. 1C). These results verified
IR induced myocardial injury in the rats in the study.
Proteomics data overview
To investigate the mechanisms in the spinal cord
that respond to myocardial injury, the present study employed a
comparative quantitative proteomics approach, based on the stable
isotope dimethy labeling strategy, to study the alterations in the
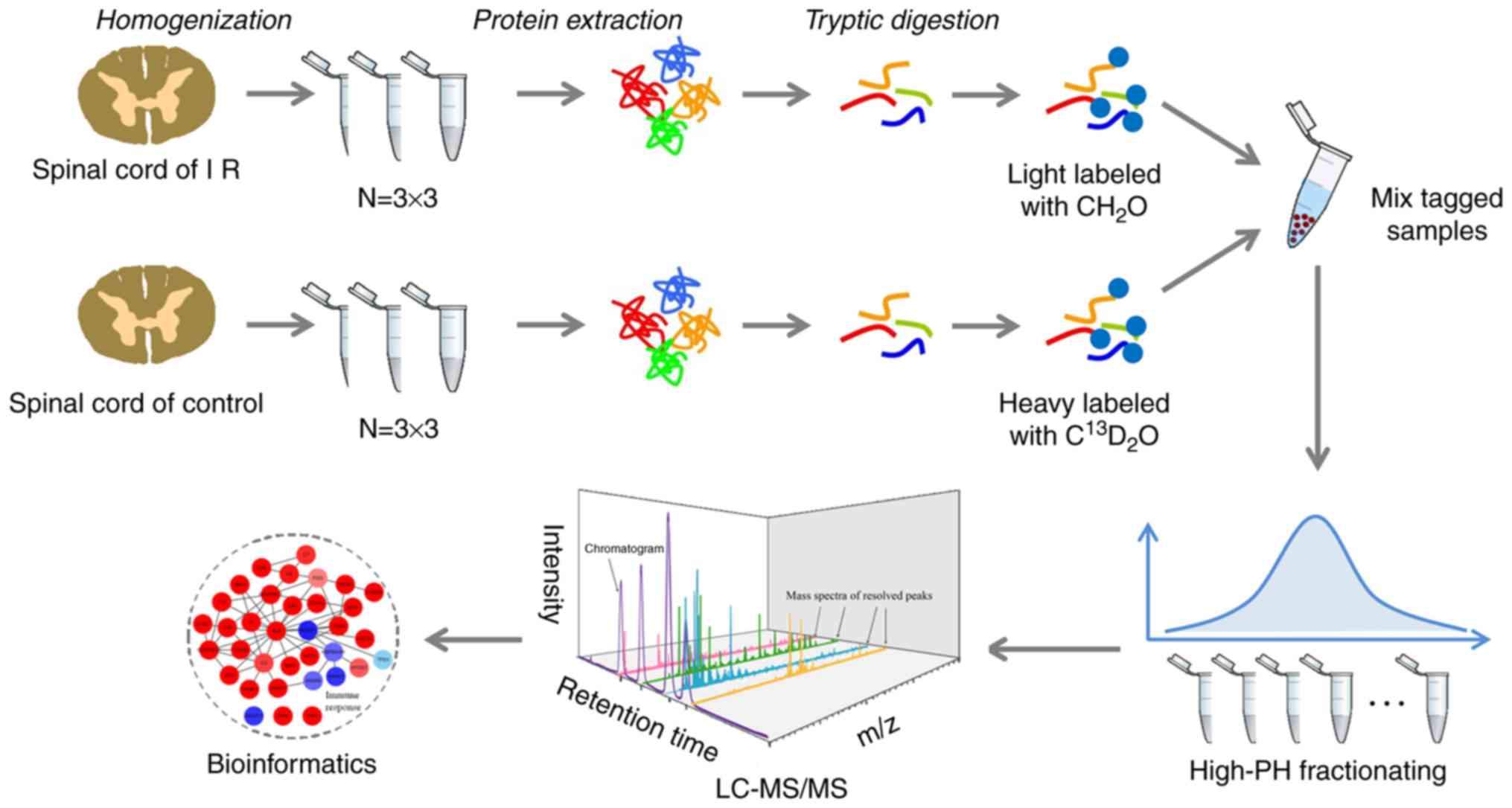
spinal cord proteome after acute myocardial IR injury in rats. An
experimental flowchart including tissue homogenization, protein
extraction, trypsin digestion, chemical labeling and LC-MS/MS
processing is shown in Fig. 2.
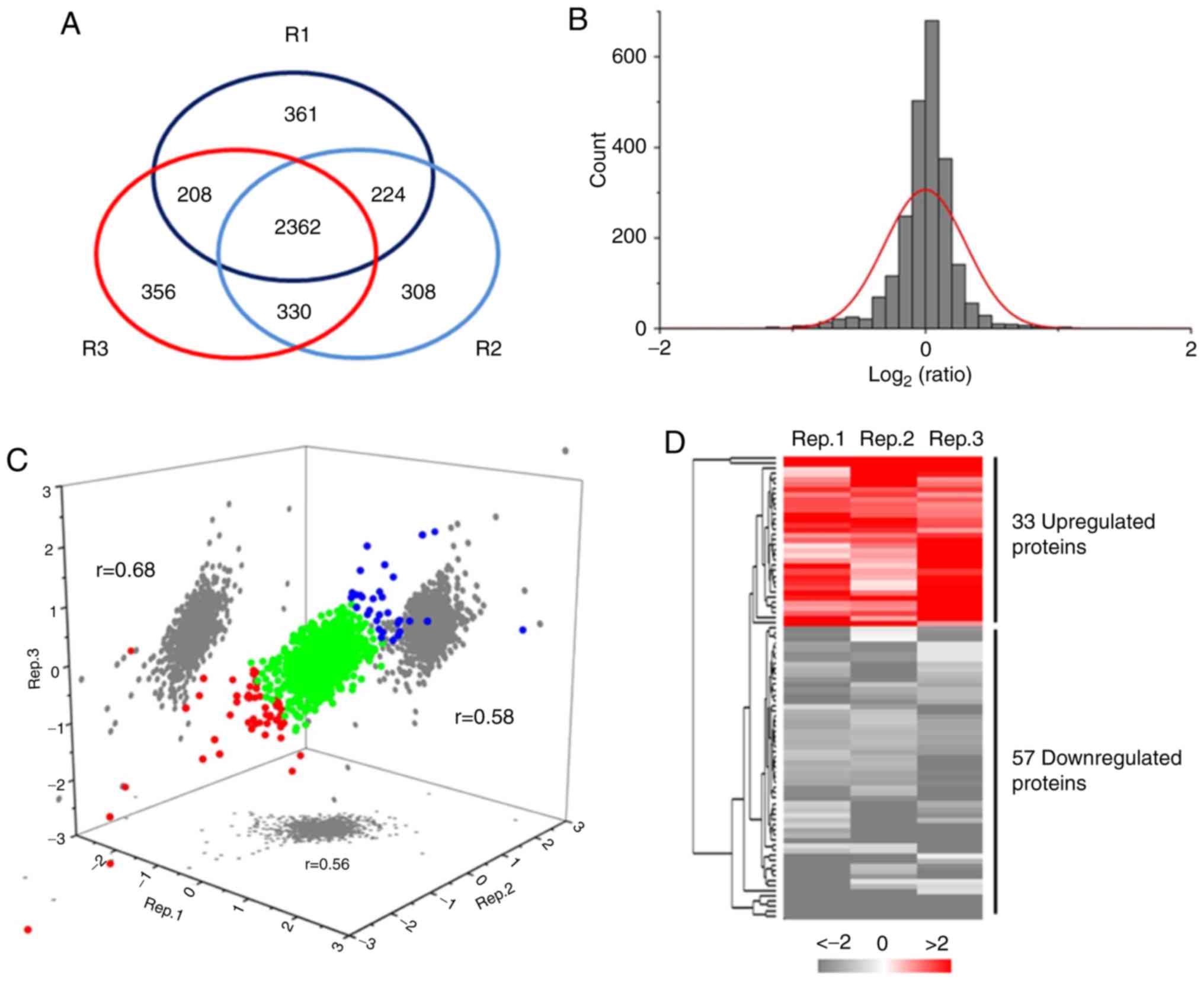
Among the three biological replicates, the present study quantified
4,149 proteins in total, in which 2,362 proteins were shared
(Fig. 3A). The gaussian
distribution of the shared quantitative data [as log2 (Ratio)] was
further analyzed, showing a reasonable ratio distribution (Fig. 3B). The Pearson correlation
coefficient analysis showed good repetitiveness among the three
biological replicates, as demonstrated by r=0.68, 0.58 and
0.56 (Fig. 3C). All the
quantified proteins are summarized in Table SI.
A total of 90 proteins are differentially
expressed in the spinal cord after IR injury
To identify the differentially expressed proteins in
the spinal cord that are regulated by myocardial injury, a
filtering rule was set as previously reported (41,45), to screen those differentially
expressed proteins: Proteins with 50% change of ratio in each
replicate, or P<0.05 in each replicate and additionally 50%
change of average ratio, were considered to be differentially
expressed. Among the whole quantitative dataset, 33 spinal cord
proteins were found to be upregulated in IR rats, whereas 57
proteins were downregu-lated (Fig.
3C and D). All the differentially expressed proteins are
summarized in Table SII.
To ascertain the functional representation of these
differentially expressed proteins, the data were further analyzed
based on the gene ontology term enrichment (Fig. S1). The present study found the
terms of biological process including axon projection,
neurofilaments assembly, microtubule based transport, amino acid
metabolism and cell adhesion were enriched among the differentia
proteins (Fig. S1A). Molecular
function terms including NADPH activity and protein binding and
cellular component terms including spiceosomal complex, endocytic
vesicle and neurofilament were enriched. These results indicate
diverse cellular functions were involved in the spinal cord after
myocardial injury.
Protein network regulated by IR
injury
Considering the possible regulatory network that
exists among the differentially expressed proteins, the present
study performed protein interaction network based on the STRING
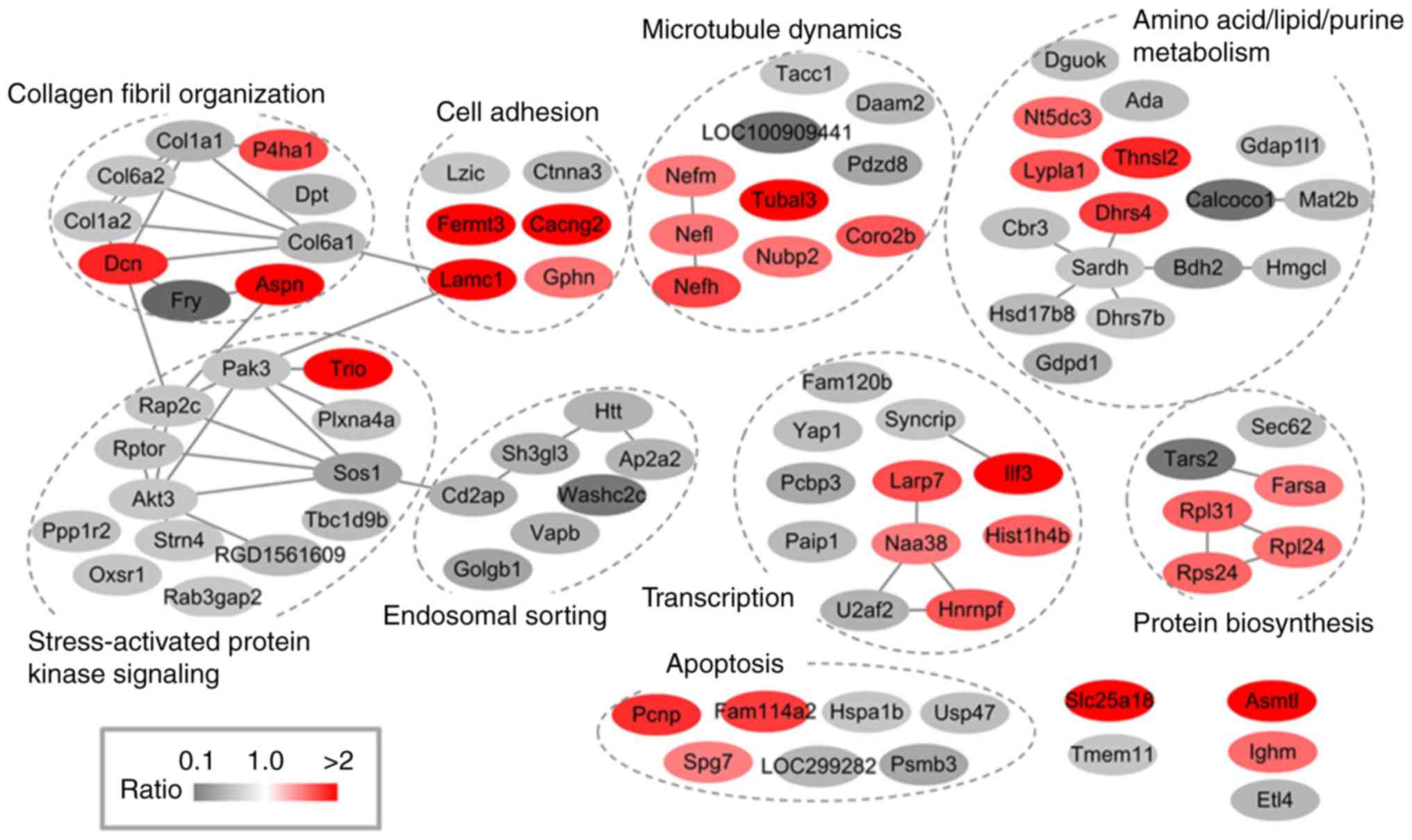
database and Uniprot annotations. As shown in Fig. 4, the differentially expressed
proteins constitute multiple functional clusters, including
collagen fibril organization, cell junction, microtubule dynamics,
amino acid/lipid/purine metabolism, stress-activated protein kinase
signaling, endosomal sorting, transcription, protein biosynthesis
and apoptosis. Several co-regulations possibly exist among these
clusters, as the present study found proteins of stress-activated
protein kinase signaling and endosomal sorting showed a consistent
down-regulatory tendency in IR rats. While upregulatory proteins
were mainly found in clusters of cell junction, microtubule
dynamics, transcription and protein biosynthesis. These results
indicate a complicated response to myocardial injury in the spinal
cord.
Alternative evidence supporting the MS
findings
To cross-check the reliability of quantitative
proteomics, several protein expression levels were further assessed
using conventional western blotting and quantitative PCR. A total
of five proteins involved in stress-activated protein kinase
signaling (Sos1 and Rptor), cell junction (Gphn), collagen fibril
organization [Dcn (Decorin)] and microtubule dynamics [Coro2b
(Corin2b)], in the regulated protein network, were selected for
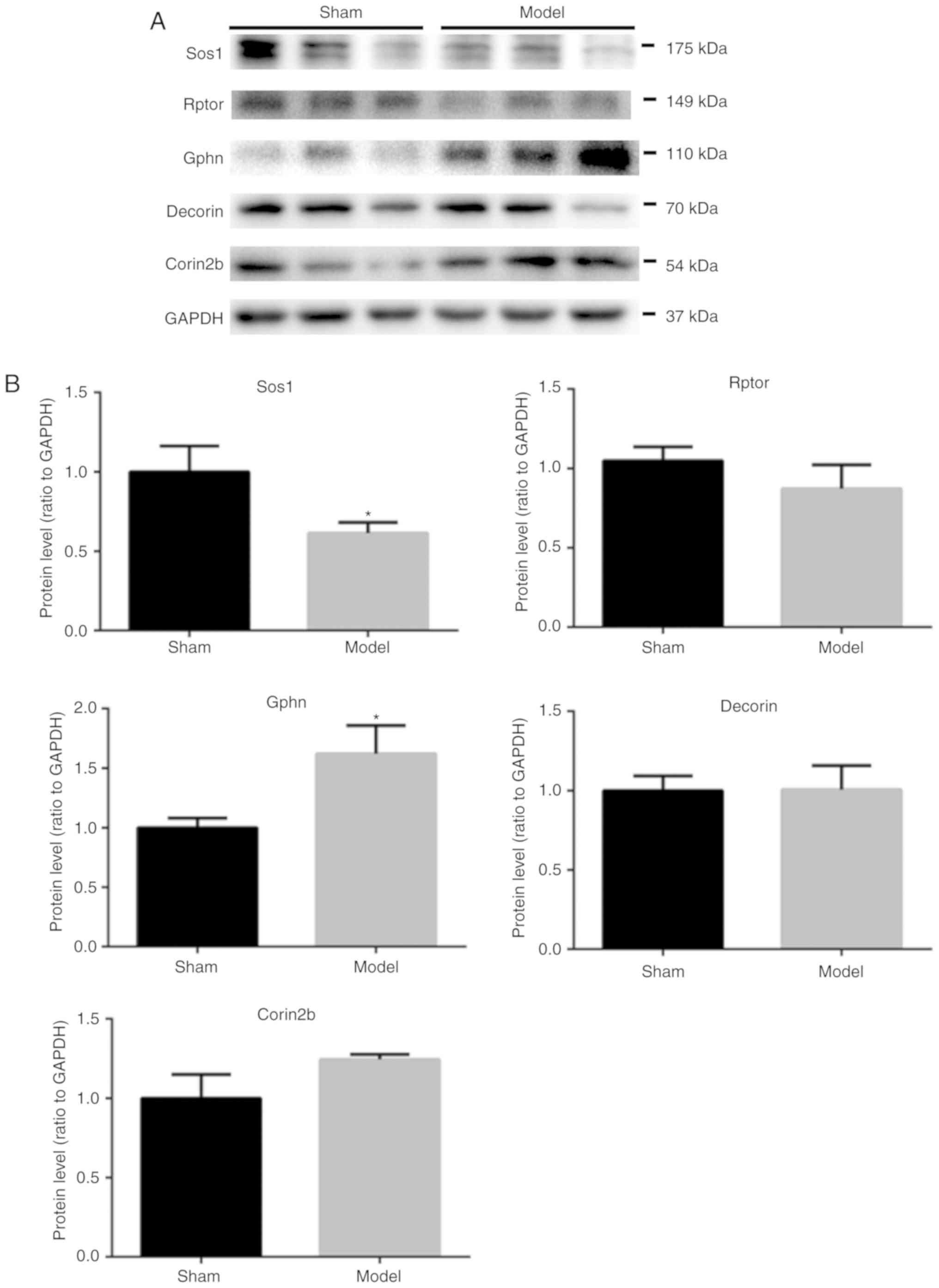
western blot analysis (Fig. 5).
The relative gray value compared with GAPDH of Sos1, Rptor, Gphn,
Dcn and Corin2b were 0.6136±0.06747 (n=6), 0.8721±0.1504 (n=4),
1.622±0.2363 (n=5), 1.006±0.1513 (n=5) and 1.245±0.03118 (n=5),
respectively. The authors found most of the analyzed proteins were
consistent with the MS results except the protein Dcn, which almost
did not change in the western blotting results.
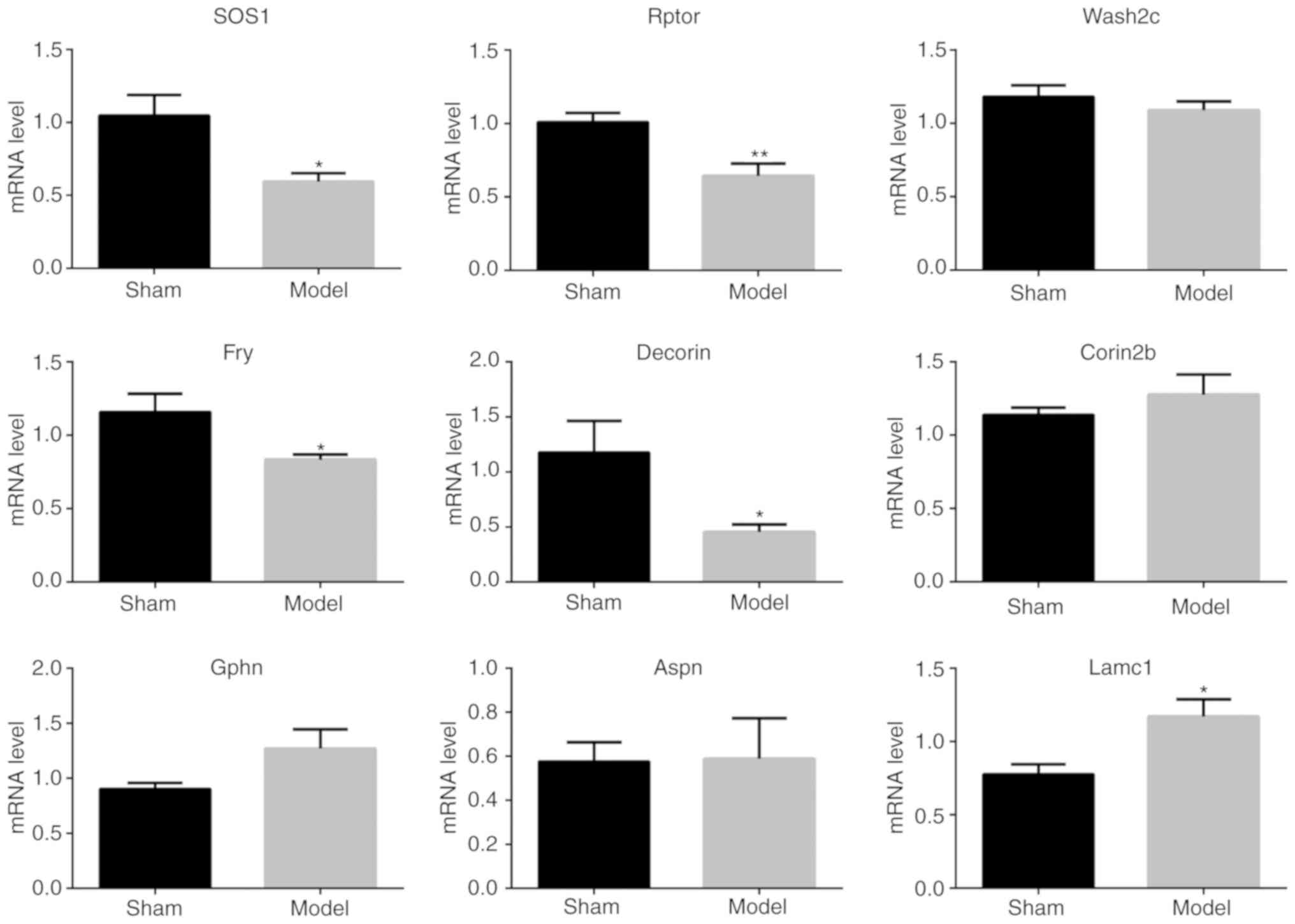
Moreover, these five genes and another four genes
(Wash2c, Fry, Aspn and Lamc1) were further analyzed by quantitative
PCR. As shown in Fig. 6, the
results showed that most of the quantitative ratios were consistent
with the MS results. These results provide alternative evidences
supporting the MS findings.
Discussion
The thoracic spinal cord plays an important role in
the regulation of myocardial ischemia-reperfusion injury. By
exploring the altered characteristics at three levels
(metabolomics, tran-scriptomics and proteomics) in the thoracic
spinal cord under myocardial IR injury, the mechanism of
heart-spinal cord neural crosstalk can be further elucidated, which
provides a new perspective for clinical intervention alleviating
myocardial IR injury by spinal nerve mechanisms in the future.
The present study attempted to systematically
profile the proteome-wide alterations of the spinal cord after
myocardial IR injury by quantitative proteomics and further
analyzed the possible regulatory mechanisms among the various
pathways, and protein-interaction networks. The myocardial IR
injury rat model first established by cross clamping the LAD for
30-min ischemia and followed by reperfusion for 2 h, as reported
previously (47,48), which showed a significant
histopathological and functional myocardial injury. Then using the
stable isotope dimethyl labeling quantitative proteomics strategy,
a total of 2,362 shared proteins with a good distribution and
correlation were successfully quantified. Among these proteins, 33
were identified to be upregulated and 57 downregulated proteins in
the spinal cord after myocardial IR injury, which are involved in
various biological processes, molecular function and cellular
components. Based on these proteins, the spinal cord protein
interaction network regulated by IR injury including apoptosis,
microtubule dynamics, stress-activated signaling and cellular
metabolism was further established. The cellular protein networks
and pathways in spinal cord associated with the myocardial I/R
injury are discussed in more detail bellow.
Spinal cord injury especially in the second phase
induces cell death, which occurs via apoptosis and autophagy
(49,50). As evolutionarily conserved
catabolic processes, apoptosis and neuronal autophagy remove those
unwanted cytosolic proteins and damaged organelles through the
autophagosome/lysosomal pathway (49). The phosphoinositide 3-kinase
(PI3K)/protein kinase B (Akt)/mammalian target of rapamycin (mTOR)
signaling is reported to play important roles after neuronal injury
and can induce apoptosis through the mitochondrial pathway
(51,52).
In the present study, proteins of the GTPase that
regulates signaling in the spinal cord were identified after
myocardial IR injury, including Rap2c, Akt3, Rptor, Sos1 and Pak3
and Strn4, which were all found to be consistently decreased.
Previous studies have reported that Rap2c expression was lower in
SCII and under hypoxic conditions, and the over-expression of Rap2c
could reverse the cell apoptosis induced by hypoxia (53). Akt3 proteins were reported to be
degraded as early as 1 h after stroke during brain injury (54). Rptor, as an associated regulatory
protein of mTOR complex 1, was found to be decreased in the present
study, implying a possible downregulation of mTOR, as suggested by
a previous study (55).
Cerebral ischemia induced a cascade of events that
may disrupt membrane trafficking pathways including Golgi
apparatus-late endosome-lysosome axis, which are important for
supplying lysosomal enzymes for cellular apoptosis and autophagy
processes (56). Massive buildup
of damaged Golgi, transport vesicles and late endosomes takes place
over time in neurons destined to die after transient cerebral
ischemia or cardiac arrest (57,58).
In the present study, several membrane trafficking
related proteins, including Cd2ap, endophilin-A3 (Sh3gl3),
huntingtin (Htt), Ap2a3, Wahc2c and Golgb1, were found to all
decrease after myocardial IR injury. Cd2ap serves as an adaptor
protein and participates in cellular apoptosis via the PI3K/Akt
signaling pathway (59). Htt
functions as a scaffold for selective macroautophagy (60) and was found to be degraded after
ischemic injury, it interacts with Sh3gl3 to function in
microtubule-based endocytotic processes (61,62). Additionally, cerebral ischemia was
reported to induce mitochondrial membrane permeabilization
(56). The present study
identified an upregulated protein Spg7, with proteolytic activity
involved in the formation and regulation of the mitochondrial
permeability transition pore, which was also reported to be
regulated during cerebral ischemic injury and treatment (63), and the abnormal expression of
which could cause paraplegias (64).
Moreover, microtubule dynamics play important roles
in autophagy and apoptotic processes after spinal cord injury,
including intracellular trafficking and extracellular matrix
remodeling (65,66). The present study found the spinal
cord neurofilament proteins Nefl, Nefm, Nefh and protein Tubal3,
Nubp2 and Coro2a were upregulated after myocardial IR injury. These
results suggested that the microtubule mediated intracellular
trafficking in spinal cord were regulated by myocardial IR
injury.
After neuron injury, the extracellular matrix (ECM)
is degraded and the composition changes, and some molecules become
aberrantly expressed or cleaved into bioactive fragments (67). In this study, the collagen type I
Col1a1, Col1a2 and collagen type VI Col6a1, Col6a2 were found to be
decreased, and the laminin subunit 1 Lamc1 was increased. The
prolyl-4-hydroxylase P4ha1, a key cellular oxygen sensor and a
regulatory enzyme in the maturation of collagens (68), was found to be increased.
Moreover, the collagen binding protein Dcn and Aspn both were
increased. Dcn is a proteoglycan constituent of the ECM reported to
possess powerful anti-inflammation properties in cardiovascular
diseases (69). These results
suggested that a possible degradation of the collagen component in
ECM of the spinal cord and the collagen metabolism may be involved
in the regulation in the spinal cord in response to myocardial IR
injury in rats.
Additionally, two neuronal receptor regulatory
proteins Cacng2 and Gphn were identified that were both upregulated
in the spinal cord of myocardial IR injured rats. Cacng2, also
called as stagazin or TRAP-2, is required for inflammation
associated AMPA receptor plasticity and involved in nociception
within the lamina of the spinal cord (70). Gphn is a scaffold protein
responsible for the traffic and synaptic anchoring of
GABAA receptors (71).
These results possibly implied special roles for the spinal cord
neuronal receptor during myocardial IR injury.
Cellular metabolism including amino acid and lipid
metabolism, and protein biosynthesis were also found to be involved
in the regulation in the spinal cord after myocardial IR injury.
The present study demonstrated that sarcosine dehydrogenase and
several of its interacting proteins (Bdh2, Dhrs7b, Hsd17b8 and
Cbr3) were downregulated after myocardial IR injury. Moreover, it
was also found that some transcription related proteins are
regulated in the spinal cord response to myocardial IR injury,
including the increased Naa38, larp7 and Hnrnpf. However, the
detailed mechanism of these proteins mostly remain rarely
investigated.
In conclusion the present study investigated the
alterations in the spinal cord that respond to myocardial IR injury
in rats using a quantitative proteomics approach. A total of nine
differentially expressed spinal cord proteins were identified in
the myocardial IR injured rats and the regulated protein networks
were further established. The results demonstrated that myocardial
IR injury induced regulation of various biological processes in the
spinal cord including apoptosis and autophagy, cellular membrane
trafficking, extracellular matrix remodeling and some other related
biological processes. The present study may provide basis for
further detailed clarification of the spinal cord regulatory
mechanisms in response to myocardial IR injury.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of P.R. China (grant nos. 81670240 and
81873467 to HX), the National Natural Science Foundation of Hubei
Province (grant no. 2016CFB625 to HX), the Medical innovation
project in Fujian Province (grant no. 2017-CX-48 to SL) and the Key
Research and Development Project of Hainan Province of China (grant
no. ZDYF2018115 to DW).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HBX and HLZ conceived and designed the study. SYL,
QW and ZXL performed the surgical procedures. YJL, QY and ZGH
participated in the experimental design. SYL, QY and ZXL performed
the experiments. DZW and SYL analyzed the data. HBX and HLZ wrote
the manuscript and all authors contributed to the final manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethical Committee of Tongji Hospital, Tongji Medical College,
Huazhong University of Science and Technology (grant no.
TJ-A20150804).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rahman W, Suzuki R, Webber M, Hunt SP and
Dickenson AH: Depletion of endogenous spinal 5-HT attenuates the
behavioural hypersensitivity to mechanical and cooling stimuli
induced by spinal nerve ligation. Pain. 123:264–274. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dogrul A, Ossipov MH and Porreca F:
Differential mediation of descending pain facilitation and
inhibition by spinal 5HT-3 and 5HT-7 receptors. Brain Res.
1280:52–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Suzuki R, Rygh LJ and Dickenson AH: Bad
news from the brain: Descending 5-HT pathways that control spinal
pain processing. Trends Pharmacol Sci. 25:613–617. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sasaki M, Obata H, Kawahara K, Saito S and
Goto F: Peripheral 5-HT2A receptor antagonism attenuates primary
thermal hyperalgesia and secondary mechanical allodynia after
thermal injury in rats. Pain. 122:130–136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kember G, Ardell JL, Shivkumar K and
Armour JA: Recurrent myocardial infarction: Mechanisms of
free-floating adaptation and autonomic derangement in networked
cardiac neural control. PLoS One. 12:pp. e01801942017, View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kember G, Armour JA and Zamir M: Neural
control hierarchy of the heart has not evolved to deal with
myocardial ischemia. Physiol Genomics. 45:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pan HL, Chen SR, Scicli GM and Carretero
OA: Cardiac interstitial bradykinin release during ischemia is
enhanced by ischemic preconditioning. Am J Physiol Heart Circ
Physiol. 279:H116–H121. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan HL, Longhurst JC, Eisenach JC and Chen
SR: Role of protons in activation of cardiac sympathetic C-fibre
afferents during ischemia in cats. J Physiol. 518:857–866. 1999.
View Article : Google Scholar
|
|
9
|
Wolfrum S, Nienstedt J, Heidbreder M,
Schneider K, Dominiak P and Dendorfer A: Calcitonin gene related
peptide mediates cardioprotection by remote preconditioning. Regul
Pept. 127:217–224. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Szallasi A and Blumberg PM: Vanilloid
(Capsaicin) receptors and mechanisms. Pharmacol Rev. 51:159–212.
1999.PubMed/NCBI
|
|
11
|
Blair RW, Weber RN and Foreman RD:
Responses of thoracic spinothalamic neurons to intracardiac
injection of bradykinin in the monkey. Circ Res. 51:83–94. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuo DC, Oravitz JJ and Degroat WC: Tracing
of afferent and efferent pathways in the left inferior cardiac
nerve of the cat using retrograde and transganglionic transport of
horseradish peroxidase. Brain Res. 321:111–118. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
White JC: Cardiac pain: Anatomic pathways
and physiologic mechanisms. Circulation. 16:644–655. 1957.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pan HL and Chen SR: Myocardial ischemia
recruits mechanically insensitive cardiac sympathetic afferents in
cats. J Neurophysiol. 87:660–668. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding X, Ardell JL, Hua F, McAuley RJ,
Sutherly K, Daniel JJ and Williams CA: Modulation of cardiac
ischemia-sensitive afferent neuron signaling by preemptive C2
spinal cord stimulation: Effect on substance P release from rat
spinal cord. Am J Physiol Regul Integr Comp Physiol. 294:R93–R101.
2008. View Article : Google Scholar
|
|
16
|
Foreman RD: Mechanisms of cardiac pain.
Annu Rev Physiol. 61:143–167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hua F, Ardell JL and Williams CA: Left
vagal stimulation induces dynorphin release and suppresses
substance P release from the rat thoracic spinal cord during
cardiac ischemia. Am J Physiol Regul Integr Comp Physiol.
287:R1468–R1477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Steagall RJ, Sipe AL, Williams CA, Joyner
WL and Singh K: Substance P release in response to cardiac ischemia
from rat thoracic spinal dorsal horn is mediated by TRPV1.
Neuroscience. 214:106–119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kang YM, Yang ZM, Ma Y, Lei JH, Yan N, Su
YK and Francis J: TNF-alpha contributes to cardiac nociception in
myocardial infarction. Circulation (81st Annual Scientific Session
of the American-Heart-Association). 118:pp. S296. 2008
|
|
20
|
Ding X, Mountain DJ, Subramanian V, Singh
K and Williams CA: The effect of high cervical spinal cord
stimulation on the expression of SP, NK-1 and TRPV1 mRNAs during
cardiac ischemia in rat. Neurosci Lett. 424:139–144. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gourine A and Gourine AV: Neural
mechanisms of cardioprotection. Physiology (Bethesda). 29:133–140.
2014.
|
|
22
|
Evonuk KS, Prabhu SD, Young ME and DeSilva
TM: Myocardial ischemia/reperfusion impairs neurogenesis and
hippocampal-dependent learning and memory. Brain Behav Immun.
61:266–273. 2017. View Article : Google Scholar :
|
|
23
|
Dou M, Ma Z, Cheng X, Zou G, Xu Y, Huang
C, Xiong W, He S and Zhang Y: Intrathecal lentivirus-mediated RNA
interference targeting nerve growth factor attenuates myocardial
ischaemia-reperfusion injury in rat. Br J Anaesth. Aug 2–2019.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan XC, Li ZX, Wu DZ, Li SY, Xiang HB and
Song YT: Mapping changes of whole brain blood flow in rats with
myocardial ischemia/reperfusion injury assessed by positron
emission tomography. Curr Med Sci. 39:653–657. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Singh H, Merry AF, Ruygrok P and Ruttley
A: Treatment of recurrent chest pain in a heart transplant
recipient using spinal cord stimulation. Anaesth Intensive Care.
36:242–244. 2008.PubMed/NCBI
|
|
26
|
Southerland EM, Milhorn DM, Foreman RD,
Linderoth B, DeJongste MJ, Armour JA, Subramanian V, Singh M, Singh
K and Ardell JL: Preemptive, but not reactive, spinal cord
stimulation mitigates transient ischemia-induced myocardial
infarction via cardiac adrenergic neurons. Am J Physiol Heart Circ
Physiol. 292:H311–H317. 2007. View Article : Google Scholar
|
|
27
|
Jiang L, Hu J, He S, Zhang L and Zhang Y:
Spinal neuronal NOS signaling contributes to morphine
cardioprotection in ischemia reperfusion injury in rats. J
Pharmacol Exp Ther. 358:450–456. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu Y, Hu J, Zhang Y and Dong C: Spinal
neuronal NOS activation mediates intrathecal fentanyl
preconditioning induced remote cardioprotection in rats. Int
Immunopharmacol. 19:127–131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu Y, Hu J, Zhang Y, Dong CS and Wong GT:
Remote intra-thecal morphine preconditioning confers
cardioprotection via spinal cord nitric oxide/cyclic guanosine
monophosphate/protein kinase G pathway. J Surg Res. 193:43–51.
2015. View Article : Google Scholar
|
|
30
|
Iwamoto T, Bai XJ and Downey HF:
Preconditioning with supply-demand imbalance limits infarct size in
dog heart. Cardiovasc Res. 27:2071–2076. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang C, Wang J, Wang N, Du F, Xiong W,
Qian J, Zhong K, Cai A, Xu S, Huang J, et al: Effect of myocardial
ischemic preconditioning on ischemia-reperfusion
stimulation-induced activation in rat thoracic spinal cord with
functional MRI. Int J Cardiol. 285:59–64. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu BW, Li ZX, He ZG, Liu C, Xiong J and
Xiang HB: Altered expression of target genes of spinal cord in
different itch models compared with capsaicin assessed by RT-qPCR
validation. Oncotarget. 8:74423–74433. 2017.PubMed/NCBI
|
|
33
|
Chen M, Li ZX, Wang Q and Xiang HB:
Altered expression of differential genes in thoracic spinal cord
involved in experimental cholestatic itch mouse model. Curr Med
Sci. 38:679–683. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Q, Li ZX, Liu BW, He ZG, Liu C, Chen
M, Liu SG, Wu WZ and Xiang HB: Altered expression of differential
gene and lncRNA in the lower thoracic spinal cord on different time
courses of experimental obstructive jaundice model accompanied with
altered peripheral nociception in rats. Oncotarget.
8:106098–106112. 2017.PubMed/NCBI
|
|
35
|
Liu T, He Z, Tian X, Kamal GM, Li Z, Liu
Z, Liu H, Xu F, Wang J and Xiang H: Specific patterns of spinal
metabolites underlying alpha-Me-5-HT-evoked pruritus compared with
histamine and capsaicin assessed by proton nuclear magnetic
resonance spectroscopy. Biochim Biophys Acta Mol Basis Dis.
1863.1222–1230. 2017.
|
|
36
|
He ZG, Liu BW, Li ZX, Liu C and Xiang HB:
Altered expression profiling of spinal genes modulated by compound
48/80 in a mouse itch model. J Anesth Perioper Med. 4:220–224.
2017. View Article : Google Scholar
|
|
37
|
Wang Q, He ZG, Li ZX, Li SY, Chen YL, Feng
MH, Hong QX and Xiang HB: Bioinformatics analysis of gene
expression profile data to screen key genes involved in cardiac
ischemia-reperfusion injury. Int J Clin Exp Med. 11:4955–4966.
2018.
|
|
38
|
Wang Q, Li ZX, Li YJ, He ZG, Chen YL, Feng
MH, Li SY, Wu DZ and Xiang HB: Identification of lncRNA and mRNA
expression profiles in rat spinal cords at various time-points
following cardiac ischemia/reperfusion. Int J Mol Med.
43:2361–2375. 2019.PubMed/NCBI
|
|
39
|
Wang Q, Li ZX, Li YJ, Manyande A, Li SY,
Feng MH, Wu DZ and Xiang HB: Alterations in amino acid levels and
metabolite ratio of spinal cord in rat with myocardial
ischemia-reperfusion injury by proton magnetic resonance
spectroscopy. Am J Transl Res. 11:3101–3108. 2019.PubMed/NCBI
|
|
40
|
Zeng HL, Yu FL, Zhang Z, Yang Q, Jin S, He
X, Chen X, Shen Y, Cheng L, Guo L and Xu F: Quantitative proteomics
study of host response to virulent and attenuated pseudorabies
virus infection in mouse brain. Biochim Biophys Acta Proteins
Proteom. 1866.307–315. 2018.
|
|
41
|
Zeng HL, Yang Q, Du H, Li H, Shen Y, Liu
T, Chen X, Kamal GM, Guan Q, Cheng L, et al: Proteomics and
metabolomics analysis of hepatic mitochondrial metabolism in
alcohol-preferring and non-preferring rats. Oncotarget.
8:102020–102032. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Murry CE, Jennings RB and Reimer KA:
Preconditioning with ischemia: A delay of lethal cell injury in
ischemic myocardium. Circulation. 74:1124–1136. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hickman DL and Johnson SW: Evaluation of
the aesthetics of physical methods of euthanasia of anesthetized
rats. J Am Assoc Lab Anim Sci. 50:695–701. 2011.
|
|
44
|
Zeng HL, Rao X, Zhang LK, Zhao X, Zhang
WP, Wang J, Xu F and Guo L: Quantitative proteomics reveals
olfactory input-dependent alterations in the mouse olfactory bulb
proteome. J Proteomics. 109:125–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang DS, Zeng HL, Li R, Huo B, Su YS,
Fang J, Yang Q, Liu LG, Hu M, Cheng C, et al: Aberrant epicardial
adipose tissue extracellular matrix remodeling in patients with
severe ischemic cardiomyopathy: Insight from comparative
quantitative proteomics. Sci Rep. 7:437872017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
47
|
Foley LS, Fullerton DA, Bennett DT,
Freeman KA, Mares J, Bell MT, Cleveland JC Jr, Weyant MJ, Meng X
and Puskas F: Reece TB. Spinal cord ischemia-reperfusion injury
induces erythropoietin receptor expression. Ann Thorac Surg.
100:41–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y, Pang QJ, Liu JT, Wu HH and Tao DY:
Down-regulated miR-448 relieves spinal cord ischemia/reperfusion
injury by up-regulating SIRT1. Braz J Med Biol Res. 51:pp.
e73192018, View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Balsam LB: Spinal cord
ischemia-reperfusion injury: MicroRNAs and mitophagy at a
crossroads. J Thorac Cardiovasc Surg. 154:1509–1510. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Beattie MS, Farooqui AA and Bresnahan JC:
Review of current evidence for apoptosis after spinal cord injury.
J Neurotrauma. 17:915–925. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Z, Zhou L, Zheng X, Chen G, Pan R, Li
J and Liu W: Autophagy protects against PI3K/Akt/mTOR-mediated
apoptosis of spinal cord neurons after mechanical injury. Neurosci
Lett. 656:158–164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jung SY, Kim DY, Yune TY, Shin DH, Baek SB
and Kim CJ: Treadmill exercise reduces spinal cord injury-induced
apoptosis by activating the PI3K/Akt pathway in rats. Exp Ther Med.
7:587–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Qiao Y, Peng C, Li J, Wu D and Wang X:
Spinal cord ischemia-reperfusion causes damage of neurocyte by
inhibiting RAP2C. Neurol Res. 39:877–884. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xie R, Cheng M, Li M, Xiong X, Daadi M,
Sapolsky RM and Zhao H: Akt isoforms differentially protect against
stroke-induced neuronal injury by regulating mTOR activities. J
Cereb Blood Flow Metab. 33:1875–1885. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hwang JY, Gertner M, Pontarelli F,
Court-Vazquez B, Bennett MV, Ofengeim D and Zukin RS: Global
ischemia induces lysosomal-mediated degradation of mTOR and
activation of autophagy in hippocampal neurons destined to die.
Cell Death Differ. 24:317–329. 2017. View Article : Google Scholar :
|
|
56
|
Yuan D, Liu C and Hu B: Dysfunction of
membrane trafficking leads to ischemia-reperfusion injury after
transient cerebral ischemia. Transl Stroke Res. 9:215–222. 2018.
View Article : Google Scholar :
|
|
57
|
Liu CL, Ge P, Zhang F and Hu BR:
Co-translational protein aggregation after transient cerebral
ischemia. Neuroscience. 134:1273–1284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang F, Liu CL and Hu BR: Irreversible
aggregation of protein synthesis machinery after focal brain
ischemia. J Neurochem. 98:102–112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ren Q and You Yu S: CD2-associated protein
participates in podocyte apoptosis via PI3K/Akt signaling pathway.
J Recept Signal Transduct Res. 36:288–291. 2016. View Article : Google Scholar
|
|
60
|
Rui YN, Xu Z, Patel B, Chen Z, Chen D,
Tito A, David G, Sun Y, Stimming EF, Bellen HJ, et al: Huntingtin
functions as a scaffold for selective macroautophagy. Nat Cell
Biol. 17:262–275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ralser M, Nonhoff U, Albrecht M, Lengauer
T, Wanker EE, Lehrach H and Krobitsch S: Ataxin-2 and huntingtin
interact with endophilin-A complexes to function in
plastin-associated pathways. Hum Mol Genet. 14:2893–2909. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hughes AC, Errington R, Fricker-Gates R
and Jones L: Endophilin A3 forms filamentous structures that
colocalise with microtubules but not with actin filaments. Brain
Res Mol Brain Res. 128:182–192. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Choi TM, Yun M, Lee JK, Park JT, Park MS
and Kim HS: Proteomic analysis of a rat cerebral ischemic injury
model after human cerebral endothelial cell transplantation. J
Korean Neurosurg Soc. 59:544–550. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Thal DR, Züchner S, Gierer S, Schulte C,
Schöls L, Schüle R and Synofzik M: Abnormal paraplegin expression
in swollen neurites, τ- and α-synuclein pathology in a case of
hereditary spastic paraplegia SPG7 with an Ala510Val mutation. Int
J Mol Sci. 16:25050–25066. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
He M, Ding Y, Chu C, Tang J, Xiao Q and
Luo ZG: Autophagy induction stabilizes microtubules and promotes
axon regeneration after spinal cord injury. Proc Natl Acad Sci USA.
113:11324–11329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hellal F, Hurtado A, Ruschel J, Flynn KC,
Laskowski CJ, Umlauf M, Kapitein LC, Strikis D, Lemmon V, Bixby J,
et al: Microtubule stabilization reduces scarring and causes axon
regeneration after spinal cord injury. Science. 331:928–931. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gaudet AD and Popovich PG: Extracellular
matrix regulation of inflammation in the healthy and injured spinal
cord. Exp Neurol. 258:24–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Myllyharju J: Prolyl 4-hydroxylases,
master regulators of the hypoxia response. Acta Physiol (Oxf).
208:148–165. 2013. View Article : Google Scholar
|
|
69
|
Vu TT, Marquez J, Le LT, Nguyen ATT, Kim
HK and Han J: The role of decorin in cardiovascular diseases: More
than just a decoration. Free Radic Res. 52:1210–1219. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sullivan SJ, Farrant M and Cull-Candy SG:
TARP γ-2 Is required for inflammation-associated AMPA receptor
plasticity within Lamina II of the spinal cord dorsal horn. J
Neurosci. 37:6007–6020. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Costa JT, Mele M, Baptista MS, Gomes JR,
Ruscher K, Nobre RJ, de Almeida LP, Wieloch T and Duarte CB:
Gephyrin cleavage in in vitro brain ischemia decreases GABAA
receptor clustering and contributes to neuronal death. Mol
Neurobiol. 53:3513–3527. 2016. View Article : Google Scholar
|