Introduction
Garlic (Allium sativum L.) is typically used
for culinary purposes (1). Garlic
extracts, including diallyl disulfide (DADS), diallyl trisulfide
and garlic polysaccharide (GP) and other associated compounds,
exhibit a wide range of pharmacological and biochemical activities,
including the inhibition of cancer progression, antibacterial and
immunomodulatory activity, hypoglycemic effects and antioxidant
effects (2-4). As a bioactive component of garlic,
DADS exhibits similar biochemical activities. It has been reported
that DADS may reverse the epithelial-to-mesenchymal transition in
lung cancer and gastric cancer cells (5,6).
DADS decreased the proliferation and invasion of osteosarcoma cells
by upregulating miR-134 expression (7) and prevented cigarette smoke-induced
airway inflammation by inhibiting the phosphorylation of
extracellular signal-regulated kinase and the expression of matrix
metalloproteinase-9 in lung tissues (8). DADS may also exhibit epigenetic
regulation by exerting histone deacetylase-inhibitory activity
(9,10).
In recent years, numerous studies have demonstrated
an association between altered gut microbiota and a number of
diseases, ranging from obesity and inflammatory diseases to
behavioral and physiological abnormalities (11,12). Gut microbiota serve an important
role in the development of these diseases as it can mediate
environmental changes in the intestinal immune system (13). Various environmental factors,
including diet and medicine, can result in compositional and
functional changes to the gut microbiota. Filocamo et al
(14) evaluated the effect of
garlic powder on the viability of representative gut bacteria in
vitro, and the results indicated that garlic intake had the
potential to temporarily modulate the gut microbiota. Wang et
al (15) reported that GP
could be used to protect the liver against fibrosis through
modulating lipid peroxidation and oxidative stress, regulating the
transforming growth factor-β1 and tumor necrosis factor-α signaling
pathways, and may be used to treat alcoholic liver fibrosis (ALF)
and ALF-induced gut microbiota dysbiosis.
Although it has been reported that DADS has
anti-cancer, anti-inflammatory and anti-oxidative effects, its
effects on the gut microbiome have not yet been determined. In the
present study, C57BL/6 mice were treated with DADS and a high fat
diet (HFD), and the effects of DADS on the liver and the intestines
were determined. The association between DADS, fatty liver, HFD and
gut microbiota was investigated, which may encourage further
studies that focus on the roles and underlying molecular mechanisms
of DADS on the development of fatty liver.
Materials and methods
Animals and tissue collection
All experimental procedures in the present study
were performed in accordance with the guidelines of the Ethics
Committee of Guangdong Pharmaceutical University. The animal
experiments were approved by the Committee on Laboratory Animal
Care and Use of Guangdong Pharmaceutical University (Guangzhou,
China). A total of 60 male C57BL/6 mice (5 weeks of age; weight,
19.92±1.05 g) obtained from the Laboratory Animal Center of
Guangdong Province, were housed in a specific pathogen-free mouse
facility, at 25°C, 60-65% humidity, 12 h light/12 h dark cycle,
with free access to food and water. The mice were randomly divided
into 6 groups and each group contained 10 mice. Group 1 (NFD-CON)
were fed a control normal food diet (NFD); group 2 (NFD-AL) were
fed with a NFD and treated with a low dose of DADS; group 3
(NFD-AH) were fed with a NFD and treated with a high dose of DADS;
group 4 (HFD-CON) were fed with a high fat diet (HFD); group 5
(HFD-AL) were fed with a HFD and treated with a low dose of DADS;
and group 6 (HFD-AH) were fed with a HFD and treated with a high
dose of DADS. The concentration of DADS used in the low-dose and
high-dose treated mice were 10 and 20 mg/kg (weight), respectively.
The mice of groups 2, 3, 5 and 6 were treated with DADS by
intragastric administration once every day, and were sacrificed
after 8 weeks. The human equivalent doses (HED) were calculated by
multiplying the mouse doses by 0.081 as previously described
(16). HED were 0.81 and 1.62
mg/kg (weight) respectively, and these doses could be achieved
through a regular diet. The dosage of DADS was calculated based on
the most recently recorded body weight of the mice, as previously
described (8,17).
16S recombinant (r)DNA gene analysis
Fecal bacterial DNA extraction, the 16S rDNA gene
PCR amplification and sequencing, and the 16S rDNA gene analysis
were performed by Gene Denovo Biotechnology Company (Guangzhou,
China). The experimental procedures were performed as previously
described (18). The sequencing
data were analyzed using Quantitative Insights into Microbial
Ecology (QIIME; version 1.9.1; http://qiime.org)
(19). The 16S rDNA gene
sequences were assigned to operational taxonomic units (OTUs) using
Uparse (version 9.2.64_i86linux32; http://www.drive5.com/uparse) (20) with a threshold of 97% pair-wise
identity, and classified taxonomically using the Ribosomal Database
Project classifier (version 2.2; http://rdp.cme.msu.edu/classifier/classifier.jsp)
(21). The abundance of bacteria
was calculated using the method of Ace and Chao (22), and the diversity of bacteria was
estimated using the method of Shannon and Simpson (23). Principal coordinates analysis
(Vegan; version 2.5-4; https://cran.r-project.org/web/packages/vegan) was
performed in order to present the differences between the gut
microbial communities of different groups. The Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway analysis was performed
http://www.kegg.jp/kegg/kegg1.html.
Linear Discriminant Analysis Effect Size (LEFse) was used to
identify the specific bacterial taxa of different groups (24).
Assays of the serum lipid profile
The mice were adequately anesthetized with ether for
blood collection prior to sacrifice. The anesthesia method was
approved by the Ethics Committee of Guangdong Pharmaceutical
University, and the pinch reflex was monitored to ensure full
anesthesia. After 1 ml blood collection, the mice were sacrificed
by cervical dislocation. The concentrations of serum non-esterified
free fatty acids (NEFA), high density lipoprotein-cholesterol
(HDL-C), low density lipoprotein-cholesterol (LDL-C), total
cholesterol (TC), triglyceride (TG), alanine aminotransferase (ALT)
and aspartate aminotransferase (AST), together with liver TC and TG
levels were measured according to the manufacturer's protocols for
each kit. Determination kits for NEFA, TC, TG, ALT and AST were
from Nanjing Jiancheng Bioengineering Institute, and determination
kits for HDL-C and LDL-C were from Shanghai Rongsheng Biotechnology
Co., Ltd. Fasting blood glucose (FBG) was determined by accu-chek
(Roche Diagnostics).
Hematoxylin and eosin and oil-red O
staining
Mouse liver and intestine tissues were fixed in 4%
paraformaldehyde at 4°C overnight for hematoxylin and eosin
staining and oil-red O staining. H&E staining was performed on
4-μm-thick sections, stained with hematoxylin for 3 min and
eosin for 20 sec at room temperature. Oil-red O staining was
performed on 7-μm-thick frozen sections, stained with
oil-red O for 10 min at room temperature. The images were captured
with a PerkinElmer Automated Quantitative Pathology System
(PerkinElmer, Inc.).
Reverse transcription-quantitative
(RT-qPCR)
Total RNA of mice liver was extracted from samples
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and were subjected to RT using a PrimeScript™ RT
Reagent kit (Takara Bio, Inc.). The RT conditions were: 37°C for 15
min and 85°C for 5 sec. The sequences of the primers used in the
present study are listed in Table
SI (Sangon Biotech (Shanghai) Co., Ltd., China). RT-qPCR was
performed using a SYBR Premix Ex Taq kit (Takara Bio, Inc.) and a
PikoReal PCR system (Thermo Fisher Scientific, Inc.). The
thermocycling conditions were: 95°C for 30 sec; followed by 40
cycles of 95°C for 5 sec, 60°C for 20 sec and 65°C for 15 sec.
GAPDH was used as the reference gene.
Western blotting
The livers of the mice were collected, lysed by
Radio-Immunoprecipitation Assay lysis buffer (Dalian Meilun
Biotechnology Co., Ltd.) and centrifuged at 13,680 x g, at 4°C for
30 min, and the supernatant was frozen at -80°C. The concentration
of the protein was determined using BCA, equal amounts of protein
(30 μg) were separated via SDS-PAGE (12% gel) and
transferred to a PVDF membrane. The PVDF membrane was blocked with
5% skimmed milk for 1 h at room temperature, and incubated
overnight with primary antibodies at 4°C. Subsequently, after
incubation at room temperature for 1 h with horseradish
peroxidase-labeled antibody, the signals were visualized using
enhanced chemiluminescence reagent. The primary antibodies
included: Mouse anti-hepatic lipase (1:1,000; cat. no. sc-21740;
Santa Cruz), mouse anti-HNF4α (1:5,000; cat. no. ab41898; Abcam),
rabbit anti-ABCG5 (1:3,000; cat. no. ab87116; Abcam), rabbit
anti-CD36 (1:1,000; cat. no. ab133625; Abcam), rabbit anti-FASN
(1:1,000; cat. no. 3180; Cell Signaling Technology, Inc.), rabbit
anti-HMGCoAR (1:3,000; cat. no. ab174830; Abcam), mouse anti-SCD1
(1:1,000; cat. no. ab19862; Abcam), and mouse anti-GAPDH (1:2,000;
cat. no. ab8245; Abcam). Secondary antibodies included HRP-donkey
anti-mouse IgG (1:2,000; cat. no. ab150105; Abcam) and HRP-goat
anti-rabbit IgG (1:2,000; cat. no. os0701; Earthox Life Sciences).
The Lane 1D software (version 5.1.0.0; SageCreation) was used for
quantification of western blotting bands.
Statistical analysis
Statistical differences were determined using SPSS
software (version 23.0; IBM Corp). One-way ANOVA with a least
significance difference post hoc test for equal variances, or
one-way ANOVA with Games-Howell for unequal variances was used to
determine the difference between different groups. The data are
presented as the mean ± standard deviation. Each experiment was
repeated at least 3 times. P<0.05 was considered to indicate a
statistically significant difference.
Results
Low-dose treatment with DADS results in
fat deposition in the liver of mice on a NFD
There was no significant difference in the body
weight of the three groups of mice fed with a NFD; however, the
body weight of the groups treated with DADS was increased in the
HFD groups compared with the control group of HFD (HFD-AL vs.
HFD-CON, P<0.05; HFD-AH vs. HFD-CON, P<0.01; Fig. 1A). The FBG of the HFD mice treated
with a low dose of DADS was elevated compared with the control
group of mice fed with a NFD (P<0.05; Fig. 1B). Serum NEFA levels were
increased in the low-dose DADS group of mice fed with a NFD and in
the HFD groups (HFD-CON and HFD-AL vs. NFD-CON, P<0.01; HFD-AH
and NFD-AL vs. NFD-CON, P<0.001; Fig. 1C). Serum TG levels of the HFD
groups treated with low and high doses of DADS were also increased
(HFD-AL vs. NFD-CON, P<0.01; HFD-AH vs. NFD-CON, P<0.001;
Fig. 1D). Serum TC, HDL-C and
LDL-C of the NFD group treated with low-dose DADS and the HFD
groups were increased (HFD-CON vs. NFD-CON, P<0.001; HFD-AL vs.
NFD-CON, P<0.01; NFD-AL vs. NFD-CON, P<0.05; Fig. 1E. HFD-CON, HFD-AL and HFD-AH vs.
NFD-CON, P<0.001; NFD-AL vs. NFD-CON, P<0.05; Fig. 1F. HFD-CON vs. NFD-CON, P<0.01;
HFD-AL and HFD-AH vs. NFD-CON, P<0.001; NFD-AL vs. NFD-CON,
P=0.068; Fig. 1G). The TG and TC
levels of the liver were determined, and the results revealed that
the liver TG and TC levels of the NFD group treated with a low-dose
of DADS were significantly increased, and in the HFD groups, only
the HFD mice treated with a low-dose of DADS exhibited similar
results (HFD-AL vs. NFD-CON, P<0.01; NFD-AL vs. NFD-CON,
P<0.001; Fig. 1H. HFD-AL vs.
NFD-CON, P<0.05; NFD-AL vs. NFD-CON, P<0.01; Fig. 1I). There were no differences in
the food intake, serum AST and ALT levels among the 6 groups
(Fig. S1A-C).
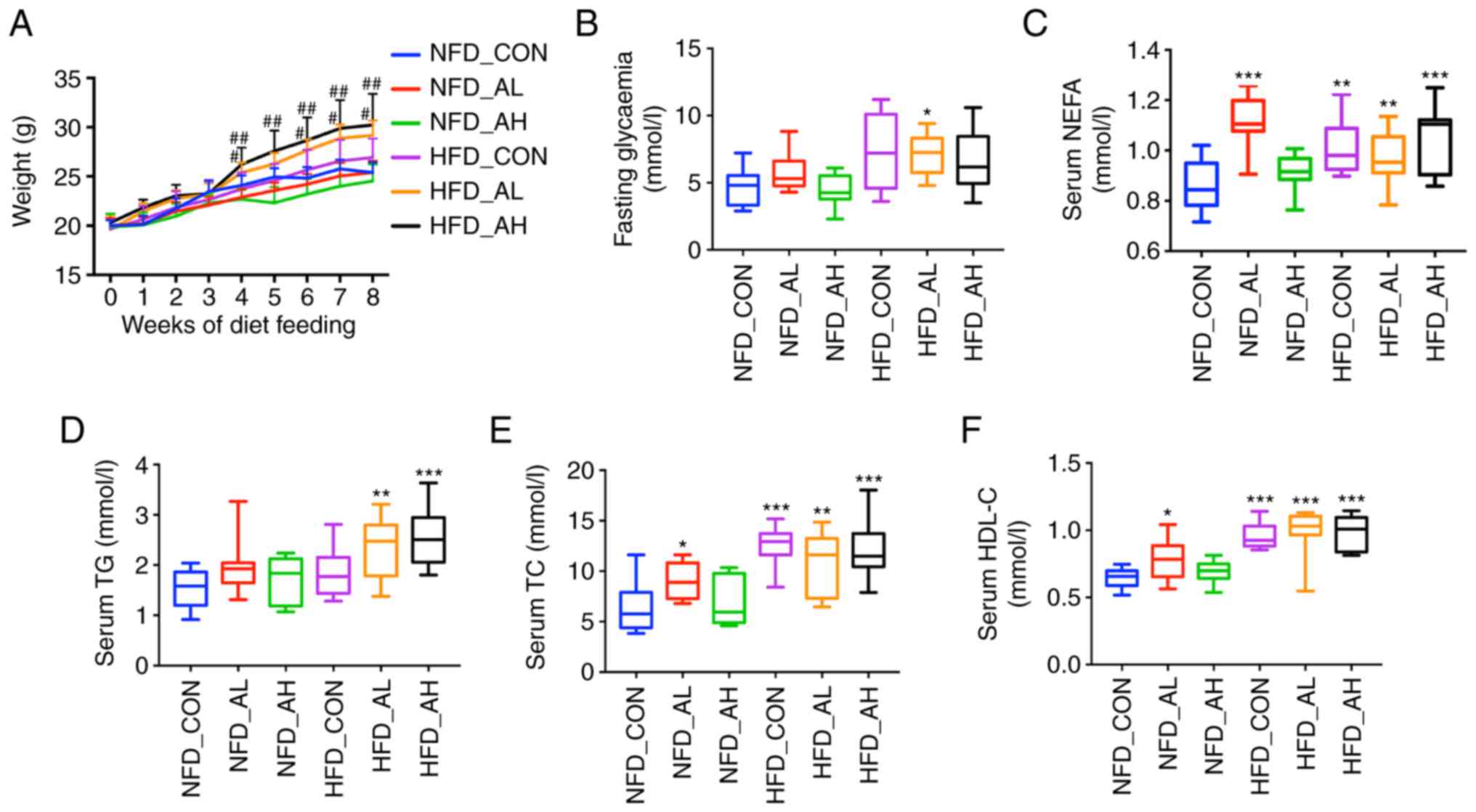 | Figure 1Effect of DADS on lipid metabolism.
(A) Effect of DADS and diet groups on the weight of the mice. (B)
Effect of DADS on FBG. (C) Effect of DADS on serum NEFA. (D) Effect
of DADS on serum TG. (E-G) Effect of DADS on serum TC, HDL-C and
LDL-C. (H and I) Effect of DADS on liver TG and TC. (J) Hematoxylin
and eosin staining of the liver of the 6 groups of mice. (K)
Oil-red O staining of the 6 groups of mice. Scale bar, 80
μm. #P<0.05, ##P<0.01 vs.
HFD-CON; *P<0.05, **P<0.01,
***P<0.001 vs. NFD-CON. NFD-CON, control mice fed
with NFD; NFD-AL, mice fed with NFD and treated with a low dose of
DADS; NFD-AH, mice fed with NFD and treated with a high dose of
DADS; HFD-CON, the control mice fed with HFD; HFD-AL, mice fed with
HFD and treated with a low dose of DADS; HFD-AH, mice fed with HFD
and treated with a high dose of DADS. NFD, normal food diet; HFD,
high fat diet; DADS, diallyl disulfide; NEFA, non-esterified free
fatty acids; TG, triglyceride; TC, total cholesterol; HDL-C, high
density lipoprotein-cholesterol; LDL-C, low density
lipoprotein-cholesterol; FBG, fasting blood glucose. |
The hematoxylin and eosin staining results
demonstrated fat deposition in the liver of the NFD groups treated
with low and high doses of DADS, similar to that in the HFD groups
(Fig. 1J). It seemed that DADS
could induce fatty liver development in the mice fed with a NFD,
and oil-red O staining was used to further visualize this. From the
images obtained from the oil-red O staining, the results were in
agreement with the hematoxylin and eosin staining, particularly in
the NFD group treated with a low-dose of DADS, which demonstrated
notable fat drops in the liver (Fig.
1K). The hematoxylin and eosin staining of different parts of
the intestine in the 6 groups exhibited no noticeable differences
(Fig. S1D).
Expression patterns of lipid
metabolism-associated genes in the liver of mice treated with
DADS
The expression levels of lipid metabolism-associated
genes were measured using RT-qPCR. Fig. 2A presents the expression levels of
lipin 1, hepatic lipase, patatin-like phospholipase
domain-containing protein 2 and peroxisome proliferator-activated
receptor γ coactivator-1α, which are associated with fatty acid
oxidation. The expression levels of hepatic lipase were
significantly decreased in the HFD groups and in the NFD group
treated with a low-dose of DADS (P<0.001). The expression levels
of PNPLA2 were decreased in the HFD groups treated with low and
high doses of DADS (HFD-AL vs. NFD-CON, P<0.05; HFD-AH vs.
NFD-CON, P<0.01). Fig. 2B
demonstrates that the expression levels of HNF4α and FXR, which are
associated with the conversion of cholesterol to bile acid, and the
expression of HNF4α and FXR, were significantly decreased in the
HFD groups and in the NFD group treated with a low-dose of DADS.
The increase in the expression levels of CYP7A1 in the HFD-CON and
HFD-AH groups were not statistically significant when compared with
the NFD-CON group (P=0.16 and P=0.118 respectively, Fig. 2B). Fig. 2C presents the expression of Abcg5,
CD36 and LDLR, which are associated with the transport of lipids.
The expression of Abcg5 was significantly decreased in the HFD
groups and in the NFD group treated with a low-dose of DADS
(NFD-AL, HFD-CON and HFD-AH vs. NFD-CON, P<0.05; HFD-AL vs.
NFD-CON, P<0.01), and the expression of CD36 was increased in
the HFD group treated with a high-dose of DADS (P<0.05).
Fig. 2D presents the expression
of ACCα1, ACCβ1, Fasn, DGAT1, DGAT2, ELOVL5, SREBP-1, SREBP-2,
HMGCoAR, PPARγ and SCD1, which are associated with lipid synthesis.
The expression levels of ACCα1, DGAT1, SREBP-2 and PPARγ were
significantly decreased in the HFD groups and in the NFD group
treated with a low-dose of DADS (for ACCα1, HFD-CON and HFD-AH vs.
NFD-CON, P<0.001; NFD-AL and HFD-AL vs. NFD-CON, P<0.01. For
DGAT1, HFD-CON, HFD-AL and HFD-AH vs. NFD-CON, P<0.01; NFD-AL
vs. NFD-CON, P<0.05. For SREBP-2, P<0.001. For PPARγ, HFD-AL
and HFD-AH vs. NFD-CON, P<0.01; HFD-CON and NFD-AL vs. NFD-CON,
P<0.05), and the expression levels of Fasn, SREBP-1 and HMGCoAR
were decreased in the HFD groups and in the NFD groups treated with
low and high-doses of DADS compared with the NFD control (for Fasn,
NFD-AL and HFD-AH vs. NFD-CON, P<0.01; NFD-AH, HFD-CON and
HFD-AL vs. NFD-CON, P<0.05. For SREBP-1, NFD-AL vs. NFD-CON,
P<0.01; NFD-AH, HFD-CON, HFD-AL and HFD-AH vs. NFD-CON,
P<0.05. For HMGCoAR and NFD-AH vs. NFD-CON, P<0.05; NFD-AL,
HFD-CON, HFD-AL and HFD-AH vs. NFD-CON, P<0.01). Fig. 2E presents the expression of FGF4α,
FGF15 and FGF21, which are associated with insulin resistance (IR).
The expression levels of FGF4α and FGF15 were decreased in the HFD
groups (P<0.05). The expression pattern of lipid
metabolism-associated genes in the liver of the NFD groups is
presented in Fig. S2.
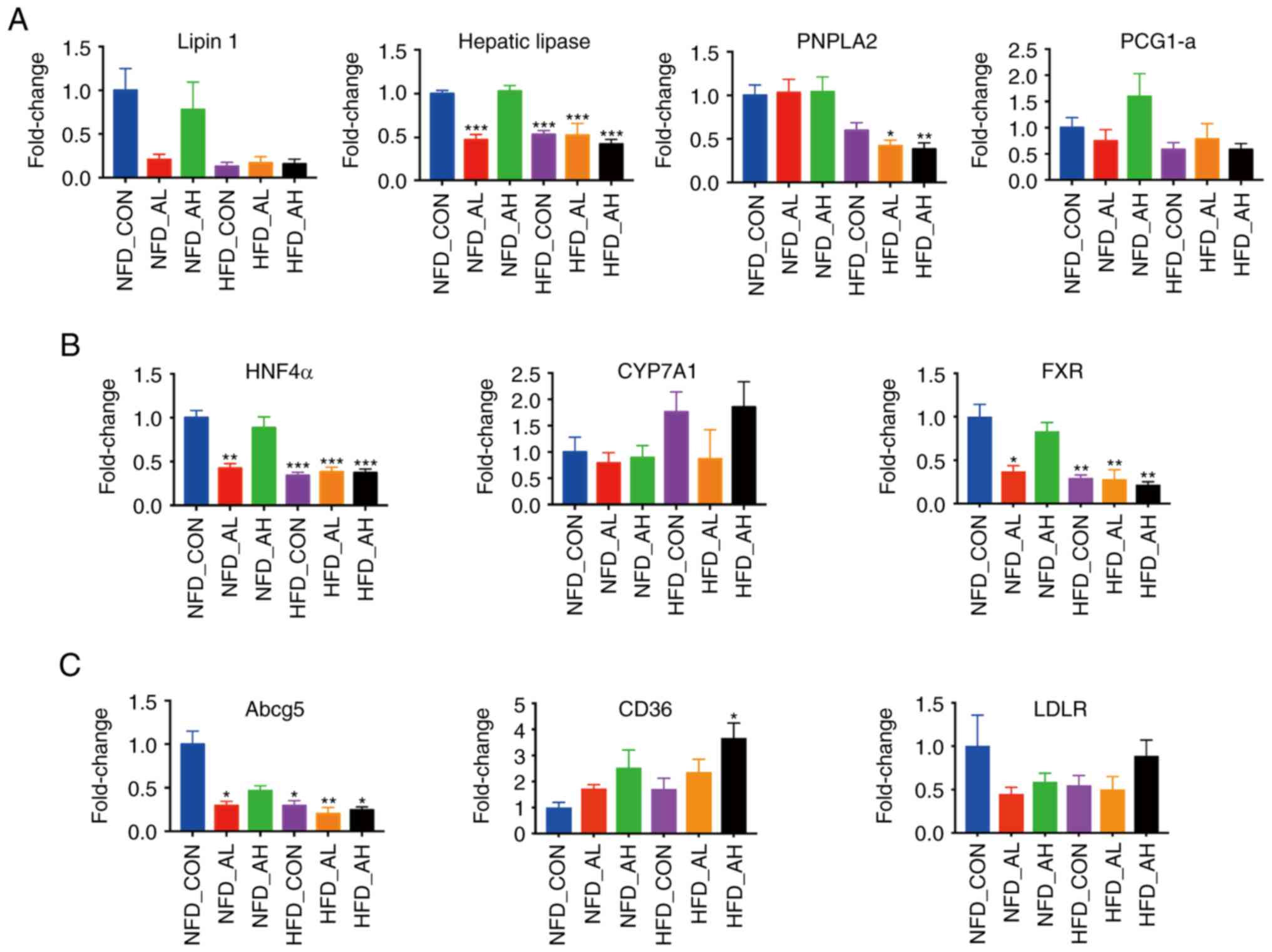 | Figure 2Expression of lipid
metabolism-associated genes in the liver of the 6 groups. (A) mRNA
expression levels of lipin 1, hepatic lipase, PNPLA2 and PGC-1α.
(B) mRNA expression levels of HNF4α, CYP7A1 and FXR. (C) mRNA
expression levels of Abcg5, CD36 and LDLR. (D) mRNA expression
levels of ACCα1, ACCβ1, Fasn, DGAT1, DGAT2, ELOVL5, SREBP-1,
SREBP-2, HMGCoAR, PPARγ and SCD1. (E) mRNA expression levels of
FGF4α, FGF15 and FGF21. *P<0.05,
**P<0.01, ***P<0.001 vs. NFD-CON.
NFD-CON, control mice fed with NFD; NFD-AL, mice fed with NFD and
treated with a low dose of DADS; NFD-AH, mice fed with NFD and
treated with a high dose of DADS; HFD-CON, control mice fed with
HFD; HFD-AL, mice fed with HFD and treated with a low dose of DADS;
HFD-AH, mice fed with HFD and treated with a high dose of DADS.
NFD, normal food diet; HFD, high fat diet; DADS, diallyl disulfide;
PNPLA2, patatin-like phospholipase domain-containing protein 2;
PGC-1α, peroxisome proliferator-activated receptor γ
coactivator-1α; HNF4α, hepatocyte nuclear factor 4α; CYP7A1,
cholesterol 7- alpha hydroxylase gene; FXR, farnesoid X receptor;
Abcg5, adenosine triphosphate-binding cassette subfamily G members
5; LDLR, low density lipoprotein receptor; ACCα1, acetyl-CoA
carboxylase α1; ACCβ1, acetyl-CoA carboxylase β1; Fasn, fatty acid
synthase; DGAT1, diacylglycerol acyltransferase 1; DGAT2,
diacylglycerol acyltransferase 2; ELOVL5, elongases of very
long-chain fatty acids 5; SREBP-1, sterol regulatory
element-binding protein-1; SREBP-2, sterol regulatory
element-binding protein-2; HMGCoAR, hydroxy-methyl-glutaryl
coen-zyme A reductase; PPARγ, peroxisome proliferator-activated
receptor γ; SCD1, Stearoyl-CoA desaturase-1; FGF4α, fibroblast
growth factor 4α; FGF15, fibroblast growth factor 15; FGF21,
fibroblast growth factor 21. |
The protein expression levels of hepatic lipase,
HNF4α, ABCG5, CD36, Fasn, HMGCoAR, SCD1 in the NFD groups was
determined using western blotting (Fig. 3A-B). The results revealed that the
protein expression levels of hepatic lipase was significantly
decreased in the NFD group treated with a low-dose of DADS compared
with the control and high-dose groups (P<0.01; Fig. 3B); the expression levels of
HMGCoAR protein was significantly decreased in the low and
high-dose treated groups (P<0.001 for NFD-AL group; P<0.01
for NFD-AH group; Fig. 3B); the
expression levels of HNF4α, ABCG5 and Fasn were markedly decreased
in the low and high-dose treated groups compared with the control
(Fig. 3B); the expression levels
of CD36 and SCD1 protein were notably increased in the low and
high-dose treated groups (Fig.
3B). The expression at the protein level was concomitant with
the mRNA expression levels.
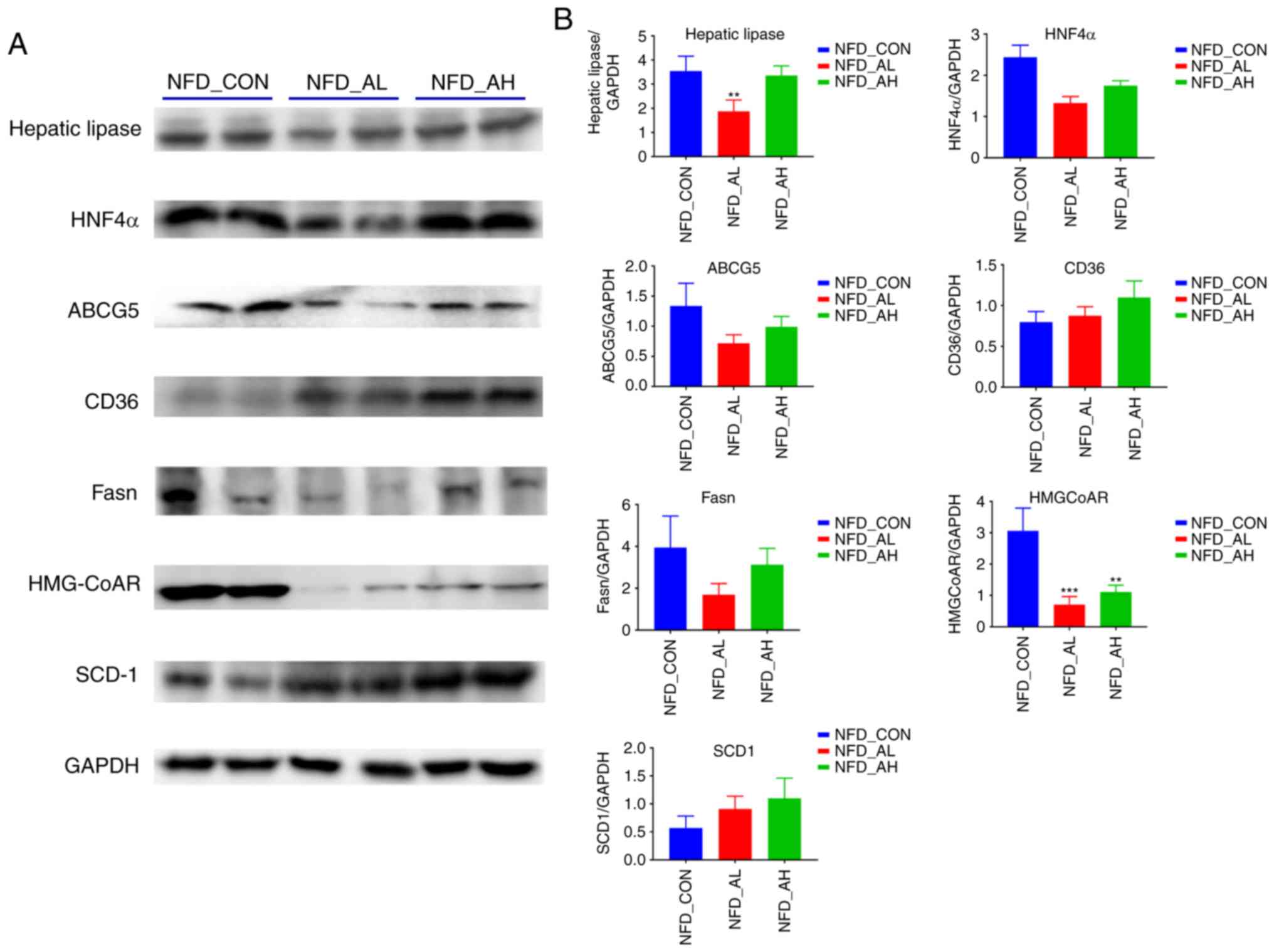 | Figure 3Protein expression of lipid
metabolism-associated genes in the liver of the NFD groups. (A) The
expression of hepatic lipase, HNF4α, ABCG5, CD36, FASN, HMGCoAR,
SCD-1 proteins was detected by western blotting. (B) The relative
protein levels of hepatic lipase, HNF4α, ABCG5, CD36, FASN, HMGCoAR
and SCD-1. The results are presented as the mean ± standard
deviation. n=6. **P<0.01, ***P<0.001
vs. NFD-CON. NFD-CON, control fed with NFD; NFD-AL, mice fed with
NFD and treated with a low dose of DADS; NFD-AH, mice fed with NFD
and treated with a high dose of DADS. NFD, normal food diet; DADS,
diallyl disulfide; HNF4α, hepatocyte nuclear factor 4α; ABCG5,
adenosine triphosphate-binding cassette subfamily G members 5;
FASN, fatty acid synthase; HMGCoAR, hydroxy-methyl-glutaryl
coenzyme A reductase; SCD1, Stearoyl-CoA desaturase-1. |
Overview of the 16S rDNA gene
analysis
In order to characterize the gut microbiota of the
mice treated with DADS, fecal samples were collected, DNA was
purified and amplified, and 16S rRNA gene tags were sequenced. The
total tags and operational taxonomic units (OTUs) of each sample
are presented in Table SII. The
abundance of bacteria was calculated by the method of Ace and Chao
(22), and the diversity of
bacteria was estimated using the method of Shannon and Simpson
(23). All the indices are
summarized in Table SII.
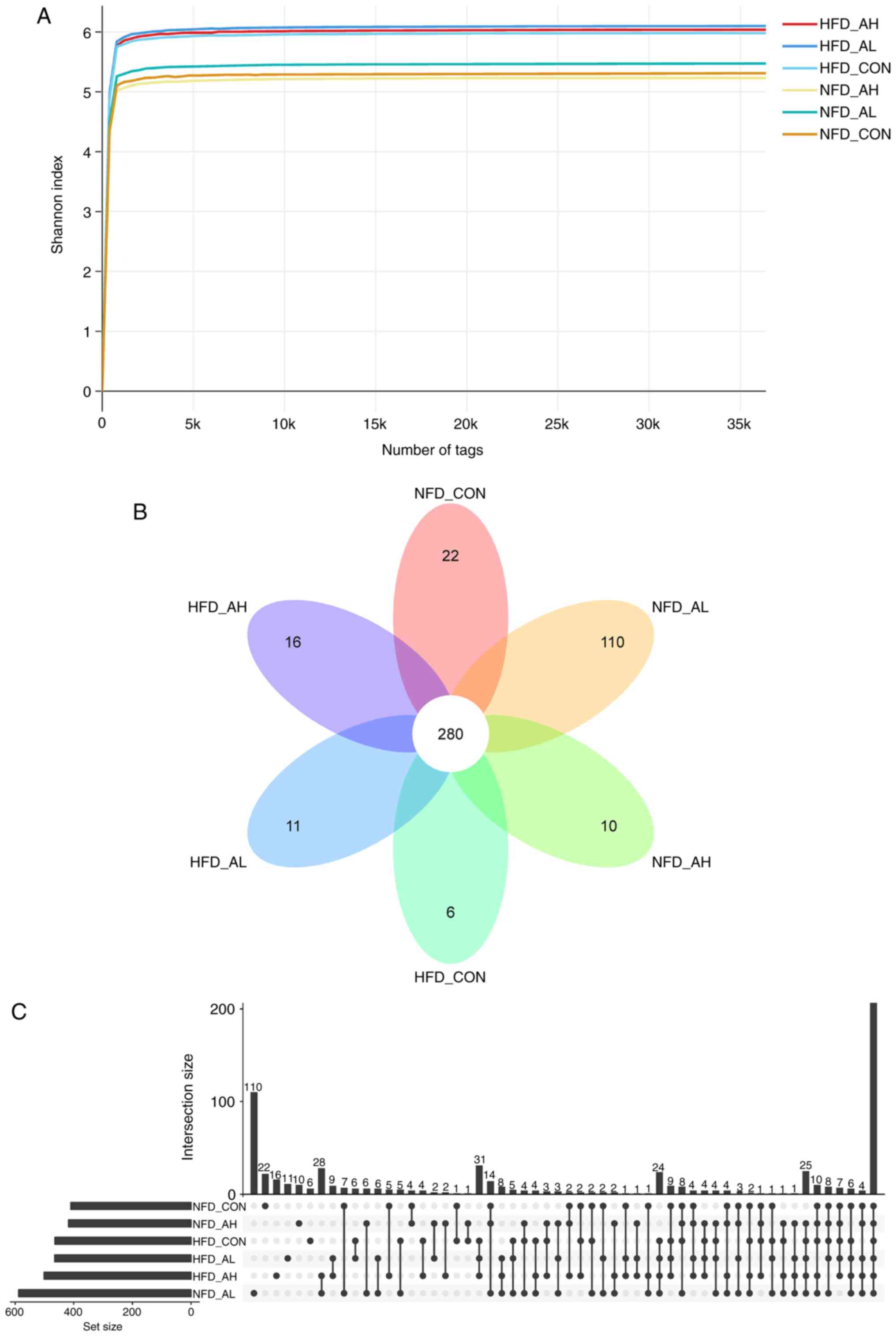
Fig. 4A demonstrates that the
Shannon rarefaction curves for each group reached a saturation
plateau, suggesting enough sequence coverage of samples to describe
the composition of bacteria. The Venn diagram and the histogram of
the six different groups demonstrated that there were 280 common
OTUs in the six groups of mice (Fig.
4B and C). The Shannon rarefaction curves and the Venn diagram
of the NFD groups and the HFD groups are presented in Fig. S3.
Effect of DADS on the relative
composition and diversity of gut microbial communities of mice
All sequences were classified from phylum to
species, and the three NFD groups exhibited notably different
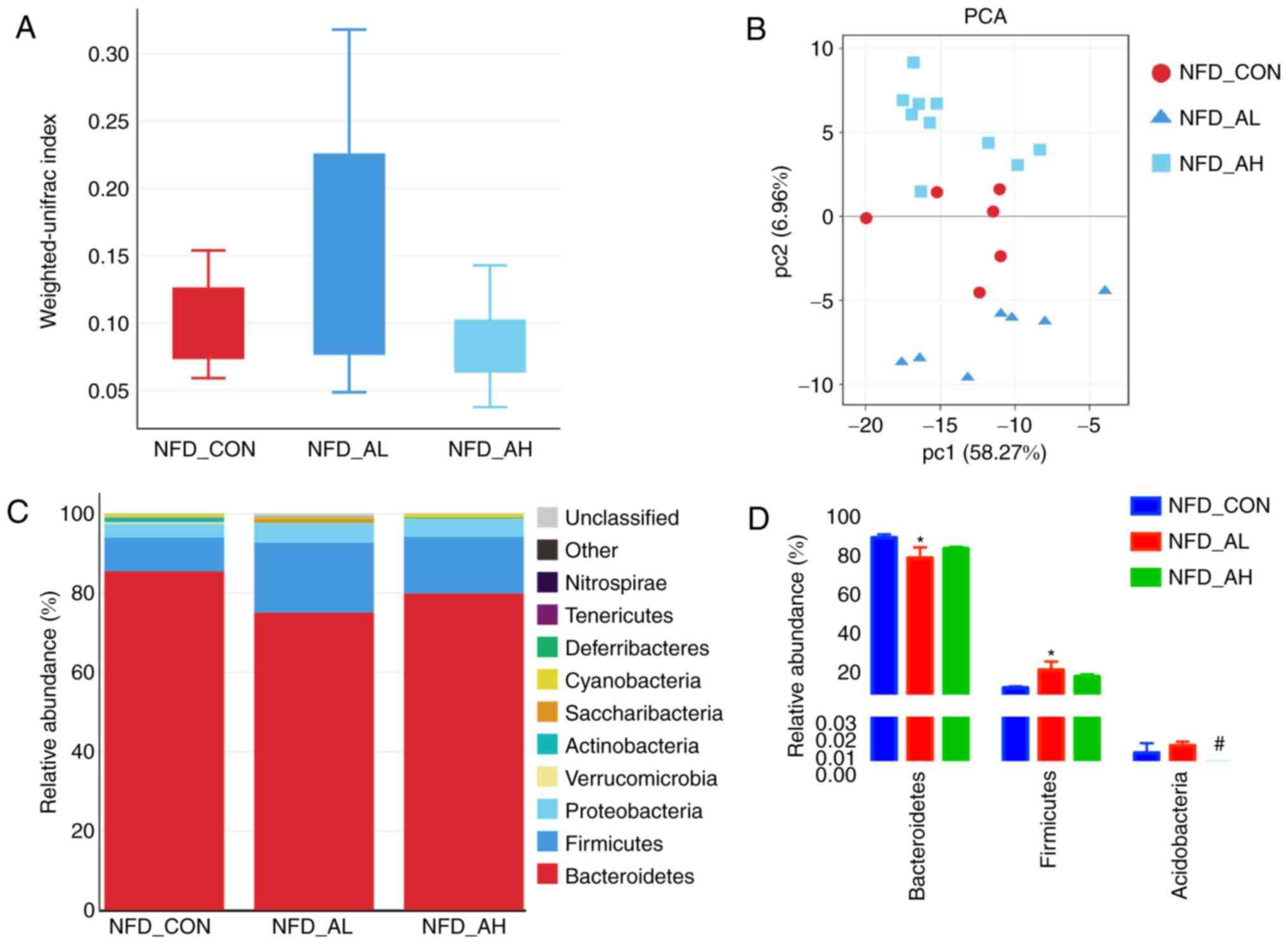
taxonomic compositions. Fig. 5A
presents the Unifrac analyses of the three groups of mice fed with
a NFD, and the results revealed notable differences among the three
groups. A PCoA was also performed, and the results revealed that
the control, low-dose DADS and high-dose groups could be
distinguished (Fig. 5B),
indicating the differences between the three gut microbial
communities, suggesting that DADS altered the bacterial community
of the mice, and the alteration depended on the dose received. The
Unifrac analyses of the six groups are presented in Fig. S4A and B.
The taxonomic compositions of the bacterial phylum
in NFD groups are presented in Fig.
5C. The majority of the samples exhibited high percentages of
Bacteroidetes, Firmicutes and Proteobacteria,
and the low-dose DADS treated group had decreased
Bacteroidetes and increased Firmicutes levels
compared with the control and high-dose groups (Fig. 5C and D). The taxonomic
compositions of the six groups, including the HFD and NFD groups,
on phylum and order level are presented in Fig. S4C-F.
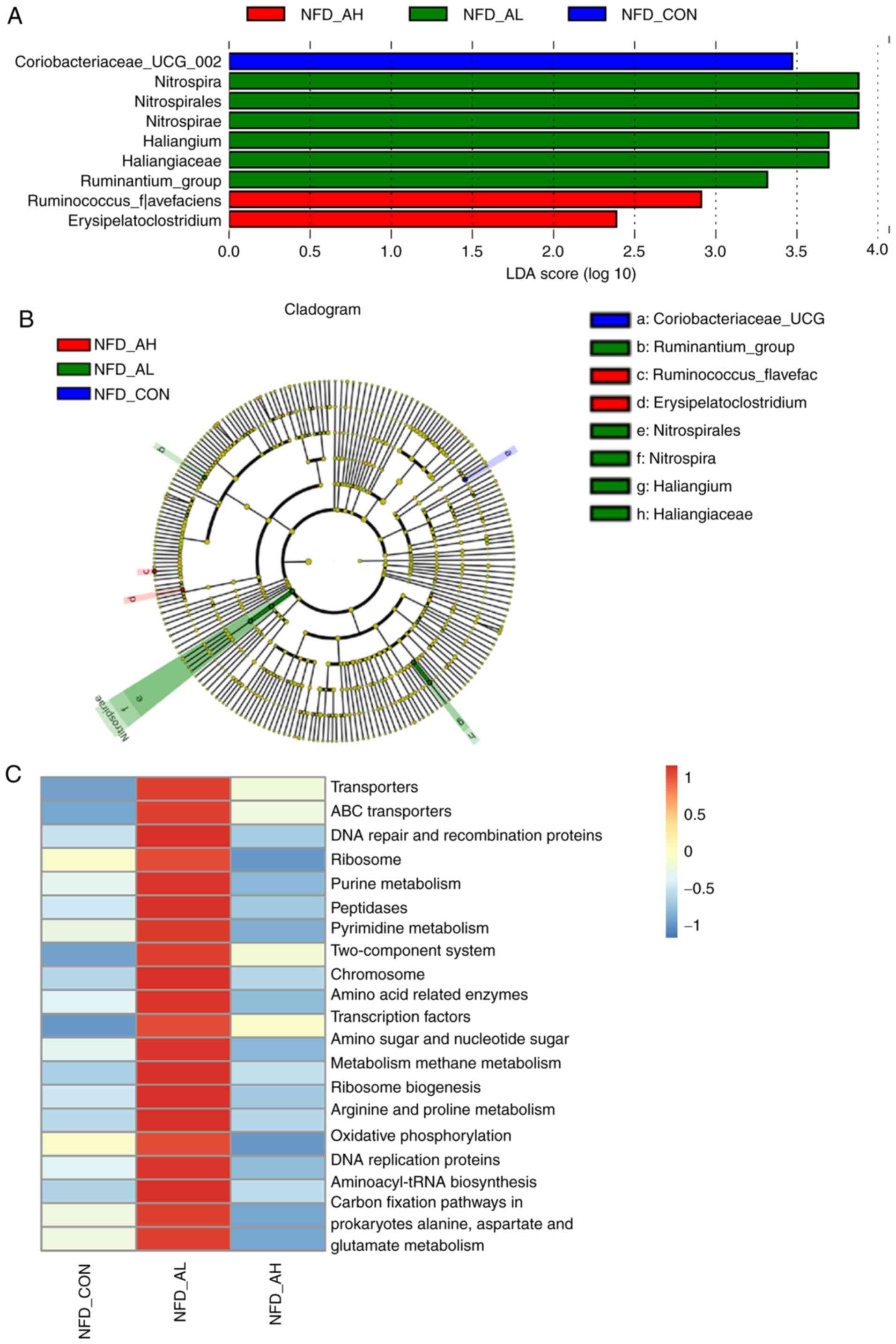
In order to identify the specific bacterial taxa
associated with DADS, the gut microbiota in the NFD groups were
compared using LEFse analysis. Fig.
6A and B demonstrates the specific and predominant bacteria of
the 3 groups, revealing that Coriobacteriaceae-UCG was
predominant in the control group; Ruminococcus-flavefac and
Erysipelatoclostridium were predominant in the high-dose
group; and Nitrospira, Nitrospirales,
Nitrospirae, Haliangium, Haliangiaceae and
Ruminantium-group were specific to the low-dose group. These
different taxa may be used as distinguishing biomarkers. The
different bacterial taxa in the HFD groups are presented in
Fig. S5A and B.
For the NFD groups, the results of the KEGG pathway
analysis demonstrated that the top 20 pathways, including 'purine
metabolism', 'pyrimidine metabolism', 'amino sugar and nucleotide
sugar metabolism', were significantly upregulated in the mice
treated with a low-dose of DADS compared with the control group and
the mice treated with a high-dose of DADS (Fig. 6C). The full results of the KEGG
pathway analysis of the six groups is presented in Fig. S5C. The top 19 pathways that were
significantly upregulated in the HFD groups were: 'Transporters',
'ABC transporters', 'DNA repair and recombination proteins',
'Ribosome', 'Purine metabolism', 'Peptidases', 'Pyrimidine
metabolism', 'Two-component system', 'Chromosome', 'Amino
acid-related enzymes', 'Transcription factors', 'Amino sugar and
nucleotide sugar metabolism', 'Methane metabolism', 'Ribosome
Biogenesis', 'Arginine and proline metabolism', 'DNA replication
proteins', 'Aminoacyl-tRNA biosynthesis', 'Carbon fixation pathways
in prokaryotes', 'Alanine, aspartate and glutamate metabolism',
compared with the NFD groups (Fig.
S5C). In Fig. S5C, the top
20 pathways of the NFD group treated with a low-dose of DADS were
also upregulated, particularly the 'Oxidative phos-phorylation
pathway' compared with the NFD control group and the NFD group
treated with a high-dose of DADS. The top 19 pathways in the HFD
group treated with a low-dose of DADS were not significantly
upregulated compared with the HFD control or HFD groups treated
with a high-dose of DADS, and the 'Oxidative phosphorylation
pathway' was significantly downregulated in the HFD group treated
with a low-dose of DADS (Fig.
S5C). Fig. S5D and E
indicate an association between fat level, liver TG and TC, and the
bacterial taxa.
Discussion
Although DADS has been demonstrated to serve a
variety of biological and pharmaceutical activities, its effects on
the gut microbiota have not yet been determined. In the present
study, C57BL/6 mice were treated with DADS and fed with a HFD, and
the effects of DADS on the gut microbiota and important organs such
as the liver and intestine were determined.
Based on the results of hematoxylin and eosin
staining, it was revealed that there was a considerable increase in
the number of fatty drops in the liver of mice treated with DADS,
similar to the liver of mice fed with a HFD. The result of the
oil-red O staining confirmed this result, and fat deposition in the
liver was more notable in the mice treated with a low-dose of DADS
compared with mice treated with a high-dose of DADS. In addition,
DADS increased the body weight of mice fed with HFD. These results
were unexpected, as previous studies have demonstrated that
components from garlic had anti-obesity, anti-diabetic,
hypolipidemic properties, and cardioprotective and hepatoprotective
functions (25-29). Serum NEFA, TC, HDL-C and LDL-C
were increased in the NFD group treated with a low-dose of DADS,
and in the HFD groups. High levels of LDL has long been recognized
as an important factor for abnormal lipid metabolism. Shi et
al (30) reported that HDL-C
levels were also significantly associated with atherosclerosis
resulting from HFD. Changes in serum TC, LDL-C and HDL-C in the
present study corroborated the results of Shi et al
(30), suggesting that the
functions of HDL-C require further study.
Based on the results of the serum tests and
histochemical staining, the expression pattern of lipid-metabolism
associated genes in the liver was measured using RT-qPCR and
western blotting. The results demonstrated that the expression of
hepatic lipase, HNF4α, FXR, Abcg5, ACCα1, DGAT1, SREBP-2 and PPARγ
were significantly decreased in the HFD groups and in the NFD group
treated with a low-dose of DADS, whereas the expression of CD36 was
increased in the HFD group treated with a high-dose of DADS, as
well as in the NFD group treated with a low-dose of DADS. The
expression of Fasn, SREBP-1 and HMGCoAR were decreased in the HFD
groups and in the NFD groups treated with low or high-doses of DADS
compared with the NFD control group. In the NFD groups, the
expression of Lipin 1 was significantly decreased in the group
treated with a low-dose of DADS compared with the control and the
high-dose groups. Lipin-1 is a mammalian phosphatidic acid
phosphatase (PAP), which is required for lipid synthesis through
its function as a PAP enzyme (31,32). Lipin1 also functions as a
transcriptional coactivator of lipid metabolism genes, which forms
a physical complex with PGC-1α and PPARα to control the expression
of genes involved in fatty acid oxidation and mitochondrial
metabolism (32). Lipase is an
enzyme that breaks down triglycerides into free fatty acids and
glycerol, and it has previously been reported that lower hepatic
lipase activity may be associated with higher concentrations of LDL
triglycerides, thus increasing cardiovascular risk (33). The results of the present study
revealed that a low dose of DADS decreased the expression levels of
hepatic lipase in the liver of mice, promoting the formation of
fatty liver. HNF4α and FXR serve key roles in the regulation of
bile acids (BA) (34), and the
results of the present study revealed that a low-dose of DADS could
decrease the expression of HNF4α and FXR in the liver of mice. As
HNF4α regulates the expression of CYP7A1 directly, a significant
decrease of HNF4α may decrease the production of BA, which worsened
fat deposition in the liver. Fatty acid translocase CD36 (FAT/CD36)
regulates the uptake and intracellular transport of long-chain
fatty acids in different cell types, and it has been reported that
the upregulation of hepatic CD36 is significantly associated with
IR and increased steatosis in patients with fatty liver (35), consistent with the results of the
present study. DGAT1, DGAT2, SREBP-1, SREBP-2, HMGcoAR serve
important roles in lipid synthesis (36-39), and the results of the present
study demonstrated that DADS downregulated the expression of these
genes. Another important gene in lipogenesis is stearoyl-CoA
desaturase-1 (SCD1), which catalyzes the rate-limiting reaction of
the synthesis of monounsaturated fatty acids, and it has been
reported that elevated SCD1 activity has been implicated in a
number of disorders, including obesity, diabetes and
atherosclerosis (40). de
Fourmestraux et al (41)
indicated that increased expression of Scd-1 and the other
lipogenic liver genes may be a critical step in the development of
obesity when mice were fed with a HFD. The results of the present
study revealed that the expression of SCD1 protein was
significantly increased in the NFD groups treated with low and
high-doses of DADS, suggesting that SCD1 may serve an important
role in the development of fatty liver caused by DADS. Fibroblast
growth factor (FGF)-21 and FGF19 (in mice, FGF15) exhibit opposite
changes in expression levels in obesity (42,43), and the results of the present
study revealed that the expression of FGF15 was decreased, whereas
expression of FGF21 was increased in the liver of mice treated with
a low-dose of DADS (although the change was not significant),
indicating that a low-dose of DADS may induce fat deposition in the
liver.
Recently, focus has been placed on the gut
microbiota after it was demonstrated that alterations in the gut
microbiota were associated with a wide range of diseases, including
obesity, diabetes, cardiovascular diseases and tumors (19,44-46). In the present study, the effects
of DADS on gut microbiota were determined by performing a 16S rDNA
gene analysis, and the results revealed that the NFD group treated
with a low-dose of DADS decreased Bacteroidetes and
increased Firmicutes compared with the control and high-dose
groups at the phyla taxonomic level. Turnbaugh et al
(47) reported that obesity was
associated with changes in the relative composition of the two
dominant bacterial divisions, Bacteroidetes and
Firmicutes, by comparing the gut microbiota of genetically
obese mice and their lean littermates, as well as those of obese
and lean human volunteers. It was revealed that the obese
individuals had higher levels of Firmicutes and lower levels
of Bacteroidetes (47).
Islam et al (48) fed rats
supplemented with different concentrations of cholic acid, and
revealed that rats demonstrated increased Firmicutes levels
accompanied by decreased Bacteroidetes levels, similar to
that of the obesity-associated gut microbiome induced by high-fat
diets. Recently, Zheng et al (49) evaluated the effects of dietary
fat-induced BA changes on the shaping of the gut microbial
composition in mice, and revealed that a HFD could induce the
secretion of BA and the concentration and species of BA in
intestines increased rapidly, which resulted in an obesity-type gut
microbiota. The results of the present study demonstrated that a
low-dose of DADS decreased Bacteroidetes levels and
increased Firmicutes levels, similar to the gut microbiota
composition of the HFD mice, and these results confirmed the
phenotype of the mice treated with a low-dose of DADS that
developed fatty liver. The results of the RT-qPCR in the present
study demonstrated that the expression of HNF4α and FXR was
decreased in the liver of the mice treated with a low-dose of DADS,
and thus, it was concluded that a low-dose of DADS may alter the
ratio of Firmicutes to Bacteroidetes by regulating
the expression of BA secretion and metabolism-associated genes,
which was in agreement with the results of the study by Zheng et
al (49), as they
demonstrated that the changes in the concentration and species of
BA in the intestine occurred earlier than the change in the gut
microbiota.
Gut microbiota provide the enzymatic and metabolic
pathways involved in food digestion, xenobiotic metabolism and
production of molecules such as amino acids, short-chain fatty
acids and metabolites, which are essential for glycolysis, the
tricarboxylic acid cycle, oxidative phosphorylation, and amino acid
and fatty acid metabolism (50).
In the present study, HFD upregulated the pathways of
'Transporters', 'ABC transporters', 'DNA repair and recombination
proteins', 'Ribosome', 'Purine metabolism', 'Peptidases',
'Pyrimidine metabolism', 'Two-component system', 'Chromosome',
'Amino acid related enzymes', 'Transcription factors', 'Amino sugar
and nucleotide sugar metabolism', 'Methane metabolism', 'Ribosome
Biogenesis', 'Arginine and proline metabolism', 'DNA replication
proteins', 'Aminoacyl-tRNA biosynthesis', 'Carbon fixation pathways
in prokaryotes', 'Alanine, aspartate and glutamate metabolism'.
Similarly, the top 20 pathways of the NFD group treated with a
low-dose of DADS were also upregulated compared with the NFD
control group and the NFD group treated with a high-dose of DADS,
indicating that a low-dose of DADS may affect the gut microbiota
and impact the pathways involved in protein and amino acid
metabolism and synthesis, lipid transport, oxidative
phosphorylation and carbon metabolism. The effects of a low-dose of
DADS on the structure of gut microbiota and pathways were similar
to a HFD. It has been reported that colonocyte metabolism is
directed towards oxidative phosphorylation during homeostasis of
microbiota, maintaining a gut flora environment dominated by
obligate anaerobic bacteria (51). Alterations of the metabolism of
the colonic epithelium may increase epithelial oxygenation,
resulting in an expansion of facultative anaerobic bacteria, which
is a marker for dysbiosis in the colon (51). The results of the present study
revealed that the oxidative phosphorylation pathway was
significantly upregulated in the NFD group treated with a low-dose
of DADS compared with the NFD control and NFD groups treated with a
high-dose of DADS, but was significantly downregulated in the HFD
group treated with a low-dose of DADS compared with the HFD control
and the HFD groups treated with a high-dose of DADS. Therefore, the
effects of DADS on metabolism and gut microbiota depend on dose and
fat content of the food, and the underlying molecular mechanisms
require further study.
In the present study, the phenotypes of mice fed
with low and high-doses of DADS had similarities and differences.
In the HFD groups, the body weight of mice treated with low and
high doses of DADS was higher than the other groups. The levels of
TC and TG were higher in the liver of mice treated with a low-dose
of DADS in both the NFD and HFD groups. The results of the
hematoxylin and eosin and oil-red O staining demonstrated that both
low and high doses of DADS resulted in fat deposition in the liver
of mice in the NFD and HFD groups, but the NFD group treated with a
low-dose of DADS exhibited more severe instances of fatty liver.
According to the results of the RT-qPCR, the expression patterns of
the majority of the lipid metabolism-associated genes in the liver
of the NFD mice treated with a low-dose of DADS was similar to that
of the HFD groups. A low-dose of DADS had similar effects on lipid
metabolism to a HFD and a larger effect than a high-dose of
DADS.
Taken together, the results of the present study
identified a notable phenomenon in mice fed with a normal diet.
Treatment with DADS resulted in fatty liver development and an
increased ratio of Firmicutes to Bacteroidetes in gut
microbiota, similar to the phenotypes of mice fed with HFD. In
addition, DADS could increase the body weight of mice fed with HFD.
It was concluded that DADS may exert its effects on lipid
metabolism and gut microbiota through regulation of the expression
of genes associated with lipogenesis and lipid metabolism.
Supplementary Data
Acknowledgments
The authors would like to thank Dr Shenghua Piao
from Guangdong Metabolic Disease Research Center of Integrated
Chinese and Western Medicine, Guangdong Pharmaceutical University
(Guangzhou, China) for helping with the language in the
article.
Funding
This study was supported by the National Natural
Science Foundation of China (grant. nos. 81803912 and 31671520),
the Science and Technology Project of Guangdong Province (grant.
nos. 2016B050501003, and 2017B050504005), the Scientific Research
Project of the Administration of Traditional Chinese Medicine of
Guangdong Province (grant. no. 20182079), the Characteristic
Innovation Project (Natural Science) of the Education Department of
Guangdong Province and the 'Innovation Strong School Project' of
Guangdong Pharmaceutical University (grant. no. 2017KTSCX102) and
the Science and Technology Project of Yue-Xiu District of Guangzhou
(grant. no. 2018-WS-011).
Availability of data and materials
The datasets generated and analyzed during the
present study are available from the corresponding author upon
reasonable request.
Authors' contributions
ZL and JG and YY designed the study and conceived
the report. YY analyzed and interpretated the results, wrote the
first draft of the manuscript and revised it critically. FY, HW,
MH, SLi, QW, CY, XZ, SLiu, YLei, LY, GC and YLiu performed the
experiments. SLiu and YLei created the figures. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The animal experiments were approved by The
Committee on Laboratory Animal Care and Use of Guangdong
Pharmaceutical University (Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nam H, Jung H, Kim Y, Kim B, Kim KH, Park
SJ and Suh JG: Aged black garlic extract regulates lipid metabolism
by inhibiting lipogenesis and promoting lipolysis in mature 3T3-L1
adipocytes. Food Sci Biotechnol. 27:575–579. 2017.
|
|
2
|
Li Z, Le W and Cui Z: A novel therapeutic
anticancer property of raw garlic extract via injection but not
ingestion. Cell Death Discov. 4:1082018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dwivedi VP, Bhattacharya D, Singh M,
Bhaskar A, Kumar S, Fatima S, Sobia P, Kaer LV and Das G: Allicin
enhances antimicrobial activity of macrophages during Mycobacterium
tuberculosis infection. J Ethnopharmacol. 243:1116342019.
View Article : Google Scholar
|
|
4
|
Behrouj H, Ziamajidi N, Abbasalipourkabir
R, Goodarzi MT and Saidijam M: Hypoglycemic and antioxidant effects
of oral administration of garlic extract in the livers of type 1
diabetic rats. J Basic Clin Physiol Pharmacol. 30:245–250. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Das B and Sinha D: Diallyl disulphide
suppresses the cannonical Wnt signaling pathway and reverses the
fibronectin-induced epithelial mesenchymal transition of A549 lung
cancer cells. Food Funct. 10:191–202. 2019. View Article : Google Scholar
|
|
6
|
Su B, Su J, Zeng Y, Ding E, Liu F, Tan T,
Xia H, Wu YH, Zeng X, Ling H, et al: Diallyl disulfide inhibits
TGF-β1-induced upregulation of Rac1 and β-catenin in
epithelial-mesenchymal transition and tumor growth of gastric
cancer. Oncol Rep. 39:2797–2806. 2018.PubMed/NCBI
|
|
7
|
Li Y, Wang Z, Li J and Sang X: Diallyl
disulfide suppresses FOXM1-mediated proliferation and invasion in
osteosarcoma by upregulating miR-134. J Cell Biochem. Nov 1. pp.
2018Epub ahead of print.
|
|
8
|
Ko JW, Jeong SH, Kwon HJ, Shin NR, Seo YS,
Kim JC, Shin IS and Kim JS: Preventive effect of garlic oil and its
organosulfur component diallyl-disulfide on cigarette smoke-induced
airway inflammation in mice. Nutrients. 10:E16592018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh AK, Bishayee A and Pandey AK:
Targeting histone deacetylases with natural and synthetic agents:
An emerging anticancer strategy. Nutrients. 10:E7312018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Y, Chi Z, Gao R and Lei Z: The roles
of natural compounds in epigenetics. Nat Product Commun.
13:1067–1072. 2018.
|
|
11
|
Schroeder BO and Bäckhed F: Signals from
the gut microbiota to distant organs in physiology and disease. Nat
Med. 22:1079–1089. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gart E, Souto Lima E, Schuren F, de Ruiter
CGF, Attema J, Verschuren L, Keijer J, Salic K, Morrison MC and
Kleemann R: Diet-independent correlations between bacteria and
dysfunction of gut, adipose tissue, and liver: A comprehensive
microbiota analysis in feces and mucosa of the ileum and colon in
obese mice with NAFLD. Int J Mol Sci. 20:E12018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hrncir T, Hrncirova L, Kverka M and
Tlaskalova-Hogenova H: The role of gut microbiota in intestinal and
liver diseases. Lab Anim. 53:271–280. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Filocamo A, Nueno-Palop C, Bisignano C,
Mandalari G and Narbad A: Effect of garlic powder on the growth of
commensal bacteria from the gastrointestinal tract. Phytomedicine.
19:707–711. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Guan M, Zhao X and Li X: Effects
of garlic polysaccha-ride on alcoholic liver fibrosis and
intestinal microflora in mice. Pharm Biol. 56:325–332. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nair AB and Jacob S: A simple practice
guide for dose conversion between animals and human. J Basic Clin
Pharm. 7:27–31. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai YS, Chen WC, Ho CT, Lu KH, Lin SH,
Tseng HC, Lin SY and Sheen LY: Garlic essential oil protects
against obesity-triggered nonalcoholic fatty liver disease through
modulation of lipid metabolism and oxidative stress. J Agric Food
Chem. 62:5897–5906. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang YH, Lei ZL, Huang L, Yang F, Zhang N,
Yuan J, Li K, Chen J and Zhang J: Antitumor ability of berberine
accompanied by modulation of gut microbiome in sarcoma-180
tumor-bearing mice. Int J Pharmacol. 14:460–470. 2018. View Article : Google Scholar
|
|
19
|
Caporaso JG, Kuczynski J, Stombaugh J,
Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich
JK, Gordon JI, et al: QIIME allows analysis of high-throughput
community sequencing data. Nat Methods. 7:335–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edgar RC: UPARSE: Highly accurate OTU
sequences from microbial amplicon reads. Nat Methods. 10:996–998.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Q, Garrity GM, Tiedje JM and Cole JR:
Naive Bayesian classifier for rapid assignment of rRNA sequences
into the new bacterial taxonomy. Appl Environ Microbiol.
73:5261–5267. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kemp PF and Aller JY: Bacterial diversity
in aquatic and other environments: What 16S rDNA libraries can tell
us. FEMS Microbiol Ecol. 47:161–177. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frosini BV: Descriptive measures of
ecological diversity. Environmetrics, in Encyclopedia of Life
Support Systems (EOLSS). Developed under the Auspices of the UNESCO
Eolss Publishers; Oxford: 2004, http://www.eolss.net/ebooks/sample%20chapters/c02/e4-26-01-01.pdf
|
|
24
|
Segata N, Izard J, Waldron L, Gevers D,
Miropolsky L, Garrett WS and Huttenhower C: Metagenomic biomarker
discovery and explanation. Genome Biol. 12:R602011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bae J, Kumazoe M, Fujimura Y and Tachibana
H: Diallyl disulfide potentiates anti-obesity effect of green tea
in high-fat/high-sucrose diet-induced obesity. J Nutr Biochem.
64:152–161. 2019. View Article : Google Scholar
|
|
26
|
Ryu JH and Kang D: Physicochemical
properties, biological activity, health benefits, and general
limitations of aged black garlic: A review. Molecules. 22:E9192017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hosseini A and Hosseinzadeh H: A review on
the effects of Allium sativum (Garlic) in metabolic syndrome. J
Endocrinol Invest. 38:1147–1157. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Elkayam A, Mirelman D, Peleg E, Wilchek M,
Miron T, Rabinkov A, Oron-Herman M and Rosenthal T: The effects of
allicin on weight in fructose-induced hyperinsulinemic,
hyper-lipidemic, hypertensive rats. Am J Hypertens.
16:1053–1056
|
|
29
|
Lee CG, Rhee DK, Kim BO, Um SH and Pyo S:
Allicin induces beige-like adipocytes via KLF15 signal cascade. J
Nutr Biochem. 64:13–24. 2019. View Article : Google Scholar
|
|
30
|
Shi Q, Hornsby PJ, Meng Q, Vandeberg JF
and Vandeberg JL: Longitudinal analysis of short-term high-fat diet
on endothelial senescence in baboons. Am J Cardiovasc Dis.
3:107–119. 2013.PubMed/NCBI
|
|
31
|
You M, Jogasuria A, Lee K, Wu J, Zhang Y,
Lee YK and Sadana P: Signal transduction mechanisms of alcoholic
fatty liver disease: Emerging role of Lipin-1. Curr Mol Pharmacol.
10:226–236. 2017. View Article : Google Scholar
|
|
32
|
Chen Y, Rui BB, Tang LY and Hu CM: Lipin
family proteins-key regulators in lipid metabolism. Ann Nutr Metab.
66:10–18. 2015. View Article : Google Scholar
|
|
33
|
Silbernagel G, Scharnagl H, Kleber ME,
Delgado G, Stojakovic T, Laaksonen R, Erdmann J, Rankinen T,
Bouchard C, Landmesser U, et al: LDL triglycerides, hepatic lipase
activity, and coronary artery disease: An epidemiologic and
Mendelian randomization study. Atherosclerosis. 282:37–44. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Matye D, Nguyen N, Zhang Y and Li
T: HNF4α regulates CSAD to couple hepatic taurine production to
bile acid synthesis in mice. Gene Expr. 18:187–196. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miquilena-Colina ME, Lima-Cabello E,
Sánchez-Campos S, García-Mediavilla MV, Fernández-Bermejo M,
Lozano-Rodríguez T, Vargas-Castrillón J, Buqué X, Ochoa B,
Aspichueta P, et al: Hepatic fatty acid translocase CD36
upregulation is associated with insulin resistance,
hyperinsulinaemia and increased steatosis in non-alcoholic
steatohepatitis and chronic hepatitis C. Gut. 60:1394–1402. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Seo YJ, Lee K, Song JH, Chei S and Lee BY:
Ishige okamurae extract suppresses obesity and hepatic steatosis in
high fat diet-induced obese mice. Nutrients. 10:E18022018.
View Article : Google Scholar
|
|
37
|
Bozic M, Guzmán C, Benet M, Sánchez-Campos
S, García-Monzón C, Gari E, Gatius S, Valdivielso JM and Jover R:
Hepatocyte vitamin D receptor regulates lipid metabolism and
mediates experimental diet-induced steatosis. J Hepatol.
65:748–757. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang F, Sun W, Chen J, Jiang L, Yang P,
Huang Y, Gong A, Liu S and Ma S: SREBP-2, a new target of
metformin? . Drug Des Devel Ther. 12:4163–4170. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sakr HF, Hussein AM, Eid EA and AlKhateeb
M: Possible mechanisms underlying fatty liver in a rat model of
male hypogonadism: A protective role for testosterone. Steroids.
135:21–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu J, Cinar R, Xiong K, Godlewski G,
Jourdan T, Lin Y, Ntambi JM and Kunos G: Monounsaturated fatty
acids generated via stearoyl CoA desaturase-1 are endogenous
inhibitors of fatty acid amide hydrolase. Proc Natl Acad Sci USA.
110:18832–188327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
de Fourmestraux V, Neubauer H, Poussin C,
Farmer P, Falquet L, Burcelin R, Delorenzi M and Thorens B:
Transcript profiling suggests that differential metabolic
adaptation of mice to a high fat diet is associated with changes in
liver to muscle lipid fluxes. J Biol Chem. 279:50743–50753. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gallego-Escuredo JM, Gómez-Ambrosi J,
Catalan V, Domingo P, Giralt M, Frühbeck G and Villarroya F:
Opposite alterations in FGF21 and FGF19 levels and disturbed
expression of the receptor machinery for endocrine FGFs in obese
patients. Int J Obes (Lond). 39:121–129. 2015. View Article : Google Scholar
|
|
43
|
Zhang F, Yu L, Lin X, Cheng P, He L, Li X,
Lu X, Tan Y, Yang H, Cai L and Zhang C: Minireview: Roles of
fibroblast growth factors 19 and 21 in metabolic regulation and
chronic diseases. Mol Endocrinol. 29:1400–1413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gérard C and Vidal H: Impact of gut
microbiota on host glycemic control. Front Endocrinol (Lausanne).
10:pp. 292019, View Article : Google Scholar
|
|
45
|
Halawa MR, El-Salam MA, Mostafa BM and
Sallout SS: The gut microbiome, lactobacillus acidophilus; relation
with type 2 diabetes mellitus. Curr Diabetes Rev. Feb 6–2019, Epub
ahead of print. View Article : Google Scholar
|
|
46
|
Ai D, Pan H, Han R, Li X, Liu G, Xia LC,
et al: Using decision tree aggregation with random forest model to
identify gut microbes associated with colorectal cancer. Genes
(Basel). 10. pp. E1122019, View Article : Google Scholar
|
|
47
|
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini
V, Mardis ER and Gordon JI: An obesity-associated gut microbiome
with increased capacity for energy harvest. Nature. 444:1027–1031.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Islam KB, Fukiya S, Hagio M, Fujii N,
Ishizuka S, Ooka T, Ogura Y, Hayashi T and Yokota A: Bile acid is a
host factor that regulates the composition of the cecal microbiota
in rats. Gastroenterology. 141:1773–1781. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zheng X, Huang F, Zhao A, Lei S, Zhang Y,
Xie G, Chen T, Qu C, Rajani C, Dong B, et al: Bile acid is a
significant host factor shaping the gut microbiome of diet-induced
obese mice. BMC Biol. 15:1202017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Belizário JE, Faintuch J and
Garay-Malpartida M: Gut micro-biome dysbiosis and immunometabolism:
New frontiers for treatment of metabolic diseases. Mediators
Inflamm. 2018.2037838:2018.
|
|
51
|
Litvak Y, Byndloss MX, Bäumler AJ, et al:
Colonocyte metabolism shapes the gut microbiota. Science. 362:pp.
eaat90762018, View Article : Google Scholar : PubMed/NCBI
|