Introduction
Long QT syndrome (LQTs) is an inherited cardiac
arrhythmia syndrome that is associated with syncope and sudden
death (1-3). It has been shown that mutations
exist in 15 genes encoding either ion channels or structural
proteins associated with LQTs (4-7). A
previous study revealed that nonsense mutations, which introduce
premature termination codons (PTCs) into the gene of interest,
account for ~10% of all mutations (8). When a nonsense mutation occurs in
the human etheragogorelated (HERG) gene, which encodes the
α-subunit of the rapid delayed rectifier potassium current channel
(IKr), it may lead to type 2 LQTs.
Aminoglycosides have been demonstrated to have the
ability of read-through on nonsense mutations to produce
full-length functional proteins in genetic disease models (9-14).
Howard et al (11) showed
for the first time that G418 suppressed PTCs to restore the
function of mutant proteins in transiently expressed cDNAs that
contained mutations in the cystic fibrosis transmembrane
conductance regulator gene in mammalian cells. Our published
previously study also demonstrated that G418 and gentamicin
partially restored the HERG protein in a dosedependent manner in
293 cells expressing R1014X mutant (15).
The importance of the local sequence context in
determining how efficiently aminoglycosides rescue nonsense
mutations has been established previously in disease models
(16-18). It appears that different stop
codons facilitate the termination process with differing
efficiencies. The efficiency with which termination may be
suppressed can also be influenced by the local sequence context in
the immediate vicinity of the stop codons, and for the majority of
the disease models, the strongest bias has been found at the
position of the base immediately following the stop codon (i.e.,
the +4 nucleotide). However, at present, how the sequence context
may influence the efficiency of aminoglycosides in terms of
rescuing HERG nonsense mutants in mammalian cells remains to be
fully elucidated. In an attempt to answer this question, the
present study aimed to examine the susceptibility of different
termination codons, and the local nucleotide sequence surrounding
them, in the process of rescuing HERG nonsense mutants by
aminoglycosides in 293 cells.
Materials and methods
HERG mutant cDNA constructs
Wildtype (WT) HERG cDNA cloned into the pcDNA3.1
vector was provided by Dr Zhengfeng Zhou (Oregon Health and Science
University, Portland, OR, USA). The HERG nonsense mutations R1014X
and W927X, containing different stop codons and +4 nucleotides,
were generated in the pcDNA3.1 WT HERG plasmid using a polymerase
chain reaction (PCR) based mutagenesis method. PCR was performed as
follows: 95°C for 2 min, 94°C for 20 sec, 55°C for 10 sec, 68°C for
2.5 min for 18 cycles, 68°C for 5 min. Briefly, the plasmid
containing WT HERG cDNA was purified using Endo-Free Plasmid
Purification kit (Qiagen, Inc.) according to the manufacture's
protocol, and used as the PCR template. The primers with the
different mutation sites were added (sequences of primers are shown
in Table I), respectively.
Amplification was performed using Fast Alteration DNA Polymerase,
and a mutated plasmid with a gap was obtained. The methylated
template plasmid was digested with DpnI, leaving the mutant
plasmid only. Following transfer to the recipient Escherichia
coli bacteria, the gap in the mutant plasmid was repaired. The
mutant plasmid was obtained and the sequences of the mutants were
subsequently verified using ABI PRISM 377 automated sequencer.
 | Table IPrimer sequences of the site-directed
mutants. |
Table I
Primer sequences of the site-directed
mutants.
| Site-directed
mutant | Primer
sequence |
|---|
| R1014X-TGAT | F:
5′-GTACCAGGAGCTCCCTTGATGCCCCGCCCCC-3′ |
| R:
5′-GGGGGCGGGGCATCAAGGGAGCTCCTGGTAC-3′ |
| R1014X-TAAT | F:
5′-GTACCAGGAGCTCCCTTAATGCCCCGCCCCC-3′ |
| R:
5′-GGGGGCGGGGCATTAAGGGAGCTCCTGGTAC-3′ |
| R1014X-TAGT | F:
5′-GTACCAGGAGCTCCCTTAGTGCCCCGCCCCC-3′ |
| R:
5′-GGGGGCGGGGCACTAAGGGAGCTCCTGGTAC-3′ |
| R1014X-TGAC | F:
5′-GTACCAGGAGCTCCCTTGACGCCCCGCCCCC-3′ |
| R:
5′-GGGGGCGGGGCGTCAAGGGAGCTCCTGGTAC-3′ |
| R1014X-TGAG | F:
5′-GTACCAGGAGCTCCCTTGAGGCCCCGCCCCC-3′ |
| R:
5′-GGGGGCGGGGCCTCAAGGGAGCTCCTGGTAC-3′ |
| R1014X-TGAA | F:
5′-GTACCAGGAGCTCCCTTGAAGCCCCGCCCCC-3′ |
| R:
5′-GGGGGCGGGGCTTCAAGGGAGCTCCTGGTAC-3′ |
| W927X-TGAG | F:
5′-CGGGGGGGCCGTGAGGGGAGAGCCCG-3′ |
| R:
5′-CGGGCTCTCCCCTCACGGCCCCCCCG-3′ |
| W927X-TAAG | F:
5′-CGGGGGGGCCGTAAGGGGAGAGCCCG-3′ |
| R:
5′-CGGGCTCTCCCCTTACGGCCCCCCCG-3′ |
| W927X-TAGG | F:
5′-CGGGGGGGCCGTAGGGGGAGAGCCCG-3′ |
| R:
5′-CGGGCTCTCCCCCTACGGCCCCCCCG-3′ |
Cell culture and transfection
The 293 cells (American Type Culture Collection)
were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.), 1% glutamine, 100
IU/ml penicillin and 100 µg/ml streptomycin at 37°C in an
incubator in a humidified atmosphere of 5% CO2/95% air.
Transfections were performed with Effectene®
transfection reagent (Qiagen, Inc.) according to the manufacturer's
protocol. The 293 cells were transiently transfected either with
0.4 µg WT or an equal quantity of mutant cDNA. The green
fluorescent protein (GFP) gene (0.2 µg) was cotransfected as
an indicator during patch-clamp recording. The transfection
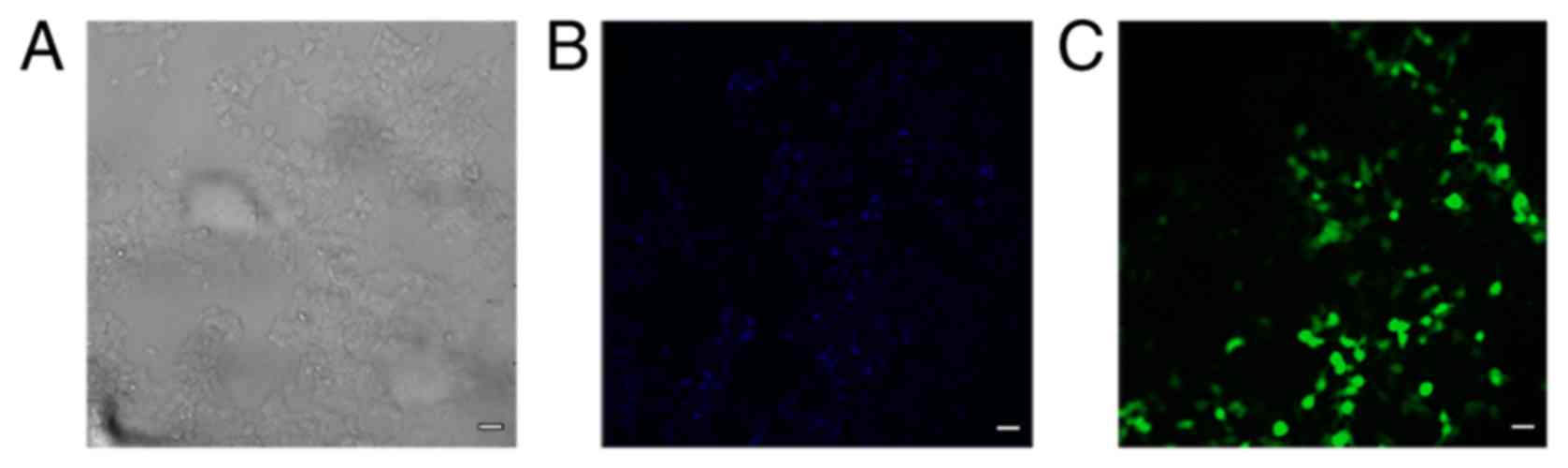
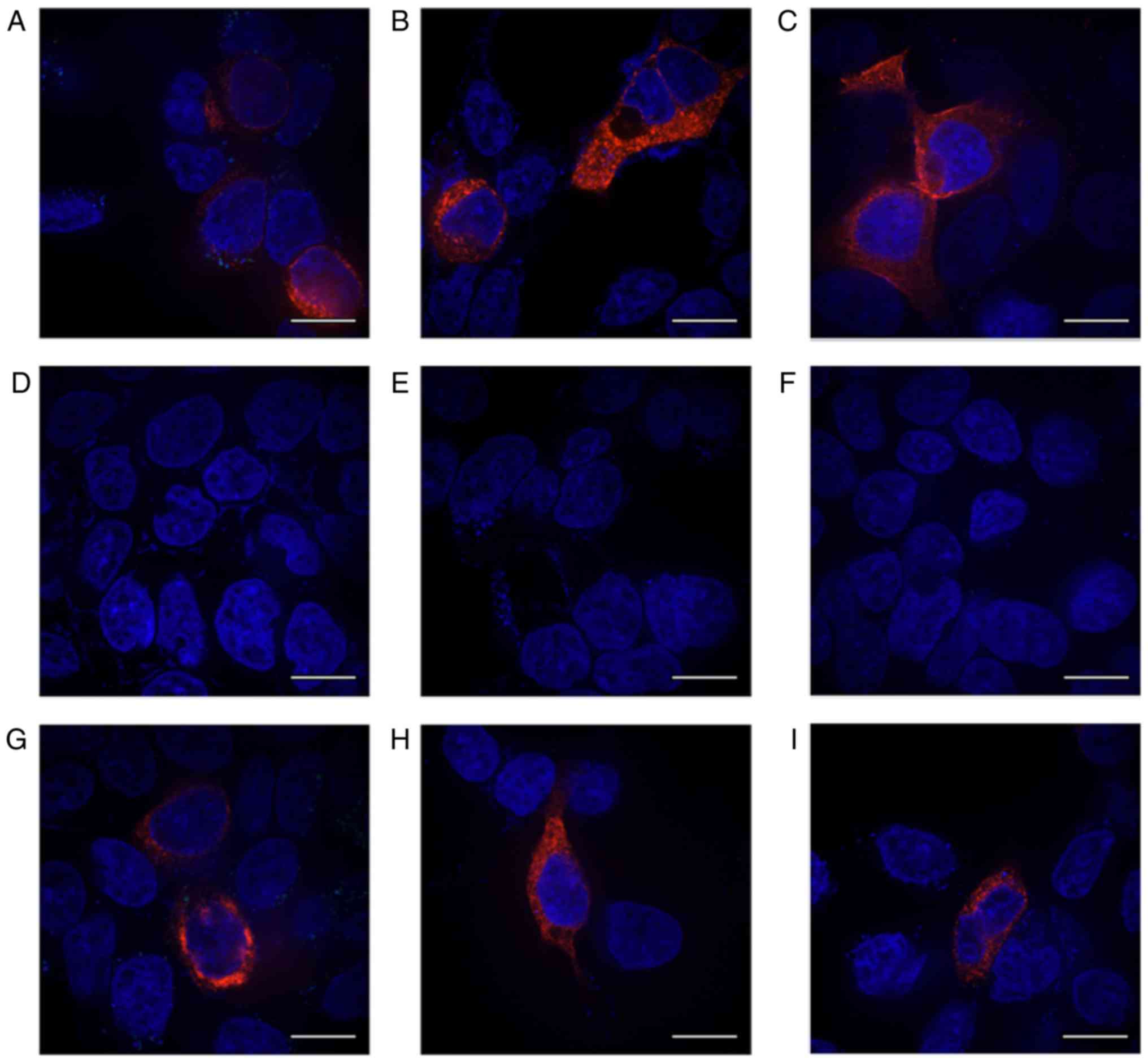
efficiency was ~65% (Fig. 1A-C).
Pharmacological rescue was examined 24 h after adding G418 (400
µg/ml) into DMEM.
Immunofluorescence and confocal
imaging
An immunofluo-rescence method was used to observe
the protein expression of HERG prior to and following drug
treatment. The 293 cells were washed three times with PBS and were
subsequently soaked in 4% paraformaldehyde for 30 min at room
temperature, followed by washing with PBS a further three times.
Following permeabilization with 0.5% Triton X-100 for 20 min, the
cells were incubated in antisera against the N- or C-terminal
antibody of HERG protein (Alomone, cat. nos. APC-062 and APC-109,
respectively; 1:400 dilution), initially at 37°C for 1 h, and then
at 4°C overnight. Subsequently, the cells were washed three times
in PBS and incubated with antirabbit Cy3-conjugated secondary
antibody (Beyotime Institute of Biotechnology, P0183; 1:500
dilution) at 37°C for 30 min, followed by three washes with PBS.
Finally, cells were incubated with DAPI (Haoxin Biotech, Inc. or
Beyotime Institute of Biotechnology) for 5 min and then were washed
three times in PBS. Confocal images were captured using a confocal
laser scanning microscope (Zeiss LSM510; Zeiss GmbH, Jena, Germany)
and analyzed using Volocity® Demo software (version 6.0;
PerkinElmer, Inc.).
Patch-clamp recording
The wholecell patch-clamp method was used in
experiments to record the HERG current. The bathing solution
contained the following: 136 mM NaCl, 5.4 mM KCl, 0.33 mM
NaH2PO4.2H2O, 1.8 mM
CaCl2, 1 mM MgCl2.6H2O, 10 mM
glucose and 10 mM HEPES. The internal pipette solution contained
the following: 140 mM KCl, 0.5 mM MgCl2.6H2O,
5 mM HEPES, 2 mM EGTA and 4 mM K2ATP. The HERG current
was activated by a depolarizing pulse (for 4 sec) to 50 mV from the
holding potential of −80 mV. The tail current of the HERG channel
was recorded following repolarization to −50 mV for 4 sec. All
patch-clamp experiments were performed at room temperature
(22±1°C).
Statistical analysis
All results are expressed as the mean ± SEM, and the
data were analyzed using SPSS software, version 17.0 (SPSS, Inc.,
Chicago, IL, USA). For statistical analyses, Student's t-test was
performed for comparisons between two data groups. For comparisons
of data among more than two groups, one-way analysis of variance
was used, followed by Bonferroni post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Construction of HERG nonsense mutants
containing different stop codons and different downstream
nucleotides
The present study aimed to investigate whether
different types of stop codon and the downstream nucleotide
context, can influence the ability of aminoglycosides to
efficiently suppress PTCs. To accomplish this, R1014X and W927X
mutants, containing TGA, TAG and TAA, were constructed using a
sitedirected mutagenesis technique. Based on the R1014X-TGAT
mutant, R1014X-TGAA, R1014X-TGAC and R1014X-TGAG mutants were also
constructed for the present study (Fig. 2A and B).
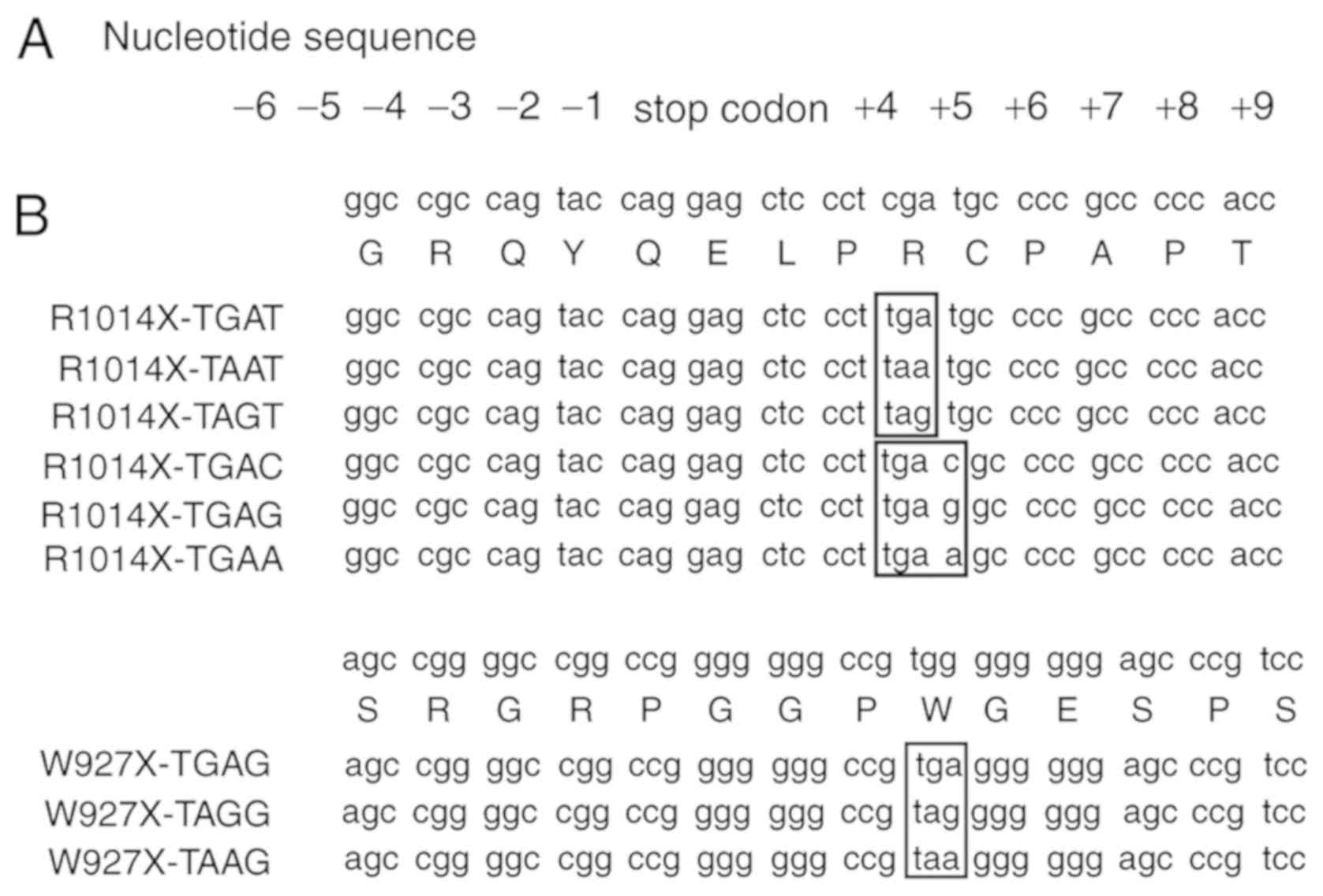 | Figure 2R1014X and W927X mutants. (A)
Nucleotide sequence surrounding the stop codon. (B) R1014X and
W927X mutants containing different stop codons (TGA, TAA and TAG
within boxes) and different +4 nucleotides immediately downstream
of the stop codon, TGA, are shown. The single-letter abbreviations
of the amino acids are as follows: A, alanine; R, arginine; C,
cysteine; Q, glutamine; E, glutamic acid; G, glycine; L, leucine;
P, proline; S, serine; T, threonine; W, tryptophan; Y,
tyrosine. |
Effect of PTCs on the G418-mediated
suppression of the R1014X and W927X mutants
Using an immunofluorescence method, the protein
expression of HERG in 293 cells transfected with the respective
R1014X mutants was detected. Protein expression was detected in the
control group (transfection reagent only) and the HERG-WT group
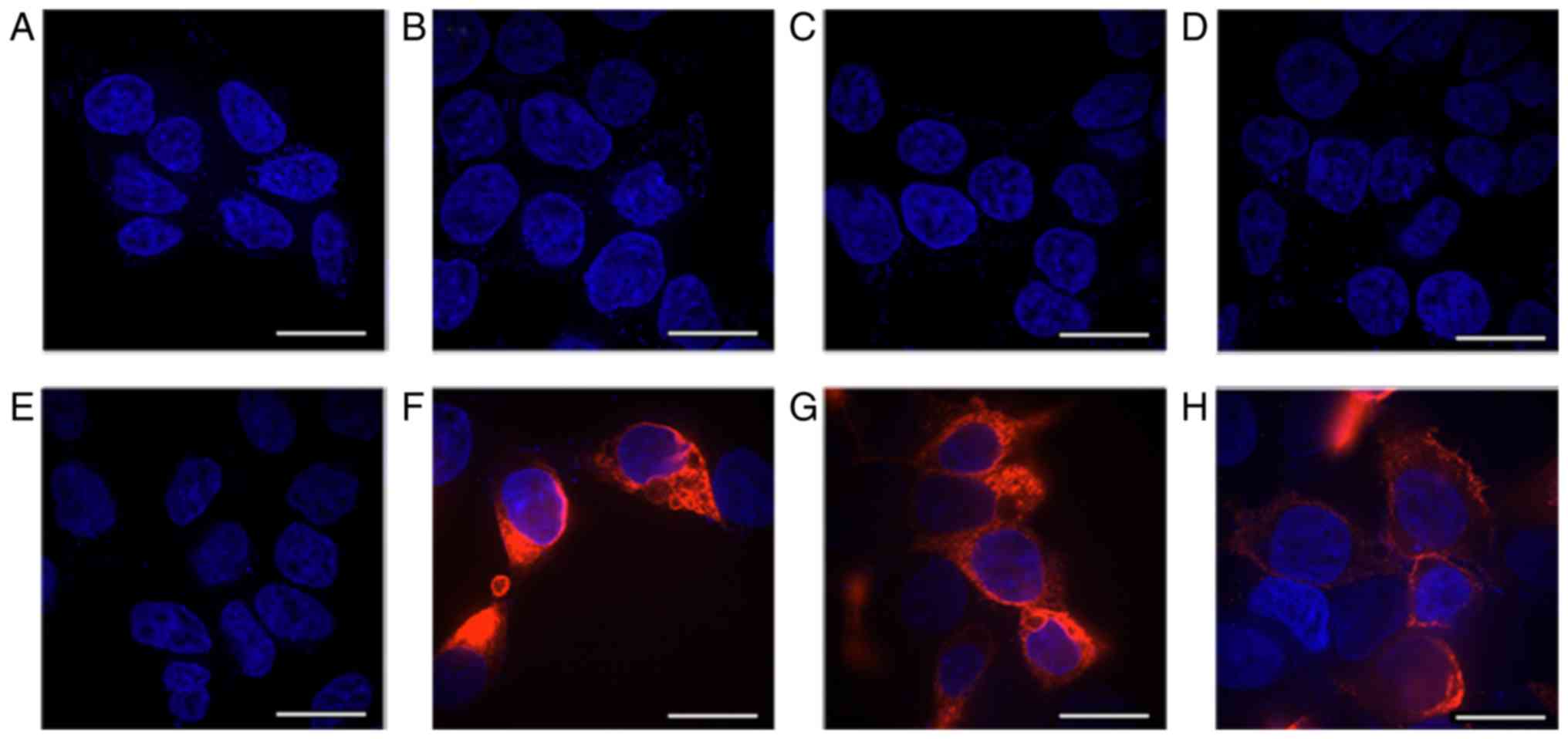
prior to and following G418 treatment in 293 cells (Fig. 3). No red fluorescence was detected
in the control group with or without G418 treatment (Fig. 3A-D). In the WT group, red
fluorescence was detected when antibody against the N-terminal
(Fig. 3F), or C-terminal
(Fig. 3G) were used prior to G418
treatment, but not when no antibody was added (Fig. 3E). Following treatment with G418,
red fluorescence was also detected following addition of the
anti-C-terminal antibody (Fig.
3H). An antibody raised against the N-terminal sequence of HERG
protein was incubated with the protein prior to the addition of
G418. As shown in Fig. 4A-C, red
fluorescence was detected in all three groups of cells (i.e.,
transfected with R1014X-TGA, R1014X-TAA or R1014X-TAG mutant,
respectively). By contrast, no red fluorescence was detected when
an antibody raised against the C-terminus of the HERG protein was
used (Fig. 4D-F). However,
following treatment with G418 (400 µg/ml), red fluorescence
was observed in each group following incubation with the
anti-C-terminal antibody (Fig.
4G-I).
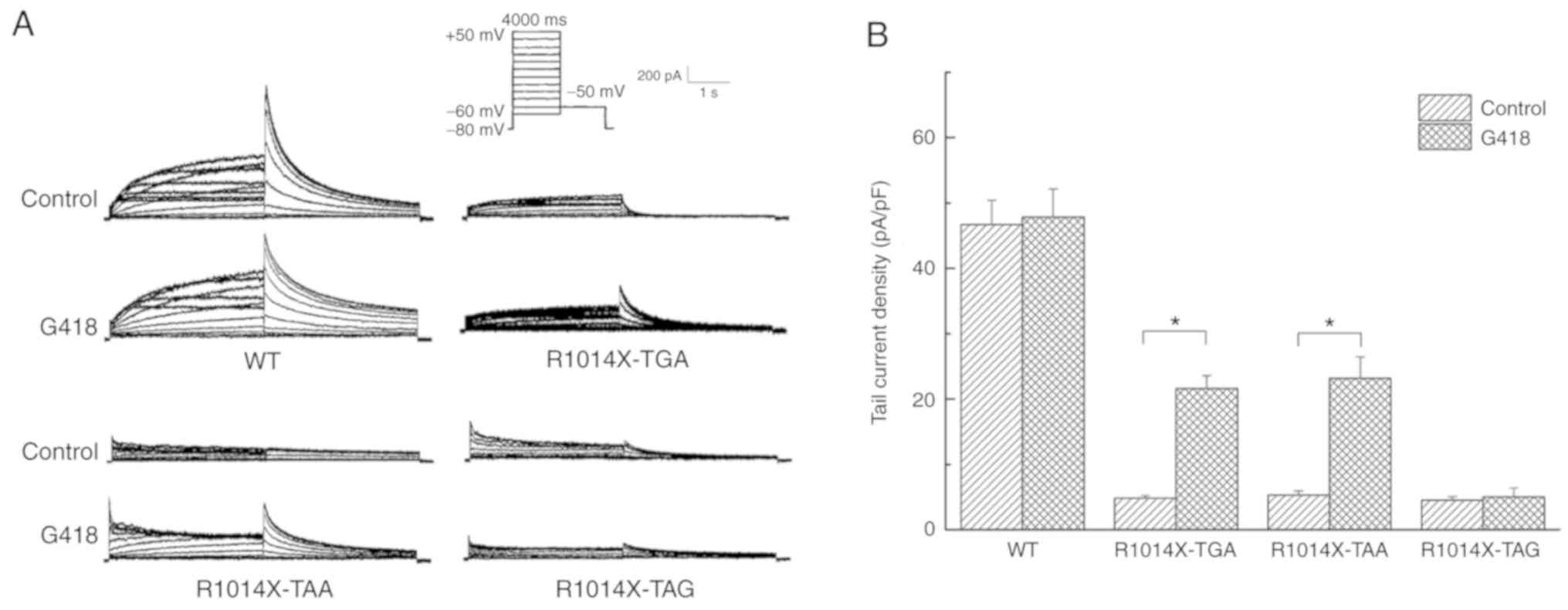
Ionic currents were also recorded for the 293 cells
transiently transfected with WT cDNA and R1014X mutant containing
the different stop codons (Fig.
5A). The mean peak tail current densities of R1014X-TGA
(4.82±0.42 pA/pF; n=8), R1014X-TAA (5.31±0.62 pA/pF; n=6) and
R1014X-TAG (4.51±0.68 pA/pF; n=6) were all lower compared with
those of the WT channel (46.69±3.74 pA/pF; n=6; P<0.05).
However, no significant differences were observed among these three
mutants. An increase in the peak tail current was only identified
with the R1014X-TGA (21.64±1.94 pA/pF; n=6; P<0.05) and
R1014X-TAA (23.19±3.27 pA/pF; n=6; P<0.05) mutants, and not with
the R1014X-TAG mutant, subsequent to the rescuing effect of G418
(Fig. 5B).
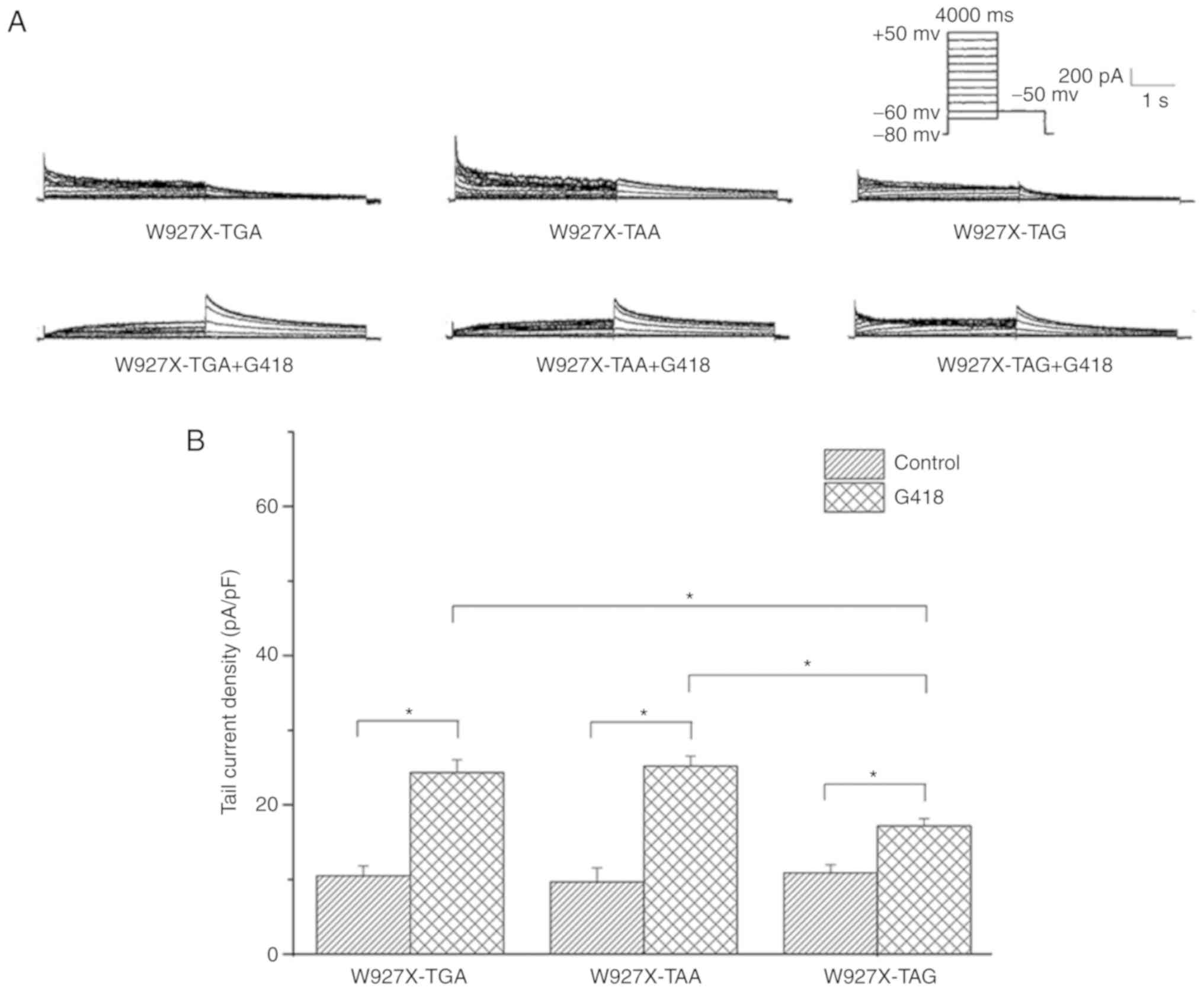
To further validate these phenomena, the same
treatment was applied to W927X mutants, which featured the PTCs of
TGA, TAA and TAG, respectively. The immunofluorescence results
(Fig. 6A-I) revealed that red
fluorescence was detected in all three mutants bearing the
different types of PTC following the administration of G418 using
the antibody raised against the C-terminus (Fig. 6). The patchclamp results suggested
that the mean peak tail electric current density was increased in
293 cells transfected with W927X-TGA, W927X-TAA and W927X-TAG
following treatment with G418. However, the increase in current
density with W927X-TAG was smaller compared with that with the
other two mutants (Fig. 7A and B;
P<0.05).
Role of the tetranucleotide termination
signal on the rescuing effect of G418
To observe the impact of the local sequence context
surrounding the PTC, the predominant focus of the present study was
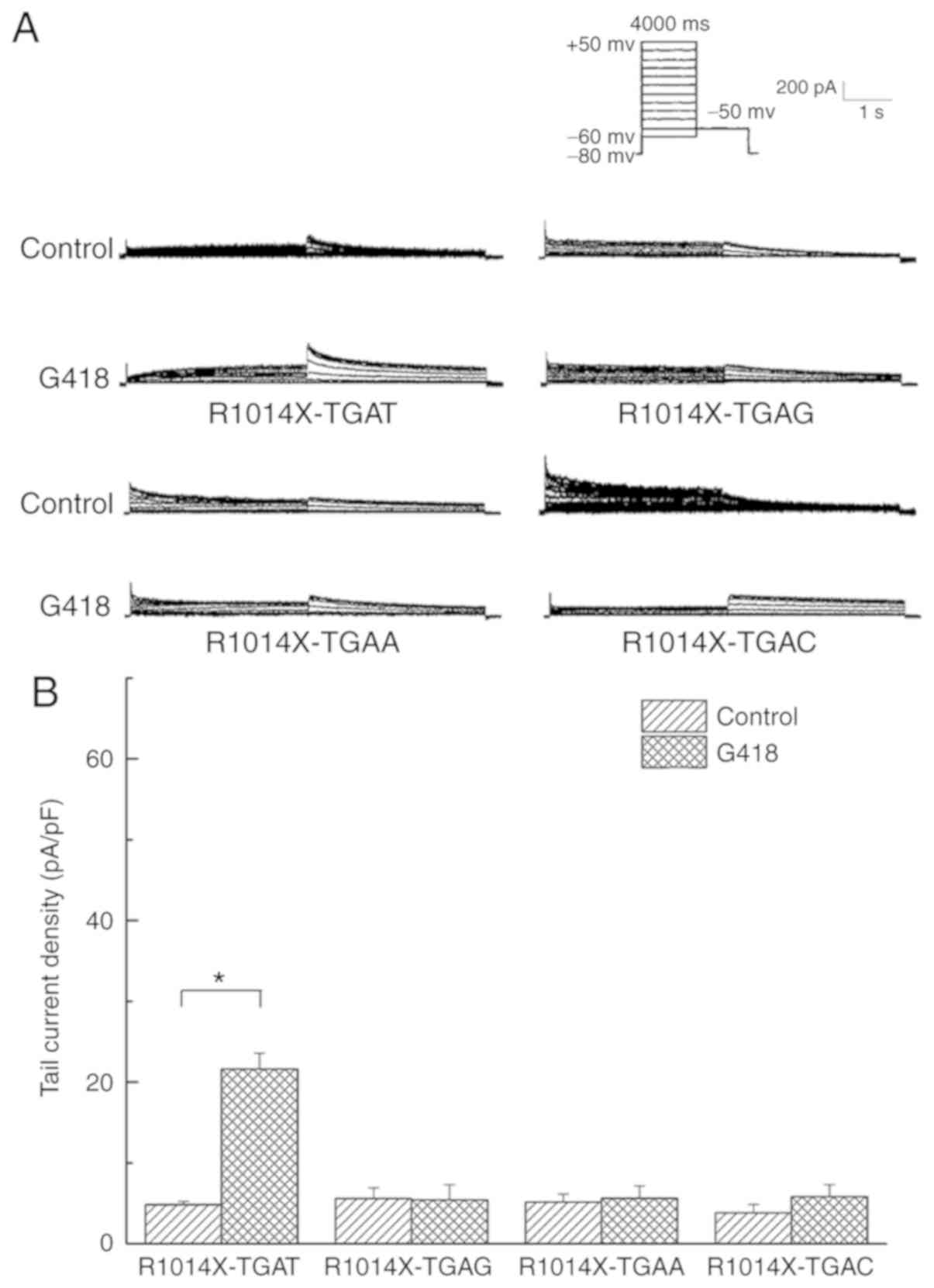
on the +4 nucleotide, and based on the R1014X-TGAT mutant,
R1014X-TGAC, R1014X-TGAG and R1014X-TGAA mutants were constructed.
Prior to the addition of G418, the N-terminal antibody was used to
observe the protein expression of HERG among the groups. Using the
immunofluorescence method, red fluorescent protein was detected in
each group (Fig. 8A-C). However,
when the Cterminal antibody was used, no red fluorescence was
detected (Fig. 8D-F). The
functional expression of HERG protein in each group without drug
treatment was subsequently tested by patch-clamp recording. On
comparing the peak tail current densities, no significant
differences were identified among the groups (5.58±1.33, 5.16±0.99
and 3.80±1.08 pA/pF for the R1014X-TGAC, R1014X-TGAG and
R1014X-TGAA mutants, respectively; Fig. 9A and B).
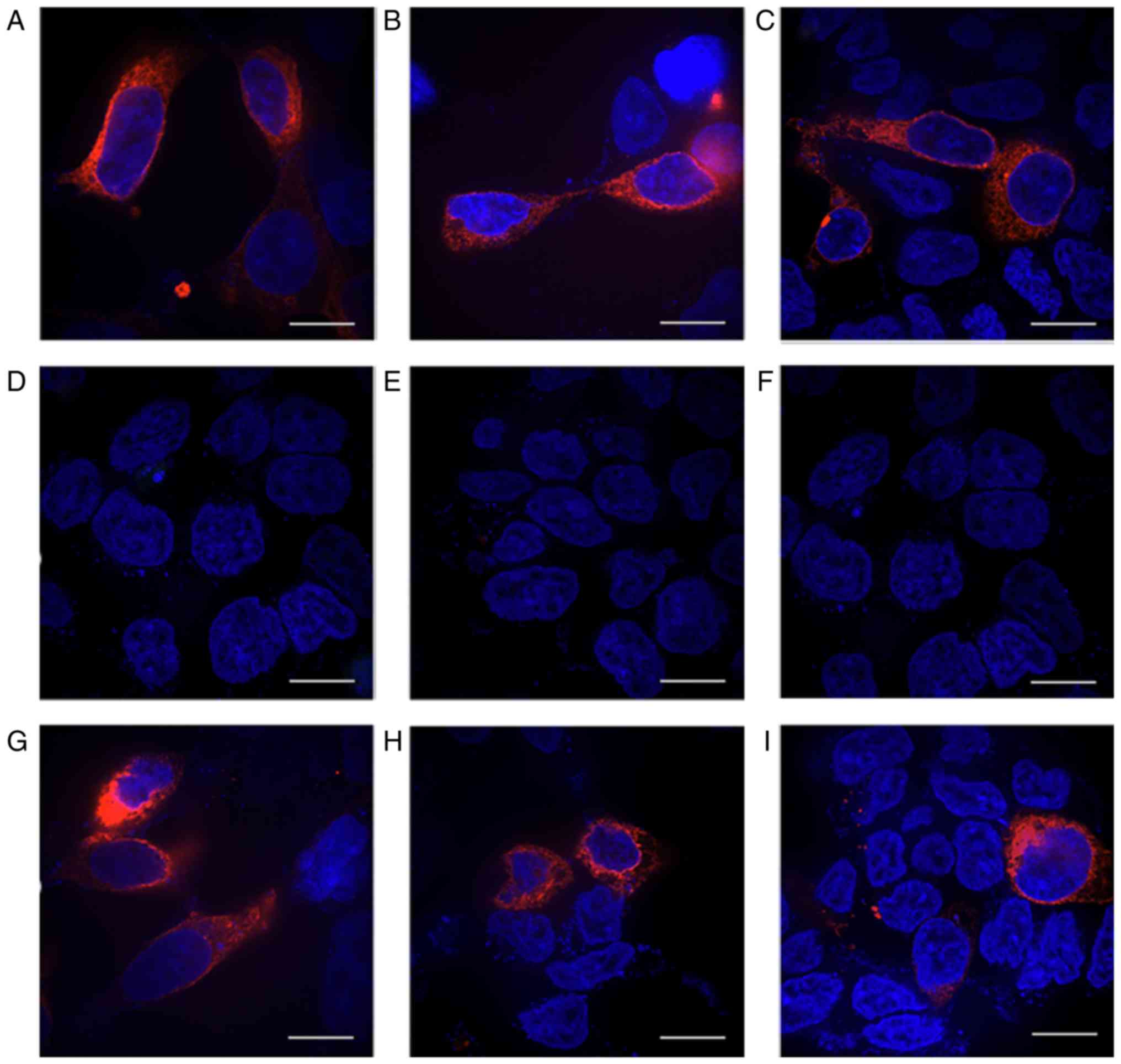
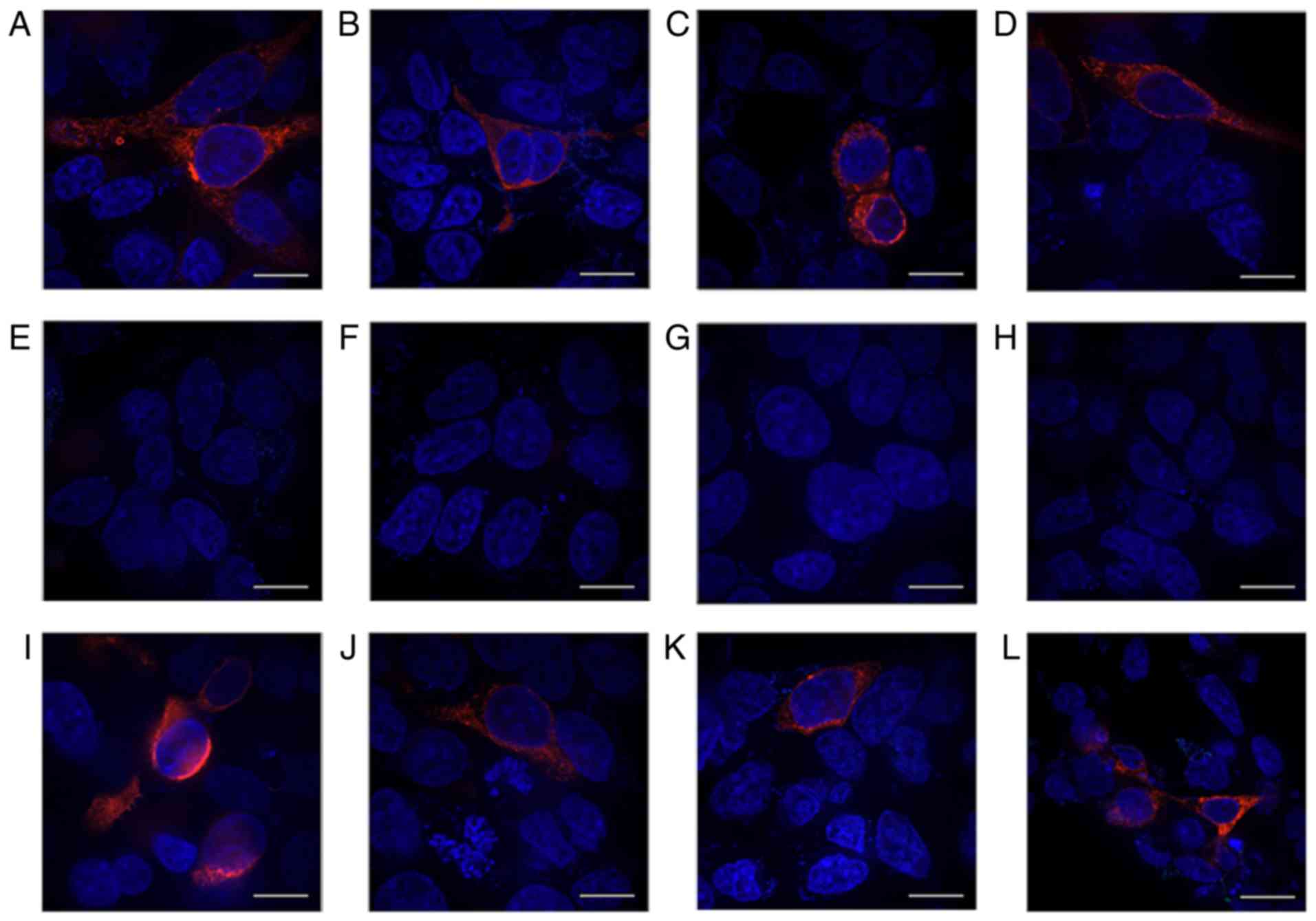 | Figure 8Protein expression in cells
transfected with HERG-R1014X mutant containing different +4
nucleotides downstream of the stop codon TGA before and after G418
treatment. Antibody against the N-terminus of the protein was used
before G418 was added to mutants with the +4 nucleotides (A) T, (B)
G, (C) A and (D) C, respectively. Antibody against the C-terminus
of the protein was used before mutants with the +4 nucleotides (E)
T, (F) G, (G) A and (H) C were treated with G418. (I-L) Following
treatment with G418, anti-C-terminal antibody was used for the (I)
T, (J) G, (K) A and (L) C mutants. Red fluorescence represents
expression of the HERG protein. Scale bar, 100 µm. HERG,
human ether-a-go-go-related. |
Following G418 treatment, red fluorescence was
detected in all groups using the C-terminal antibody (Fig. 8G-I). For the group transfected
with R1014X-TGAT, the peak tail current density was increased in
the cells treated with G418 (22.96±1.75 pA/pF; n=5; P<0.05)
compared with that in the untreated cells (4.54±0.51 pA/pF; n=6),
although this was still lower compared with that of the WT channel
(47.83±4.31 pA/pF; n=5). In the other groups transfected with
R1014X-TGAC, R1014X-TGAG or R1014X-TGAA, the tail current density
did not alter significantly upon treatment with G418 (5.58±1.33 vs.
5.45±1.84 pA/pF, 5.16±0.99 vs. 5.65±1.50 pA/pF and 3.80±1.08 vs.
5.84±1.46 pA/pF, respectively; all P>0.05; Fig. 9A and B).
Discussion
In the present study, to investigate how stop codons
and the +4 nucleotide may influence the efficiency of G418 in
rescuing nonsense mutations of the HERG gene, the possible
impacting factors were examined, such as the type of PTC and the
nucleotide sequences immediately downstream of them. It was
revealed that the stop codons TGA and TAA were rescued more easily
by the aminoglycoside compared with TAG. In addition, the +4
nucleotide influenced the function of HERG protein following the
rescuing effect of the aminoglycoside. This is the first time, to
the best of our knowledge, that the effect of the intrinsic
sequence on the nonsense mutation rescue of the HERG gene has been
observed. The HERG gene was a particularly good candidate for PTC
suppression, as it has a higher frequency of nonsense mutations
than other genes associated with arrhythmia and sudden death, the
majority of which are inactivated by missense mutations.
The read-through effect on nonsense mutations to
produce full-length functional proteins has been demonstrated for
aminoglycosides in numerous genetic disease models, including
Duchenne muscular dystrophy (DMD) and ocular genetic diseases
(19,20). In our previous study, the
expression of WT and R1014X mutant in 293 cells was confirmed by
western blotting and PCR following the transfection of
corresponding plasmids, respectively. It was also demonstrated that
G418 and gentamicin partially restored the HERG-like currents in
293 cells expressing W927X and R1014X mutants (15). It has also been reported that the
type of stop codon can influence the rescuing efficiency of an
aminoglycoside. Gentamicin and G418 enabled aminoacyl-tRNA to read
through UGA more easily than other PTCs in disease models
associated with APC or p53 gene nonsense mutations (18,21). In clinical trials associated with
DMD, upon administration of gentamicin, the level of the myotubes
was upregulated more markedly in patients with the PTC of UGA,
followed by UAA and UAG (22). In
the present study, the results confirmed that G418 suppressed UGA
and UAA, and the IKr current was restored more readily for these
PTCs than for UAG in HERG nonsense mutations.
The context of the stop codon and its surrounding
sequences influence the efficiency of translation termination, as
has been reported in previous studies (23,24). However, the mechanism remains
unclear. Aminoglycoside antibiotics can exert their effect by
binding to the A site of ribosomal rRNA via hydrogen bonds, thus
altering the conformation of ribosomal rRNA, mRNA and
aminoacyl-tRNA. This effect reduces the fidelity of the translation
process, causing similar aminoacyl-tRNAs to enter the A site. When
an amino is added to the stop codon site, the read-through effect
occurs (25).
It has been reported that, when WT AUGG was mutated
to AUAG, AUGA and AUAA in nonsense mutation models, the ability to
bind to ribosomal and aminoacyl transfer RNA was 0.1, 3.2 and 0.3
times higher compared with AUGG, respectively (25,26). This means that, among the three
stop codons, UGA was more likely to bind to the aminoacyl-tRNA in
the ribosome, inhibiting the termination of translation and causing
the protein to continue to synthesize. Therefore, G418 may induce
more efficient translational read-through and produce sufficient
full-length protein in the nonsense mutation with stop codon UGA or
UAA. Additional investigations are required to obtain further
understanding of the termination of translation at normal and
premature stop codons. It is important to characterize the identity
of the amino acid inserted by the treatment of G418 in HERG
nonsense mutations with different stop codons.
Nonsense-mediated decay (NMD) is an RNA quality
control mechanism that selectively degrades mRNA harboring PTCs.
NMD eliminates abnormal mRNA transcripts harboring PTCs, thereby
preventing the production of truncated proteins that often have
dominant-negative effects (27).
However, the role of NMD in the majority of eukaryotic cells
requires at least one intron downstream of the PTCs and the
corresponding splicing process. Pre-mRNA splicing results in the
deposition of a multi-protein complex, known as the
exon-junction-complex (EJC), 20-24 nt upstream of each exon-exon
junction. The EJCs are displaced by the ribosome during the pioneer
round of translation. If translation terminates at a PTC that is
located 50-55 nt upstream of an exon-exon junction, the downstream
EJC serves as a binding platform for NMD factors that trigger NMD.
If no intron exists, the mRNA can escape NMD. In the present study,
cDNA of the HERG gene containing exons was used to rule out the
interference of NMD on the experimental results.
The efficiency of translation termination is also
affected by upstream and downstream sequences around the PTC,
particularly by the +4 nucleotide (28,29). When an adenine nucleotide is
located at this position, the highest rescuing efficiency may be
detected, followed by a pyrimidine nucleotide at the same position
(30-32). Therefore, it was hypothesized that
the local sequence context featuring the +4 nucleotide is also
involved in the process of translation termination, in addition to
the PTC. Previous studies on DMD and other disease models have
confirmed the important influence of the +4 nucleotide (33,34). In a QX (N) expression system, it
was revealed that the stop codon had a close association with the
+4 nucleotide (35). Gentamicin
and other aminoglycosides were observed to have different rescuing
efficiencies with different types of nucleotide at the +4 position.
When the stop codon was TGA, the effectiveness of the +4 nucleotide
was in the order of C > T, A > G (35).
The results of the present study suggested that the
+4 nucleotide downstream of the stop codon had a significant impact
on the functional expression of HERG protein following the rescue
of readthrough by the drug. When the original base T was mutated to
C, G or A, respectively, red fluorescence remained apparent
following the addition of an antibody against the C-terminus of
HERG following G418 treatment, indicating expression of the intact
HERG protein. However, the peak tail current did not alter
significantly following the administration of G418. Amino acid
sequence analysis predicted that nucleotide mutation results in the
original-site cysteine (corresponding to the nucleotide sequence,
tgc) being mutated to arginine (corresponding to nucleotide
sequence, cgc), serine (corresponding to nucleotide sequence, agc)
or glycine (corresponding to the codon, ggc). Therefore, a mutation
at the +4 nucleotide base position altered the amino acid sequence
coded by the nucleotides downstream of the stop codon upon
intervention with G418. In this manner, no functional integral
protein can be produced. Notably, these results differed from
previous results obtained in other disease models (31,34). Due to the lack of suitable
technology, it is not possible to analyze and predict the amino
acid that is inserted into the site of the PTC. However, the
results of our previous study demonstrated that certain HERG
nonsense mutants, such as R863X, E698X and W412X, positioned close
to the Nterminus, were not rescued upon treatment with an
amino-glycoside (15); therefore,
the amino acid may differ from the original amino acid upon rescue
with G418. If the mutation is located in the domain of the HERG
gene that is most critical for protein structure, the conformation
of the HERG protein may be altered upon rescue with an
aminoglycoside, leading to its function not being restored.
However, further investigations are required in order to elucidate
the specific mechanism.
In conclusion, the present study demonstrated that
the type of PTC, and the context of the nucleotides downstream of
it, may determine the pharmacological rescue efficiency of the HERG
gene. TGA and TAA were more easily rescued by the aminoglycoside
compared with TAG. Furthermore, the +4 nucleotide was found to
influence the function of the HERG protein following the rescuing
effect of the aminoglycoside. By identifying the type of stop codon
and the following sequence, clinicians can establish patients who
are more sensitive to nonsense mutation rescue therapy among those
with LQT2 in the future. Pacemaker implantation, which is expensive
and traumatic, may be avoided in these individuals. However, these
findings highlight the fact that the mechanism of
aminoglycosidemediated suppression in mammalian cells remains to be
fully elucidated.
In the present study, GFP and the target gene were
not placed in the same plasmid. Therefore, it was only possible to
demonstrate successful transfection of the target gene through the
expression of GFP indirectly. However, by recording the HERG
current and using an antibody against the target gene, the
expression of the target gene was confirmed. Additionally,
transfection of the 293 cells in the experiments was performed with
Effectene transfection reagent according to the manufacturer's
instructions. After 24 h, Pharmacological rescue was assessed by
adding G418 (400 µg/ml) into DMEM for another 24 h. As there
was a period of only 48 h after transfection, it was not possible
to examine the stability of HERG protein in different groups. This
is an important question and it is sensible to examine the
stability of HERG protein with long-tern drug administration in
nonsense mutation animal models.
Acknowledgments
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81070150) and the Beijing
Municipal Natural Science Foundation (grant no. 7192105).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
HY performed the patch-clamp experiment, analyzed
data and was a major contributor in writing the manuscript. YM
performed PCR and immunofluorescence experiments and contributed to
writing the manuscript. SZ and CT performed cell culture and the
transfection experiment. FW, NL and QL analyzed and interpreted the
data collected from patch-clamp experiments. YJ contributed to
improvements in the manuscript and provided financial support for
the study. JP made substantial contributions to conception and
design and provided financial support for the study. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li G, Shi R, Wu J, Han W, Zhang A, Cheng
G, Xue X and Sun C: Association of the hERG mutation with longQT
syndrome type 2, syncope and epilepsy. Mol Med Rep. 13:2467–2475.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Czosek RJ, Kaltman JR, Cassedy AE, Shah
MJ, Vetter VL, Tanel RE, Wernovksy G, Wray J and Marino BS: Quality
of life of pediatric patients with long QT syndrome. Am J Cardiol.
117:605–610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kanters JK, Yuan L, Hedley PL, Stoevring
B, Jons C, Bloch Thomsen PE, Grunnet M, Christiansen M and
Jespersen T: Flecainide provocation reveals concealed brugada
syndrome in a long QT syndrome family with a novel L1786Q mutation
in SCN5A. Circ J. 78:1136–1143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Splawski I, Shen J, Timothy KW, Lehmann
MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent
GM and Keating MT: Spectrum of mutations in long-QT syndrome genes.
KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 102:1178–1185.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marsman RF, Barc J, Beekman L, Alders M,
Dooijes D, van den Wijngaard A, Ratbi I, Sefiani A, Bhuiyan ZA,
Wilde AA and Bezzina CR: A mutation in CALM1 encoding calmodulin in
familial idiopathic ventricular fibrillation in childhood and
adolescence. J Am Coll Cardiol. 63:259–266. 2014. View Article : Google Scholar
|
|
6
|
Mototani H, Iida A, Nakamura Y and Ikegawa
S: Identification of sequence polymorphisms in CALM2 and analysis
of association with hip osteoarthritis in a Japanese population. J
Bone Miner Metab. 28:547–553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ortiz-Bonnin B, Rinne S, Moss R, Streit
AK, Scharf M, Richter K, Stöber A, Pfeufer A, Seemann G, Kääb S, et
al: Electrophysiological characterization of a large set of novel
variants in the SCN5A-gene: Identification of novel LQTS3 and BrS
mutations. Pflugers Arch. 468:1375–1387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rowe SM and Clancy JP: Pharmaceuticals
targeting nonsense mutations in genetic diseases: Progress in
development. Biodrugs. 23:165–174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Floquet C, Hatin I, Rousset JP and Bidou
L: Statistical analysis of readthrough levels for nonsense
mutations in mammalian cells reveals a major determinant of
response to gentamicin. PLoS Genet. 8:e10026082012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brendel C, Belakhov V, Werner H, Wegener
E, Gärtner J, Nudelman I, Baasov T and Huppke P: Readthrough of
nonsense mutations in Rett syndrome: Evaluation of novel
aminoglyco-sides and generation of a new mouse model. J Mol Med
(Berl). 89:389–398. 2011. View Article : Google Scholar
|
|
11
|
Howard M, Frizzell RA and Bedwell DM:
Aminoglycoside antibiotics restore CFTR function by overcoming
premature stop mutations. Nat Med. 2:467–469. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Baradaran-Heravi A, Balgi AD, Zimmerman C,
Choi K, Shidmoossavee FS, Tan JS, Bergeaud C, Krause A, Flibotte S,
Shimizu Y, et al: Novel small molecules potentiate premature
termination codon readthrough by aminoglycosides. Nucleic Acids
Res. 44:6583–6598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McHugh DR, Steele MS, Valerio DM, Miron A,
Mann RJ, LePage DF, Conlon RA, Cotton CU, Drumm ML and Hodges CA: A
G542X cystic fibrosis mouse model for examining nonsense mutation
directed therapies. PLoS One. 13:e1995732018. View Article : Google Scholar
|
|
14
|
Bolze F, Mocek S, Zimmermann A and
Klingenspor M: Aminoglycosides, but not PTC124 (Ataluren), rescue
nonsense mutations in the leptin receptor and in luciferase
reporter genes. Sci Rep. 7:10202017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu H, Liu X, Huang J, Zhang Y, Hu R and Pu
J: Comparison of read-through effects of aminoglycosides and PTC124
on rescuing nonsense mutations of HERG gene associated with long QT
syndrome. Int J Mol Med. 33:729–735. 2014. View Article : Google Scholar
|
|
16
|
Lee HL and Dougherty JP: Pharmaceutical
therapies to recode nonsense mutations in inherited diseases.
Pharmacol Ther. 136:227–266. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Floquet C, Rousset JP and Bidou L:
Readthrough of premature termination codons in the adenomatous
polyposis coli gene restores its biological activity in human
cancer cells. PLoS One. 6:e241252011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zilberberg A, Lahav L and Rosin-Arbesfeld
R: Restoration of APC gene function in colorectal cancer cells by
aminoglycoside- and macrolide-induced read-through of premature
termination codons. Gut. 59:496–507. 2010. View Article : Google Scholar
|
|
19
|
Wang X and Gregory-Evans CY: Nonsense
suppression therapies in ocular genetic diseases. Cell Mol Life
Sci. 72:1931–1938. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
LucaDe A, Nico B, Rolland JF, Cozzoli A,
Burdi R, Mangieri D, Giannuzzi V, Liantonio A, Cippone V, De Bellis
M, et al: Gentamicin treatment in exercised mdx mice:
Identification of dystrophin-sensitive pathways and evaluation of
efficacy in work-loaded dystrophic muscle. Neurobiol Dis.
32:243–253. 2008. View Article : Google Scholar
|
|
21
|
Floquet C, Deforges J, Rousset JP and
Bidou L: Rescue of non-sense mutated p53 tumor suppressor gene by
aminoglyco-sides. Nucleic Acids Res. 39:3350–3362. 2011. View Article : Google Scholar
|
|
22
|
Malik V, Rodino-Klapac LR, Viollet L, Wall
C, King W, Al-Dahhak R, Lewis S, Shilling CJ, Kota J,
Serrano-Munuera C, et al: Gentamicin-induced readthrough of stop
codons in Duchenne muscular dystrophy. Ann Neurol. 67:771–780.
2010.PubMed/NCBI
|
|
23
|
Cridge AG, Crowe-McAuliffe C, Mathew SF
and Tate WP: Eukaryotic translational termination efficiency is
influenced by the 3′ nucleotides within the ribosomal mRNA channel.
Nucleic Acids Res. 46:1927–1944. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Frolova L, Le Goff X, Zhouravleva G,
Davydova E, Philippe M and Kisselev L: Eukaryotic polypeptide chain
release factor eRF3 is an eRF1- and ribosome-dependent guanosine
triphos-phatase. RNA. 2:334–341. 1994.
|
|
25
|
Moazed D and Noller HF: Interaction of
antibiotics with functional sites in 16S ribosomal RNA. Nature.
327:389–394. 1987. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Engelberg-Kulka H: UGA suppression by
normal tRNA Trp in Escherichia coli: Codon context effects. Nucleic
Acids Res. 9:983–991. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gong Q, Stump MR and Zhou Z: Position of
premature termination codons determines susceptibility of hERG
mutations to nonsense-mediated mRNA decay in long QT syndrome.
Gene. 539:190–197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brown CM, Dalphin ME, Stockwell PA and
Tate WP: The translational termination signal database. Nucleic
Acids Res. 21:3119–3123. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Brown CM, Stockwell PA, Trotman CN and
Tate WP: Sequence analysis suggests that tetra-nucleotides signal
the termination of protein synthesis in eukaryotes. Nucleic Acids
Res. 18:6339–6345. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bonetti B, Fu L, Moon J and Bedwell DM:
The efficiency of translation termination is determined by a
synergistic interplay between upstream and downstream sequences in
Saccharomyces cerevisiae. J Mol Biol. 251:334–345. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McCaughan KK, Brown CM, Dalphin ME, Berry
MJ and Tate WP: Translational termination efficiency in mammals is
influenced by the base following the stop codon. Proc Natl Acad Sci
USA. 92:5431–5435. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tate WP, Poole ES, Horsfield JA, Mannering
SA, Brown CM, Moffat JG, Dalphin ME, McCaughan KK, Major LL and
Wilson DN: Translational termination efficiency in both bacteria
and mammals is regulated by the base following the stop codon.
Biochem Cell Biol. 73:1095–1103. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Manuvakhova M, Keeling K and Bedwell DM:
Aminoglycoside antibiotics mediate context-dependent suppression of
termination codons in a mammalian translation system. RNA.
6:1044–1055. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Howard MT, Shirts BH, Petros LM, Flanigan
KM, Gesteland RF and Atkins JF: Sequence specificity of
aminoglycoside-induced stop codon read-through: Potential
implications for treatment of Duchenne muscular dystrophy. Ann
Neurol. 48:164–169. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Keeling KM and Bedwell DM: Clinically
relevant aminoglyco-sides can suppress disease-associated premature
stop mutations in the IDUA and P53 cDNAs in a mammalian translation
system. J Mol Med (Berl). 80:367–376. 2002. View Article : Google Scholar
|