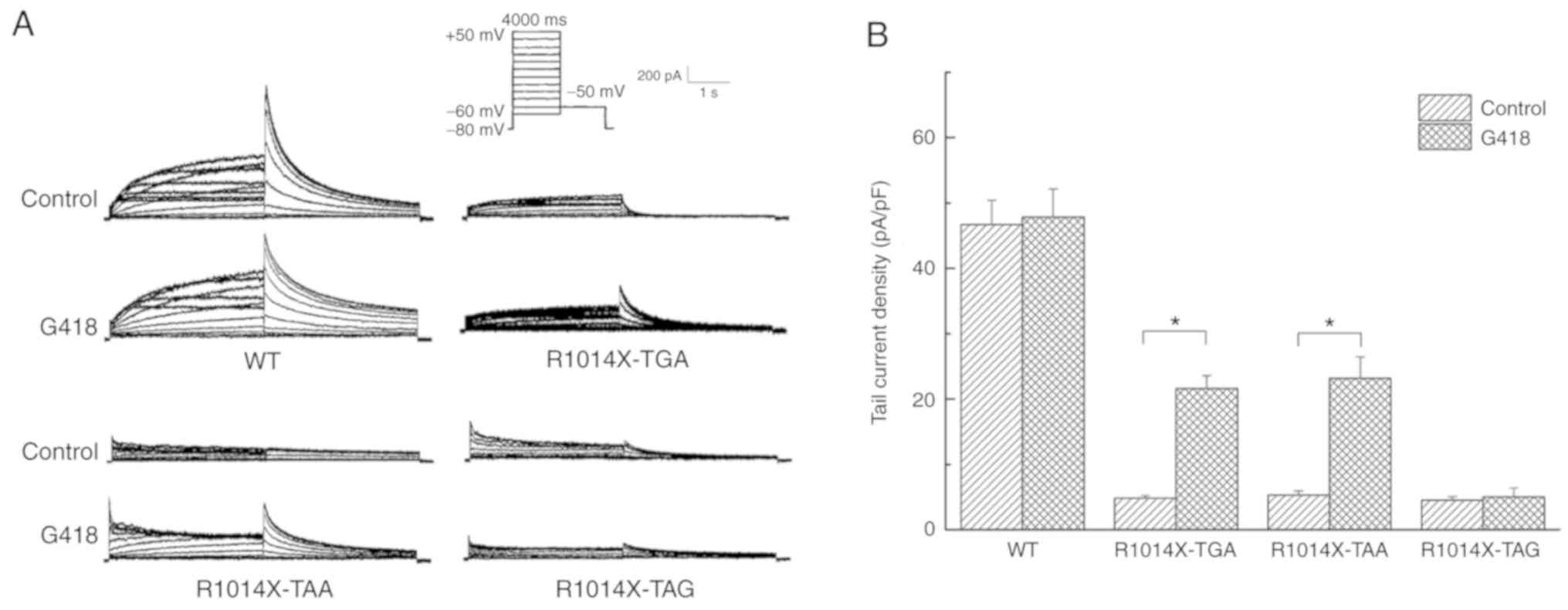
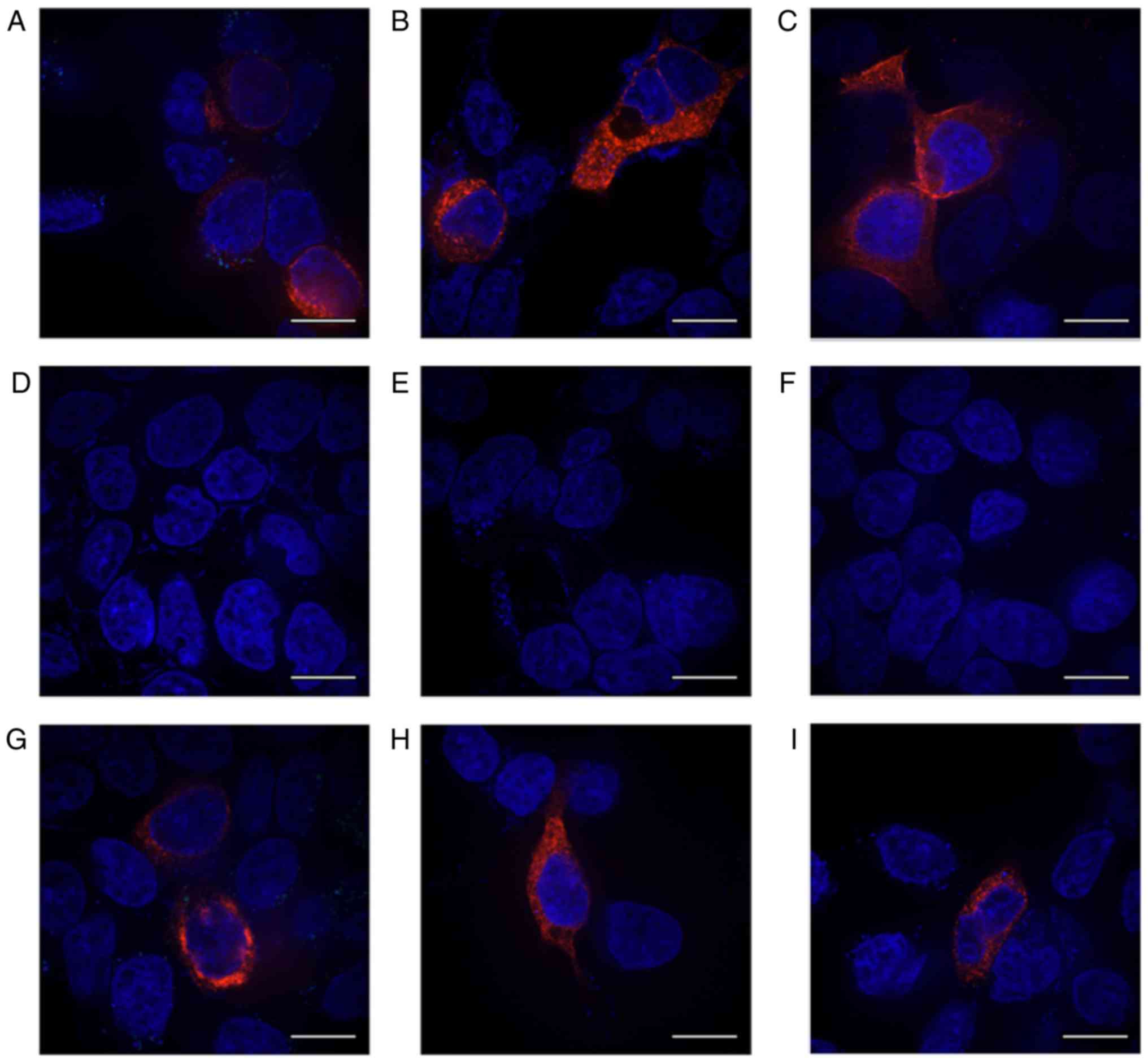
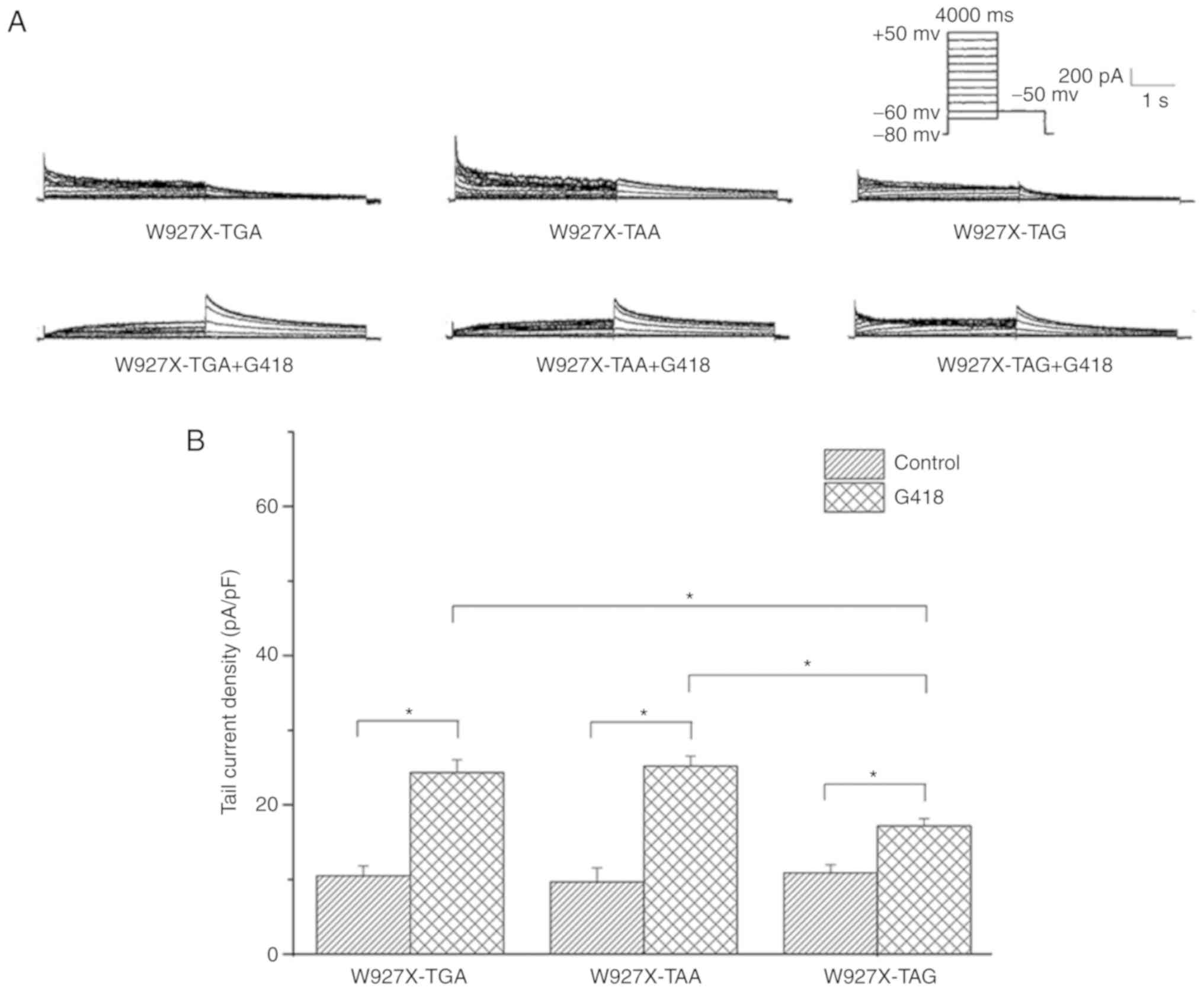
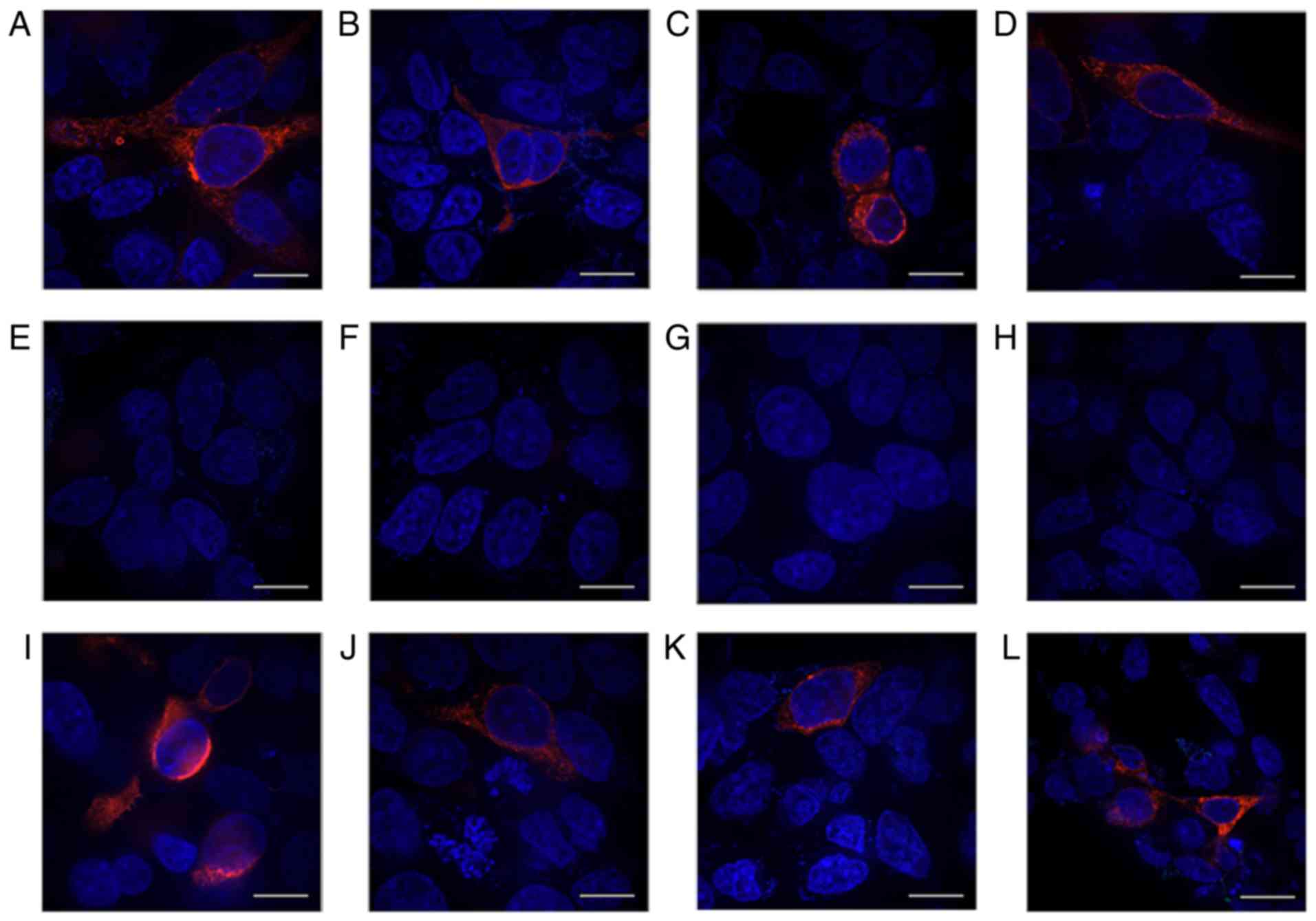
|
1
|
Li G, Shi R, Wu J, Han W, Zhang A, Cheng
G, Xue X and Sun C: Association of the hERG mutation with longQT
syndrome type 2, syncope and epilepsy. Mol Med Rep. 13:2467–2475.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Czosek RJ, Kaltman JR, Cassedy AE, Shah
MJ, Vetter VL, Tanel RE, Wernovksy G, Wray J and Marino BS: Quality
of life of pediatric patients with long QT syndrome. Am J Cardiol.
117:605–610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kanters JK, Yuan L, Hedley PL, Stoevring
B, Jons C, Bloch Thomsen PE, Grunnet M, Christiansen M and
Jespersen T: Flecainide provocation reveals concealed brugada
syndrome in a long QT syndrome family with a novel L1786Q mutation
in SCN5A. Circ J. 78:1136–1143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Splawski I, Shen J, Timothy KW, Lehmann
MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent
GM and Keating MT: Spectrum of mutations in long-QT syndrome genes.
KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 102:1178–1185.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marsman RF, Barc J, Beekman L, Alders M,
Dooijes D, van den Wijngaard A, Ratbi I, Sefiani A, Bhuiyan ZA,
Wilde AA and Bezzina CR: A mutation in CALM1 encoding calmodulin in
familial idiopathic ventricular fibrillation in childhood and
adolescence. J Am Coll Cardiol. 63:259–266. 2014. View Article : Google Scholar
|
|
6
|
Mototani H, Iida A, Nakamura Y and Ikegawa
S: Identification of sequence polymorphisms in CALM2 and analysis
of association with hip osteoarthritis in a Japanese population. J
Bone Miner Metab. 28:547–553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ortiz-Bonnin B, Rinne S, Moss R, Streit
AK, Scharf M, Richter K, Stöber A, Pfeufer A, Seemann G, Kääb S, et
al: Electrophysiological characterization of a large set of novel
variants in the SCN5A-gene: Identification of novel LQTS3 and BrS
mutations. Pflugers Arch. 468:1375–1387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rowe SM and Clancy JP: Pharmaceuticals
targeting nonsense mutations in genetic diseases: Progress in
development. Biodrugs. 23:165–174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Floquet C, Hatin I, Rousset JP and Bidou
L: Statistical analysis of readthrough levels for nonsense
mutations in mammalian cells reveals a major determinant of
response to gentamicin. PLoS Genet. 8:e10026082012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brendel C, Belakhov V, Werner H, Wegener
E, Gärtner J, Nudelman I, Baasov T and Huppke P: Readthrough of
nonsense mutations in Rett syndrome: Evaluation of novel
aminoglyco-sides and generation of a new mouse model. J Mol Med
(Berl). 89:389–398. 2011. View Article : Google Scholar
|
|
11
|
Howard M, Frizzell RA and Bedwell DM:
Aminoglycoside antibiotics restore CFTR function by overcoming
premature stop mutations. Nat Med. 2:467–469. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Baradaran-Heravi A, Balgi AD, Zimmerman C,
Choi K, Shidmoossavee FS, Tan JS, Bergeaud C, Krause A, Flibotte S,
Shimizu Y, et al: Novel small molecules potentiate premature
termination codon readthrough by aminoglycosides. Nucleic Acids
Res. 44:6583–6598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McHugh DR, Steele MS, Valerio DM, Miron A,
Mann RJ, LePage DF, Conlon RA, Cotton CU, Drumm ML and Hodges CA: A
G542X cystic fibrosis mouse model for examining nonsense mutation
directed therapies. PLoS One. 13:e1995732018. View Article : Google Scholar
|
|
14
|
Bolze F, Mocek S, Zimmermann A and
Klingenspor M: Aminoglycosides, but not PTC124 (Ataluren), rescue
nonsense mutations in the leptin receptor and in luciferase
reporter genes. Sci Rep. 7:10202017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu H, Liu X, Huang J, Zhang Y, Hu R and Pu
J: Comparison of read-through effects of aminoglycosides and PTC124
on rescuing nonsense mutations of HERG gene associated with long QT
syndrome. Int J Mol Med. 33:729–735. 2014. View Article : Google Scholar
|
|
16
|
Lee HL and Dougherty JP: Pharmaceutical
therapies to recode nonsense mutations in inherited diseases.
Pharmacol Ther. 136:227–266. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Floquet C, Rousset JP and Bidou L:
Readthrough of premature termination codons in the adenomatous
polyposis coli gene restores its biological activity in human
cancer cells. PLoS One. 6:e241252011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zilberberg A, Lahav L and Rosin-Arbesfeld
R: Restoration of APC gene function in colorectal cancer cells by
aminoglycoside- and macrolide-induced read-through of premature
termination codons. Gut. 59:496–507. 2010. View Article : Google Scholar
|
|
19
|
Wang X and Gregory-Evans CY: Nonsense
suppression therapies in ocular genetic diseases. Cell Mol Life
Sci. 72:1931–1938. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
LucaDe A, Nico B, Rolland JF, Cozzoli A,
Burdi R, Mangieri D, Giannuzzi V, Liantonio A, Cippone V, De Bellis
M, et al: Gentamicin treatment in exercised mdx mice:
Identification of dystrophin-sensitive pathways and evaluation of
efficacy in work-loaded dystrophic muscle. Neurobiol Dis.
32:243–253. 2008. View Article : Google Scholar
|
|
21
|
Floquet C, Deforges J, Rousset JP and
Bidou L: Rescue of non-sense mutated p53 tumor suppressor gene by
aminoglyco-sides. Nucleic Acids Res. 39:3350–3362. 2011. View Article : Google Scholar
|
|
22
|
Malik V, Rodino-Klapac LR, Viollet L, Wall
C, King W, Al-Dahhak R, Lewis S, Shilling CJ, Kota J,
Serrano-Munuera C, et al: Gentamicin-induced readthrough of stop
codons in Duchenne muscular dystrophy. Ann Neurol. 67:771–780.
2010.PubMed/NCBI
|
|
23
|
Cridge AG, Crowe-McAuliffe C, Mathew SF
and Tate WP: Eukaryotic translational termination efficiency is
influenced by the 3′ nucleotides within the ribosomal mRNA channel.
Nucleic Acids Res. 46:1927–1944. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Frolova L, Le Goff X, Zhouravleva G,
Davydova E, Philippe M and Kisselev L: Eukaryotic polypeptide chain
release factor eRF3 is an eRF1- and ribosome-dependent guanosine
triphos-phatase. RNA. 2:334–341. 1994.
|
|
25
|
Moazed D and Noller HF: Interaction of
antibiotics with functional sites in 16S ribosomal RNA. Nature.
327:389–394. 1987. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Engelberg-Kulka H: UGA suppression by
normal tRNA Trp in Escherichia coli: Codon context effects. Nucleic
Acids Res. 9:983–991. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gong Q, Stump MR and Zhou Z: Position of
premature termination codons determines susceptibility of hERG
mutations to nonsense-mediated mRNA decay in long QT syndrome.
Gene. 539:190–197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brown CM, Dalphin ME, Stockwell PA and
Tate WP: The translational termination signal database. Nucleic
Acids Res. 21:3119–3123. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Brown CM, Stockwell PA, Trotman CN and
Tate WP: Sequence analysis suggests that tetra-nucleotides signal
the termination of protein synthesis in eukaryotes. Nucleic Acids
Res. 18:6339–6345. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bonetti B, Fu L, Moon J and Bedwell DM:
The efficiency of translation termination is determined by a
synergistic interplay between upstream and downstream sequences in
Saccharomyces cerevisiae. J Mol Biol. 251:334–345. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McCaughan KK, Brown CM, Dalphin ME, Berry
MJ and Tate WP: Translational termination efficiency in mammals is
influenced by the base following the stop codon. Proc Natl Acad Sci
USA. 92:5431–5435. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tate WP, Poole ES, Horsfield JA, Mannering
SA, Brown CM, Moffat JG, Dalphin ME, McCaughan KK, Major LL and
Wilson DN: Translational termination efficiency in both bacteria
and mammals is regulated by the base following the stop codon.
Biochem Cell Biol. 73:1095–1103. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Manuvakhova M, Keeling K and Bedwell DM:
Aminoglycoside antibiotics mediate context-dependent suppression of
termination codons in a mammalian translation system. RNA.
6:1044–1055. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Howard MT, Shirts BH, Petros LM, Flanigan
KM, Gesteland RF and Atkins JF: Sequence specificity of
aminoglycoside-induced stop codon read-through: Potential
implications for treatment of Duchenne muscular dystrophy. Ann
Neurol. 48:164–169. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Keeling KM and Bedwell DM: Clinically
relevant aminoglyco-sides can suppress disease-associated premature
stop mutations in the IDUA and P53 cDNAs in a mammalian translation
system. J Mol Med (Berl). 80:367–376. 2002. View Article : Google Scholar
|