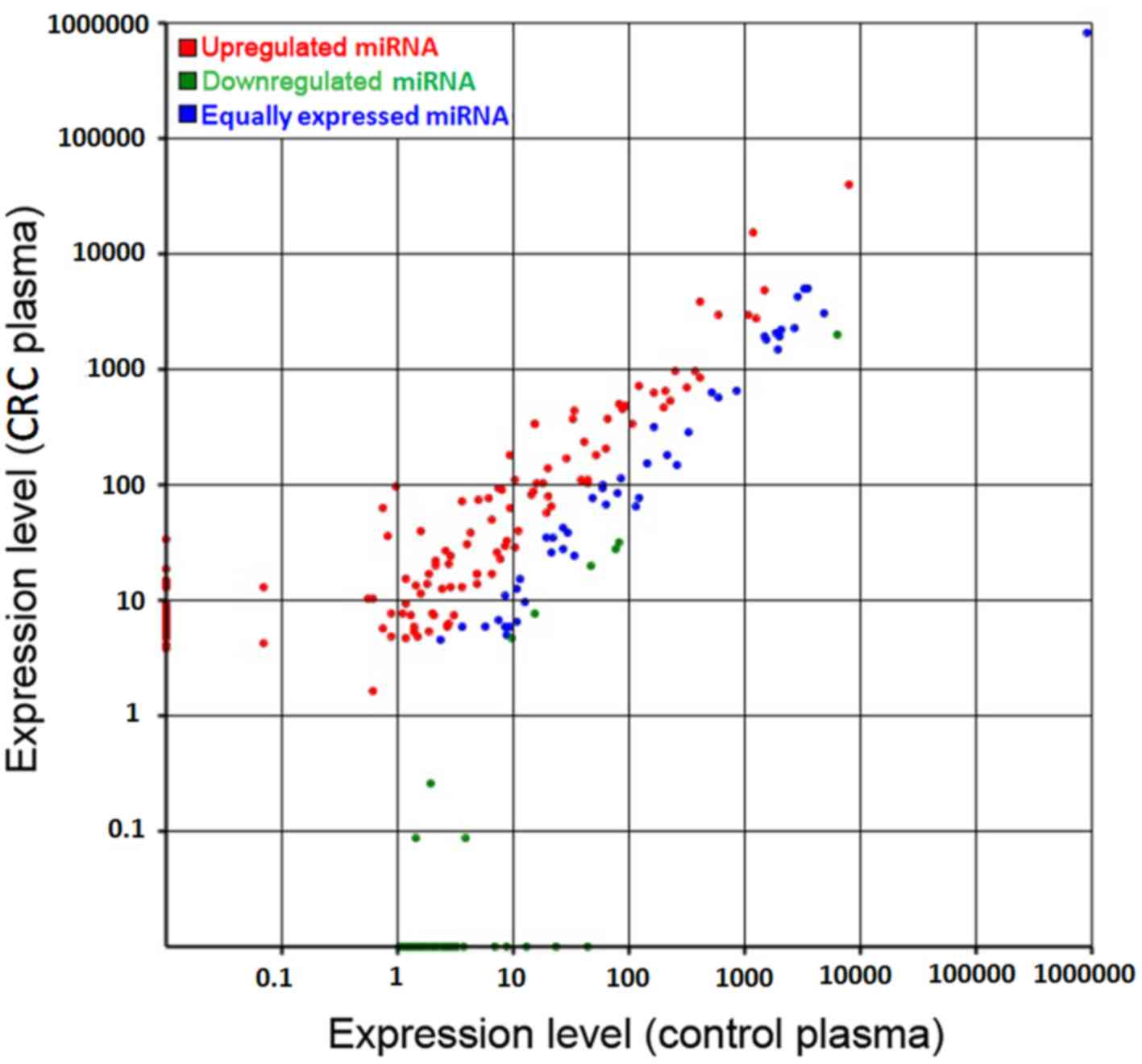
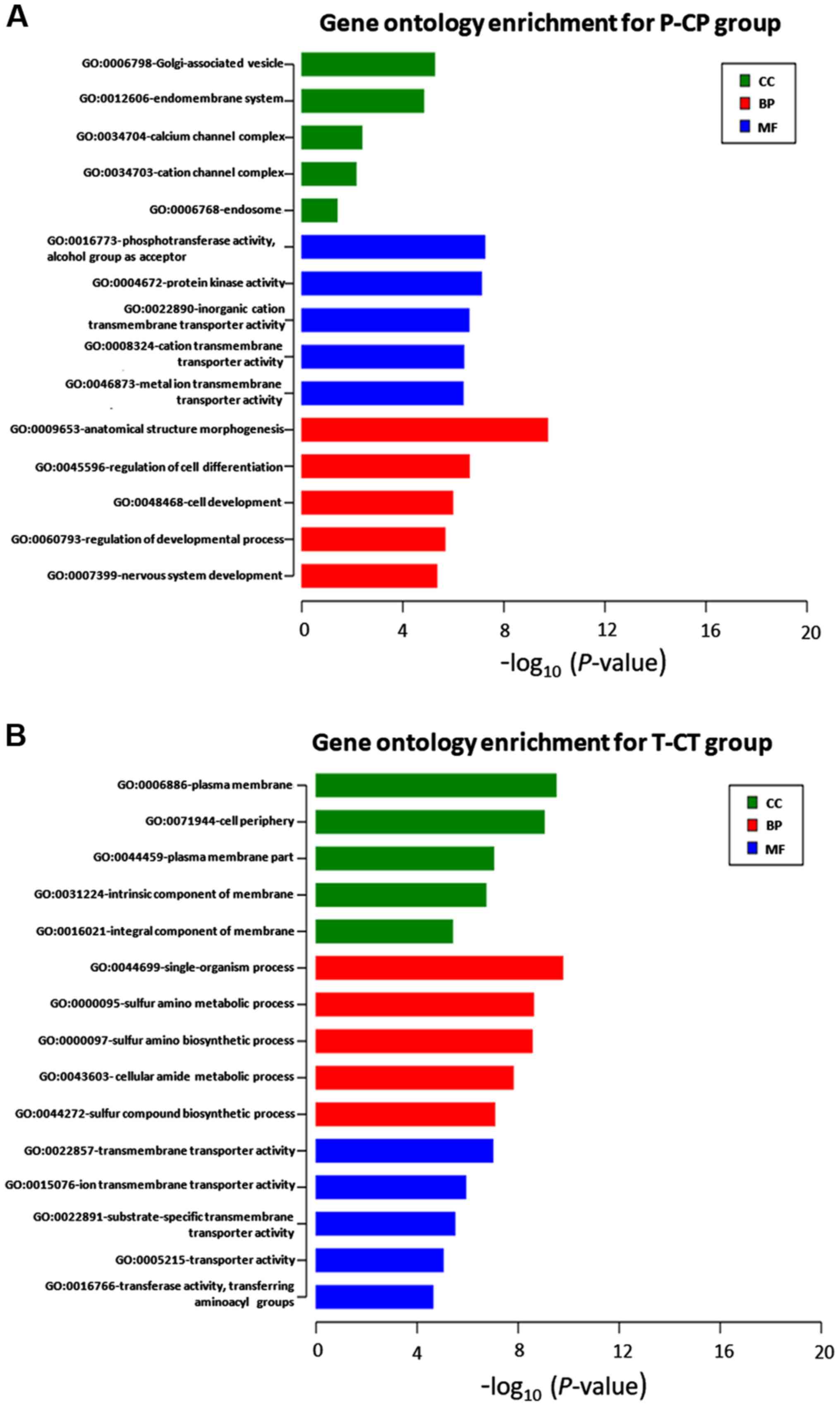
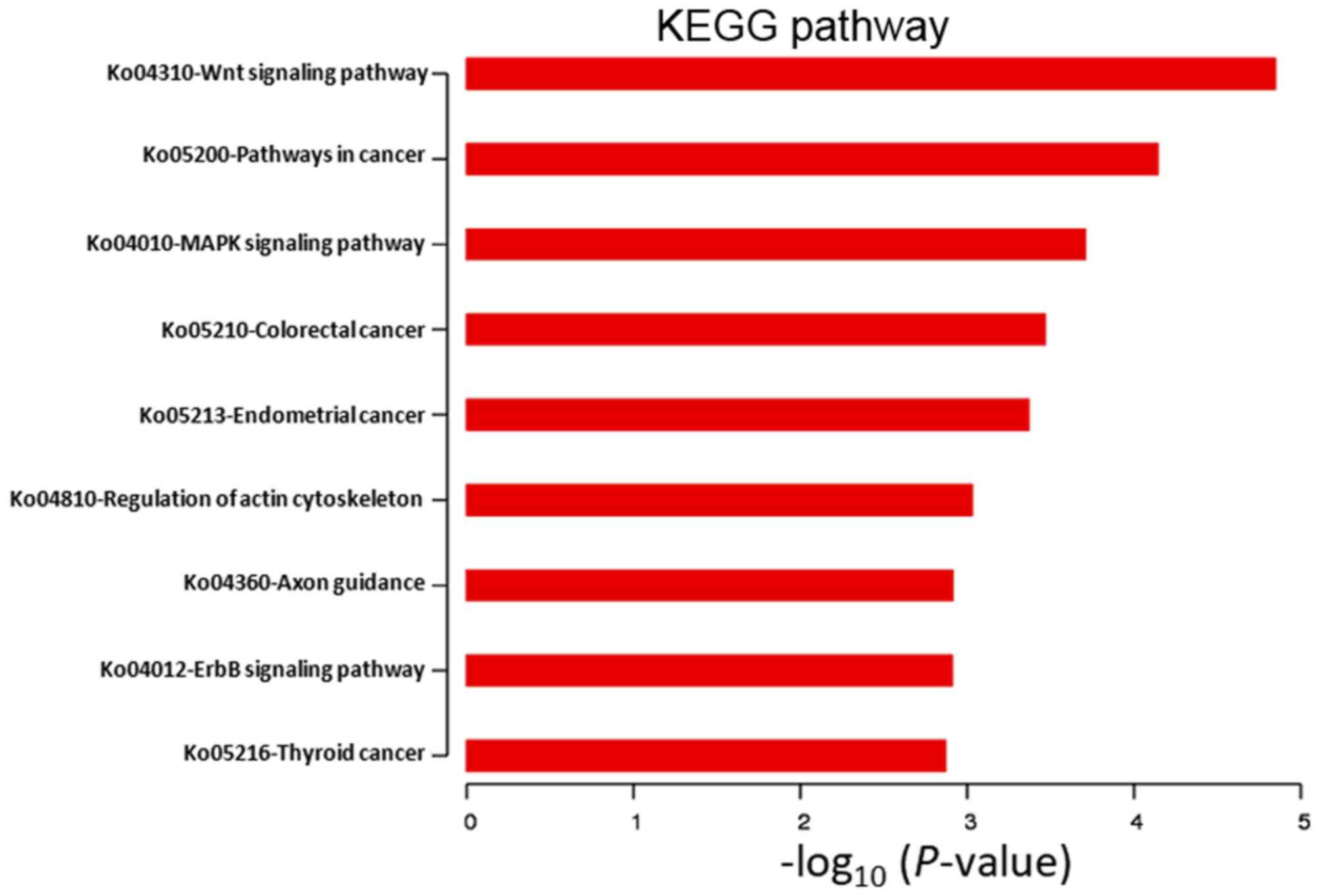
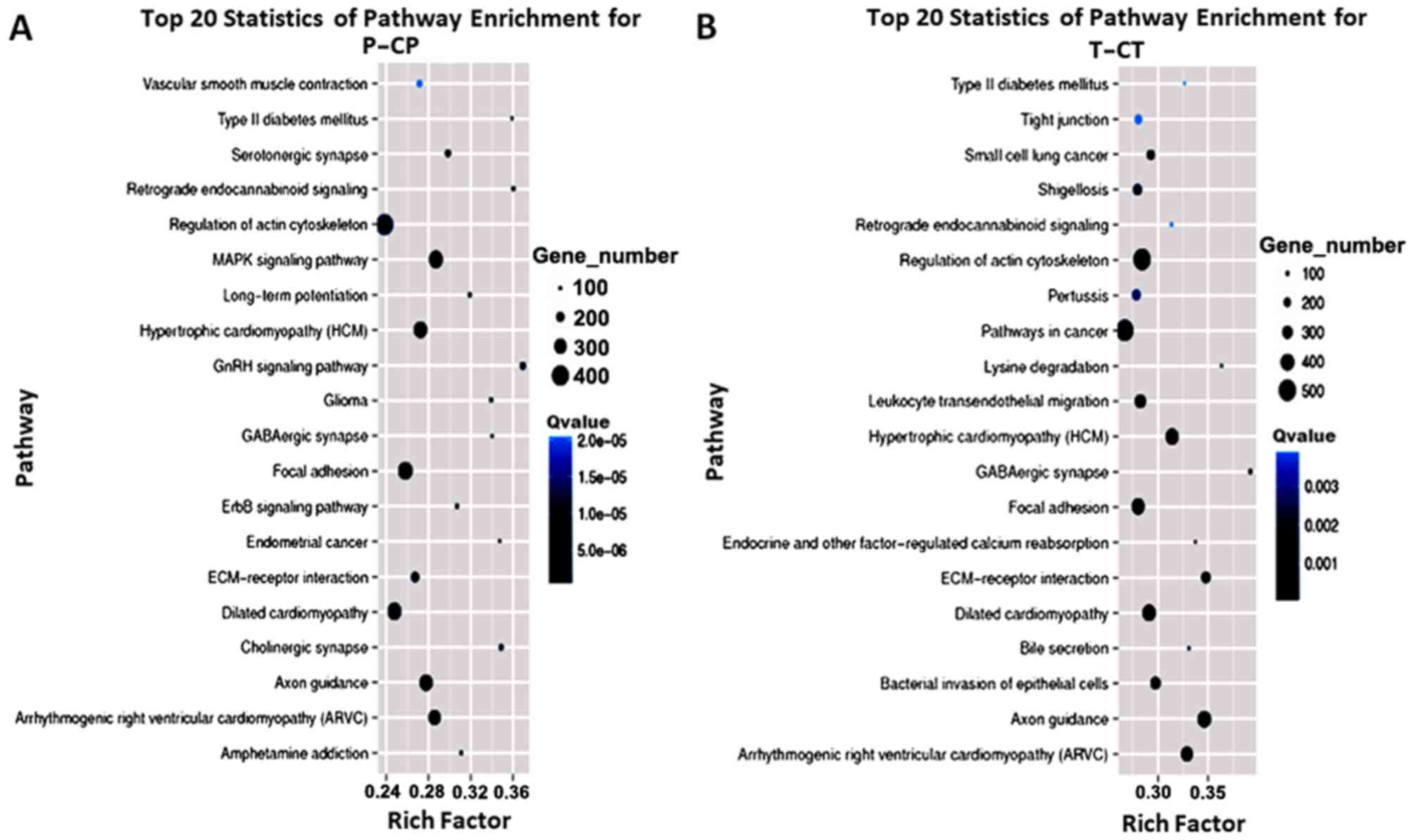
|
1
|
The International Agency for Research on
Cancer (IARC), Global Cancer Observatory, World Health Organization
(WHO): Report on the latest estimates on the global burden of
cancer. Date of release. September 12–2018, Geneva, Switzerland:
https://www.iarc.fr/wp-content/uploads/2018/09/pr263_E.pdf.
Accessed December 13, 2018.
|
|
2
|
Al-Sheikh YA, Ghneim HK, Softa KI,
Al-Jobran AA, Al-Obeed O, Mohamed MA, Abdulla M and Aboul-Soud MA:
Expression profiling of selected microRNA signatures in plasma and
tissues of Saudi colorectal cancer patients by qPCR. Oncol Lett.
11:1406–1412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ibrahim EM, Zeeneldin AA, El-Khodary TR,
Al-Gahmi AM and Bin Sadiq BM: Past, present and future of
colorectal cancer in the Kingdom of Saudi Arabia. Saudi J
Gastroenterol. 14:178–182. 2008. View Article : Google Scholar
|
|
4
|
Kumar R, Price TJ, Beeke C, Jain K, Patel
G, Padbury R, Young GP, Roder D, Townsend A, Bishnoi S, et al:
Colorectal cancer survival: An analysis of patients with metastatic
disease synchronous and metachronous with the primary tumor. Clin
Colorectal Cancer. 13:87–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
O'Connell JB, Maggard MA and Ko CY: Colon
cancer survival rates with the new American Joint Committee on
Cancer sixth edition staging. J Natl Cancer Inst. 96:1420–1425.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Herranz H and Cohen SM: MicroRNAs and gene
regulatory networks: Managing the impact of noise in biological
systems. Genes Dev. 24:1339–1344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
10
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating micro- RNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar
|
|
11
|
Hannafon BN, Sebastiani P, de las Morenas
A, Lu J and Rosenberg CL: Expression of microRNA and their gene
targets are dysregulated in preinvasive breast cancer. Breast
Cancer Res. 13:R242011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miko E, Czimmerer Z, Csánky E, Boros G,
Buslig J, Dezso B and Scholtz B: Differentially expressed microRNAs
in small cell lung cancer. Exp Lung Res. 35:646–664. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Akao Y, Nakagawa Y and Naoe T:
MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 26:311–320.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bandrés E, Cubedo E, Agirre X, Malumbres
R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, et
al: Identification by real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:29–38. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Michael MZ, O' Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
16
|
Peng J, Xie Z, Cheng L, Zhang Y, Chen J,
Yu H, Li Z and Kang H: Paired design study by real-time PCR:
miR-378* and miR-145 are potent early diagnostic biomarkers of
human colorectal cancer. BMC Cancer. 15:158–165. 2015. View Article : Google Scholar
|
|
17
|
Schepeler T, Reinert JT, Ostenfeld MS,
Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ,
Kruhøffer M, Laurberg S, et al: Diagnostic and prognostic microRNAs
in stage II colon cancer. Cancer Res. 68:6416–6424. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Waldman SA and Terzic A: MicroRNA
signatures as diagnostic and therapeutic targets. Clin Chem.
54:943–944. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsang JC and Lo YM: Circulating nucleic
acids in plasma/serum. Pathology. 39:197–207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang Z, Huang D, Ni S, Peng Z, Sheng W
and Du X: Plasma microRNAs are promising novel biomarkers for early
detection of colorectal cancer. Int J Cancer. 127:118–126. 2010.
View Article : Google Scholar
|
|
24
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu X, Xu X, Pan B, He B, Chen X, Zeng K,
Xu M, Pan Y, Sun H, Xu T, et al: Circulating miR-1290 and miR-320d
as novel diagnostic biomarkers of human colorectal cancer. J
Cancer. 10:43–50. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Motoyama K, Inoue H, Takatsuno Y, Tanaka
F, Mimori K, Uetake H, Sugihara K and Mori M: Over- and
under-expressed microRNAs in human colorectal cancer. Int J Oncol.
34:1069–1075. 2009.PubMed/NCBI
|
|
27
|
Wang X, Chen L, Jin H, Wang S, Zhang Y,
Tang X and Tang G: Screening miRNAs for early diagnosis of
colorectal cancer by small RNA deep sequencing and evaluation in a
Chinese patient population. Onco Targets Ther. 9:1159–1166.
2016.PubMed/NCBI
|
|
28
|
UICC: TNM Classification of Malignant
Tumours. Sobin LH and Wittekind Ch: 6th edition. 2006
|
|
29
|
Kirschner MB, Kao SC, Edelman JJ,
Armstrong NJ, Vallely MP, van Zandwijk N and Reid G: Haemolysis
during sample preparation alters microRNA content of plasma. PLoS
One. 6:e241452011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jeong SY, Chessin DB, Schrag D, Riedel E,
Wong WD and Guillem JG: Re: Colon cancer survival rates with the
new American Joint Committee on Cancer sixth edition staging. J
Natl Cancer Inst. 97:1705–1707. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Launoy G, Smith TC, Duffy SW and Bouvier
V: Colorectal cancer mass-screening: Estimation of faecal occult
blood test sensitivity, taking into account cancer mean sojourn
time. Int J Cancer. 73:220–224. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hassan C, Pickhardt PJ, Laghi A, Kim DH,
Zullo A, Iafrate F, Di Giulio L and Morini S: Computed tomographic
colonography to screen for colorectal cancer, extracolonic cancer,
and aortic aneurysm: Model simulation with cost-effectiveness
analysis. Arch Intern Med. 168:696–705. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Carpelan-Holmström M, Louhimo J, Stenman
UH, Alfthan H and Haglund C: CEA, CA 19-9 and CA 72-4 improve the
diagnostic accuracy in gastrointestinal cancers. Anticancer Res.
22:2311–2316. 2002.PubMed/NCBI
|
|
34
|
Wang J, Li J, Shen J, Wang C, Yang L and
Zhang X: Micro-RNA-182 downregulates metastasis suppressor 1 and
contributes to metastasis of hepatocellular carcinoma. BMC Cancer.
12:2272012. View Article : Google Scholar
|
|
35
|
Xu X, Ayub B, Liu Z, Serna VA, Qiang W,
Liu Y, Hernando E, Zabludoff S, Kurita T, Kong B, et al:
Anti-miR182 reduces ovarian cancer burden, invasion, and
metastasis: An in vivo study in orthotopic xenografts of nude mice.
Mol Cancer Ther. 13:1729–1739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li P, Sheng C, Huang L, Zhang H, Huang L,
Cheng Z and Zhu Q: MiR-183/-96/-182 cluster is up-regulated in most
breast cancers and increases cell proliferation and migration.
Breast Cancer Res. 16:4732014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Segura MF, Hanniford D, Menendez S, Reavie
L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A,
Bogunovic D, et al: Aberrant miR-182 expression promotes melanoma
metastasis by repressing FOXO3 and microphthalmia-associated
transcription factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang
Z, Wang X, Lin Y, Mao Y, et al: Downregulation of microRNA-182-5p
contributes to renal cell carcinoma proliferation via activating
the AKT/FOXO3a signaling pathway. Mol Cancer. 13:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Li H, Cui L, Feng J and Fan Q:
MicroRNA-182 suppresses clear cell renal cell carcinoma migration
and invasion by targeting IGF1R. Neoplasma. 63:717–725. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kouri FM, Ritner C and Stegh AH: miRNA-182
and the regulation of the glioblastoma phenotype - toward
miRNA-based precision therapeutics. Cell Cycle. 14:3794–3800. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kouri FM, Hurley LA, Daniel WL, Day ES,
Hua Y, Hao L, Peng CY, Merkel TJ, Queisser MA, Ritner C, et al:
miR-182 integrates apoptosis, growth, and differentiation programs
in glioblastoma. Genes Dev. 29:732–745. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qin J, Luo M, Qian H and Chen W:
Upregulated miR-182 increases drug resistance in cisplatin-treated
HCC cell by regulating TP53INP1. Gene. 538:342–347. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar :
|
|
44
|
Knight T and Irving JA: Ras/Raf/MEK/ERK
pathway activation in childhood acute lymphoblastic leukemia and
its therapeutic targeting. Front Oncol. 4:1602014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Santarpia L, Lippman SL and El-Naggar AK:
Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy.
Expert Opin Ther Targets. 16:103–119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chappell WH, Steelman LS, Long JM, Kempf
RC, Abrams SL, Franklin RA, Bäsecke J, Stivala F, Donia M, Fagone
P, et al: Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors:
Rationale and importance to inhibiting these pathways in human
health. Oncotarget. 2:135–164. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Slattery ML, Mullany LE, Sakoda LC, Wolff
RK, Samowitz WS and Herrick JS: The MAPK-signaling pathway in
colorectal cancer: Dysregulated genes and their association with
micro-RNAs. Cancer Inform. 17:11769351187665222018. View Article : Google Scholar
|