Introduction
Extracellular matrices (ECMs) provide cell
microenvironments with several secreted molecules, such as
collagens, glycoproteins, glycosaminoglycans (GAGs) and
proteoglycans. Proteoglycans regulate cell behaviors by binding GAG
chains or through direct protein-protein interactions via the core
protein (1). The structure of
small leucine-rich proteoglycans (SLRPs) consists of a small
protein core with various GAG side chains, and SLRPs are grouped
into five classes based on common structural properties (1,2).
Osteoadherin, also termed osteomodulin (the Omd gene
product), was originally isolated from guanidine extracts of bovine
bone, and is a 47-kDa keratan sulfate proteoglycan of the class II
subfamily of SLRPs (2,3). Several SLRPs are ECM components;
however, osteoadherin is the only member that is restricted to
mineralized tissues, such as bones and teeth (4-6).
In bone tissues, Omd is expressed by fetal and adult
osteoblasts, and its product osteoadherin can reach up to 400
µg/g wet weight in bovine bone (3).
As with numerous SLRPs, osteoadherin binds a number
of ECM components; in particular, it binds fibrillar collagens to
stabilize fibrillar tissue frameworks (7,8).
Leucine-rich repeat (LRR) motifs comprise 6-10 repeats of ~20-29
amino acids with conserved leucine spacing, and are folded into
structures with one β-sheet and one α-helix. LRRs participate in a
wide range of biological processes, and interact directly with
collagen, heparin-binding proteins and cell surface receptors to
block ligand binding (9).
Osteoadherin contains six closely spaced tyrosine sulfate residues
in its N-terminal region and two in its C-terminal region (9,10).
Tyrosine sulfate domains bind heparin-binding proteins, such as
basic growth factor-2 and thrombospondin I (10). Osteoadherin also contains large
numbers of acidic amino acid residues in its N-terminal region
(3), which may bind to
hydroxyapatite in calcified tissues and basic cluster motifs of
heparin-binding proteins and growth factors (9,11).
SLRPs also carry GAG chains that are covalently attached to the
core protein, and can modulate biological functions depending on
the nature of GAG chains. Through these interactions, SLRPs are
involved in the initial triggering of multiple cellular responses
in various tissues (2). However,
it is unclear whether osteoadherin in bone tissues serves roles in
the regulation of osteoblast growth, apoptosis, and
differentiation.
Bone morphogenetic proteins (BMPs) regulate
differentiation and proliferation in various cell types, including
osteoblasts (12). BMP2
reportedly triggers osteoblast differentiation and upregulates
several genes that encode osteoblast phenotype-related proteins
in vitro (13). In the
intracellular BMP2 signaling pathway, Smad1 is phosphorylated
directly by type I receptors for BMP2 and is transported into the
nucleus in complexes with Smad4 (14). This complex interacts with the
regulatory elements of target genes and regulates their expression.
To date, little is understood of how Omd expression is
regulated in response to BMP2 during osteoblast
differentiation.
Apoptosis can be initiated by various stimuli and is
directed by a family of cysteine proteases known as caspases. At
the molecular level, caspase activation plays a central role in the
execution of apoptosis. To the best of our knowledge, so far, 14
mammalian caspases have been identified, three of which (caspase-3,
-6 and -7) are the effector caspases that coordinate the execution
phase of apoptosis by degrading multiple substrates, including the
structural and regulatory proteins in the cell nucleus, cytoplasm
and cytoskeleton (15).
The present study determined the Omd
expression levels during osteoblast differentiation and
investigated the effects of overexpression and knockdown of
osteoadherin in osteoblast cells. Overexpression of Omd
increased the numbers of viable cells and decreased the activity of
caspase 3/7 in MC3T3-E1 cells. By contrast, knockdown of Omd
decreased viable cell numbers and increased caspase 3/7 activity in
these cells. Omd overexpression also reduced the expression
of CCN family 2 (CCN2). Thus, it can be concluded that osteoadherin
regulates apoptosis and growth in osteoblast cells.
Materials and methods
Reagents
Recombinant human BMP2 was kindly supplied by
Astellas Pharma Ltd. Dexamethasone and 3-isobutyl-1-meth-ylxanthine
(IBMX) were purchased from Sigma-Aldrich; Merck KGaA.
Cell cultures
MC3T3-E1 mouse osteoblast cells and C2C12 myoblast
cells were obtained from RIKEN BioResource Center and were cultured
in α-minimal essential medium (α-MEM; Sigma-Aldrich; Merck KGaA)
containing 100 µg/ml kanamycin (Meiji Seika Kaisha, Ltd.)
and 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA) at 37°C
under a humidified atmosphere containing 5% CO2, as
described previously (16).
MC3T3-E1 cells were cultured and differentiated in α-MEM containing
10% FBS, 10 mM β-glycerophosphate (Tokyo Chemical Industry Co.,
Ltd.) and 50 µg/ml ascorbic acid (Wako Pure Chemical
Industries, Ltd.) for 1-3 weeks. Pre-adipocyte 3T3-L1 cells were
purchased from DS Pharma Biomedical Co., Ltd. and grown in
Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich; Merck
KGaA) supplemented with 10% FBS at 37°C until confluence. At 2 days
after confluence (day 0), differentiation was induced in adipocyte
cultures by adding 500 µM IBMX, 10 µg/ml insulin
(Cell Science and Technology Institute, Inc.) and 1 µM
dexamethasone to the basal medium. On day 3, media were replaced
with adipogenic medium comprising DMEM supplemented with 10% FBS
and 10 µg/ml insulin, which was changed every 2 days
thereafter until analysis. C2C12 cells were seeded into culture
dishes, and then cultured in media containing 50 or 200 ng/ml of
BMP2 for 6-24 h. Cells of the mouse stromal cell line ST2 (RIKEN
BioResource Center) were obtained and cultured as described
previously (17). Similarly,
bEnd.3 mouse vascular endothelial cells (American Type Culture
Collection) (18) were cultured
in DMEM supplemented with 10% FBS as described previously (19) and culture media were changed every
3-4 days.
Reverse transcription
(RT)-semi-quantitative PCR (qPCR)
Total RNA was extracted from cells using ISOGEN
(Nippon Gene Co., Ltd.) and complementary DNA was synthesized with
the Omniscript RT kit (Qiagen GmbH) using an oligo(dT)15
primer (1 µM) at 37°C for 30 min. semi-qPCR analyses were
performed with Taq DNA polymerase (Qiagen GmbH) as described
previously (20). The specific
primer sequences for each gene are listed in Table I. Each reaction consisted of an
initial denaturation at 94°C for 3 min followed by three-step
cycling: Denaturation at 94°C for 30 sec, annealing at a
temperature optimized for each primer pair for 30 sec, and
extension at 72°C for 40 sec. After the requisite number of cycles
(25-35 cycles), reactions underwent a final extension at 72°C for 5
min. To accommodate the differences in RNA quantities, expression
levels were normalized to those of the housekeeping gene GAPDH.
Amplification products were separated using electrophoresis on 2%
agarose gels and visualized by SYBR Green staining followed by UV
light illumination. The relative intensity of the gel bands was
measured using ImageJ software (version 1.52a; National Institutes
of Health), and results were normalized to the GAPDH mRNA
level.
 | Table IPrimers used for reverse
transcription-semi-quantitative PCR. |
Table I
Primers used for reverse
transcription-semi-quantitative PCR.
| Specificity | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) | Predicted size,
bp |
|---|
| Omd |
ATGGTGTATTCGCTAAACTTTCAA |
AAGATGATTATAGCAGAGGTCAAG | 194 |
| Bgn |
TGCCACCGCCATTGTTCCATC |
GTTTGTGGAGGGACTGGCTAGA | 486 |
| Dcn |
ATGGCAGTCTGGCCAATGTT |
GCCGTCTGAGGGTTACTTGT | 308 |
| Fmod |
CATGATCTGCACCCCTTCTT |
AATGCAGAGGAAGCCAGGTC | 300 |
| Lum |
CCTGAGGAATAACCAAATCGAC |
AGACCAGCAGGCAGCTTGCTCA | 393 |
| GAPDH |
TCCACCACCCTGTTGCTGTA |
ACCACAGTCCATGCCATCAC | 452 |
RT-qPCR
Total RNA was extracted from cells using ISOGEN
(Nippon Gene Co., Ltd.) and complementary DNA was synthesized with
High-Capacity cDNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) at 37°C for 60 min. RT-qPCR
analyses were performed using the assay-on-demand TaqMan probes
(Mm00449589_m1 for Omd, Mm00650798g1 for Npy1r,
Mm00475834_m1 for Alp, Mm03413826_mH for Bglap,
Mm01192932_g1 for Ccn2, and Mm99999915_g1 for GAPDH;
Applied Biosystems; Thermo Fisher Scientific, Inc.) and the
StepOne® real time PCR system, according to the
manufacturer's protocol, as described previously (21). The qPCR thermocycling conditions
included an initial denaturation of 95°C for 10 min followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min. Relative gene
expression levels were quantified using the comparative
2-ΔΔCq method (22)
using GAPDH expression as an endogenous control.
Western blot analysis
MC3T3-E1 cells were transfected according to the
experimental conditions and then collected at specific time points,
washed in ice-cold PBS, and suspended in CelLytic-M Mammalian cell
lysis/extraction reagent (Sigma-Aldrich; Merck KGaA) plus a
protease inhibitor (Complete mini; Roche Diagnostics). Whole-cell
extracts were quantified using a Protein assay reagent (Bio-Rad
Laboratories, Inc.) and ~30 µg protein/lane was separated
using 10% SDS-PAGE followed by transfer to a polyvinylidene
fluoride membrane (EMD Millipore). Blocking was performed with 0.3%
skim milk (BD Biosciences) dissolved in TBS with 0.05% Tween-20
(TBST) for 1 h at room temperature. The membrane was probed with
antibodies against osteoadherin (rabbit polyclonal anti-human
polyclonal antibody; 1:2,000; cat. no. NBP2-19626; Novus
Biologicals, LLC) or β-actin (rabbit polyclonal antibody; 1:2,000;
cat. no. 20536-1-AP, ProteinTech Group, Inc.) for 1 h at room
temperature. The membrane was subsequently washed three times and
incubated with anti-mouse horseradish peroxidase-linked antibody
(1:20,000; cat. no. NA934; GE Healthcare Life Sciences) in TBST for
1 h at room temperature. Following three additional washes,
chemiluminescence detection was performed with ECL Prime Western
Blotting Detection Reagent (GE Healthcare Life Sciences) and a LAS
1000 image analyzer, according to the manufacturer's
instructions.
Reporter constructs and assays of
luciferase activity
Luciferase reporter plasmids for the murine
Omd promoter were generated as follows. The 1774-bp
Omd promoter fragment (-1,609 to +165) (chromosome 13; MGI:
1350918) was isolated from mouse genomic DNA using PCR and was
subcloned into the pGL4.12 vector (Promega Corporation) to generate
the luciferase reporter plasmid pOmdluc. The nucleotide sequences
of promoter regions were verified by sequencing. The Smad
expression plasmids encoding wild-type Smad4 and constitutively
active Smad1 (DVD) were provided by Dr Katagiri (Saitama Medical
University, Saitama, Japan) (23). For the reporter assay, C2C12 cells
were plated 24 h prior to transfection at a density of
1×105 cells/well (24-well plate) and cultured in α-MEM
supplemented with 10% FBS. Transfection of plasmid DNA into cells
was performed using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Luciferase reporter assays were
performed as described previously (20). Briefly, cells were lysed with a
PicaGene cell culture lysis reagent Luc (Wako Pure Chemical
Industries, Ltd.; cat. no. 300-04351) and firefly luciferase
activities were measured using a Mini Lumat LB 9506 luminometer
(Berthold Technologies). Luciferase activity was normalized by
comparison with β-galactosidase activity and fold-changes in the
luciferase activities of transfected cells were calculated on the
basis of those in control MC3T3-E1 cells that were transfected with
pcDNA3.
Transfection of small interfering RNAs
(siRNAs)
MC3T3-E1 cells were transfected with 2.5 nM Silencer
Select pre-designed siRNAs for osteoadherin (siOmd; Thermo Fisher
Scientific Inc.; cat. nos. s77492 and s77494), the Y1 receptor
(siY1; Thermo Fisher Scientific, Inc.; cat. no. s70765) or Silencer
negative control siRNA no. 1 (siCont; Thermo Fisher Scientific,
Inc.; cat. no. 4390843) using Lipofectamine RNAiMAX (Invitrogen;
Thermo Fisher Scientific, Inc.) as previously described (16). Total RNA was extracted after 2 or
6 days for the determination of mRNA level. Total protein was
extracted after 2 days for western blot analysis.
Generation of plasmid construct and
overexpression of Omd
Omd overexpression plasmid was generated as
follows and was subsequently designated pOmd. Total RNA was
extracted from MC3T3-E1 cells using ISOGEN (Nippon Gene Co., Ltd.).
Mouse Omd complementary DNA was reverse-transcribed from RNA
using Omniscript RT kit (Qiagen), and then amplified using primers
that flank the mouse Omd open reading frame (forward, 5′-CCA
GCC CGA GGA CAA GAA AA-3′ and reverse, 5′-GGC TTT ATG GAG GCA TAA
ATG TGT-3′) and PrimeSTAR Max DNA polymerase (Clontech
Laboratories, Inc.), according to the manufacturer's instructions.
PCR products were electrophoresed on 1% agarose gels and were
purified and subcloned into KpnI and XbaI restriction
sites of the pcDNA3 vector (Invitrogen; Thermo Fisher Scientific,
Inc.) using In-Fusion HD cloning kit (Clontech Laboratories, Inc.;
cat. no. 639648), according to the manufacturer's protocol.
Individual clones of transformed Escherichia coli were
isolated from agar plates and nucleotide sequences of each plasmid
were confirmed by DNA sequencing. MC3T3-E1 cells were transfected
in 24-well plates with 0.1 µg pOmd or empty vector (pcDNA3)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol.
Alkaline phosphatase (ALP) staining
analyses
MC3T3-E1 cells were plated onto 24-well plates at a
density of 1.7×104/ml. Following 24 h, cells were
transfected in 24-well plates with 0.1 µg pOmd or pcDNA3 and
were cultured in α-MEM for 14 days. ALP staining was performed as
previously described (20).
Briefly, cells were rinsed in PBS, fixed in 10% formalin at room
temperature for 30 min, rinsed again in PBS, and then incubated at
room temperature for 1 h in 300 µl aliquots of solution
containing 0.15 mg/ml 5-bromo-4 chloro-3-indolylphosphate, 0.3
mg/ml nitro-blue tetrazolium (Wako Pure Chemical Industries, Ltd.),
0.1 M Tris-HCl (pH 9.0), 0.01 N NaOH and 0.05 mM MgCl2.
Images were captured using an iPhone7 smartphone (Apple
Corporation). The same settings were applied to all experimental
samples.
Quantitation of cell viability
Cell counting kit-8 (Dojindo Molecular Technologies,
Inc.; cat no. CK04-01) were used to evaluate cell viability. In
these experiments, 10-µl aliquots of the WST-8 substrate
[5-mM 2-(2-methoxy-4-nitrophenyl)-3-(4-nitr
ophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt]
were added to each well. After incubation for 1 h at 37°C, optical
densities were measured at a wavelength of 450 nm using a
microplate absorbance reader. Data are expressed as fold changes of
transfected samples relative to cell numbers in samples transfected
with control siRNA or pcDNA3.
Measurements of caspase 3/7 activity
The enzyme activities of caspase 3/7 were determined
using a caspase colorimetric assay (Caspase-Glo 3/7 assay system;
Promega Corporation) as described previously (21). Briefly, for each reaction, cells
were lysed and incubated with luminogenic substrates containing the
Asp-Glu-Val-Asp sequence, which is cleaved by activated caspase
3/7. Following incubation at room temperature for 1 h, luminescence
was quantified using a Mini Lumat LB 9506 luminometer (Berthold
Technologies). Fold changes in caspase 3/7 activity in transfected
cells were calculated on the basis of those in control siRNA or
pcDNA3 transfected MC3T3-E1 cells.
Statistical analysis
Data are expressed as the mean ± standard deviation
of three to six independent experiments. Comparison of two groups
was performed with a Student's t-test using Microsoft Excel version
2016 (Microsoft Corporation). Comparisons of multiple groups were
performed using one-way analysis of variance using Microsoft Excel
version 2016 (Microsoft Corporation) followed by Dunnett's multiple
comparison test using software provided by Osaka University, Osaka,
Japan (http://www.gen-info.osaka-u.ac.jp/MEPHAS/dunnett-e.html).
P<0.05 was considered to indicate a statistically significant
difference.
Results
SLRP mRNA expression levels in cultured
cells
To evaluate the roles of osteoadherin in mesenchymal
cells, Omd mRNA expression levels were determined in various
mesenchymal cell cultures and compared with those of other SLRPs,
including biglycan, decorin, fibromodulin, and lumican, using
RT-semi-qPCR analyses. Omd mRNA was expressed in MC3T3-E1
cells (osteoblasts) but not in C2C12 myoblasts, adipocyte
differentiated 3T3-L1 cells, ST2 bone marrow stromal cells or
bEnd.3 endothelial cells (Fig.
1A). By contrast, biglycan (Bgn), decorin (Dcn)
and fibromodulin (Fmod) mRNA expression was detected in
MC3T3-E1, C2C12 and 3T3-L1 cells. Meanwhile, lumican (Lum)
mRNA was expressed only in MC3T3-E1 and 3T3-L1 cells. Analysis of
the Omd and Bgn mRNA/GAPDH mRNA ratio in
various mesenchymal cell cultures is presented in Fig. 1B. These data indicate that
Omd expression varies during osteoblast differentiation. To
determine whether Omd expression reflects the stages of
osteoblast differentiation, MC3T3-E1 cells were cultured in
differentiation medium for 1-3 weeks and expression levels of
Omd mRNA were determined using RT-qPCR. Omd mRNA
expression was significantly increased from 1 week and increased
further over the culture period of 3 weeks in differentiating
osteoblast cells (Fig. 1C).
Protein levels were also assayed using western blot analysis and
were identified to be increased during in vitro osteoblastic
differentiation in MC3T3-E1 cells (Fig. 1D).
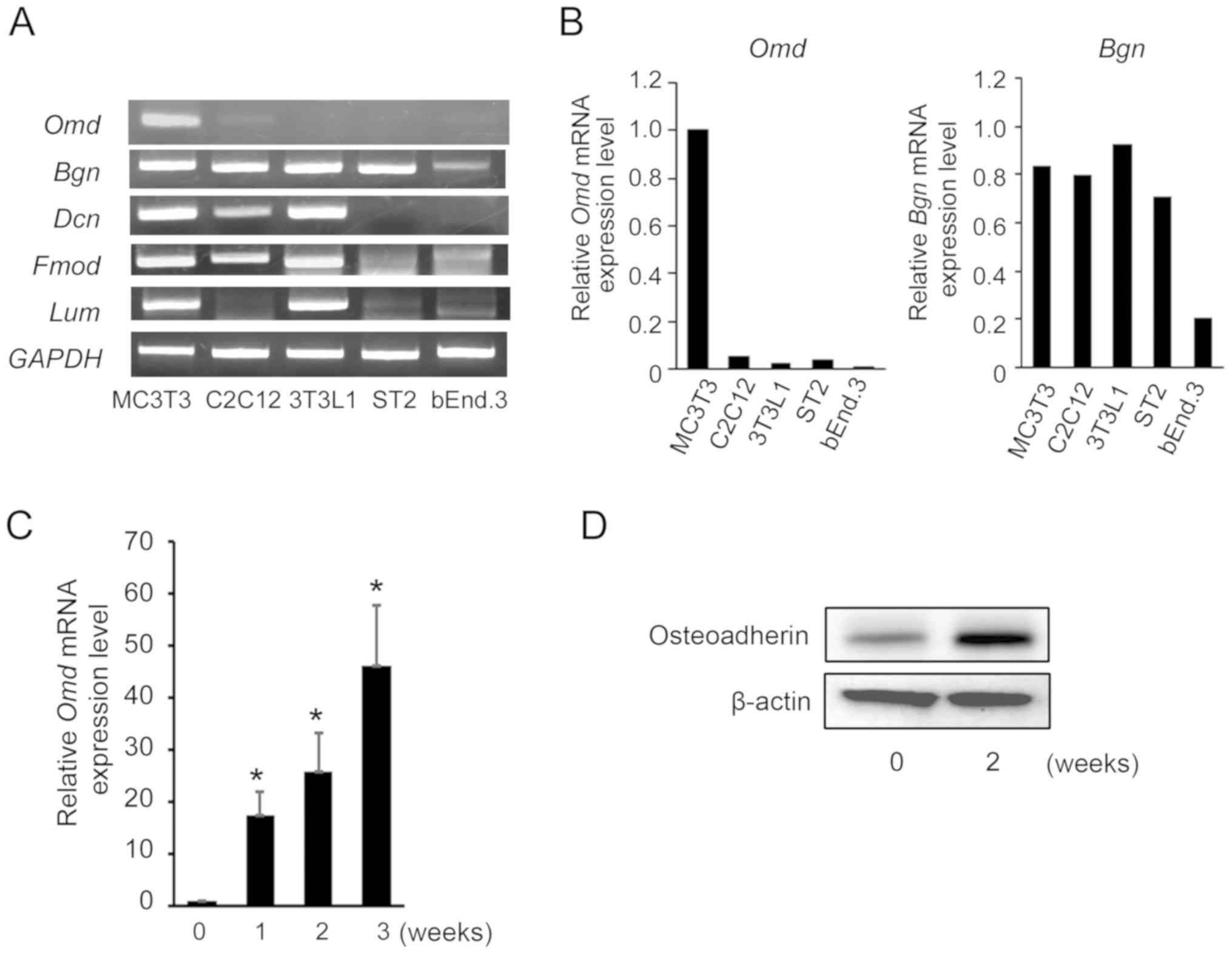 | Figure 1SLRP mRNA expression levels under
various culture conditions. (A) Total RNA was extracted from
MC3T3-E1, 3T3-L1, C2C12, ST2 and bEnd.3 cells, and mRNA expression
levels of the SLRPs Omd, Dcn, Bgn, Fmod and
Lum were determined using RT-semi-qPCR analyses. Equal
loading of cDNA samples was confirmed by amplification of
GAPDH cDNA. The presented data are representative of three
experiments with similar results. (B) mRNA expression levels of
Omd and Bgn normalized to GAPDH. (C) Confluent
MC3T3-E1 cells were incubated in differentiation media for 1-3
weeks. Total RNA was extracted from cells and Omd mRNA
expression was determined using RT-qPCR. mRNA expression levels
were normalized to those of GAPDH, and are presented as fold
changes relative to the expression levels in control cells. (D)
Confluent MC3T3-E1 cells were incubated in differentiation media
for 2 weeks. Total protein was extracted from MC3T3-E1 cells and
the levels of Omd protein were determined by western blot analysis.
β-actin was used as an endogenous control. Data are presented as
the mean ± standard deviation of separate experiments performed in
triplicate. *P<0.05 vs. week 0. SLRP, small
leucine-rich proteoglycan; Omd, osteoadherin; Dcn,
decorin; Bgn, biglycan; Fmod, fibromodulin;
Lum, lumican; RT, reverse transcription; qPCR, quantitative
PCR; cDNA, complementary DNA. |
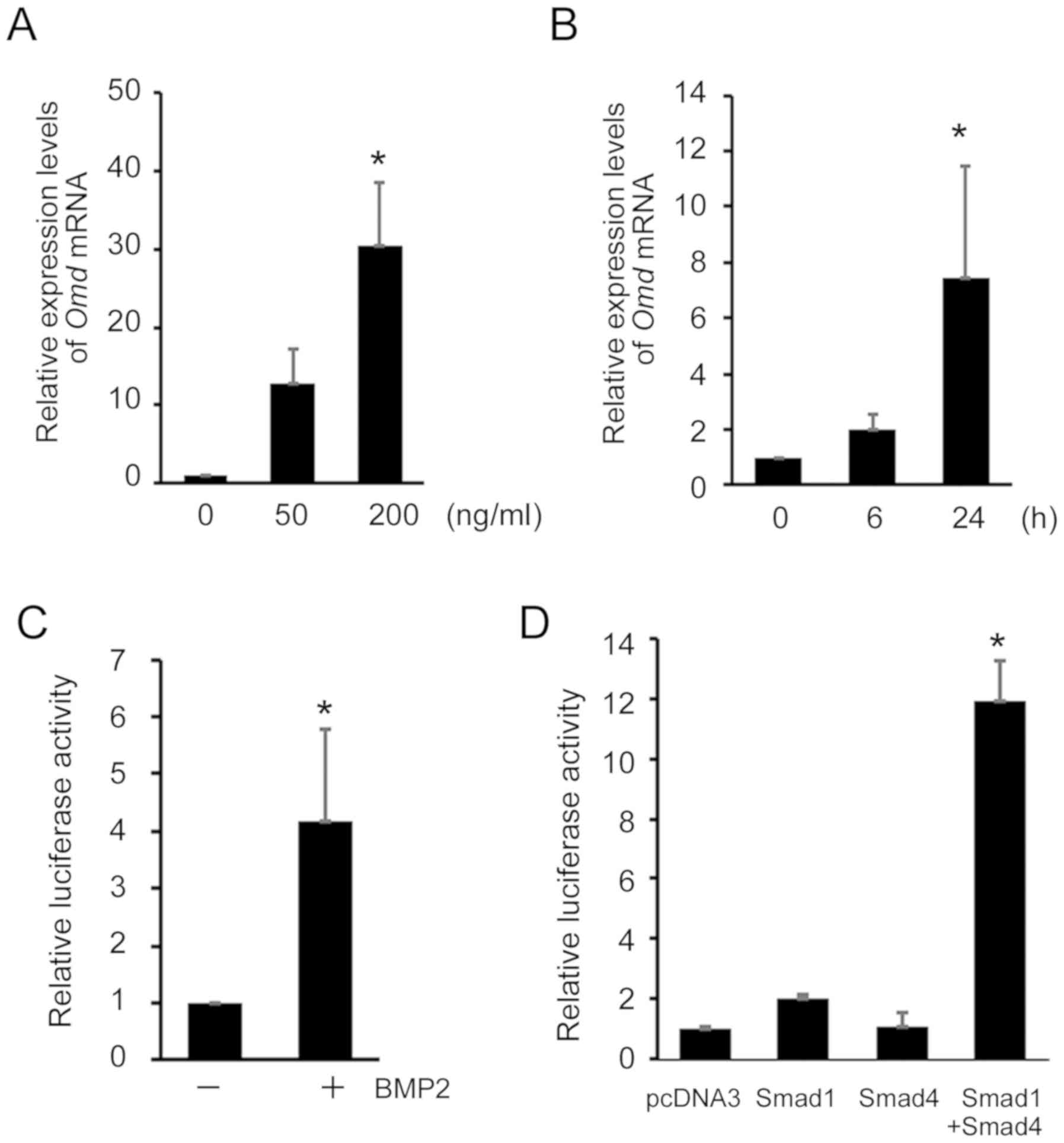
BMP2 induces Omd expression in C2C12
cells and regulates Omd promoter activity via Smad signaling
To further investigate the roles of osteoadherin in
osteoblast differentiation, experiments were performed with C2C12
cells, which are well-characterized model cells that differentiate
into myotubes or osteoblasts by the presence of BMP2 (13). In C2C12 cells, Omd mRNA
expression was not detected using RT-semi-qPCR analyses (Fig. 1A). However, in subsequent RT-qPCR
analyses, Omd mRNA expression was increased >10-fold
after treatment with 50 ng/ml BMP2. A higher BMP2 concentration
increased the expression of Omd mRNA to >30-fold at 200
ng/ml (Fig. 2A). In time course
analyses of BMP2-induced Omd mRNA expression, Omd
mRNA expression increased after 6 h, and a significant
time-dependent increase in mRNA level was observed at 24 h
(Fig. 2B). These findings
indicate that BMP2 induces Omd expression in these
cells.
To elucidate the mechanisms by which BMP2 signaling
activates Omd transcription, the present study cloned an
~1.7 kb-pair mouse genomic DNA fragment corresponding to the
5'-flanking promoter region of the mouse Omd gene. To
determine responsiveness to BMP2 signaling, this Omd
promoter region was ligated into a luciferase reporter expression
vector (pOmd-luc), and luciferase activity was observed in
BMP2-treated C2C12 cells (Fig.
2C). Transient transfection of these cells with pOmd-luc and
co-transfection with Smad1 (DVD) and Smad4 expression plasmids
resulted in a significant increase in luciferase activity (Fig. 2D). However, co-transfection with
Smad1 (DVD) and Smad4 expression plasmids did not alter pOmd-luc
activity in MC3T3-E1 cells (data not shown).
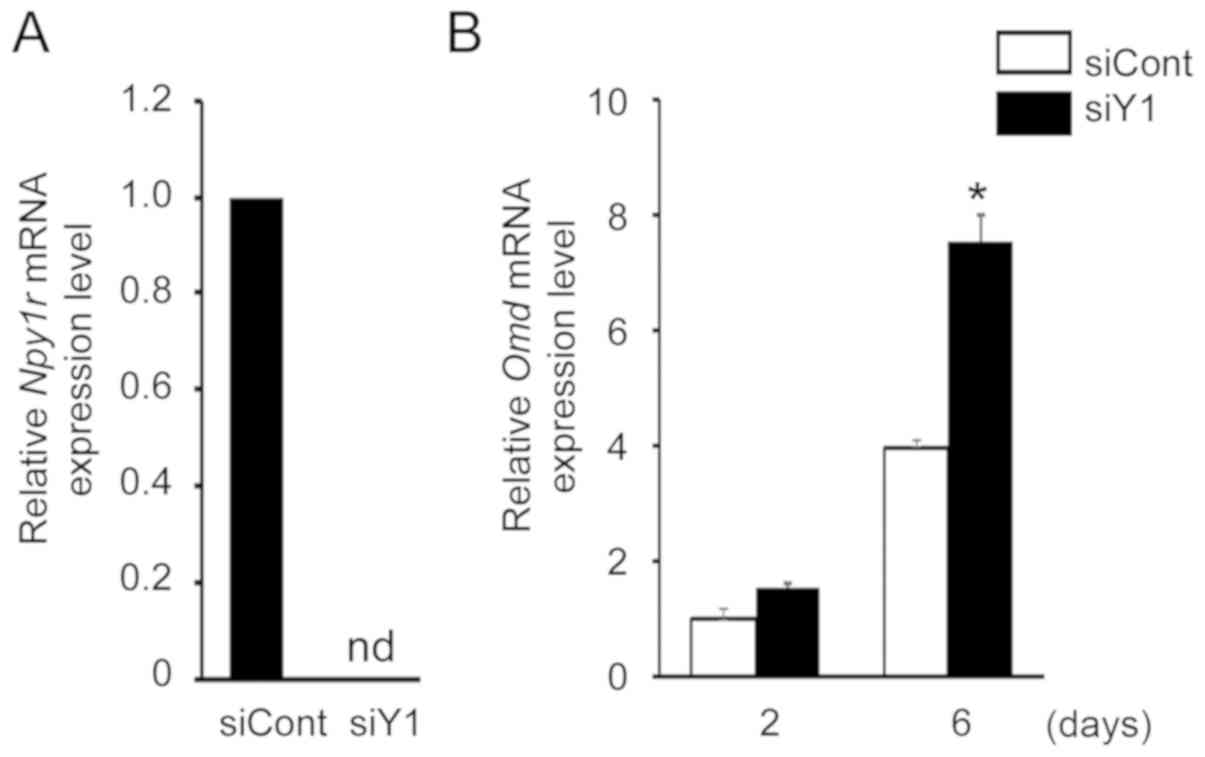
Knockdown of Y1 receptor upregulates Omd
mRNA expression in MC3T3-E1 cells
Several studies have reported that neuropeptide Y
and its Y1 receptor are directly involved in osteoblast regulation
(24-26). Previously, we demonstrated that
knockdown of the Y1 receptor using siRNA promotes osteoblast
differentiation (16). Therefore,
the present study examined the effects of Y1 receptor inhibition on
Omd mRNA expression. Following transfection of MC3T3-E1
cells with siRNA for the Y1 receptor, Y1 receptor (Npy1r)
mRNA expression level was decreased to undetectable levels,
confirming that the siRNA was effective at silencing endogenous Y1
receptor expression (Fig. 3A).
Under these conditions, Omd mRNA expression was
significantly increased (Fig.
3B), indicating that Omd expression is Y1 receptor
dependent and that the NPY signaling pathway regulates Omd
expression in MC3T3-E1 cells.
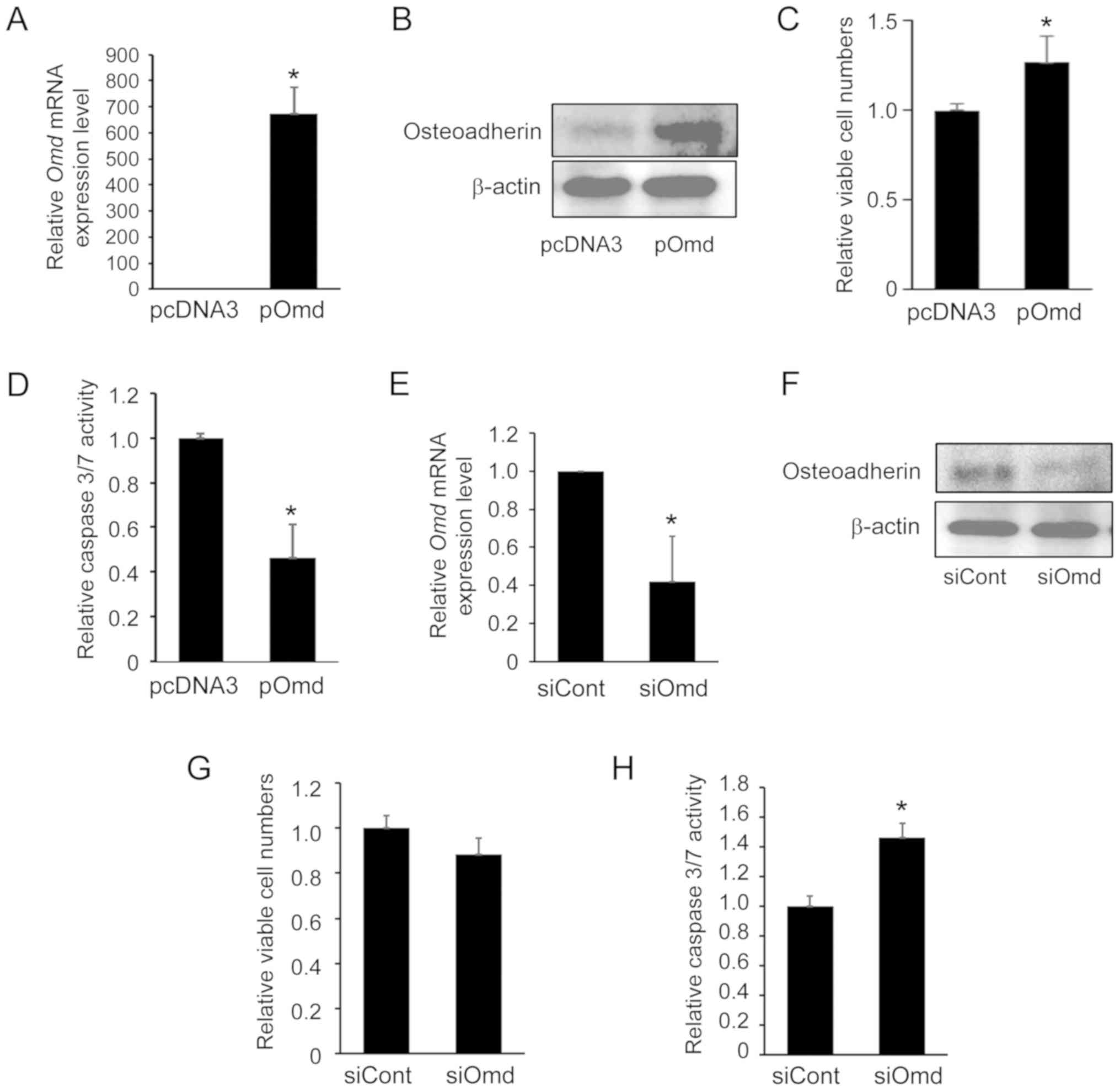
Effects of overexpression or knockdown of
Omd on viability and apoptosis in MC3T3-E1 cells
To determine the biological relevance of Omd
regulation in osteoblast differentiation, Omd was
significantly overexpressed in MC3T3-E1 cells by transfecting cells
with the pOmd expression plasmid (Fig. 4A). Western blot analysis
demonstrated that osteoadherin protein was overexpressed in
pOmd-transfected MC3T3-E1 cells (Fig.
4B). Viability was significantly increased in these cells
compared with those transfected with control plasmid (Fig. 4C). Caspase 3/7 activity, which is
associated with apoptosis (15),
was also significantly decreased following transfection with pOmd
(Fig. 4D), which further
indicates that Omd is involved in the regulation of
apoptosis in MC3T3-E1 cells.
In further experiments, the effects of
Omd-knockdown was examined using RNA interference in
MC3T3-E1 cells. Following transfection with Omd siRNA,
Omd mRNA expression was significantly decreased (Fig. 4E). As presented in Fig. 4F, cells transfected with
Omd siRNA exhibited a reduction in osteoadherin expression
compared with cells transfected with siCont. Viable cell numbers
were decreased compared with those in control siRNA-transfected
cells, although statistical significance between control siRNA- and
Omd siRNA-transfected cells was not confirmed (Fig. 4G). By contrast, caspase 3/7
activity was significantly increased following exposure to
Omd siRNA (Fig. 4H). These
results indicate that cell survival/death is Omd-dependent
and that osteoadherin may regulate cell growth and apoptosis in
osteoblast cells.
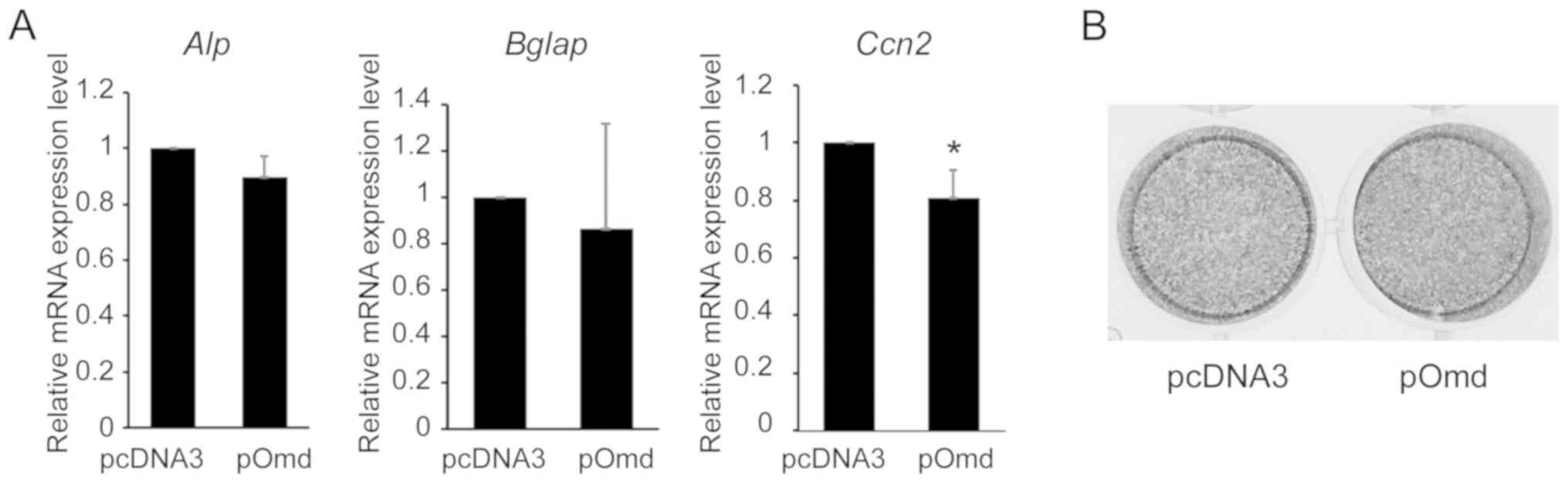
Effects of Omd overexpression on the mRNA
expression of specific osteoblast differentiation genes in MC3T3-E1
cells
To investigate the effects of osteoadherin in
osteoblast differentiation, the present study examined the effects
on genes with characterized roles in osteoblast differentiation.
Following transfection with Omd siRNA, Alp mRNA
expression level and ALP activities were not affected in MC3T3-E1
cells (data not shown). Although Alp and Bglap mRNA
expression levels were not significantly altered by Omd
overexpression, Ccn2 expression was significantly reduced
(Fig. 5A). However, as ALP
activity was not affected by Omd overexpression on day 14
(Fig. 5B), it can be concluded
that osteoblast-related gene expression is not dependent on
Omd in MC3T3-E1 cells.
Discussion
SLRP family members are secreted into the ECM after
synthesis by the cells of connective tissues. These proteoglycans
have core proteins with LRRs and are important structural
components of the ECM. Accordingly, numerous SLRPs have been
reported to bind multiple ECM constituents, particularly fibrillar
collagens, and thereby stabilize tissue frameworks (7,8).
SLRPs include five classes of structurally related proteoglycans.
Among them, osteoadherin, fibromodulin, keratocan and lumican are
class II SLRPs (2), and
osteoadherin has highly restricted expression in cells of
mineralized tissues (4,6). In the present study, BMP2 enhanced
Omd expression in C2C12 cells. Y1R inhibition also enhanced
Omd expression, suggesting specific functions of
osteoadherin in bone homeostasis and osteoblast differentiation.
The class I SLPRs biglycan and decorin are also expressed in
osteoblasts and are present in bone ECM (2,27),
and their affinity for bone apatite has been demonstrated by
chromatographic purification with hydroxyapatite columns (28). These observations suggest
significant roles of these SLPRs in the regulation of
mineralization (28).
Osteoadherin also interacts with calcium phosphate mineral
(29), suggesting that
osteoadherin contributes to biomineralization and the control of
mineral crystal nucleation, growth, and maturation.
Although few previous reports show regulation of
Omd expression by growth factors and signaling molecules,
Rehn et al (30) reported
that transforming growth factor-β (TGF-β) down-regulates Omd
expression in osteoblasts. The present study demonstrated that BMP2
enhances Omd gene expression and activates the Omd
gene promoter. Upon activation by BMP2, Smad1/Smad4 complexes
regulate the transcription of various target genes, and among these
Id1, Tlx-2 and Mix.2 reportedly respond to BMP
signaling via the Smad-binding motifs on their promoters (23,31,32). Several DNA binding motifs for
Smads, including GTCT, have been identified (33). The present analyses indicate the
presence of transcriptional machinery that is sensitive to BMP2
signaling and regulates transcription through interactions with the
Omd promoter.
SLRPs have multiple complex roles. Class II SLRPs
contain charged KS chains in their LRRs and N termini with multiple
sulfated tyrosine residues that contribute to the anionic
properties of these proteoglycans. The GAG KS is reportedly present
in several tissues, including bone, cartilage, reproductive and
neural tissues (1,2). KS binds the growth factors
fibroblast growth factor 2, TGF-β and sonic hedgehog with high
affinity (34,35), suggesting roles of KS
proteoglycans in the regulation of growth factor activities and
morphogen gradient formation. Decorin has been characterized as a
proteoglycan with roles in the control of cell growth (2), as indicated by binding and
inhibition of TGF-β. Biglycan also interacts with TGF-β and
modulates its activities (2). In
addition, decorin was reported to interact with epidermal growth
factor receptor and vascular endothelial growth factor receptor,
and consequently regulate the cell cycle (2,9,36).
The KS of osteoadherin interacts with fibronectin to promote
osteoblast attachment in vitro (3). Furthermore, the tyrosine
sulfate-rich domains of osteoadherin have been shown to bind motifs
of basic clusters in a variety of heparin-binding proteins,
including some with bioactive properties (10). These studies suggest that
osteoadherin binds various growth factors, growth factor receptors
and matrix proteins that regulate apoptosis and cell growth via
cell cycle- and proliferation-related mechanisms.
CCN2, which is widely considered a connective tissue
growth factor, is a cysteine-rich ECM protein that acts as an
anabolic growth factor to regulate cell functions (37,38). Ccn2 mRNA expression also
induces apoptosis in human aortic smooth muscle cells (39), breast cancer cells (40) and mouse osteocytes (41) by down-regulating anti-apoptotic
genes. In accordance with these observations, Omd
overexpression reduced Ccn2 expression in MC3T3-E1 cells in
the present study, suggesting an apoptotic mechanism involving CCN2
that is regulated by osteoadherin expression in osteoblasts.
In conclusion, the present study, to the best of our
knowledge, is the first to show that osteoadherin regulates
apoptosis and proliferation in osteoblast cells, and thus
established a molecular relationship between osteoblast-derived
SLRP osteoadherin and caspase 3/7 activity in osteoblasts. However,
the mechanism of regulation on caspase 3/7 activity by osteoadherin
remains unclear. Further investigations are required to identify
molecular mechanisms of osteoadherin and associated signaling
molecules.
Acknowledgments
Not applicable.
Funding
The present study was supported in part by a
research grant from the Japan Society for the Promotion of Science
Grants-in-aid for Scientific Research (grant no. 17K1163707).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
EH and MT conceived and designed the experiments. EH
performed the experiments. EH and TF analyzed the data. EH and MT
wrote the manuscript. All authors read and approved the final
manuscript for publication.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Heinegård D: Fell-Muir Lecture:
Proteoglycans and more-from molecules to biology. Int J Exp Pathol.
90:575–586. 2009. View Article : Google Scholar
|
|
2
|
Iozzo RV and Schaefer L: Proteoglycan form
and function: A comprehensive nomenclature of proteoglycans. Matrix
Biol. 42:11–55. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wendel M, Sommarin Y and Heinegård D: Bone
matrix proteins: Isolation and characterization of a novel
cell-binding keratan sulfate proteoglycan (osteoadherin) from
bovine bone. J Cell Biol. 141:839–847. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sommarin Y, Wendel M, Shen Z, Hellman U
and Heinegârd D: Osteoadherin, a cell-binding keratan sulfate
proteoglycan in bone, belongs to the family of leucine-rich repeat
proteins of the extracellular matrix. J Biol Chem. 273:16723–16729.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Buchaille R, Couble ML, Magloire H and
Bleicher F: Expression of the small leucine-rich proteoglycan
osteoadherin/osteo-modulin in human dental pulp and developing rat
teeth. Bone. 27:265–270. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nikdin H, Olsson ML, Hultenby K and Sugars
RV: Osteoadherin accumulates in the predentin towards the
mineralization front in the developing tooth. PLoS One.
7:e315252012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sjöberg AP, Manderson GA, Mörgelin M, Day
AJ, Heinegård D and Blom AM: Short leucine-rich glycoproteins of
the extracellular matrix display diverse patterns of complement
interaction and activation. Mol Immunol. 46:830–839. 2009.
View Article : Google Scholar :
|
|
8
|
Kalamajski S and Oldberg A: The role of
small leucine-rich proteoglycans in collagen fibrillogenesis.
Matrix Biol. 29:248–253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schaefer L and Iozzo RV: Biological
functions of the small leucine-rich proteoglycans: From genetics to
signal transduction. J Biol Chem. 283:21305–21309. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tillgren V, Onnerfjord P, Haglund L and
Heinegård D: The tyrosine sulfate-rich domains of the LRR proteins
fibromodulin and osteoadherin bind motifs of basic clusters in a
variety of heparin-binding proteins, including bioactive factors. J
Biol Chem. 284:28543–28553. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rehn AP, Cerny R, Sugars RV, Kaukua N and
Wendel M: Osteoadherin is upregulated by mature osteoblasts and
enhances their in vitro differentiation and mineralization. Calcif
Tissue Int. 82:454–464. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Canalis E, Economides AN and Gazzerro E:
Bone morphogenetic proteins, their antagonists, and the skeleton.
Endocr Rev. 24:218–235. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen G, Deng C and Li YP: TGF-β and BMP
signaling in osteoblast differentiation and bone formation. Int J
Biol Sci. 8:272–288. 2012. View Article : Google Scholar :
|
|
14
|
Katagiri T and Tsukamoto S: The unique
activity of bone morphogenetic proteins in bone: A critical role of
the Smad signaling pathway. Biol Chem. 394:703–714. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Van Opdenbosch N and Lamkanfi M: Caspases
in cell death, inflammation, and disease. Immunity. 50:1352–1364.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yahara M, Tei K and Tamura M: Inhibition
of neuropeptide Y Y1 receptor induces osteoblast differentiation in
MC3T3-E1 cells. Mol Med Rep. 16:2779–2784. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tamura M, Sato MM and Nashimoto M:
Regulation of CXCL12 expression by canonical Wnt signaling in bone
marrow stromal cells. Int J Biochem Cell Biol. 43:760–767. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Montesano R, Pepper MS, Möhle-Steinlein U,
Risau W, Wagner EF and Orci L: Increased proteolytic activity is
responsible for the aberrant morphogenetic behavior of endothelial
cells expressing the middle T oncogene. Cell. 62:435–445. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsuji-Tamura K and Ogawa M: Dual
inhibition of mTORC1 and mTORC2 perturbs cytoskeletal organization
and impairs endothelial cell elongation. Biochem Biophys Res
Commun. 497:326–331. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakashima A, Katagiri T and Tamura M:
Cross-talk between Wnt and bone morphogenetic protein 2 (BMP-2)
signaling in differentiation pathway of C2C12 myoblasts. J Biol
Chem. 280:37660–37668. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iizuka S, Oridate N, Nashimoto M, Fukuda S
and Tamura M: Growth inhibition of head and neck squamous cell
carcinoma cells by sgRNA targeting the cyclin D1 mRNA based on TRUE
gene silencing. PLoS One. 9:e1141212014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Nojima J, Kanomata K, Takada Y, Fukuda T,
Kokabu S, Ohte S, Takada T, Tsukui T, Yamamoto TS, Sasanuma H, et
al: Dual roles of smad proteins in the conversion from myoblasts to
osteoblastic cells by bone morphogenetic proteins. J Biol Chem.
285:15577–15586. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Teixeira L, Sousa DM, Nunes AF, Sousa MM,
Herzog H and Lamghari M: NPY revealed as a critical modulator of
osteoblast function in vitro: New insights into the role of Y1 and
Y2 receptors. J Cell Biochem. 107:908–916. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khor EC and Baldock P: The NPY system and
its neural and neuroendocrine regulation of bone. Curr Osteoporos
Rep. 10:160–168. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kurebayashi N, Sato M, Fujisawa T,
Fukushima K and Tamura M: Regulation of neuropeptide Y Y1 receptor
expression by bone morphogenetic protein 2 in C2C12 myoblasts.
Biochem Biophys Res Commun. 439:506–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nikitovic D, Aggelidakis J, Young MF,
Iozzo RV, Karamanos NK and Tzanakakis GN: The biology of small
leucine-rich proteogly-cans in bone pathophysiology. J Biol Chem.
287:33926–33933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boskey AL, Spevak L, Doty SB and Rosenberg
L: Effects of bone CS-proteoglycans, DS-decorin, and DS-biglycan on
hydroxyapatite formation in a gelatin gel. Calcif Tissue Int.
61:298–305. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou HY: Proteomic analysis of
hydroxyapatite interaction proteins in bone. Ann N Y Acad Sci.
1116:323–326. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rehn AP, Chalk AM and Wendel M:
Differential regulation of osteoadherin (OSAD) by TGF-beta1 and
BMP-2. Biochem Biophys Res Commun. 349:1057–1064. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang SJ, Hoodless PA, Lu Z, Breitman ML,
McInnes RR, Wrana JL and Buchwald M: The Tlx-2 homeobox gene is a
downstream target of BMP signalling and is required for mouse
mesoderm development. Development. 125:1877–1887. 1998.PubMed/NCBI
|
|
32
|
Dennler S, Itoh S, Vivien D, ten Dijke P,
Huet S and Gauthier JM: Direct binding of Smad3 and Smad4 to
critical TGF beta-inducible elements in the promoter of human
plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091–3100.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zawel L, Dai JL, Buckhaults P, Zhou S,
Kinzler KW, Vogelstein B and Kern SE: Human Smad3 and Smad4 are
sequence-specific transcription activators. Mol Cell. 1:611–617.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schaefer L and Schaefer RM: Proteoglycans:
From structural compounds to signaling molecules. Cell Tissue Res.
339:237–246. 2010. View Article : Google Scholar
|
|
35
|
Weyers A, Yang B, Solakyildirim K, Yee V,
Li L, Zhang F and Linhardt RJ: Isolation of bovine corneal keratan
sulfate and its growth factor and morphogen binding. FEBS J.
280:2285–2293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Buraschi S, Neill T, Goyal A, Poluzzi C,
Smythies J, Owens RT, Schaefer L, Torres A and Iozzo RV: Decorin
causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci
USA. 110:E2582–E2591. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen CC and Lau LF: Functions and
mechanisms of action of CCN matricellular proteins. Int J Biochem
Cell Biol. 41:771–783. 2009. View Article : Google Scholar :
|
|
38
|
Kubota S and Takigawa M: Cellular and
molecular actions of CCN2/CTGF and its role under physiological and
pathological conditions. Clin Sci (Lond). 128:181–196. 2015.
View Article : Google Scholar
|
|
39
|
Hishikawa K, Oemar BS, Tanner FC, Nakaki
T, Fujii T and Lüscher TF: Overexpression of connective tissue
growth factor gene induces apoptosis in human aortic smooth muscle
cells. Circulation. 100:2108–2112. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hishikawa K, Oemar BS, Tanner FC, Nakaki
T, Lüscher TF and Fujii T: Connective tissue growth factor induces
apoptosis in human breast cancer cell line MCF-7. J Biol Chem.
274:37461–37466. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sakai Y, Balam TA, Kuroda S, Tamamura N,
Fukunaga T, Takigawa M and Takano-Yamamoto T: CTGF and apoptosis in
mouse osteocytes induced by tooth movement. J Dent Res. 88:345–350.
2009. View Article : Google Scholar : PubMed/NCBI
|