NOTCH1, NOTCH2, NOTCH3 and NOTCH4 are cell surface
receptors that transduce juxtacrine signals of delta-like canonical
Notch ligand (DLL)1, DLL3, DLL4, jagged canonical Notch ligand
(JAG)1 and JAG2 from adjacent cells (1-3).
Germline mutations in the NOTCH1, NOTCH2 and
NOTCH3 genes cause Adams-Oliver syndrome, Alagille syndrome
and cerebral autosomal dominant arteriopathy with subcortical
infarcts and leukoencephalopathy, respectively (4), and DLL4-NOTCH3 signaling in human
vascular organoids induces basement membrane thickening and drives
vasculopathy in the diabetic microenvironment (5). By contrast, somatic alterations in
the genes encoding Notch signaling components drive various types
of human cancer, such as breast cancer, small-cell lung cancer
(SCLC) and T-cell acute lymphoblastic leukemia (T-ALL) (6-9).
Notch signaling dysregulation is involved in a variety of
pathologies, including cancer and non-cancerous diseases.
Small-molecule inhibitors, antagonistic monoclonal
antibodies (mAbs), antibody-drug conjugates (ADCs), bispecific
antibodies or biologics (bsAbs) and chimeric antigen
receptor-modified T cells (CAR-Ts) targeting Notch signaling
components have been developed as investigational anti-cancer drugs
(10-12). The safety, tolerability and
anti-tumor effects of these compounds have been studied in clinical
trials; however, Notch-targeted therapeutics are not yet approved
for the treatment of patients with cancer. Here, Notch signaling in
the tumor microenvironment and Notch-targeted therapeutics are
reviewed, and perspectives on Notch-related precision oncology are
discussed with emphases on biologics, clinical sequencing and
explainable artificial intelligence.
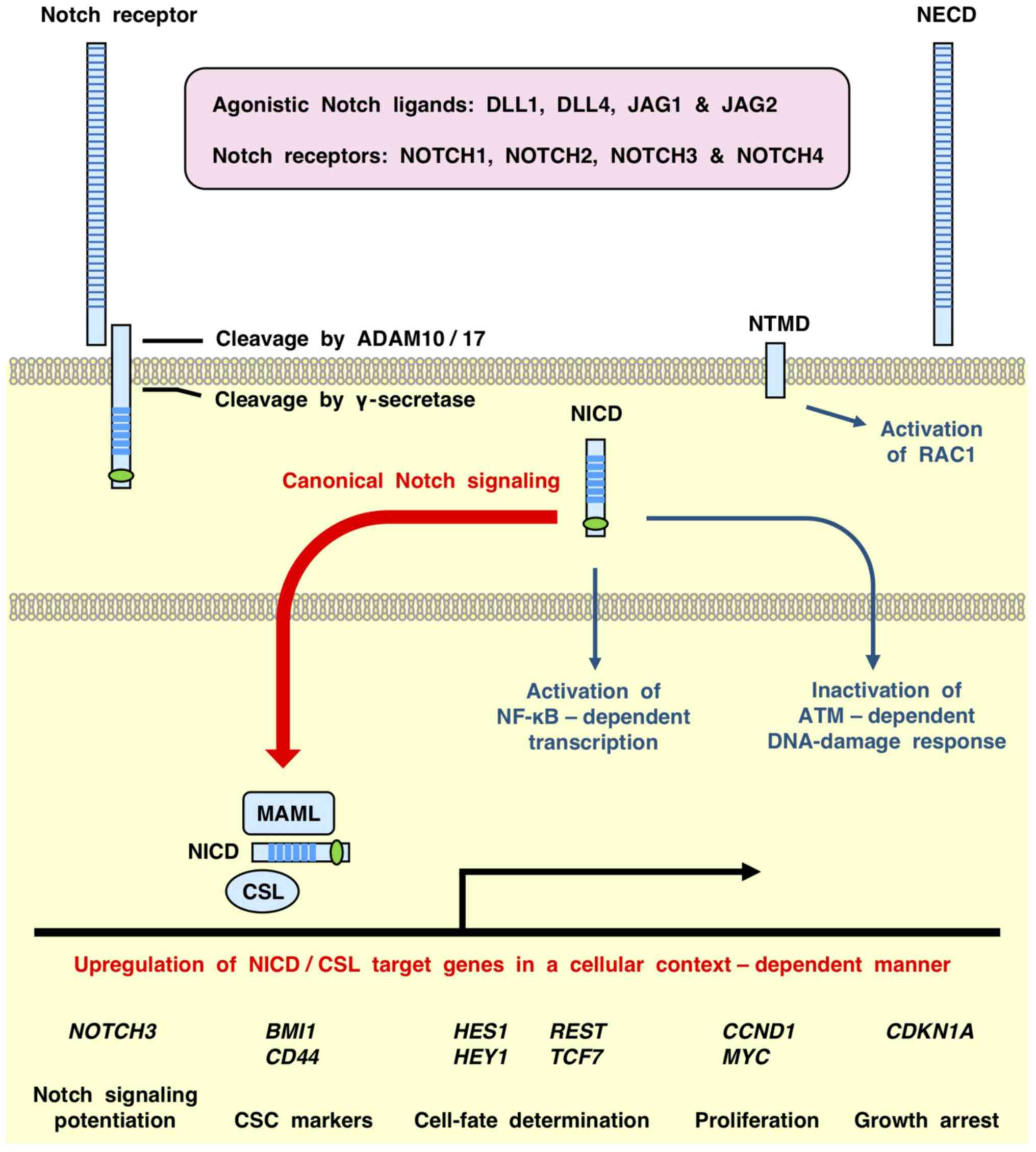
Interactions with DLL/JAG agonistic ligands trigger
sequential proteolytic cleavage of Notch receptors by disintegrin
and metalloproteinase domain-containing protein (ADAM)10/17 and
γ-secretase (2,6,18,19), which generates the following: i)
Notch extracellular domain; ii) Notch transmembrane domain (NTMD);
and iii) Notch intracellular domain (NICD) (Fig. 1). The NICD is then translocated to
the nucleus and associates with CBF1-suppressor of hairless-LAG1
(CSL) and mastermind like proteins (MAML1, MAML2 or MAML3) to
activate the transcription of target genes. NICD/CSL-dependent
transcription of Notch target genes is defined as the canonical
Notch signaling cascade (20),
whereas CSL-independent cellular responses, such as NICD-dependent
activation of NF-κB (21),
NICD-dependent inhibition of serine-protein kinase ATM (22) and NTMD-dependent activation of
Ras-related C3 botulinum toxin substrate 1 (RAC1) (23), are defined as non-canonical Notch
signaling cascades (Fig. 1).
NICDs undergo posttranslational modifications such
as phosphorylation, ubiquitination and PARylation. Cyclin-dependent
kinase (CDK)8-dependent phosphorylation of the NOTCH1 intracellular
domain (NICD1) within the intracellular proline-, glutamate-,
serine- and threonine-rich region leads to F-box/WD
repeat-containing protein 7 (FBXW7)-mediated ubiquitination and
proteasomal degradation (24,25), whereas ubiquitin carboxyl-terminal
hydrolase 7-mediated deubiquitination stabilizes NOTCH1 receptors
(26). SRC-dependent
phosphorylation of NICD1 within the intracellular ankyrin repeat
region represses Notch signaling through blockade of the NICD1-MAML
interaction and degradation of NICD1 (27). AKT-dependent phosphorylation of
NICD4 at S1495, S1847, S1865 and S1917 tethers NICD4 in the
cytoplasm and represses NICD4-dependent transcription (28). MDM2-dependent NICD4 ubiquitination
and E3 ubiquitin-protein ligase LNX (NUMB)-dependent NICD1
ubiquitination degrade NICDs and attenuate Notch signaling
(29,30), whereas MDM2-dependent NICD1
ubiquitination does not degrade NICD1 and activates Notch signaling
(31). Poly [ADP-ribose]
polymerase tankyrase-1 (TNKS) PARylates NOTCH1, NOTCH2 and NOTCH3,
and TNKS-dependent PARylation of NOTCH2 is required for nuclear
translocation of the NICD (32).
Posttranslational modifications of NICDs modulate their stability
and intracellular localization to fine-tune intracellular Notch
signaling.
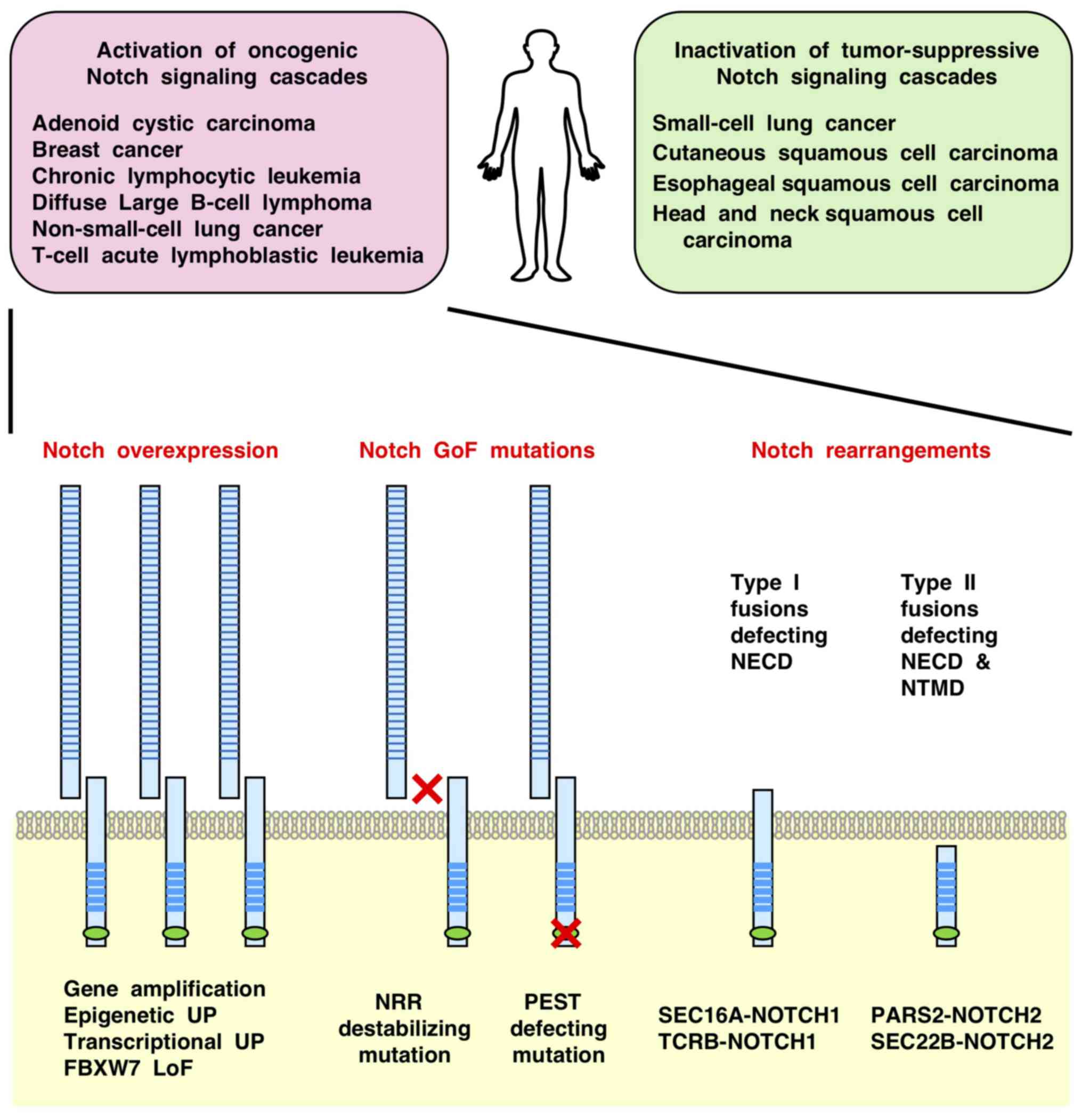
Transcriptional or epigenetic alterations also
dysregulate Notch signaling in the absence of genetic alterations
in the Notch signaling components (Fig. 2). Oncogenic Notch signaling is
reinforced due to NOTCH3 upregulation through ETS-related
transcription factor ELF3-dependent transcription in
KRAS-mutant lung adenocarcinoma (73); JAG1 upregulation through
CpG hypomethylation in renal cell carcinoma (74); and upregulation of JAG1,
MAML2, NOTCH1, NOTCH2 and NOTCH3,
partially through increased histone H3K27 acetylation, in
neuroblastoma (75).
Tumor-suppressive Notch signaling is inactivated in Ewing's sarcoma
due to repression of JAG1 by RNA binding protein EWS-friend
leukemia integration 1 transcription factor fusion protein
(76) and repression of
NOTCH1 and REST through decreased H3K27 acetylation
in SCLC (77).
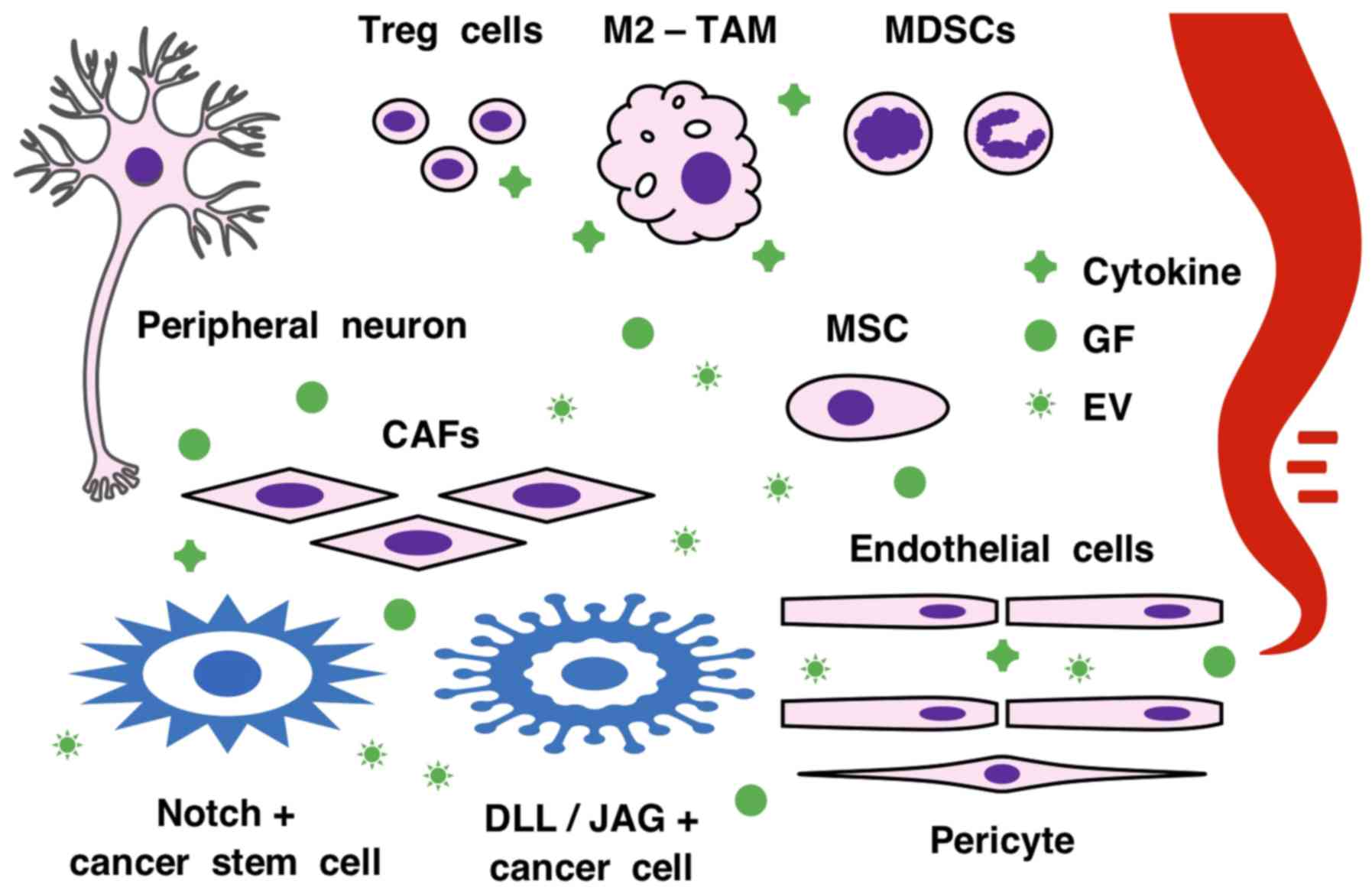
The tumor microenvironment comprises a heterogeneous
population of cancer cells, cancer-associated fibroblasts (CAFs),
endothelial cells, mesenchymal stem/stromal cells (MSCs),
pericytes, peripheral neurons and immune cells (90-92) (Fig.
3). Single-cell RNA sequencing (scRNAseq) revealed seven
subgroups of fibroblasts, six subgroups of endothelial cells and 30
subgroups of immune cells in NSCLC (93), and four subtypes of
cancer-associated fibroblasts in mouse mammary tumors (94). Cancerous and non-cancerous cells
communicate via growth factors, cytokines and extracellular
vesicles for paracrine signaling, and via membrane-type
ligand/receptor pairs for juxtacrine signaling (3,95-97). These intercellular communications
turn the anti-tumor microenvironment into a pro-tumor
microenvironment through 'omics reprogramming' (98), which includes epigenetic changes
(99), epithelial-to-mesenchymal
transition (100), immunoediting
(101) and vascular remodeling
(102).
Tumor angiogenesis is characterized by excessive
endothelial sprouting from preexisting blood vessels, which leads
to overgrowth of randomly organized and leaky tumor vessels
(124-126). Vascular endothelial growth
factor (VEGFA) signaling through VEGF receptor 2 (VEGFR2) (KDR) and
neuropilin-1 (NRP1) receptors on endothelial tip cells drives
vascular sprouting and DLL4 upregulation, and DLL4 signaling
through Notch receptors on endothelial stalk cells restricts
angiogenic sprouting and proliferation through downregulation of
VEGFR2 and NRP1 (127,128). By contrast, Notch signaling
induces JAG1 upregulation to antagonize the DLL4-dependent 'stalk'
phenotype, and promote endothelial sprouting and proliferation
(129,130). NICD1-dependent Notch signaling
activation in endothelial cells promotes lung metastasis (131), but that in hepatic endothelial
cells represses liver metastasis (132). Thus, Notch signaling regulates
tumor angiogenesis and metastasis in a context-dependent
manner.
Notch signals are involved in the development and
homeostasis of immune cells: JAG1-Notch, DLL4-Notch1 and
DLL1-Notch2 signals promote the self-renewal of long-term
hematopoietic stem cells, differentiation of early T-lymphocyte
progenitors and differentiation of marginal zone B lymphocytes,
respectively (133,134); DLL1/4 and JAG1/2 signals induce
the differentiation of naïve T lymphocytes into Th1 and Th2 cells,
respectively (135,136); DLL1 and JAG1 signals promote the
differentiation of tumor-associated macrophages (TAMs) into M1- and
M2-like phenotypes, respectively (137,138); DLL1 or JAG1 on MSCs and JAG2 on
hematopoietic progenitor cells induce the expansion of regulatory T
(Treg) cells (139-141); and DLL4 on dendritic cells
promotes Treg differentiation (142). By contrast, Notch-related
immunological reprogramming in the tumor microenvironment may be
more complex; scRNAseq revealed 20 subsets of T lymphocytes,
including circulating Treg cells, non-cancerous tissue-infiltrating
Treg cells and cancerous tissue-infiltrating Treg cells (143). For example, Notch-mediated
immune regulation in the hypoxic tumor microenvironment is
potentiated by the interaction between NICD and hypoxia-induced
hypoxia inducible factor-1α, and is modulated by the crosstalk with
the FGF, Hedgehog, transforming growth factor (TGF)-β, VEGF and WNT
signaling cascades (102,124,125,144-147).
Notch1 signaling elicited immune evasion through TGF-β upregulation
and accumulation of myeloid-derived suppressor cells (MDSCs) and
Treg cells in a mouse xenograft model with B16 melanoma cells
(148), and through upregulation
of cytotoxic T-lymphocyte protein 4, lymphocyte activation gene 3
protein, programmed cell death protein 1 and hepatitis A virus
cellular receptor 2, and accumulation of MDSCs, TAMs and Treg
cells, in an engineered mouse model of HNSCC (149).
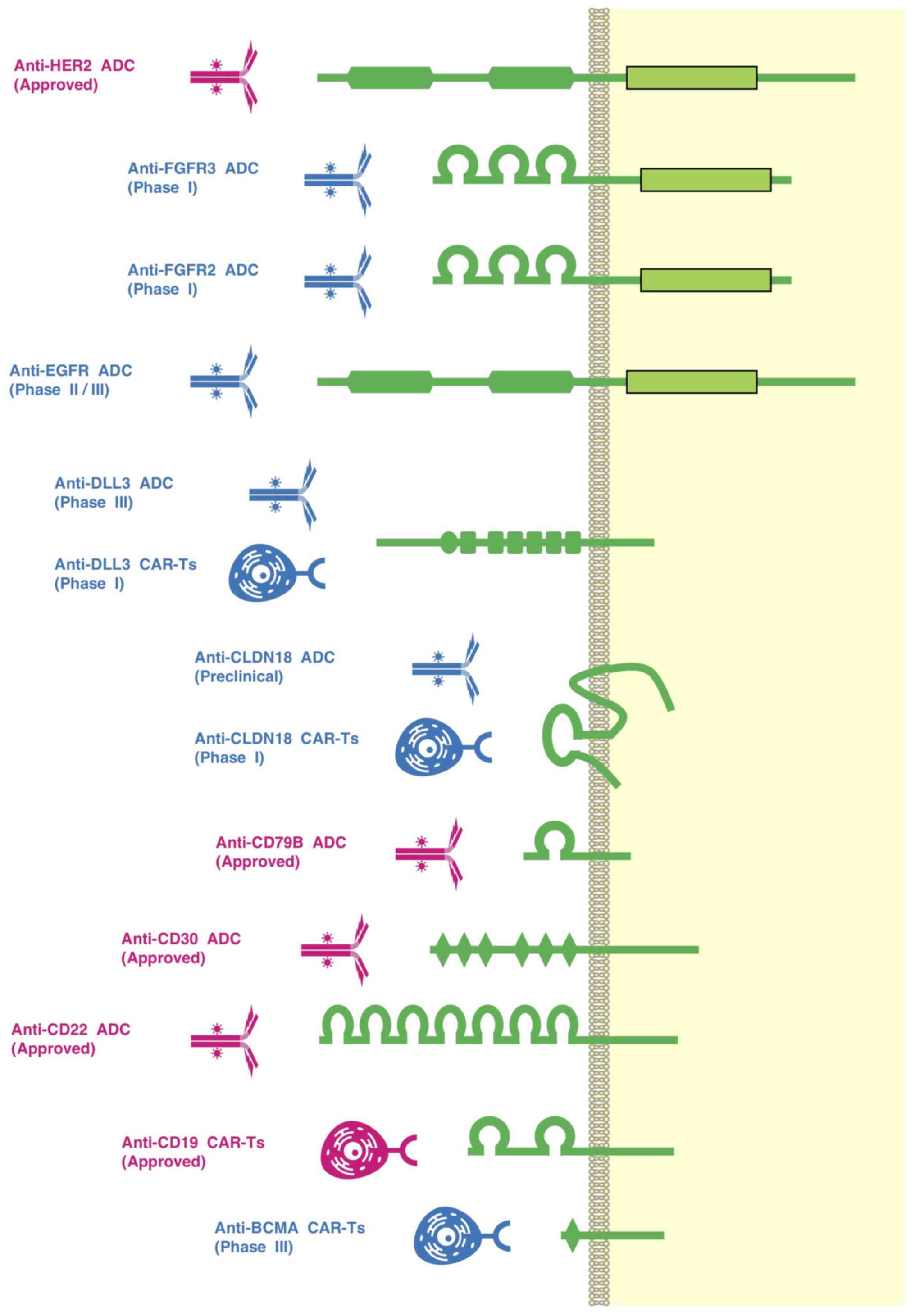
Investigational drugs that target Notch signaling
cascades are classified as follows: i) Small-molecule γ-secretase
inhibitors that block the final step of ligand-induced processing
of Notch receptors; ii) biologics, including mAbs, ADCs, bsAbs and
CAR-Ts, that bind to the extracellular region of Notch ligands or
receptors; iii) ADAM17 inhibitors that block the initial step of
ligand-induced processing of Notch receptors; and iv) NICD
protein-protein-interaction inhibitors that block the
NICD-dependent transcription of Notch target genes (Table I).
Antibody drugs that can selectively block Notch
ligands or receptors have been predicted to be an optimal choice
for cancer therapy compared with γ-secretase inhibitors for
pan-Notch signaling blockade. Anti-DLL4 mAbs (demcizumab,
enoticumab and MEDI0639) (160-162), an anti-NOTCH1 mAb
(brontictuzumab) (163) and an
anti-NOTCH2/3 mAb (tarextumab) (164,165) have been investigated in phase I
clinical trials for the treatment of patients with cancer (Table I), and were relatively well
tolerated with common adverse effects, including diarrhea, fatigue
and nausea. However, because DLL4-NOTCH signaling in endothelial
cells (127,128) and DLL4-NOTCH3 signaling in
pericytes (5) mediate
cardiovascular homeostasis, anti-DLL4 and anti-NOTCH2/3 mAbs elicit
cardiovascular toxicities, such as hypertension, acute myocardial
infarction, left ventricular dysfunction and peripheral edema. The
ORRs of monotherapy with anti-DLL4, anti-NOTCH1 and anti-NOTCH2/3
mAbs were <5% (160-165).
Repression of targeted antigens owing to the
intratu-moral heterogeneity and omics reprogramming of tumor cells
is a common mechanism of resistance to ADCs and CAR-Ts (98,196,197). Clinical trials of ADCs in
patients with solid tumors have produced disappointing results,
owing to a narrow therapeutic window and unavoidable therapeutic
resistance or recurrence (Tables
II and III). Recruitment of
new patients for the randomized phase III clinical trial of Rova-T
in patients with SCLC (registration no. NCT03061812) was halted
owing to shorter overall survival times in the Rova-T treatment
group than in the topotecan treatment group (12). LoF NOTCH1 mutations that
decrease DLL3 dependence to suppress Notch signaling might lead to
intrinsic resistance to Rova-T, whereas trans-differentiation from
DLL3-high SCLC to DLL3-low SCLC or NSCLC might elicit acquired
resistance to Rova-T. To enhance the clinical benefits of Rova-T in
patients with SCLC, the mechanisms of resistance and biomarkers of
responders should be elucidated by monitoring DLL3 expression,
NOTCH mutations and tumor phenotypes before, during and
after Rova-T therapy.
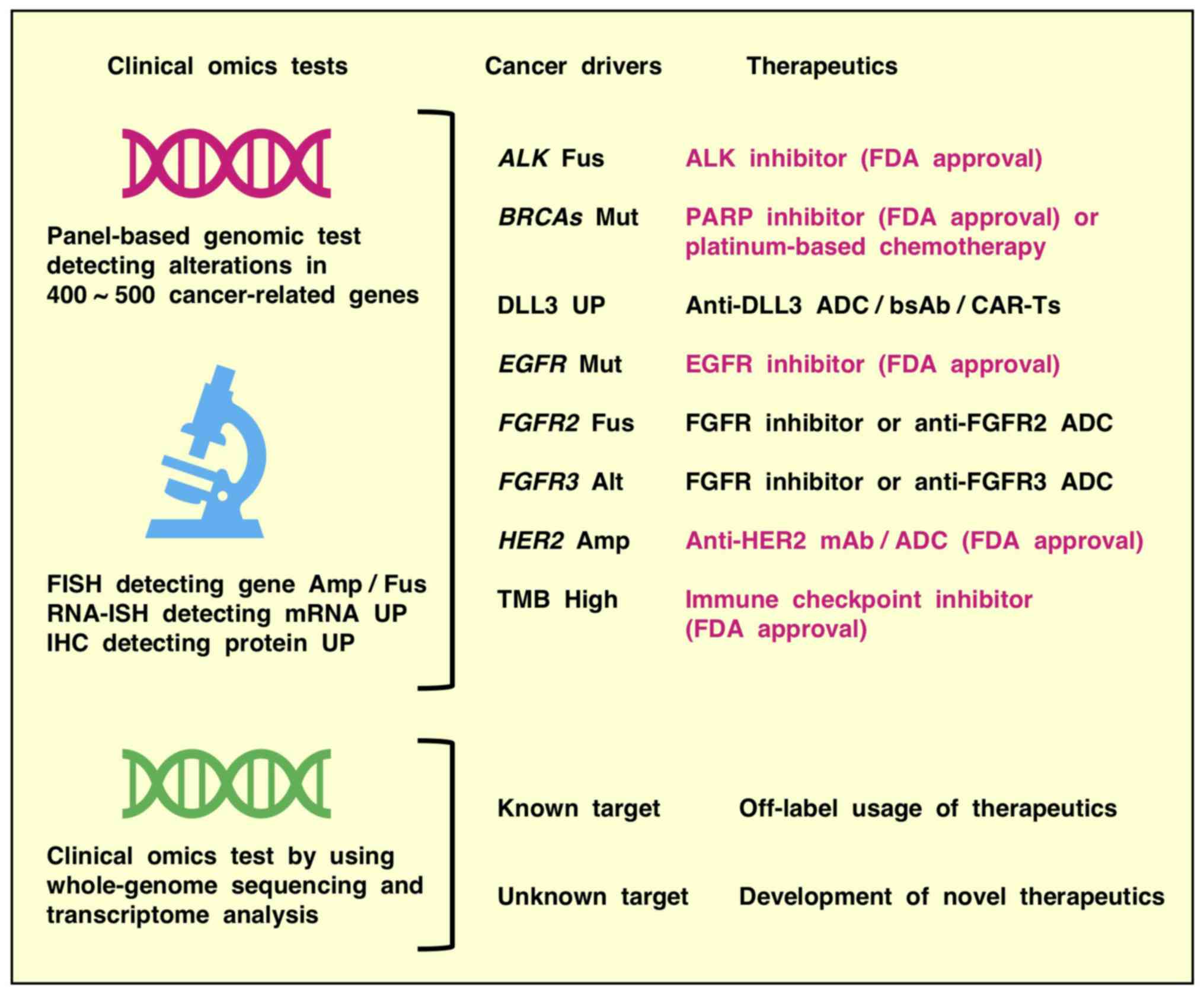
Clinical genomic tests using panel-based
next-generation sequencing are utilized to match approved marketed
drugs or investigational drugs to cancer patients in clinical
trials in the era of precision oncology (198-200) (Fig. 5). These up-to-date genomic tests,
which detect alterations in 400-500 cancer-related genes, but not
out-of-date genomic tests, which detect many fewer cancer-related
genes, can be reliably applied to diagnose tumor mutational
burden-high cancers that predict responders to immune checkpoint
inhibitors and non-responders to EGFR inhibitors (201-204). By contrast, because of their
optimization for the major genetic alterations in various human
cancer types, panel-based genomic tests cannot detect rare genetic
alterations, promoter/enhancer mutations and epigenetic alterations
that elicit aberrant activation of Notch and other oncogenic
signaling pathways. Genomic tests that detect GoF mutations in the
NOTCH1, NOTCH2, NOTCH3 and NOTCH4
genes, as well as mRNA in situ hybridization and
immunohistochemical analyses that detect overexpression of Notch
family receptors, would enhance the benefits of Notch pathway
inhibitors, such as blocking mAbs and γ-secretase inhibitors,
through successful positive selection of putative responders.
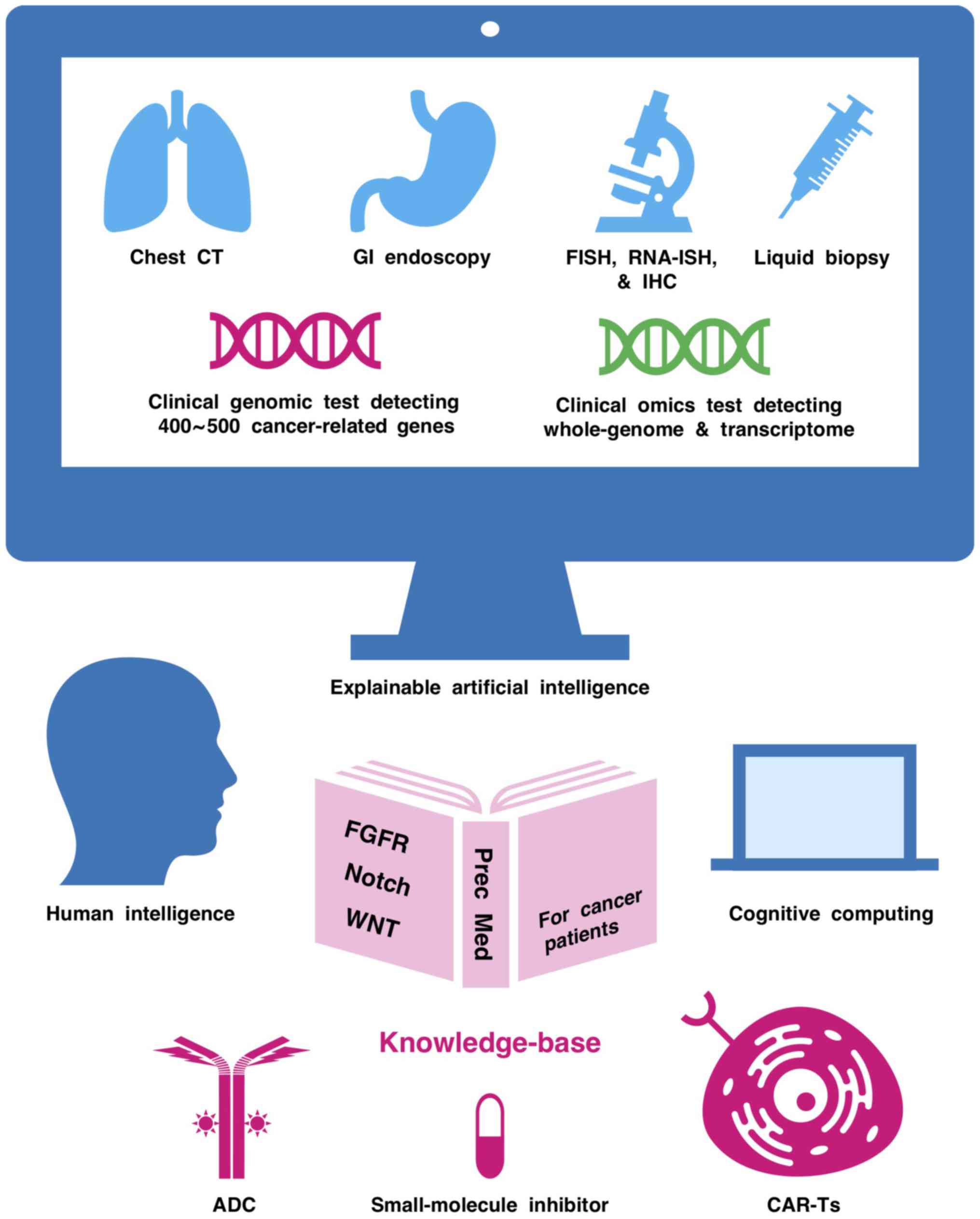
Whole-genome sequencing, as well as wholeexome
sequencing plus transcriptome analysis, is applied for the
exploration of unknown cancer drivers, and the development of novel
therapeutics for known but intractable targets with the aid of
human intelligence, cognitive computing and artificial intelligence
in basic and translational oncology (205-208). Moreover, artificial intelligence
is also applied for computer-aided diagnostic approaches (209,210), such as chest computed tomography
(211), dermoscopy (212), gastrointestinal endoscopy
(213), mammography (214) and histopathological diagnosis
(215-218). To avoid the lack of transparency
associated with black box artificial intelligence based on deep
learning technologies, the development of explainable artificial
intelligence is necessary (219). Construction of a Notch-related
knowledge base via human intelligence, explainable artificial
intelligence, and cognitive computing based on natural language
processing and text mining (Fig.
6) would promote the clinical application of Notch-targeted
therapeutics in the era of omics-based precision medicine.
This study was supported in part by a grant-in-aid
from Masaru Katoh's Fund for the Knowledge-Base Project.
Not applicable.
MasukoK and MasaruK contributed to the conception
of the study, performed the literature search and wrote the
manuscript. MasukoK prepared the tables. MasaruK prepared the
figures. All the authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Guruharsha KG, Kankel MW and
Artavanis-Tsakonas S: The Notch signalling system: Recent insights
into the complexity of a conserved pathway. Nat Rev Genet.
13:654–666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray SJ: Notch signalling in context. Nat
Rev Mol Cell Biol. 17:722–735. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meurette O and Mehlen P: Notch signaling
in the tumor micro-environment. Cancer Cell. 34:536–548. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siebel C and Lendahl U: Notch signaling in
development, tissue homeostasis, and disease. Physiol Rev.
97:1235–1294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wimmer RA, Leopoldi A, Aichinger M, Wick
N, Hantusch B, Novatchkova M, Taubenschmid J, Hämmerle M, Esk C,
Bagley JA, et al: Human blood vessel organoids as a model of
diabetic vasculopathy. Nature. 565:505–510. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ranganathan P, Weaver KL and Capobianco
AJ: Notch signalling in solid tumours: A little bit of everything
but not all the time. Nat Rev Cancer. 11:338–351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ntziachristos P, Lim JS, Sage J and
Aifantis I: From fly wings to targeted cancer therapies: A
centennial for notch signaling. Cancer Cell. 25:318–334. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aster JC, Pear WS and Blacklow SC: The
varied roles of Notch in cancer. Annu Rev Pathol. 12:245–275. 2017.
View Article : Google Scholar
|
|
9
|
Nowell CS and Radtke F: Notch as a tumour
suppressor. Nat Rev Cancer. 17:145–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Espinoza I and Miele L: Notch inhibitors
for cancer treatment. Pharmacol Ther. 139:95–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takebe N, Miele L, Harris PJ, Jeong W,
Bando H, Kahn M, Yang SX and Ivy SP: Targeting Notch, Hedgehog, and
Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin
Oncol. 12:445–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Owen DH, Giffin MJ, Bailis JM, Smit MD,
Carbone DP and He K: DLL3: An emerging target in small cell lung
cancer. J Hematol Oncol. 12:612019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
D'Souza B, Miyamoto A and Weinmaster G:
The many facets of Notch ligands. Oncogene. 27:5148–5167. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kangsamaksin T, Murtomaki A, Kofler NM,
Cuervo H, Chaudhri RA, Tattersall IW, Rosenstiel PE, Shawber C and
Kitajewski J: NOTCH decoys that selectively block DLL/NOTCH or
JAG/NOTCH disrupt angiogenesis by unique mechanisms to inhibit
tumor growth. Cancer Discov. 5:182–197. 2015. View Article : Google Scholar
|
|
15
|
Kakuda S and Haltiwanger RS: Deciphering
the Fringe-mediated Notch code: Identification of activating and
inhibiting sites allowing discrimination between ligands. Dev Cell.
40:193–201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nandagopal N, Santat LA, LeBon L, Sprinzak
D, Bronner ME and Elowitz MB: Dynamic ligand discrimination in the
Notch signaling pathway. Cell. 172:869–880.e19. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sjöqvist M and Andersson ER: Do as I say,
Not(ch) as I do: Lateral control of cell fate. Dev Biol. 447:58–70.
2019. View Article : Google Scholar
|
|
18
|
Lambrecht BN, Vanderkerken M and Hammad H:
The emerging role of ADAM metalloproteinases in immunity. Nat Rev
Immunol. 18:745–758. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang G, Zhou R, Zhou Q, Guo X, Yan C, Ke
M, Lei J and Shi Y: Structural basis of Notch recognition by human
γ-secretase. Nature. 565:192–197. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kopan R and Ilagan MX: The canonical Notch
signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vilimas T, Mascarenhas J, Palomero T,
Mandal M, Buonamici S, Meng F, Thompson B, Spaulding C, Macaroun S,
Alegre ML, et al: Targeting the NF-kappaB signaling pathway in
Notch1-induced T-cell leukemia. Nat Med. 13:70–77. 2007. View Article : Google Scholar
|
|
22
|
Vermezovic J, Adamowicz M, Santarpia L,
Rustighi A, Forcato M, Lucano C, Massimiliano L, Costanzo V,
Bicciato S, Del Sal G and d'Adda di Fagagna F: Notch is a direct
negative regulator of the DNA-damage response. Nat Struct Mol Biol.
22:417–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Polacheck WJ, Kutys ML, Yang J, Eyckmans
J, Wu Y, Vasavada H, Hirschi KK and Chen CS: A non-canonical Notch
complex regulates adherens junctions and vascular barrier function.
Nature. 552:258–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
O'Neil J, Grim J, Strack P, Rao S,
Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters
R, et al: FBW7 mutations in leukemic cells mediate NOTCH pathway
activation and resistance to γ-secretase inhibitors. J Exp Med.
204:1813–1824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Liu P, Inuzuka H and Wei W: Roles
of F-box proteins in cancer. Nat Rev Cancer. 14:233–247. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shan H, Li X, Xiao X, Dai Y, Huang J, Song
J, Liu M, Yang L, Lei H, Tong Y, et al: USP7 deubiquitinates and
stabilizes NOTCH1 in T-cell acute lymphoblastic leukemia. Signal
Transduct Target Ther. 3:292018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
LaFoya B, Munroe JA, Pu X and Albig AR:
Src kinase phosphorylates Notch1 to inhibit MAML binding. Sci Rep.
8:155152018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ramakrishnan G, Davaakhuu G, Chung WC, Zhu
H, Rana A, Filipovic A, Green AR, Atfi A, Pannuti A, Miele L and
Tzivion G: AKT and 143-3 regulate Notch4 nuclear localization. Sci
Rep. 5:87822015. View Article : Google Scholar
|
|
29
|
Sun Y, Klauzinska M, Lake RJ, Lee JM,
Santopietro S, Raafat A, Salomon D, Callahan R and
Artavanis-Tsakonas S: Trp53 regulates Notch 4 signaling through
Mdm2. J Cell Sci. 124:1067–1076. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McGill MA and McGlade CJ: Mammalian Numb
proteins promote Notch1 receptor ubiquitination and degradation of
the Notch1 intracellular domain. J Biol Chem. 278:23196–23203.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pettersson S, Sczaniecka M, McLaren L,
Russell F, Gladstone K, Hupp T and Wallace M: Non-degradative
ubiquitination of the Notch1 receptor by the E3 ligase MDM2
activates the Notch signalling pathway. Biochem J. 450:523–536.
2013. View Article : Google Scholar
|
|
32
|
Bhardwaj A, Yang Y, Ueberheide B and Smith
S: Whole proteome analysis of human tankyrase knockout cells
reveals targets of tankyrase-mediated degradation. Nat Commun.
8:22142017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schaller MA, Logue H, Mukherjee S, Lindell
DM, Coelho AL, Lincoln P, Carson WF IV, Ito T, Cavassani KA,
Chensue SW, et al: Delta-like 4 differentially regulates murine CD4
T cell expansion via BMI1. PLoS One. 5:e121722010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
López-Arribillaga E, Rodilla V,
Pellegrinet L, Guiu J, Iglesias M, Roman AC, Gutarra S, González S,
Muñoz-Cánoves P, Fernández- Salguero P, et al: Bmi1 regulates
murine intestinal stem cell proliferation and self-renewal
downstream of Notch. Development. 142:41–50. 2015. View Article : Google Scholar
|
|
35
|
Ronchini C and Capobianco AJ: Induction of
cyclin D1 transcription and CDK2 activity by Notch(ic): Implication
for cell cycle disruption in transformation by Notch(ic). Mol Cell
Biol. 21:5925–5934. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tanis KQ, Podtelezhnikov AA, Blackman SC,
Hing J, Railkar RA, Lunceford J, Klappenbach JA, Wei B, Harman A,
Camargo LM, et al: An accessible pharmacodynamic transcriptional
biomarker for Notch target engagement. Clin Pharmacol Ther.
99:370–380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
García-Peydró M, Fuentes P, Mosquera M,
García-León MJ, Alcain J, Rodríguez A, García de Miguel P, Menéndez
P, Weijer K, Spits H, et al: The NOTCH1/CD44 axis drives
pathogenesis in a T cell acute lymphoblastic leukemia model. J Clin
Invest. 128:2802–2818. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rangarajan A, Talora C, Okuyama R, Nicolas
M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et
al: Notch signaling is a direct determinant of keratinocyte growth
arrest and entry into differentiation. EMBO J. 20:3427–3436. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Procopio MG, Laszlo C, Al Labban D, Kim
DE, Bordignon P, Jo SH, Goruppi S, Menietti E, Ostano P, Ala U, et
al: Combined CSL and p53 downregulation promotes cancer-associated
fibroblast activation. Nat Cell Biol. 17:1193–1204. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jarriault S, Le Bail O, Hirsinger E,
Pourquié O, Logeat F, Strong CF, Brou C, Seidah NG and Isra l A:
Delta-1 activation of Notch-1 signaling results in HES-1
transactivation. Mol Cell Biol. 18:7423–7431. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lim JS, Ibaseta A, Fischer MM, Cancilla B,
O'Young G, Cristea S, Luca VC, Yang D, Jahchan NS, Hamard C, et al:
Intratumoural heterogeneity generated by Notch signalling promotes
small-cell lung cancer. Nature. 545:360–364. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Stoeck A, Lejnine S, Truong A, Pan L, Wang
H, Zang C, Yuan J, Ware C, MacLean J, Garrett-Engele PW, et al:
Discovery of biomarkers predictive of GSI response in
triple-negative breast cancer and adenoid cystic carcinoma. Cancer
Discov. 4:1154–1167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Maier MM and Gessler M: Comparative
analysis of the human and mouse Hey1 promoter: Hey genes are new
Notch target genes. Biochem Biophys Res Commun. 275:652–660. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Weng AP, Millholland JM, Yashiro-Ohtani Y,
Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H,
Tobias J, et al: c-Myc is an important direct target of Notch1 in
T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev.
20:2096–2109. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gekas C, D'Altri T, Aligué R, González J,
Espinosa L and Bigas A: β-Catenin is required for T-cell leukemia
initiation and MYC transcription downstream of Notch1. Leukemia.
30:2002–2010. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tottone L, Zhdanovskaya N, Carmona Pestaña
Á, Zampieri M, Simeoni F, Lazzari S, Ruocco V, Pelullo M, Caiafa P,
Felli MP, et al: Histone modifications drive aberrant Notch3
expression/activity and growth in T-ALL. Front Oncol. 9:1982019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pirot P, van Grunsven LA, Marine JC,
Huylebroeck D and Bellefroid EJ: Direct regulation of the Nrarp
gene promoter by the Notch signaling pathway. Biochem Biophys Res
Commun. 322:526–534. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wakabayashi N, Skoko JJ, Chartoumpekis DV,
Kimura S, Slocum SL, Noda K, Palliyaguru DL, Fujimuro M, Boley PA,
Tanaka Y, et al: Notch-Nrf2 axis: Regulation of Nrf2 gene
expression and cytoprotection by Notch signaling. Mol Cell Biol.
34:653–663. 2014. View Article : Google Scholar :
|
|
49
|
VanDussen KL, Carulli AJ, Keeley TM, Patel
SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud
Å, et al: Notch signaling modulates proliferation and
differentiation of intestinal crypt base columnar stem cells.
Development. 139:488–497. 2012. View Article : Google Scholar :
|
|
50
|
Weber BN, Chi AW, Chavez A, Yashiro-Ohtani
Y, Yang Q, Shestova O and Bhandoola A: A critical role for TCF-1 in
T-lineage specification and differentiation. Nature. 476:63–68.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Germar K, Dose M, Konstantinou T, Zhang J,
Wang H, Lobry C, Arnett KL, Blacklow SC, Aifantis I, Aster JC and
Gounari F: T-cell factor 1 is a gatekeeper for T-cell specification
in response to Notch signaling. Proc Natl Acad Sci USA.
108:20060–20065. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bray SJ and Gomez-Lamarca M: Notch after
cleavage. Curr Opin Cell Biol. 51:103–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen B, Jiang L, Zhong ML, Li JF, Li BS,
Peng LJ, Dai YT, Cui BW, Yan TQ, Zhang WN, et al: Identification of
fusion genes and characterization of transcriptome features in
T-cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA.
115:373–378. 2018. View Article : Google Scholar
|
|
54
|
Sanchez-Vega F, Mina M, Armenia J, Chatila
WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia
S, et al: Oncogenic signaling pathways in The Cancer Genome Atlas.
Cell. 173:321–337.e10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Weng AP, Ferrando AA, Lee W, Morris JP IV,
Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT and Aster
JC: Activating mutations of NOTCH1 in human T cell acute
lympho-blastic leukemia. Science. 306:269–271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Palomero T, Barnes KC, Real PJ, Glade
Bender JL, Sulis ML, Murty VV, Colovai AI, Balbin M and Ferrando
AA: CUTLL1, a novel human T-cell lymphoma cell line with t(7;9)
rearrangement, aberrant NOTCH1 activation and high sensitivity to
gamma-secretase inhibitors. Leukemia. 20:1279–1287. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Bie De J, Demeyer S, Alberti-Servera L,
Geerdens E, Segers H, Broux M, De Keersmaecker K, Michaux L,
Vandenberghe P, Voet T, et al: Single-cell sequencing reveals the
origin and the order of mutation acquisition in T-cell acute
lymphoblastic leukemia. Leukemia. 32:1358–1369. 2018. View Article : Google Scholar
|
|
58
|
Puente XS, Pinyol M, Quesada V, Conde L,
Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz
M, et al: Whole-genome sequencing identifies recurrent mutations in
chronic lymphocytic leukaemia. Nature. 475:101–105. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fabbri G and Dalla-Favera R: The molecular
pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer.
16:145–162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Karube K, Enjuanes A, Dlouhy I, Jares P,
Martin-Garcia D, Nadeu F, Ordóñez GR, Rovira J, Clot G, Royo C, et
al: Integrating genomic alterations in diffuse large B-cell
lymphoma identifies new relevant pathways and potential therapeutic
targets. Leukemia. 32:675–684. 2018. View Article : Google Scholar :
|
|
61
|
González-Rincón J, Méndez M, Gómez S,
García JF, Martín P, Bellas C, Pedrosa L, Rodríguez-Pinilla SM,
Camacho FI, Quero C, et al: Unraveling transformation of follicular
lymphoma to diffuse large B-cell lymphoma. PLoS One.
14:e02128132019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kridel R, Meissner B, Rogic S, Boyle M,
Telenius A, Woolcock B, Gunawardana J, Jenkins C, Cochrane C,
Ben-Neriah S, et al: Whole transcriptome sequencing reveals
recurrent NOTCH1 mutations in mantle cell lymphoma. Blood.
119:1963–1971. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Robinson DR, Kalyana-Sundaram S, Wu YM,
Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso
CS, et al: Functionally recurrent rearrangements of the MAST kinase
and Notch gene families in breast cancer. Nat Med. 17:1646–1651.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang K, Zhang Q, Li D, Ching K, Zhang C,
Zheng X, Ozeck M, Shi S, Li X, Wang H, et al: PEST domain mutations
in Notch receptors comprise an oncogenic driver segment in
triple-negative breast cancer sensitive to a γ-secretase inhibitor.
Clin Cancer Res. 21:1487–1496. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Robinson DR, Wu YM, Lonigro RJ, Vats P,
Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, et
al: Integrative clinical genomics of metastatic cancer. Nature.
548:297–303. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Westhoff B, Colaluca IN, D'Ario G,
Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G,
Viale G, et al: Alterations of the Notch pathway in lung cancer.
Proc Natl Acad Sci USA. 106:22293–22298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang NJ, Sanborn Z, Arnett KL, Bayston LJ,
Liao W, Proby CM, Leigh IM, Collisson EA, Gordon PB, Jakkula L, et
al: Loss-of-function mutations in Notch receptors in cutaneous and
lung squamous cell carcinoma. Proc Natl Acad Sci USA.
108:17761–17766. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Agrawal N, Frederick MJ, Pickering CR,
Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et
al: Exome sequencing of head and neck squamous cell carcinoma
reveals inactivating mutations in NOTCH1. Science. 333:1154–1157.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Stransky N, Egloff AM, Tward AD, Kostic
AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C,
McKenna A, et al: The mutational landscape of head and neck
squamous cell carcinoma. Science. 333:1157–1160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Martincorena I, Fowler JC, Wabik A, Lawson
ARJ, Abascal F, Hall MWJ, Cagan A, Murai K, Mahbubani K, Stratton
MR, et al: Somatic mutant clones colonize the human esophagus with
age. Science. 362:911–917. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
George J, Lim JS, Jang SJ, Cun Y, Ozretić
L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al:
Comprehensive genomic profiles of small cell lung cancer. Nature.
524:47–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ali SA, Justilien V, Jamieson L, Murray NR
and Fields AP: Protein kinase Cι drives a NOTCH3-dependent
stem-like phenotype in mutant KRAS lung adenocarcinoma. Cancer
Cell. 29:367–378. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Bhagat TD, Zou Y, Huang S, Park J, Palmer
MB, Hu C, Li W, Shenoy N, Giricz O, Choudhary G, et al: Notch
pathway is activated via genetic and epigenetic alterations and is
a therapeutic target in clear cell renal cancer. J Biol Chem.
292:837–846. 2017. View Article : Google Scholar :
|
|
75
|
van Groningen T, Akogul N, Westerhout EM,
Chan A, Hasselt NE, Zwijnenburg DA, Broekmans M, Stroeken P,
Haneveld F, Hooijer GKJ, et al: A NOTCH feed-forward loop drives
reprogramming from adrenergic to mesenchymal state in
neuroblastoma. Nat Commun. 10:15302019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ban J, Bennani-Baiti IM, Kauer M, Schaefer
KL, Poremba C, Jug G, Schwentner R, Smrzka O, Muehlbacher K, Aryee
DN and Kovar H: EWS-FLI1 suppresses NOTCH-activated p53 in Ewing's
sarcoma. Cancer Res. 68:7100–7109. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Augert A, Eastwood E, Ibrahim AH, Wu N,
Grunblatt E, Basom R, Liggitt D, Eaton KD, Martins R, Poirier JT,
et al: Targeting NOTCH activation in small cell lung cancer through
LSD1 inhibition. Sci Signal. 12:pii: eaau2922. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Song Y, Zhang Y, Jiang H, Zhu Y, Liu L,
Feng W, Yang L, Wang Y and Li M: Activation of Notch3 promotes
pulmonary arterial smooth muscle cells proliferation via
Hes1/p27Kip1 signaling pathway. FEBS Open Bio. 5:656–660. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Maraver A, Fernández-Marcos PJ, Herranz D,
Muñoz-Martin M, Gomez-Lopez G, Cañamero M, Mulero F, Megías D,
Sanchez-Carbayo M, Shen J, et al: Therapeutic effect of γ-secretase
inhibition in KrasG12V-driven non-small cell lung carcinoma by
derepression of DUSP1 and inhibition of ERK. Cancer Cell.
22:222–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Palomero T, Sulis ML, Cortina M, Real PJ,
Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, et
al: Mutational loss of PTEN induces resistance to NOTCH1 inhibition
in T-cell leukemia. Nat Med. 13:1203–1210. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhou X, Smith AJ, Waterhouse A, Blin G,
Malaguti M, Lin CY, Osorno R, Chambers I and Lowell S: Hes1
desynchronizes differentiation of pluripotent cells by modulating
STAT3 activity. Stem Cells. 31:1511–1122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Weng MT, Tsao PN, Lin HL, Tung CC, Change
MC, Chang YT, Wong JM and Wei SC: Hes1 increases the invasion
ability of colorectal cancer cells via the STAT3-MMP14 pathway.
PLoS One. 10:e01443222015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Jin S, Mutvei AP, Chivukula IV, Andersson
ER, Ramsköld D, Sandberg R, Lee KL, Kronqvist P, Mamaeva V, Ostling
P, et al: Non-canonical Notch signaling activates IL-6/JAK/STAT
signaling in breast tumor cells and is controlled by p53 and
IKKα/IKKβ. Oncogene. 32:4892–4902. 2013. View Article : Google Scholar
|
|
84
|
Schreck KC, Taylor P, Marchionni L,
Gopalakrishnan V, Bar EE, Gaiano N and Eberhart CG: The Notch
target Hes1 directly modulates Gli1 expression and Hedgehog
signaling: A potential mechanism of therapeutic resistance. Clin
Cancer Res. 16:6060–6070. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Bonyadi Rad E, Hammerlindl H, Wels C,
Popper U, Ravindran Menon D, Breiteneder H, Kitzwoegerer M, Hafner
C, Herlyn M, Bergler H and Schaider H: Notch4 signaling induces a
mesenchymal-epithelial-like transition in melanoma cells to
suppress malignant behaviors. Cancer Res. 76:1690–1697. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang D, Xu J, Liu B, He X, Zhou L, Hu X,
Qiao F, Zhang A, Xu X, Zhang H, et al: IL6 blockade potentiates the
anti-tumor effects of γ-secretase inhibitors in Notch3-expressing
breast cancer. Cell Death Differ. 25:330–339. 2018. View Article : Google Scholar
|
|
87
|
Hartman BH, Reh TA and Bermingham-McDonogh
O: Notch signaling specifies prosensory domains via lateral
induction in the developing mammalian inner ear. Proc Natl Acad Sci
USA. 107:15792–15797. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Petrovic J, Formosa-Jordan P,
Luna-Escalante JC, Abelló G, Ibañes M, Neves J and Giraldez F:
Ligand-dependent Notch signaling strength orchestrates lateral
induction and lateral inhibition in the developing inner ear.
Development. 141:2313–2324. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sun W, Gaykalova DA, Ochs MF, Mambo E,
Arnaoutakis D, Liu Y, Loyo M, Agrawal N, Howard J, Li R, et al:
Activation of the NOTCH pathway in head and neck cancer. Cancer
Res. 74:1091–1104. 2014. View Article : Google Scholar :
|
|
90
|
Turley SJ, Cremasco V and Astarita JL:
Immunological hallmarks of stromal cells in the tumour
microenvironment. Nat Rev Immunol. 15:669–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Valkenburg KC, de Groot AE and Pienta KJ:
Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin
Oncol. 15:366–381. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Östman A and Corvigno S: Microvascular
mural cells in cancer. Trends Cancer. 4:838–848. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lambrechts D, Wauters E, Boeckx B, Aibar
S, Nittner D, Burton O, Bassez A, Decaluwé H, Pircher A, Van den
Eynde K, et al: Phenotype molding of stromal cells in the lung
tumor microenvironment. Nat Med. 24:1277–1289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Bartoschek M, Oskolkov N, Bocci M, Lövrot
J, Larsson C, Sommarin M, Madsen CD, Lindgren D, Pekar G, Karlsson
G, et al: Spatially and functionally distinct subclasses of breast
cancer-associated fibroblasts revealed by single cell RNA
sequencing. Nat Commun. 9:51502018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Altorki NK, Markowitz GJ, Gao D, Port JL,
Saxena A, Stiles B, McGraw T and Mittal V: The lung
microenvironment: An important regulator of tumour growth and
metastasis. Nat Rev Cancer. 19:9–31. 2019. View Article : Google Scholar :
|
|
97
|
Wang Z and Zöller M: Exosomes, metastases,
and the miracle of cancer stem cell markers. Cancer Metastasis Rev.
38:259–295. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Katoh M: Canonical and non-canonical WNT
signaling in cancer stem cells and their niches: Cellular
heterogeneity, omics reprogramming, targeted therapy and tumor
plasticity (Review). Int J Oncol. 51:1357–1369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Dotto GP: Multifocal epithelial tumors and
field cancerization: Stroma as a primary determinant. J Clin
Invest. 124:1446–1453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Schreiber RD, Old LJ and Smyth MJ: Cancer
immunoediting: Integrating immunity's roles in cancer suppression
and promotion. Science. 331:1565–1570. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Fukumura D, Kloepper J, Amoozgar Z, Duda
DG and Jain RK: Enhancing cancer immunotherapy using
antiangiogenics: Opportunities and challenges. Nat Rev Clin Oncol.
15:325–340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Robbins J, Blondel BJ, Gallahan D and
Callahan R: Mouse mammary tumor gene int-3: A member of the Notch
gene family transforms mammary epithelial cells. J Virol.
66:2594–2599. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Peters G, Lee AE and Dickson C: Concerted
activation of two potential proto-oncogenes in carcinomas induced
by mouse mammary tumour virus. Nature. 320:628–631. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Shackleford GM, MacArthur CA, Kwan HC and
Varmus HE: Mouse mammary tumor virus infection accelerates mammary
carcinogenesis in Wnt-1 transgenic mice by insertional activation
of int-2/Fgf-3 and hst/Fgf-4. Proc Natl Acad Sci USA. 90:740–744.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Katoh M: WNT and FGF gene clusters
(review). Int J Oncol. 21:1269–1273. 2002.PubMed/NCBI
|
|
107
|
Lowther W, Wiley K, Smith GH and Callahan
R: A new common integration site, Int7, for the mouse mammary tumor
virus in mouse mammary tumors identifies a gene whose product has
furin-like and thrombospondin-like sequences. J Virol.
79:10093–10096. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Theodorou V, Kimm MA, Boer M, Wessels L,
Theelen W, Jonkers J and Hilkens J: MMTV insertional mutagenesis
identifies genes, gene families and pathways involved in mammary
cancer. Nat Genet. 39:759–769. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar :
|
|
110
|
Morgan RG, Mortensson E and Williams AC:
Targeting LGR5 in colorectal cancer: Therapeutic gold or too
plastic. Br J Cancer. 118:1410–1418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Estrach S, Ambler CA, Lo Celso C, Hozumi K
and Watt FM: Jagged 1 is a beta-catenin target gene required for
ectopic hair follicle formation in adult epidermis. Development.
133:4427–4438. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ma L, Wang Y, Hui Y, Du Y, Chen Z, Feng H,
Zhang S, Li N, Song J, Fang Y, et al: WNT/NOTCH pathway is
essential for the maintenance and expansion of human MGE
progenitors. Stem Cell Reports. 12:934–949. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Boulter L, Govaere O, Bird TG, Radulescu
S, Ramachandran P, Pellicoro A, Ridgway RA, Seo SS, Spee B, Van
Rooijen N, et al: Macrophage-derived Wnt opposes Notch signaling to
specify hepatic progenitor cell fate in chronic liver disease. Nat
Med. 18:572–579. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Högström J, Heino S, Kallio P, Lähde M,
Leppänen VM, Balboa D, Wiener Z and Alitalo K: Transcription factor
PROX1 suppresses Notch pathway activation via the nucleosome
remod-eling and deacetylase complex in colorectal cancer stem-like
cells. Cancer Res. 78:5820–5832. 2018.
|
|
115
|
Phng LK, Potente M, Leslie JD, Babbage J,
Nyqvist D, Lobov I, Ondr JK, Rao S, Lang RA, Thurston G and
Gerhardt H: Nrarp coordinates endothelial Notch and Wnt signaling
to control vessel density in angiogenesis. Dev Cell. 16:70–82.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Liu W, Li H, Hong SH, Piszczek GP, Chen W
and Rodgers GP: Olfactomedin 4 deletion induces colon
adenocarcinoma in ApcMin/+ mice. Oncogene. 35:5237–5247.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Giancotti FG: Mechanisms governing
metastatic dormancy and reactivation. Cell. 155:750–764. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Katoh M: Multi-layered prevention and
treatment of chronic inflammation, organ fibrosis and cancer
associated with canonical WNT/β-catenin signaling activation
(Review). Int J Mol Med. 42:713–725. 2018.PubMed/NCBI
|
|
119
|
Dimri GP, Martinez JL, Jacobs JJ, Keblusek
P, Itahana K, Van Lohuizen M, Campisi J, Wazer DE and Band V: The
Bmi-1 oncogene induces telomerase activity and immortalizes human
mammary epithelial cells. Cancer Res. 62:4736–4745. 2002.PubMed/NCBI
|
|
120
|
De Jaime-Soguero A, Aulicino F, Ertaylan
G, Griego A, Cerrato A, Tallam A, Del Sol A, Cosma MP and Lluis F:
Wnt/Tcf1 pathway restricts embryonic stem cell cycle through
activation of the Ink4/Arf locus. PLoS Genet. 13:e10066822017.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Srinivasan T, Walters J, Bu P, Than EB,
Tung KL, Chen KY, Panarelli N, Milsom J, Augenlicht L, Lipkin SM
and Shen X: NOTCH signaling regulates asymmetric cell fate of fast-
and slow-cycling colon cancer-initiating cells. Cancer Res.
76:3411–3421. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Germann M, Xu H, Malaterre J, Sampurno S,
Huyghe M, Cheasley D, Fre S and Ramsay RG: Tripartite interactions
between Wnt signaling, Notch and Myb for stem/progenitor cell
functions during intestinal tumorigenesis. Stem Cell Res.
13:355–366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Mourao L, Jacquemin G, Huyghe M, Nawrocki
WJ, Menssouri N, Servant N and Fre S: Lineage tracing of
Notch1-expressing cells in intestinal tumours reveals a distinct
population of cancer stem cells. Sci Rep. 9:8882019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Jain RK: Normalizing tumor
microenvironment to treat cancer: Bench to bedside to biomarkers. J
Clin Oncol. 31:2205–2218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
De Palma M, Biziato D and Petrova TV:
Microenvironmental regulation of tumour angiogenesis. Nat Rev
Cancer. 17:457–474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Yunus M, Jansson PJ, Kovacevic Z,
Kalinowski DS and Richardson DR: Tumor-induced neoangiogenesis and
receptor tyrosine kinases-Mechanisms and strategies for acquired
resistance. Biochim Biophys Acta Gen Subj. 1863:1217–1225. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Potente M, Gerhardt H and Carmeliet P:
Basic and therapeutic aspects of angiogenesis. Cell. 146:873–887.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Goel HL and Mercurio AM: VEGF targets the
tumour cell. Nat Rev Cancer. 13:871–882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Bridges E, Oon CE and Harris A: Notch
regulation of tumor angiogenesis. Future Oncol. 7:569–588. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Simons M, Gordon E and Claesson-Welsh L:
Mechanisms and regulation of endothelial VEGF receptor signalling.
Nat Rev Mol Cell Biol. 17:611–625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wieland E, Rodriguez-Vita J, Liebler SS,
Mogler C, Moll I, Herberich SE, Espinet E, Herpel E, Menuchin A,
Chang-Claude J, et al: Endothelial Notch1 activity facilitates
metastasis. Cancer Cell. 31:355–367. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wohlfeil SA, Häfele V, Dietsch B,
Schledzewski K, Winkler M, Zierow J, Leibing T, Mohammadi MM,
Heineke J, Sticht C, et al: Hepatic endothelial Notch activation
protects against liver metastasis by regulating endothelial-tumor
cell adhesion independent of angiocrine signaling. Cancer Res.
79:598–610. 2019. View Article : Google Scholar
|
|
133
|
Radtke F, MacDonald HR and
Tacchini-Cottier F: Regulation of innate and adaptive immunity by
Notch. Nat Rev Immunol. 13:427–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Lobry C, Oh P, Mansour MR, Look AT and
Aifantis I: Notch signaling: Switching an oncogene to a tumor
suppressor. Blood. 123:2451–2459. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Amsen D, Helbig C and Backer RA: Notch in
T cell differentiation: all things considered. Trends Immunol.
36:802–814. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Charbonnier LM, Wang S, Georgiev P, Sefik
E and Chatila TA: Control of peripheral tolerance by regulatory T
cell-intrinsic Notch signaling. Nat Immunol. 16:1162–1173. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Wang YC, He F, Feng F, Liu XW, Dong GY,
Qin HY, Hu XB, Zheng MH, Liang L, Feng L, et al: Notch signaling
determines the M1 versus M2 polarization of macrophages in
antitumor immune responses. Cancer Res. 70:4840–4849. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Liu H, Wang J, Zhang M, Xuan Q, Wang Z,
Lian X and Zhang Q: Jagged1 promotes aromatase inhibitor resistance
by modulating tumor-associated macrophage differentiation in breast
cancer patients. Breast Cancer Res Treat. 166:95–107. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Rashedi I, Gómez-Aristizábal A, Wang XH,
Viswanathan S and Keating A: TLR3 or TLR4 activation enhances
mesenchymal stromal cell-mediated Treg induction via Notch
signaling. Stem Cells. 35:265–275. 2017. View Article : Google Scholar
|
|
140
|
Cahill EF, Tobin LM, Carty F, Mahon BP and
English K: Jagged-1 is required for the expansion of CD4+ CD25+
FoxP3+ regulatory T cells and tolerogenic dendritic cells by murine
mesenchymal stromal cells. Stem Cell Res Ther. 6:192015. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Kared H, Adle-Biassette H, Foïs E, Masson
A, Bach JF, Chatenoud L, Schneider E and Zavala F:
Jagged2-expressing hematopoietic progenitors promote regulatory T
cell expansion in the periphery through Notch signaling. Immunity.
25:823–834. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Ting HA, de Almeida Nagata D, Rasky AJ,
Malinczak CA, Maillard IP, Schaller MA and Lukacs NW: Notch ligand
Delta-like 4 induces epigenetic regulation of Treg cell
differentiation and function in viral infection. Mucosal Immunol.
11:1524–1536. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Zhang L, Yu X, Zheng L, Zhang Y, Li Y,
Fang Q, Gao R, Kang B, Zhang Q, Huang JY, et al: Lineage tracking
reveals dynamic relationships of T cells in colorectal cancer.
Nature. 564:268–272. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Gustafsson MV, Zheng X, Pereira T, Gradin
K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U and
Bondesson M: Hypoxia requires Notch signaling to maintain the
undifferentiated cell state. Dev Cell. 9:617–628. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Katoh M: FGFR inhibitors: Effects on
cancer cells, tumor microenvironment and whole-body homeostasis
(Review). Int J Mol Med. 38:3–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Grazioli P, Felli MP, Screpanti I and
Campese AF: The mazy case of Notch and immunoregulatory cells. J
Leukoc Biol. 102:361–368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Katoh M: Genomic testing, tumor
microenvironment and targeted therapy of Hedgehog-related human
cancers. Clin Sci (Lond). 133:953–970. 2019. View Article : Google Scholar
|
|
148
|
Yang Z, Qi Y, Lai N, Zhang J, Chen Z, Liu
M, Zhang W, Luo R and Kang S: Notch1 signaling in melanoma cells
promoted tumor-induced immunosuppression via upregulation of
TGF-β1. J Exp Clin Cancer Res. 37:12018. View Article : Google Scholar
|
|
149
|
Mao L, Zhao ZL, Yu GT, Wu L, Deng WW, Li
YC, Liu JF, Bu LL, Liu B, Kulkarni AB, et al: γ-Secretase inhibitor
reduces immunosuppressive cells and enhances tumour immunity in
head and neck squamous cell carcinoma. Int J Cancer. 142:999–1009.
2018. View Article : Google Scholar
|
|
150
|
El-Khoueiry AB, Desai J, Iyer SP, Gadgeel
SM, Ramalingam SS, Horn L, LoRusso P, Bajaj G, Kollia G, Qi Z, et
al: A phase I study of AL101, a pan-NOTCH inhibitor, in patients
(pts) with locally advanced or metastatic solid tumors. J Clin
Oncol. 36(15 Suppl): S25152018. View Article : Google Scholar
|
|
151
|
Massard C, Azaro A, Soria JC, Lassen U, Le
Tourneau C, Sarker D, Smith C, Ohnmacht U, Oakley G, Patel BKR, et
al: First-in-human study of LY3039478, an oral Notch signaling
inhibitor in advanced or metastatic cancer. Ann Oncol.
29:1911–1917. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Habets RA, de Bock CE, Serneels L,
Lodewijckx I, Verbeke D, Nittner D, Narlawar R, Demeyer S, Dooley
J, Liston A, et al: Safe targeting of T cell acute lymphoblastic
leukemia by pathology-specific NOTCH inhibition. Sci Transl Med.
11:pii: eaau6246. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Messersmith WA, Shapiro GI, Cleary JM,
Jimeno A, Dasari A, Huang B, Shaik MN, Cesari R, Zheng X, Reynolds
JM, et al: A phase I, dose-finding study in patients with advanced
solid malignancies of the oral γ-secretase inhibitor PF-03084014.
Clin Cancer Res. 21:60–67. 2015. View Article : Google Scholar
|
|
154
|
Kummar S, O'Sullivan Coyne G, Do KT,
Turkbey B, Meltzer PS, Polley E, Choyke PL, Meehan R, Vilimas R,
Horneffer Y, et al: Clinical activity of the γ-secretase inhibitor
PF-03084014 in adults with desmoid tumors (aggressive
fibromatosis). J Clin Oncol. 35:1561–1569. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Tolcher AW, Messersmith WA, Mikulski SM,
Papadopoulos KP, Kwak EL, Gibbon DG, Patnaik A, Falchook GS, Dasari
A, Shapiro GI, et al: Phase I study of RO4929097, a gamma
secre-tase inhibitor of Notch signaling, in patients with
refractory metastatic or locally advanced solid tumors. J Clin
Oncol. 30:2348–2353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Strosberg JR, Yeatman T, Weber J, Coppola
D, Schell MJ, Han G, Almhanna K, Kim R, Valone T, Jump H and
Sullivan D: A phase II study of RO4929097 in metastatic colorectal
cancer. Eur J Cancer. 48:997–1003. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Jordan NV, Bardia A, Wittner BS, Benes C,
Ligorio M, Zheng Y, Yu M, Sundaresan TK, Licausi JA, Desai R, et
al: HER2 expression identifies dynamic functional states within
circulating breast cancer cells. Nature. 537:102–106. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Diluvio G, Del Gaudio F, Giuli MV,
Franciosa G, Giuliani E, Palermo R, Besharat ZM, Pignataro MG,
Vacca A, d'Amati G, et al: NOTCH3 inactivation increases triple
negative breast cancer sensitivity to gefitinib by promoting EGFR
tyrosine dephosphorylation and its intracellular arrest.
Oncogenesis. 7:422018. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Yao J, Qian C, Shu T, Zhang X, Zhao Z and
Liang Y: Combination treatment of PD98059 and DAPT in gastric
cancer through induction of apoptosis and downregulation of
WNT/β-catenin. Cancer Biol Ther. 14:833–839. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Smith DC, Eisenberg PD, Manikhas G, Chugh
R, Gubens MA, Stagg RJ, Kapoun AM, Xu L, Dupont J and Sikic B: A
phase I dose escalation and expansion study of the anticancer stem
cell agent demcizumab (anti-DLL4) in patients with previously
treated solid tumors. Clin Cancer Res. 20:6295–6303. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Chiorean EG, LoRusso P, Strother RM,
Diamond JR, Younger A, Messersmith WA, Adriaens L, Liu L, Kao RJ,
DiCioccio AT, et al: A phase I first-in-human study of enoticumab
(REGN421), a fully human Delta-like ligand 4 (DLL4) monoclonal
antibody, in patients with advanced solid tumors. Clin Cancer Res.
21:2695–2703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Falchook GS, Dowlati A, Naing A, Gribbin
MJ, Jenkins DW, Chang LL, Lai DW and Smith DC: Phase I study of
MEDI0639 in patients with advanced solid tumors. J Clin Oncol.
33(15 Suppl): S30242015. View Article : Google Scholar
|
|
163
|
Ferrarotto R, Eckhardt G, Patnaik A,
LoRusso P, Faoro L, Heymach JV, Kapoun AM, Xu L and Munster P: A
phase I dose-escalation and dose-expansion study of brontictuzumab
in subjects with selected solid tumors. Ann Oncol. 29:1561–1568.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Yen WC, Fischer MM, Axelrod F, Bond C,
Cain J, Cancilla B, Henner WR, Meisner R, Sato A, Shah J, et al:
Targeting Notch signaling with a Notch2/Notch3 antagonist
(tarextumab) inhibits tumor growth and decreases tumor-initiating
cell frequency. Clin Cancer Res. 21:2084–2095. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Smith DC, Chugh R, Patnaik A, Papadopoulos
KP, Wang M, Kapoun AM, Xu L, Dupont J, Stagg RJ and Tolcher A: A
phase 1 dose escalation and expansion study of Tarextumab
(OMP-59R5) in patients with solid tumors. Invest New Drugs.
37:722–730. 2019. View Article : Google Scholar :
|
|
166
|
Beck A, Goetsch L, Dumontet C and Corvaïa
N: Strategies and challenges for the next generation of
antibody-drug conjugates. Nat Rev Drug Discov. 16:315–337. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Lambert JM and Berkenblit A: Antibody-Drug
conjugates for cancer treatment. Annu Rev Med. 69:191–207. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Carter PJ and Lazar GA: Next generation
antibody drugs: Pursuit of the 'high-hanging fruit'. Nat Rev Drug
Discov. 17:197–223. 2018. View Article : Google Scholar
|
|
169
|
June CH, O'Connor RS, Kawalekar OU,
Ghassemi S and Milone MC: CAR T cell immunotherapy for human
cancer. Science. 359:1361–1365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Saunders LR, Bankovich AJ, Anderson WC,
Aujay MA, Bheddah S, Black K, Desai R, Escarpe PA, Hampl J, Laysang
A, et al: A DLL3-targeted antibody-drug conjugate eradicates
high-grade pulmonary neuroendocrine tumor-initiating cells in vivo.
Sci Transl Med. 7:302ra1362015. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Rudin CM, Pietanza MC, Bauer TM, Ready N,
Morgensztern D, Glisson BS, Byers LA, Johnson ML, Burris HA III,
Robert F, et al: Rovalpituzumab tesirine, a DLL3-targeted
antibody-drug conjugate, in recurrent small-cell lung cancer: A
first-in-human, first-in-class, open-label, phase 1 study. Lancet
Oncol. 18:42–51. 2017. View Article : Google Scholar :
|
|
172
|
Carbone DP, Morgensztern D, Le Moulec S,
Santana-Davila R, Ready N, Hann CL, Glisson BS, Dowlati A, Rudin
CM, Lally S, et al: Efficacy and safety of rovalpituzumab tesirine
in patients With DLL3-expressing, ≥ 3rd line small cell lung
cancer: Results from the phase 2 TRINITY study. J Clin Oncol. 36(15
Suppl): S85072018. View Article : Google Scholar
|
|
173
|
Rosen LS, Wesolowski R, Baffa R, Liao KH,
Hua SY, Gibson BL, Pirie-Shepherd S and Tolcher AW: A phase I,
dose-escalation study of PF-06650808, an anti-Notch3 antibody-drug
conjugate, in patients with breast cancer and other advanced solid
tumors. Invest New Drugs. Mar 18–2019.Epub ahead of print.
PubMed/NCBI
|
|
174
|
Smit MAD, Borghaei H, TOwonikoko TK,
Hummel HD, Johnson ML, Champiat S, Salgia R, Udagawa H, Boyer MJ
and Govindan R: Phase 1 study of AMG 757, a half-life extended
bispecific T cell engager (BiTE) antibody construct targeting DLL3,
in patients with small cell lung cancer (SCLC). J Clin Oncol. 37(15
Suppl): TPS85772019.
|
|
175
|
Li Y, Hickson JA, Ambrosi DJ, Haasch DL,
Foster-Duke KD, Eaton LJ, DiGiammarino EL, Panchal SC, Jiang F,
Mudd SR, et al: ABT-165, a dual variable domain immunoglobulin
(DVD-Ig) targeting DLL4 and VEGF, demonstrates superior efficacy
and favorable safety profiles in preclinical models. Mol Cancer
Ther. 17:1039–1050. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Jimeno A, Moore KN, Gordon M, Chugh R,
Diamond JR, Aljumaily R, Mendelson D, Kapoun AM, Xu L, Stagg R and
Smith DC: A first-in-human phase 1a study of the bispecific
anti-DLL4/anti-VEGF antibody navicixizumab (OMP-305B83) in patients
with previously treated solid tumors. Invest New Drugs. 37:461–472.
2019. View Article : Google Scholar
|
|
177
|
Hu S, Fu W, Li T, Yuan Q, Wang F, Lv G, Lv
Y, Fan X, Shen Y, Lin F, et al: Antagonism of EGFR and Notch limits
resistance to EGFR inhibitors and radiation by decreasing
tumor-initiating cell frequency. Sci Transl Med. 9:pii: eaag0339.
2017. View Article : Google Scholar
|
|
178
|
Fu W, Lei C, Yu Y, Liu S, Li T, Lin F, Fan
X, Shen Y, Ding M, Tang Y, et al: EGFR/Notch antagonists enhance
the response to inhibitors of the PI3K-Akt pathway by decreasing
tumor-initiating cell frequency. Clin Cancer Res. 25:2835–2847.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Byers LA, Chiappori A and Smit MAD: Phase
1 study of AMG 119, a chimeric antigen receptor (CAR) T cell
therapy targeting DLL3, in patients with relapsed/refractory small
cell lung cancer (SCLC). J Clin Oncol. 37(15 Suppl):
TPS85762019.
|
|
180
|
Puca L, Gavyert K, Sailer V, Conteduca V,
Dardenne E, Sigouros M, Isse K, Kearney M, Vosoughi A, Fernandez L,
et al: Delta-like protein 3 expression and therapeutic targeting in
neuroendocrine prostate cancer. Sci Transl Med. 11:pii: eaav0891.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Locke FL, Ghobadi A, Jacobson CA, Miklos
DB, Lekakis LJ, Oluwole OO, Lin Y, Braunschweig I, Hill BT,
Timmerman JM, et al: Long-term safety and activity of axicabtagene
ciloleucel in refractory large B-cell lymphoma (ZUMA-1): A
single-arm, multicentre, phase 12 trial. Lancet Oncol. 20:31–42.
2019. View Article : Google Scholar
|
|
182
|
Schuster SJ, Bishop MR, Tam CS, Waller EK,
Borchmann P, McGuirk JP, Jäger U, Jaglowski S, Andreadis C, Westin
JR, et al: Tisagenlecleucel in adult relapsed or refractory diffuse
large B-cell lymphoma. N Engl J Med. 380:45–56. 2019. View Article : Google Scholar
|
|
183
|
Kantarjian HM, DeAngelo DJ, Stelljes M,
Martinelli G, Liedtke M, Stock W, Gökbuget N, O'Brien S, Wang K,
Wang T, et al: Inotuzumab ozogamicin versus standard therapy for
acute lymphoblastic leukemia. N Engl J Med. 375:740–753. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Horwitz S, O'Connor OA, Pro B, Illidge T,
Fanale M, Advani R, Bartlett NL, Christensen JH, Morschhauser F,
Domingo-Domenech E, et al: Brentuximab vedotin with chemotherapy
for CD30-positive peripheral T-cell lymphoma (ECHELON-2): A global,
double-blind, randomised, phase 3 trial. Lancet. 393:229–240. 2019.
View Article : Google Scholar :
|
|
185
|
Tilly H, Morschhauser F, Bartlett NL,
Mehta A, Salles G, Haioun C, Munoz J, Chen AI, Kolibaba K, Lu D, et
al: Polatuzumab vedotin in combination with immunochemotherapy in
patients with previously untreated diffuse large B-cell lymphoma:
An open-label, non-randomised, phase 1b-2 study. Lancet Oncol.
20:998–1010. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Verma S, Miles D, Gianni L, Krop IE,
Welslau M, Baselga J, Pegram M, Oh DY, Diéras V, Guardino E, et al:
Trastuzumab emtansine for HER2-positive advanced breast cancer. N
Engl J Med. 367:1783–1791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Doi T, Shitara K, Naito Y, Shimomura A,
Fujiwara Y, Yonemori K, Shimizu C, Shimoi T, Kuboki Y, Matsubara N,
et al: Safety, pharmacokinetics, and antitumour activity of
trastuzumab deruxtecan (DS-8201), a HER2-targeting antibody-drug
conjugate, in patients with advanced breast and gastric or
gastro-oesophageal tumours: A phase 1 dose-escalation study. Lancet
Oncol. 18:1512–1522. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Banerji U, van Herpen CML, Saura C,
Thistlethwaite F, Lord S, Moreno V, Macpherson IR, Boni V, Rolfo C,
de Vries EGE, et al: Trastuzumab duocarmazine in locally advanced
and metastatic solid tumours and HER2-expressing breast cancer: A
phase 1 dose-escalation and dose-expansion study. Lancet Oncol.
20:1124–1135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Lassman AB, van den Bent MJ, Gan HK,
Reardon DA, Kumthekar P, Butowski N, Lwin Z, Mikkelsen T, Nabors
LB, Papadopoulos KP, et al: Safety and efficacy of depatuxizumab
mafodotin + temozolomide in patients with EGFR-amplified, recurrent
glioblastoma: Results from an international phase I multicenter
trial. Neuro Oncol. 21:106–114. 2019. View Article : Google Scholar
|
|
190
|
Moore KN, Martin LP, O'Malley DM,
Matulonis UA, Konner JA, Perez RP, Bauer TM, Ruiz-Soto R and Birrer
MJ: Safety and activity of mirvetuximab soravtansine (IMGN853), a
folate receptor alpha-targeting antibody-drug conjugate, in
platinum-resistant ovarian, fallopian tube, or primary peritoneal
cancer: A phase I expansion study. J Clin Oncol. 35:1112–1118.
2017. View Article : Google Scholar
|
|
191
|
Challita-Eid PM, Satpayev D, Yang P, An Z,
Morrison K, Shostak Y, Raitano A, Nadell R, Liu W, Lortie DR, et
al: Enfortumab vedotin antibody-drug conjugate targeting Nectin-4
is a highly potent therapeutic agent in multiple preclinical cancer
models. Cancer Res. 76:3003–3013. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Bardia A, Mayer IA, Vahdat LT, Tolaney SM,
Isakoff SJ, Diamond JR, O'Shaughnessy J, Moroose RL, Santin AD,
Abramson VG, et al: Sacituzumab govitecanhziy in refractory
metastatic triple-negative breast cancer. N Engl J Med.
380:741–751. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Raje N, Berdeja J, Lin Y, Siegel D,
Jagannath S, Madduri D, Liedtke M, Rosenblatt J, Maus MV, Turka A,
et al: Anti-BCMA CAR T-cell therapy bb2121 in relapsed or
refractory multiple myeloma. N Engl J Med. 380:1726–1737. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Merlino G, Fiascarelli A, Bigioni M,
Bressan A, Carrisi C, Bellarosa D, Salerno M, Bugianesi R, Manno R,
Bernadó Morales C, et al: MEN1309/OBT076, a first-in-class
antibody-drug conjugate targeting CD205 in solid tumors. Mol Cancer
Ther. 18:1533–1543. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Zhan X, Wang B, Li Z, Li J, Wang H, Chen
L, Jiang H, Wu M, Xiao J, Peng X, et al: Phase I trial of Claudin
18.2-specific chimeric antigen receptor T cells for advanced
gastric and pancreatic adenocarcinoma. J Clin Oncol. 37(15 Suppl):
S25092019. View Article : Google Scholar
|
|
196
|
García-Alonso S, Ocaña A and Pandiella A:
Resistance to antibody-drug conjugates. Cancer Res. 78:2159–2165.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Shah NN and Fry TJ: Mechanisms of
resistance to CAR T cell therapy. Nat Rev Clin Oncol. 16:372–385.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Singal G, Miller PG, Agarwala V, Li G,
Kaushik G, Backenroth D, Gossai A, Frampton GM, Torres AZ, Lehnert
EM, et al: Association of patient characteristics and tumor
genomics with clinical outcomes among patients with non-small cell
lung cancer using a clinicogenomic database. JAMA. 321:1391–1399.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Samstein RM, Lee CH, Shoushtari AN,
Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ,
Omuro A, et al: Tumor mutational load predicts survival after
immunotherapy across multiple cancer types. Nat Genet. 51:202–206.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Rubinstein JC, Nicolson NG and Ahuja N:
Next-generation sequencing in the management of gastric and
esophageal cancers. Surg Clin North Am. 99:511–527. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Hirsch FR, Suda K, Wiens J and Bunn PA Jr:
New and emerging targeted treatments in advanced non-small-cell
lung cancer. Lancet. 388:1012–1024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Rizvi H, Sanchez-Vega F, La K, Chatila W,
Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N,
et al: Molecular determinants of response to anti-programmed cell
death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in
patients with non-small-cell lung cancer profiled with targeted
next-generation sequencing. J Clin Oncol. 36:633–641. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Offin M, Rizvi H, Tenet M, Ni A,
Sanchez-Vega F, Li BT, Drilon A, Kris MG, Rudin CM, Schultz N, et
al: Tumor mutation burden and efficacy of EGFR-tyrosine kinase
inhibitors in patients with EGFR-mutant lung cancers. Clin Cancer
Res. 25:1063–1069. 2019. View Article : Google Scholar
|
|
204
|
Buchhalter I, Rempel E, Endris V, Allgäuer
M, Neumann O, Volckmar AL, Kirchner M, Leichsenring J, Lier A, von
Winterfeld M, et al: Size matters: Dissecting key parameters for
panel-based tumor mutational burden analysis. Int J Cancer.
144:848–858. 2019. View Article : Google Scholar
|
|
205
|
Schneider G: Automating drug discovery.
Nat Rev Drug Discov. 17:97–113. 2018. View Article : Google Scholar
|
|
206
|
Katoh M: Fibroblast growth factor
receptors as treatment targets in clinical oncology. Nat Rev Clin
Oncol. 16:105–122. 2019. View Article : Google Scholar
|
|
207
|
Somashekhar SP, Sepúlveda MJ, Puglielli S,
Norden AD, Shortliffe EH, Rohit Kumar C, Rauthan A, Arun Kumar N,
Patil P, Rhee K, et al: Watson for oncology and breast cancer
treatment recommendations: Agreement with an expert
multidisciplinary tumor board. Ann Oncol. 29:418–423. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Paley S and Karp PD: The MultiOmics
explainer: Explaining omics results in the context of a
pathway/genome database. BMC Bioinformatics. 20:3992019. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM,
Chang YC, Huang CS, Shen D and Chen CM: Computer-aided diagnosis
with deep learning architecture: Applications to breast lesions in
US images and pulmonary nodules in CT scans. Sci Rep. 6:244542016.
View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Esteva A, Kuprel B, Novoa RA, Ko J,
Swetter SM, Blau HM and Thrun S: Dermatologist-level classification
of skin cancer with deep neural networks. Nature. 542:115–118.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Ardila D, Kiraly AP, Bharadwaj S, Choi B,
Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, et al:
End-to-end lung cancer screening with three-dimensional deep
learning on low-dose chest computed tomography. Nat Med.
25:954–961. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Dascalu A and David EO: Skin cancer
detection by deep learning and sound analysis algorithms: A
prospective clinical study of an elementary dermoscope.
EBioMedicine. 43:107–113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Mori Y, Kudo SE, Misawa M, Saito Y,
Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y,
et al: Real-time use of artificial intelligence in identification
of diminutive polyps during colonoscopy: A prospective study. Ann
Intern Med. 169:357–366. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Aboutalib SS, Mohamed AA, Berg WA, Zuley
ML, Sumkin JH and Wu S: Deep learning to distinguish recalled but
benign mammography images in breast cancer screening. Clin Cancer
Res. 24:5902–5909. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Coudray N, Ocampo PS, Sakellaropoulos T,
Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N and Tsirigos
A: Classification and mutation prediction from non-small cell lung
cancer histopathology images using deep learning. Nat Med.
24:1559–1567. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Kather JN, Pearson AT, Halama N, Jäger D,
Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, et al:
Deep learning can predict microsatellite instability directly from
histology in gastrointestinal cancer. Nat Med. 25:1054–1056. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Campanella G, Hanna MG, Geneslaw L,
Miraflor A, Werneck Krauss, Silva V, Busam KJ, Brogi E, Reuter VE,
Klimstra DS and Fuchs TJ: Clinical-grade computational pathology
using weakly supervised deep learning on whole slide images. Nat
Med. 25:1301–1309. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Hekler A, Utikal JS, Enk AH, Solass W,
Schmitt M, Klode J, Schadendorf D, Sondermann W, Franklin C,
Bestvater F, et al: Deep learning outperformed 11 pathologists in
the classification of histopathological melanoma images. Eur J
Cancer. 118:91–96. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Topol EJ: High-performance medicine: The
convergence of human and artificial intelligence. Nat Med.
25:44–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Zheng H, Bae Y, Kasimir-Bauer S, Tang R,
Chen J, Ren G, Yuan M, Esposito M, Li W, Wei Y, et al: Therapeutic
antibody targeting tumor- and osteoblastic niche-derived Jagged1
sensitizes bone metastasis to chemotherapy. Cancer Cell.
32:731–747.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Li DD, Zhao CH, Ding HW, Wu Q, Ren TS,
Wang J, Chen CQ and Zhao QC: A novel inhibitor of ADAM17 sensitizes
colorectal cancer cells to 5-Fluorouracil by reversing Notch and
epithelial-mesenchymal transition in vitro and in vivo. Cell
Prolif. 51:e124802018. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Weber D, Lehal R, Frismantas V, Bourquin
J, Bauer M, Murone M and Radtke F: 411P-Pharmacological activity of
CB-103-an oral pan-NOTCH inhibitor with a novel mode of action. Ann
Oncol. 28(Suppl 5): v122–v141. 2017. View Article : Google Scholar
|
|
223
|
Moellering RE, Cornejo M, Davis TN, Del
Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL
and Bradner JE: Direct inhibition of the NOTCH transcription factor
complex. Nature. 462:182–188. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Sano R, Krytska K, Larmour CE, Raman P,
Martinez D, Ligon GF, Lillquist JS, Cucchi U, Orsini P, Rizzi S, et
al: An antibody-drug conjugate directed to the ALK receptor
demonstrates efficacy in preclinical models of neuroblastoma. Sci
Transl Med. 11:pii: eaau9732. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Boshuizen J, Koopman LA, Krijgsman O,
Shahrabi A, van den Heuvel EG, Ligtenberg MA, Vredevoogd DW, Kemper
K, Kuilman T, Song JY, et al: Cooperative targeting of melanoma
heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK
inhibitors. Nat Med. 24:203–212. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Tao Y, Wang R, Lai Q, Wu M, Wang Y, Jiang
X, Zeng L, Zhou S, Li Z, Yang T, et al: Targeting of DDR1 with
antibody-drug conjugates has antitumor effects in a mouse model of
colon carcinoma. Mol Oncol. 13:1855–1873. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Sommer A, Kopitz C, Schatz CA, Nising CF,
Mahlert C, Lerchen HG, Stelte-Ludwig B, Hammer S, Greven S,
Schuhmacher J, et al: Preclinical efficacy of the auristatin-based
antibody-drug conjugate BAY 1187982 for the treatment of
FGFR2-positive solid tumors. Cancer Res. 76:6331–6339. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Surguladze D, Pennello A, Ren X, Mack T,
Rigby A, Balderes P, Navarro E, Amaladas N, Eastman S, Topper M, et
al: LY3076226, a novel anti-FGFR3 antibody drug conjugate exhibits
potent and durable anti-tumor activity in tumor models harboring
FGFR3 mutations or fusions. Cancer Res. 79(13 Suppl):
S48352019.
|
|
229
|
Rudra-Ganguly N, Challita-Eid PM, Lowe C,
Mattie M, Moon SJ, Mendelsohn BA, Leavitt M, Virata C, A Verlinsky
A, Capo L, et al: AGS62P1, a novel site-specific antibody drug
conjugate targeting FLT3 exhibits potent anti-tumor activity
regardless of FLT3 kinase activation status. Cancer Res. 76(14
Suppl): S5742016.
|
|
230
|
Avilés P, Domínguez JM, Guillén MJ,
Muñoz-Alonso MJ, Mateo C, Rodriguez-Acebes R, Molina-Guijarro JM,
Francesch A, Martínez-Leal JF, Munt S, et al: MI130004, a novel
antibody-drug conjugate combining trastuzumab with a molecule of
marine origin, shows outstanding in iivo activity against
HER2-expressing tumors. Mol Cancer Ther. 17:786–794. 2018.
View Article : Google Scholar
|
|
231
|
Koganemaru S, Kuboki Y, Koga Y, Kojima T,
Yamauchi M, Maeda N, Kagari T, Hirotani K, Yasunaga M, Matsumura Y
and Doi T: U3-1402, a novel HER3-targeting antibody-drug conjugate,
for the treatment of colorectal cancer. Mol Cancer Ther.
18:2043–2050. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Abrams T, Connor A, Fanton C, Cohen SB,
Huber T, Miller K, Hong EE, Niu X, Kline J, Ison-Dugenny M, et al:
Preclinical antitumor activity of a novel anti-c-KIT antibody-drug
conjugate against mutant and wild-type c-KIT-positive solid tumors.
Clin Cancer Res. 24:4297–4308. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Strickler JH, Weekes CD, Nemunaitis J,
Ramanathan RK, Heist RS, Morgensztern D, Angevin E, Bauer TM, Yue
H, Motwani M, et al: First-in-human phase I, dose-escalation and
-expansion study of telisotuzumab vedotin, an antibody-drug
conjugate targeting c-Met, in patients with advanced solid tumors.
J Clin Oncol. 36:3298–3306. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Sachdev JC, Maitland ML, Sharma M, Moreno
V, Boni V, Kummar S, Stringer-Reasor EM, Forero-Torres A, Lakhani
NJ, Gibson B, et al: PF-06647020 (PF-7020), an antibody-drug
conjugate (ADC) targeting protein tyrosine kinase 7 (PTK7), in
patients (pts) with advanced solid tumors: Results of a phase I
dose escalation and expansion study. J Clin Oncol. 36(15-Suppl):
S55652018. View Article : Google Scholar
|
|
235
|
Nguyen M, Miyakawa S, Kato J, Mori T, Arai
T, Armanini M, Gelmon K, Yerushalmi R, Leung S, Gao D, et al:
Preclinical efficacy and safety assessment of an antibody-drug
conjugate targeting the c-RET proto-oncogene for breast carcinoma.
Clin Cancer Res. 21:5552–5562. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Yao HP, Feng L, Suthe SR, Chen LH, Weng
TH, Hu CY, Jun ES, Wu ZG, Wang WL, Kim SC, et al: Therapeutic
efficacy, pharmacokinetic profiles, and toxicological activities of
humanized antibody-drug conjugate Zt/g4-MMAE targeting RON receptor
tyrosine kinase for cancer therapy. J Immunother Cancer. 7:752019.
View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Berger C, Sommermeyer D, Hudecek M, Berger
M, Balakrishnan A, Paszkiewicz PJ, Kosasih PL, Rader C and Riddell
SR: Safety of targeting ROR1 in primates with chimeric antigen
receptor-modified T cells. Cancer Immunol Res. 3:206–216. 2015.
View Article : Google Scholar :
|
|
238
|
Trudel S, Lendvai N, Popat R, Voorhees PM,
Reeves B, Libby EN, Richardson PG, Hoos A, Gupta I, Bragulat V, et
al: Antibody-drug conjugate, GSK2857916, in relapsed/refractory
multiple myeloma: An update on safety and efficacy from dose
expansion phase I study. Blood Cancer J. 9:372019. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Godwin CD, Gale RP and Walter RB:
Gemtuzumab ozogamicin in acute myeloid leukemia. Leukemia.
31:1855–1868. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Socinski MA, Kaye FJ, Spigel DR, Kudrik
FJ, Ponce S, Ellis PM, Majem M, Lorigan P, Gandhi L, Gutierrez ME,
et al: Phase 1/2 study of the CD56-targeting antibody-drug
conjugate lorvotuzumab mertansine (IMGN901) in combination with
carboplatin/etoposide in small-cell lung cancer patients with
extensive-stage disease. Clin Lung Cancer. 18:68–76.e2. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
241
|
de Bono JS, Concin N, Hong DS,
Thistlethwaite FC, Machiels JP, Arkenau HT, Plummer R, Jones RH,
Nielsen D, Windfeld K, et al: Tisotumab vedotin in patients with
advanced or metastatic solid tumours (InnovaTV 201): A
first-in-human, multicentre, phase 1-2 trial. Lancet Oncol.
20:383–393. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Dotan E, Cohen SJ, Starodub AN, Lieu CH,
Messersmith WA, Simpson PS, Guarino MJ, Marshall JL, Goldberg RM,
Hecht JR, et al: Phase I/II trial of labetuzumab govitecan
(anti-CEACAM5/SN-38 antibody-drug conjugate) in patients with
refractory or relapsing metastatic colorectal cancer. J Clin Oncol.
35:3338–3346. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Zhu G, Foletti D, Liu X, Ding S, Melton
Witt J, Hasa-Moreno A, Rickert M, Holz C, Aschenbrenner L, Yang AH,
et al: Targeting CLDN18.2 by CD3 bispecific and ADC modalities for
the treatments of gastric and pancreatic cancer. Sci Rep.
9:84202019. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Bhakta S, Crocker LM, Chen Y, Hazen M,
Schutten MM, Li D, Kuijl C, Ohri R, Zhong F, Poon KA, et al: An
anti-GDNF family receptor alpha 1 (GFRA1) antibody-drug conjugate
for the treatment of hormone receptor-positive breast cancer. Mol
Cancer Ther. 17:638–649. 2018. View Article : Google Scholar
|
|
245
|
Ott PA, Pavlick AC, Johnson DB, Hart LL,
Infante JR, Luke JJ, Lutzky J, Rothschild NE, Spitler LE, Cowey CL,
et al: A phase 2 study of glembatumumab vedotin, an antibody-drug
conjugate targeting glycoprotein NMB, in patients with advanced
melanoma. Cancer. 125:1113–1123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Gong X, Azhdarinia A, Ghosh SC, Xiong W,
An Z, Liu Q and Carmon KS: LGR5-targeted antibody-drug conjugate
eradicates gastrointestinal tumors and prevents recurrence. Mol
Cancer Ther. 15:1580–1590. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Purcell JW, Tanlimco SG, Hickson J, Fox M,
Sho M, Durkin L, Uziel T, Powers R, Foster K, McGonigal T, et al:
LRRC15 is a novel mesenchymal protein and stromal target for
antibody-drug conjugates. Cancer Res. 78:4059–4072. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Willuda J, Linden L, Lerchen HG, Kopitz C,
Stelte-Ludwig B, Pena C, Lange C, Golfier S, Kneip C, Carrigan PE,
et al: Preclinical antitumor efficacy of BAY 1129980-a novel
auristatin-based anti-C4.4A (LYPD3) antibody-drug conjugate for the
treatment of non-small cell lung cancer. Mol Cancer Ther.
16:893–904. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Golfier S, Kopitz C, Kahnert A, Heisler I,
Schatz CA, Stelte-Ludwig B, Mayer-Bartschmid A, Unterschemmann K,
Bruder S, Linden L, et al: Anetumab ravtansine: A novel
mesothelin-targeting antibody-drug conjugate cures tumors with
heterogeneous target expression favored by bystander effect. Mol
Cancer Ther. 13:1537–1548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Banerjee S, Oza AM, Birrer MJ, Hamilton
EP, Hasan J, Leary A, Moore KN, Mackowiak-Matejczyk B, Pikiel J,
Ray-Coquard I, et al: Anti-NaPi2b antibody-drug conjugate
lifastuzumab vedotin (DNIB0600A) compared with pegylated liposomal
doxorubicin in patients with platinum-resistant ovarian cancer in a
randomized, open-label, phase II study. Ann Oncol. 29:917–923.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Sussman D, Smith LM, Anderson ME, Duniho
S, Hunter JH, Kostner H, Miyamoto JB, Nesterova A, Westendorf L,
Van Epps HA, et al: SGN-LIV1A: A novel antibody-drug conjugate
targeting LIV-1 for the treatment of metastatic breast cancer. Mol
Cancer Ther. 13:2991–3000. 2014. View Article : Google Scholar : PubMed/NCBI
|