Introduction
The global burden of rheumatic heart disease (RHD)
has decreased in recent decades, but the prevalence and mortality
in low- and middle-income countries remain high. RHD affects >30
million people each year and results in >300,000 deaths annually
(1). The pathogenesis of RHD
results from streptococcus-induced autoimmunity, but the specific
pathophysiology and molecular mechanisms remain unknown (2,3).
MicroRNAs (miRNAs/miRs) are small noncoding RNAs
that regulate posttranscriptional gene expression by binding to 3′
untranslated regions (3′UTRs). The roles played by miRNAs in
regulating inflammation, immunity and fibrosis have been confirmed,
but the role of miR-155-5p in RHD remains unclear. The tyrosine
phosphorylation of signal transducer and activator of transcription
3 (STAT3) is associated with malignancy (4) and rheumatoid arthritis (5-7).
The phosphorylation of STAT3 is negatively regulated by suppressor
of cytokine signaling 1 (SOCS1) in atherogenesis (8) and rheumatoid arthritis (9). Sphingosine-1-phosphate receptor 1
(S1PR1), a sphingosine-1-phosphate (S1P) receptor, has been shown
to be a factor in inflammation, autoimmunity and anti-fibrotic
function. The authors previously reported low expression of S1PR1
and activation of STAT3 phosphorylation in the RHD rat model
(10), but the specific
mechanisms of these findings remain unclear.
The purpose of the current study was to determine
the roles of miR-155/S1PR1 and the miR-155/SOCS1/STAT3 signaling
pathways in RHD-induced valvular damage. A rat model of RHD was
established by injecting formaldehyde-inactivated Group A
streptococci and complete Freund's adjuvant, and the expression of
miR-155-5p in the heart was inhibited by injecting recombinant
adeno-associated virus. After inhibition of miR-155-5p, the valvar
damage was detected. The relationships between miR-155-5p, and
S1PR1 and SOCS1 were investigated, and the expression of S1PR1,
SOCS1, STAT3, interleukin (IL)-6 and IL-17 was detected. The
findings suggested that the S1PR1 and SOCS1 genes are targets of
miR-155-5p, that the miR-155/S1PR1 and miR-155/SOCS1/STAT3 pathways
are major components of RHD-induced valvular damage, and that the
inhibition of miR-155 could alleviate the progression of valvular
damage.
Materials and methods
Animals
An experimental RHD animal model was established as
described previously (10,11).
Female Lewis rats were purchased at 8 weeks of age (150-180 g) from
Beijing Vital River Animal Technology, Co., Ltd. The acclimation
period for the rats was 5 days. The rats were bred in a specific
pathogen-free animal laboratory at the Center of Animal Experiments
of Guangxi Medical University. The rats were housed at 23±2°C with
a 12-h light/dark cycle and were allowed unrestricted cage activity
and unlimited access to water and standard chow. All animal
experimental procedures were performed according to the ethical
guidelines for the care and use of laboratory animals and were
approved by the Medical Ethics Committee of the First Affiliated
Hospital of Guangxi Medical University (grant no.
2019-KY-E-009).
Antigen preparation
Group A streptococci (GAS, ATCC19615; American Type
Culture Collection) were cultured in brain heart infusion fluid
medium (Guangdong Huankai Microbial Sci. & Tech. Co., Ltd.) at
37°C for 24 h, collected and then washed with normal saline (NS).
After harvesting, the GAS were inactivated by 10% neutral formalin
for 12 h. The formaldehyde-inactivated GAS were washed and
resuspended in sterile NS adjusted to a concentration of
4.0×1011 CFU/ml. Suspensions were fully emulsified using
sonic breaking (Sonics & Materials, Inc.) to create antigen A.
Antigen B was prepared using a GAS suspension with an equal volume
of complete Freund's adjuvant (Sigma-Aldrich, Merck KGaA).
Immunization of Lewis rats
A total of 20, 8-week-old female Lewis rats were
randomly divided into two groups: A control group and an RHD group.
Briefly, the RHD rat model was established by injecting
formaldehyde-inactivated GAS and complete Freund's adjuvant.
Patients with RHD can develop carditis, valve damage, fibrosis and
calcification (12). According to
this, rats with valvular damage and fibrosis were considered as
successfully established models (13). More specifically, rats in the RHD
group were injected into the hind foot pad with 0.2 ml antigen B on
day 0. After one week of rest, the rats were injected
subcutaneously in the abdomen with 0.5 ml antigen B on days 7, 14,
21 and 28, followed by antigen A on days 35, 42, 49 and 56. The
rats in the control group were injected with NS following the same
protocol. A total of 1.5 ml blood was collected via tail veins
without extra anaesthesia on day 63. All the rats were sacrificed
by intraperitoneal injection of pentobarbital sodium (150 mg/kg) on
day 63; lack of heartbeat and breathing for >5 min were
considered to indicate animal death. Body weight loss of >15%
with a decreased ability to consume food and water was used as
humane endpoint.
Histochemistry
Samples were taken from valves in every group and
fixed in 4% paraformaldehyde for 24 h at 4°C. Then, the tissues
were decalcified and embedded in paraffin blocks. The blocks were
sliced at 5 µm for hematoxylin and eosin (H&E) staining
and Sirius red staining. For H&E staining, the sections were
stained with hematoxylin for 4-10 min at room temperature and then
eosin for 0.5-2 min at room temperature. The images were captured
using a BX43 light microscope (Olympus Corporation). For Sirius red
staining, sections were stained with Sirius red solution for 1 h at
room temperature. The images were captured using a BX43 confocal
microscope (magnification, ×100; Olympus Corporation).
Immunohistochemistry
The valve tissues stained for S1PR1 (1:80; cat. no.
ab77076; Abcam), STAT3 (1:75; cat. no. ab69153; Abcam),
phosphorylated (p-)STAT3 (1:70; cat. no. ab76315; Abcam), IL-6
(1:65; cat. no. ab9324; Abcam) and IL-17 (1:90; cat. no. ab214588;
Abcam) were analyzed by immunohistochemistry. Sections (5
µm) of formalin-fixed paraffin-embedded tissue specimens
were blocked in 5% bovine serum albumin (BSA; Beijing Solarbio
Science & Technology Co., Ltd.) solution for 1 h at room
temperature after deparaffinization and rehydration. The sections
were added to hydrogen peroxide for 20 min at 25°C to eliminate
endogenous peroxidase and washed using phosphate-buffered saline.
Sections were incubated for 12 h at 4°C with the primary antibodies
mentioned above and then incubated with anti-rabbit horseradish
peroxidase (HRP)-conjugated (1:10; cat. no. PV-6001; OriGene
Technologies, Inc.) or anti-mouse HRP-conjugated (1:10; cat. no.
PV-6002; OriGene Technologies, Inc.) secondary antibodies for 30
min at room temperature. Diaminobenzidine (DAB) was used as an
enhancement factor for color development. After restaining with
hematoxylin and dehydrating, the images were captured using a BX43
light microscope (Olympus Corporation). Brownish yellow staining
detected by microscopy indicated positive expression. Quantitative
assessments were performed based on the methods described by
Friedrichs et al (14).
Briefly, five high-power field (magnification, ×400) images were
randomly selected and the immunoreactive score and positive cell
percentage were used to describe the expression levels. Each test
was performed in triplicate.
RT-qPCR
Total RNA was extracted from valves and serum
exosomes using the TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The RNA concentration was measured using a NanoDrop™ 2000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). A total of 0.5 µg of total RNA was reverse
transcribed into cDNA using the PrimeScript RT Reagent kit (Takara
Bio, Inc.) for mRNA. The reverse transcription conditions were as
follows: 37°C for 15 min and 85°C for 5 sec. A total of 1 µg
of total RNA or serum exosome RNA was reverse-transcribed using the
Mir-X™ miRNA First-Strand Synthesis kit (Takara Bio, Inc.). The
reverse transcription conditions were as follows: 37°C for 60 min
and 85°C for 5 min. RT-qPCR was performed using TB Green Premix Ex
Taq II (Takara Bio, Inc.) in a StepOne system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the following thermocycling
conditions: 95°C for 30 min, followed by 40 cycles at 95°C for 5
sec and 60°C for 30 sec. The primers used are listed in Table I. The mRNA and miRNA expression
levels were normalized to β-actin and U6 expression, respectively,
using the 2−ΔΔCq method (15). Samples were measured in three
independent replicates.
 | Table ISequences of primers used in reverse
transcription-quantitative PCR. |
Table I
Sequences of primers used in reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′-3′) |
|---|
| miR-155-5p | Forward:
ACGCGTTAATGCTAATTGTGATAGGGGT |
| U6 | Forward:
GGAACGATACAGAGAAGATTAGC |
| Reverse:
TGGAACGCTTCACGAATTTGCG |
| STAT3 | Forward:
TTTGAGACAGAGGTGTACCACCAAG |
| Reverse:
ACCACAGGATTGATGCCCAAG |
| S1PR1 | Forward:
GCTTCATCACTCACTACCCTAGCA |
| Reverse:
TTCTCCCTTCCCTCCCTCTC |
| SOCS1 | Forward:
CACGCACTTCCGCACATTCC |
| Reverse:
TCCAGCAGCTCGAAGAGGGA |
| Col3a1 | Forward:
ACTTCTGGTCCTCCTGGTCTGC |
| Reverse:
CGCCTGGCTCACCCTTTTCAC |
| FSP1 | Forward:
TGGGGAGAAGGACAGACGAAGC |
| Reverse:
TGGCAATGCAGGACAGGAAGAC |
| β-actin | Forward:
GGAGATTACTGCCCTGGCTCCTA |
| Reverse:
GACTCATCGTACTCCTGCTTGCTG |
Western blotting
Samples of valve tissues were treated with RIPA
lysis buffer (Sangon Biotech Co., Ltd.) and were centrifuged at
12,000 × g at 4°C for 15 min to obtain the supernatant, according
to the manufacturer's protocol. The protein concentration was
subsequently quantified with a bicinchoninic acid protein assay
(Sangon Biotech Co., Ltd.). Protein lysates were mixed with 4X
loading buffer (Beijing Solarbio Science & Technology Co.,
Ltd.) and boiled at 95°C for 5 min. Equal amounts of protein (20
µg) were separated by 10% SDS-PAGE at 60 V for 30 min and
120 V for 70 min by a blotting system (Bio-Rad Laboratories, Inc.).
The separated protein was electrotransferred to 0.22 µm PVDF
membranes (EMD Millipore) at a constant 300 mA for 90 min. After
electrotransfer, the membranes were blocked with 3% BSA blocking
solution (Sangon Biotech Co., Ltd.) for 1 h at room temperature and
then probed with the following antibodies for 12 h at 4°C: S1PR1
(1:3,000; cat. no. ab125074; Abcam), SOCS1 (1:1,000; cat. no. 3950;
Cell Signaling Technology, Inc.), STAT3 (1:1,000; cat. no. ab68153;
Abcam), p-STAT3 (1:1,000; cat. no. 9145; Cell Signaling Technology,
Inc.) and β-tubulin (1:3,000; cat. no. 10068-1-AP; ProteinTech
Group, Inc.). The membranes were incubated with HRP-conjugated
secondary antibody (10,000; cat. no. ab6721; Abcam) for 1 h at room
temperature. All samples were measured in triplicate. The
expression of S1PR1, STAT3 and p-STAT3 was quantified by
normalizing to β-tubulin using ImageJ software (1.51j, National
Institute of Health).
Isolation and identification of
exosomes
Exosomes were isolated from serum using the ExoQuick
ULTRA Isolation kit (System Biosciences) according to the
manufacturer's protocol. Western blotting was used to detect the
exosome-specific membrane markers cluster of differentiation (CD)9
(cat. no. ab92726; Abcam; 1:1,000) and CD63 (cat. no. ab108950;
Abcam; 1:1,000). Nanoparticle tracking analysis (NTA) was used to
measure both the size and concentration of exosomes with a ZetaView
PMX120 system (Particle Metrix). Transmission electron microscopy
(TEM; Tecnai G2 spirit, FEI) was used to observe the morphology of
exosomes at 80 kV after staining with 2% uranyl acetate for 1 min
at room temperature. All experiments were performed in
triplicate.
Dual luciferase assay
The potential relationships between miR-155-5p and
SOCS1 and S1PR1 were predicted by TargetScan 7.2 (http://www.targetscan.org). The 293T cells were
obtained from the Cell Bank of Type Culture Collection of Chinese
Academy of Sciences. The cells were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.)
in an incubator at 37°C and 5% CO2. Both the wild-type
(WT) and the mutant-type (MUT) 3′-UTR of S1PR1 and SOCS1 mRNA were
amplified by PCR and inserted into psiCHECK-2 dual-luciferase
reporter plasmids downstream of the Renilla luciferase gene
(Promega Corporation). The 0.16 µg S1PR1 WT 3′-UTR, S1PR1
MUT 3′-UTR or negative control (NC) 3′-UTR plasmids were
cotransfected with 5 pmol miR-155-5p mimics (5′-UUA AUG CUA AUU GUG
AUA GGG GU-3′) or miR-NC mimics (5′-UUC UCC GAA CGU GUC ACG UdT
dT-3′) into 293T cells using X-tremeGENE™ HP DNA Transfection
Reagent (Roche Diagnostics). The 0.16 µg SOCS1 WT 3′-UTR or
SOCS1 MUT 3′-UTR were cotransfected with 5 pmol miR-155-5p mimics
or miR-NC mimics into 293T cells. After 48 h, the luciferase
activities were tested using a Dual-Luciferase Reporter Assay
system (Promega Corporation), Renilla luciferase activity
was normalized to firefly luciferase activity; experiments were
performed in triplicate.
In vivo gene therapy
Recombinant adeno-associated virus (serotype 9)
vectors carrying a rat miR-155-5p (MIMAT0030409) inhibition
sequence with a c-TNT promoter (AAV-miR155-inhibitor; Han
Biomedical, Inc.) were used. An AAV-control was used as a negative
control. A total of 24 female Lewis rats were randomly divided into
four groups: Control group (n=6), RHD group (n=6), RHD+AAV-control
group (AAV-control; n=6) and RHD+AAV-miR155-inhibitor group
(AAV-miR155-inhibitor; n=6). Each rat in the AAV-control and
AAV-miR155-inhibitor group was given a single injection of
2.5×1011 viral genome particles (AAV-control or AAV-
miR155-inhibitor, diluted in 200 µl normal saline) via the
tail vein. A total of 3 weeks after the injection, RHD models were
established in the RHD group, the AAV-control group and the
AAV-miR155-inhibitor group following the procedure mentioned
above.
ELISA
To study the cytokine levels in the serum, rat IL-6
and IL-17 were measured using ELISA kits according to the
manufacturer's protocol (cat. nos. E04640r and E07451r; Cusabio).
All samples were measured in triplicate.
Statistical analysis
All data are expressed as the mean ± standard
deviation for at least three separate experiments. Statistical
analysis was performed using SPSS software 16.0 (SPSS, Inc.). A
Student's t-test was used to analyze the differences between two
groups. One-way analysis of variance with Tukey's post hoc test was
used to analyze the differences among four groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Pathological examination of the RHD
model
H&E staining showed that the rats in the RHD
group presented with acute valvulitis. Diffuse infiltration of
inflammatory cells, thickened valves, inflammatory exudate and
fibrosis were observed under microscopy. None of these changes were
found in the control group (Fig.
1A).
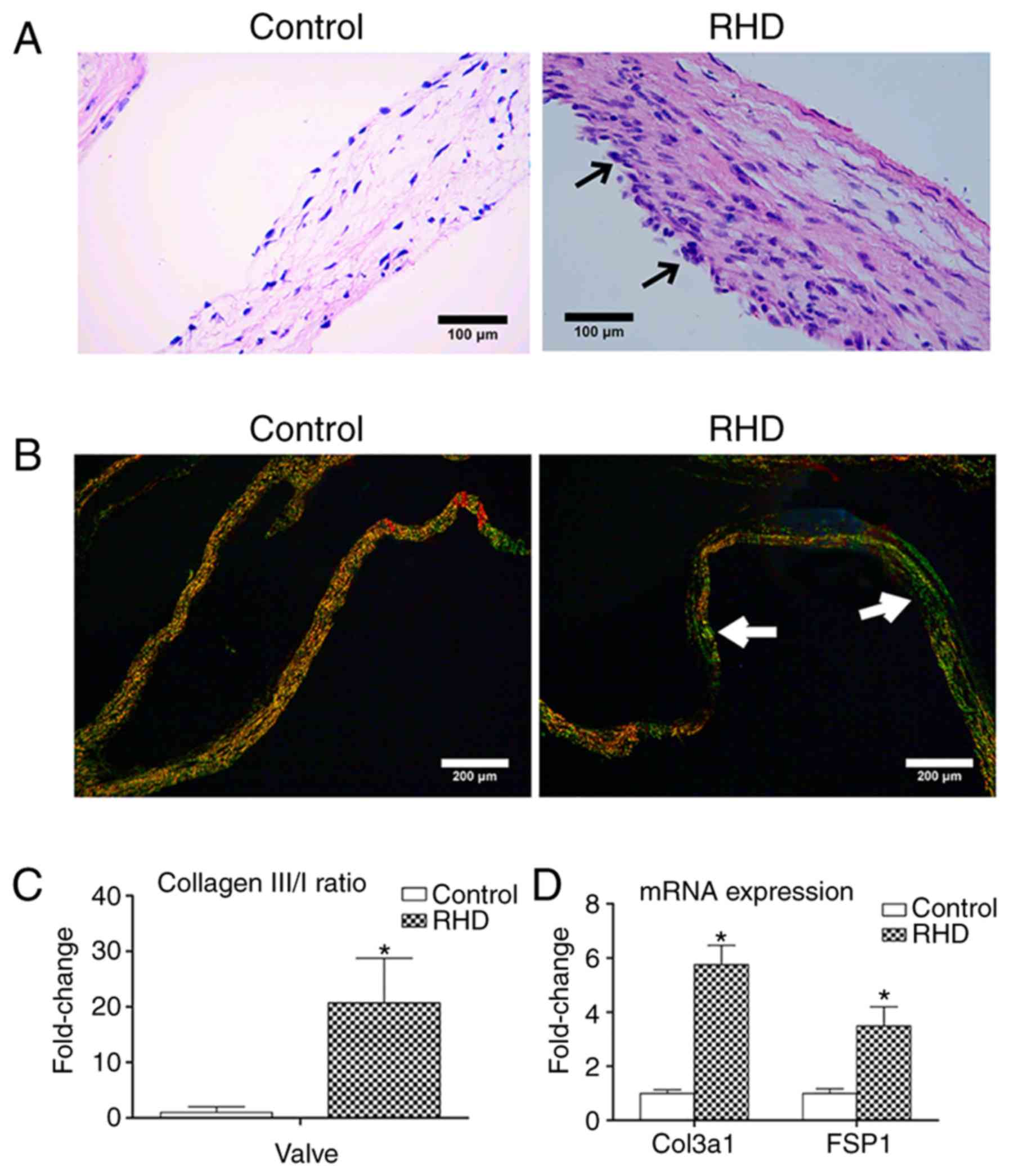 | Figure 1H&E, Sirius red staining and
fibrosis molecular markers of valve tissues. (A) H&E staining;
RHD-induced valvular damage was observed in RHD group;
magnification, ×400; scale bar: 100 µm; the arrows represent
inflammatory cells and inflammatory exudate (B). Sirius red
staining, RHD-induced fibrosis was observed in RHD group;
magnification, ×100; scale bar: 200 µm; the arrows represent
COL3 (green fibers with weak birefringence). (C) The COL3/1 ratio
of valves in the RHD group was higher than that in the control
group. (D) Relative expression of Col3a1 and FSP1 in that RHD group
were increased compared with those in the control group. Data are
shown as the mean ± standard deviation; *P<0.05 vs.
the control group. H&E, hematoxylin and eosin staining; COL3,
collagen fiber type 3; COL1, collagen fiber type 1; Col3a1,
collagen type III α1 chain; FSP1, fibroblast-specific protein 1;
RHD, rheumatic heart disease. |
The types of collagen fibers can be distinguished by
Sirius red staining with polarized light. Collagen fiber type 1
(COL1) appeared as close-packed yellow and red fibers with obvious
birefringence, and collagen fiber type 3 (COL3) appeared as loosely
arranged green fibers with weak birefringence. The relative
increase in COL3 and decrease in COL1 were calculated by the
COL3/COL1 (COL3/1) ratio. The COL3/1 ratio of valves in the RHD
group was significantly 20.32-fold higher than that in the control
group (P<0.05; Fig. 1B and C).
A previous study indicated that COL1 is the major type of collagen
in valves without fibrosis (16).
With the progression of fibrosis, the proportion of COL3 increases
gradually. The significant increase in the COL3/1 ratio therefore
confirmed the initiation of fibrosis in the valves and
myocardium.
The mRNA expression levels of collagen type III α1
chain (Col3a1) and fibroblast-specific protein 1 (FSP1) were used
as fibrosis molecular markers. The mRNA levels of Col3a1 and FSP1
in the RHD group were significantly increased compared with the
control group (P<0.05; Fig.
1D).
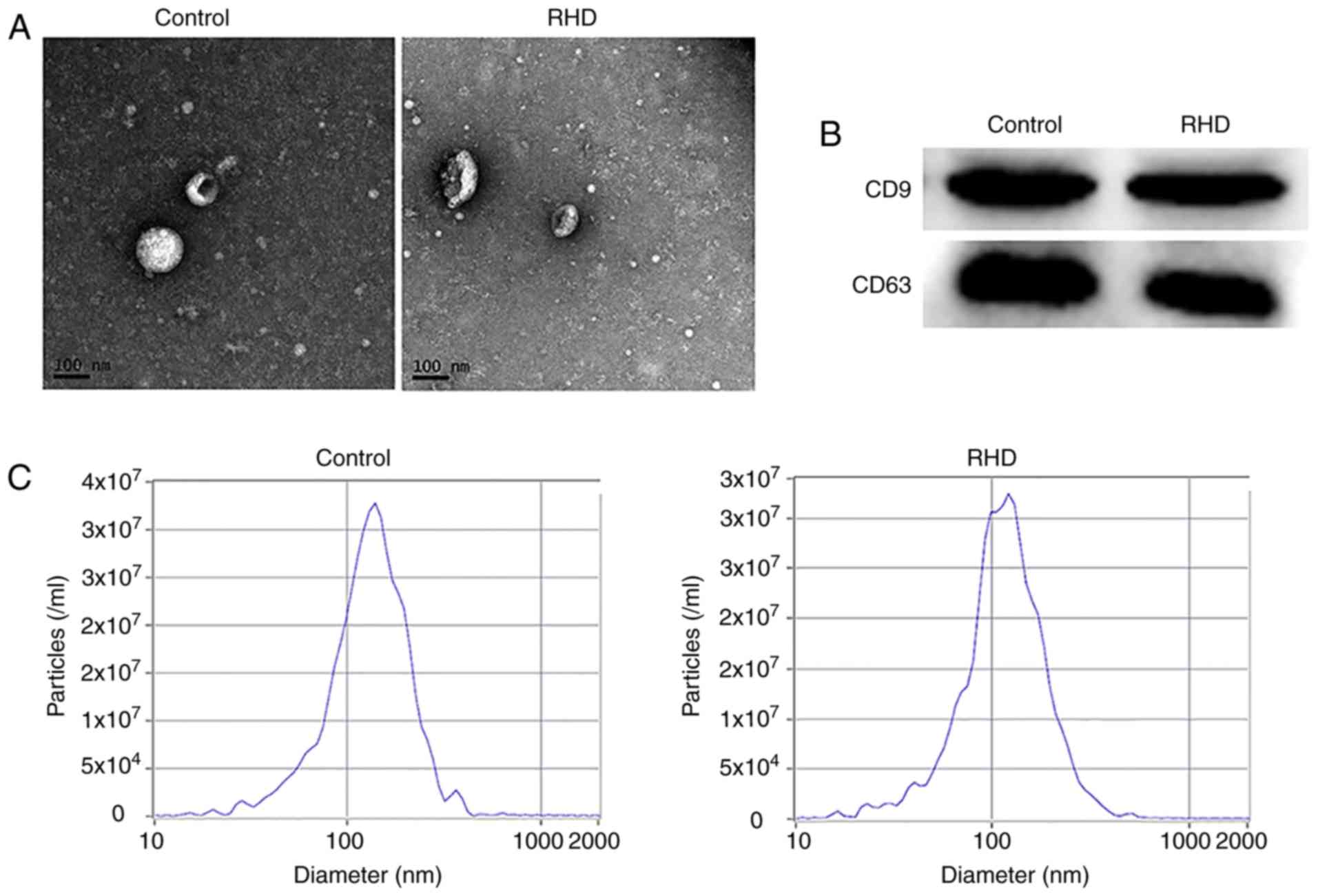
Identification of serum exosomes
Exosomes were isolated from the serum and observed
using TEM. Their morphology was round vesicles with double
membranes (Fig. 2A). Specific
membrane markers CD9 and CD63 were detected in the exosomes by
western blotting (Fig. 2B). The
NTA results demonstrated that the peak diameters were 117.2 nm (RHD
group) and 137.1 nm (control group) and the concentrations were
3.2×107/ml (RHD group) and 3.3×107/ml
(control group; Fig. 2C).
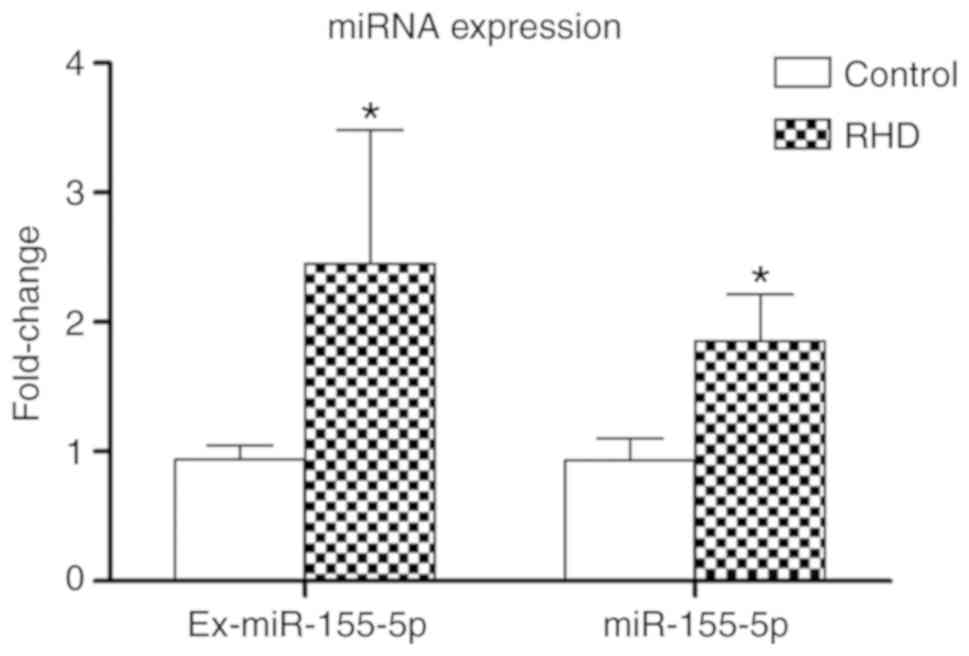
miR-155-5p is upregulated in serum
exosomes and valve tissues in the RHD model
RT-qPCR results showed that the amount of miR-155-5p
extracted from the serum exosomes (ex-miR155-5p) and valves of the
RHD group were significantly 2.61-fold and 1.99-fold higher than in
the control group, respectively (P<0.05; Fig. 3). These results demonstrate a
potential relationship between miR-155-5p and RHD.
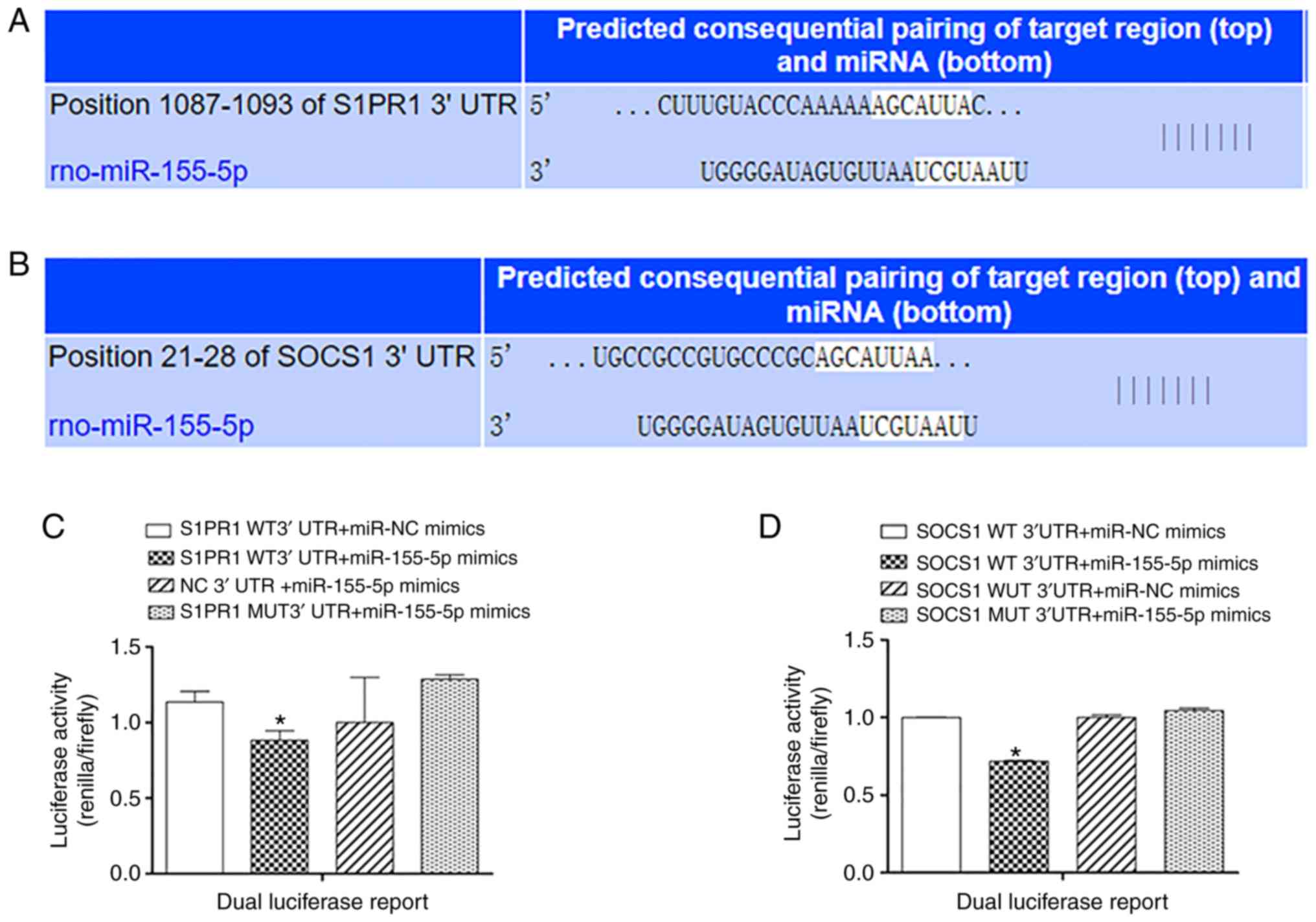
miR-155-5p directly targets S1PR1
The TargetScan results predicted that the S1PR1 gene
could be a potential target of miR-155-5p (Fig. 4A). A dual luciferase assay was
established to verify this prediction. When the miR-155-5p mimics
were cotransfected with S1PR1 WT 3′ UTR, the relative luciferase
activity (Renilla/firefly) was significantly decreased
compared with S1PR1 WT 3′ UTR cotransfected with miR-NC mimics
(P<0.05). The same effect did not occur when miR-155-5p was
overexpressed in the S1PR1 MUT 3′ UTR (Fig. 4C).
miR-155-5p directly targets SOCS1
The TargetScan results predicted that the SOCS1 gene
could be a potential target of miR-155-5p (Fig. 4B). A dual luciferase assay was
established to verify this prediction. When the miR-155-5p mimics
were cotransfected with SOCS1 WT 3′ UTR, the relative luciferase
activity (Renilla/firefly) was significantly decreased
compared with when SOCS1 WT 3′ UTR was cotransfected with miR-NC
mimics (P<0.05). The same effect did not occur when miR-155-5p
or miR-NC were cotransfected with the SOCS1 MUT 3′ UTR (Fig. 4D).
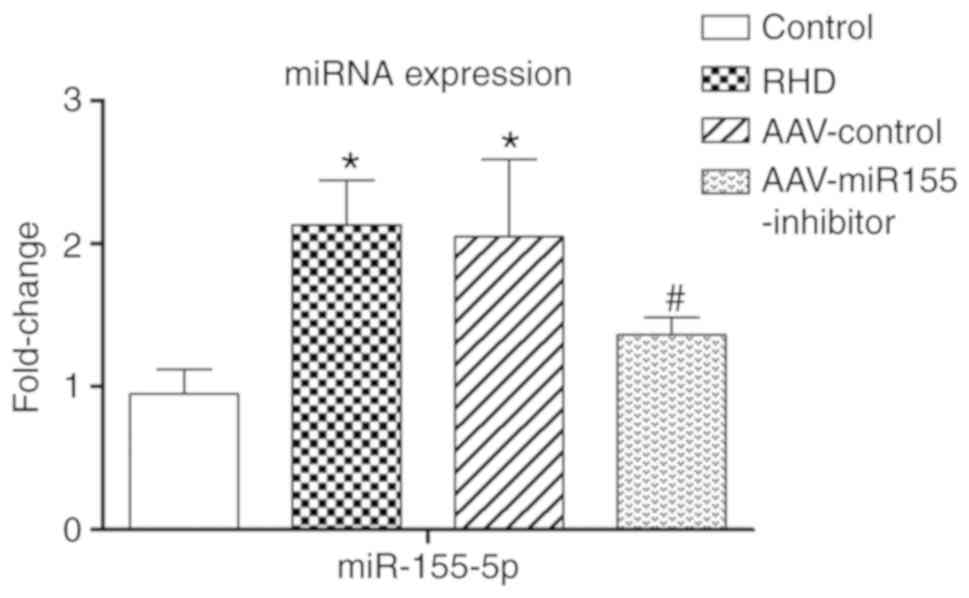
AAV-miR155-inhibitor pretreatment
attenuates valvular damage
To determine whether miR-155-5p participates in
RHD-induced valvular damage, the cardiac-specific
AAV-miR155-inhibitor was injected via the rat tail vein. RT-qPCR
results showed that the amount of miR-155-5p in the
AAV-miR155-inhibitor group was significantly 1.56-fold lower than
in the RHD group after a 9-week modeling procedure (P<0.05;
Fig. 5). Reduced inflammatory
cell infiltration and inflammatory exudate were also observed
(Fig. 6A). Sirius red staining
results showed that both valves in the AAV-miR155-inhibitor group
had less fibrosis compared with the RHD group (Fig. 6B). The COL3/1 ratio in the
AAV-miR155-inhibitor group was significantly 2.36-fold lower than
that in the RHD group (P<0.05; Fig. 6C). The mRNA expression of Col3a1
and FSP1 in the AAV-miR155-inhibitor group was significantly
decreased compared with the RHD group (P<0.05; Fig. 6C). Thus, these findings indicate
that miR-155-5p is a negative regulatory factor of RHD-induced
valvular damage.
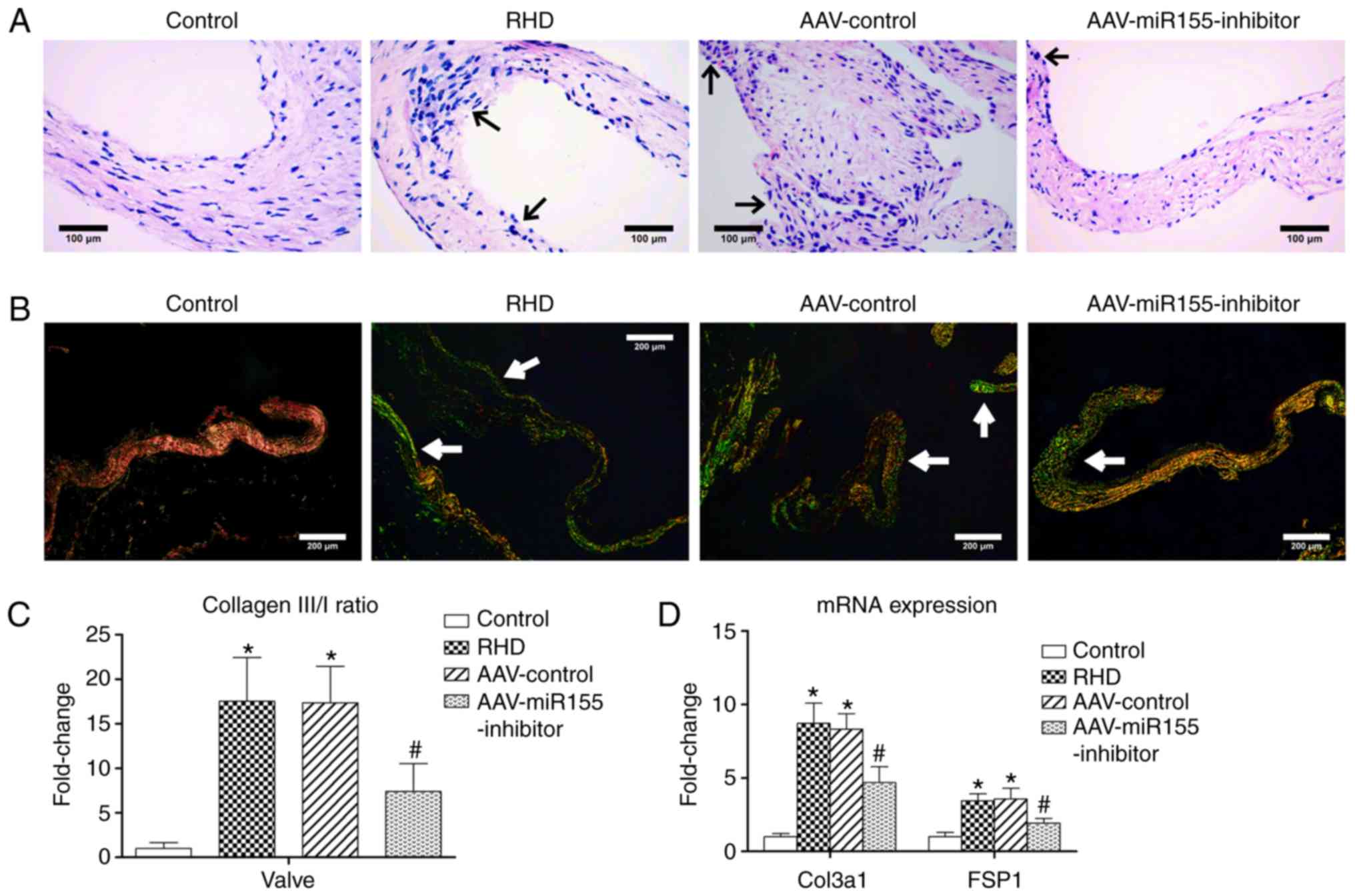 | Figure 6H&E, Sirius red staining and
fibrosis molecular markers of four groups. (A) H&E, reduced
inflammatory cell infiltration and inflammatory exudate were also
observed after AAV-injection; magnification, ×400; scale bar: 100
µm; the arrows represent inflammatory cells and inflammatory
exudate. (B) Sirius red staining, less fibrosis was observed after
AAV-injection; magnification, ×100; scale bar: 200 µm, the
arrows represent COL3 (green fibers with weak birefringence). (C)
COL3/COL1 ratio in AAV-miR155-inhibitor group was lower than that
in RHD group. (D) The relative expression of Col3a1 and FSP1 in the
AAV-miR155-inhibitor group was lower than that in the RHD group.
Data are shown as the mean ± standard deviation;
*P<0.05 vs. the control group. #P<0.05
vs. the RHD group. H&E, hematoxylin and eosin staining; COL3,
collagen fiber type 3; COL1, collagen fiber type 1; Col3a1,
collagen type III α1 chain; FSP1, fibroblast-specific protein 1;
RHD, rheumatic heart disease; miR, microRNA. |
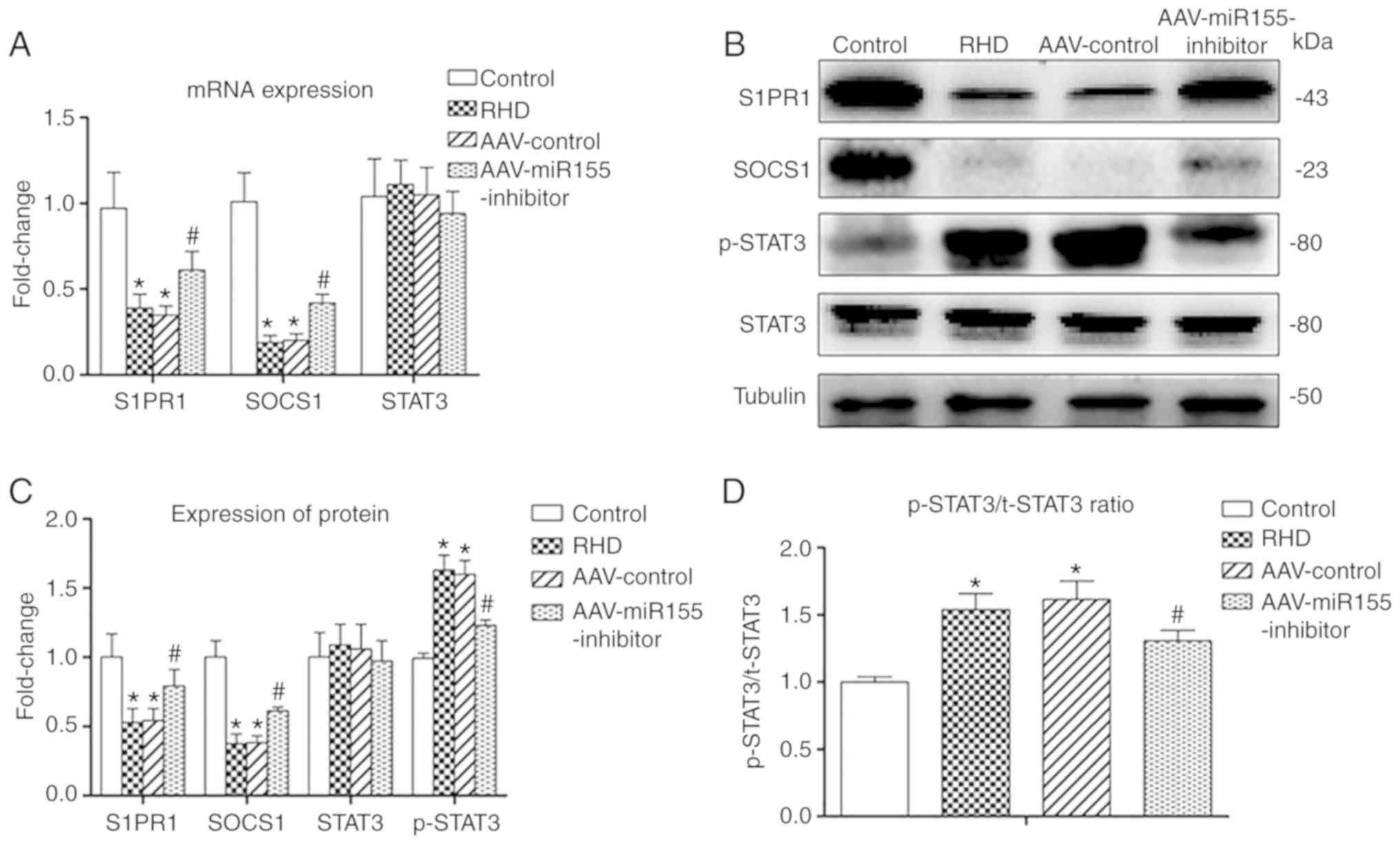
mRNA levels of S1PR1, SOCS1 and
STAT3
The mRNA levels of S1PR1 and SOCS1 in the RHD group
and the AAV-control group were significantly decreased compared
with in the control group (P<0.05), and there was no significant
difference between the two groups. The mRNA levels of S1PR1 and
SOCS1 in the AAV-miR155-inhibitor group were significantly
increased compared with those in the RHD group (P<0.05). There
were no differences between the four groups in the mRNA level of
STAT3 (Fig. 6A).
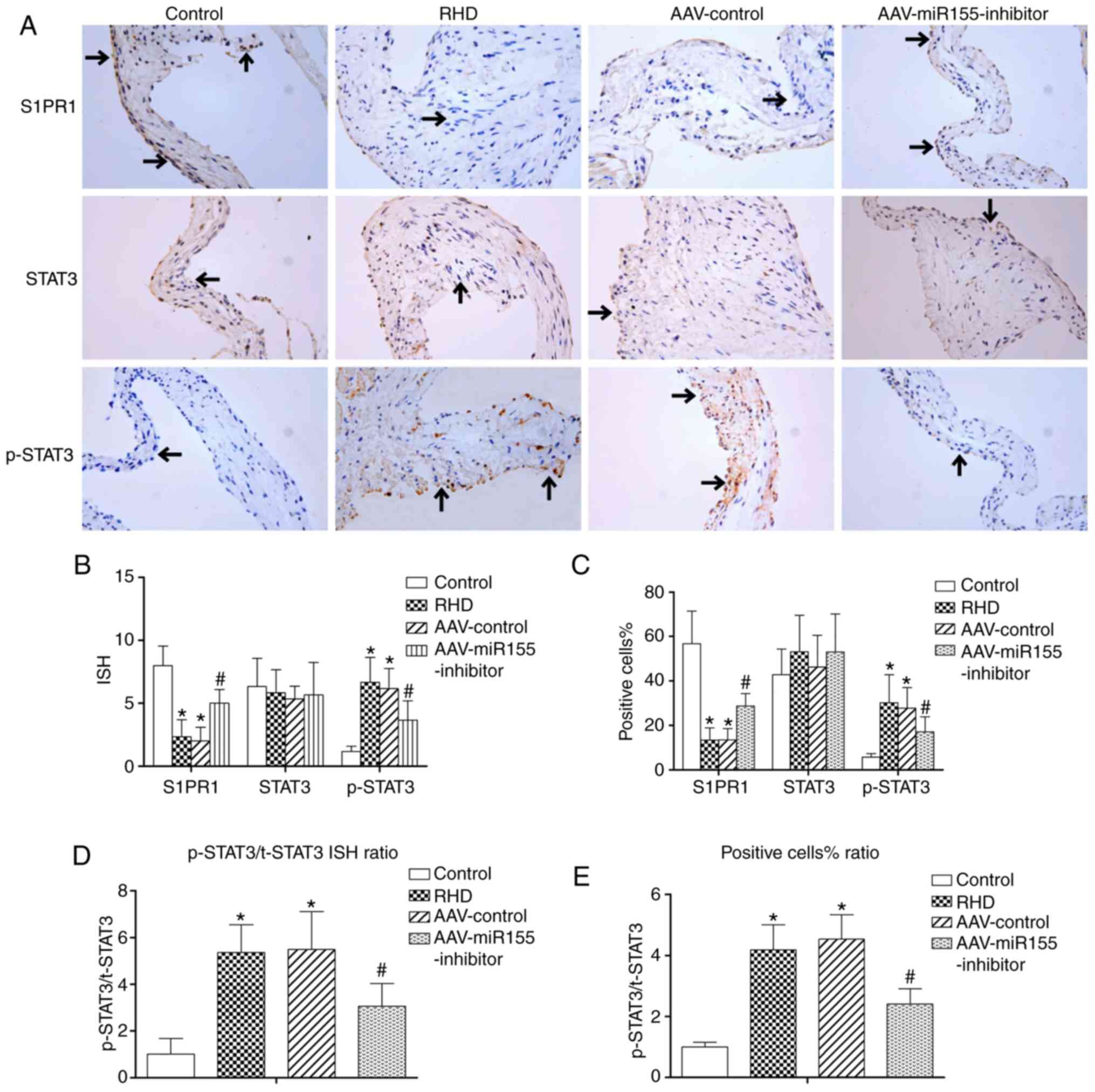
Protein levels of S1PR1, SOCS1, STAT3 and
p-STAT3
Western blotting and immunohistochemistry showed
significant downregulation of S1PR1 and SOCS1 in the RHD and
AAV-control groups compared with the control group (P<0.05), and
there was no significant difference between the RHD and AAV-control
groups. The protein levels of p-STAT3, p-STAT3/t-STAT3 protein
ratio, p-STAT3/t-STAT3 ISH ratio and p-STAT3/t-STAT3 positive cell%
ratio in the RHD and AAV-control groups were significantly
increased compared with those in the control group (P<0.05), and
there was no significant difference between the RHD and AAV-control
groups. The protein levels of STAT3 in the four groups were
similar. Nevertheless, the AAV-miR155-inhibitor pretreatment
significantly enhanced the S1PR1 and SOCS1 protein levels compared
with the RHD group (P<0.05) and significantly reduced the
p-STAT3 protein expression, p-STAT3/t-STAT3 protein ratio,
p-STAT3/t-STAT3 ISH ratio and p-STAT3/ t-STAT3 positive cell%
ratio. (P<0.05; Figs. 7B-D,
and 8A-E).
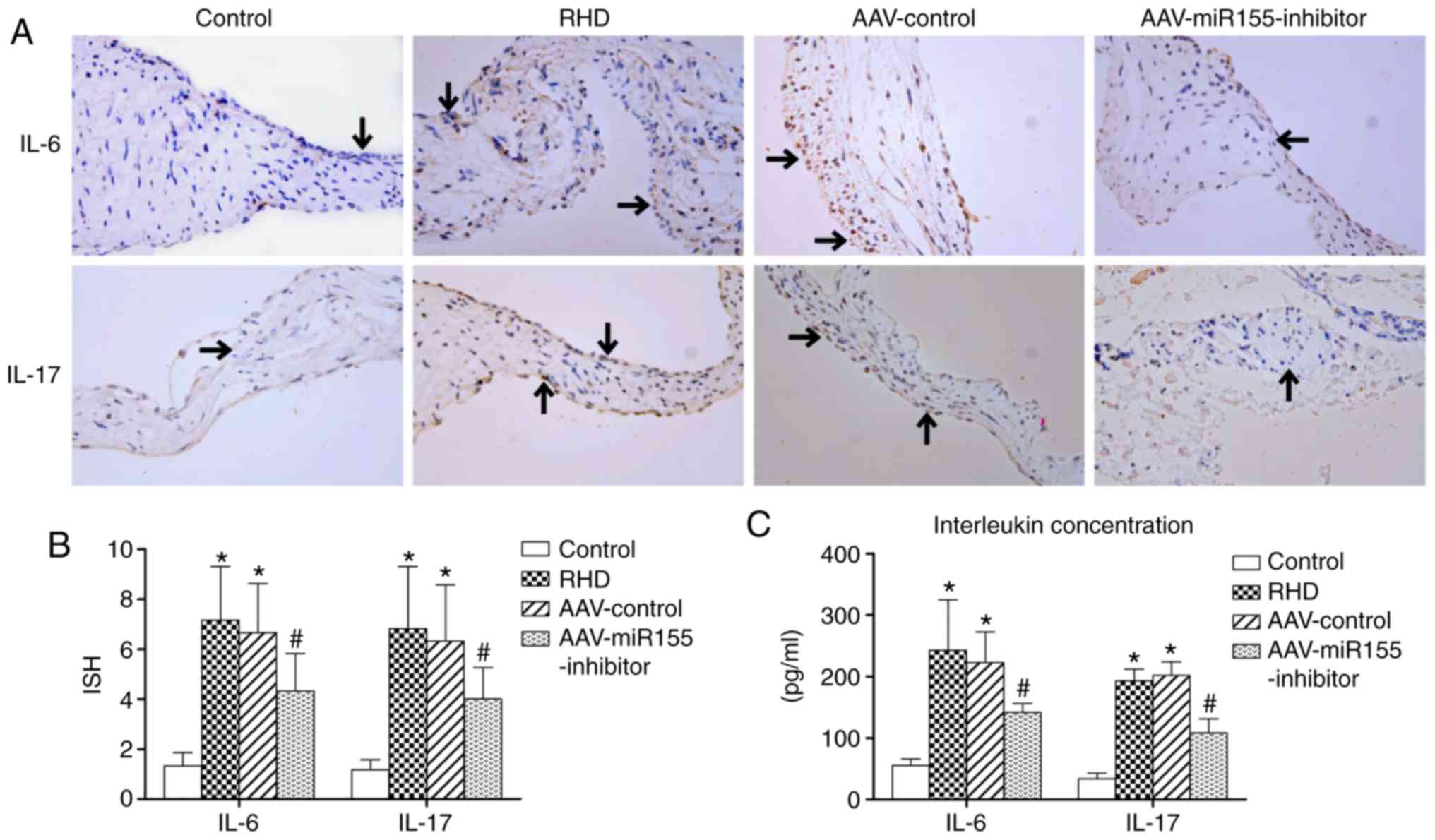
AAV-miR155-inhibitor pretreatment
attenuates the expression of IL-6 and IL-17
Immunohistochemistry and ELISA showed that the
expression levels of IL-6 and IL-17 in the RHD and AAV-control
groups were significantly increase compared with in the control
group (P<0.05). The AAV-miR155-inhibitor pretreatment also
significantly attenuated the expression of IL-6 and IL-17 in the
valve tissues and serum (P<0.05; Fig. 9).
Discussion
RHD affects >30 million people and kills 300,000
annually, but the specific pathophysiology and molecular mechanisms
of this disease remain unknown. In recent decades, miRNAs have been
shown to function in regulating various cellular pathways by
targeting different genes. Several studies have reported that the
expression of miRNAs is altered in patients with chronic RHD,
including miR-205-3p, miR-1183 and miR-101. These miRNAs regulate
the progression of RHD by targeting different pathways, such as
IL-1β and TLR2 (17-19). The mechanism of action of
miR-155-5p in RHD is still unclear, but scholars have reported that
miR-155 is involved in autoimmunity, inflammation and fibrosis
(20-22). Studies have shown that miR-155
plays a crucial role in rheumatoid arthritis and osteoarthritis
(23). Inhibition of miR-155
attenuated the development of an abdominal aortic aneurysm by
regulating macrophage inflammation (24). The paracrine activity of
miR-155-enriched exosomes regulated fibroblast proliferation and
inflammation in cardiac injury (22). Additionally, the inhibition of
miR-155 attenuated inflammation after myocardial infarction through
the SOCS1/nuclear factor-κB pathway (25). Knockout of miR-155 reduced the
infarct size, inflammation and attenuated collagen deposition in
acute myocardial infarction by targeting p53-inducible nuclear
protein 1 (26). In the current
study, the high expression of miR-155-5p in the RHD rat model was
confirmed by RT-qPCR. With the inhibition of miR-155-5p induced by
AAV pretreatment, decreased inflammation and fibrosis were observed
in the valves. The miRNAs in exosomes, such as miR-let-7b (27) and miR-19a-3p (28), played a critical role in
inflammation, autoimmunity and fibrosis. A high expression of
miR-155-5p in serum exosomes was also observed, suggesting that
exosomes might participate in RHD-induced valvular damage via the
transfer of miR-155-5p. These results confirmed that miR-155-5p
accelerated the progression of valvular damage and that the
inhibition of miR-155-5p could alleviate this damage.
S1PR1, a G protein-coupled receptor, is one of the
receptors for sphingosine 1-phosphate (S1P). The various functions
of S1PR1 have been verified in recent decades. S1PR1 is involved in
inflammation, autoimmunity and anti-fibrotic functions. FTY-720, a
S1PR1 agonist, can significantly reduce joint destruction and
inflammation in rheumatoid arthritis mice with osteoporosis
(29). A study showed that the
expression of S1PR1 in RA patients was significantly lower than in
healthy control patients (30).
The present authors have previously reported the downregulation of
S1PR1 in an RHD rat model (10).
In the current study, the dual luciferase assays confirmed that
miR-155-5p directly targeted S1PR1 by binding to its 3′ UTR in
vitro. With the inhibition of miR-155-5p in the rat valves, the
expression of S1PR1 was upregulated, as demonstrated by RT-qPCR,
western blotting and immunohistochemistry in vivo. These
results suggest that activation of the miR-155-5p/S1PR1 pathway
promoted RHD-induced valvular damage and inhibition of this pathway
attenuated this damage.
The STAT3 pathway plays a pivotal role in cancer
inflammation and anti-tumor immunity (31,32). Various previous studies have shown
high expression of p-STAT3 in rheumatoid arthritis (5-7)
and the STAT3 pathway was activated in permanent atrial
fibrillation patients with rheumatic heart disease (33). The present study showed activation
of the STAT3 pathway in an RHD rat model and the expression of the
p-STAT3 protein was decreased with the inhibition of miR-155-5p.
These results suggest a correlation between STAT3 activation and
miR-155 upregulation, but the specific function of miR-155 in
regulating the STAT3 signal pathway in RHD is unclear. Scholars
have reported that SOCS1 is downregulated by miR-155 in non-small
cell lung cancer (34), atopic
dermatitis (35), ulcerative
colitis (36), chronic
neuropathic pain (37) and
atherosclerosis (38). PDCD4 was
reported to be regulated by miR155 via the SOCS1-STAT3 signaling
pathway (8). The promotion of T
helper 17 (Th17) cell differentiation induced by activation of the
STAT3 pathway could be inhibited by an increase in SOCS1 in
rheumatoid arthritis (9). In the
present study, the dual luciferase assay confirmed that miR-155-5p
directly targeted SOCS1 by binding to its 3′ UTR region in
vitro. With the inhibition of miR-155-5p induced by AAV
injection, the expression of SOCS1 was increased in vivo, as
shown by RT-qPCR and western blotting. These results suggest that
miR-155-5p regulated the SOCS1/STAT3 pathway and that the
activation of the miR-155-5p/SOCS1/STAT3 pathway promoted
RHD-induced valvular damage. Inhibition of this pathway could
attenuate the progression of this damage.
Studies have demonstrated the pivotal role of IL-6
in immune system activation and inflammation in cancer (39,40) and autoimmune diseases (41,42). Liu et al (43) reported that the expression of IL-6
and TNF-α was attenuated in miR-155-inhibited RA fibroblast-like
synoviocytes. The IL-6/STAT3 axis is a key factor that regulates
numerous autoimmune diseases (44). In the current study, the high
expression of IL-6 in the valves and serum was detected by
immunohisto-chemistry and ELISA. With the inhibition of miR-155-5p,
the expression of IL-6 in valves and serum decreased. Consistent
with these results, the upregulation of IL-6 induced by the
upregulation of miR-155 also participated in the activation of the
STAT3 signal pathway. This miR-155-5p/IL-6/STAT3 pathway also
promoted RHD-induced valvular damage and inhibition of this pathway
alleviated the progression of valvular damage.
One study reported that the serum level of IL-17 was
higher in rheumatic mitral stenosis patients (45) and the biological function of
proinflammation in rheumatic disease has been confirmed by numerous
scholars (46,47). miR-155 promotes the development of
Th17 cell and Th1 cell subsets (21). Studies have reported the essential
roles of miR-155 in the immune response to Streptococcus
pneumoniae (48) and Th17
cell differentiation (35). The
authors previously reported that Th17 cell-associated cytokines
were significantly higher in patients with RHD, including IL-17 and
IL-21 (11). In the present
study, the high expression of IL-17 in serum and valve tissue was
suppressed by the downregulation of miR-155-5p. Consistent with
this finding, the present data suggested that miR-155-5p promoted
Th17 cell differentiation and participated in the progression of
RHD.
However, some important limitations should be
mentioned in this study. Firstly, valvular inflammation and
fibrosis after upregulating of miR-155-5p were not detected.
Secondly, experiments in cell lines were not performed, which would
provide another layer of tests for the present study. Thirdly, the
expression of miR-155-5p in serum exosomes after AAV-injection and
differential expressions of other miRNAs and proteins in serum
exosomes after valvular damage were not detected, it would be
valuable to measure the expression in an RHD rat model in the
future. In addition, the relative mRNA expression of FSP1 and
Col3a1 was used as fibrosis molecular markers, but the protein
expression was not detected.
The current study clarified that miR-155-5p in serum
exosome and valve tissues was upregulated in a GAS-induced RHD rat
model. Further study showed that miR-155-5p regulated the progress
of RHD-induced valvular damage by regulating multiple signaling
pathways. miR-155-5p directly suppressed S1PR1 expression and
miR-155-5p also directly suppressed the expression of SOCS1 and
then activated STAT3 phosphorylation. miR-155-5p promoted the
expression of IL-6 and then activated STAT3 phosphorylation.
miR-155-5p also promoted the differentiation of Th17 cells. Thus,
miR-155-5p played a critical role in RHD progression.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81660069), Guangxi
Key Laboratory Base of Precision Medicine in Cardio-cerebrovascular
Disease Control (grant no. 17-259-85) and Prevention and Guangxi
Clinical Research Center for Cardio-cerebrovascular Diseases (grant
no. AD17129014).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZZ and FH conceived and designed the study. AC and
JW participated in the experimental design. AC, JW, CL and SX
performed the experiments. AC, BL and YW analyzed the data. AC
wrote the manuscript and all authors contributed to the final
manuscript.
Ethics approval and consent to
participate
Protocols involving animals were approved by the
Medical Ethics Committee of the First Affiliated Hospital of
Guangxi Medical University (permit no. 2016-KY-201).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Watkins DA, Johnson CO, Colquhoun SM,
Karthikeyan G, Beaton A, Bukhman G, Forouzanfar MH, Longenecker CT,
Mayosi BM, Mensah GA, et al: Global, regional, and national burden
of rheumatic heart disease, 1990-2015. N Engl J Med. 377:713–722.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marijon E, Mirabel M, Celermajer DS and
Jouven X: Rheumatic heart disease. Lancet. 379:953–964. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guilherme L and Kalil J: Rheumatic fever
and rheumatic heart disease: Cellular mechanisms leading autoimmune
reactivity and disease. J Clin Immunol. 30:17–23. 2010. View Article : Google Scholar
|
|
4
|
Turkson J and Jove R: STAT proteins: Novel
molecular targets for cancer drug discovery. Oncogene.
19:6613–6626. 2000. View Article : Google Scholar
|
|
5
|
Gao W, McCormick J, Connolly M, Balogh E,
Veale DJ and Fearon U: Hypoxia and STAT3 signalling interactions
regulate pro-inflammatory pathways in rheumatoid arthritis. Ann
Rheum Dis. 74:1275–1283. 2015. View Article : Google Scholar
|
|
6
|
Liu J, Fei D, Xing J and Du J:
MicroRNA-29a inhibits proliferation and induces apoptosis in
rheumatoid arthritis fibroblast-like synoviocytes by repressing
STAT3. Biomed Pharmacother. 96:173–181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee SH, Kim EK, Kwon JE, Lee JK, Lee D,
Kim SY, Seo HB, Na HS, Jung K, Kwok SK, et al: Ssu72 attenuates
autoimmune arthritis via targeting of STAT3 signaling and Th17
activation. Sci Rep. 7:55062017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye J, Guo R, Shi Y, Qi F, Guo C and Yang
L: miR-155 regulated inflammation response by the SOCS1-STAT3-PDCD4
axis in atherogenesis. Mediators Inflamm. 2016:80601822016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jahreis S, Kuhn S, Madaj AM, Bauer M and
Polte T: Mold metabolites drive rheumatoid arthritis in mice via
promotion of IFN-gamma- and IL-17-producing T cells. Food Chem
Toxicol. 109:405–413. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu XD, Zeng ZY, Gong DP, Wen JL and Huang
F: Potential involvement of S1PR1/STAT3 signaling pathway in
cardiac valve damage due to rheumatic heart disease. Biotech
Histochem. 94:398–403. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wen Y, Zeng Z, Gui C, Li L and Li W:
Changes in the expression of Th17 cell-associated cytokines in the
development of rheumatic heart disease. Cardiovasc Pathol.
24:382–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Teker E, Akadam-Teker AB, Ozturk O, Eronat
AP, Yalin K, Golcuk SE and Bugra Z: Association between the
interferon gamma 874 T/A polymorphism and the severity of valvular
damage in patients with rheumatic heart disease. Biochem Genet.
56:225–234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gorton D, Govan B, Olive C and Ketheesan
N: B- and T-cell responses in group a streptococcus M-protein- or
Peptide-induced experimental carditis. Infect Immun. 77:2177–2183.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Friedrichs K, Gluba S, Eidtmann H and
Jonat W: Overexpression of p53 and prognosis in breast cancer.
Cancer. 72:3641–3647. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Purushothaman KR, Purushothaman M,
Turnbull IC, Adams DH, Anyanwu A, Krishnan P, Kini A, Sharma SK,
O'Connor WN and Moreno PR: Association of altered collagen content
and lysyl oxidase expression in degenerative mitral valve disease.
Cardiovasc Pathol. 29:11–18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu Q, Sun Y, Duan Y, Li B, Xia J, Yu S and
Zhang G: Comprehensive microRNA profiling reveals potential
augmentation of the IL1 pathway in rheumatic heart valve disease.
BMC Cardiovasc Disord. 18:532018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li N, Lian J, Zhao S, Zheng D, Yang X,
Huang X, Shi X, Sun L, Zhou Q, Shi H, et al: Detection of
differentially expressed MicroRNAs in rheumatic heart disease:
miR-1183 and miR-1299 as potential diagnostic biomarkers. Biomed
Res Int. 2015:5245192015.PubMed/NCBI
|
|
19
|
Dong H, Sun Y, Shan F, Sun Q and Yang B:
Down-regulation of miR-101 contributes to rheumatic heart disease
through up-regulating TLR2. Med Sci Monit. 21:1500–1506. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thai TH, Calado DP, Casola S, Ansel KM,
Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et
al: Regulation of the germinal center response by microRNA-155.
Science. 316:604–608. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
O'Connell RM, Kahn D, Gibson WS, Round JL,
Scholz RL, Chaudhuri AA, Kahn ME, Rao DS and Baltimore D:
MicroRNA-155 promotes autoimmune inflammation by enhancing
inflammatory T cell development. Immunity. 33:607–619. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang C, Zhang C, Liu L, A X, Chen B, Li Y
and Du J: Macrophage-derived mir-155-containing exosomes suppress
fibroblast proliferation and promote fibroblast inflammation during
cardiac injury. Mol Ther. 25:192–204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kriegsmann M, Randau TM, Gravius S,
Lisenko K, Altmann C, Arens N and Kriegsmann J: Expression of
miR-146a, miR-155, and miR-223 in formalin-fixed paraffin-embedded
synovial tissues of patients with rheumatoid arthritis and
osteoarthritis. Virchows Arch. 469:93–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Z, Liang K, Zou G, Chen X, Shi S,
Wang G, Zhang K, Li K and Zhai S: Inhibition of miR-155 attenuates
abdominal aortic aneurysm in mice by regulating macrophage-mediated
inflammation. Biosci Rep. 38:BSR201714322018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu J, Huang CX, Rao PP, Cao GQ, Zhang Y,
Zhou JP, Zhu LY, Liu MX and Zhang GG: MicroRNA-155 inhibition
attenuates endoplasmic reticulum stress-induced cardiomyocyte
apoptosis following myocardial infarction via reducing macrophage
inflammation. Eur J Pharmacol. 857:1724492019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He W, Huang H, Xie Q, Wang Z, Fan Y, Kong
B, Huang D and Xiao Y: MiR-155 knockout in fibroblasts improves
cardiac remodeling by targeting tumor protein p53-inducible nuclear
protein 1. J Cardiovasc Pharmacol Ther. 21:423–435. 2016.
View Article : Google Scholar
|
|
27
|
Kim SJ, Chen Z, Essani AB, Elshabrawy HA,
Volin MV, Volkov S, Swedler W, Arami S, Sweiss N and Shahrara S:
Identification of a novel toll-like receptor 7 endogenous ligand in
rheumatoid arthritis synovial fluid that can provoke arthritic
joint inflammation. Arthritis Rheumatol. 68:1099–1110. 2016.
|
|
28
|
Gollmann-Tepeköylü C, Pölzl L, Graber M,
Hirsch J, Nägele F, Lobenwein D, Hess MW, Blumer MJ, Kirchmair E,
Zipperle J, et al: miR-19a-3p containing exosomes improve function
of ischemic myocardium upon shock wave therapy. Cardiovasc Res. Aug
13–2019.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kikuta J, Iwai K, Saeki Y and Ishii M:
S1P-targeted therapy for elderly rheumatoid arthritis patients with
osteoporosis. Rheumatol Int. 31:967–969. 2011. View Article : Google Scholar
|
|
30
|
Choi HS, Kim KH, Jin S, Kim J, Yoo I, Pack
SP, Ha UH, Park TW, Choi SA, Yuk SH, et al: Decreased expression of
sphingosine-1-phosphate receptor 1 in the blood leukocyte of
rheumatoid arthritis patients. Immune Netw. 18:e392018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu H, Kortylewski M and Pardoll D:
Crosstalk between cancer and immune cells: Role of STAT3 in the
tumour microenvironment. Nat Rev Immunol. 7:41–51. 2007. View Article : Google Scholar
|
|
33
|
Xue XD, Huang JH and Wang HS: Angiotensin
II activates signal transducers and activators of transcription 3
via Rac1 in the atrial tissue in permanent atrial fibrillation
patients with rheumatic heart disease. Cell Biochem Biophys.
71:205–213. 2015. View Article : Google Scholar
|
|
34
|
Xue X, Liu Y, Wang Y, Meng M, Wang K, Zang
X, Zhao S, Sun X, Cui L, Pan L and Liu S: miR-21 and miR-155
promote non-small cell lung cancer progression by downregulating
SOCS1, SOCS6, and PTEN. Oncotarget. 7:84508–84519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma L, Xue HB, Wang F, Shu CM and Zhang JH:
MicroRNA-155 may be involved in the pathogenesis of atopic
dermatitis by modulating the differentiation and function of T
helper type 17 (Th17) cells. Clin Exp Immunol. 181:142–149. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pathak S, Grillo AR, Scarpa M, Brun P,
D'Incà R, Nai L, Banerjee A, Cavallo D, Barzon L, Palù G, et al:
miR-155 modulates the inflammatory phenotype of intestinal
myofibroblasts by targeting SOCS1 in ulcerative colitis. Exp Mol
Med. 47:e1642015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tan Y, Yang J, Xiang K, Tan Q and Guo Q:
Suppression of microRNA-155 attenuates neuropathic pain by
regulating SOCS1 signalling pathway. Neurochem Res. 40:550–560.
2015. View Article : Google Scholar
|
|
38
|
Yang Y, Yang L, Liang X and Zhu G:
MicroRNA-155 promotes atherosclerosis inflammation via targeting
SOCS1. Cell Physiol Biochem. 36:1371–1381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Scheller J, Chalaris A, Schmidt-Arras D
and Rose-John S: The pro- and anti-inflammatory properties of the
cytokine interleukin-6. Biochim Biophys Acta. 1813:878–888. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hodge DR, Hurt EM and Farrar WL: The role
of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer.
41:2502–2512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Narazaki M, Tanaka T and Kishimoto T: The
role and therapeutic targeting of IL-6 in rheumatoid arthritis.
Expert Rev Clin Immunol. 13:535–551. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tanaka T, Narazaki M and Kishimoto T: IL-6
in inflammation, immunity, and disease. Cold Spring Harb Perspect
Biol. 6:a0162952014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu N, Feng X, Wang W, Zhao X and Li X:
Paeonol protects against TNF-α-induced proliferation and cytokine
release of rheumatoid arthritis fibroblast-like synoviocytes by
upregulating FOXO3 through inhibition of miR-155 expression.
Inflamm Res. 66:603–610. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Camporeale A and Poli V: IL-6, IL-17 and
STAT3: A holy trinity in auto-immunity? Front Biosci (Landmark Ed).
17:2306–2326. 2012. View
Article : Google Scholar
|
|
45
|
Bilik MZ, Kaplan I, Polat N, Akil MA,
Akyüz A, Acet H, Yüksel M, İnci Ü, Kayan F and Toprak N: Serum
levels of IL-17 and IL-23 in patients with rheumatic mitral
stenosis. Medicine (Baltimore). 95:e35622016. View Article : Google Scholar
|
|
46
|
Volin MV and Shahrara S: Role of TH-17
cells in rheumatic and other autoimmune diseases. Rheumatology
(Sunnyvale). 1:21692011. View Article : Google Scholar
|
|
47
|
Al-Saadany HM, Hussein MS, Gaber RA and
Zaytoun HA: Th-17 cells and serum IL-17 in rheumatoid arthritis
patients: Correlation with disease activity and severity. The
Egyptian Rheumatologist. 38:1–7. 2016. View Article : Google Scholar
|
|
48
|
Verschoor CP, Dorrington MG, Novakowski
KE, Kaiser J, Radford K, Nair P, Anipindi V, Kaushic C, Surette MG
and Bowdish DM: MicroRNA-155 is required for clearance of
Streptococcus pneumoniae from the nasopharynx. Infect Immun.
82:4824–4833. 2014. View Article : Google Scholar : PubMed/NCBI
|