Introduction
In recent years, the study of sleep disorders in
critically ill patients has drawn increasing attention (1). In these patients, the amount of
sleep is reduced and the pattern of sleep is fragmented without
following the normal circadian rhythm. In addition, these patients
have a higher proportion of sleep in the N1-N2 stages, whereas the
amount of rapid eye movement (REM) in these patients is decreased.
Since REM and sleep in the N3 stage play an important role in a
wide range of physiological functions involving the immune,
respiratory, endocrine, cardiovascular and central nervous systems,
the lack of REM and N3 sleep in patients in the intensive care unit
(ICU) significantly affects their recovery (1). For example, inadequate sleep has
been shown to cause post-traumatic stress disorder, non-invasive
ventilation failure and delirium (2-4).
It is known that α2 agonists can result in a loss of
consciousness, a loss of righting reflex (LORR), presumed surrogate
and sedation (5). On the other
hand, the activity of locus coeruleus (LC) neurons is decreased
during sleep and begins to increases before an individual wakes up,
indicating that these neurons are associated with consciousness
(6). Notably, although the
selective stimulation of LC neurons induces waking, their
inhibition cannot help sleep (7).
In addition, in mice lacking the ability to produce noradrenalin
(NA), an agonist of the adra2a (α2A) receptor, the administration
of dexmedetomi-dine can still result in LORR (5). Nevertheless, in the absence of its
endogenous ligand, the response of α2A-receptors can become
hypersensitized in the long-term, making it difficult to interpret
the results (5).
As a class of small and non-coding RNAs containing
19-25 nucleotides, microRNAs (miRNAs or miRs) have been shown to
mediate the expression of a wide range of genes both in animals and
plants (8). In addition, a number
of miRNAs are involved in various cellular processes, such as
metabolism, stress response, stem cell renewal, embryonic
development, cell proliferation, apoptosis and differentiation
(9). Given these critical
functions of miRNAs, it is reasonable to expect that the
abnormality of miRNA pathways and miRNA synthesis can result in
several human diseases, including obesity, cardiovascular diseases,
cancer, psoriasis, schizophrenia, chronic hepatitis, acquired
immune deficiency syndrome (AIDS) and diabetes (10-15). In addition, single-nucleotide
polymorphisms (SNPs) in the 3′untranslated region (3′UTR) of genes
can alter the binding of miRNAs and affect the expression of target
proteins, thus influencing the pathogenesis of a number of diseases
(16). A previous study on
healthy subjects demonstrated that a 3′UTR polymorphism in
adrenoceptor alpha 2A (ADRA2A) mediates the onset of autonomic
responses upon environmental and physiological stimulation, while
the polymorphisms of ADRA2A can also be used to predict the
responses to induced pain (17).
In addition, the correlation between rs3750625, a 3′UTR SNP in
ADRA2A, and acute musculoskeletal pain (MSP) has been evaluated
among individuals experiencing sexual assault and motor vehicle
collision (MVC) (18,19). Furthermore, results of
bioinformatics analyses have suggested that rs3750625 is located in
the seed binding region of miR-34a, which is known to mediate
stress and pain (20).
ADRA2A has been reported to be associated with sleep
quality (21). Since miR-34a acts
as a direct regulator of ARDA2A, the presence of rs3750625 has been
shown to interrupt the interaction between miR-34a and ADRA2A
(18,22). Therefore, in this study, we
evaluated the regulatory asociation between miR-34a and ARDA2A, as
well as the association between rs3750625 and sleep quality in
patients in the ICU.
Materials and methods
Collection of blood samples
The Human Research Ethics Committees of Dongguan
Houjie Hospital approved this research. Written informed consent
was obtained from all patients or their first-degree relatives
prior to the onset of this research. The research process was in
conformity with the latest version of Declaration of Helsinki. In
this study, peripheral blood samples were collected from 38
subjects (the ethnicity of all participants was Han; sex: Female,
18 male, 20 mean age, 50±0.6 years old) enrolled from patients who
stay in ICU for at least 3 days. The inclusion criteria were the
following: i) An age between 18 and 65 years; ii) a stay in the ICU
of at least 3 days, but the use of mechanical ventilation not
required; iii) stable hemodynamic parameters without sedative
treatment. The exclusion criteria were the following: i) Patients
diagnosed with hypoxic-ischemic encephalopathy; ii) patients
diagnosed with psychiatric disorders; iii) patients diagnosed with
hemodynamic instability; iv) patients diagnosed with severe
abnormalities in the cardiac conduction system; v) patients
diagnosed with ventricular dysfunction; vi) patients diagnosed with
liver failure and a bilirubin level >100 mmol/l; vii) patients
diagnosed with neurological injury or diseases that may influence
sleep quality; viii) patients with a scale of Glasgow Coma score of
<11; and ix) patients with an acute physiology score portion of
the Acute physiology and Chronic Health evaluation II (APACHe II)
score of >15.
Genotyping
Genomic DNA was isolated from abdomen tissue samples
using a DNA extraction kit (Shunhua Bioengineer Co. Ltd.) and
amplified using PCR. An ExoSAP-IT purification kit (USB) was
utilized to purify the PCR products, which were then sent to a core
facility for sequencing.
Sleep quality measurement
During the entire course of this study, the light in
the ward was maintained at a minimum level so as to not affect the
sleep of the subjects. For subjects in the 'no sedation' group of
this study, they had to be removed from the study if they suffered
from sleep disorders and required sedation. For all the remaining
subjects, a measurement of bispectral index (BIS) was carried out
by recording their sleep time and sleep depth continuously using a
Covidien BIS VISTA monitor. In addition, the level of sedation (if
applicable) and the depth of sleep were graded based on the BIS
index, which scored EEG signals from 0 to 100. A score of 65-85
indicated a patient was in sedation or sleep, while a score of 85
to 100 suggested the patient was awake. In addition, the Richmond
Agitation-Sedation Scale (RASS) score (23), the duration of consciousness and
sleep, blood pressure, heart rate, respiratory rate, oxyhemoglobin
saturation, and the duration of total sleep of each subject were
examined every 2 h. The ratio between the duration of effective
sleep and the duration of total sleep was defined as sleep
efficiency.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific) was utilized to extract total RNA from HCN-1A and U251
cells and blood samples. The quality of the RNA was determined by
measuring its absorbance with a NanoDrop 1000 spectrophotometer
(Thermo Fisher Scientific). A Sprint Power Script Pre Primed Single
Shots kit (Clontech) was utilized to synthesize first-strand ADRA2A
cDNA from total RNA using the following reaction conditions: 5
cycles of 30 sec at 94°C, 30 sec at 60°C and 60 sec at 68°C. A
TaqMan hsa-miR-34a amplification kit (Applied Biosystems) was used
to amplify the cDNA, which was then measured on an Applied
Biosystems Real-Time PCR System (Applied Biosystems). The real-time
PCR reaction mixture contained 4.0 µl (a total of 40 ng) of
ADRA2A cDNA, 10 µl of 2X gene expression master mix, 1
µl of 20X Gene Expression Assay buffer and 5 µl of
RNase-free water. The 3′UTR of ADRA2A was amplified with primers,
including ATG ATG CTC GAG ACT CAG AAA CCC GGG CGC (forward) and ATG
ATG AAT TCC CAT AAA ATC AGA TGT TCC CAG AG (reverse). GAPDH and U6
were used as internal controls to normalize the relative expression
of miR-34a and ADRA2A, respectively. The reactive expression of
miR-34a and ADRA2A mRNA was calculated using the 2-ΔΔCq
method (24). Each experiment was
carried out at least 3 times.
Cells, cell culture and transfection
The HCN-1A and U251 cell lines were obtained from
the American Type Culture Collection (ATCC). For transfection
experiments, the cells were seeded in 24-well plates and cultured
for 24 h at 37°C and 5% CO2 in a RPMI medium containing
10% heat-inactivated fetal bovine serum (FBS), 50 U/ml
streptomycin/penicillin and 2 mM glutamine (Euroclone). When the
cells reached 50-80% confluence, they were transfected with 30 nM
miR-34a precursors, ADRA2A siRNA or a scramble control (RiboBio)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific).
Subsequent experiments were accordingly performed after 48 h of
transfection. Each experiment was performed at least 3 times.
Luciferase assay
The 3′-UTR of ADRA2A, which was found to contain the
putative binding site of miR-34a by use of online database miRDB
(http://mirdb.org/), was amplified using PCR.
Subsequently, the PCR products (the major allele ADRA2A 3′UTR) were
cloned into a pGL3 vector (Promega) and the correct sequence of the
construct was confirmed by direct sequencing. In addition, a
specific single-base mutation in ADRA2A 3′UTR (the minor allele
ADRA2A 3′UTR), as well as a mutant ADRA2A 3′UTR was produced using
a QuikChange® Lightning Site-Directed Mutagenesis kit
(Agilent Technologies) and primers, including GCT AAG GGC AGC ACT
GCC TGC CCT C (forward) and GAG GGC AGG CAG TGC TGC CCT TAG C
(reverse). Subsequently, both minor allele ADRA2A 3′UTR and mutant
ADRA2A 3′UTR were inserted into pGL3 vectors (Promega),
respectively, and the correct sequences of these constructs were
also confirmed by direct sequencing. During the subsequent
luciferase assay, HCN-1A and U251 cells were co-transfected with
0.4 µg of Firefly luciferase reporter constructs containing
the major, minor or mutant ADRA2A 3′UTR, 0.02 µg of control
vector containing a Renilla luciferase (Promega), and
various concentrations of miR-34a precursors using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific). At 48 h
post-transfection, a Dual-Luciferase Reporter Assay System
(Promega) was utilized to determine the luciferase activity of
Firefly luciferase, which was then normalized to the luciferase
activity of Renilla luciferase to obtain the final results.
All tests were performed in triplicate.
Western blot analysis
RIPA buffer (Invitrogen; Thermo Fisher Scientific)
supplemented with a protease inhibitor cocktail and phosphatase
(Roche Diagnostics) was used to extract the total protein from
HCN-1A and U251 cells. The lysate was then boiled for 10 min in
2-mercaptoethanol and 10% SDS-PAGE was used to separate the
proteins. Subsequently, the proteins were transferred onto a
nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ) at
90V for 2 h. In the next step, the membrane was blocked at room
temperature for 60 min in TBS (Tris-buffered saline) containing
Tween-20 (TBST) and 5% non-fat milk, followed by incubation at 37°C
for 120 min with goat anti-ADRA2A (ab45871, 1:5,000 dilution,
Abcam) or anti-β-actin primary antibodies (4970s, 1:9,000 dilution,
Cell Signaling Technologies). Subsequently, the membrane was washed
and incubated at room temperature for 2 h with horseradish
peroxidase (HRP)-conjugated anti-goat secondary antibodies (ab6721,
1:13,000 dilution, Abcam). A Western Lightning Enhanced
Chemiluminescence solution (Perkin Elmer) was used to detect the
protein signals, which were acquired and analyzed using ImageJ
software (NIH) to obtain the density of each protein band. All
experiments were performed 3 times.
Statistical analysis
The data were analyzed by non-parametric tests and
the significance of different variables was compared using one-way
ANOVA, followed by Dunnett's test as a post hoc test. All
continuous variables were presented as medians and a P-value of
<0.05 was considered to indicate a statistically significant
difference. All statistical analyses were carried out using SPSS
18.0 software (SPSS, Inc.).
Results
rs3750625 is located in the seed region
of ADRA2A containing a putative binding site for miR-34a
According to the results of in silicon analysis by
the utilization of miRdSNP (http://mirdsnp.ccr.buffalo.edu/), rs3750625 was
located in the seed region of ADRA2A containing a putative binding
site for miR-34a. In addition, A allele, the minor allele in
rs3750625 polymorphism, could increase the binding affinity between
ADRA2A and miR-34a by creating an A-U base pair. Furthermore, the
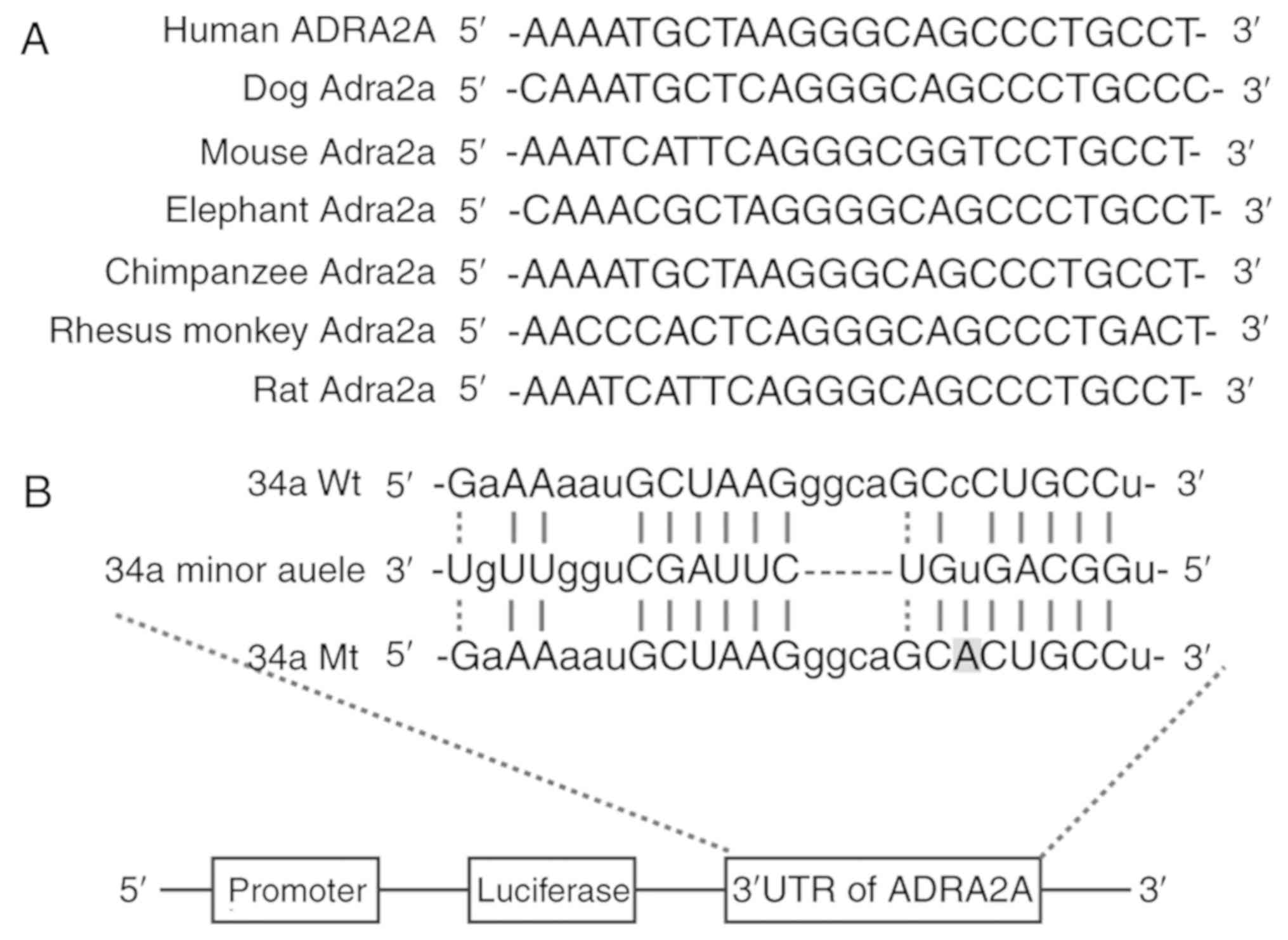
'seed sequence' in the 3′UTR of ADRA2A was highly conserved among
different species (Fig. 1A),
indicating that such sequence may play a very important role. As
shown in Fig. 1B, the binding of
miR-34a to ADRA2A was successful when the major allele of ADRA2A
was present; however, the mutation in ADRA2A 3′UTR hindered such
binding.
rs3750625 allele affects miR-34a
binding
According to our results from an online search upon
miRDB, miR-34a could directly target ADRA2A 3′UTR (Fig. 1B). To further confirm the
interaction between miR-34a and ADRA2A, the major, minor or mutant
3′UTR of ADRA2A was inserted into a pGL3 vector immediately
downstream of the luciferase reporter gene (Fig. 1B), followed by the co-transfection
of HCN-1A and U251 cells with these luciferase reporter constructs
and various concentrations of the miR-34a mimics (20 fmol, 60 fmol
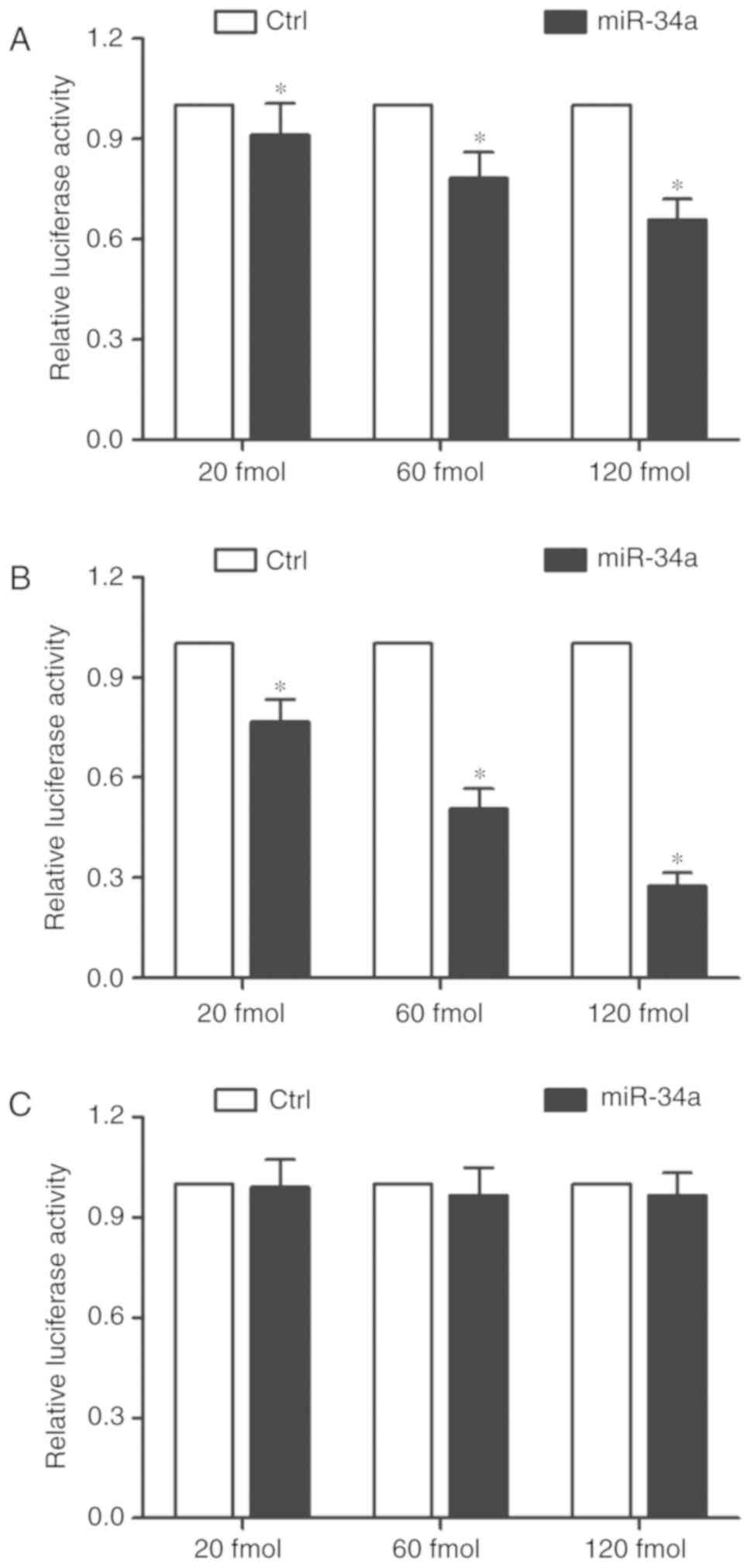
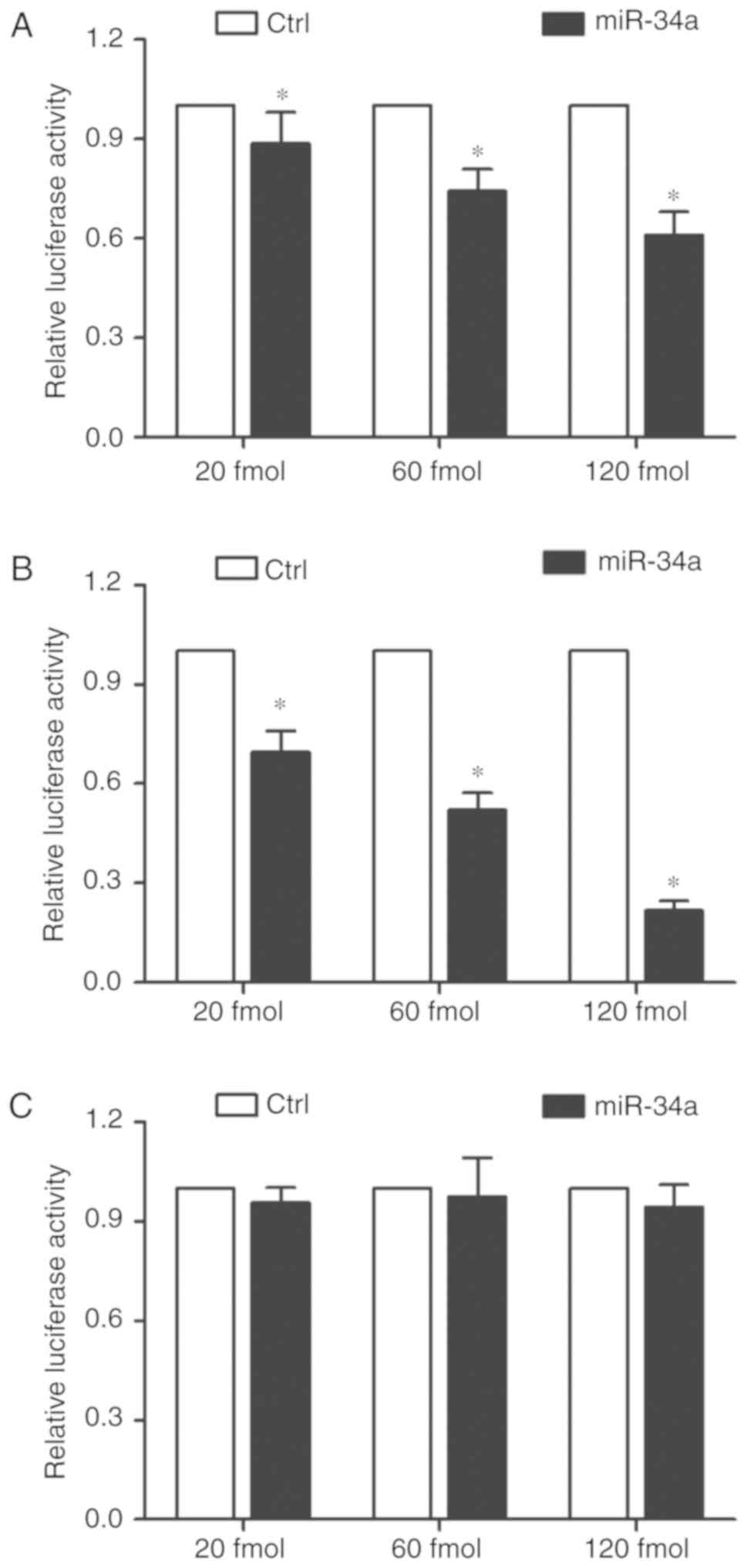
and 120 fmol). As shown in Figs.
2A and 3A, transfection with
miR-34a mimics reduced the luciferase activity of the cells
transfected with the major ADRA2A 3′UTR, and such a decrease in
luciferase activity was dose-dependent. Furthermore, the luciferase
activity in the cells transfected with the major ADRA2A 3′UTR
decreased in a stepwise manner at an increasing concentration of
miR-34a mimics (Figs. 2B and
3B). Notably, when the
concentration of miR-34a remained the same, the inhibitory effect
of miR-34a on the luciferase activity of the cells transfected with
the minor ADRA2A 3′UTR was much stronger than that of the cells
transfected with the major ADRA2A 3′UTR. As expected, miR-34a
mimics exerted no effect on the luciferase activity of the cells
transfected with the mutant ADRA2A 3′UTR (Figs. 2C and 3C), suggesting that the A allele located
in rs3750625 increased the binding affinity between ADRA2A and
miR-34a, thus reducing the expression of ADRA2A.
miR-34a suppresses ADRA2A expression
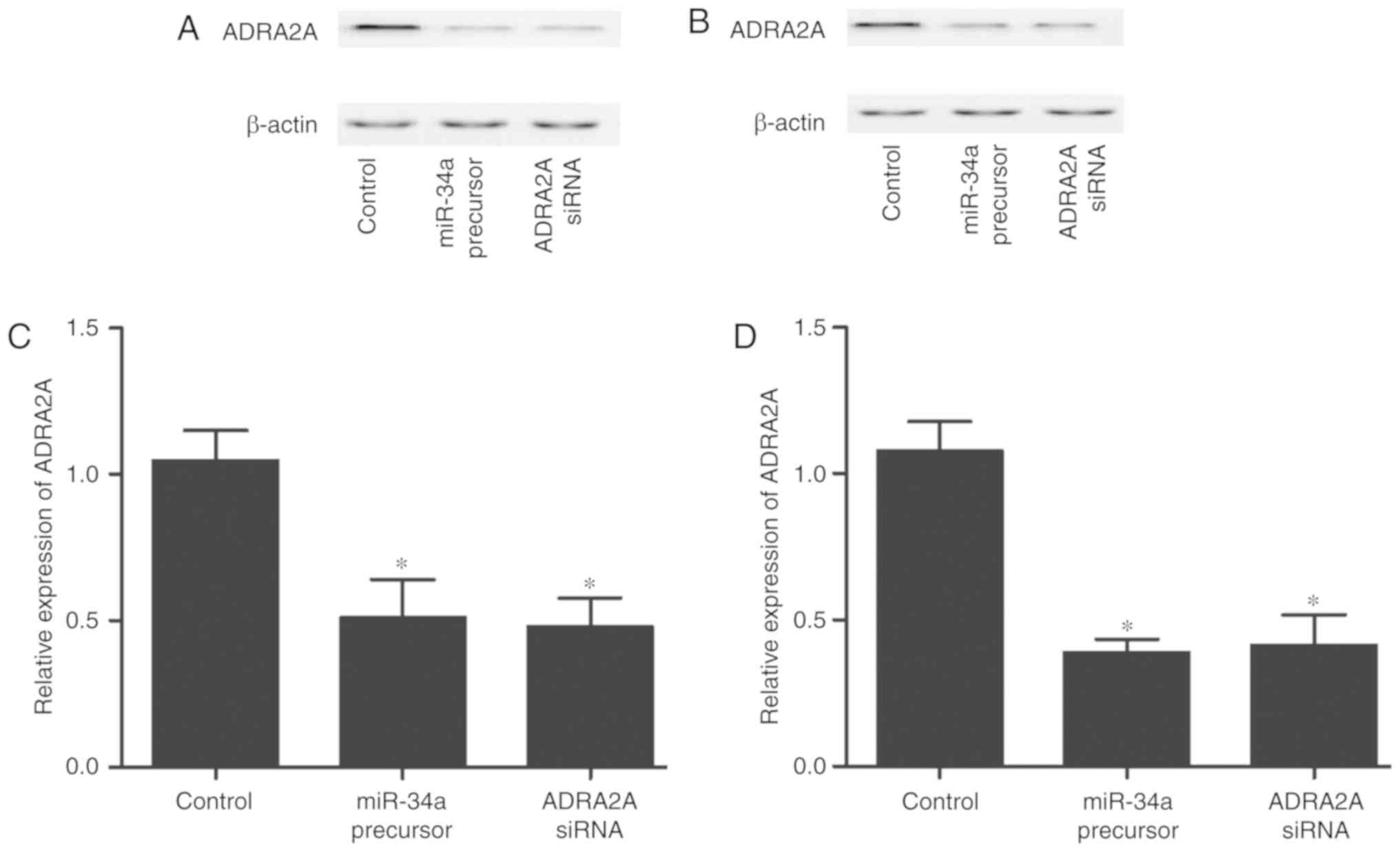
To further confirm whether miR-34a inhibits ADRA2A
expression, the HCN-1A and U251 cells were transfected with miR-34a
precursor or ADRA2A siRNA, as shown in Fig. 4. Following transfection with
miR-34a precursor or ADRA2A siRNA, both the HCN-1A (Fig. 4A and C) and U251 (Fig. 4B and D) cells exhibited a
decreased protein and mRNA expression of ADRA2A.
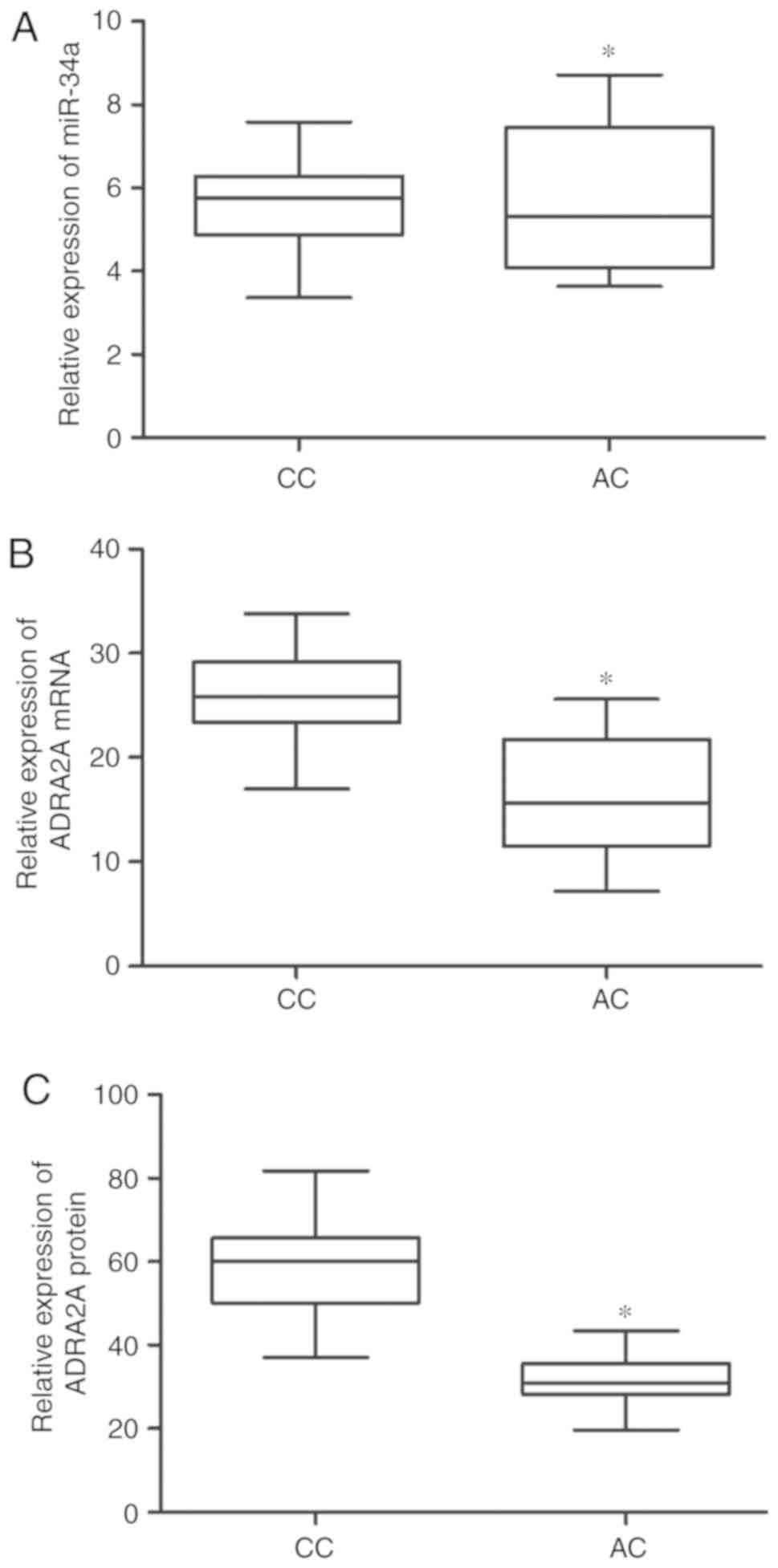
miR-34a and ADRA2A are differentially
expressed in the different groups
The 38 subjects in this study were divided into 2
groups, i.e., the CC (n=23) and AC (n=15) groups, according to
their genotype of rs3750625. Subsequently, RT-qPCR and western blot
analysis were performed to measure the levels of miR-34a, ADRA2A
mRNA and ADRA2A protein in these 2 groups. As shown in Fig. 5A, the level of miR-34a in the CC
group was similar to that in the AC group; however, the protein and
mRNA level of ADRA2A in the CC group was much higher than that in
the AC group (Fig. 5B and C).
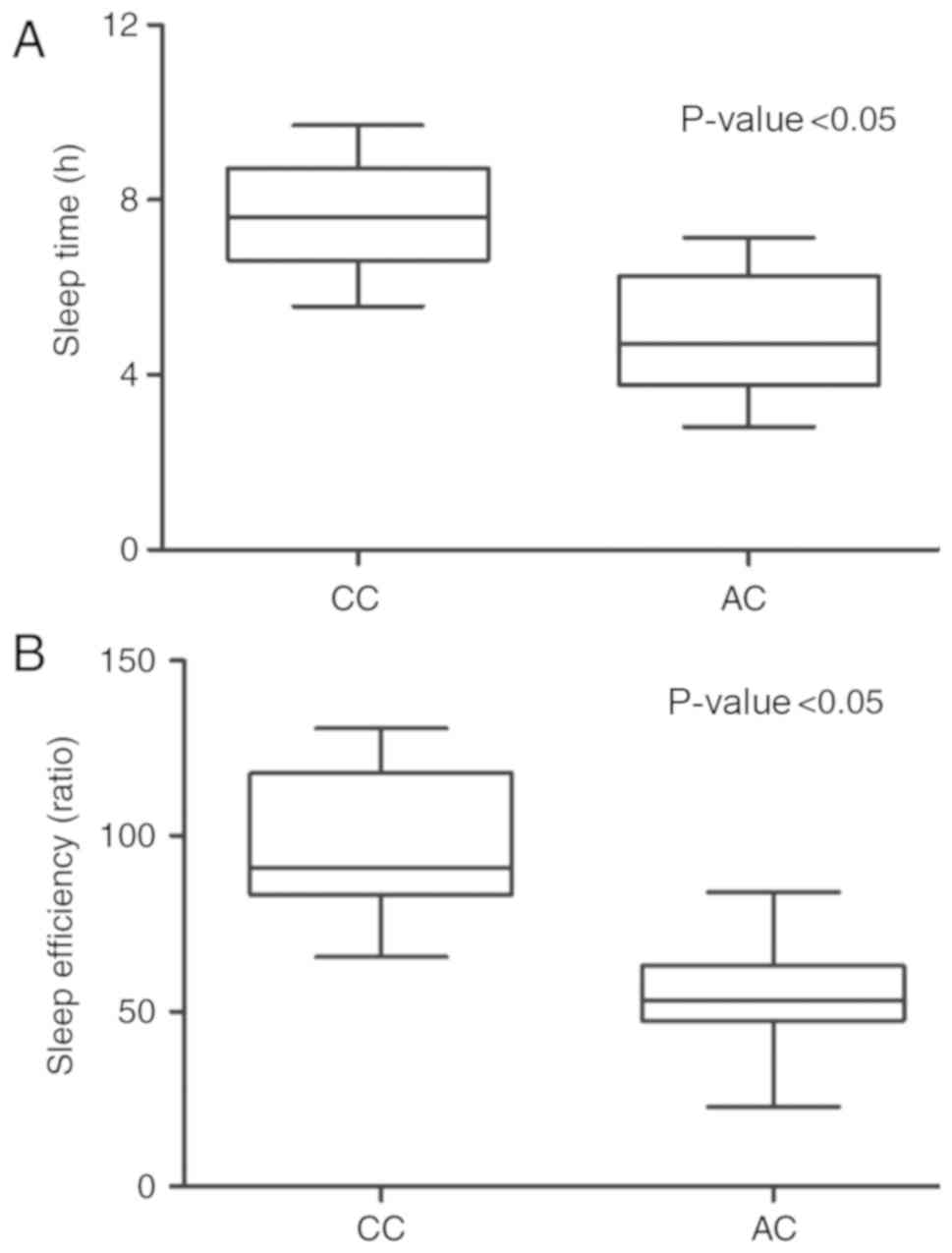
Determination of sleep quality in
different groups
The duration of sleep and the depth of sleep in the
2 groups were continuously monitored and recorded using a Covidien
BIS VISTA machine. As shown in Fig.
6, sleep time (Fig. 6A) and
sleep efficiency (Fig. 6B) in the
CC group were much higher than those in the AC group, indicating
that ADRA2A rs3750625 polymorphism was associated with the sleep
quality of patients in the ICU.
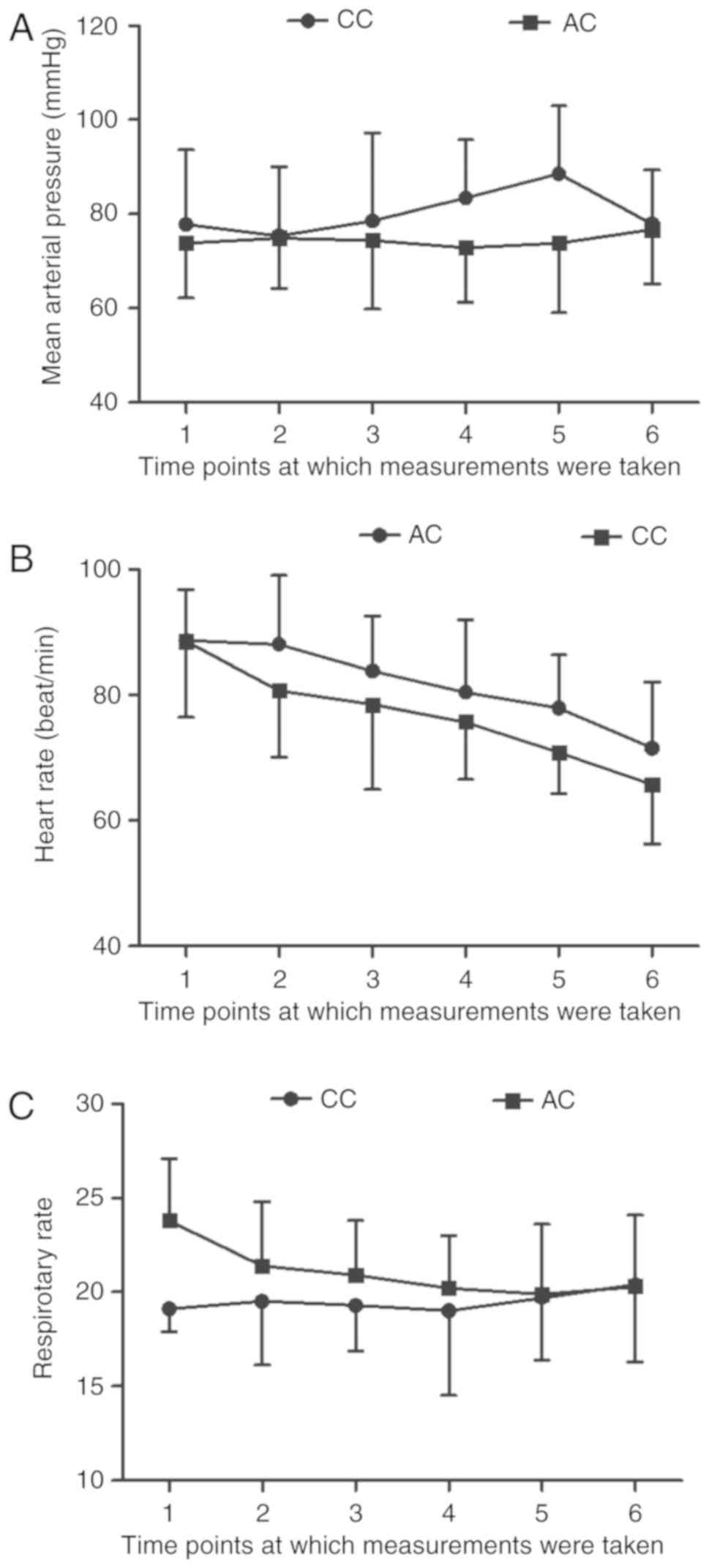
Mean arterial pressure (MAP), heart rate
and respiratory rate in the different groups
Compared to the baseline values, the value of MAP In
the AC group was similar to that in the CC group. In fact, the MAP
values in both AC and CC groups remained stable from 22:00 to
07:00, measured every 1.5 h (Fig.
7A). In addition, a lower heart rate was observed among
patients in the CC group as compared to those in the AC group
(Fig. 7B). Furthermore, compared
to the baseline values, the subjects in the CC group exhibited a
significantly reduced heart rate tendency (Fig. 7B). As shown in Fig. 7C, no obvious difference was
observed between the AC and CC groups in terms of respiratory
rate.
Discussion
Poor sleep is very common among patients in the ICU
and hence, sleep quality is considered an important aspect of
intensive care (25). In fact,
patients in the ICU spend approximately 50% of their total sleeping
time during the daytime, with a significantly reduced or missing
circadian rhythm (26).
Furthermore, inadequate sleep may cause extended hospitalization,
as well as an extended stay in the ICU. In severe cases, inadequate
sleep can trigger long-term cognitive impairment and delirium
(27). Therefore, sedation is
critical for a successful treatment of patients in the ICU as it
can reduce pain, relieve anxiety and decrease stress.
The adrenergic neurons in the brain are closely
related to the regulation of sleep cycles (27). For example, the activation of α1
and β receptors can induce neuronal excitation, while the induction
of inhibitory α2A receptors by agonists, including dexmedetomidine,
xylazine or clonidine, can cause hypnosis (28,29). Furthermore, the knockdown or
deletion of the ADRA2A gene, which encodes the α2A
adrenergic receptor, can reduce the capability of dexmedetomidine
to trigger LORR (30). Since the
location of ADRA2A expression in the brain is highly
restricted, it is possible to not only identify which downstream
pathways are used by dexmedetomidine to trigger sleep, but also
determine which pathways can inhibit sleep, thus allowing us to
gain insight into natural sleep circuitry (31,32).
The ADRA2A gene is found in chromosome
10q24-q26 and is located in a linkage region to T2DM (33). It was shown previously that
ADRA2A is involved in the function of β cells (34). Animal models have been used to
study the function of miRNAs in the regulation of sleep. For
example, sleep deprivation can lead to a significant change in
miRNA profiles in rat adipose tissues and brain, and such a change
is independent of the plasma level of corticosterone (35). In addition, sleep in experimental
animals can be altered by the cortical or intraventricular
injection of miRNAs and specific anti-miRNAs into the brain, as
suggested by the slow wave activity on electroencephalographic
(EEG) (36). The role of miRNAs
in the control of sleep quality may be mediated by their ability to
regulate the expression of sleep associated genes. Recently, the
full 3′UTR of ADRA2A was cloned into a luciferase reporter plasmid
downstream of a Firefly luciferase reporter gene, and the
luciferase assay results revealed a potential location for miR-34a
binding, which in turn led to the reduction in luciferase activity
in a dose-dependent manner (18,37). In this study, we searched an
online database and found that miR-34a could directly bind to the
3′UTR of ADRA2A. In addition, a single site or a 'seed sequence' in
ADRA2A 3′UTR were mutated in this study to generate minor or mutant
ADRA2A 3′UTR, which were then inserted into pGL3 vectors to
generate different luciferase constructs. Furthermore, the
above-mentioned luciferase constructs were transfected into the
HCN-1A and U251 cells, and the results revealed that miR-34a
reduced the luciferase activity of the cells transfected with the
major or minor ADRA2A 3′UTR in a dose-dependent manner. As
expected, miR-34a had no effect on the luciferase activity of the
cells transfected with the mutant ADRA2A 3′UTR.
It was previously suggested that, following sexual
assault- or MVC-associated trauma, subjects harboring the rs3750625
minor allele may show a 1-point increase (on a scale of 0-10)
during acute MSP. This may be caused by a more efficient binding
between ADRA2A 3′UTR and miR-34a in the presence of the minor
allele. Such results are consistent with the role of miR-34a in the
regulation of stress, indicating that the symptoms of stress may be
caused by the activation of physiologic systems that also promote
miR-34a expression and increase the effect of rs3750625 on stress
(18,38). In this study, we recruited 38
patients in the ICU and divided them into 2 groups, i.e., the CC
(n=32) and AC (n=6) groups, according to their genotypes of
rs3750625. The results revealed that miR-34a expression in the CC
group was comparable to that in the AC group, whereas the mRNA and
protein level of ADRA2A was significantly higher in the CC group
compared to that in the AA group. In addition, we found that the
sleep time and sleep efficiency in the CC group were much higher
than those in the AC group. Furthermore, we revealed that the MAP
value in both the AC and CC groups remained stable from 22:00 to
08:00, and their respiratory rates were also similar. However, a
lower heart rate was observed in the patients from the CC group.
Donello et al previously demonstrated that animals displayed
acute hyperalgesia upon sound exposure if ADRA2A was absent
(39). It has also been shown
that, in individuals harboring the minor allele of rs3750625, an
increased level of miR-34a binding, a reduced level of ADRA2A
transcripts, and an increased level of acute MSP were observed in
those suffering from trauma (18). rs3750625 has been previously shown
to play a role in the postprandial gastric volume of obese or
overweight European-Americans (22). It has also been demonstrated that
rs3750625 is close to or within the seed region of 23 miRNAs,
suggesting that rs3750625 may increase the expression of ADRA2A by
affecting miRNA binding (18). In
a recent study, Linnstaedt et al demonstrated that rs3750625
further decreased ADRA2A expression by increasing miR-34a binding
affinity, and hence reduced the risk of gestational diabetes
mellitus (GDM) by increasing the release of insulin (18).
There are limitations to this study: Firstly, the
sample size of this study was relatively small, and further studies
with a larger sample size are warranted to confirm the conclusions
of this study; secondly, further functional studies are warranted
to confirm the conclusions of this study.
Taken together, it can be concluded that the SNP
rs3750625 in the 3′UTR of ADRA2A affects the sleep quality of
patients in the ICU in by increasing the binding of miR-34a to
ADRA2A. In brief, the presence of the rs3750625 minor allele in the
3′UTR of ADRA2A increases the binding between miR-34a and ADRA2A, a
direct target of miR-34a. As a result, by further inhibiting the
expression of ADRA2A, the presence of the rs3750625 minor allele
can affect the sleep quality of patients in the ICU.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
QH and JZ were involved in the conception and design
of the study. GC, YH and JL were involved in the literature
collection. QH, GC, YD, SZ, XC, QX and LX were involved in data
collection. YH, JL, QN and JZ were involved in data analysis and
processing. QH, GC and JZ were involved in manuscript preparation.
YH, JL, YD, SZ, XC, QX, QN and LX were involved in the final
proofreading of the manuscript. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
The Human Research Ethics Committees of Dongguan
Houjie Hospital approved this research. Written informed consent
was obtained from all patients or their first-degree relatives
prior to the onset of this research. The research process was in
conformity with the latest version of Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Pisani MA, Friese RS, Gehlbach BK, Schwab
RJ, Weinhouse GL and Jones SF: Sleep in the intensive care unit. Am
J Respir Crit Care Med. 191:731–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weinhouse GL, Schwab RJ, Watson PL, Patil
N, Vaccaro B, Pandharipande P and Ely EW: Bench-to-bedside review:
Delirium in ICU patients-importance of sleep deprivation. Crit
Care. 13:2342009. View
Article : Google Scholar
|
|
3
|
Roche Campo F, Drouot X, Thille AW, Galia
F, Cabello B, d'Ortho MP and Brochard L: Poor sleep quality is
associated with late noninvasive ventilation failure in patients
with acute hypercapnic respiratory failure. Crit Care Med.
38:477–485. 2010. View Article : Google Scholar
|
|
4
|
Wulff K, Gatti S, Wettstein JG and Foster
RG: Sleep and circadian rhythm disruption in psychiatric and
neurodegenerative disease. Nat Rev Neurosci. 11:589–599. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanders RD and Maze M: Noradrenergic
trespass in anesthetic and sedative states. Anesthesiology.
117:945–947. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takahashi K, Kayama Y, Lin JS and Sakai K:
Locus coeru-leus neuronal activity during the sleep-waking cycle in
mice. Neuroscience. 169:1115–1126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carter ME, Yizhar O, Chikahisa S, Nguyen
H, Adamantidis A, Nishino S, Deisseroth K and de Lecea L: Tuning
arousal with optogenetic modulation of locus coeruleus neurons. Nat
Neurosci. 13:1526–1533. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karp X and Ambros V: Developmental
biology. Encountering microRNAs in cell fate signaling. Science.
310:1288–1289. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weiler J, Hunziker J and Hall J:
Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs
implicated in human disease. Gene Ther. 13:496–502. 2006.
View Article : Google Scholar
|
|
11
|
Latronico MV, Catalucci D and Condorelli
G: Emerging role of microRNAs in cardiovascular biology. Circ Res.
101:1225–1236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lynam-Lennon N, Maher SG and Reynolds JV:
The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos
Soc. 84:55–71. 2009. View Article : Google Scholar
|
|
13
|
Sonkoly E, Wei T, Janson PC, Saaf A,
Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B,
Scheynius A, et al: MicroRNAs: Novel regulators involved in the
pathogenesis of psoriasis. PLoS One. 2:e6102007. View Article : Google Scholar
|
|
14
|
Hansen T, Olsen L, Lindow M, Jakobsen KD,
Ullum H, Jonsson E, Andreassen OA, Djurovic S, Melle I, Agartz I,
et al: Brain expressed microRNAs implicated in schizophrenia
etiology. PLoS One. 2:e8732007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murakami Y, Kubo S, Tamori A, Itami S,
Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T and Taguchi YH:
Comprehensive analysis of transcriptome and metabolome analysis in
Intrahepatic Cholangiocarcinoma and Hepatocellular Carcinoma. Sci
Rep. 5:162942015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deveci M, Catalyurek UV and Toland AE:
mrSNP: Software to detect SNP effects on microRNA binding. BMC
Bioinformatics. 15:732014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Finley JC Jr, O'Leary M, Wester D,
MacKenzie S, Shepard N, Farrow S and Lockette W: A genetic
polymorphism of the alpha2-adrenergic receptor increases autonomic
responses to stress. J Appl Physiol. 96:2231–2239. 1985. View Article : Google Scholar
|
|
18
|
Linnstaedt SD, Walker MG, Riker KD, Nyland
JE, Hu J, Rossi C, Swor RA, Jones JS, Diatchenko L, Bortsov AV, et
al: Genetic variant rs3750625 in the 3'UTR of ADRA2A affects
stress-dependent acute pain severity after trauma and alters a
microRNA-34a regulatory site. Pain. 158:230–239. 2017. View Article : Google Scholar :
|
|
19
|
Kohli U, Muszkat M, Sofowora GG, Harris
PA, Friedman EA, Dupont WD, Scheinin M, Wood AJ, Stein CM and
Kurnik D: Effects of variation in the human alpha2A- and
alpha2C-adre-noceptor genes on cognitive tasks and pain perception.
Eur J Pain. 14:154–159. 2010. View Article : Google Scholar
|
|
20
|
Haramati S, Navon I, Issler O, Ezra-Nevo
G, Gil S, Zwang R, Hornstein E and Chen A: MicroRNA as repressors
of stress-induced anxiety: The case of amygdalar miR-34. J
Neurosci. 31:14191–14203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gelegen C, Gent TC, Ferretti V, Zhang Z,
Yustos R, Lan F, Yang Q, Overington DW, Vyssotski AL, van Lith HA,
et al: Staying awake-a genetic region that hinders α2 adrenergic
receptor agonist-induced sleep. Eur J Neurosci. 40:2311–2319. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Papathanasopoulos A, Camilleri M, Carlson
PJ, Vella A, Nord SJ, Burton DD, Odunsi ST and Zinsmeister AR: A
preliminary candidate genotype-intermediate phenotype study of
satiation and gastric motor function in obesity. Obesity (Silver
Spring). 18:1201–1211. 2010. View Article : Google Scholar
|
|
23
|
Sessler CN, Gosnell MS, Grap MJ, Brophy
GM, O'Neal PV, Keane KA, Tesoro EP and Elswick RK: The richmond
agitation-sedation scale: Validity and reliability in adult
intensive care unit patients. Am J Respir Crit Care Med.
166:1338–1344. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
25
|
Freedman NS, Gazendam J, Levan L, Pack AI
and Schwab RJ: Abnormal sleep/wake cycles and the effect of
environmental noise on sleep disruption in the intensive care unit.
Am J Respir Crit Care Med. 163:451–457. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parthasarathy S and Tobin MJ: Sleep in the
intensive care unit. Intensive Care Med. 30:197–206. 2004.
View Article : Google Scholar
|
|
27
|
Saper CB, Fuller PM, Pedersen NP, Lu J and
Scammell TE: Sleep state switching. Neuron. 68:1023–1042. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schmeichel BE and Berridge CW:
Wake-promoting actions of noradrenergic α1- and beta-receptors
within the lateral hypotha-lamic area. Eur J Neurosci. 37:891–900.
2013. View Article : Google Scholar
|
|
29
|
Drew GM, Gower AJ and Marriott AS: Alpha
2-adrenoceptors mediate clonidine-induced sedation in the rat. Br J
Pharmacol. 67:133–141. 1979.PubMed/NCBI
|
|
30
|
Lakhlani PP, MacMillan LB, Guo TZ, McCool
BA, Lovinger DM, Maze M and Limbird LE: Substitution of a mutant
alpha2a-adren-ergic receptor via 'hit and run' gene targeting
reveals the role of this subtype in sedative, analgesic, and
anesthetic-sparing responses in vivo. Proc Natl Acad Sci USA.
94:9950–9955. 1997. View Article : Google Scholar
|
|
31
|
Franks NP: General anaesthesia: From
molecular targets to neuronal pathways of sleep and arousal. Nat
Rev Neurosci. 9:370–386. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nicholas AP, Pieribone V and Hokfelt T:
Distributions of mRNAs for alpha-2 adrenergic receptor subtypes in
rat brain: An in situ hybridization study. J Comp Neurol.
328:575–594. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duggirala R, Blangero J, Almasy L, Dyer
TD, Williams KL, Leach RJ, O'Connell P and Stern MP: Linkage of
type 2 diabetes mellitus and of age at onset to a genetic location
on chromosome 10q in Mexican Americans. Am J Hum Genet.
64:1127–1140. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Boesgaard TW, Grarup N, Jorgensen T,
Borch-Johnsen K, Hansen T and Pedersen O; Meta-Analysis of Glucose
and Insulin-Related Trait Consortium (MAGIC): Variants at
DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with
reduced glucose-stimulated beta cell function in middle-aged Danish
people. Diabetologia. 53:1647–1655. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu KX, Chen GP, Lin PL, Huang JC, Lin X,
Qi JC and Lin QC: Detection and analysis of apoptosis- and
autophagy-related miRNAs of mouse vascular endothelial cells in
chronic intermittent hypoxia model. Life Sci. 193:194–199. 2018.
View Article : Google Scholar
|
|
36
|
Borbely S, Vilagi I, Haraszti Z, Szalontai
O, Hajnik T, Toth A and Detari L: Sleep deprivation decreases
neuronal excitability and responsiveness in rats both in vivo and
ex vivo. Brain Res Bull. 137:166–177. 2018. View Article : Google Scholar
|
|
37
|
Gottwein E and Cullen BR: A human
herpesvirus microRNA inhibits p21 expression and attenuates
p21-mediated cell cycle arrest. J Virol. 84:5229–5237. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dias BG, Goodman JV, Ahluwalia R, Easton
AE, Andero R and Ressler KJ: Amygdala-dependent fear memory
consolidation via miR-34a and Notch signaling. Neuron. 83:906–918.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Donello JE, Guan Y, Tian M, Cheevers CV,
Alcantara M, Cabrera S, Raja SN and Gil DW: A peripheral
adrenoceptor-mediated sympathetic mechanism can transform
stress-induced analgesia into hyperalgesia. Anesthesiology.
114:1403–1416. 2011. View Article : Google Scholar : PubMed/NCBI
|