Introduction
It is estimated that 1.8 million new colorectal
cancer cases and 881,000 colorectal cancer-associated mortalities
occurred worldwide in 2018 (1).
Colorectal cancer is responsible for ~10% of mortalities associated
with cancer and exhibits the second highest cancer mortality rate
worldwide (1). Conventional
surgery, chemotherapy and radiotherapy are commonly used for the
treatment of patients with colorectal cancer (2). Recently, targeted therapy against
vascular endothelial growth factor and epidermal growth factor
receptor in combination with chemotherapy has been demonstrated to
prolong the overall survival of patients (3). The identification of novel oncogenes
and tumor suppressors may aid in the investigation and development
of new agents to improve the prognosis of patients with colorectal
cancer in the future.
MicroRNAs (miRNAs) are characterized as small (18-25
nucleotides), single-stranded non-coding RNA molecules. In recent
years, accumulating evidence has demonstrated that miRNAs are
important regulators of the majority of physiological processes in
mammal cells (4). The seed region
of miRNAs can form a complementary base pair with the
3′-untranslated region (3′UTR) of target gene mRNAs, leading to
mRNA degradation or translation inhibition (5). The dysregulation of miRNAs has been
reported to be associated with the initiation and development of a
number of cancer types (6). In
colorectal cancer, a number of miRNAs, including miR-17-5p and
miR-21, have been revealed to be key drivers of tumorigenesis
(7,8). Furthermore, miR-1290 and miR-139-3p
are markedly decreased and increased in colorectal tumors,
respectively, and can serve as biomarkers for patients with
colorectal cancer (9,10). Although numerous differentially
expressed miRNAs have been identified via RNA sequencing and
microarray (11,12), the roles and function of the
majority of these molecules have not yet been determined. Through
the retrieval and analysis of previously published microarray data
(11), miR-3651 has been
identified to be significantly upregulated in colon tumors
(11). The prognostic value of
miR-3651 has previously been evaluated in esophageal squamous cell
and oral squamous cell carcinomas (13,14). However, the role and molecular
mechanism of this miRNA remain unclear in colorectal cancer.
T-box transcription factor 1 (TBX1) is known to be a
developmentally important transcription factor, which has a 180-200
amino acid conserved DNA-binding domain, namely the T-box (15). The expression of TBX1 is essential
for the proliferation and differentiation of heart progenitor
cells, and is important for the development of the heart, inner
ear, sweat gland, teeth and thyroid (16-19). TBX1 has also been identified as a
novel tumor suppressor in thyroid cancer and basal cell carcinoma
(20,21). As a transcription factor, TBX1
interacts with chromatin remodeling complexes to control the
expression of numerous genes (22). TBX1 has also been reported to
control the activity of phosphoinositide-3 kinase/protein kinase B
(PI3K/AKT) and mitogen-activated protein kinase/extracellular
signal-regulated kinase (MAPK/ERK) signaling in thyroid cancer
cells (20). The downregulation
of TBX1 in tumor cells is due to promoter methylation and aberrant
miRNA expression (20,21).
In the present study, a published miRNA expression
profile of colon tumors and normal tissues was analyzed, in
combination with reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) examination of collected specimens. It was
observed that miR-3651 was significantly upregulated in the colon
tumor tissues compared with the normal tissues. In vitro
assays further revealed that downregulation of miR-3651 inhibited
the proliferation and induced the apoptosis of colorectal cancer
cells. Furthermore, it was demonstrated that miR-3651 targeted TBX1
to control the PI3K/AKT and MAPK/ERK signaling pathways.
Collectively, the data of the current study demonstrated the
clinical relevance of miR-3651 in colorectal carcinogenesis.
Materials and methods
Sample collection
Normal and tumor tissues were collected from 40
patients who underwent surgery at the China-Japan Union Hospital of
Jilin University (Changchun, China) between July 2016 and January
2018. The patient characteristics were recorded following
enrollment and included the following: Age, sex, tumor size, degree
of tumor differentiation and tumor-node-metastasis (TNM) stage
(23). No patients received
chemotherapy or radiotherapy prior to surgery. The experiments were
performed under the supervision of the Ethics Committee of the
China-Japan Union Hospital of Jilin University. Written informed
consent was acquired from all participants. Tissues were
immediately stored in liquid nitrogen prior to subjecting to RNA
extraction.
Cell culture and transfection
A normal colonic mucosal cell line (FHC) and two
human colorectal cancer cell lines (HCT116 and HT29) were purchased
from the American Type Culture Collection and used within 6 months
of receipt. The cells were cultured in Dulbecco's modified Eagle
medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.). Cell culture was performed in a humidified incubator with 5%
CO2 at 37°C. miR-3651 mimic (5′-AUG GCA CUG GUA GAA UUC
ACU G-3′), miR-negative control (NC) mimic (5′-CAG UAC UUU UGU GUA
GUA CAA-3′), miR-3651 inhibitor (5′-CAG UGA AUU CUA CCA GUG CCA
U-3′) and miR-NC inhibitor (5′-UUC UCC GAA CGU GUC ACG U-3′) were
synthesized and purchased from Suzhou GenePharma Co., Ltd. The
mimics and inhibitors (50 nM) were transfected into HCT116 and HT29
ells using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer′s protocol.
TBX1 siRNA (5′-UAU UUC UCG CUA UCU UUG CGU-3′) and control siRNA
(5′-UUC UCC GAA CGU GUC ACG U-3′) were products of Shanghai
GenePharma Co., Ltd. siRNAs (25 nM) were transfected into
1×105 cells in each well of
24-well plates with Lipofectamine RNAiMax (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol. A
total of 2 days after transfection, the transfection efficiency was
determined by reverse transcription-quantitative PCR (RT-qPCR).
Bioinformatics analysis
The data of GSE115513 (11) (containing miRNA expression data of
381 normal colonic mucosa, 51 colon adenoma and 411 colorectal
carcinoma) were downloaded from the Gene Expression Omnibus (GEO)
database (https://www.ncbi.nlm.nih.gov/geoprofiles/). The data
were subsequently analyzed with GEO2R, which was provided by the
GEO database. The potential target genes of miR-3651 were predicted
with TargetScan software v.7.2 (http://www.targetscan.org/vert_72/).
RNA isolation and RT-qPCR
Total RNA was isolated from the cells and tissues
using an RNeasy Mini kit (Qiagen GmbH), according to the
manufacturer's protocol. The concentration of RNA was then measured
using a NanoDrop 2000 system (Thermo Fisher Scientific, Inc.).
Next, RNA was reverse transcribed into first-stranded cDNA using a
PrimeScript™ RT reagent kit (Takara Bio, Inc.). Subsequently, the
qPCR reaction was performed using TB Green® Fast qPCR
Mix (Takara Bio, Inc.) on a CFX-96 Realtime system (Bio-Rad
Laboratories, Inc.). The qPCR conditions included two steps: Step
1: 95°C for 30 sec; and step 2: 35 cycles of 95°C for 15 sec and
60°C for 10 sec. GAPDH and U6 served as the internal controls for
mRNA and miRNA detection, respectively. The relative gene
expression was calculated using the 2−ΔΔCq method
(24). The primer sequences used
in qPCR were as follows: Stem-loop primer, 5′-CTC AAC TGG TGT CGT
GGA GTC GGC AAT TCA GTT GAG TCA TGT AC-3′; miR-3651 forward, 5′-GCC
GAG CAT AGC CCG GTC GC-3′, and reverse, 5′-CTC AAC TGG TGT CGT
GGA-3′; U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′, and reverse,
5′-AAC GCT TCA CGA ATT TGC GT-3′; TBX1 forward, 5′-ACG ACA ACG GCC
ACA TTA TTC-3′, and reverse, 5′-CCT CGG CAT ATT TCT CGC TAT CT-3′;
GAPDH forward, 5′-ACA ACT TTG GTA TCG TGG AAG G-3′, and reverse,
5′-GCC ATC ACG CCA CAG TTT C-3′.
Protein extraction and western blot
analysis
Protein lysates were prepared using a RIPA lysis
buffer, and the concentration of lysates was calculated using a BCA
Protein Assay kit (both from Thermo Fisher Scientific, Inc.). A
total of 20 µg protein was then loaded onto an 8% SDS-PAGE
gel and separated using electrophoresis. Proteins were then
transferred to a PVDF membrane and blocked with 5% non-fat milk at
room temperature for 1 h. Subsequently, the membrane was incubated
in a solution containing primary antibodies at room temperature for
1 h, followed by incubation with secondary antibodies at room
temperature for 1 h. The primary antibodies used in western blot
analysis were as follows: p-AKT (cat. no. 4060; 1:2,000), AKT (cat.
no. 4685; 1:2,000), p-ERK1/2 (cat. no. 4370; 1:2,000) and ERK1/2
(cat. no. 4695; 1:2,000) purchased from Cell Signaling Technology,
Inc.; TBX1 antibody (cat. no. ab109313; 1:500) obtained from Abcam;
GAPDH (cat. no. AMAB91152; 1:10,000) from Sigma-Aldrich (Merck
KGaA); and B-cell lymphoma 2 (Bcl2; cat. no. sc-130307; 1:200)
Bcl2-associated X protein (Bax; cat. no. sc-7480; 1:200) antibodies
from Santa Cruz Biotechnology, Inc. Mouse (cat. no. ab97040;
1:10,000) and rabbit (cat. no. ab7090; 1:10,000) IgG secondary
antibodies were purchased from Abcam. The blots were developed
using a Pierce™ ECL Western Blotting Substrate (Thermo Fisher
Scientific, Inc.). ImageJ V.1.6.0 (National Institutes of Health)
was used to quantify the protein expression of all blots.
Cell proliferation assay
The cell proliferation ability was detected by Cell
Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). Briefly, a total of 10,000 HCT116 or HT29 cells
were seeded into each well of 96-well plates. On the following day,
cells were transfected with miR-3651 inhibitor or miR-NC inhibitor,
and maintained for 3 days. At 0, 1, 2 and 3 days after
transfection, 10 µl CCK-8 solution was added into each well
and sustained for a further 2 h. The medium was then transferred to
a new 96-well plate, and the absorbance at 450 nM was detected to
reflect the cell number.
Cell apoptosis assay
The cell apoptosis rate was determined using a Dead
Cell Apoptosis kit with Annexin-V Alexa Fluor™ 488 and propidium
iodide (PI; Invitrogen; Thermo Fisher Scientific, Inc.). Briefly,
cells were transfected for 48 h, and then harvested and suspended
in 1X Annexin-binding buffer. Next, PI and Annexin V were added
into the cell suspension and incubated for 15 min at room
temperature. Following incubation, 400 µl 1X Annexin-binding
buffer was added and mixed. The samples were immediately subjected
to flow cytometry analysis. A positive PI and Annexin V result
indicated that cells were at the late apoptosis stage, while a
negative PI and positive Annexin V result indicated cells at the
early apoptosis stage.
Dual-luciferase reporter assay
The TBX1 3′UTR was amplified from HCT116 cDNA and
ligated into a pGL3-basic plasmid. Mutations were introduced into
the TBX1 3′UTR using a QuikChange Site-Directed Mutagenesis kit
(Agilent Technologies, Inc., Santa Clara, CA, USA). In order to
perform dual-luciferase reporter assay, 2×105 HCT116 or HT29 cells/well were seeded
into 24-well plates, and transfected with miR-3651 mimic or miR-NC
mimic in combination with pGL3-TBX1 3′UTR-wild-type (WT) or
3′UTR-mutant (Mut). After 48 h, the relative luciferase activity of
each group was detected using a Dual-Luciferase Reporter system
(Promega Corporation, Madison, WI, USA) on a Synergy H1 microplate
reader (BioTek Instruments, Inc.; Agilent Technologies, Inc).
Statistical analysis
The data were analyzed with GraphPad Prism version
6.0 (GraphPad Software, San Diego, CA, USA), and are presented as
the mean ± standard deviation. Comparisons between two groups were
performed by Student's t-test. Comparison between three groups were
performed using one-way analysis of variance, followed by
Newman-Keuls post hoc test. The correlation between miRNA and mRNA
expression was analyzed using Pearson correlation analysis. All
experiments were performed at least three times. P<0.05 was
considered to indicate a statistically significant difference.
Results
Overexpression of miR-3651 in colorectal
cancer tissues and cells
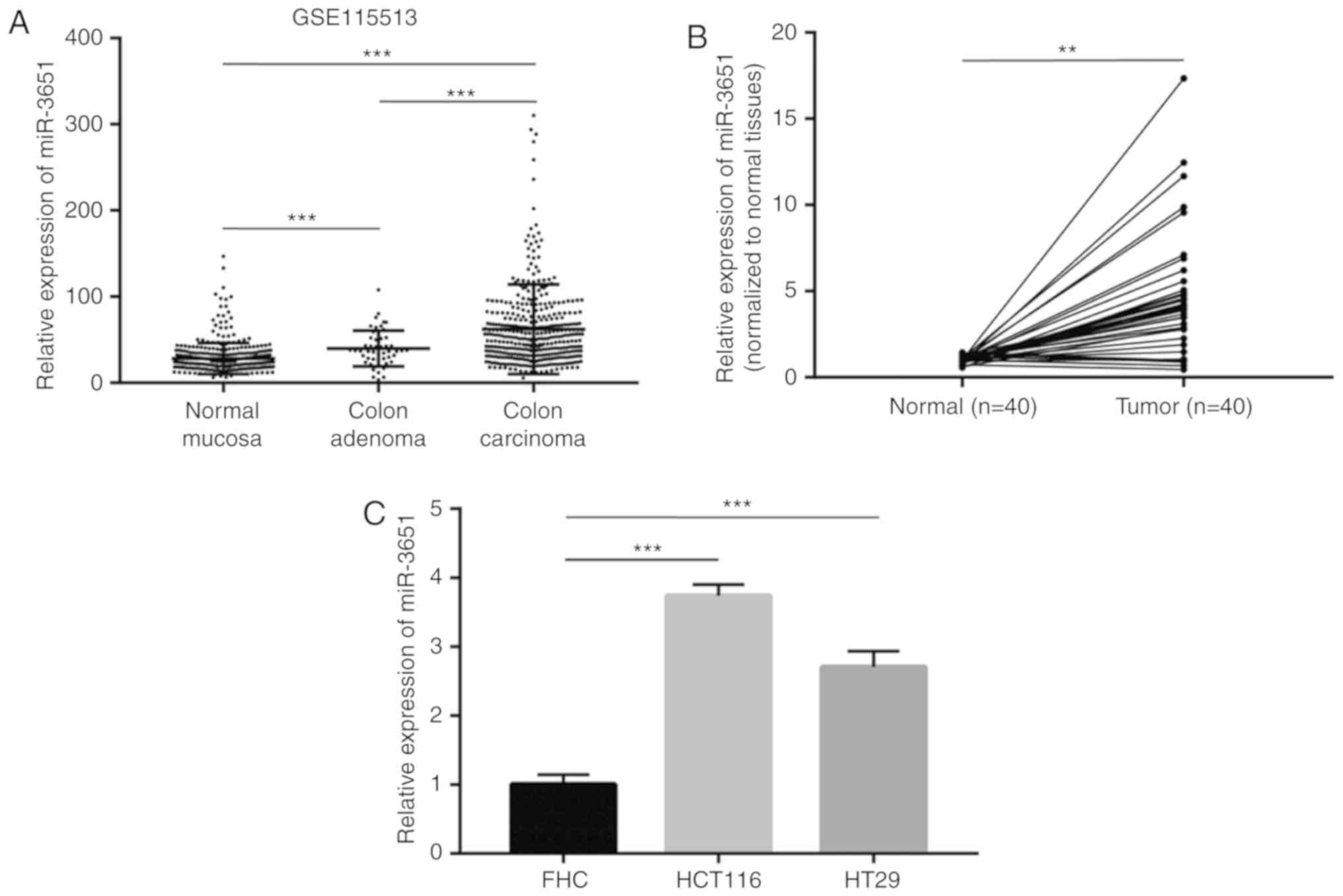
In order to identify the potential oncogenic miRNAs
in colorectal cancer, microarray data on the expression of miRNAs
in 381 normal colonic mucosa, 51 colon adenoma and 411 colorectal
carcinoma samples were collected from the Gene Expression Omnibus
database (series GSE115513). miRNAs that were differentially
expressed between colon adenoma and normal mucosa were screened.
The expression of these miRNAs in colorectal carcinoma and adenoma
was subsequently compared. According to the results of this
screening strategy, miR-3651 was revealed to be significantly
overexpressed in colon adenoma and colorectal carcinoma compared
with the normal colonic mucosa (Fig.
1A).
For validation of the elevation of miR-3651 observed
in the microarray data, 40 pairs of normal and tumor tissues were
collected from patients with colorectal cancer. Using RT-qPCR,
miR-3651 expression was detected in the paired tissues and
normalized to the normal tissue expression. Consistent with the
published microarray data, the miR-3651 levels were found to be
increased in 85% (34/40) of patients (Fig. 1B). The association between
miR-3651 expression and the clinicopathological features of
patients was further analyzed (Table
I). It was observed that high miR-3651 expression was
significantly associated with high TNM stage in patients with
colorectal cancer (Table I). In
addition, miR-3651 expression was subsequently detected in the
immortalized colon normal cells (FHC) and two colorectal cancer
cell lines (HCT116 and HT29). The results indicated that miR-3651
was increased by 3-fold in HCT116 and HT29 cancer cells as compared
with that in normal FHC cells (Fig.
1C).
 | Table IAssociation between miR-3651
expression and the clinicopathological features of patients with
colorectal cancer. |
Table I
Association between miR-3651
expression and the clinicopathological features of patients with
colorectal cancer.
| Clinicopathological
feature | miR-3651 expression
(n)
| P-value |
|---|
| High | Low |
|---|
| Age | | | 0.99 |
| ≥60 years | 11 | 12 | |
| <60 years | 9 | 8 | |
| Sex | | | 0.69 |
| Male | 15 | 17 | |
| Female | 5 | 3 | |
| TNM stage | | | <0.05 |
| I | 1 | 4 | |
| II | 1 | 5 | |
| III-IV | 18 | 11 | |
| Tumor size | | | 0.48 |
| ≥15 cm | 7 | 4 | |
| <15 cm | 13 | 16 | |
|
Differentiation | | | 0.99 |
| Poor | 5 | 4 | |
| High | 15 | 16 | |
Downregulation of miR-3651 inhibits
colorectal cancer cell proliferation via induction of cell
apoptosis
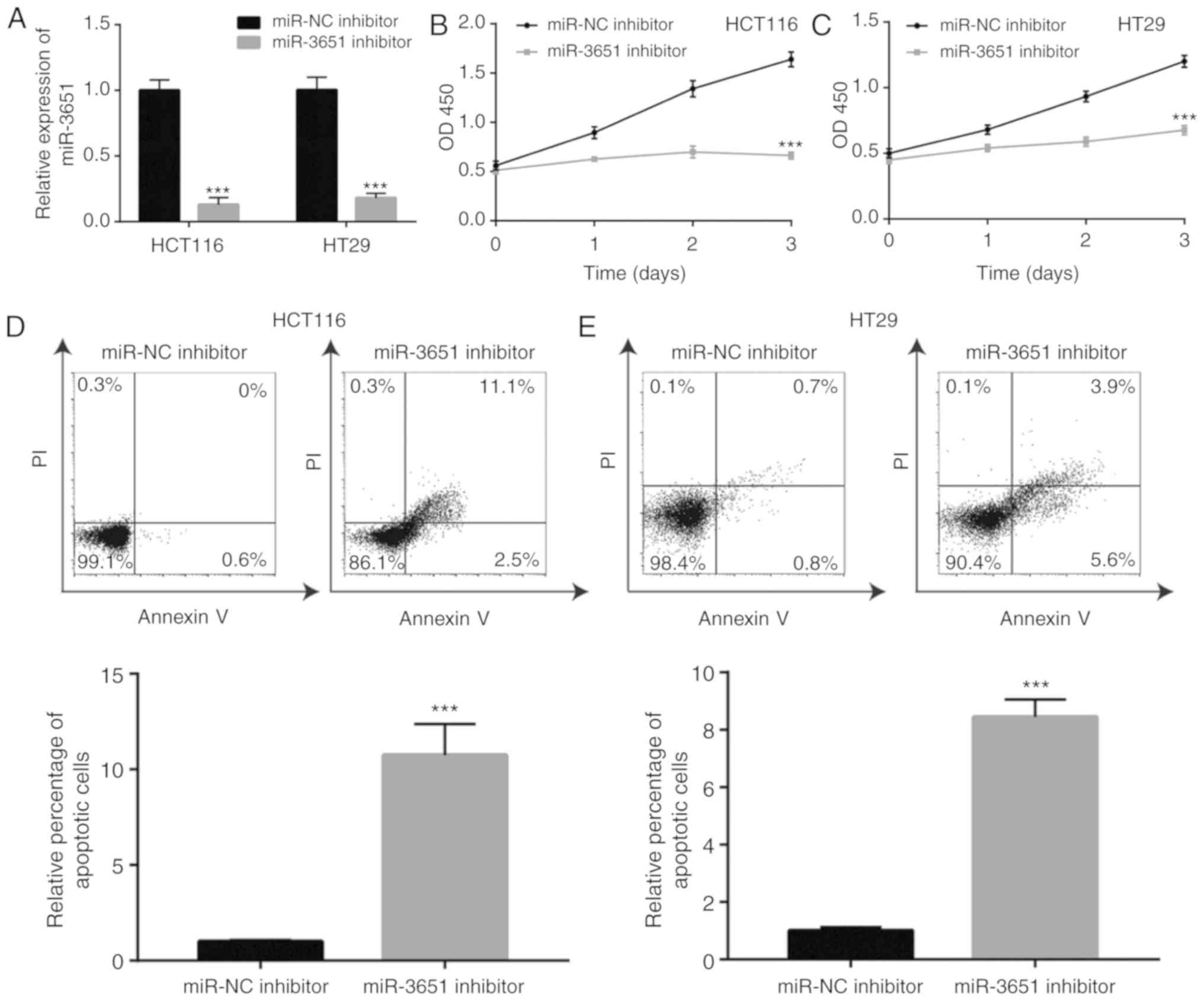
The current study then further assessed the
potential role of miR-3651 in colorectal cancer cells. A miR-3651
inhibitor or miR-NC inhibitor was transfected into colorectal
cancer cells, and the cell proliferation was detected. Transfection
with miR-3651 inhibitor induced an approximately 5-fold decrease in
miR-3651 expression in HCT116 and HT29 cells (Fig. 2A). In the cell proliferation
assay, the downregulation of miR-3651 significantly repressed cell
proliferation during the first 2 days of culture, with a
significant reduction in proliferation observed on day 3 in HCT116
cells (Fig. 2B). Similarly, the
downregulation of miR-3651 inhibited cell proliferation in HT29
cells (Fig. 2C). To examine
whether the cell growth arrest was associated with cell apoptosis,
flow cytometry was used to detect cell apoptosis rate in colorectal
cancer cells treated with miR-3651 inhibitor. The downregulation of
miR-3651 induced significant apoptosis in HCT116 cells (Fig. 2D), with similar results also
observed in HT29 cells (Fig. 2E).
These data indicated that miR-3651 inhibition reduced colorectal
cancer cell proliferation and promoted cell apoptosis.
Downregulation of miR-3651 deactivates
PI3K/AKT and MAPK/ERK signaling in colorectal cancer cells
The PI3K/AKT and MAPK/ERK signaling pathways have
been reported to be involved in colorectal cancer cell
proliferation and survival (20).
The results of western blot analysis demonstrated that the
downregulation of miR-3651 significantly decreased the ratio of
p-AKT/total (T)-AKT and p-ERK1/2/ T-ERK1/2, but did not alter the
expression levels of T-AKT and T-ERK1/2 in HCT116 cells, indicating
the deactivation of PI3K/AKT and MAPK/ERK signaling (Fig. 3A and B). A similar effect of
miR-3651 downregulation on the ratio of p-AKT/T-AKT and
p-ERK1/2/T-ERK1/2 was also observed in HT29 cells (Fig. 3C and D). Since the PI3K/AKT and
MAPK/ERK pathways regulate cell apoptosis by controlling the
expression of pro-apoptotic and anti-apoptotic proteins in cells
(25,26), the levels of such proteins were
subsequently investigated in the current study. The results
revealed that the expression of the anti-apoptotic protein Bcl2 was
significantly decreased, whereas the expression of the
pro-apoptotic protein Bax was significantly increased in HCT116 and
HT29 cells transfected with miR-3651 inhibitor (Fig. 3E and F). Taken together, these
results indicated that miR-3651 may be associated with mediating
the PI3K/AKT and MAPK/ERK pathways to sustain colorectal cancer
cell growth.
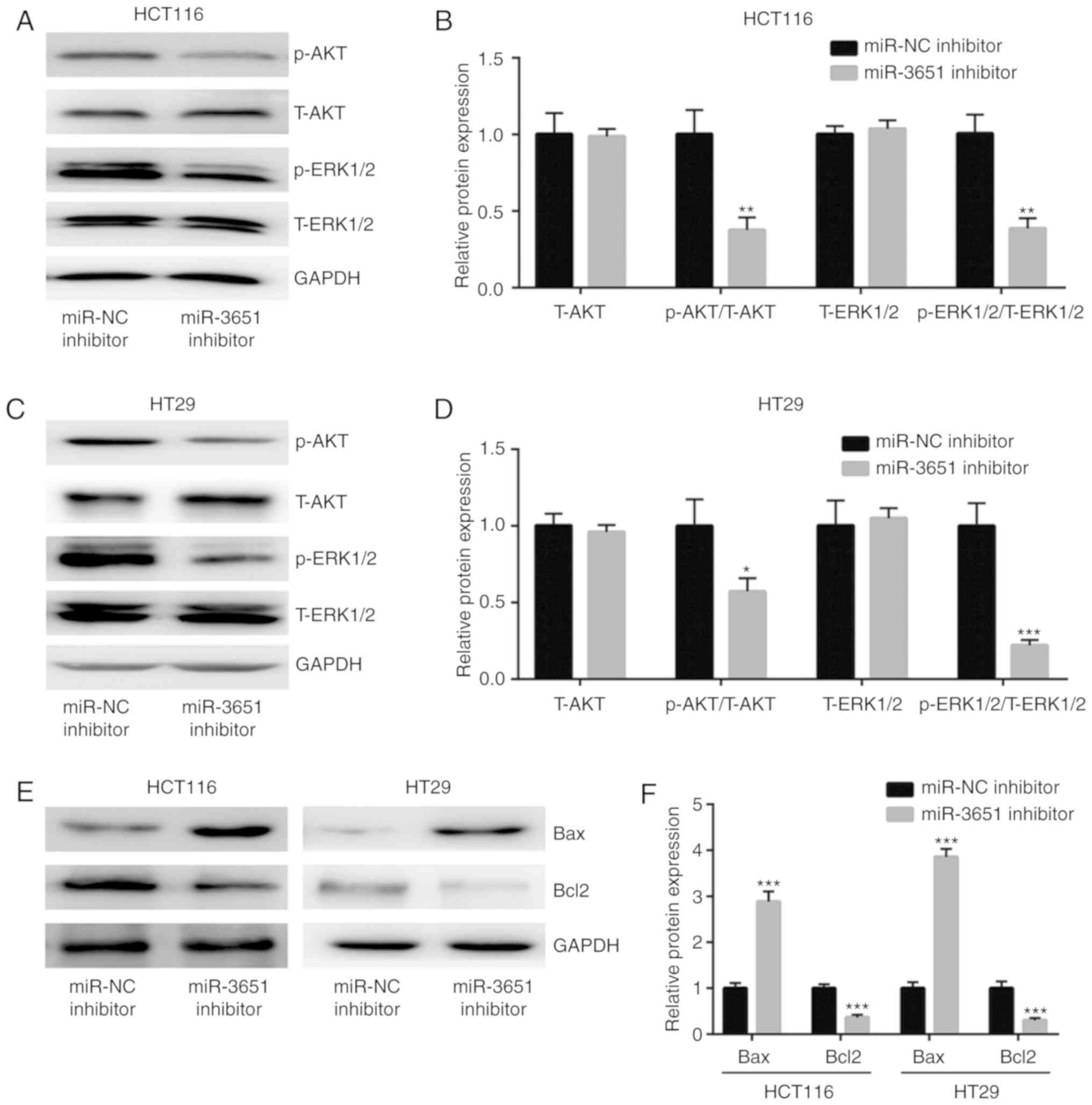 | Figure 3miR-3651 activated the PI3K/AKT and
MAPK/ERK signaling pathways in colorectal cancer cells. (A) Western
blots and (B) quantified protein expression results revealed that
miR-3651 downregulation decreased the ratio of p-AKT/T-AKT and
p-ERK1/2/T-ERK1/2 in HCT116 cells. (C) Western blots and (D)
quantified protein expression results revealed that miR-3651
downregulation decreased the ratio of p-AKT/T-AKT and
p-ERK1/2/T-ERK1/2 in HT29 cells. (E) Western blots and (F)
quantified results of Bcl2 and Bax protein expression indicated
that miR-3651 downregulation elevated Bax and decreased Bcl2 levels
in HCT116 and HT29 cells. *P<0.05,
**P<0.01 and ***P<0.001. miR, microRNA;
NC, negative control; PI3K, phosphoinositide-3 kinase; AKT, protein
kinase B; MAPK, mitogen-activated protein kinase; ERK,
extracellular signal-regulated kinase; p-, phosphorylated; T-,
total; Bcl2, B-cell lymphoma 2; Bax, Bcl2-associated X protein. |
miR-3651 directly represses TBX1
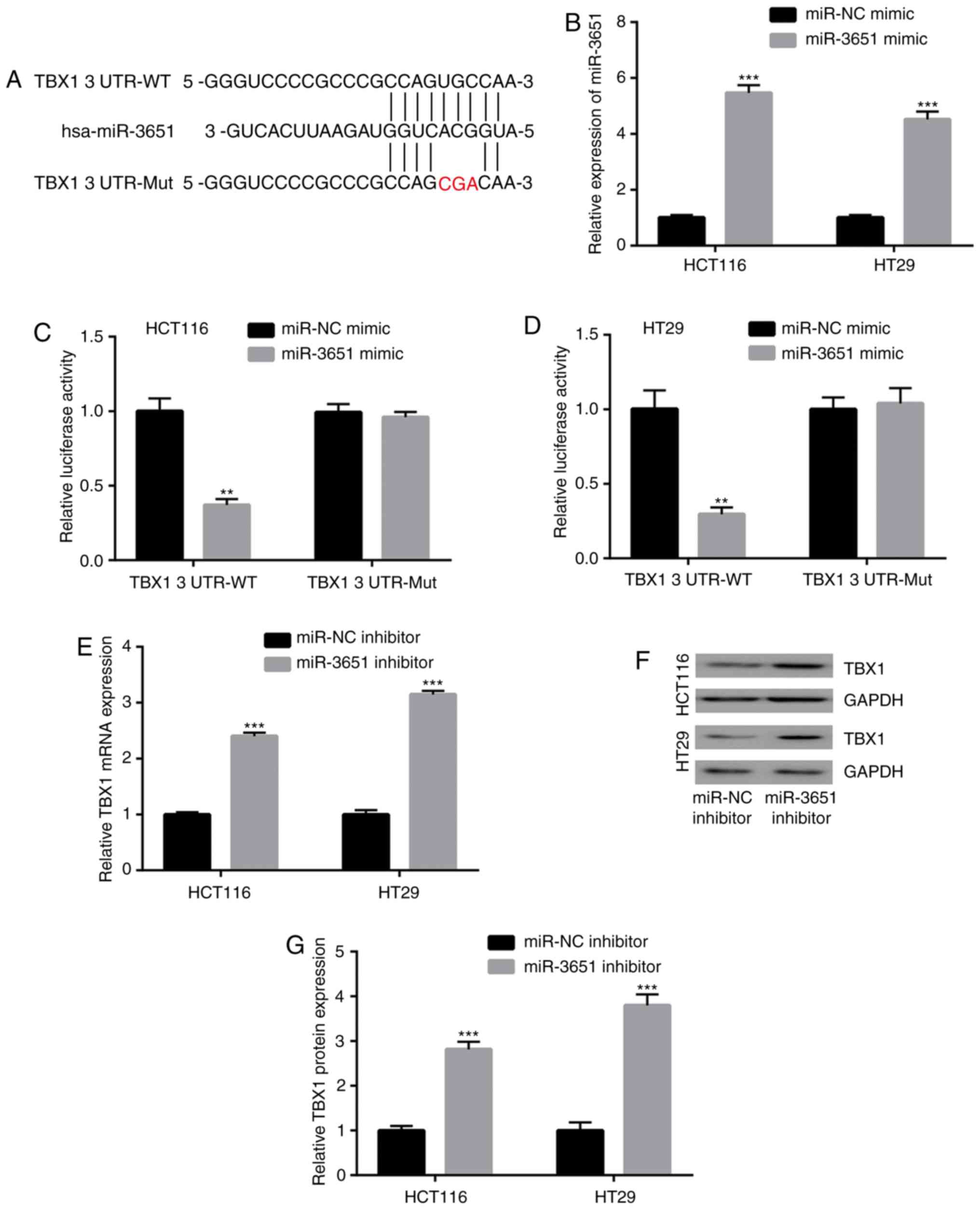
expression in colorectal cancer cells
miRNAs function through binding to the 3′UTR of
target gene mRNAs to inhibit the target gene expression (5). In the present study, the online tool
TargetScan was used to predict potential target genes of miR-3651.
Through a literature review, TBX1 was identified as a tumor
suppressor in colorectal cancer among the predicted target genes of
miR-3651, and the 3′UTR of TBX1 was observed to complementary bind
to miR-3651 (Fig. 4A). To explore
whether miR-3651 directly regulates TBX1 expression, miR-3651 mimic
was transfected in HCT116 and HT29 cells to upregulate miR-3651
expression (Fig. 4B). The
dual-luciferase reporter assay indicated that the overexpression of
miR-3651 significantly decreased the relative luciferase activity
in HCT116 cells co-transfected with TBX1 3′UTR-WT (Fig. 4C). Overexpression of miR-3651 had
a similar inhibitory effect on the relative luciferase activity of
HT29 cells co-transfected with TBX1 3′UTR-WT (Fig. 4D). However, miR-3651
overexpression did not alter the luciferase activity of cells
transfected with TBX1 3′UTR-Mut in the two cell lines. Furthermore,
RT-qPCR demonstrated that the downregulation of miR-3651
significantly elevated TBX1 mRNA levels in HCT116 and HT29 cells
(Fig. 4E). Western blot analysis
also indicated the elevation of TBX1 protein expression in HCT116
and HT29 cells (Fig. 4F and G).
These data suggested that TBX1 is a target gene of miR-3651 in
colorectal cancer cells.
miR-3651 regulates PI3K/AKT and MAPK/ERK
pathways via targeting TBX1 in colorectal cancer cells
TBX1 has previously been reported to be a regulator
of the PI3K/AKT and MAPK/ERK signaling pathways (20). To assess whether TBX1 was
associated with the regulation of these signaling pathways via
miR-3651, TBX1 expression was silenced via transfection of TBX1
small interfering RNA (siRNA) in HCT116 cells (Fig. 5A and B). The results revealed that
TBX1 silencing reversed the miR-3651 downregulation-induced
deactivation of the PI3K/AKT and MAPK/ERK signaling pathways
(Fig. 5C and D).
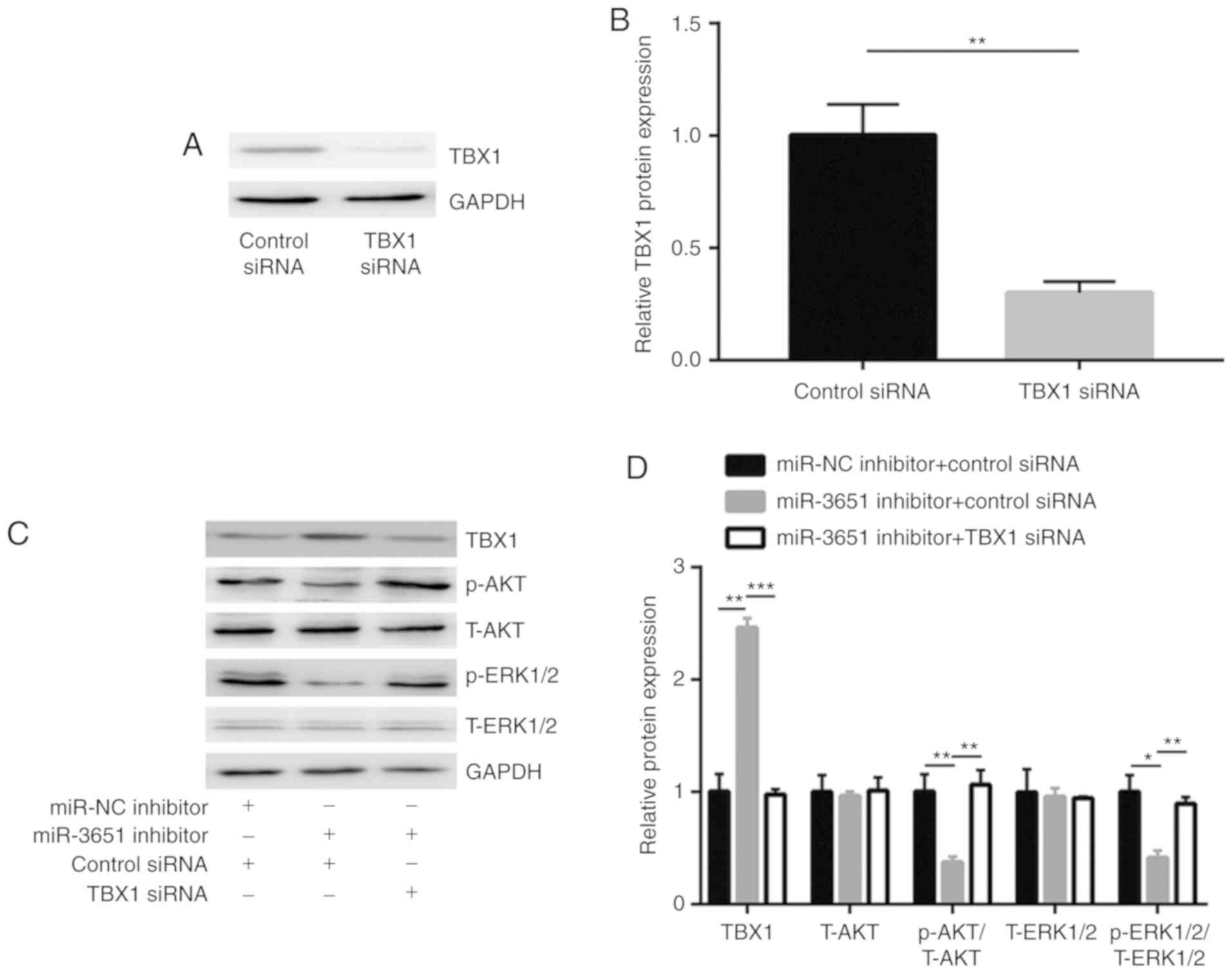 | Figure 5miR-3651 regulated PI3K/AKT and
MAPK/ERK pathways via repression of TBX1. (A) Western blots and (B)
quantified results, showing that transfection with TBX1 siRNA
decreased TBX1 protein expression in HCT116 cells. (C) Western
blots and (D) quantified protein expression levels, indicating that
miR-3651 downregulation in HCT116 cells increased TBX1 level, and
decreased the ratio of p-AKT/T-AKT and p-ERK1/2/T-ERK1/2, which was
then reversed by TBX1 silencing. *P<0.05,
**P<0.01 and ***P<0.001. miR, microRNA;
TBX1, T-box transcription factor 1; siRNA, small interfering RNA;
NC, negative control; PI3K, phosphoinositide-3 kinase; AKT, protein
kinase B; MAPK, mitogen-activated protein kinase; ERK,
extracellular signal-regulated kinase; p-, phosphorylated; T-,
total. |
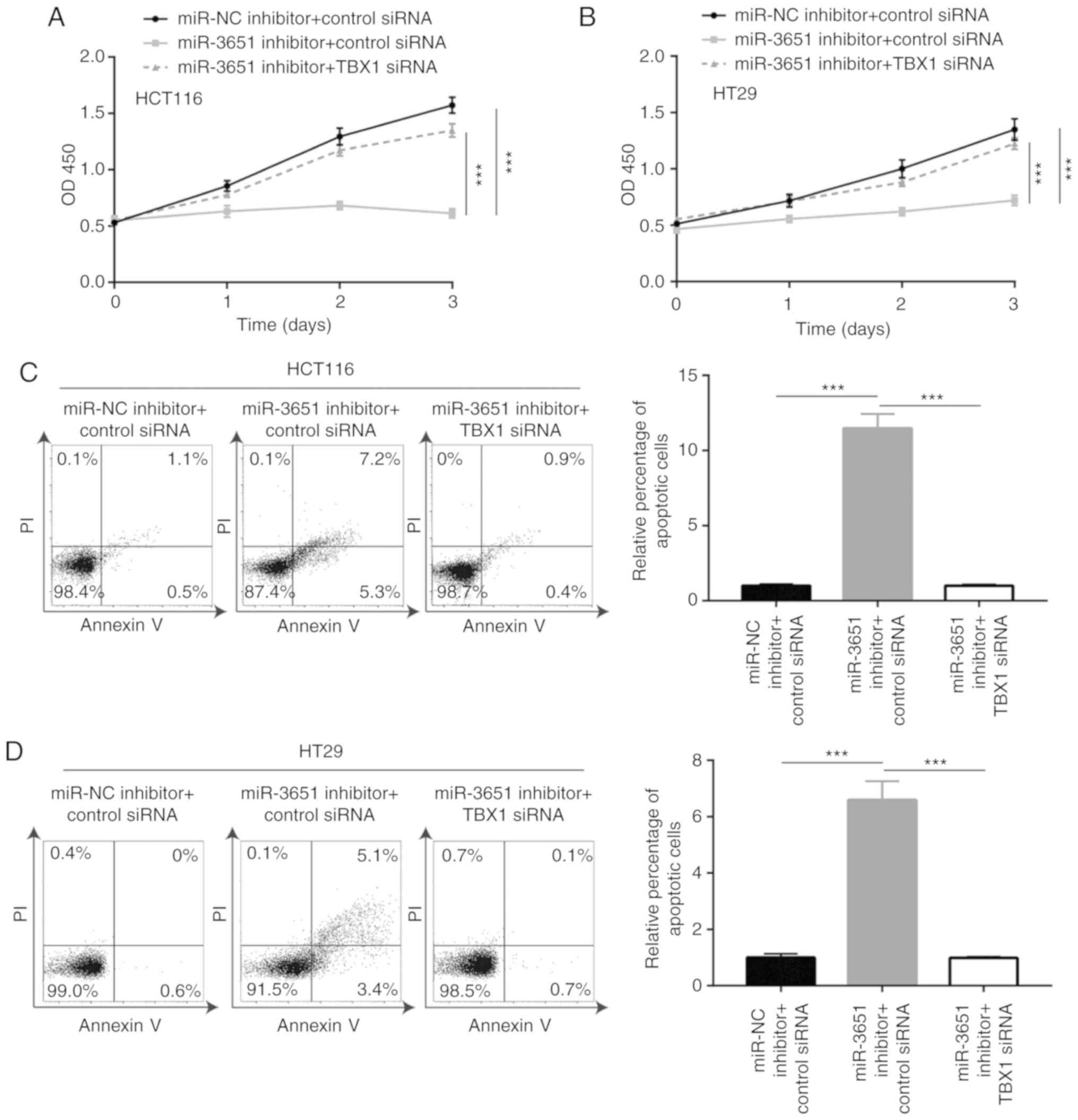
miR-3651 promotes colorectal cancer cell
proliferation and inhibits cell apoptosis via targeting TBX1
As miR-3651 directly repressed TBX1 expression to
activate the PI3K/AKT and MAPK/ERK signaling pathways, it was then
hypoth-esized that TBX1 is essential for miR-3651 function during
colorectal cancer cell proliferation. According to the cell
proliferation assay results, the silencing of TBX1 reversed the
miR-3651 downregulation-induced growth arrest in HCT116 cells
(Fig. 6A). Consistent with the
results observed in HCT116 cells, TBX1 siRNA also reversed the
miR-3651 downregulation-induced growth arrest in HT29 cells
(Fig. 6B). Furthermore, TBX1
silencing attenuated the elevation of apoptosis that was induced by
the miR-3651 inhibitor in HCT116 and HT29 cells (Fig. 6C and D). These results
collectively confirmed that miR-3651 regulated colorectal cancer
cell proliferation by directly targeting TBX1.
miR-3651 expression is negatively
correlated with TBX1 mRNA levels in colorectal tumor tissues
To further investigate the clinical relevance of
miR-3651 and TBX1, TBX1 mRNA expression was detected in the 40
pairs of normal and tumor tissues obtained from patients with
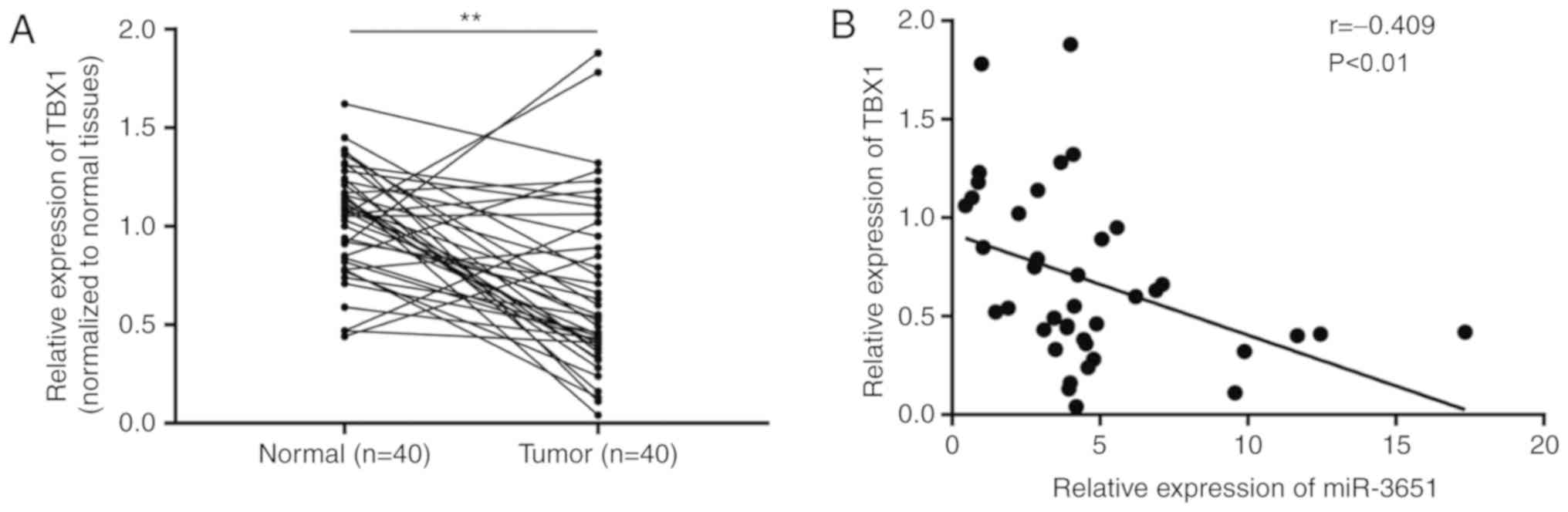
colorectal cancer. TBX1 was down-regulated in 77% (31/40) of
tumors, as compared with the paired normal tissues (Fig. 7A). In addition, Pearson
correlation analysis revealed that the miR-3651 expression was
negatively correlated with TBX1 mRNA expression in colorectal
tumors (Fig. 7B).
Discussion
Accumulating evidence has indicated that miRNAs
serve a role in colorectal cancer progression (27). For instance, microarray analysis
of tumors from 12 patients revealed that miR-224 was overexpressed
in colorectal adenoma and cancer, and that it regulated KRAS, AKT
and ERK signaling activity in colorectal cancer cells (28). Furthermore, in a large cohort of
1,893 carcinoma and normal paired samples, a number of upregulated
miRNAs (including miR-204-5p and miR-10a-5p) and downregulated
miRNAs (including miR-378a-5p and miR-145-5p) were identified in
colorectal cancer (11). Although
certain of these have been experimentally verified (29), the role of the majority of
differentially expressed miRNAs remains largely undetermined.
miR-3651 has been reported to be a prognostic biomarker in
esophageal squamous cell, oral squamous cell and hepatocellular
carcinomas (13,14,30). In another independent study that
was based on microarray analysis, miR-3651 was observed to be most
significantly upregulated in tumors from 51 patients with
colorectal cancer, and these results were confirmed using RT-qPCR
(31). In the current study,
bioinformatics analysis revealed high expression of miR-3651 in
colorectal cancer. Subsequently, the overexpression of miR-3651 in
tumor tissues obtained from 40 patients with colorectal cancer was
further validated. It was also observed that the downregulation of
miR-3651 inhibited the proliferation and induced the apoptosis of
colorectal cancer cells. The data gained in the present study
suggested a pro-survival and pro-proliferation role of miR-3651 in
colorectal cancer.
The MAPK/ERK and PI3K/AKT signaling pathways are
well-studied in cancer cells (32,33). In a variety of cancer types,
including colorectal cancer, the overactivation of MAPK/ERK and
PI3K/AKT pathways has been observed, and indicated to be important
for cell proliferation and resistance to apoptosis (34-36). In the present study, miR-3651
downregulation was observed to decrease p-ERK1/2 and p-AKT
expression levels, demonstrating that miR-3651 mediated the
activation of MAPK/ERK and PI3K/AKT pathways in colorectal cancer
cells. The inhibition of hyperactivated MAPK/ERK and PI3K/AKT
pathways has been demonstrated to lead to cancer cell growth arrest
and cell apoptosis (37,38). The current data suggested that
miR-3651 may regulate colorectal cancer cell proliferation and
apoptosis via the MAPK/ERK and PI3K/AKT pathways.
TBX1 is a transcription factor that mediates key
gene expression during development (22), and is a well-documented tumor
suppressor in a number of cancer types (20). TBX1 has been revealed to be
frequently decreased in papillary thyroid cancer tissues and
thyroid cancer cell lines due to the hypermethylation of the gene
promoter (20). In thyroid cancer
cells, TBX1 regulated cell cycle progression and the expression of
apoptosis-associated genes via the MAPK/ERK and PI3K/AKT pathways,
while its overexpression inhibited cancer cell proliferation,
migration and invasion accompanied with cell cycle arrest and
increased cell apoptosis (20).
Another study reported that TBX1 was decreased in the tumor tissues
of patients with basal cell carcinoma, and that it inhibited cell
proliferation and cell cycle progression (21). Furthermore, the overexpression of
TBX1 in a mouse spindle cell carcinoma cell line was associated
with cell growth inhibition and cell cycle arrest (39). In the current study, TBX1 was
predicted to be a target gene of miR-3651 using TargetScan
analysis. Similar to its function in thyroid cancer, it was
demonstrated that TBX1 also inactivated the MAPK/ERK and PI3K/AKT
pathways in colorectal cancer cells. Additionally, miR-3651
regulated the MAPK/ERK and PI3K/AKT pathways via the downregulation
of TBX1. TBX1 was previously reported to be a target gene of
miR-17-92 cluster, miR-182 and miR-451a in different cell types
(21,40,41). In the present study, the results
of the dual-luciferase assay also indicated that TBX1 was a target
gene of miR-3651 in colorectal cancer cells. Furthermore, miR-3651
exerted its pro-proliferation function by targeting TBX1 in
colorectal cancer cells. These results collectively demonstrated
the oncogenic potential of miR-3651 in colorectal cancer.
In conclusion, the current study indicated that
miR-3651 directly targeted TBX1 to activate the MAPK/ERK and
PI3K/AKT pathways, facilitated cell proliferation and inhibited
cell apoptosis in colorectal cancer cells. These results provided
an insight into the role of miR-3651 in colorectal cancer and the
potential application of this miRNA in colorectal cancer
therapy.
Funding
The present study was approved by the National
Natural Science Foundation of China (grant no. 81872323) and the
Science and Technology Department of Jilin Province (grant no.
820190303149SF).
Availability of data and materials
The data are available under special request.
Authors' contributions
CL, YG and YL performed clinical sample collection.
DD designed and supervised the study. DD, YL, CL and YG performed
the data acquisition and analysis. YL and DD prepared, edited and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent prior
to participation in the study. The Ethic Committee of China-Japan
Union Hospital of Jilin University supervised the protocol of the
experiments and approved the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hansen RM: Systemic therapy of colorectal
cancer. Wis Med J. 87:27–29. 1988.PubMed/NCBI
|
|
3
|
Moriarity A, O'Sullivan J, Kennedy J,
Mehigan B and McCormick P: Current targeted therapies in the
treatment of advanced colorectal cancer: A review. Ther Adv Med
Oncol. 8:276–293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yates L, Norbury C and Gilbert RC: The
long and short of MicroRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rasnic R, Linial N and Linial M: Enhancing
identification of cancer types via lowly-expressed microRNAs.
Nucleic Acids Res. 45:5048–5060. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma Y, Zhang P, Wang F, Zhang H, Yang Y,
Shi C, Xia Y, Peng J, Liu W, Yang Z and Qin H: Elevated oncofoetal
miR-17-5p expression regulates colorectal cancer progression by
repressing its target gene P130. Nat Commun. 3:12912012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
9
|
Imaoka H, Toiyama Y, Fujikawa H, Hiro J,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, et al:
Circulating microRNA-1290 as a novel diagnostic and prognostic
biomarker in human colorectal cancer. Ann Oncol. 27:1879–1886.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ng L, Wan TM, Man JH, Chow AK, Iyer D,
Chen G, Yau TC, Lo OS, Foo DC, Poon JT, et al: Identification of
serum miR-139-3p as a non-invasive biomarker for colorectal cancer.
Oncotarget. 8:27393–27400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Slattery ML, Herrick JS, Pellatt DF,
Stevens JR, Mullany LE, Wolff E, Hoffman MD, Samowitz WS and Wolff
RK: MicroRNA profiles in colorectal carcinomas, adenomas and normal
colonic mucosa: Variations in miRNA expression and disease
progression. Carcinogenesis. 37:245–261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang G, Li J, Sun B, Li S, Lü L, Wang Y,
Chen B and Xiao Z: Deep sequencing reveals complex mechanisms of
microRNA deregulation in colorectal cancer. Int J Oncol.
45:603–610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang C, Guan S, Chen X, Liu B, Liu F, Han
L, Un Nesa E, Song Q, Bao C, Wang X and Cheng Y: Clinical potential
of miR-3651 as a novel prognostic biomarker for esophageal squamous
cell cancer. Biochem Biophys Res Commun. 465:30–34. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ries J, Vairaktaris E, Agaimy A, Kintopp
R, Baran C, Neukam FW and Nkenke E: miR-186, miR-3651 and miR-494:
Potential biomarkers for oral squamous cell carcinoma extracted
from whole blood. Oncol Rep. 31:1429–1436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lindsay EA, Vitelli F, Su H, Morishima M,
Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler
PJ, et al: Tbx1 haploinsufficieny in the DiGeorge syndrome region
causes aortic arch defects in mice. Nature. 410:97–101. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen L, Fulcoli FG, Tang S and Baldini A:
Tbx1 regulates proliferation and differentiation of multipotent
heart progenitors. Circ Res. 105:842–851. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu H, Viola A, Zhang Z, Gerken CP,
Lindsay-Illingworth EA and Baldini A: Tbx1 regulates population,
proliferation and cell fate determination of otic epithelial cells.
Dev Biol. 302:670–682. 2007. View Article : Google Scholar :
|
|
18
|
Botchkarev VA and Fessing MY: Edar
signaling in the control of hair follicle development. J Investig
Dermatol Symp Proc. 10:247–251. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ryan AK, Goodship JA, Wilson DI, Philip N,
Levy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B,
Prieur M, et al: Spectrum of clinical features associated with
interstitial chromosome 22q11 deletions: A European collaborative
study. J Med Genet. 34:798–804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang N, Li Y, Wei J, Pu J, Liu R, Yang Q,
Guan H, Shi B, Hou P and Ji M: TBX1 functions as a tumor suppressor
in thyroid cancer through inhibiting the activities of the PI3K/AKT
and MAPK/ERK pathways. Thyroid. 29:378–394. 2019. View Article : Google Scholar
|
|
21
|
Sun H and Jiang P: MicroRNA-451a acts as
tumor suppressor in cutaneous basal cell carcinoma. Mol Genet
Genomic Med. 6:1001–1009. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hari DM, Leung AM, Lee JH, Sim MS, Vuong
B, Chiu CG and Bilchik AJ: AJCC cancer staging manual 7th edition
criteria for colon cancer: Do the complex modifications. J Am Coll
Surg. 217:181–190. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu C, Chen X, Wang Q, Xu X and Xu B: TNFα
promotes glio-blastoma A172 cell mitochondrial apoptosis via
augmenting mitochondrial fission and repression of MAPK-ERK-YAP
signaling pathways. Onco Targets Ther. 11:7213–7227. 2018.
View Article : Google Scholar :
|
|
25
|
Baldini A, Fulcoli FG and Illingworth E:
Tbx1: Transcriptional and developmental functions. Curr Top Dev
Biol. 122:223–243. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Moridikia A, Mirzaei H, Sahebkar A and
Salimian J: MicroRNAs: Potential candidates for diagnosis and
treatment of colorectal cancer. J Cell Physiol. 233:901–913. 2018.
View Article : Google Scholar
|
|
28
|
Amankwatia EB, Chakravarty P, Carey FA,
Weidlich S, Steele RJ, Munro AJ, Wolf CR and Smith G: MicroRNA-224
is associated with colorectal cancer progression and response to
5-fluorouracil-based chemotherapy by KRAS-dependent and
-independent mechanisms. Br J Cancer. 112:1480–1490. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li S, Wu X, Xu Y, Wu S, Li Z, Chen R,
Huang N, Zhu Z and Xu X: miR-145 suppresses colorectal cancer cell
migration and invasion by targeting an ETS-related gene. Oncol Rep.
36:1917–1926. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu HR, Huang RZ, Yu XN, Shi X,
Bilegsaikhan E, Guo HY, Song GQ, Weng SQ, Dong L, Janssen HLA, et
al: Microarray expression profiling of microRNAs reveals potential
biomarkers for hepatocellular carcinoma. Tohoku J Exp Med.
245:89–98. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Della Vittoria Scarpati G, Calura E, Di
Marino M, Romualdi C, Beltrame L, Malapelle U, Troncone G, De
Stefano A, Pepe S, De Placido S, et al: Analysis of differential
miRNA expression in primary tumor and stroma of colorectal cancer
patients. Biomed Res Int. 2014:8409212014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Burotto M, Chiou VL, Lee JM and Kohn EC:
The MAPK pathway across different malignancies: A new perspective.
Cancer. 120:3446–3456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Araujo WM, Robbs BK, Bastos LG, de
Souza WF, Vidal FC, Viola JP and Morgado-Diaz JA: PTEN
overexpression cooperates with lithium to reduce the malignancy and
to increase cell death by apoptosis via PI3K/AKT suppression in
colorectal cancer cells. J Cell Biochem. 117:458–469. 2016.
View Article : Google Scholar
|
|
35
|
Tian XQ, Guo FF, Sun DF, Wang YC, Yang L,
Chen SL, Hong J and Fang JY: Downregulation of ZNF278 arrests the
cell cycle and decreases the proliferation of colorectal cancer
cells via inhibition of the ERK/MAPK pathway. Oncol Rep.
38:3685–3692. 2017.PubMed/NCBI
|
|
36
|
Zhou Q, Chen J, Feng J, Xu Y, Zheng W and
Wang J: SOSTDC1 inhibits follicular thyroid cancer cell
proliferation, migration, and EMT via suppressing PI3K/Akt and
MAPK/Erk signaling pathways. Mol Cell Biochem. 435:87–95. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vakana E, Pratt S, Blosser W, Dowless M,
Simpson N, Yuan XJ, Jaken S, Manro J, Stephens J, Zhang Y, et al:
LY3009120, a panRAF inhibitor, has significant anti-tumor activity
in BRAF and KRAS mutant preclinical models of colorectal cancer.
Oncotarget. 8:9251–9266. 2017. View Article : Google Scholar :
|
|
38
|
Soo HC, Chung FF, Lim KH, Yap VA, Bradshaw
TD, Hii LW, Tan SH, See SJ, Tan YF, Leong CO and Mai CW:
Cudraflavone C induces tumor-specific apoptosis in colorectal
cancer cells through inhibition of the phosphoinositide 3-kinase
(PI3K)-AKT pathway. PLoS One. 12:e01705512017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Trempus CS, Wei SJ, Humble MM, Dang H,
Bortner CD, Sifre MI, Kissling GE, Sunman JA, Akiyama SK, Roberts
JD, et al: A novel role for the T-box transcription factor Tbx1 as
a negative regulator of tumor cell growth in mice. Mol Carcinog.
50:981–991. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang J, Bai Y, Li H, Greene SB, Klysik E,
Yu W, Schwartz RJ, Williams TJ and Martin JF: MicroRNA-17-92, a
direct Ap-2α transcriptional target, modulates T-box factor
activity in orofacial clefting. PLoS Genet. 9:e10037852013.
View Article : Google Scholar
|
|
41
|
Wang XR, Zhang XM, Du J and Jiang H:
MicroRNA-182 regulates otocyst-derived cell differentiation and
targets T-box1 gene. Hear Res. 286:55–63. 2012. View Article : Google Scholar : PubMed/NCBI
|