Introduction
Liver cancer is a malignant tumor type that
seriously endangers human life and health, and its morbidity and
mortality rates continue to rise faster compared with those of
other cancer types (1). China is
a high-risk area for liver cancer, accounting for ~50% of new
patients and liver cancer-associated mortalities globally each year
(2). The occurrence and
development of liver cancer are known to utilize a complex
evolutionary process involving the accumulation of multiple
factors, stages and gene variations (3). Identifying the genes that serve key
functions in the development of liver cancer and the associated
molecular mechanisms are prerequisites for the treatment of liver
cancer. To identify the key genes affecting the development of
liver cancer, a previous study proposed a research strategy based
on the cross-species screening of early-stage disease, and CDC25A
was revealed to be highly expressed in liver cancer (4).
As an important member of the CDC25 family, CDC25A
is a bispecific protein phosphatase that hydrolyses tyrosine and
serine/threonine residues for dephosphorylation (5). CDC25A activates the
cyclincyclin-dependent kinase complex, which promotes the
transition of the G1/S and G2/M phases and is an important target
for DNA damage response (6,7).
CDC25A, which has the characteristics of a proto-oncogene, is known
to be highly expressed in various malignant tumor types, including
lung cancer (8), breast cancer
(9) and esophageal cancer
(10), and is closely associated
with poor patient prognosis. To further investigate the molecular
mechanism of CDC25A in liver cancer, an Affymetrix human gene
expression profiling chip was used to analyze the differentially
expressed genes in HepG2 cells with the CDC25A gene silenced. Based
on ingenuity pathway analysis, signaling pathways were revealed to
be enriched among the genes with significant differential
expression, and the interleukin (IL)-6 signaling pathway was
revealed to be significantly inhibited.
IL-6 is a cytokine produced by various cells and
serves important functions in information transmittance, immune
cell activation and regulation, T and B cell activation,
proliferation and differentiation and inflammatory responses
(11). Previously, one study
demonstrated that IL-6 is closely associated with the occurrence,
development and metastasis of various malignant tumor types, which
may be associated with its antiapoptotic and proangiogenic effects
(12). IL-6 has been confirmed to
affect liver cancer proliferation and serves an important function
in the development and recurrence of liver cancer (13). IL-6 is a major inducer of signal
transducer and activator of transcription 3 (STAT3) phosphorylation
and activation. IL-6 binds to glycoprotein 130 and induces Janus
kinase phosphorylation, which results in the phosphorylation of
Tyr705 in STAT3, the promotion of STAT3 binding to DNA and the
activation of certain genes downstream of STAT3 to promote the
development of liver cancer (14,15).
Nuclear factor-κB (NF-κB), an important
transcriptional regulator, participating in the bodily inflammatory
response, the immune response, cell apoptosis, differentiation,
regeneration and other important physiological and pathological
processes. NF-κB is an important molecular marker linking the
inflammatory response with tumors (16,17). Activated NF-κβ is involved in cell
proliferation, increases the expression of inflammation-associated
factors, inhibits apoptosis and promotes the malignant
transformation, invasion and metastasis of a tumor (18,19). One study confirmed that NF-κB
serves an important function in the induction of IL-6 expression
(20). Interestingly, CDC25A has
been revealed to positively regulate NF-κB, inhibit apoptosis and
promote tumor cell survival (21). Therefore, the present study
hypothesized that CDC25A may regulate the expression of IL-6
through the NF-κB gene, further affecting the occurrence and
development of liver cancer.
The present study investigated the molecular
mechanism of CDC25A in the development of liver cancer, and the
results may provide a novel theoretical basis for the treatment of
liver cancer based on the utilization of CDC25A as a target.
Materials and methods
Cell culture
HepG2 cells (human liver cancer cell line) were
purchased from the Shanghai Cell Biology Institute and cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a 5% CO2 incubator.
Cell transfection
Lentiviral particles [LV-short hairpin RNA
(shRNA)-CDC25A] for silencing the CDC25A gene and negative control
empty lentiviral particles (LV-shRNA-NC) were obtained from
Shanghai GeneChem Co., Ltd., and the lentiviral interference
sequences were 5′-CAT GCA CCA CGA GGA CTT T-3′ and 5′-TTC TCC GAA
CGT GTC ACG T-3′, respectively. The two lentiviruses contained a
green fluorescent protein (GFP) marker. Cells were divided into the
following three groups: LV-shRNA-CDC25A-transfected cells
(CDC25A-shRNA group), LV-shRNA-NC-transfected cells (CDC25A-NC
group) and untransfected cells (control group). There were 3
replicate wells per group. Cells in the logarithmic growth phase
were plated into 6-well plates at a density of 5×104/ml
in 2 ml per well, containing the virus particles, polybrene (a
final concentration of 5 g/ml; Shanghai GeneChem Co., Ltd.) and the
enhanced infection solution (ENi.s). Once the cells reached 20-30%
confluence, they were transfected at a multiplicity of infection of
30. The GFP positive expression rate was observed at 24, 48 and 72
h after infection under a fluorescence inverted microscope (Olympus
Corporation). The cell transfection efficiencies were detected by
RT-qPCR and western blot analyses.
Cell proliferation assay
MTT Cell Proliferation and Cytotoxicity Assay kit
(Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China) was used to detect cell proliferation, according to the
manufacturers protocol. The cell suspension was seeded in a 96-well
plate at a density of 2×104/ml (100 µl total).
Each group was placed in 5 wells. The cells were cultured at 37°C
in a 5% CO2 incubator. On days 1, 2, 3, 4 and 5, cells
were incubated with 10 µl 5 mg/ml MTT solution for 4 h at
37°C. Next, 100 µl dimethyl sulfoxide was added to each
well, and the plates were shaken for 5-10 min. The absorbance at
490 nm was analyzed using a microplate reader (Thermo Fisher
Scientific, Inc.) to plot the growth curve.
Cell migration assay
Cell migration was conducted in a Transwell chamber.
Subsequent to resuspending the cells in RPMI-1640 medium, 100
µl cell suspension (1×105 cells) was added to the
upper chamber, and 30% FBS in RPMI-1640 medium was added to the
lower chamber. The plate was placed at 37°C in a 5% CO2
incubator for 24 h. Next, the cells that did not pass through the
polycarbonate membrane were removed with a cotton swab, and the
cells on the lower surface were stained with 1% Giemsa (Beijing
Solarbio Science & Technology Co., Ltd.) at room temperature
for 20 min, rinsed with phosphate buffered saline (PBS), air-dried,
and counted under an inverted microscope (Olympus Corporation).
Cell cycle assay
Single-cell suspensions from the CDC25A-shRNA
CDC25A-NC and control groups (1×106 cells/ml in ice-cold
PBS) were prepared, and then subsequently fixed with 70% ethanol at
4°C for 2 h. Subsequent to treatment with RNase A (10 µg/ml)
for 30 min at 37°C, the cells were resuspended in 500 µl
propidium iodide solution. Cell cycle distribution was analyzed by
a FACS Calibur flow cytometer (BD Biosciences).
Gene chip screening
Gene chip screening was performed as described in
our previous study (22).
Construction of the liver cancer
xenograft model
A total of 30 female BALB/c nude mice (weighing
18-24 g and aged 4-6 weeks) were purchased from Beijing Vital River
Laboratory Animal Technology Co., Ltd. (Beijing, China). The study
was ethically approved and supervised by the Ethics Committee of
Guangxi Medical University Affiliated Tumor Hospital (approval no.
LW2019045). The mice were housed in a specific pathogen-free animal
room at 22-24°C with a humidity of 55-70% and a 12/12 h light/dark
cycle at the Experimental Animal Center of Guangxi Medical
University, and all animals were provided ad libitum access
to standard laboratory feed and water. Their health and behavior
were monitored every 5 days. The nude mice were randomly divided
into the following three groups (n=10): The knockdown (KD), NC and
control groups. The KD group was HepG2 cells transfected with
LV-shRNA-CDC25A, the NC group was HepG2 cells transfected with
LV-shRNA-NC, and the control group was untreated cells. All cells
were resus-pended at a concentration of 1×107/ml, and
cell suspensions (200 µl) were subcutaneously injected into
the right axilla (0.5 cm) of the nude mice. The tumor volume was
observed and recorded every 7 days. When the animals exhibited a
loss of appetite and inability to eat, clinical symptoms of severe
loss of organ function or when the tumor was observed to be
ulcerated, infected or necrotic, the animals were euthanized.
Following 35 days, the nude mice were euthanized by cervical
dislocation, then the breathing, corneal reflex and heartbeat were
observed to determine euthanasia. The criteria for verifying animal
mortality are no breathing, no heartbeat and no corneal reflex. The
liver cancer xenograft tumors were then harvested. The tumor volume
formula used was as follows: Volume
(mm3)=width2 (mm2) x
length(mm)/2.
Hematoxylin and eosin (H&E)
staining
Xenograft tumors were fixed with 4% paraformaldehyde
at room temperature for 24 h, embedded in paraffin and serially
sectioned at a thickness of 5 µm. The sections were dewaxed
and stained with H&E using a staining kit (cat. no. c0105;
Beyotime Institute of Biotechnology), and visualized using a phase
contrast microscope (Olympus Corporation) at a magnification of
×200.
RNA extraction and RT-qPCR
Total RNA was extracted from the liver cancer cells
and xenograft tumors using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.), and cDNA was synthesized using the PrimeScript
RT Reagent kit (Takara Bio, Inc.), according to the manufacturer's
protocol. qPCR was performed with a qTOWER3G PCR
instrument (Analytik Jena AG) and SYBR® Premix Ex Taq™
(Takara Bio, Inc.), according to the manufacturer's protocol. The
reaction conditions were as follows: Predenaturation at 95°C for 30
sec, denaturation at 95°C for 5 sec and annealing at 60°C for 30
sec for a total of 40 cycles. The results obtained were analyzed
using the 2−ΔΔCq method (23). The primers used were as follows:
CDC25A forward, 5′-TTC CTC TTT TTA CAC CCC AGT CA-3′ and reverse,
5′-TCG GTT GTC AAG GTT TGT AGT TC-3′; IL-6 forward, 5′-ACT CAC CTC
TTC AGA ACG AAT TG-3′ and reverse, 5′-CCA TCT TTG GAA GGT TCA GGT
TG-3′; IL-1β forward, 5′-GCC AGT GAA ATG ATG GCT TAT T-3′ and
reverse, 5′-AGG AGC ACT TCA TCT GTT TAG G-3′; NIK forward, 5′-AGG
AGA AGA CGC CGC CA C TG-3′ and reverse, 5′-TGC CTC GGA GCC TTC CTT
GG-3′; NF-κB forward, 5′-CCC ACG AGC TTG TAG GAA AGG-3′ and
reverse, 5′-GGA TTC CCA GGT TCT GGA AAC-3′; GAPDH forward, 5′-TGA
CTT CAA CAG CGA CAC CCA-3′ and reverse, 5′-CAC CCT GTT GCT GTA GCC
AAA-3′.
Western blot analysis
The cells and xenograft tumors were lysed with
RIPA-phenylmethylsulfonyl fluoride buffer (cat. no. P0013K;
Beyotime Institute of Biotechnology) to extract the total protein,
and the protein concentration was determined using the BCA method.
Protein buffer was added to the samples and denatured at 100°C.
Each well was loaded with 50 µg proteins. Then, the samples
were electrophoresed on a 10% SDS-PAGE gel at 60V for 150 min,
transferred to a 0.22-µm-pore size poly-vinyline difluoride
(PVDF) membrane, and blocked with 5% skimmed milk at room
temperature for 2 h. The membranes were washed and incubated while
shaking overnight at 4°C with anti- CDC25A (1:5,000; rabbit; cat.
no. ab202485; Abcam), anti-IL-6 (1:1,000; rabbit; cat. no.
ab233706; Abcam), anti-IL-1β (1:1,500; rabbit; cat. no. ab2105;
Abcam), anti-NIK (1:1,000; rabbit; cat. no. ab155583; Abcam),
anti-NF-κB (1:1,000; rabbit; cat. no. ab32536; Abcam), anti-β-actin
(1:1,000; rabbit; cat. no. ab8227; Abcam) and anti-GAPDH (1:1,000;
mouse; cat. no. ab8245) antibodies. Subsequent to washing, the
membranes were incubated with the appropriate horseradish
peroxidase-conjugated secondary antibody (1:1,000; goat anti-rabbit
or anti-mouse antibody; cat. nos. A0208 and A0216, respectively;
Beyotime Institute of Biotechnology) for 1 h at room temperature.
The PVDF membranes were scanned with an enhanced chemiluminescence
detection system ChemiDoc MP (Bio-Rad Laboratories, Inc.) and the
densitometry was performed using Image Lab software (version 3.0;
Bio-Rad Laboratories, Inc.).
Immunohistochemistry
The sections at a thickness of 5 µm were
dewaxed, immersed in sodium citrate buffer, incubated at 100°C for
2 min in an autoclave, and then rinsed 3 times with PBS to retrieve
the antigens. Then, the sections were placed in 3%
H2O2 and incubated at room temperature for 25
min in the dark to block the endogenous peroxide. The sections were
sequentially incubated overnight with rabbit anti-human CDC25A
(1:2,000; cat. no. GB11283; Wuhan Servicebio Technology Co., Ltd.)
and rabbit anti-human IL-6 (1:600; cat. no. GB11117; Wuhan
Servicebio Technology Co., Ltd.) antibodies at 4°C, and then with a
goat anti-rabbit immunoglobulin G solution (1:200; cat. no. G23303;
Wuhan Servicebio Technology Co., Ltd.) for 50 min at room
temperature. The immunocomplexes were visualized using
3,3′-diaminoben-zidine and a light microscope (Olympus Corporation)
at a magnification of ×200. Two blinded pathologists independently
assessed all specimens. The staining intensity was scored as
follows: 0 (negative); 1 (weak); 2 (moderate); and 3 (strong). The
positive range scores were defined as follows: 0 (0-20%); 1
(21-50%); 2 (51-80%); and 3 (81-100%). The final score was obtained
by multiplying the intensity score and the positive range score,
and a score ³4 was regarded as high expression.
Statistical analysis
Analyses were performed using SPSS 19.0 statistical
software (IBM Corp.). Differences among the CDC25A-shRNA, CDC25A-NC
and control groups or among the KD, NC and control groups were
analyzed using one-way analysis of variance followed by
Student-Newman-Keuls. Correlational analysis was performed with
Pearson's correlation coefficient. Measurement data are expressed
as the mean ± standard deviation. P<0.05 was considered to
indicate a statistically significant difference.
Results
Efficiency of lentiviral infection in
HepG2 cells
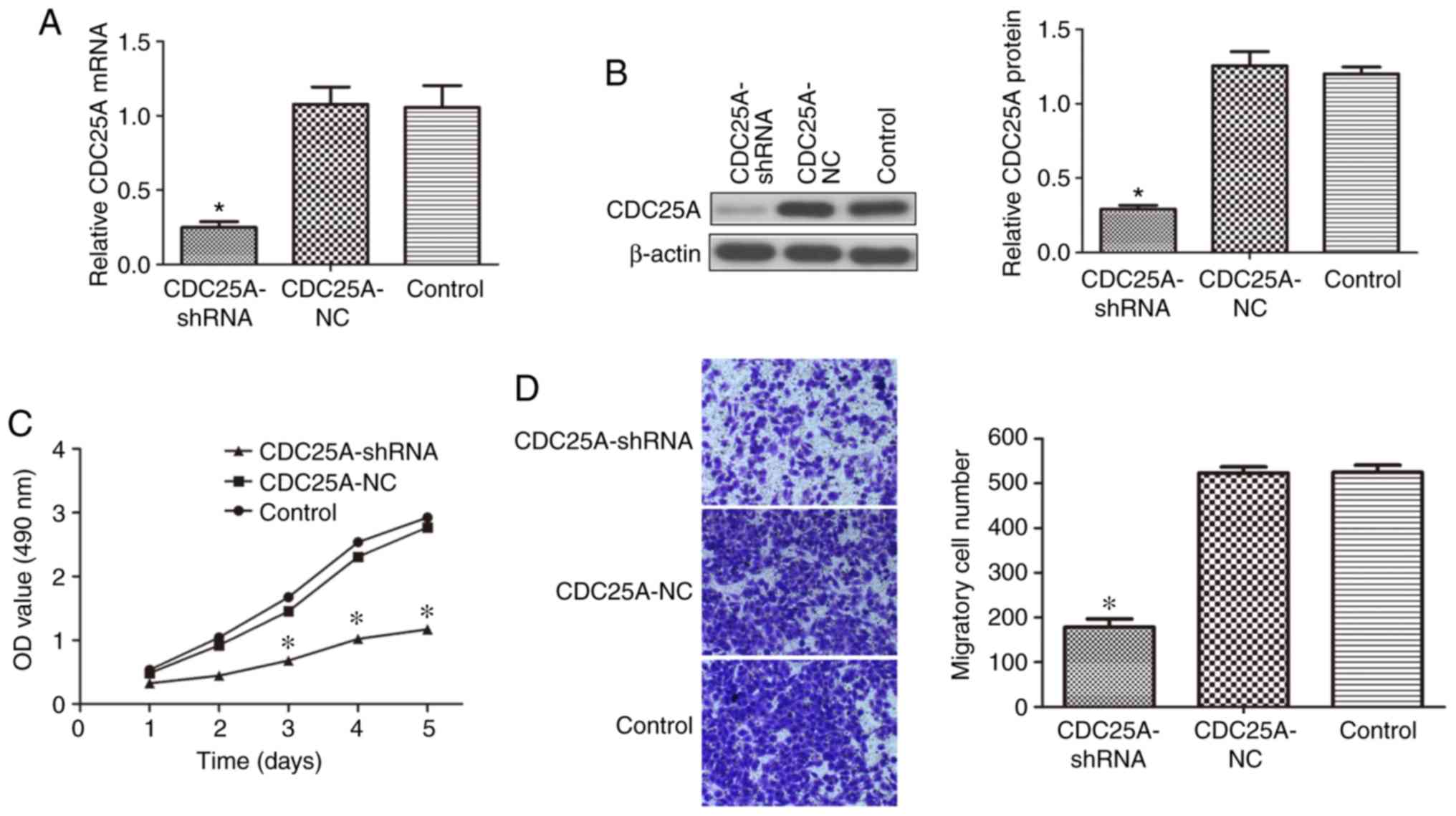
The expression of GFP was detected after 48 h.
Following transfection, the mRNA and protein expression levels of
CDC25A in the CDC25A-shRNA group were significantly lower compared
with those in the CDC25A-NC and control groups (P<0.05; Fig. 1A and B).
Effect of silencing CDC25A on liver
cancer cell growth
The effect of silencing CDC25A on the growth of
HepG2 cells was confirmed by an MTT assay, revealing that the
proliferation of HepG2 cells was significantly decreased in
LV-shRNA-CDC25A transfected cells compared with the CDC25A-NC and
control groups (P<0.05; Fig.
1C).
Effect of silencing CDC25A on liver
cancer cells migration
Migration experiments revealed that significantly
fewer cells passed through the membrane in the CDC25A-shRNA group
compared with in the CDC25A-NC and control groups (P<0.05;
Fig. 1D), and no significant
difference was observed between the CDC25A-NC and control
groups.
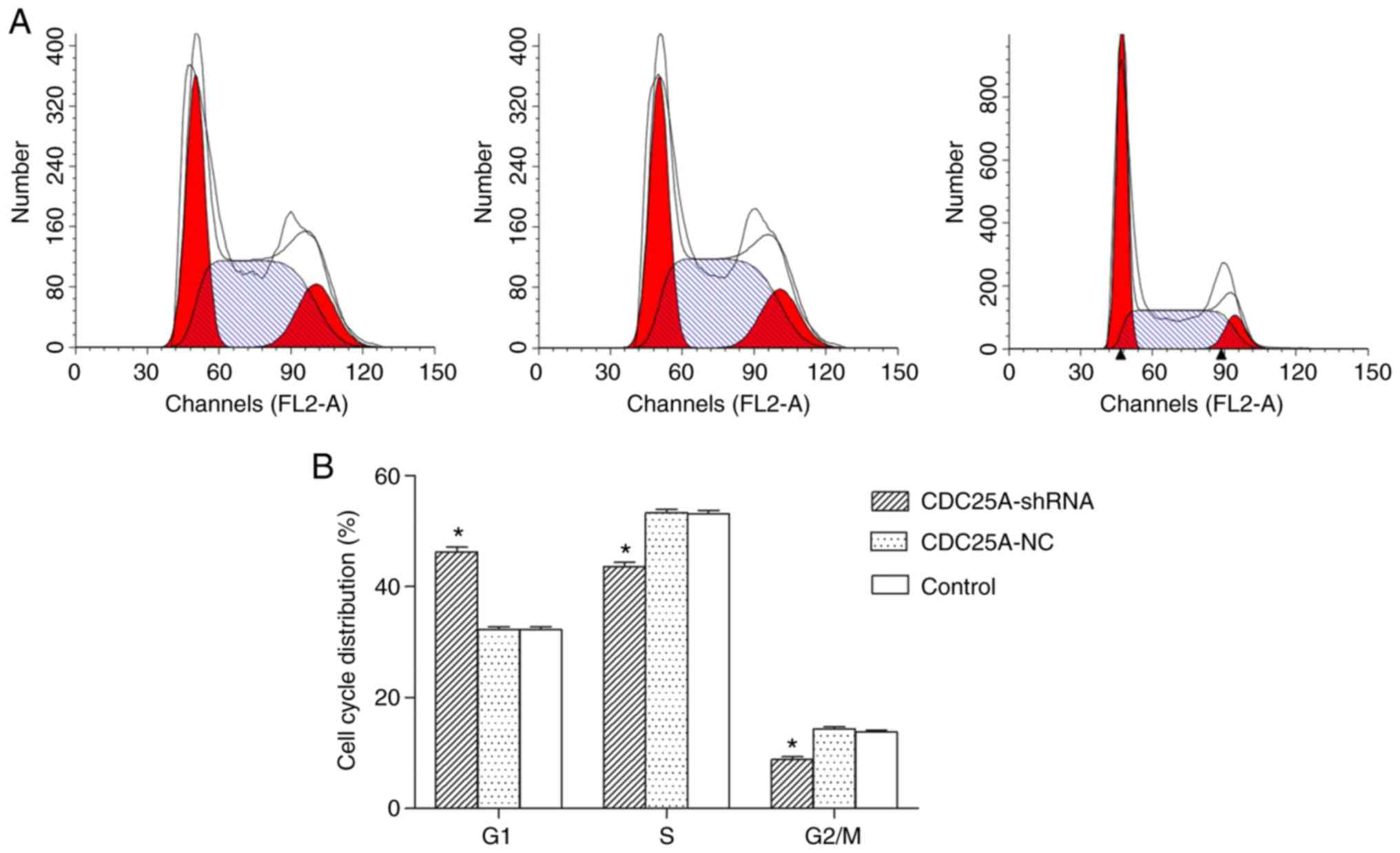
Effect of silencing CDC25A on liver
cancer cell cycle
As presented in Fig.
2A and B, the percentage of cells in the G1 phase was
significantly increased, accompanied by a significant decrease in S
and G2 phases in the CDC25A-shRNA group in comparison with the
CDC25A-NC and control groups (P<0.05).
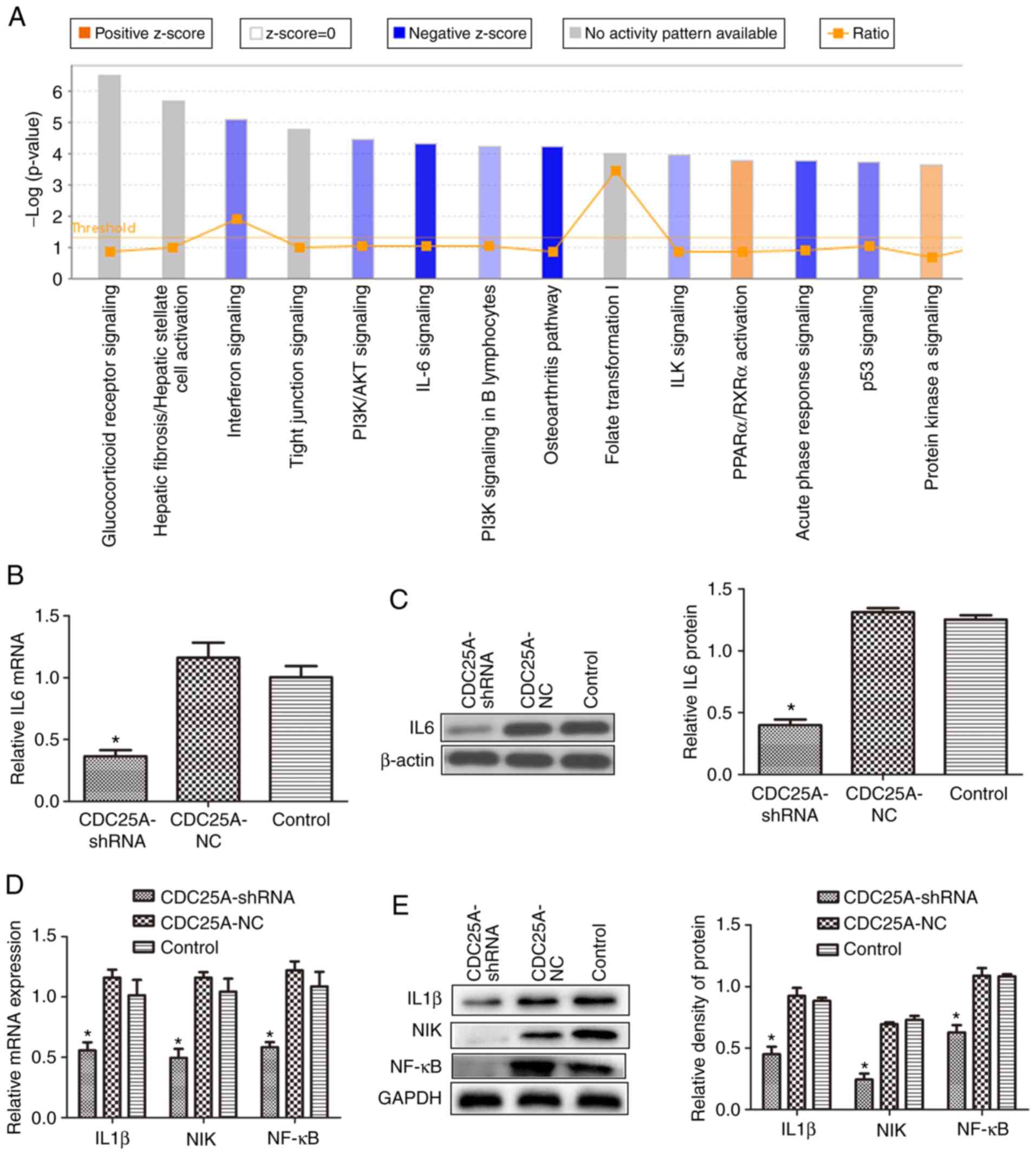
Effect of silencing CDC25A on IL6
expression in HepG2 cells
The differentially expressed genes in HepG2 cells in
which the CDC25A gene was silenced mainly involved interferon
signaling, the phosphoinositide-3-kinase/protein kinase B signaling
pathway, the IL-6 signaling pathway, the P53 signaling pathway and
the cell cycle pathway (Fig. 3A),
and the IL-6 signaling pathway exhibited the most significant
inhibition. These results were verified by RT-qPCR and western blot
analyses. As presented in Fig. 3B and
C, subsequent to silencing the CDC25A gene, the mRNA and
protein expression levels of IL-6 in the CDC25A-shRNA group were
significantly lower compared with those in the CDC25A-NC and
control groups (P<0.05). These results suggest that CDC25A may
affect the expression of IL-6 in HepG2 cells.
Effect of silencing CDC25A on the
IL-1β/NIK/NF-κB signaling axis in HepG2 cells
Gene chip screening was performed as described in
our previous study (22). The
differentially expressed genes in HepG2 cells with the CDC25A gene
silenced revealed that the expression of IL-1β, NIK and NF-κB,
which are IL-6 pathway-associated genes, were significantly
downregulated (P<0.001; Table
I). RT-qPCR analysis revealed that the mRNA expression levels
of IL-1β, NIK and NF-κB in the CDC25A-shRNA group were
significantly lower compared with those in the CDC25A-NC and
control groups (P<0.05). In addition, similar results were
obtained regarding protein expression levels (P<0.05; Fig. 3D and E).
 | Table IGenes with differential expression
following the downregulation of cell division cycle 25A
expression. |
Table I
Genes with differential expression
following the downregulation of cell division cycle 25A
expression.
| Gene symbol | Gene title | Fold change | Regulation | P-value |
|---|
| IL-6 | Interleukin-6 | −5.769 | Down | <0.001 |
| IL-1β | Interleukin-1β | −1.666 | Down | <0.001 |
| NIK | Mitogen-activated
protein kinase kinase kinase kinase 4 | −1.547 | Down | <0.001 |
| NF-κB | Nuclear
factor-κB | −1.743 | Down | <0.001 |
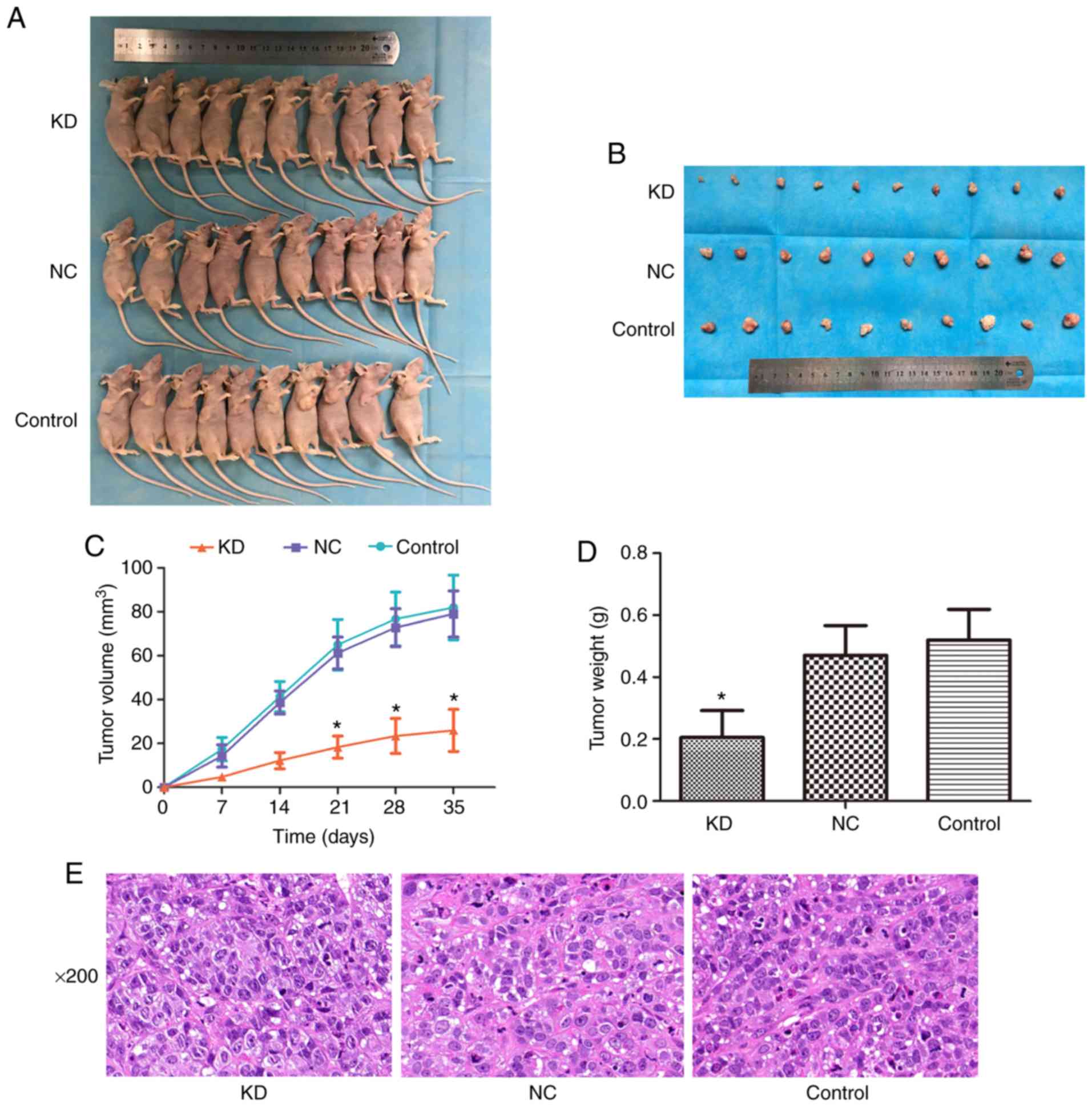
Effect of silencing the CDC25A gene on
liver cancer xenograft growth
On the seventh day following inoculation,
transplanted tumor masses appeared in the nude mice at all
inoculation sites. The width and length of each xenograft tumor
were measured every 7 days, the tumor volume was calculated and the
growth curve was plotted. On the 35th day, the nude mice were
sacrificed (Fig. 4A) and the
xenograft tumors were harvested (Fig.
4B). As presented in Fig. 4C,
the tumor volume in the KD group was significantly smaller compared
with that in the NC or control group from day 21 onwards
(P<0.05). Correspondingly, the tumor weight in the KD group was
significantly decreased (P<0.05; Fig. 4D). Finally, Fig. 4E presents the results of the
H&E staining and shows the xenografts were liver cancer
tissues, indicating that the liver cancer xenograft models were
successfully constructed. These results indicate that silencing the
CDC25A gene may inhibit the growth of liver cancer xenografts.
CDC25A affects liver cancer growth by
targeting IL-6 through the IL-1β/NIK/NF-κB signaling axis
The expression levels of CDC25A and IL6
pathway-associated molecules (including IL-1β, NIK, NF-κB and IL-6)
in the liver cancer xenografts were evaluated by RT-qPCR, western
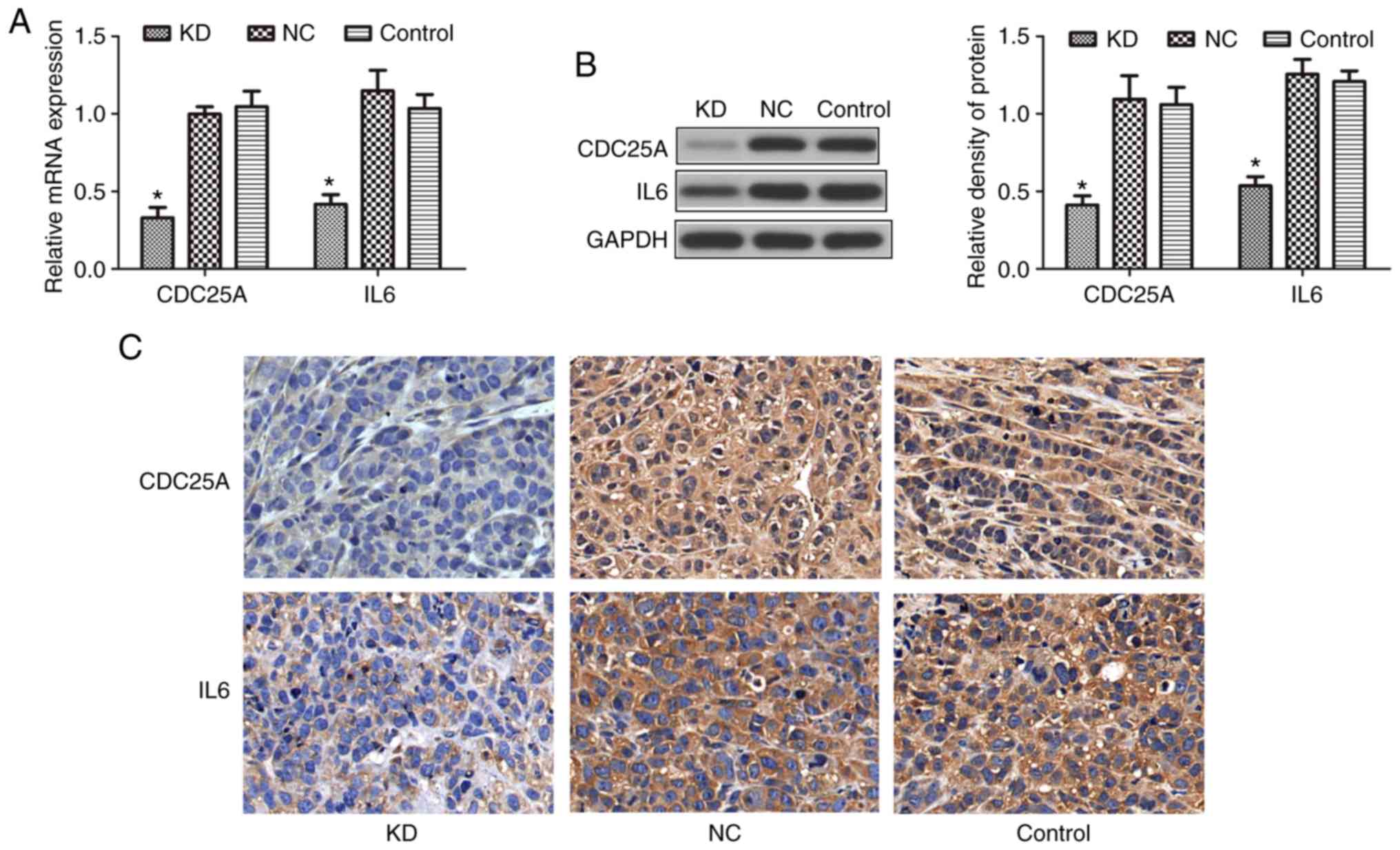
blot analysis and immunohistochemistry. As presented in Fig. 5A and B, the mRNA and protein
expression levels of CDC25A and IL-6 were significantly reduced in
the KD group compared with the NC or control groups (P<0.05).
Further immunohistochemistry analysis revealed that the CDC25A and
IL-6 proteins were strongly expressed in the NC and control groups,
but their expression levels were lower in the KD group (Fig. 5C). A significant positive
correlation was observed between the expression levels of CDC25A
and IL-6 in the xenograft tissue samples (R=0.669, P<0.05; data
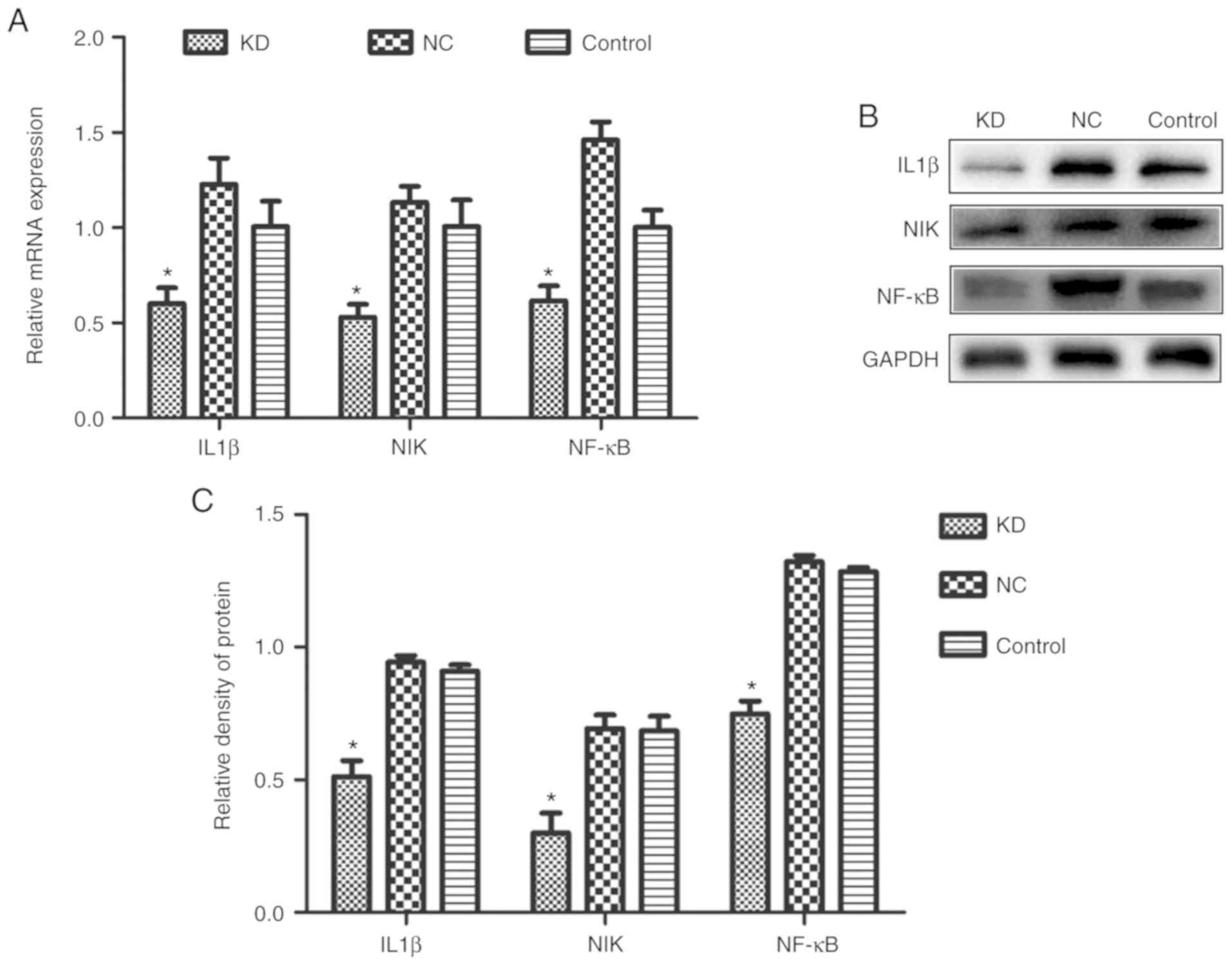
not shown). The mRNA and protein expression levels of IL-1β, NIK
and NF-κB in the KD group were significantly lower compared with
those in the NC or control group (P<0.05; Fig. 6A-C). These results indicate that
CDC25A overexpression in liver cancer may positively regulate IL6
expression by regulating the IL-1β/NIK/NF-κB signaling axis,
thereby promoting the growth of liver cancer.
Discussion
CDC25A is a bispecific protein phosphatase
consisting of 524 amino acid residues that contains an N-terminal
regulatory domain and a C-terminal catalytic domain (24). CDC25A has been revealed to promote
cell cycle progression, and the overexpression of CDC25A causes
abnormal cell cycle regulation and results in tumorigenesis
(25). In addition, previous
studies have revealed that CDC25A serves key functions in
apoptosis, cell metabolism and tumor cell metastasis (4,26).
Overexpressed CDC25A may interact with tumor-associated factors
including mitogen-activated protein kinase kinase kinase 5, NF-κB,
STAT3, NIMA related kinase 11, pyruvate kinase M1/2 and forkhead
box O1 to promote tumor progression (27-29). CDC25A is overexpressed in liver
cancer (30), which is positively
associated with clinicopathological parameters including portal
vein thrombosis, extrahepatic metastasis and tumor differentiation
in patients with liver cancer (31).
In the present study, lentiviral-mediated shRNA
transfection was used to silence the expression of CDC25A in HepG2
cells, and the treated cells were inoculated subcutaneously into
nude mice. Silencing the CDC25A gene significantly inhibited the
growth of the xenograft tumors (P<0.05), indicating that CDC25A
is a key gene in the development of liver cancer. Xu et al
(32) produced a study that used
CDC25A antisense to inhibit CDC25A in liver cancer cells, and
revealed that cell growth, invasion and cell cycle were inhibited,
which was consistent with the results of the present study.
IL-6 is a multifunctional cytokine that has been
associated with multiple tumor types, including breast cancer
(33), lung cancer (34) and ovarian cancer (35). IL-6 may promote the development of
tumor types by mediating various signaling pathways, with the
induction of STAT3 phosphorylation being the most common mechanism.
The IL-6/STAT3 signaling transduction pathway is critical for the
development and progression of malignant tumor types (36). IL6-mediated STAT3 activation may
substantially upregulate the expression of a number of genes
associated with tumor cell proliferation, apoptosis (BCL2 like 1,
MCL1 apoptosis regulator, BCL2 family member, survivin and P53),
the hypoxia response, metastasis and angiogenesis and may
downregulate the expression of proapoptotic genes (37-39); this activation also serves a key
function in the proliferation, apoptosis, invasion and metastasis
of liver cancer cells (40,41). The present screened the
differentially expressed genes in HepG2 cells with silenced CDC25A
using a gene chip and revealed that the IL-6 signaling pathway was
the most significantly inhibited pathway. The expression of IL-6 in
HepG2 cells was confirmed to be significantly decreased subsequent
to silencing the CDC25A gene (P<0.05). The same results were
obtained in vivo, and a significant positive correlation was
observed between CDC25A and IL-6 expression in xenograft tumors
(P<0.05). These results suggest that CDC25A may promote the
development of liver cancer by regulating the expression of IL-6.
Subsequently, the present study investigated how CDC25A regulates
IL-6.
NF-κB is well known to inhibit apoptosis and promote
tumor cell survival via various mechanisms. NF-κB, an important
transcription factor that links inflammation and tumorigenesis,
regulates the expression of various proinflammatory cytokines,
including IL-6, in cancer cells and promotes tumor cell
proliferation (42). Targeting
the inhibition of NF-κB may downregulate the expression of IL-6
(43).
Inflammatory cytokines, including IL-1, IL-6 and
IL-8, may activate the STAT3/NF-κB pathway in tumor cells, and
these pathways stimulate further cytokine production, resulting in
a positive feedback loop (44).
IL-1β is an inflammatory factor that is involved in the NF-κB cell
signaling pathway. IL-1β binds its receptor, IL-1 receptor, and
exerts its effects on the NF-κB kinase NIK, which activates the Iκβ
kinase complex and activates NF-κB (45,46). IL-1β may be induced by NF-κB
activation, while IL-1β activates an auto-crine signal loop that
upregulates the NF-κB signal (47,48). The activated NF-κB then further
promotes the expression of downstream targets IL-6 and IL-8
(49). This positive feedback
loop promotes angiogenesis, tumor growth and metastasis. Gene chip
analysis revealed that the IL-1β/NIK/NF-κB/IL-6 signaling axis was
significantly downregulated in HepG2 cells subsequent to knocking
out the CDC25A gene. The present study confirmed that the
expression levels of IL-6, IL-1β, NIK and NF-κB were consistent
with the chip results, and these results were also confirmed in
vivo. Subsequent to silencing the CDC25A gene, the expression
levels of IL-1β, NIK and NF-κB in the xenograft tumors were
significantly decreased (P<0.05). The present study confirmed
that CDC25A may regulate the expression of IL-6 by regulating the
IL-1β/NIK/NF-κB signaling axis, thereby promoting the growth of
liver cancer.
In summary, the present study revealed that CDC25A
may positively regulate IL-6 through the IL-1β/NIK/NF-κB signaling
axis, thereby promoting the proliferation and growth of liver
cancer cells, and these results provide novel insight into the
function and mechanism of CDC25A in liver cancer.
Funding
The present study was supported by the National
Science Foundation of China (grant no. 30960428), the Guangxi
Natural Science Foundation (grant no. 2017GXNSFBA198003) and the
Basic Ability Enhancement Program for Young and Middle-aged
Teachers of Guangxi (grant no. 2017KY0102).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JC designed and guided the experiments. SC designed
and conducted the experiments, analyzed the data and drafted the
manuscript. YT designed the experiments and revised the manuscript
critically for important intellectual content. CY, KL and XH
contributed to the analysis and interpretation of data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was ethically approved by the
Affiliated Tumor Hospital of Guangxi Medical University Ethics
Committees (Guangxi, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
Acknowledgments
The authors would like to thank the Department of
Research, Affiliated Tumor Hospital of Guangxi Medical University
and the Experimental Animal Center of Guangxi Medical University
for providing laboratories in which to perform the experiments.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamashita T and Kaneko S: Liver cancer.
Rinsho byori. 64:787–796. 2016.In Japanese. PubMed/NCBI
|
|
4
|
Lu X, Sun W, Tang Y, Zhu L, Li Y, Ou C,
Yang C, Su J, Luo C, Hu Y and Cao J: Identification of key genes in
hepatocellular carcinoma and validation of the candidate gene,
cdc25a, using gene set enrichment analysis, meta-analysis and
cross-species comparison. Mol Med Rep. 13:1172–1178. 2016.
View Article : Google Scholar :
|
|
5
|
Shen T and Huang S: The role of Cdc25A in
the regulation of cell proliferation and apoptosis. Anticancer
Agents Med Chem. 12:631–639. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harbour JW, Luo RX, Dei Santi A, Postigo
AA and Dean DC: Cdk phosphorylation triggers sequential
intramolecular interactions that progressively block Rb functions
as cells move through G1. Cell. 98:859–869. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sur S and Agrawal DK: Phosphatases and
kinases regulating CDC25 activity in the cell cycle: Clinical
implications of CDC25 overexpression and potential treatment
strategies. Mol Cell Biochem. 416:1–14. 2016. View Article : Google Scholar
|
|
8
|
Li H, Jiang M, Cui M, Feng G, Dong J, Li
Y, Xiao H and Fan S: MiR-365 enhances the radiosensitivity of
non-small cell lung cancer cells through targeting CDC25A. Biochem
Biophys Res Commun. 512:392–398. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qin H and Liu W: MicroRNA-99a-5p
suppresses breast cancer progression and cell-cycle pathway through
downregulating CDC25A. J Cell Physiol. 234:3526–3537. 2019.
View Article : Google Scholar
|
|
10
|
Luo A, Zhou X, Shi X, Zhao Y, Men Y, Chang
X, Chen H, Ding F, Li Y, Su D, et al: Exosome-derived miR-339-5p
mediates radio-sensitivity by targeting Cdc25A in locally advanced
esophageal squamous cell carcinoma. Oncogene. 38:4990–5006. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ataie-Kachoie P, Pourgholami MH,
Richardson DR and Morris DL: Gene of the month: Interleukin 6
(IL-6). J Clin Pathol. 67:932–937. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sato Y, Goto Y, Narita N and Hoon DS:
Cancer cells expressing toll-like receptors and the tumor
microenvironment. Cancer Microenviron. 2(Suppl 1): S205–S214. 2009.
View Article : Google Scholar
|
|
13
|
Yuan FJ, Zhou YS, Wei Y, Zou C, Chen L,
Huang L and Liu Z: Increased expression of IL-6 mRNA in
hepatocellular carcinoma cell lines correlates with biological
characteristics. Asian Pac J Cancer Prev. 12:3361–3365.
2011.PubMed/NCBI
|
|
14
|
Schmidt-Arras D and Rose-John S: IL-6
pathway in the liver: From physiopathology to therapy. J Hepatol.
64:1403–1415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou M, Yang H, Learned RM, Tian H and
Ling L: Non-cell-autonomous activation of IL-6/STAT3 signaling
mediates FGF19-driven hepatocarcinogenesis. Nat Commun.
8:154332017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rayet B and Gelinas C: Aberrant rel/nfkb
genes and activity in human cancer. Oncogene. 18:6938–6947. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yin L and Yu X: Arsenic-induced apoptosis
in the p53-proficient and p53-deficient cells through differential
modulation of NFkB pathway. Food Chem Toxicol. 118:849–860. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sheng ML, Xu GL, Zhang CH, Jia WD, Ren WH,
Liu WB, Zhou T, Wang YC, Lu ZL, Liu WF, et al: Aberrant estrogen
receptor alpha expression correlates with hepatocellular carcinoma
metastasis and its mechanisms. Hepatogastroenterology. 61:146–150.
2014.PubMed/NCBI
|
|
19
|
Xu H, Wei Y, Zhang Y, Xu Y, Li F, Liu J,
Zhang W, Han X, Tan R and Shen P: Oestrogen attenuates tumour
progression in hepatocellular carcinoma. J Pathol. 228:216–229.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hoesel B and Schmid JA: The complexity of
NF-κB signaling in inflammation and cancer. Mol Cancer. 12:862013.
View Article : Google Scholar
|
|
21
|
Hong HY, Choi J, Cho YW and Kim BC: Cdc25A
promotes cell survival by stimulating NF-κB activity through IκB-α
phosphorylation and destabilization. Biochem Biophys Res Commun.
420:293–296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He P: Screening of differentially
expressed genes in liver cancer HepG2 cells after silencing CDC25A
gene. Guangxi Med Univ. 2018.
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Källström H, Lindqvist A, Pospisil V,
Lundgren A and Rosenthal CK: Cdc25A localisation and shuttling:
Characterisation of sequences mediating nuclear export and import.
Exp Cell Res. 303:89–100. 2005. View Article : Google Scholar
|
|
25
|
Dozier C, Mazzolini L, Cénac C, Froment C,
Burlet-Schiltz O, Besson A and Manenti S: CyclinD-CDK4/6 complexes
phosphorylate CDC25A and regulate its stability. Oncogene.
36:3781–3788. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Z, Kar S and Carr BI: Cdc25A protein
phosphatase: A therapeutic target for liver cancer therapies.
Anticancer Agents Med Chem. 8:863–871. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zou X, Tsutsui T, Ray D, Blomquist JF,
Ichijo H, Ucker DS and Kiyokawa H: The cell cycle-regulatory CDC25A
phosphatase inhibits apoptosis signal-regulating kinase 1. Mol Cell
Biol. 21:4818–4828. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liang J, Cao R, Zhang Y, Xia Y, Zheng Y,
Li X, Wang L, Yang W and Lu Z: PKM2 dephosphorylation by Cdc25A
promotes the Warburg effect and tumorigenesis. Nat Commun.
7:124312016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Feng X, Wu Z, Wu Y, Hankey W, Prior TW, Li
L, Ganju RK, Shen R and Zou X: Cdc25A regulates matrix
metalloprotease 1 through Foxo1 and mediates metastasis of breast
cancer cells. Mol Cell Biol. 31:3457–3471. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang XQ, Zhu YQ, Lui KS, Cai Q, Lu P and
Poon RT: Aberrant Polo-like kinase 1-Cdc25A pathway in metastatic
hepatocellular carcinoma. Clin Cancer Res. 14:6813–6820. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu X, Yamamoto H, Sakon M, Yasui M, Ngan
CY, Fukunaga H, Morita T, Ogawa M, Nagano H, Nakamori S, et al:
Overexpression of CDC25A phosphatase is associated with hypergrowth
activity and poor prognosis of human hepatocellular carcinomas.
Clin Cancer Res. 9:1764–1772. 2003.PubMed/NCBI
|
|
32
|
Xu X, Yamamoto H, Liu G, Ito Y, Ngan CY,
Kondo M, Nagano H, Dono K, Sekimoto M and Monden M: CDC25A
inhibition suppresses the growth and invasion of human
hepatocellular carcinoma cells. Int J Mol Med. 21:145–152.
2008.PubMed/NCBI
|
|
33
|
Fu S and Lin J: Blocking interleukin-6 and
interleukin-8 signaling inhibits cell viability, colony-forming
activity, and cell migration in human triple-negative breast cancer
and pancreatic cancer cells. Anticancer Res. 38:6271–6279. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang Q, Zhang Z, Liao Y, Liu C, Fan S,
Wei X, Ai B and Xiong J: 17β-estradiol upregulates IL6 expression
through the ERβ pathway to promote lung adenocarcinoma progression.
J Exp Clin Cancer Res. 37:1332018. View Article : Google Scholar
|
|
35
|
Wang Y, Zong X, Mitra S, Mitra AK, Matei D
and Nephew KP: IL-6 mediates platinum-induced enrichment of ovarian
cancer stem cells. JCI Insight. 3:pii: 122360. 2018. View Article : Google Scholar
|
|
36
|
Yin Z, Ma T, Lin Y, Lu X, Zhang C, Chen S
and Jian Z: IL-6/STAT3 pathway intermediates M1/M2 macrophage
polarization during the development of hepatocellular carcinoma. J
Cell Biochem. 119:9419–9432. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bournazou E and Bromberg J: Targeting the
tumor microenvironment: JAK-STAT3 signaling. JAKSTAT.
2:e238282013.PubMed/NCBI
|
|
38
|
Rokavec M, Wu W and Luo JL: IL6-mediated
suppression of miR-200c directs constitutive activation of
inflammatory signaling circuit driving transformation and
tumorigenesis. Mol Cell. 45:777–789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Al Zaid Siddiquee K and Turkson J: STAT3
as a target for inducing apoptosis in solid and hematological
tumors. Cell Res. 18:254–267. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Subramaniam A, Shanmugam MK, Ong TH, Li F,
Perumal E, Chen L, Vali S, Abbasi T, Kapoor S, Ahn KS, et al:
Emodin inhibits growth and induces apoptosis in an orthotopic
hepato-cellular carcinoma model by blocking activation of STAT3. Br
J Pharmacol. 170:807–821. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma H, Yan D, Wang Y, Shi W, Liu T, Zhao C,
Huo S, Duan J, Tao J, Zhai M, et al: Bazedoxifene exhibits growth
suppressive activity by targeting interleukin-6/glycoprotein
130/signal transducer and activator of transcription 3 signaling in
hepatocellular carcinoma. Cancer Sci. 110:950–961. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiang M, Birkbak NJ, Vafaizadeh V, Walker
SR, Yeh JE, Liu S, Kroll Y, Boldin M, Taganov K, Groner B, et al:
STAT3 induction of miR-146b forms a feedback loop to inhibit the
NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes.
Sci Signal. 7:ra112014. View Article : Google Scholar
|
|
43
|
Duan XH, Li H, Han XW, Ren JZ, Li FY, Ju
SG, Chen PF and Kuang DL: Upregulation of IL-6 is involved in
moderate hyperthermia induced proliferation and invasion of
hepatocellular carcinoma cells. Eur J Pharmacol. 833:230–236. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Korkaya H, Liu S and Wicha MS: Regulation
of cancer stem cells by cytokine networks: Attacking cancer's
inflammatory roots. Clin Cancer Res. 17:6125–6129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stylianou E and Saklatvala J:
Interleukin-1. Int J Biochem Cell Biol. 30:1075–1079. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu R, Chen B, Jia X, Qiu Y, Liu M, Huang
C, Feng J and Wu Q: Interleukin-1β influences functional
regeneration following nerve injury in mice through nuclear
factor-κB signaling pathway. Immunology. 156:235–248. 2019.
View Article : Google Scholar
|
|
47
|
Sun K, Xu L, Jing Y, Jing Y, Han Z, Chen
X, Cai C, Zhao P, Zhao X and Yang L: Autophagy-deficient Kupffer
cells promote tumorigenesis by enhancing
mtROS-NF-κB-IL1α/β-dependent inflammation and fibrosis during the
preneoplastic stage of hepatocarcinogenesis. Cancer Lett.
388:198–207. 2017. View Article : Google Scholar
|
|
48
|
Nomura A, Gupta VK, Dauer P, Sharma NS,
Dudeja V, Merchant N, Saluja AK and Banerjee S: NFκB-mediated
inva-siveness in CD133+ pancreatic TICs is regulated by autocrine
and paracrine activation of IL1 signaling. Mol Cancer Res.
16:162–172. 2018. View Article : Google Scholar
|
|
49
|
Voronov E, Shouval DS, Krelin Y, Cagnano
E, Benharroch D, Iwakura Y, Dinarello CA and Apte RN: IL-1 is
required for tumor invasiveness and angiogenesis. Proc Natl Acad
Sci USA. 100:2645–2650. 2003. View Article : Google Scholar : PubMed/NCBI
|