Introduction
Cardiac hypertrophy is characterized by the abnormal
enlargement of the heart muscle, which occurs as a result of
increased myocyte size and non-muscle cell proliferation (1,2).
Cardiac hypertrophy occurs in response to hemodynamic overload and
it may predict future coronary artery disease and heart failure
(1). Cardiac hypertrophy is a
complex process that occurs at the cellular and molecular levels,
and involves imbalance of the local autocrine/paracrine network and
circulating biologically active mediators (3). To date, several cell-derived factors
have been shown to improve cardiac function and have intensively
been studied as potential pharmacological targets to prevent and
reverse cardiac hypertrophy-associated diseases (3-5).
Secretoneurin (SN) is a 33-amino acid neuropeptide
derived from a member of the chromogranin/secretogranin family,
secretogranin-II (6). SN is
considered a novel biomarker for cardiovascular diseases including
ischemic heart disease and heart failure (7-12).
In addition, SN demonstrates a protective function in myocardial
ischemia/reperfusion injury in experimental animal models (6,13).
However, little is known regarding the regulation of SN in the
hypertrophic injury of cardiomyocytes. Our preliminary study
demonstrated that SN played a protective role against cardiac
hypertrophy induced by DL-isoproterenol hydrochloride (ISO) in mice
(14). Nevertheless, the
mechanism of the protective action of SN against cardiac
hypertrophy remains unclear.
Proteomics is a quantitative analysis of protein
expression in biological samples. This method is a powerful
screening technology for the global evaluation of protein
expression in complex samples. The isobaric tags for relative and
absolute quantification (iTRAQ)-labeling method is one of the most
reliable techniques that allows the quantitative analysis of
proteins based on peptide identification (15). Differential proteomics relies on
iTRAQ technology and can reflect the regulatory mechanisms
associated with pathological conditions. This approach may be
employed in a wide variety of disorders, including cancer,
cardiovascular disease and psychiatric illness (16-19). Proteomic profiling has revealed
that considerable pathophysiological changes, including altered
energy metabolism, enhanced protein synthesis, proto-oncogene
expression, elevated oxidative stress, occur during cardiac
hypertrophy (20-22). However, the majority of these
studies have only compared patients with cardiac hypertrophy and
healthy subjects (23,24), and the proteomic expression of
SN-overexpressing cardiac hypertrophic cells has not been
investigated. To the best of our knowledge, the protective
mechanism of SN on cardiovascular diseases has not been previously
examined using proteomic analysis.
Therefore, in the present study, proteins were
labeled by iTRAQ and identified by liquid chromatography-Triple
time of flight (LC-TripleTOF®) and bioinformatics
analyses. The putative target proteins and molecular pathways
associated with the protective effect of SN on ISO-induced cardiac
hypertrophy in mice were identified. The results of the present
study provide information with regard to the possible target
proteins and regulatory mechanisms of SN against cardiac
hypertrophy and support the understanding of the potential clinical
application of SN in the treatment of this disease.
Materials and methods
Materials
The protease inhibitor cocktail used was obtained
from Roche Diagnostics GmbH. The iTRAQ Reagent-8plex kit was
purchased from AB Sciex. The BCA Protein assay kit was from Thermo
Fisher Scientific, Inc., and the anti-apolipoprotein C-III (Apoc3)
antibody was purchased from Boster Biological Technology.
DL-isoproterenol hydrochloride (ISO; purity ≥98.5%) was purchased
from Sigma-Aldrich; Merck KGaA. The other chemicals used were of
analytical grade.
Adenoviral constructs
The SN expression vector was customized from
Genechem Co., Ltd., as previously described (14). A signal peptide for the secretion
of recombinant SN from transfected cells (ATG GAG TTT GGG CTG AGC
TGG CTT TTT CTT GTT GCT GCA TTA AGA GGT GTC CAG TCC) and the human
SN peptide (ACA AAT GAA ATA GTG GAG GAA CAA TAT ACT CCT CAA AGC CTT
GCT ACA TTG GAA TCT GTC TTC CAA GAG CTG GGG AAA CTG ACA GGA CCA AAC
AAC CAG) were synthesized and cloned into the vector (Ad-SN). A
similar recombinant adenovirus expressing green fluorescent protein
was used as a negative control (Ad-con). Recombinant adenoviruses
were amplified and purified as previously described (25).
Animals
A total of 40 male C57BL/6 mice (age, 8-12 weeks;
weight, 22±2 g) were purchased from the Animal Center, Health
Sciences Center, Sichuan University. Mice were housed under
standard conditions (room temperature, 20±1°C; humidity 60±10%;12-h
light/dark cycles) and given free access to standard rodent chow
and water. The experimental procedures were performed in compliance
with the Guide for the Care and Use of Laboratory Animals published
by the US National Institute of Health (publication no. 85-23,
revised 1985). The protocols were approved by the Animal Care and
Use Committee of Sichuan University.
Experimental design
The animals were randomly divided into four groups
(n=10 per group) as follows: i) The control group (G1) including
animals treated with Ad-con by myocardial injection and with saline
by subcutaneous injection; ii) the control group with high SN
levels (G2) including animals treated with Ad-SN by myocardial
injection and with saline by subcutaneous administration; iii) the
cardiac hypertrophy group (G3) including animals treated with
Ad-con by myocardial injection and with ISO by subcutaneous
administration; iv) the cardiac hypertrophy group with high SN
levels (G4), including animals treated with Ad-SN by myocardial
injection and with ISO by subcutaneous administration.
In vivo gene delivery and the mouse model
of cardiac hypertrophy
The mice were anesthetized with isoflurane (2%) in
oxygen and kept warm at 37°C. After loss of the righting reflex,
the mice were ventilated by intratracheal intubation with a rodent
ventilator (Harvard Apparatus). The chest of each mouse was opened
to expose the heart and 2×1010 particles of the Ad-con
or Ad-SN (injection volume, 50 µl) were injected into the
myocardium at five different sites, as previously described
(26,27). Subsequently, the mice received
subcutaneous injection of either ISO (5 mg/kg/day) or saline into
their back for seven days (28).
Plasma SN levels were determined by a ultrasensitive
electrochemical detection method, which was based on a
Pb2+-decorated reduced graphene oxidetetraethylene
pentamine label (12).
Echocardiography and cardiac hemodynamic
measurements
The mice were anesthetized with isoflurane (2%) in
oxygen and kept warm at 37°C. After loss of the righting reflex,
the mice were examined by transthoracic echocardiography and
subsequently hemodynamic measurements were performed following a
7-day period administration of adenoviral constructs (29). Transthoracic echocardiography was
performed with a GE Vivid 7 (GE Healthcare) and a 12-MHz imaging
transducer. The investigator was blinded to the treatments and the
complete procedure of the echocardiography was performed by the
same person. Following echocardiography, the chest of each animal
was opened and the left ventricular function was evaluated by
inserting a pressure-volume catheter 1.2 F transducer (4.5-mm
electrode spacing; serial no. 112B-B057; SCIsense Inc.) into the
left ventricle from the apex and positioning it along the cardiac
longitudinal axis (29). The
signals were recorded by an eight-channel physiological recorder
(iWorx 308; iWorx/CB Sciences, Inc.). Following hemodynamic
recording, the mice were euthanized by pentobarbital overdose (at
least 200 mg/kg, i.p.) (30).
After cessation of the heartbeat, the hearts were collected
immediately to measure the heart weight and extract the
proteins.
Protein sample preparation
The heart tissues of the animals in each group were
washed with cold PBS following drug treatment and homogenized in
cold PBS at 4°C. The cells were collected by centrifugation at
1,000 × g for 5 min at 4°C and subsequently lysed in PBS containing
protease inhibitor cocktail. The lysates were freeze-thawed three
times at −80°C followed by sonication (200 W; 30 sec) on ice and
subsequently centrifuged at 11,500 × g for 20 min at 4°C. The
supernatant was collected and stored at −80°C prior to further
analysis.
iTRAQ labeling
The samples were mixed with cold acetone (99.5%) for
2 h at -20°C and centrifuged at 3,000 × g for 5 min at 4°C to
precipitate the proteins. Following air-drying, the precipitates
were dissolved in dissolution buffer UA (8 M urea and 0.1 M
Tris-HCl; pH 8.5). The concentrations of the proteins were measured
using the Bradford method, and the proteins (200 µg per
sample) were alkylated and digested with trypsin at 37°C for 12 h.
The samples were labeled with different isobaric tagging reagents
according to the manufacturer's protocol (iTRAQ8plex reagents; AB
Sciex). A total of 3 biological replicate samples were merged in
one labeling sample, and two samples were prepared for iTRAQ
labeling. The peptides from G1-1, G1-2, G2-1, G2-2, G3-1, G3-2,
G4-1 and G4-2 were labeled with 115, 116, 117, 118, 113, 114, 119
and 121 iTRAQ reagents, respectively. The iTRAQ reagent-labeled
samples were finally pooled.
Strong cation exchange (SCX)
chromatography
The pooled samples were added to SCX buffer A (20 mM
ammonium formate; pH 10.0) and mixed well. Following centrifugation
at 11,000 × g for 5 min at 4°C, iTRAQ-labeled peptides were
separated using a HPLC 2010A system (Shimadzu Corporation) with a
Gemini-NX chromatographic column (4.6×250 mm; 5 µm; 110 Å;
Phenomenex; PN:00G-4454-E0). The column was eluted with a linear
gradient of 0-20% SCX buffer A containing 20 mM ammonium formate
for 60 min and subsequently eluted with a gradient of 20-100% SCX
buffer B containing 80% acetonitrile for an additional 60 min with
a flow rate of 0.8 ml/min at 25°C. The fractions were collected at
1 min intervals and lyophilized in a vacuum concentrator.
Reverse-phase LC-TripleTOF and mass
spectrometric analysis
The SCX fractions were resuspended in reverse phase
buffer A (98% H2O, 2% acetonitrile, 0.1% formic acid).
The peptides were captured by the reverse phase nano LC Chrom XP
C18 column (Chrom XP C18, 350 µm x 0.5 mm; 3 µm; 120
Å; Eksigent) and separated using a 60 min linear gradient of buffer
A (0.1% formic acid in 2% acetonitrile) to buffer B (0.1% formic
acid in 98% acetonitrile) at a flow rate of 300 nl/min. MS was
performed in a TripleTOF 5600 instrument (AB Sciex, LLC).The
electrospray ionization (ionspray voltage of 2.3 KV) was used to
generate positive ions and the quadrupole time-of-flight mass
spectrometer (Q-TOF-MS) was operated in an information-dependent
acquisition mode (scan range, 350-1500 m/z; accumulation time, 0.5
sec). The peptides were finally selected for MS/MS analyses.
Western blotting
The mouse heart tissues of each group were washed
with cold PBS at 4°C following drug treatment. The tissues were
homogenized and lysed in RIPA buffer (Beyotime Institute of
Biotechnology) containing the protein inhibitor cocktail. The
lysates were centrifuged at 12,000 × g for 15 min at 4°C to collect
the supernatant. The protein concentration was determined by the
BCA method. A total of 50 µg protein was separated from each
sample by SDS-PAGE on a 10% gel and transferred to polyvinylidene
fluoride membranes. The membranes were blocked in 5% non-fat milk
diluted in TBS/T buffer at 4°C for 1 h, and then immunoblotted with
antibodies against Apoc3 (1:200 in TBS/T buffer; cat. no. PB1097;
Boster Biological Technology) or β-actin (1:1,000 in TBS/T buffer;
cat. no. sc-47778; Santa Cruz Biotechnology, Inc.) at 4°C
overnight. After washing 3 times for 5 min with TBS/T, the
membranes were incubated with HRPconjugated secondary antibodies
(1:5,000 diluted in TBS/T; cat. no. ZB-2301 for antirabbit and cat.
no. ZB-2305 for antimouse; both OriGene Technologies, Inc.) for 1 h
at room temperature. The bands were visualized with enhanced
chemiluminescence reagents (Merck KGaA) using Image Lab software
(version 4.0; Bio-Rad Laboratories, Inc.).
Data processing and analysis
The MS raw data were accessed at www.iprox.org/page/PSV023.html;?url=1561949914798JAZl
with the following password: [KTuX]. The MS raw data files were
uploaded into the PeakView software (version 1.2; AB Sciex, LLC)
for peak detection, generation of peak lists of mass error
corrected peptides and database searches. Protein identification
and quantification were performed with ProteinPilot software
(version 4.0; AB SCIEX) against the uniprot-taxonomy_10090 mouse
protein database (www.uniprot.org/uniprot/?query=taxonomy:10090). All
common fixed and variable modifications of proteins were
considered. iTRAQ 8 plex (Peptide Labeled) was selected as the
quantitation mode. A total of three missed cleavages were allowed
and the enzyme specificity was selected as 'trypsin'. Mass
tolerance for the initial search was selected as '5 ppm' and the
MS/MS tolerance was set to 0.1 Da. The data were estimated using
automatic decoy searching for false discovery rate (FDR) analysis.
The relative expression levels of the proteins at different
treatment protocols were calculated using G3 as the reference. The
proteins that met the following criteria were selected for further
analysis: i) Significantly changed expression (fold-change ≥1.5
fold or ≤0.667 fold vs. control; P<0.05); ii) a minimum of three
peptides with 95% confidence; and iii) 1% global FDR. The
bioinformatics analysis ensured that all identified protein
accessions were submitted to the Database for annotation,
visualization and integrated discovery (DAVID; version 6.8;
http://david.abcc.ncifcrf.gov).
Subsequently, the default matched organism was selected as
'background to do' analysis. The Gene Ontology (GO; geneontology.org) and Kyoto Encyclopedia of Genes and
Genomes (KEGG; www.genome.jp/kegg) reports from DAVID
were downloaded to draw charts using specific in-house Perl and R
scripts. The accession of DEPs was submitted to the Search Tool for
the Retrieval of Interacting Genes/Proteins (STRING; version 10;
www.string-db.org) for the protein-protein interaction
(PPI) files. Subsequently, the PPI files and the protein expression
file were imported to Cytoscape software (version 3.2.1; www.cytoscape.org) to construct and display the PPT
network.
Statistical analysis
The results are presented as the mean ± SEM (n=6).
The comparisons were performed using GraphPad Prism software
(version 5; GraphPad Software, Inc.) with unpaired Student's t-test
and the one-way ANOVA followed by the Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
SN gene overexpression improves the left
ventricular hypertrophy and dysfunction induced by ISO
The echocardiography and cardiac hemodynamic
measurements were performed following ISO treatment in order to
evaluate the cardiac protective effect of SN (Fig. 1). Following a 7-day period of
treatment with ISO, the mice that received Ad-con injections
exhibited significant cardiac hypertrophy and cardiac dysfunction
compared with the vehicle-treated Ad-con mice. The plasma SN levels
were detected by electrochemical detection methods. Compared with
the mice that received Ad-con injections, significantly higher
plasma SN levels were observed in mice receiving Ad-SN injections
(both P<0.01; Table I). ISO
treatment also significantly increased plasma SN levels in the mice
receiving Ad-con injections (P<0.01; Table I), compared with vehicle-treated
mice receiving Ad-con injections. The data indicated a higher left
ventricle (LV) end systolic diameter (IVSd, P<0.05; Fig. 1A) and a lower ejection fraction,
as estimated by the Teichholz method (EF, P<0.01; Fig. 1B). In addition, the maximal values
of the first derivative of LV pressure (dPmax, P<0.01; Fig. 1C) and the cardiac output (CO,
P<0.05; Fig. 1D) were noted.
However, SN gene therapy partly restored these levels to
approximately 10.4% (P>0.05) lower in IVSd, and 60.8%
(P<0.01), 104.5% (P<0.01) and 101.1% (P>0.05) higher in
EF, dPmax and CO, respectively compared with the corresponding
levels noted in ISO-treated Ad-con mice. The results demonstrated
that SN played a protective role in ISO-induced cardiac
hypertrophy.
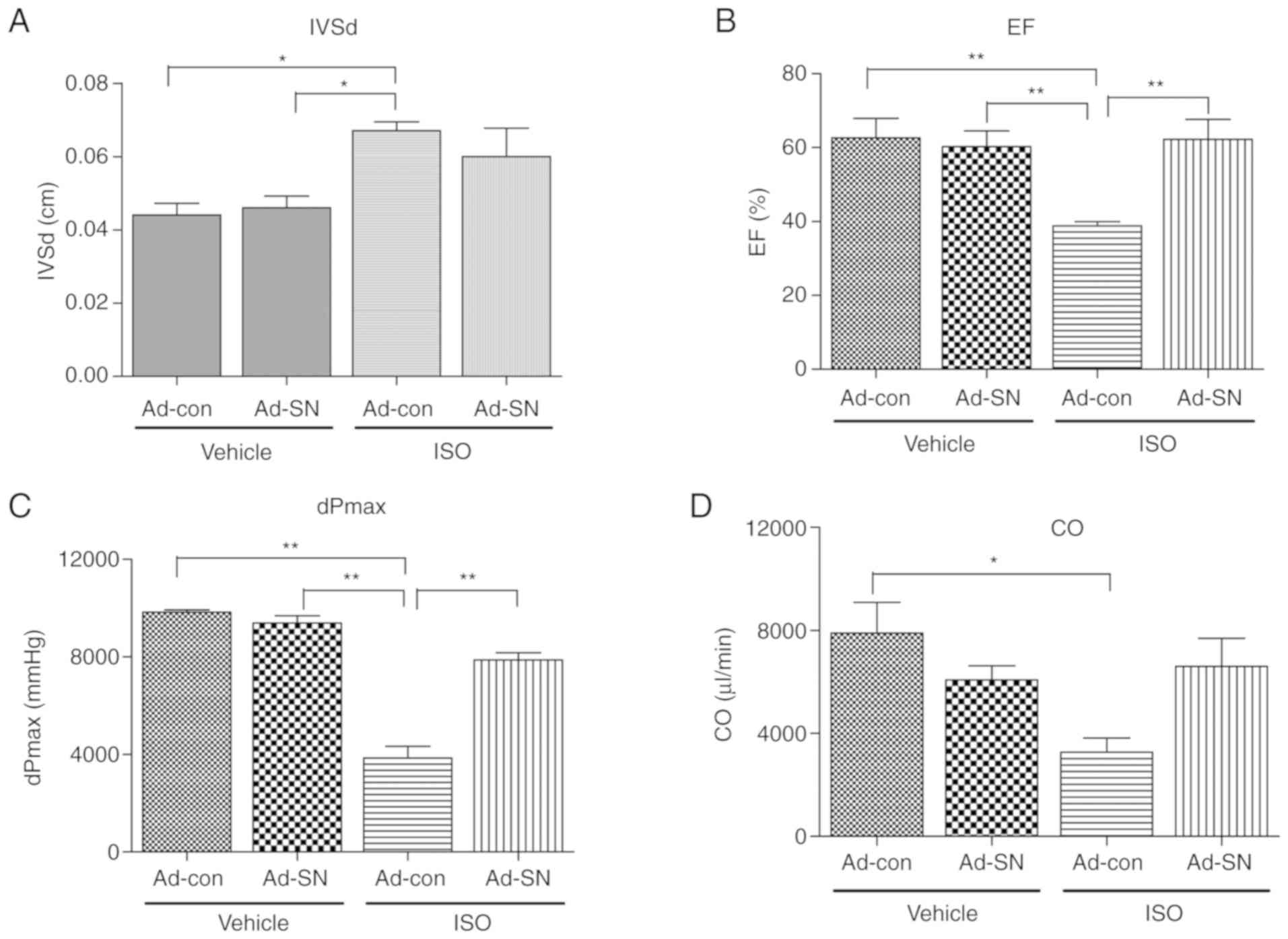 | Figure 1Echocardiography and hemodynamic
measurements. G1 mice received Ad-con intramyocardial injection and
vehicle subcutaneous injection, G2 mice received Ad-SN
intramyocardial injection and vehicle subcutaneous injection, G3
mice received Ad-con intramyocardial injection and ISO subcutaneous
injection and G4 mice received Ad-SN intramyocardial injection and
ISO subcutaneous injection. (A) IVSd of mice in G1, G2, G3 and G4;
(B) EF (Teich)% of mice in G1, G2, G3 and G4; (C) dPmax of mice in
G1, G2, G3 and G4; (D) CO of mice in G1, G2, G3 and G4. The data
are presented as the mean ± SEM (n=6/group). *P<0.05 and
**P<0.01. G1-4, groups 1-4; Ad, adenovirus; SN,
secretoneurin; ISO, isoprenaline; IVSd, left ventricle (LV) end
systolic diameter; EF (Teich)%, ejection fraction calculated by the
Teichholz method; dPmax, maximal value of the first derivative of
LV pressure; CO, cardiac output. |
 | Table IPlasma SN levels determined by
electrochemical methods in vehicle- or ISO-treated mice receiving
Ad-con or Ad-SN intramyocardial injections. |
Table I
Plasma SN levels determined by
electrochemical methods in vehicle- or ISO-treated mice receiving
Ad-con or Ad-SN intramyocardial injections.
| Group | Plasma SN level
(ng/ml) |
|---|
| 1 | 3.1±0.4 |
| 2 | 8.8±0.4a |
| 3 | 7.4±0.5a |
| 4 | 13.0±0.5b,c |
Identification of differentially
expressed proteins following SN gene injection in the ISO-induced
cardiac hypertrophy mouse model
Using iTRAQ coupled with LC TripleTOF, the present
study identified 2,044 proteins with a 1% FDR from 19,729 distinct
peptides derived from 106,837 spectra. A 1.5-fold-change of
expression cut-off value for 1,217 proteins was used as the cut-off
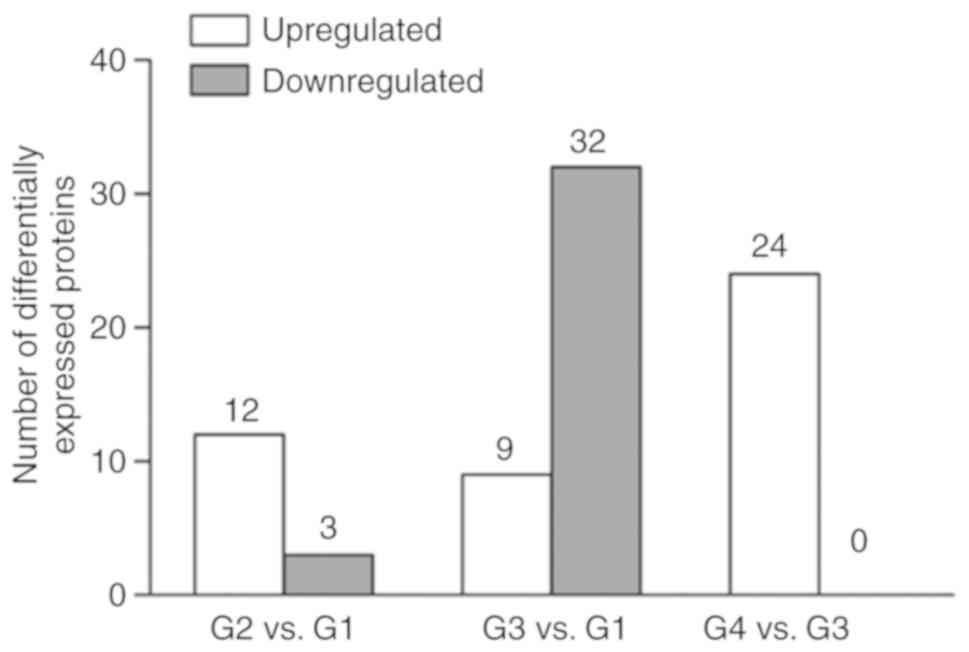
point. A total of 15 proteins indicated a significant change
(P<0.05) in their expression levels between the G2 and G1
groups, whereas 41 proteins exhibited a significant change
(P<0.05) in their expression levels between the G3 and G1
groups. A total of 24 proteins exhibited a significant change
(P<0.05) in their expression levels between the G4 and the G3
groups (Fig. 2). A total of 12
proteins exhibited common upregulated expression levels and three
proteins demonstrated common downregulated expression levels
following SN gene therapy compared with those of the control group
(G2 vs. G1; Fig. 2). In cardiac
hypertrophic animals (G3), a higher number of common proteins
exhibited differentially expression levels (9 upregulated and 32
downregulated) compared with the expression levels noted in the
control group (G1; Fig. 2).
Moreover, following SN gene therapy, 24 proteins indicated
upregulated expression compared with the corresponding proteins of
the cardiac hypertrophic group (G4 vs. G3; Fig. 2). A total of 32 downregulated
proteins were identified in the cardiac hyper-trophic group and 15
of them exhibited upregulated expression following SN gene therapy.
These proteins did not exhibit a change between the G1 and G2
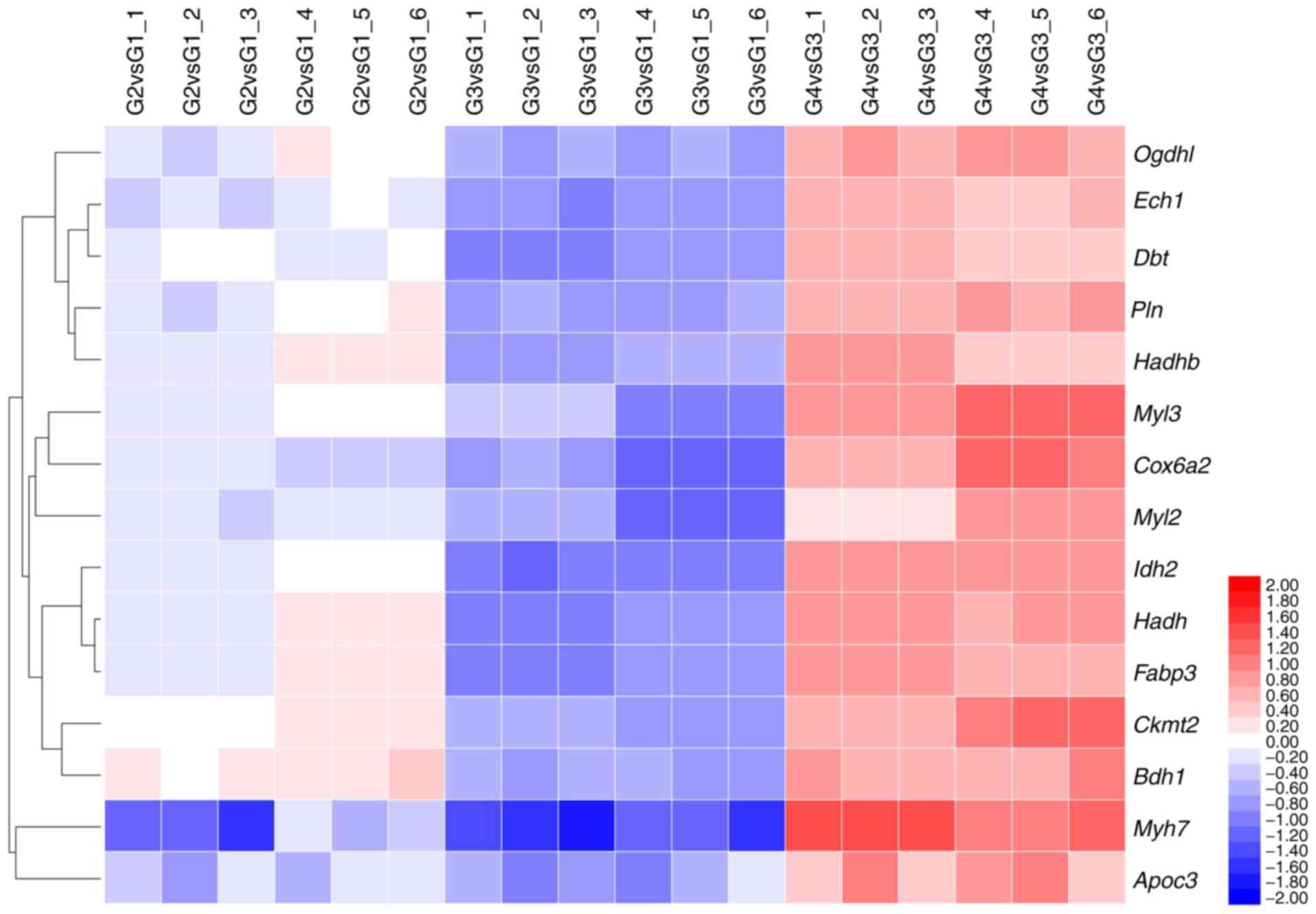
groups (Fig. 3 and Table II).
 | Table IIList of the SN-related significantly
differential proteins(n=6).a |
Table II
List of the SN-related significantly
differential proteins(n=6).a
| Protein name | Protein ID | Gene | Fold-change
|
|---|
| G2/G1 | G3/G1 | G4/G3 |
|---|
| Oxoglutarate
dehydrogenase like | B2RXT3_MOUSE | Ogdhl | 1.01±0.15 | 0.66±0.03 | 1.73±0.08 |
| Δ(3,5)-Δ(2,4)-dienoyl-CoA isomerase,
mitochondrial | ECH1_MOUSE | Ech1 | 0.93±0.06 | 0.61±0.03 | 1.57±0.09 |
| Myosin light chain
3 | MYL3_MOUSE | Myl3 | 0.97±0.04 | 0.65±0.13 | 2.21±0.38 |
| Myosin regulatory
light chain 2, ventricular/cardiac muscle isoform | MLRV_MOUSE | Myl2 | 0.89±0.03 | 0.59±0.12 | 1.57±0.39 |
| Lipoamide
acyltransferase component of branched-chain α-keto acid
dehydrogenase complex, mitochondrial | ODB2_MOUSE | Dbt | 0.99±0.02 | 0.58±0.04 | 1.52±0.12 |
| Isocitrate
dehydrogenase [NADP], mitochondrial | IDHP_MOUSE | Idh2 | 1.04±0.08 | 0.53±0.03 | 1.95±0.02 |
| Cardiac
phospholamban | PPLA_MOUSE | Pln | 1.01±0.12 | 0.64±0.04 | 1.71±0.06 |
|
Hydroxyacyl-coenzyme A dehydrogenase,
mitochondrial | HCDH_MOUSE | Hadh | 1.15±0.18 | 0.58±0.02 | 1.82±0.08 |
| Creatine kinase
S-type, mitochondrial | KCRS_MOUSE | Ckmt2 | 1.14±0.03 | 0.65±0.01 | 1.97±0.36 |
| D-β-hydroxybutyrate
dehydrogenase, mitochondrial | BDH_MOUSE | Bdh1 | 1.25±0.09 | 0.66±0.03 | 1.81±0.17 |
| Trifunctional
enzyme subunit β, mitochondrial | ECHB_MOUSE | Hadhb | 1.061±0.12 | 0.63±0.05 | 1.59±0.18 |
| Myosin, heavy
polypeptide 7, cardiac muscle, β | B2RXX9_MOUSE | Myh7 | 0.63±0.21 | 0.39±0.06 | 2.48±0.39 |
| Apolipoprotein
C-III | E9QP56_MOUSE | Apoc3 | 0.81±0.13 | 0.64±0.13 | 1.78±0.39 |
| Fatty acid binding
protein 3, muscle and heart | Q5EBJ0_MOUSE | Fabp3 | 1.03±0.15 | 0.58±0.03 | 1.83±0.14 |
| Cytochrome c
oxidase subunit 6A, mitochondrial | Q8R2L0_MOUSE | Cox6a2 | 0.89±0.04 | 0.56±0.09 | 2.11±0.45 |
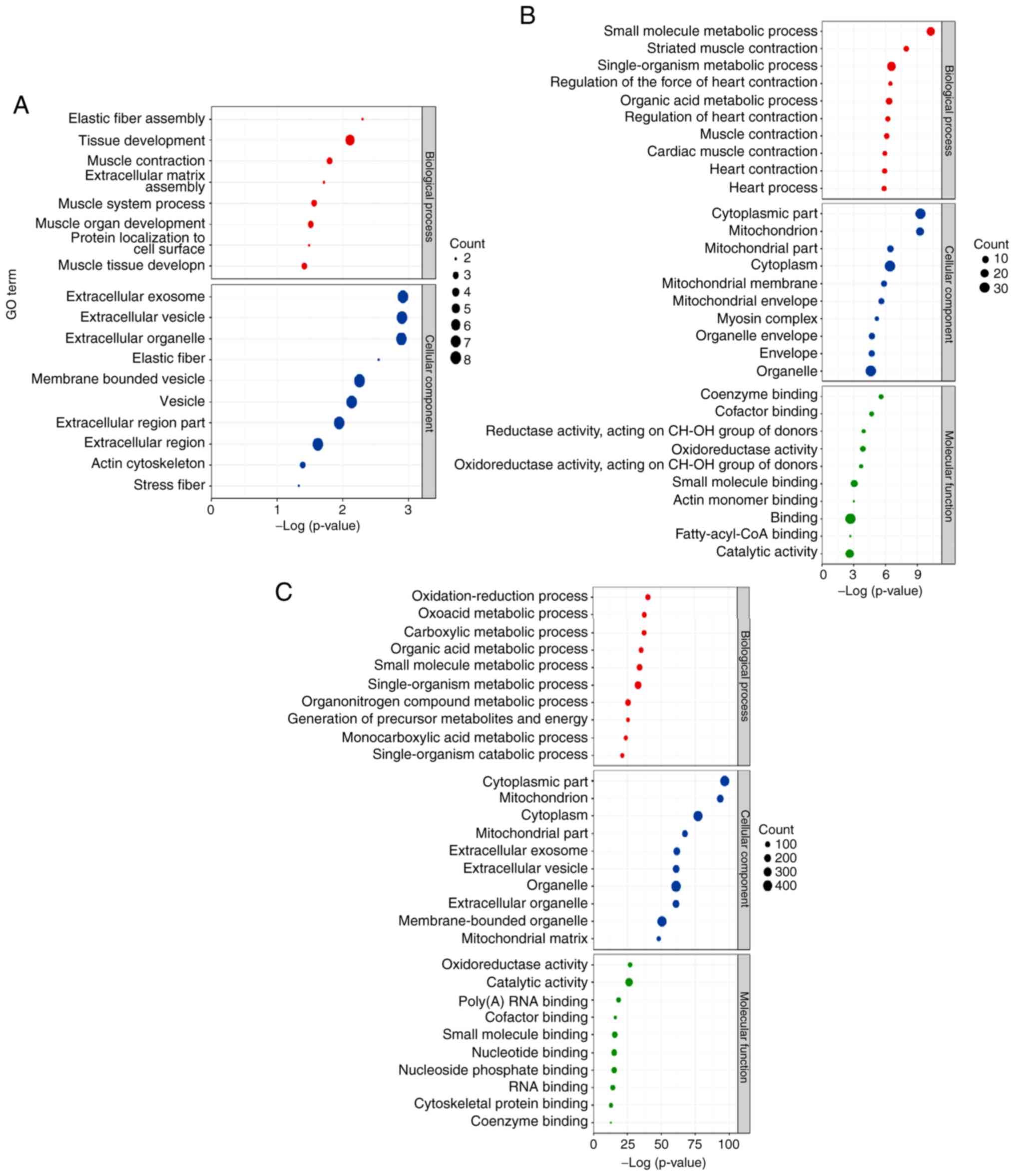
GO classification of differentially
expressed proteins
To further understand the function and role of
SN-associated proteins, GO and pathway enrichment analysis were
performed using DAVID. The top ten significantly enriched
biological processes were identified, including the cellular
component and molecular function terms (Fig. 4).
Following comparison of the biological process GO
terms corresponding to the differentially expressed proteins of the
G3 and G1 groups, seven terms were closely associated with cardiac
hypertrophy and cardiac injury ('striated muscle contraction',
'regulation of the force of heart contraction', 'regulation of
heart contraction', 'muscle contraction', 'cardiac muscle
contraction', 'heart contraction' and 'heart process'). The other
three significantly enriched biological processes were associated
with the metabolic process (Fig.
4B). The comparison of the differentially expressed proteins of
the G4 and G3 groups demonstrated that all the significantly
enriched biological processes were associated with metabolic
processes (Fig. 4C). With regard
to the cellular component, the top two terms of the differentially
expressed proteins were compared between the G3 and G1 and the G4
and G3 groups. These terms were involved in the cytoplasmic and the
mitochondrial function processes (Fig. 4B and C).
The comparison of the G2 and G1 groups revealed no
significant differences in the enriched molecular function terms of
the differentially expressed proteins (Fig. 4A). The top three terms in the
differentially expressed proteins of the G3 and the G1 groups were
'coenzyme binding', 'cofactor binding' and 'oxidoreductase activity
acting on the CH-OH group of donors' (Fig. 4B). The top three terms in the
differentially expressed proteins of the G4 and the G3 groups were
'oxidoreductase activity', 'catalytic activity' and 'poly(A) RNA
binding' (Fig. 4C).
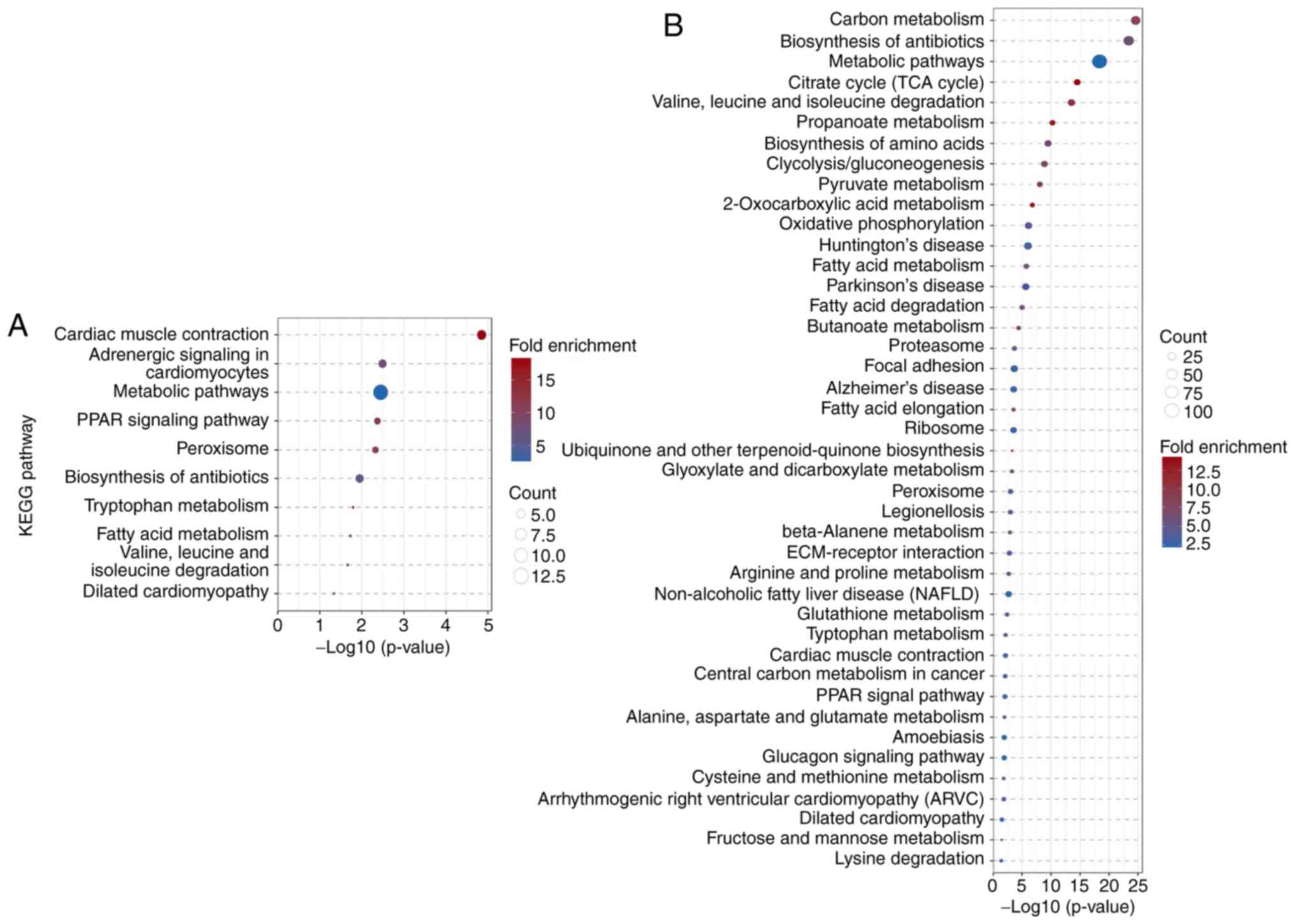
Metabolic pathways involved in
SN-mediated protection against cardiac hypertrophy in mice
KEGG pathway analysis was used to identify the
metabolic pathways of the differentially expressed proteins. The 41
differentially expressed proteins of the G3 and the G1 groups were
significantly enriched in ten signaling pathways, with 'cardiac
muscle contraction', 'adrenergic signaling in cardiomyocytes' and
'metabolic pathways' as the top three enriched pathways (Fig. 5A). The 24 differentially expressed
proteins derived from the comparison of the G4 and G3 groups were
significantly enriched in 24 signaling pathways, with 'carbon
metabolism', 'biosynthesis of antibiotics' and 'metabolic pathways'
as the top three enriched pathways (Fig. 5B). The comparison of the
differentially expressed proteins of the G2 and G1 groups did not
reveal enriched pathways.
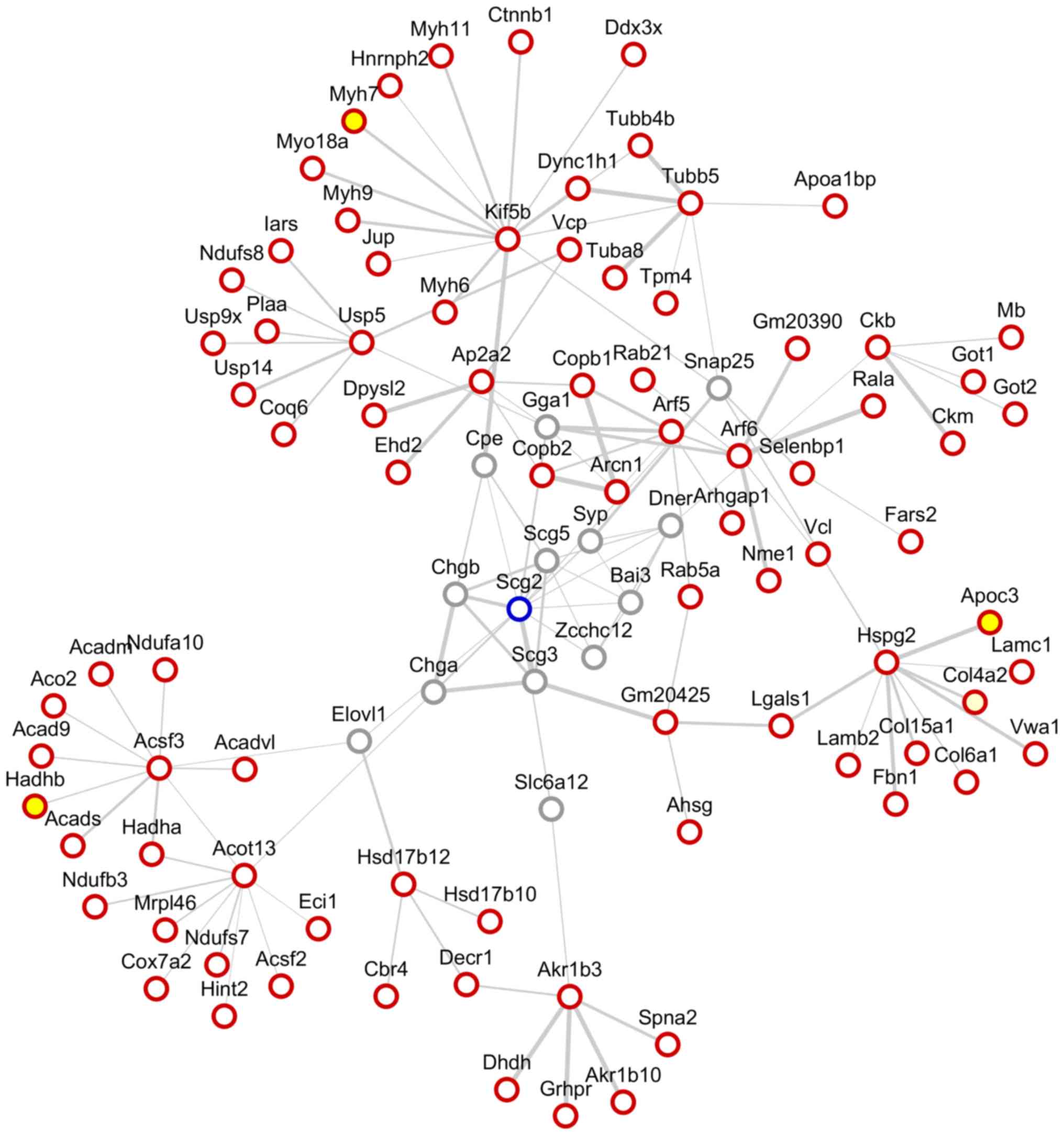
Furthermore, a protein-protein interaction network
analysis of SN was performed using the STRING website and Cytoscape
software (Fig. 6). Among the
network of the protein-protein interaction surrounding SN, the
differentially expressed proteins were identified by iTRAQ as
follows: Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme
complex subunit β (Hadhb), apolipoprotein C-III (Apoc3) and myosin
heavy polypeptide 7 cardiac muscle β (Myh7).
SN influences cardiac hypertrophy by
altering Apoc3 levels
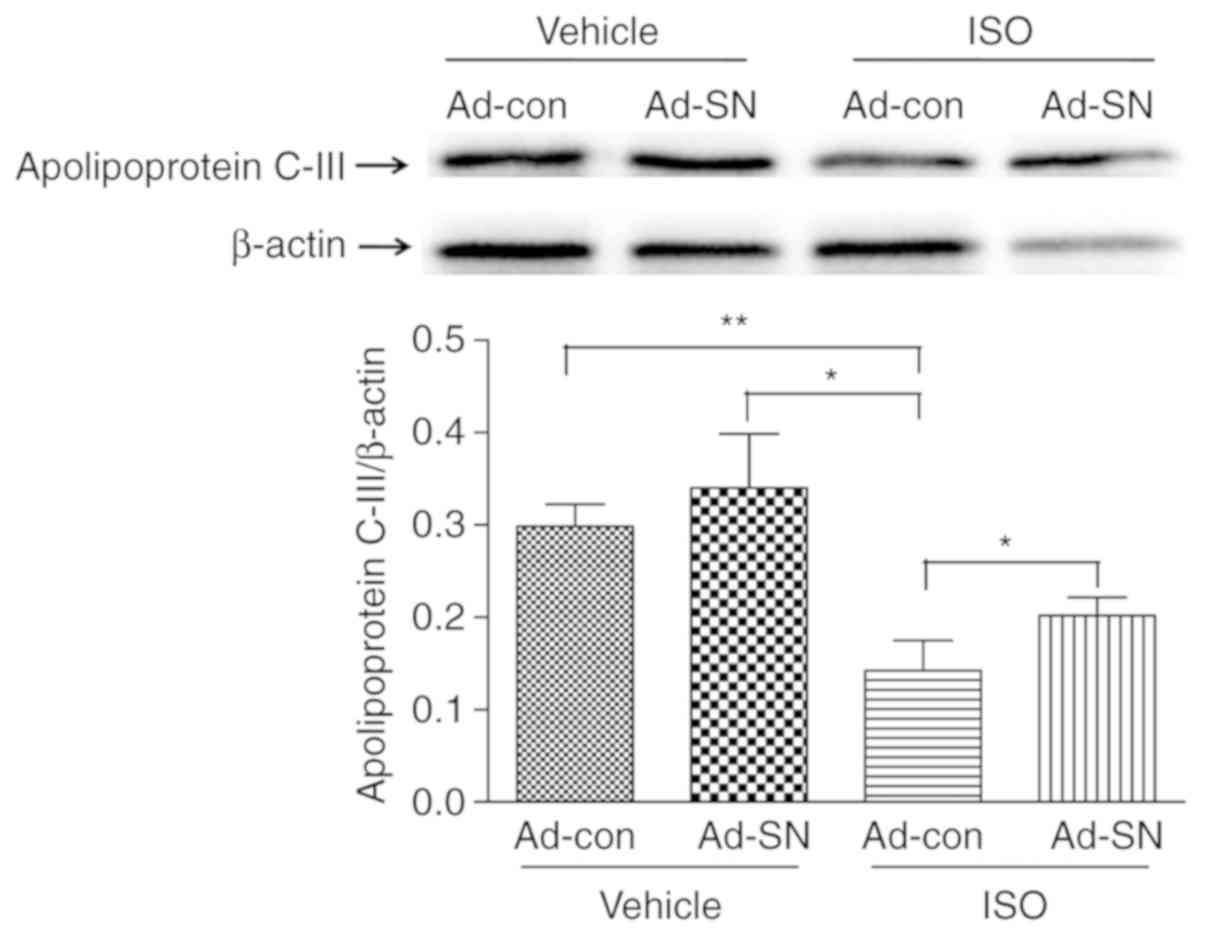
For quantitative validation of candidate proteins,
western blot analysis of Apoc3 was performed. The protein
expression levels of Apoc3 were significantly decreased in the
cardiac hypertrophy animals (Fig.
7; P<0.01). However, the comparison of the cardiac
hypertrophic group with the control group demonstrated that SN gene
therapy effectively induced Apoc3 expression by 41.8% (Fig. 7; P<0.05). The results suggested
that SN increased Apoc3 expression levels in the hypertrophic heart
tissues.
Discussion
Various forms of ventricular pressure overload can
cause cardiac hypertrophy, which is an imbalance in cardiac
homeostasis (31). In cardiac
hypertrophy, catecholamine levels are increased and adrenergic
stimulation occurs (32). During
the development of this condition, several endogenous factors are
involved (3,33). SN was initially discovered as a
regulator of neurogenic inflammation (34). However, subsequent studies
demonstrated that it could act as a potential protector against
cardiovascular events (6,13,35). SN plays a protective role in
myocardial ischemia and heart failure (36,37) by inducing angiogenesis and
postnatal vasculogenesis (13,38). However, to the best of our
knowledge, the regulation of SN in cardiac hypertrophy has not been
previously investigated. In the preliminary study conducted by our
research group, SN played a protective role on cardiac hypertrophy
induced by ISO in mice. SN gene therapy resulted in a significant
improvement in cardiac hypertrophy and cardiac dysfunction
(14). However, the mechanism of
the protective role of SN on cardiac hypertrophy was not fully
elucidated.
iTRAQ quantitative proteomic technology has been
previously used to reveal the mechanisms of diseases (39) or drug treatment (40,41). However, to the best of our
knowledge, no studies focusing on the protective role of SN on
cardiac hypertrophy have been previously reported. Therefore, in
the present study, iTRAQ coupled with a LC-TripleTOF-based
proteomic technology was used to identify and quantify the
differentially expressed candidate proteins in response to
SN-mediated inhibition of cardiac hypertrophy induced by ISO for
the first time. A total of 41 differentially expressed proteins
were observed between hypertrophic and non-hypertrophic heart
tissues in mice (G3 vs. G1). A total of 9 proteins were
upregulated, whereas 32 proteins were downregulated. Following SN
gene therapy, 24 proteins exhibited upregulated expression compared
with those noted in the hypertrophic heart with the control gene
therapy (G4 vs. G3). In iTRAQ based-comparative proteomic studies,
more upregulated differentially expressed proteins were found than
down-regulated ones (42-45). In the majority of the studies
using similar quantitative proteomic techniques, the criteria for
selecting differential expression protein were set as 'fold-change
≥1.2 fold or ≤0.83 fold vs. control' (39,40). Whereas, in the present study, the
criteria were set as 'fold change ≥1.5 fold or ≤0.667 fold vs.
control; P<0.05'. Higher criteria for selecting differential
expressed proteins were selected, as this approach was likely to
filter out a large number of downregulated differential expressed
proteins whose changes were smaller than the criteria. Therefore,
it was not difficult to infer that all differentially expressed
proteins were upregulated in the results. A total of 15 out of 32
downregulated proteins (G3 vs. G1 groups) were closely associated
with the protective effects of SN gene therapy on cardiac
hypertrophy in mice.
GO analysis of differentially expressed proteins
indicated that the significantly enriched biological processes were
cellular component and molecular function terms. The GO analysis of
the differentially expressed proteins resulting from the comparison
of the hypertrophic and the non-hypertrophic heart tissues (G3 vs.
G1) indicated the top ten significantly enriched biological
processes, of which seven terms were closely associated with
cardiac hypertrophy and cardiac injury. This result was expected,
since the samples from the hypertrophic hearts were pooled. The
other three terms were associated with metabolic processes, as
myocardial metabolic impairment is a major feature of cardiac
hypertrophy and heart failure (46-48). The effect of SN gene therapy on
the hypertrophic heart tissues (comparison of G4 and G3 groups)
resulted in the identification of significantly enriched biological
process terms that were associated with the metabolic process.
These results indicated that SN played a protective role in cardiac
hypertrophy by regulating the metabolic process in cardiomyocytes.
KEGG analysis indicated similar results to those derived from the
GO analysis, suggesting that the differentially expressed proteins
in the hypertrophic heart were significantly associated with
cardiac muscle contraction, adrenergic signaling in cardiomyocytes
and metabolic pathways, whereas following SN gene therapy, the
differentially expressed proteins were all associated with
metabolic pathways. These results suggested that the metabolic
pathways played a critical role in the protective effect of SN in
ISO-induced cardiac hypertrophy in mice.
To further investigate the mechanism of SN
regulation in the hypertrophic heart, a protein-protein interacting
network analysis of SN was performed. The network of
protein-protein interaction surrounding SN highlighted the
identification of three proteins in 50 candidate targets by iTRAQ,
which were considered to be closely associated with the protective
role of SN-gene therapy on cardiac hypertrophy in mice. These three
proteins were Hadhb, Apoc3 and Myh7. Myh7 is a slow molecular
ATPase involved in muscle contraction (49) and several studies have
demonstrated that the gene encoding for this protein is mutated in
cardiac hypertrophy, resulting in altered protein expression levels
(50-52). Hadhb is involved in the pathway of
fatty acid β-oxidation. To the best of our knowledge, the role of
this protein in cardiac hypertrophy has not yet been demonstrated.
Apoc3 is associated with high levels of triglycerides and remnant
cholesterol. Apoc3 inhibits the hydrolysis of triglyceride-rich
lipoproteins by lipoprotein lipase (53) and reduces the uptake of
triglyceride-rich lipoproteins by the liver, thereby increasing
plasma triglyceride levels (54,55). Apoc3 has been proposed as a
potential target for reducing residual cardiovascular risk
(56-58). Several studies have demonstrated
that the role of Apoc3 in ischemic cardiovascular disease is
associated with its ability to impair plasma lipoprotein
metabolism, which in turn leads to increased triacylglyceraldehyde
levels (56-58). In the present study, Apoc3 was
identified as a major protein involved in the SN-mediated
inhibition of cardiac hypertrophy. These data were derived by iTRAQ
and were further validated by western blotting, suggesting that
Apoc3 was indeed involved in the protection of SN against cardiac
hypertrophy. The results indicated significantly reduced expression
levels of Apoc3 in hypertrophic heart tissues compared with those
noted in non-hypertrophic heart tissues. However, the expression
levels of Apoc3 were significantly increased following SN gene
therapy. These results indicated the SN protective role against
cardiac hypertrophy by increasing the levels of Apoc3 expression.
However, although the expression of Apoc3 was altered, the
limitation of present study was that Apoc3 was not knocked down to
confirm its role in protecting cardiac hypertrophy by SN. Further
follow-up studies are undergoing to provide additional information
on the protective mechanism of Apoc3 on cardiac hypertrophy by
SN.
The present study utilized comparative proteomic
analysis following iTRAQ labeling to identify 15 differentially
expressed proteins in cardiac hypertrophy and SN-overexpressed
cardiac hypertrophy groups. Moreover, the protective role of SN on
cardiac hypertrophy was revealed. The majority of these proteins
were involved in cardiac muscle contraction and metabolism
pathways. A protein-protein interaction network analysis of SN
demonstrated that of the 15 differentially expressed proteins,
Hadhb, Apoc3 and Myh7 were associated with the function of SN-gene
overexpression in cardiac hypertrophy. Western blotting indicated
that Apoc3 levels were downregulated in hypertrophic heart tissues,
whereas they were upregulated follwing SN gene therapy. Further
validation of these putative target proteins and pathways is
essential in future studies in order to improve our understanding
of the regulatory mechanisms of SN in cardiac hypertrophy.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81400212 and
81370403), Chongqing Basic and Frontier Research Project (grant
nos. CSTC2015jcyjBX0053 and CSTC2018jcyjAX0126), Chongqing
Precision Medical Key Technology Research and Development and
Demonstration Projects (grant no. cstc2016shms-ztzx0042), and the
Funds for Outstanding Young Scholars in Chongqing Medical
University (grant no. CYYQ201309).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The MS raw data can be accessed at https://iprox.org/page/SSV024.html;url=1574211720733k2Jh
with password [7DSA].
Authors' contributions
HC performed the mouse model study and contributed
to manuscript writing. MW performed the iTRAQ labeling and the
LC-MS/MS analysis. WJ performed the echocardiography and cardiac
hemodynamic measurements. XL performed the bioinformatics analysis.
JZ prepared the protein samples and performed the western blotting.
CY designed the study and contributed to manuscript writing. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The protocols were approved by the Animal Care and
Use Committee of Sichuan University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Mr. Hong Yu
(Shanghai Shengzi biological technology Co., Ltd., Shanghai, China)
for assistance with the bioinformatics analysis.
References
|
1
|
Frey N and Olson EN: Cardiac hypertrophy:
The good, the bad, and the ugly. Annu Rev Physiol. 65:45–79. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oparil S: Pathogenesis of ventricular
hypertrophy. J Am Coll Cardiol. 5:57B–65B. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cingolani HE, Ennis IL, Aiello EA and
Perez NG: Role of auto-crine/paracrine mechanisms in response to
myocardial strain. Pflugers Arch. 462:29–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Doroudgar S and Glembotski CC: The
cardiokine story unfolds: Ischemic stress-induced protein secretion
in the heart. Trends Mol Med. 17:207–214. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Finckenberg P and Mervaala E: The
cardiokine story unfolds: Ischemic stress-induced protein secretion
in the heart. J Hypertens. 28(Suppl): S33–S38. 2010. View Article : Google Scholar
|
|
6
|
Albrecht-Schgoer K, Schgoer W, Holfeld J,
Theurl M, Wiedemann D, Steger C, Gupta R, Semsroth S,
Fischer-Colbrie R, Beer AG, et al: The angiogenic factor
secretoneurin induces coronary angiogenesis in a model of
myocardial infarction by stimulation of vascular endothelial growth
factor signaling in endothelial cells. Circulation. 126:2491–2501.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pertl C, Kaufmann W, Amann R, Heinemann A,
Ebeleseder K, Polansky R, Saria A and Kim S: Secretoneurin, a novel
neuropeptide, in the human dental pulp. Arch Oral Biol. 43:361–365.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ceconi C, Ferrari R, Bachetti T, Opasich
C, Volterrani M, Colombo B, Parrinello G and Corti A: Chromogranin
A in heart failure; a novel neurohumoral factor and a predictor for
mortality. Eur Heart J. 23:967–974. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rosjo H, Masson S, Latini R, Flyvbjerg A,
Milani V, La Rovere MT, Revera M, Mezzani A, Tognoni G, Tavazzi L,
et al: Prognostic value of chromogranin A in chronic heart failure:
Data from the GISSI-Heart Failure trial. Eur J Heart Fail.
12:549–556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosjo H, Husberg C, Dahl MB, Stridsberg M,
Sjaastad I, Finsen AV, Carlson CR, Oie E, Omland T and Christensen
G: Chromogranin B in heart failure: A putative cardiac biomarker
expressed in the failing myocardium. Circ Heart Fail. 3:503–511.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jansson AM, Rosjo H, Omland T, Karlsson T,
Hartford M, Flyvbjerg A and Caidahl K: Prognostic value of
circulating chromogranin A levels in acute coronary syndromes. Eur
Heart J. 30:25–32. 2009. View Article : Google Scholar :
|
|
12
|
Yuan G, Chen H, Xia C, Gao L and Yu C:
Ultrasensitive electrochemical detection of secretoneurin based on
Pb(2+)-decorated reduced graphene oxide-tetraethylene pentamine as
a label. Biosens Bioelectron. 69:95–99. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rosjo H, Stridsberg M, Florholmen G,
Stenslokken KO, Ottesen AH, Sjaastad I, Husberg C, Dahl MB, Oie E,
Louch WE, et al: Secretogranin II; a protein increased in the
myocardium and circulation in heart failure with cardioprotective
properties. PLoS One. 7:e374012012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen HL, Liu Y, Jiang W, Wang XX, Yuan GL,
Zhao YL and Yu C: Secretoneurin suppresses cardiac hypertrophy
through suppression of oxidant stress. Eur J Pharmacol. 822:13–24.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zieske LR: A perspective on the use of
iTRAQ reagent technology for protein complex and profiling studies.
J Exp Bot. 57:1501–1508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han X, Shao W, Liu Z, Fan S, Yu J, Chen J,
Qiao R, Zhou J and Xie P: iTRAQ-based quantitative analysis of
hippocampal postsynaptic density-associated proteins in a rat
chronic mild stress model of depression. Neuroscience. 298:220–292.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sharma R, Gowda H, Chavan S, Advani J,
Kelkar D, Kumar GS, Bhattacharjee M, Chaerkady R, Prasad TS, Pandey
A, et al: Proteomic signature of endothelial dysfunction identified
in the serum of acute ischemic stroke patients by the itraq-based
lc-MS approach. J Proteome Res. 14:2466–2479. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Q, Su X, Jiang X, Dong X, Fan Y,
Zhang J, Yu C, Gao W, Shi S, Jiang J, et al: iTRAQ technology-based
identification of human peripheral serum proteins associated with
depression. Neuroscience. 330:291–325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu DD, Deng DF, Li X, Wei LL, Li YY, Yang
XY, Yu W, Wang C, Jiang TT, Li ZJ, et al: Discovery and
identification of serum potential biomarkers for pulmonary
tuberculosis using iTRAQ-coupled two-dimensional LC-MS/MS.
Proteomics. 14:322–331. 2014. View Article : Google Scholar
|
|
20
|
Mohamed BA, Asif AR, Schnelle M, Qasim M,
Khadjeh S, Lbik D, Schott P, Hasenfuss G and Toischer K: Proteomic
analysis of short-term preload-induced eccentric cardiac
hypertrophy. J Transl Med. 14:1492016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mitra A, Basak T, Ahmad S, Datta K, Datta
R, Sengupta S and Sarkar S: Comparative proteome profiling during
cardiac hypertrophy and myocardial infarction reveals altered
glucose oxidation by differential activation of pyruvate
dehydrogenase e1 component subunit β. J Mol Biol. 427:2104–2120.
2015. View Article : Google Scholar
|
|
22
|
Chowdhury D, Tangutur AD, Khatua TN,
Saxena P, Banerjee SK and Bhadra MP: A proteomic view of
isoproterenol induced cardiac hypertrophy: Prohibitin identified as
a potential biomarker in rats. J Transl Med. 11:1302013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zamorano-Leon JJ, Modrego J,
Mateos-Caceres PJ, Macaya C, Martin-Fernandez B, Miana M, de las
Heras N, Cachofeiro V, Lahera V and López-Farré AJ: A proteomic
approach to determine changes in proteins involved in the
myocardial metabolism in left ventricles of spontaneously
hypertensive rats. Cell Physiol Biochem. 25:347–358. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mesaros C and Blair IA: Mass
spectrometry-based approaches to targeted quantitative proteomics
in cardiovascular disease. Clin Proteomics. 13:202016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo J, Deng ZL, Luo X, Tang N, Song WX,
Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, et al: A protocol
for rapid generation of recombinant adenoviruses using the AdEasy
system. Nat Protoc. 2:1236–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lopez-Gordo E, Kohlbrenner E, Katz MG and
Weber T: AAV vectors for efficient gene delivery to rodent hearts.
Methods Mol Biol. 1950:311–332. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Garcia-Olloqui P, Rodriguez-Madoz JR, Di
Scala M, Abizanda G, Vales A, Olague C, Iglesias-Garcia O, Larequi
E, Aguado-Alvaro LP, Ruiz-Villalba A, et al: Effect of heart
ischemia and administration route on biodistribution and
transduction efficiency of AAV9 vectors. J Tissue Eng Regen Med.
Nov 1–2019.Epub ahead of print.
|
|
28
|
Tshori S, Gilon D, Beeri R, Nechushtan H,
Kaluzhny D, Pikarsky E and Razin E: Transcription factor MITF
regulates cardiac growth and hypertrophy. J Clin Invest.
116:2673–2681. 2006. View Article : Google Scholar PubMed/NCBI
|
|
29
|
Chen H, Wang X, Tong M, Wu D, Wu S, Chen
J, Wang X, Kang Y, Tang H, Tang C and Jiang W: Intermedin
suppresses pressure overload cardiac hypertrophy through activation
of autophagy. PLoS One. 8:e647572013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang XX, Wang XL, Tong MM, Gan L, Chen H,
Wu SS, Chen JX, Li RL, Wu Y, Zhang HY, et al: SIRT6 protects
cardiomyocytes against ischemia/reperfusion injury by augmenting
FoxO3α-dependent antioxidant defense mechanisms. Basic Res Cardiol.
111:132016. View Article : Google Scholar
|
|
31
|
Kehat I and Molkentin JD: Molecular
pathways underlying cardiac remodeling during pathophysiological
stimulation. Circulation. 122:2727–2735. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zimmer HG: Catecholamine-induced cardiac
hypertrophy: Significance of proto-oncogene expression. J Mol Med
(Berl). 75:849–859. 1997. View Article : Google Scholar
|
|
33
|
Kamalov G, Bhattacharya SK and Weber KT:
Congestive heart failure: Where homeostasis begets dyshomeostasis.
J Cardiovasc Pharmacol. 56:320–328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dunzendorfer S, Schratzberger P, Reinisch
N, Kahler CM and Wiedermann CJ: Secretoneurin, a novel
neuropeptide, is a potent chemoattractant for human eosinophils.
Blood. 91:1527–1532. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kirchmair R, Marksteiner J, Troger J,
Mahata SK, Mahata M, Donnerer J, Amann R, Fischer-Colbrie R,
Winkler H and Saria A: Human and rat primary C-fibre afferents
store and release secre-toneurin, a novel neuropeptide. Eur J
Neurosci. 6:861–868. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schgoer W, Theurl M, Albrecht-Schgoer K,
Jonach V, Koller B, Lener D, Franz WM and Kirchmair R:
Secretoneurin gene therapy improves blood flow in an ischemia model
in type 1 diabetic mice by enhancing therapeutic
neovascularization. PLoS One. 8:e740292013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chan CK and Vanhoutte PM: Secretoneurin
facilitates endothelium-dependent relaxations in porcine coronary
arteries. Am J Physiol Heart Circ Physiol. 300:H1159–H1165. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schgoer W, Theurl M, Jeschke J, Beer AG,
Albrecht K, Gander R, Rong S, Vasiljevic D, Egger M, Wolf AM, et
al: Gene therapy with the angiogenic cytokine secretoneurin induces
therapeutic angiogenesis by a nitric oxide-dependent mechanism.
Circ Res. 105:994–1002. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang K Master's degree, Chen Z Master's
degree, Long L Master's degree, Tao Y Master's degree, Wu Q
Master's degree, Xiang M Master's degree, Liang Y Bachelor's
degree, Xie X Bachelor's degree, Jiang Y Master's degree, Xiao Z
Doctor's degree, et al: iTRAQ-based quantitative proteomic analysis
of differentially expressed proteins in chemoresistant
nasopharyngeal carcinoma. Cancer Biol Ther. 19:809–824. 2018.
View Article : Google Scholar
|
|
40
|
Gupta D, Mohammed M, Mekala LP,
Chintalapati S and Chintalapati VR: iTRAQ-based quantitative
proteomics reveals insights into metabolic and molecular responses
of glucose-grown cells of Rubrivivax benzoatilyticus JA2. J
Proteomics. 194:49–59. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Luo H, Yao L, Zhang Y and Li R: Liquid
chromatography-mass spectrometry-based quantitative proteomics
analysis reveals chondroprotective effects of astragaloside IV in
interleukin-1β-induced SW1353 chondrocyte-like cells. Biomed
Pharmacother. 91:796–802. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ren J, Zhao G, Sun X, Liu H, Jiang P, Chen
J, Wu Z, Peng D, Fang Y and Zhang C: Identification of plasma
biomarkers for distinguishing bipolar depression from major
depressive disorder by iTRAQ-coupled LC-MS/MS and bioinformatics
analysis. Psychoneuroendocrinology. 86:17–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li Z, Huang T, Tang M, Cheng B, Peng Y and
Zhang X: iTRAQ-based proteomics reveals key role of
gamma-amino-butyric acid (GABA) in regulating drought tolerance in
perennial creeping bentgrass (Agrostis stolonifera). Plant Physiol
Biochem. 145:216–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen C, Hao X, Geng Z and Wang Z:
ITRAQ-based quantitative proteomic analysis of MG63 in response to
HIF-1α inducers. J Proteomics. 211:1035582019. View Article : Google Scholar
|
|
45
|
Tian L, You HZ, Wu H, Wei Y, Zheng M, He
L, Liu JY, Guo SZ, Zhao Y, Zhou RL and Hu X: iTRAQ-based
quantitative proteomic analysis provides insight for molecular
mechanism of neuroticism. Clin Proteomics. 16:382019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Diguet N, Trammell SAJ, Tannous C, Deloux
R, Piquereau J, Mougenot N, Gouge A, Gressette M, Manoury B, Blanc
J, et al: Nicotinamide riboside preserves cardiac function in a
mouse model of dilated cardiomyopathy. Circulation. 137:2256–2273.
2018. View Article : Google Scholar
|
|
47
|
Huang CY, Lee FL, Peng SF, Lin KH, Chen
RJ, Ho TJ, Tsai FJ, Padma VV and Kuo WW: HSF1 phosphorylation by
ERK/GSK3 suppresses RNF126 to sustain IGF-IIR expression for
hypertension-induced cardiomyocyte hypertrophy. J Cell Physiol.
233:979–989. 2018. View Article : Google Scholar
|
|
48
|
Wang Y, Zhang Y, Ding G, May HI, Xu J,
Gillette TG, Wang H and Wang ZV: Temporal dynamics of cardiac
hypertrophic growth in response to pressure overload. Am J Physiol
Heart Circ Physiol. 313:H1119–H1129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kesidis N, Metaxas TI, Vrabas IS,
Stefanidis P, Vamvakoudis E, Christoulas K, Mandroukas A, Balasas D
and Mandroukas K: Myosin heavy chain isoform distribution in single
fibres of bodybuilders. Eur J Appl Physiol. 103:579–583. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Petropoulou E, Soltani M, Firoozabadi AD,
Namayandeh SM, Crockford J, Maroofian R and Jamshidi Y: Digenic
inheritance of mutations in the cardiac troponin (TNNT2) and
cardiac beta myosin heavy chain (MYH7) as the cause of severe
dilated cardiomyopathy. Eur J Med Genet. 60:485–488. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang L, Zuo L, Hu J, Shao H, Lei C, Qi W,
Liu Y, Miao Y, Ma X, Huang CL, et al: Dual LQT1 and HCM phenotypes
associated with tetrad heterozygous mutations in KCNQ1, MYH7,
MYLK2, and TMEM70 genes in a three-generation Chinese family.
Europace. 18:602–609. 2016. View Article : Google Scholar
|
|
52
|
Chugh S, Ouzounian M, Lu Z, Mohamed S, Li
W, Bousette N, Liu PP and Gramolini AO: Pilot study identifying
myosin heavy chain 7, desmin, insulin-like growth factor 7, and
annexin A2 as circulating biomarkers of human heart failure.
Proteomics. 13:2324–2334. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ginsberg HN, Le NA, Goldberg IJ, Gibson
JC, Rubinstein A, Wang-Iverson P, Norum R and Brown WV:
Apolipoprotein B metabolism in subjects with deficiency of
apolipoproteins CIII and AI. Evidence that apolipoprotein CIII
inhibits catabolism of triglyceride-rich lipoproteins by
lipoprotein lipase in vivo. J Clin Invest. 78:1287–1295. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ooi EM, Barrett PH, Chan DC and Watts GF:
Apolipoprotein C-III: Understanding an emerging cardiovascular risk
factor. Clin Sci (Lond). 114:611–624. 2008. View Article : Google Scholar
|
|
55
|
Clavey V, Lestavel-Delattre S, Copin C,
Bard JM and Fruchart JC: Modulation of lipoprotein B binding to the
LDL receptor by exogenous lipids and apolipoproteins CI, CII, CIII,
and E. Arterioscler Thromb Vasc Biol. 15:963–971. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wyler von Ballmoos MC, Haring B and Sacks
FM: The risk of cardiovascular events with increased apolipoprotein
CIII: A systematic review and meta-analysis. J Clin Lipidol.
9:498–510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jorgensen AB, Frikke-Schmidt R,
Nordestgaard BG and Tybjaerg-Hansen A: Loss-of-function mutations
in APOC3 and risk of ischemic vascular disease. N Engl J Med.
371:32–41. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Luo M and Peng D: The emerging role of
apolipoprotein C-III: Beyond effects on triglyceride metabolism.
Lipids Health Dis. 15:1842016. View Article : Google Scholar : PubMed/NCBI
|