Introduction
Diabetes mellitus (DM) is a major metabolic
disorder, affecting over 350 million people worldwide in 2017
(1). The increased incidence of
DM is associated with an increased incidence of complications of
diabetes, making DM one of the most important current public health
issues (2). Diabetic enteropathy
(DE) is a common complication of DM, and the majority of previous
studies have examined neuron loss, which results in dysmotility and
altered secretion within the entire gastrointestinal tract;
therefore, it has been proposed that DE should be considered a
panenteric disorder (3). However,
in patients with DM, intestinal epithelial cells (IECs) often
undergo significant changes. A number of recent studies have shown
that DM is an independent risk factor for the occurrence and
development of colorectal cancer (4,5),
supporting the hypothesis that there is an association between DM
and the abnormal proliferation of intestinal epithelial stem cells
(IESCs).
Long noncoding RNAs (lncRNAs) constitute a cluster
of transcripts that are made of >200 nucleotides, have no
protein-coding ability and are able to regulate gene expression at
the transcriptional, epigenetic, and translational levels (6,7).
These aberrantly expressed lncRNAs may be involved in epigenetic
regulatory processes, such as chromatin modification, X chromosome
silencing and genetic imprinting and in the regulation of
transcription, translation, protein activity, and RNA alternative
splicing (8,9). The lncRNA expression profiles of a
variety of tumors are significantly different. In our previous
study, the knockdown of lncRNA H19 inhibited the abnormal
differentiation of small IECs in diabetic mice (10). Metastasis associated lung
adenocarcinoma transcript 1 (MALAT1), a highly conserved lncRNA, is
expressed at high levels in the majority of cells. Previous
research suggests that MALAT1 is involved in the pathogenesis of
various human diseases, especially cancer. A number of recent
studies point to the involvement of MALAT1 in the proliferation of
cancer cells, vascular smooth muscle cells, high glucose-induced
endothelial cells and periodontal ligament stem cells (11-13). However, the role of MALAT1 in the
proliferation of IESCs in DM requires further understanding.
MicroRNAs (miRNAs) are a major class of short (~22 nucleotides)
noncoding RNAs that function to block protein translation and/or
degrade their messenger RNA targets. They bind to complementary
sequences in the 3′-untranslated regions (UTRs), 5′UTRs, and/or
coding regions of target mRNAs (11). One of the most novel functions of
lncRNAs is their ability to serve as competing endogenous RNAs
(ceRNAs), which compete with coding RNAs for shared miRNAs, thus
regulating the functions of these genes (14,15). miRNAs direct a number of important
processes that are associated with cellular growth, apoptosis,
differentiation, metabolism and the immune response (16). Furthermore, miRNAs are known to
have a pivotal role in DM (10,17). Numerous studies have reported the
roles of miR-129-5p (18-20). miR-129-5p is involved in lncRNA
Trinucleotide repeat containing adaptor 6C-antisense RNA 1-mediated
processes and regulates Unc-5 netrin receptor B in thyroid cancer
to influence cell proliferation, migration, and invasion (18). Although studies have investigated
the functions of miR-129-5p in cancer, its role in IESCs of DM
remains largely unclear. Therefore, the present study investigated
whether MALAT1 functions as a miRNA sponge by regulating the
proliferation of IESCs in DM.
IESCs are located in the intestinal crypt, which
maintains intestinal epithelial balance by regulating its own
proliferation and differentiation (18,21). The normal proliferation of IESCs
is maintained by highly specialized and well-regulated signaling
cascades. The Wnt pathway is a classical pathway in the IEC, and
this pathway acts primarily on IESCs to promote intestinal
epithelial proliferation. Additionally, deregulated Wnt signaling
is involved in the pathophysiological processes of numerous
diseases, including the occurrence and development of cancer
(22). Recent evidence suggests
that Wnt/β-catenin signaling is activated under diabetic conditions
(23) and that increased
proliferation of IECs in diabetic rats has been associated with the
accumulation of β-catenin (23).
In our previous study, the abnormal proliferation of IECs in DM
mice was observed (24); however,
the potential mechanisms underlying the association between
Wnt/β-catenin signaling pathway activation and abnormal IESC
proliferation in DM are still poorly understood.
Numerous studies on MALAT1 have examined its role in
the progression and prognosis of cancer (25-27). Despite the aforementioned
findings, the role of MALAT1 in IESCs of DM needs to be
investigated. In the present study, MALAT1 expression was
significantly elevated in the IESCs of DM mice. Furthermore, it was
demonstrated that MALAT1 acts as a 'molecular sponge' for
miR-129-5p to regulate the abnormal proliferation of IESCs via the
Wnt signaling pathway.
Materials and methods
Streptozocin (STZ)-induced DM mice
model
A total of 96 8-week-old male C57BL/6J mice (weight,
20-40 g) were obtained from the Animal Laboratory Center in The
Affiliated Hospital of Qingdao University (Qingdao, China). All
animals were maintained in a thermostatically controlled room with
a 12-h light/dark cycle. Diabetes was induced by daily
intraperitoneal injection of STZ (Sigma-Aldrich; Merck KGaA; 70
mg/kg) for 5 days (10,17,28); and the mice in the control group
received ip injections the same volume of citrate buffer (0.1 mol/l
(10,17,28). DM in the experimental mice was
defined as a fasting blood glucose ≥16.7 mmol/l for ten consecutive
days (10,17,28). Then, all mice were euthanized with
an intraperitoneal injection of ketamine/xylazine (100/10 mg/kg
body weight). The small intestines were carefully removed and
flushed with 0.1 M PBS (pH 7.4) for the isolation of primary IESCs.
All experiments with mice were approved by the Animal Care
Committee of The Affiliated Hospital of Qingdao University.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples and cell
lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Then, RT was performed with PrimeScript™ RT Master mix
(Takara Biotechnology Co., Ltd.). The RT conditions were as
follows: 42°C for 5 min and 95°C for 10 sec. qPCR was performed on
a CFX Connect™ real-time PCR detection system (Bio-Rad
Laboratories, Inc.) using SYBR® Premix Ex Taq™ (Takara
Biotechnology Co., Ltd.). The thermo-cycling conditions were as
follows: Initial denaturation at 95°C for 5 min, followed by 40
cycles of denaturation at 95°C for 15 sec and annealing/elongation
at 60°C for 30 sec. miRNA expression levels were measured using the
SYBR PrimeScript™ miRNA RT-PCR kit (Takara Biotechnology Co.,
Ltd.). miRNA samples were normalized to U6. Each experiment was
repeated six times. The primers are described in Table SI. The data
were analyzed using the 2−ΔΔCq method (29).
Bioinformatics analysis
DIANA-LncBase Predicted v.2 (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=lncBase/index)
and TargetScan 7.2 (http://www.targetscan.org/) were used to predict the
putative target genes for MALAT1 and miR-129-5p.
Culture of cell lines
CT26 cells, NIH 3T3 cells and 293T cells were
obtained from American Type Culture Collection. These cell lines
were all cultured under standard culture conditions as described
previously (10,17). Cell lines were cultured in DMEM
(Invitrogen; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin.
Cells were grown in a 95% air, 5% CO2 atmosphere at
37°C.
Primary IESC isolation and culture
Primary IESCs were isolated from the small
intestines of mice and cultured in Matrigel as described previously
(10,17,30,31).
Cell transfection
The siRNAs targeting MALAT1 were purchased from
GenePharma (si#1 and si#2; Shanghai GenePharma Co., Ltd.). CT26
cells and primary IESCs were seeded in six-well plates 1 day prior
to transfection. The siRNAs (15 nM), miRNA mimic (15 nM) and
inhibitor (15 nM) were transfected into cells according to the
Lipofectamine 3000 transfection reagent protocol (Thermo Fisher
Scientific, Inc.). The miRNA mimic (agomiR-129-5p) and inhibitor
(antagomiR-129-5p) were all purchased from Shanghai GenePharma Co.,
Ltd. The silencing efficiency was evaluated at 48 h following
transfection using RT-qPCR. Each experiment was repeated six
times.
Dual-luciferase reporter plasmid
transfection
The MALAT1 and SRY-box 9 (SOX9) wild-type (WT)
sequences with potential miR-129-5p-binding sites were amplified
from the genomic DNA of NIH 3T3 cells and cloned into the
pmiR-RB-REPORT™ plasmid (Guangzhou RiboBio Co., Ltd.). A plasmid
containing a potential miR-129-5p-binding site with a mutation was
used as the negative control (Guangzhou RiboBio Co., Ltd.).
Mutations were introduced with the KOD-plus mutagenesis kit (Toyobo
Life Science). Measurements of firefly and Renilla
luciferase activity were performed using the Dual-Luciferase
Reporter Assay system (Promega Corporation). Briefly, A total of 48
h after cell transfection, for the Dual-Luciferase Reporter assay,
the firefly luciferase reporter was measured first by adding
Luciferase Assay Reagent II to generate a stabilized luminescent
signal. After the firefly luminescence was quantified for 3 min,
the reaction was quenched, and the Renilla luciferase
reaction was simultaneously initiated by adding Stop &
Glo® reagent to the same tube. The Stop &
Glo® reagent also produced a stabilized signal from the
Renilla luciferase protein, which decayed slowly over the
course of the measurement. The luciferase efficiency was evaluated
at 2 min following Stop & Glo® reagent using
SpectraMAX Multifunctional Microplate Reader (Molecular Devices).
Each experiment was repeated six times.
Downregulating the expression of MALAT1
in vivo
A total of 120 C57BL/6J mice were randomly divided
into five groups, with 24 in each group. All mice received a tail
vein injection once a day for 3 days. The Con-NS group comprised
control mice receiving saline (0.9%; same volume as the
experimental group) injections (14,15), and the DM-NS group comprised DM
mice receiving saline (0.9%; same volume as the experimental group)
injections (14,15); the DM-siRNA (si#1 and si#2) mice
received injections of MALAT1 siRNAs (80 mg/kg body weight
(14,15), and the DM-CT mice received
injections of antagomiR-129-5p and MALAT1 siRNAs (80 mg/kg body
weight (14,15). In each group, six mice were
euthanized with an intraperitoneal injection of ketamine/xylazine
(100/10 mg/kg body weight) on day 0 (prior to injection), days 2, 4
and 6 for further study. Each experiment was repeated six
times.
Fluorescence in situ hybridization
A DIG-labeled LNA-MALAT1 probe was synthesized by
Bersinbio (Guangzhou, China.) and the probe sequences are available
upon request. In brief, a 5-mm section of paraffin-embedded tissues
was incubated with 50% methanol in PBST (0.1% Tween-20) solution
for 5 min, 30% methanol for 5 min, PBST solution for 5 min and then
with 4% paraformaldehyde in PBS solution for 20 min at room
temperature. The tissues was washed twice with PBST for 5 min at
room temperature and then followed by the treatment with proteinase
K (15 µg/ml; New England Biolabs) at 37°C for 15 min. After
being washed three times with PBS and dried with ethanol, the
section was hybridized using 30 nM LNA-MALAT1 probe at 55°C for 1
h. After three incubations with SCC buffer at 60°C for 30 min, the
samples were washed with PBST (PBS containing 0.1% Tween-20) for 15
min three times at room temperature. The section was then incubated
with anti-DIG-AP (cat. no. 11093274910; 1:300; Roche Diagnostics)
at 4°C overnight. Then, the section was stained with NBT/BCIP
(Thermo Fisher Scientific, Inc.) at 30°C for 2 h, and the reaction
was stopped with stop-buffer. When the section was dried with
ethanol, the expression of MALAT1 was determined using
diaminobenzidine solution (1:900; Boster Biological Technology) for
3 min at room temperature, and the staining intensity was observed
using a fluorescence microscope (Olympus Corporation). The staining
was quantified by counting the number of positive cells at a
magnification of ×400. Each experiment was repeated six times.
Immunohistochemistry
A 5-mm section was prepared from the
paraffin-embedded intestinal section, and hematoxylin and eosin
(H&E) staining was further used for histological analysis. In
brief, fresh tissue was fixed with 4% paraformaldehyde for 24 h at
room temperature, then dehydrated with a gradient alcohol series
(75% alcohol for 4 h, 85% alcohol for 2 h, 90% alcohol for 2 h, 95%
alcohol for 1 h), absolute ethanol II for 30 min, alcohol benzene
for 5-10 min, xylene for 5-10 min, and wax for 3 h; the wax-soaked
tissue was embedded and stored at −20°C. After the wax had
solidified, it was paraffin embedded and sliced. For
immunohistochemistry, sections were placed in 1% hydrogen peroxide
in PBS for 10 min and then placed in citrate buffer in a pressure
cooker for 45 min (Beyotime Institute of Biotechnology). Goat serum
(5-10%; Beyotime Institute of Biotechnology) was applied for 30 min
at room temperature, and then sections were incubated with
anti-BrdU (cat. no. 560210; 1:200; BD Biosciences; Becton,
Dickinson and Company,) and anti-SOX9 antibody (cat. no. 82630;
1:250; Cell Signaling Technology, Inc.) antibodies overnight at
4°C. The tissue was then incubated with EnVision+/HRP/Rb (Dako;
Agilent Technologies, Inc.) for 30 min at room temperature. Slides
were developed using 3,3-diaminobenzidine tetrahydrochloride (DAB)
and counterstained with hematoxylin. The H&E-stained sections
were imaged with a light microscope BX51 (Olympus Corporation) to
measure the length of villi and the level of cell proliferation.
The stained sections were quantified by counting the number of
positive cells at a magnification of ×400 in 10 contiguous,
well-oriented intestinal crypts by an examiner blinded to sample
identity. Each experiment was repeated six times.
Protein extraction and western
blotting
Total protein from tissues and cells was isolated in
RIPA Buffer (Thermo Fisher Scientific, Inc.) containing a protease
inhibitor cocktail (Roche Applied Science). Protein samples (40
µg/sample) were separated on 10% SDS-PAGE gels. The
separated proteins were transferred onto polyvinylidene fluoride
membranes and blocked with 5% skim milk at room temperature for 1
h. Blots were incubated at 4°C overnight with primary antibodies:
Anti-β-catenin antibody (cat. no. 8480S; 1:1,000), anti-SOX9
antibody (cat. no. 82630; 1:1,000), anti-cyclin D1 antibody (cat.
no. 55506S; 1:1,000), anti-cyclin-dependent kinase 2 (CDK2)
antibody (cat. no. 2546; 1:1,000), anti-cell division cycle 42
(CDC42) antibody (cat. no. 2466; 1:1,000), and anti-β-actin
antibody (cat. no. 4970; 1:1,000) (all from Cell Signaling
Technology, Inc.). After three washes with TBS-T, the membranes
were washed and incubated for 1 h with horseradish
peroxidase-conjugated secondary antibody (cat. no. 7074S; Cell
Signaling Technology, Inc.) at 37°C. The blots were visualized
using an enhanced chemiluminescence Ultra Western HRP Substrate kit
(cat. no. WBULS0100; EMD Millipore) and autoradiography with X-ray
film. Protein quantification was analyzed by Quantity One software
version 4.6.2 (Bio-Rad Laboratories, Inc.) and the intensity values
were normalized to β-actin. Each experiment was repeated six
times.
Statistical analysis
Results are expressed as the mean ± standard
deviation and analyzed using the statistical software package (SAS
8.0 for Windows; SAS Institute, Inc.). Comparisons between groups
were analyzed using a Student's test and multiple group comparisons
were analyzed using one-way ANOVA with Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
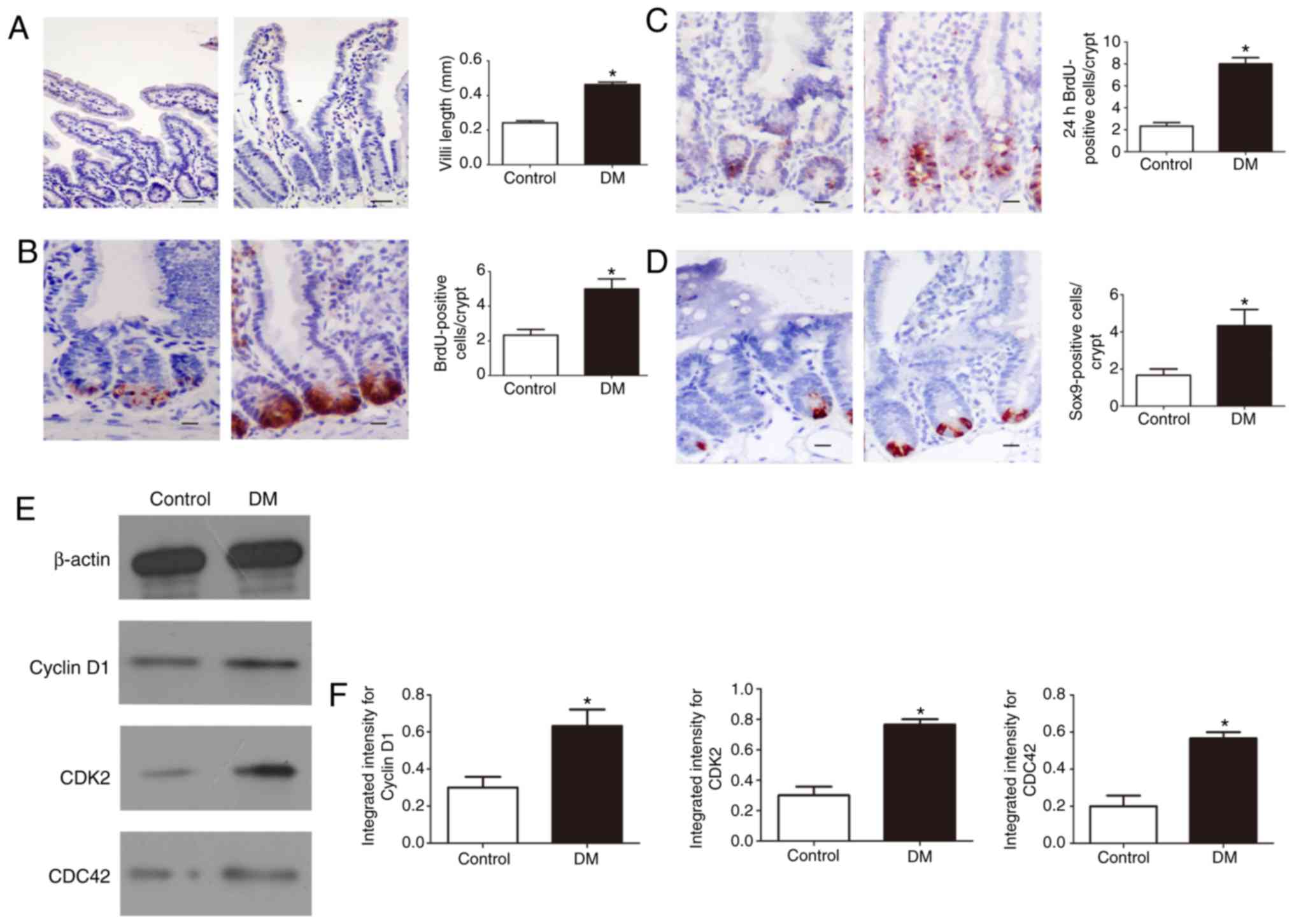
Abnormal proliferation of IECs in DM
mice
To further evaluate IEC proliferation in DM,
immunostaining was used to measure the length of the villi. The
results showed that the length of the villi was significantly
increased in DM mice compared with that in control mice (P<0.05;
Fig. 1A). Moreover, BrdU
(intraperitoneal) injection into mice was used to label the S-phase
cells of the intestinal epithelium. BrdU analysis showed that the
BrdU positivity of intestinal crypts in DM mice increased
significantly compared with that in the control group after 1 h of
BrdU injection (P<0.05; Fig.
1B). Additionally, the number of BrdU-positive cells in DM mice
increased significantly after 24 h (P<0.05; Fig. 1C). To detect a significant
increase in intestinal cell proliferation, SOX9 staining was
performed, which is a marker for the crypt cell population
containing stem and progenitor cells. The crypts in the IECs of the
DM mice contained an increased number of SOX9-positive cells
compared with those of the control mice (P<0.05; Fig. 1D). Based on the observed abnormal
proliferation of IECs, the levels of several important cell
cycle-related proteins were determined. An increased expression of
cyclin D1, CDK2, and CDC42 was observed in the IECs of the DM mice
compared with the control group (P<0.05; Fig. 1E and F). These findings suggest
that the abnormal proliferation of stem cells in the crypt may be
responsible for the elongation of villi in DM mice.
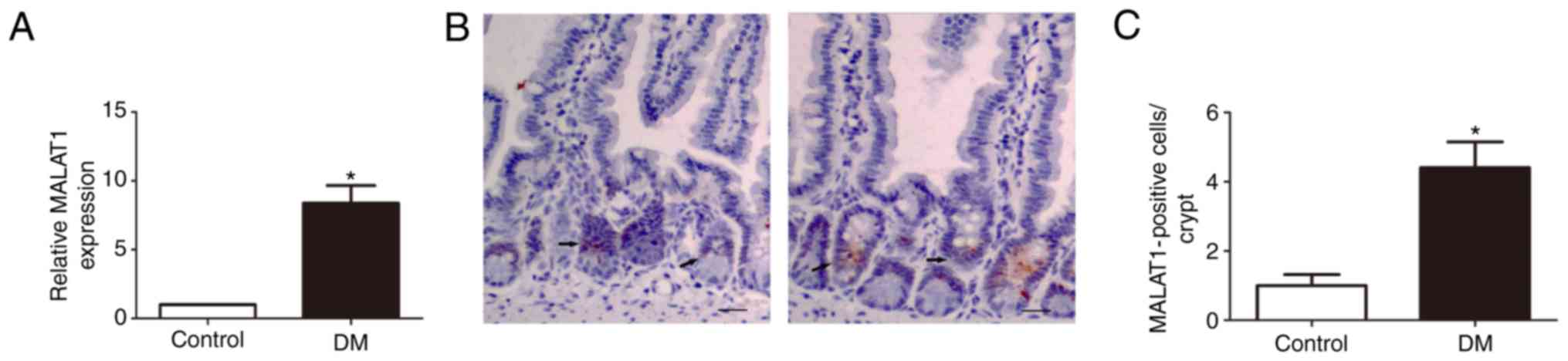
MALAT1 is upregulated in the IESCs of DM
mice
To determine whether MALAT1 is expressed in the
IESCs of DM mice, its expression profile was examined.
Interestingly, RT-qPCR analysis revealed that MALAT1 levels were
significantly upregulated in the IESCs of DM compared with normal
tissues (P<0.05; Fig. 2A).
Fluorescence in situ hybridization of a DIG-labeled
LNA-MALAT1 probe further showed that MALAT1 was predominantly
localized to the cytoplasm of the intestinal crypt and that
expression of MALAT1 in DM mice was significantly increased
(P<0.05; Fig. 2B and C).
Therefore, it was hypothesized that MALAT1, whose function is still
not fully understood, plays an important role in the abnormal
proliferation of IECs in DM mice.
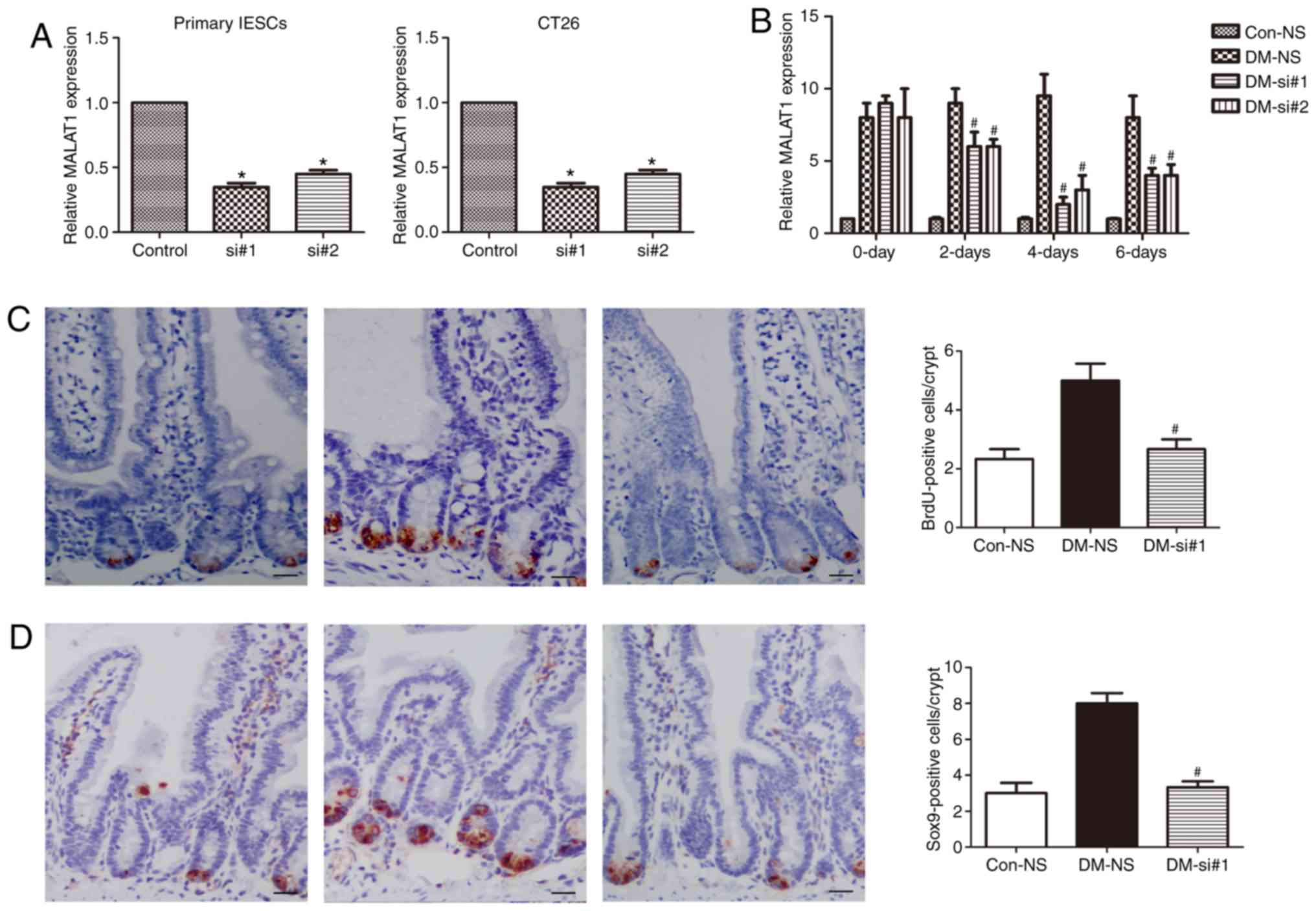
Knockdown of MALAT1 inhibits the abnormal
proliferation of IESCs in DM mice
To study the functional role of MALAT1 in the IESCs
of DM, knockdown of MALAT1 in primary IESCs and CT26 cells was
conducted. RT-qPCR showed that siRNA-mediated knockdown of MALAT1
significantly downregulated the expression of MALAT1 in both cell
lines (P<0.05; Fig. 3A).
Additionally, tail vein injections of two siRNAs against MALAT1
into DM mice were performed. MALAT1 expression in the IESCs of
DM-siRNAs mice was significantly downregulated at 2, 4 and 6 days
compared to that in DM-NS mice (P<0.05; Fig. 3B). MALAT1 expression on the 4th
day after DM-siRNA injection was similar to that in the Con-NS
mice. On the 4th day after DM-siRNA injection, BrdU was injected
into DM-siRNA mice. One hour after the BrdU injection, the increase
in BrdU positivity in the intestinal crypts was significantly
inhibited in the DM-siRNA mice (P<0.05; Fig. 3C) and close to the levels observed
in the Con-NS mice (P<0.05; Fig.
3C). Additionally, SOX9 expression was measured using
immunohistochemistry, and the expression of SOX9-positive cells in
DM-siRNA mice was significantly decreased after siRNA
administration (P<0.05; Fig.
3D). These data indicate that MALAT1 may be involved in IESC
proliferation in DM mice.
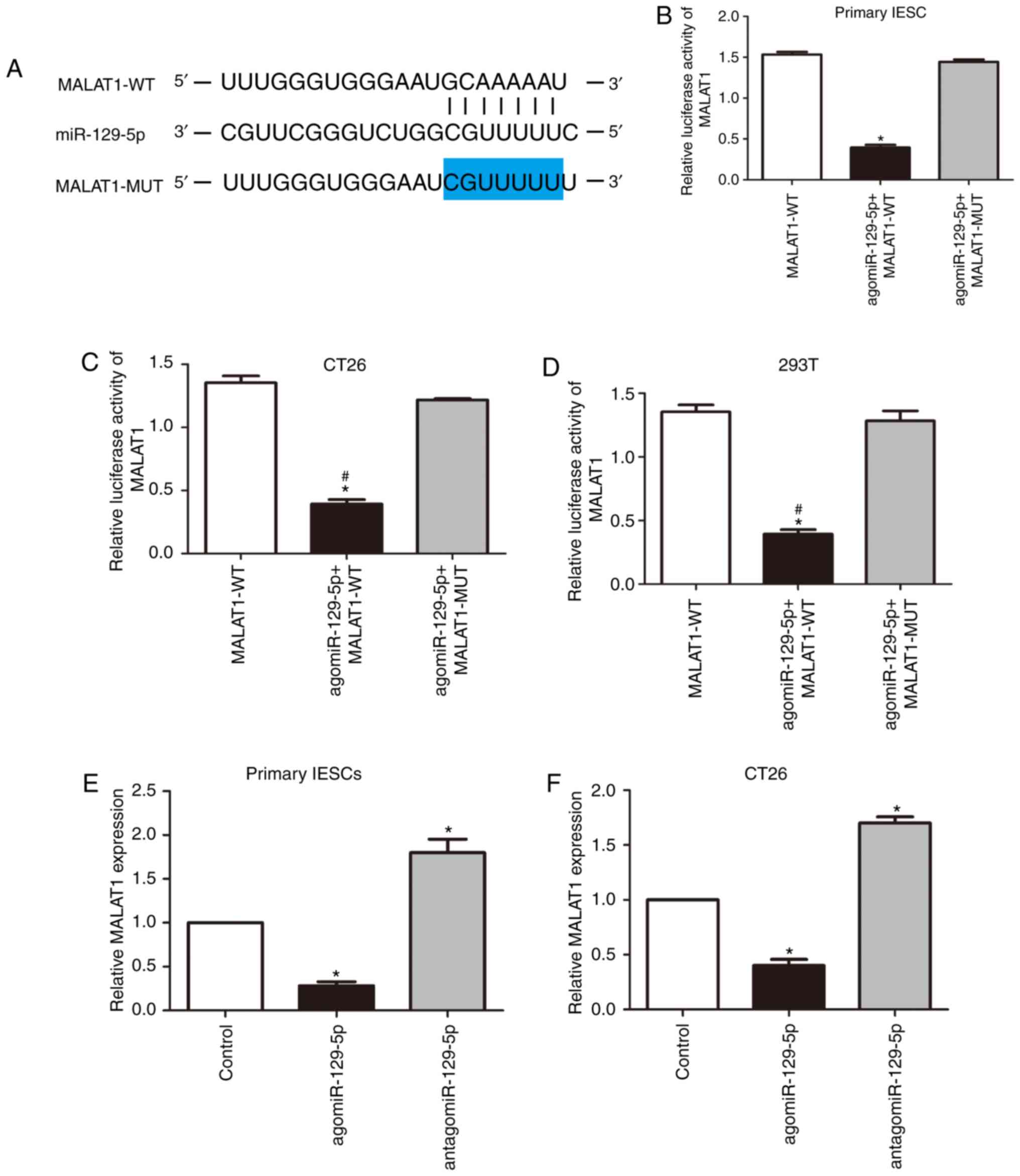
miR-129-5p directly binds and
downregulates MALAT1
To further investigate the potential mechanism by
which MALAT1 contributes to the abnormal proliferation of IESCs,
DIANA-LncBase Predicted v.2 and the TargetScan database were used
to predict a MALAT1 microRNA target and selected miR-129-5p
(P<0.05; Fig. 4A). To verify
the prediction, wild-type (MALAT1-WT) and miR-129-5p-binding-site
mutant (MALAT1-MUT) MALAT1 luciferase reporters were constructed.
As shown in Fig. 4B-D, miR-129-5p
expression significantly attenuated the luciferase activity of the
reporter with WT MALAT, but did not attenuate that of the mutant
reporter (P<0.05). To further assess the potential association
between miR-129-5p and MALAT1, primary IESCs and CT26 cells were
transfected with antagomiR-129-5p or agomiR-129-5p. It was
demonstrated that restoration of miR-129-5p significantly reduced
MALAT1 levels, whereas antagonism of miR-129-5p increased MALAT1
expression (P<0.05; Fig. 4E and
F). Taken together, these results indicate that MALAT1-mediated
promotion of IESC proliferation is partly dependent on miR-129-5p
sponging.
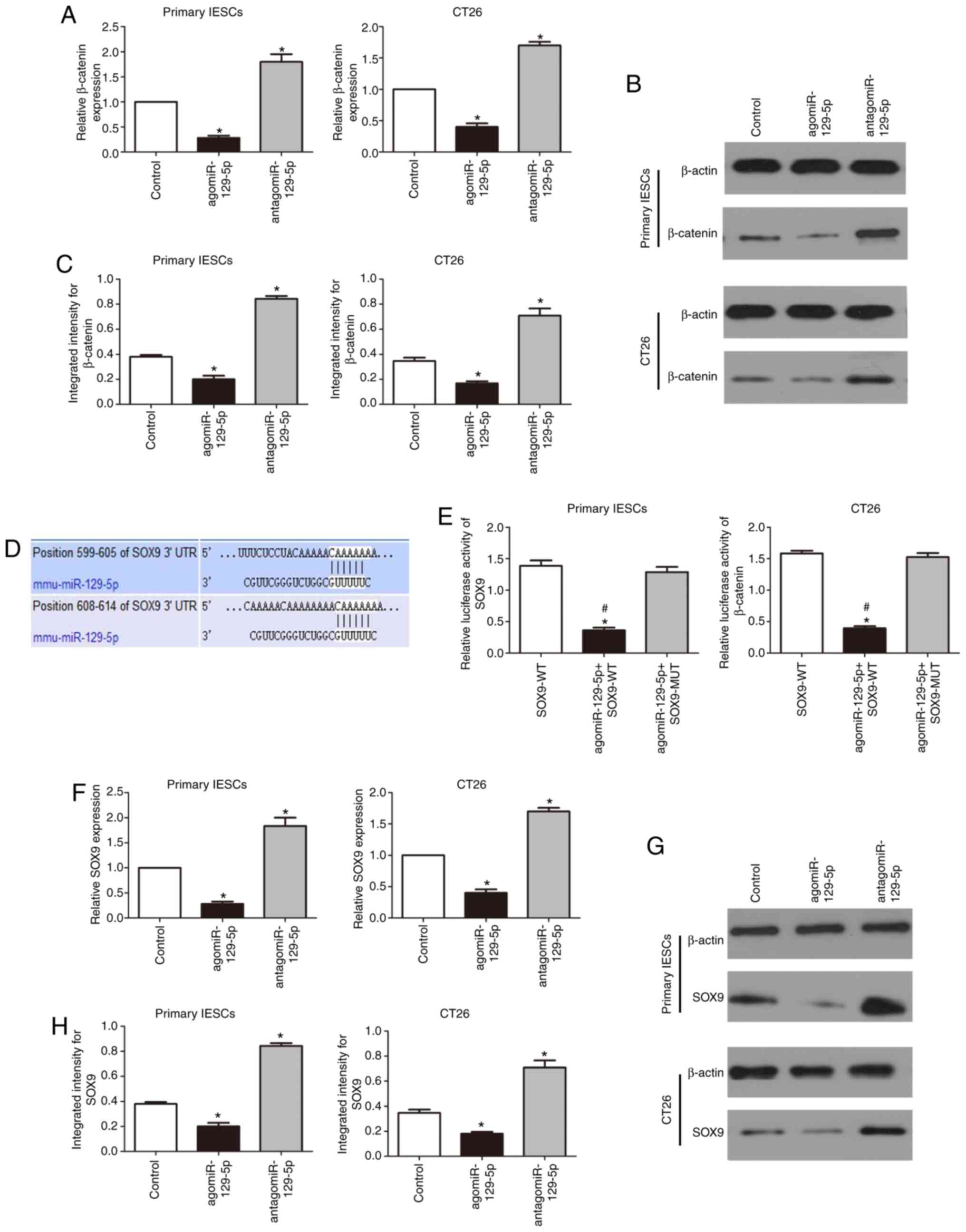
miR-129-5p regulates WNT/β-catenin
signaling by targeting SOX9
The WNT/β-catenin signaling pathway is essential for
maintaining the development and homeostasis of IECs. Overexpression
of miR-129-5p in primary IESCs and CT26 cells was performed and it
was demonstrated that β-catenin mRNA and protein expression levels
decreased. Inhibition of miR-129-5p expression resulted in the
significant upregulation of β-catenin expression levels (P<0.05;
Fig. 5A-C). SOX9 was identified
as a downstream target of miR-129-5p using the TargetScan
(http://www.targetscan.org) online
database. There are two putative SOX9 binding sites within
miR-129-5p: Regions 599-605 and 608-614 (Fig. 5D). Activity analysis demonstrated
that luciferase expression in IESCs and CT26 cells that were
cotransfected with miR-129-5p and the SOX9-WT plasmid were
significantly decreased compared with that of the cells
cotransfected with miR-129-5p and the SOX9-MUT plasmid or
transfected with the SOX9-WT plasmid alone (P<0.05; Fig. 5E). To further assess the potential
association between miR-129-5p and SOX9, cells were transfected
with antagomiR-129-5p or agomiR-129-5p. As shown in Fig. 5F, forced expression of miR-129-5p
significantly decreased SOX9 mRNA expression, while inhibition of
miR-129-5p expression significantly increased SOX9 expression at
the mRNA level (P<0.05); likewise, the protein expression of
SOX9 was significantly decreased and increased, respectively
(P<0.05; Fig. 5G and H). Our
previous study showed that SOX9 regulates WNT/β-catenin signaling
in the IESCs of diabetic mice (25). These data further showed that
miR-129-5p regulates WNT/β-catenin signaling by targeting SOX9.
MALAT1 knockdown results in the
downregulation of β-catenin expression through miR-129-5p in DM
mice
First, primary IESCs and CT26 cells were transfected
with siRNAs or cotransfected them with antagomiR-129-5p and siRNAs;
the inhibition of MALAT1 significantly suppressed SOX9 and
β-catenin expression, whereas inhibition of miR-129-5p partly
abolished the silencing effect of MALAT1 knockdown on SOX9 and
β-catenin (P<0.05; Fig. 6A-C).
Then, to investigate the biological role of MALAT1 in the abnormal
proliferation mechanism of IESCs in DM, siRNAs were subcutaneously
injected into mice. The levels of SOX9 and β-catenin protein in the
DM-siRNA mice were significantly reduced compared with those in the
DM-NS mice (P<0.05; Fig. 6D and
E) and were similar to those in the Con-NS mice. Additionally,
the expression levels of SOX9 and β-catenin proteins were higher in
DM-CT mice compared with in DM-siRNA mice (P<0.05; Fig. 6D and E) and similar to those in
DM-NS mice. More importantly, in our previous study, β-catenin was
localized to the crypts of IECs (10). These results suggest that MALAT1
can regulate WNT/β-catenin signaling by sequestering endogenous
miR-129-5p in the IESCs of DM.
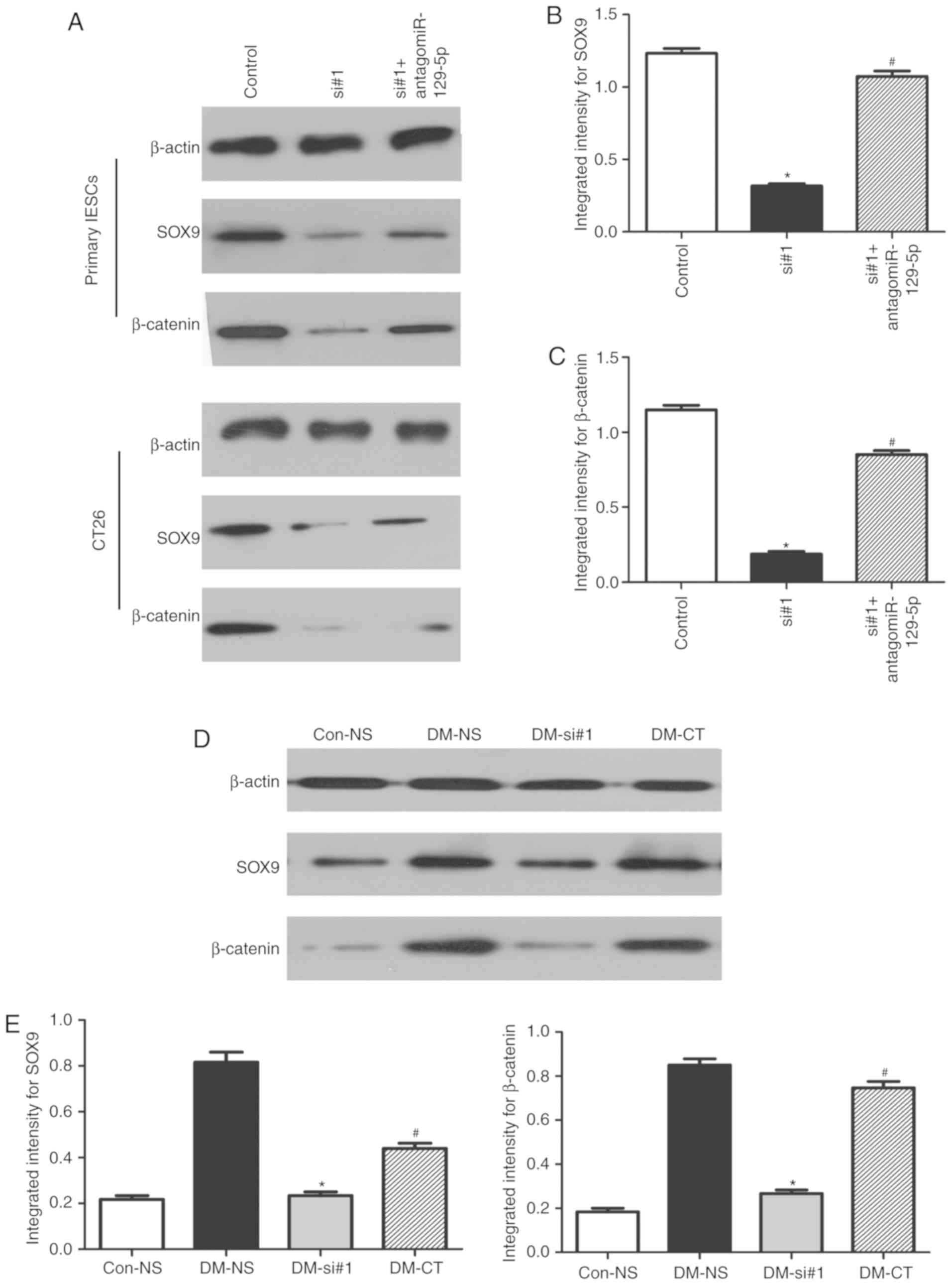 | Figure 6MALAT1 knockdown induces the
downregulation of β-catenin levels. (A) The levels of SOX9 and
β-catenin in primary IECs and CT26 after transfection with siRNAs
or co-transfection with antagomiR-129-5p and siRNAs were determined
by western blotting analysis. Quantification of (B) SOX9 and (C)
β-catenin protein levels. n=6; *P<0.05 vs. NC;
#P<0.05 vs. siRNA groups. (D) Representative image
and (E) quantification of the protein levels of miR-129-5p targets
in DM mice after tail vein injections of siRNAs or cotransfection
with antagomiR-129-5p or siRNAs. n=6; *P<0.05 vs.
DM-NS group; #P<0.05 vs. DM-siRNAs group). IESCs,
intestinal epithelial stem cells; miRNA/miR, microRNA; siRNA/si,
small interfering RNA; antagomiR, antagonist miR; SOX9, SRY-box 9;
MALAT1, metastasis associated lung adenocarcinoma transcript 1;
Con-NS, control mice receiving saline; DM-NS, DM mice receiving
saline; DM-si, DM mice receiving siRNA; DM-CT, DM mice receiving
antagomiR-129-5p and MALAT1 siRNAs. |
Discussion
As a complication of DM, DE is often present in
diabetic patients, and there is still a lack of early diagnosis and
effective treatment measures to mitigate the harmful and
potentially irreversible effects of DE on the small intestine. A
growing body of evidence indicates that abnormal expression of
lncRNAs is associated with tumorigenesis and development, and that
certain lncRNAs are associated with poor cancer prognosis (32-34). In fact, previous studies have
demonstrated that MALAT1 can cause the tumorigenesis and
development of multiple types of tumors, as well as abnormal
development of vascular smooth muscle and endothelial cells
(12,13). However, the role of MALAT1 in the
IESCs of DM is still is poorly understood. In the present study, a
pathogenic role for MALAT1 was revealed in the IESCs of DM and its
possible molecular mechanisms were elucidated. The findings
suggested that MALAT1 is a specific lncRNA that causes the abnormal
proliferation of IESCs in DM. In the present study, MALAT1
expression was significantly increased in the IESCs of DM mice.
IESCs are located in the intestinal crypt of the intestinal
epithelium and are mainly responsible for the renewal of IECs.
Fluorescence in situ hybridization of a DIG-labeled
LNA-MALAT1 probe showed that MALAT1 appears to be predominantly
localized to the crypts of IECs and to the cytoplasm of crypt
cells. Taken together, the data indicate that MALAT1 appears to
play an important role in regulating the cell fate of the IESC in
DM. Although the classical pathway for controlling IESC
proliferation has been studied, little is known about the
underlying molecular mechanisms controlling IESC proliferation
(10,17,24,35). In our previous studies, we found
an increase in the number of goblet cells, Paneth cells, and
absorptive cells and a reduction in endocrine cells in DM mice
(10,17). Immunostaining was used to show
that the length of the villi was significantly increased in DM
mice. Moreover, BrdU positivity was significantly higher in the
intestinal crypts of DM mice than in those of normal mice after 1
and 24 h. Given that IECs are highly dependent on IESC function,
whether Sox9 affects IESCs was investigated. It was observed that
the crypts in the IECs of the DM mice contained an increased number
of Sox9-positive cells compared with those of the control mice. The
precise balance between IESC proliferation and the principal
molecular determinant of IESC proliferation remains to be
elucidated. Inspired by these results, it was hypothesized that
MALAT1 may be involved in the abnormal proliferation of IESCs in
DM. Using loss-of-function methods in vivo, it was revealed
that MALAT1 plays a key role in the abnormal proliferation of IESCs
in DM. Following siRNA injections into DM mice, the abnormal
proliferation of IESCs was normalized according to the results of
BrdU assays. These results indicate that inhibiting MALAT1 may be
an effective method to prevent the abnormal proliferation of IESCs
in DM mice. However, it remained plausible that other upregulated
lncRNAs or even downregulated lncRNAs may exhibit large differences
in the DM model. Additionally, the regulatory network of lncRNAs
appears to be complex and may be highly dependent on the cellular
context. Thus, more research is needed in the future.
Recently, emerging evidence has suggested that
lncRNAs regulate miRNAs by functioning as endogenous sponges, and
it has been experimentally demonstrated that miRNA targeting
regulates the stability of lncRNAs (36,37). In accordance with the ceRNA
hypothesis, MALAT1 also functions as a decoy to reduce or eliminate
the effects of ceRNAs on their native mRNA targets. With the help
of bioinformatics analysis, MALAT1 was confirmed to harbor
miR-129-5p binding sites. To further clarify this, the association
between MALAT1 and miR-129-5p was analyzed by performing luciferase
assays. Restoration of miR-129-5p levels reduced MALAT1 levels,
whereas antagonism of miR-129-5p increased MALAT1 expression. These
studies indicate that miR-129-5p directly inhibits MALAT1
expression by directly targeting MALAT1.
As a class of nucleic acid-based molecules, miRNAs
have therapeutic potential, depending on their characteristics, to
modulate one or more gene targets within a particular signal
transduction pathway, or even one or more targets across multiple
independent pathways (38). Study
has also confirmed the important role of miRNAs in the abnormal
proliferation of cancer stem cells (39). Studies have shown that miR-129-5p
plays important roles in regulating the proliferation of various
human cancer types (40) and in
diabetes (41). However, in the
diabetic context, the mechanism of miR-129-5p actions in the
proliferation of IESCs remains unclear. In the present study,
bioinformatics analysis suggested that SOX9 was a target of
miR-129-5p. To further confirm that SOX9 was a target of
miR-129-5p, the effect of cotransfection of the miR-129-5p and
SOX9-WNT plasmids was investigated by evaluating luciferase
activity. The analysis showed that luciferase expression in cells
cotransfected with miR-129-5p and the SOX9-WT plasmid was
significantly decreased compared with that in cells cotransfected
with miR-129-5p and the SOX9-MUT plasmid or transfected with the
SOX9-WT plasmid alone. Thus, the luciferase reporter analysis
confirmed that SOX9 is a target gene of miR-129-5p. To further
confirm the association between miR-129-5p and SOX9, SOX9
expression was evaluated in primary IESCs and CT26 cells after
transfection of miR-129-5p mimic or inhibitor. It was found that
SOX9 expression was decreased at the mRNA and protein levels after
upregulation of miR-129-5p expression. In contrast, downregulation
of miR-129-5p expression increased SOX9 expression at the mRNA and
protein levels. These results confirm that miR-129-5p could
directly target SOX9 and suggest a potential mechanism for
regulating SOX9 expression. Our previous study showed that SOX9
regulates WNT/β-catenin signaling in IESCs in the diabetic context
(30).
In mammals, the WNT/β-catenin signaling pathway is
essential for maintaining IESC proliferation and IEC homeo-stasis
(42-44). The effect of MALAT1 on the
Wnt/β-catenin pathway has not been fully investigated under
diabetic conditions. To further elucidate the underlying molecular
mechanism of abnormal IESC proliferation in DM, the expression of
MALAT1 in DM was examined by transfecting siRNAs in vitro
and in vivo. After transfection of siRNA in primary IECs and
DM mice, the downregulation of MALAT1 inhibited the expression of
SOX9 and β-catenin, in a similar manner to that of the miR-129-5p
mimic. Furthermore, lowering the expression of miR-129-5p partially
offset the silencing effect of MALAT1 on these genes. Therefore,
the effect of MALAT1 on IESC proliferation in DM could be explained
in part by its function as a molecular sponge of miR-129-5p,
thereby confirming the role of MALAT1 in the regulatory mechanism
of IESC proliferation. A study has demonstrated that the
upregulation of MALAT1 is induced by activation of the
Wnt/β-catenin pathway (45).
However, there are numerous reports on MALAT1-mediated regulation
of β-catenin expression (26,46). In this study, in the context of
diabetes, the downregulation of MALAT1 was demonstrated to inhibit
the expression of SOX9 and β-catenin. We hypothesize that in the
complex signaling networks of cells, adjustments between MALAT1 and
β-catenin may occur through direct or indirect means, which may
depend on different conditions. Thus, there is a need for more
research in the diabetic context.
The results of the present study showed that IESC
proliferation depended partly on MALAT1 expression. The potential
mechanisms underlying these effects involve the regulation of the
WNT/β-catenin signaling pathways, MALAT1 actions as a molecular
sponge for miR-129-5p and MALAT1-mediated regulation of its target
gene SOX9. Furthermore, evidence for an apoptotic mechanism that
serves to govern the proliferation of IESCs in DM was provided,
offering a platform for the development of targeted
therapeutics.
Supplementary Data
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TDS conceived the study, wrote the original draft,
collected the data for the proliferation of IESCs and investigated
the SOX9-mediated WNT/β-catenin signaling pathway in mice. ZBT
collected the expression data of MALAT1 in mice. YPJ collected the
data for the number of SOX9-positive cells. TDS and ZBT provided
the resources and supervised the study. TDS and YPJ reviewed and
edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Affiliated Hospital of Qingdao University (Qingdao, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
lncRNA
|
long noncoding RNA
|
|
miRNA
|
microRNA
|
|
mRNA
|
messenger RNA
|
|
ceRNA
|
competing endogenous RNA
|
|
DM
|
diabetes mellitus
|
|
IECs
|
intestinal epithelial cells
|
|
IESCs
|
intestinal epithelial stem cells
|
|
STZ
|
streptozocin
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
UTR
|
untranslated region
|
|
DAB
|
3,3′-diaminobenzidine
tetrahydrochloride
|
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant no. 81800461).
References
|
1
|
Davies MJ, D'Alessio DA, Fradkin J, Kernan
WN, Mathieu C, Mingrone G, Rossing P, Tsapas A, Wexler DJ and Buse
JB: Management of hyperglycemia in type 2 diabetes, 2018. A
consensus report by the american diabetes association (ADA) and the
European association for the study of diabetes (EASD). Diabetes
Care. 41:2669–2701. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nickerson HD and Dutta S: Diabetic
complications: Current challenges and opportunities. J Cardiovasc
Transl Res. 5:375–379. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meldgaard T, Olesen SS, Farmer AD, Krogh
K, Wendel AA, Brock B, Drewes AM and Brock C: Diabetic enteropathy:
From molecule to mechanism-based treatment. J Diabetes Res.
2018:38273012018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Kort S, Simons C, van den Brandt PA,
Janssen-Heijnen MLG, Sanduleanu S, Masclee AAM and Weijenberg MP:
Diabetes mellitus, genetic variants in the insulin-like growth
factor pathway and colorectal cancer risk. Int J Cancer.
145:1774–1781. 2019.PubMed/NCBI
|
|
5
|
Wang X, Häring MF, Rathjen T, Lockhart SM,
Sørensen D, Ussar S, Rasmussen LM, Bertagnolli MM, Kahn CR and
Rask-Madsen C: Insulin resistance in vascular endothelial cells
promotes intestinal tumour formation. Oncogene. 36:4987–4996. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pirogov SA, Gvozdev VA and Klenov MS: Long
noncoding RNAs and stress response in the nucleolus. Cells.
8:E6682019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rafiee A, Riazi-Rad F, Havaskary M and
Nuri F: Long noncoding RNAs: Regulation, function and cancer.
Biotechnol Genet Eng Rev. 34:153–180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klingenberg M, Matsuda A, Diederichs S and
Patel T: Non-coding RNA in hepatocellular carcinoma: Mechanisms,
biomarkers and therapeutic targets. J Hepatol. 67:603–618. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Phelps M, Coss C, Wang H and Cook M:
Registered report: Coding-independent regulation of the tumor
suppressor PTEN by competing endogenous mRNAs. Elife. 5:e124702016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shan TD, Lv SY, Tian ZB, Liu XS, Liu FG
and Sun XG: Knockdown of lncRNA H19 inhibits abnormal
differentiation of small intestinal epithelial cells in diabetic
mice. J Cell Physiol. 234:837–848. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Krishnan P and Damaraju S: The challenges
and opportunities in the clinical application of noncoding RNAs:
The road map for miRNAs and piRNAs in cancer diagnostics and
prognostics. Int J Genomics. 2018:58480462018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu S, Sun H, Wang Y, Yang X, Meng Q, Yang
H, Zhu H, Tang W, Li X, Aschner M and Chen R: MALAT1 rs664589
polymorphism inhibits binding to miR-194-5p, contributing to
colorectal cancer risk, growth, and metastasis. Cancer Res.
79:5432–5441. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang D, Xu H, Wu B, Jiang S, Pan H, Wang R
and Chen J: Long noncoding RNA MALAT1 sponges miR1243p.1/KLF5 to
promote pulmonary vascular remodeling and cell cycle progression of
pulmonary artery hypertension. Int J Mol Med. 44:871–884.
2019.PubMed/NCBI
|
|
14
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thyagarajan A, Shaban A and Sahu RP:
MicroRNA-directed cancer therapies: Implications in melanoma
intervention. J Pharmacol Exp Ther. 364:1–12. 2018. View Article : Google Scholar :
|
|
17
|
Shan TD, Ouyang H, Yu T, Li JY, Huang CZ,
Yang HS, Zhong W, Xia ZS and Chen QK: miRNA-30e regulates abnormal
differentiation of small intestinal epithelial cells in diabetic
mice by downregulating Dll4 expression. Cell Prolif. 49:102–114.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hou S, Lin Q, Guan F and Lin C: lncRNA
TNRC6C-AS1 regulates UNC5B in thyroid cancer to influence cell
proliferation, migration, and invasion as a competing endogenous
RNA of miR-129-5p. J Cell Biochem. 119:8304–8316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu C, Zhang X, Chen P, Ruan X, Liu W, Li
Y, Sun C, Hou L, Yin B, Qiang B, et al: MicroRNA-129 modulates
neuronal migration by targeting Fmr1 in the developing mouse
cortex. Cell Death Dis. 10:2872019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Z, Lu J, Zeng G, Pang J, Zheng X, Feng
J and Zhang J: MiR-129-5p inhibits liver cancer growth by targeting
calcium calmodulin-dependent protein kinase IV (CAMK4). Cell Death
Dis. 10:7892019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Clevers H, Loh KM and Nusse R: Stem cell
signaling. An integral program for tissue renewal and regeneration:
Wnt signaling and stem cell control. Science. 346:12480122014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perochon J, Carroll LR and Cordero JB: Wnt
signalling in intestinal stem cells: Lessons from mice and flies.
Genes (Basel). 9:E1382018. View Article : Google Scholar
|
|
23
|
Dorfman T, Pollak Y, Sohotnik R, Coran AG,
Bejar J and Sukhotnik I: Enhanced intestinal epithelial cell
proliferation in diabetic rats correlates with beta-catenin
accumulation. J Endocrinol. 226:135–143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ouyang H, Yang HS, Yu T, Shan TD, Li JY,
Huang CZ, Zhong W, Xia ZS and Chen QK: MEK/ERK pathway activation
by insulin receptor isoform alteration is associated with the
abnormal proliferation and differentiation of intestinal epithelial
cells in diabetic mice. Mol Cell Biochem. 413:165–178. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu S, Qiu J, He G, Liang Y, Wang L, Liu C
and Pan H: lncRNA MALAT1 acts as a miR-125a-3p sponge to regulate
FOXM1 expression and promote hepatocellular carcinoma progression.
J Cancer. 10:6649–6659. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang J, Li Q, Xue B and He R: MALAT1
inhibits the Wnt/β-catenin signaling pathway in colon cancer cells
and affects cell proliferation and apoptosis. Bosn J Basic Med Sci.
Nov 15–2019.Epub ahead of print. View Article : Google Scholar
|
|
27
|
Wu X, Li R, Song Q, Zhang C, Jia R, Han Z,
Zhou L, Sui H, Liu X, Zhu H, et al: JMJD2C promotes colorectal
cancer metastasis via regulating histone methylation of
MALAT1promoter and enhancing β-catenin signaling pathway. J Exp
Clin Cancer Res. 38:4352019. View Article : Google Scholar
|
|
28
|
Brito-Casillas Y, Melian C and Wagner AM:
Study of the pathogenesis and treatment of diabetes mellitus
through animal models. Endocrinol Nutr. 63:345–353. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Huang CZ, Xu JH, Zhong W, Xia ZS, Wang SY,
Cheng D, Li JY, Wu TF, Chen QK and Yu T: Sox9 transcriptionally
regulates Wnt signaling in intestinal epithelial stem cells in
hypomethylated crypts in the diabetic state. Stem Cell Res Ther.
8:602017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gracz AD, Ramalingam S and Magness ST:
Sox9 expression marks a subset of CD24-expressing small intestine
epithelial stem cells that form organoids in vitro. Am J Physiol
Gastrointest Liver Physiol. 298:G590–G600. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dhamija S and Diederichs S: From junk to
master regulators of invasion: lncRNA functions in migration, EMT
and metastasis. Int J Cancer. 139:269–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lv SY, Shan TD, Pan XT, Tian ZB, Liu XS,
Liu FG, Sun XG, Xue HG, Li XH, Han Y, et al: The lncRNA ZEB1-AS1
sponges miR-181a-5p to promote colorectal cancer cell proliferation
by regulating Wnt/beta-catenin signaling. Cell Cycle. 17:1245–1254.
2018. View Article : Google Scholar :
|
|
34
|
Yu T, Shan TD, Li JY, Huang CZ, Wang SY,
Ouyang H, Lu XJ, Xu JH, Zhong W and Chen QK: Knockdown of linc-UFC1
suppresses proliferation and induces apoptosis of colorectal
cancer. Cell Death Dis. 7:e22282016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koren E, Yosefzon Y, Ankawa R, Soteriou D,
Jacob A, Nevelsky A, Ben-Yosef R, Bar-Sela G and Fuchs Y: ARTS
mediates apoptosis and regeneration of the intestinal stem cell
niche. Nat Commun. 9:45822018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ballantyne MD, McDonald RA and Baker AH:
lncRNA/ MicroRNA interactions in the vasculature. Clin Pharmacol
Ther. 99:494–501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Adams BD, Parsons C, Walker L, Zhang WC
and Slack FJ: Targeting noncoding RNAs in disease. J Clin Invest.
127:761–771. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Khawar MB, Mehmood R and Roohi N:
MicroRNAs: Recent insights towards their role in male infertility
and reproductive cancers. Bosn J Basic Med Sci. 19:31–42. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tian Y, Ma X, Lv C, Sheng X, Li X, Zhao R,
Song Y, Andl T, Plikus MV, Sun J, et al: Stress responsive miR-31
is a major modulator of mouse intestinal stem cells during
regeneration and tumorigenesis. Elife. 6:e295382017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shaker OG, Abdelwahed MY, Ahmed NA, Hassan
EA, Ahmed TI, Abousarie MA and Ayoub SE: Evaluation of serum long
noncoding RNA NEAT and MiR-129-5p in hepatocellular carcinoma.
IUBMB Life. 71:1571–1578. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Demirsoy İH, Ertural DY, Balci Ş, Çınkır
Ü, Sezer K, Tamer L and Aras N: Profiles of circulating MiRNAs
following metformin treatment in patients with type 2 diabetes. J
Med Biochem. 37:499–506. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mao J, Hu X, Xiao Y, Yang C, Ding Y, Hou
N, Wang J, Cheng H and Zhang X: Overnutrition stimulates intestinal
epithelium proliferation through beta-catenin signaling in obese
mice. Diabetes. 62:3736–3746. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bello SA, Torres-Gutiérrez V,
Rodríguez-Flores EJ, Toledo-Román EJ, Rodríguez N, Díaz-Díaz LM,
Vázquez-Figueroa LD, Cuesta JM, Grillo-Alvarado V, Amador A, et al:
Insights into intestinal regeneration signaling mechanisms. Dev
Biol. Feb 1–2020, Epub ahead of print. View Article : Google Scholar
|
|
44
|
Merenda A, Fenderico N and Maurice MM: Wnt
signaling in 3D: Recent advances in the applications of intestinal
organoids. Trends Cell Biol. 30:60–73. 2020. View Article : Google Scholar
|
|
45
|
Li H, Zhao Q, Chang L, Wei C, Bei H, Yin
Y, Chen M, Wang H, Liang J and Wu Y: lncRNA MALAT1 modulates ox-LDL
induced EndMT through the Wnt/β-catenin signaling pathway. Lipids
Health Dis. 18:622019. View Article : Google Scholar
|
|
46
|
Guo C and Wang X, Chen LP, Li M, Li M, Hu
YH, Ding WH and Wang X: Long non-coding RNA MALAT1 regulates
ovarian cancer cell proliferation, migration and apoptosis through
Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci.
22:3703–3712. 2018.PubMed/NCBI
|