Introduction
Sensorineural hearing loss (SNHL) is one of the
major leading causes of hearing impairment, and is typically
characterized by the degeneration of spiral ganglion neurons (SGNs)
(1). Multiple internal and
external causative factors greatly affect the development of this
disease. Currently, the clinical treatment for severe cases of SNHL
is a cochlear implant to locate an electrode array in the scala
tympani and electrically stimulate SGNs (2). However, the therapeutic efficacy is
reduced due to the continuous degeneration of SGNs (3–5)
and a gradual reduction in SGN numbers. Therefore, the urgent
investigation of the mechanisms responsible for SGN degeneration
and the development of more effective treatment methods are
required.
The development of SNHL is influenced by multiple
hereditary and developmental factors (6). Numerous genetic loci have been
reported to increase the possibility of permanent SNHL caused by
ototoxic medications or noise trauma (7). The transmembrane protease, serine 3
(TMPRSS3) gene encodes a proteolytic enzyme that belongs to the
subfamily of type II transmembrane serine proteases (8,9).
TMPRSS3 is expressed in SGNs, inner hair cells and the stria
vascularis of the cochlear duct (10). The TMPRSS3 gene plays crucial
roles in the morphological and functional maturation of the inner
ear, as well as in maintaining the contents of the endolymph and
perilymph (11,12). However, the function of the
TMPRSS3 gene in the auditory system has not yet been fully
elucidated. A previous study by the authors reported a high TMPRSS3
expression in SGNs of mouse cochleae (13). In addition, it has been
demonstrated that microRNA (miRNA or miR)-204-5p suppresses the
survival of cochlear SGNs in vitro by directly targeting the
TMPRSS3 gene (14). However, the
upstream regulatory mechanisms of miR-204-5p have not yet been
fully elucidated.
Long non-coding RNAs (lncRNAs) are a type of RNA
molecules that are unable to encode proteins, with a length of
>200 nucleotides (nts) (15).
lncRNAs have been defined as RNA molecules functioning as either
primary or spliced transcripts, which do not match with the known
small RNAs or some structural RNAs (16). lncRNAs have been reported to
regulate gene expression through multiple mechanisms, including
transcription, post-transcription and chromatin modification
(16,17). lncRNAs can regulate gene
transcription by co-activating transcription factors, interacting
with RNA binding proteins (RBPs) or repressing the promoters of
targeted genes (18–20). The present study demonstrated that
lncRNA EBLN3P promotes the recovery of the function of impaired
SGNs by competitively binding to miR-204-5p and regulating TMPRSS3
expression.
Materials and methods
Establishment of mouse model with
kanamycin sulfate-induced hearing impairment
Specific pathogen-free mice (male, 2 months old,
weighing 25–35 g), were purchased from the Department of
Experimental Animal Science of Central South University. Their
auditory brainstem response (ABR) threshold was normal, and middle
and inner ear diseases were excluded. The mice were randomly
divided into 2 groups as follows: An experimental group and a
control group. The mice in the experimental group were
intramuscularly injected with 500 mg/kg/day kanamycin sulfate
(040810OG; Ameresco, Inc.) for 14 days, while those in the control
group were injected with an equal volume of normal saline for 14
days, as previously described (21). For each group at the beginning, 8
mice in total were used for the whole experimental procedure. All
the mice were supplied with water and food ad libitum and
allowed to acclimatize to their environment for a period of 7 days
prior to being used in the corresponding experiment. Humane
endpoints were applied for all mice in the experiments. The care of
the laboratory animals and animal experimentation were performed in
accordance with animal ethics guidelines and approved by the Animal
Care and Use Committee at Xiangya School of Medicine from Central
South University. Humane endpoints were used and mice displaying
severe illness were euthanized prior to the end of the experiments
to minimize suffering and distress. The health and behavior of the
mice were monitored twice per day. The criteria for determining
when the animals should be euthanized included weight loss, loss of
appetite, lack of feeding or drinking, weakness, ruffled fur, signs
of severe organ system dysfunction and not responsive to treatment.
The duration of the experiment was approximately 14 days. In the
process of the experiment, one mouse from the experimental group
was found to have sustained weight loss (18% weight loss was
observed) and also exhibited a loss of appetite; this this mouse
was euthanized by CO2 exposure according to the IACUC
policy. The optimal flow rate was set 20% of the chamber volume per
minute until the mouse became unconscious. At the completion of all
the in vivo experiments, 6 mice from each group were used
for quantification, and the remaining 3 mice from the 2 groups were
euthanized by CO2 exposure as described above. The death
of the mice was verified by monitoring cardiac cessation and
respiratory arrest. All the best efforts were made to minimize the
suffering and distress of the animals.
ABR test
The 6 mice from each group mentioned above, which
were used for quantification were also used for the ABR test.
Anesthesia was performed by an intraperitoneal injection of 2%
sodium pentobarbital (40 mg/kg, prepared with physiological
saline), as previously described (22). After 5–10 min, the stinging and
blink reflexes of the mice disappeared, and the breathing slowed
down, thereby indicating that the state of anesthesia was moderate
and the subsequent experiment could be initiated (23). The moderately anesthetized mice
were fixed on an experimental bench in prone position, and their
body temperature was maintained at a constant level during the
experiment. The placement of three electrodes to detect ABR was
carried out as follows: The recording electrode was placed
subcutaneously in the middle of the cranial crest; the referred
electrode was placed under the skin of the mastoid of the ear to be
measured; and the ground wire was placed under the skin of the
papillary part of the contralateral ear. The experimental mice, the
electrodes, the front signal amplifier of the ABR detection
instrument and the sound stimulator were placed in the acoustic-
and electric-shielded room, while the other instruments were placed
outside the shielded room.
The Smart-EP function option of the SmartEP3.91USBez
software (Intelligent Hearing Systems) stimulates the tone-pip
stimuli generated by the high-frequency sound transducer, and is
sent to the external auditory canal of the experimental animal by
0.5 cm through a plastic tube of ~2-mm diameter. The output signal
is input to the computer via a bio-signal pre-amplifier and is
monitored by a computer monitor. A tone-pip of 8 kHz frequency was
selected as the stimulation sound; the stimulation rate was 39.10
counts/sec; the stimulation duration was 5 cycles; and the rise and
fall times were 2 cycles. A trapezoidal envelope was used; the
recording analysis time was 16.0 msec, and the number of
superpositions was 1,024. Tone-pip starts from 90 dB of sound
pressure level (dBSPL) and is first decremented by 10 dBSPL and
then by 5 dBSPL. The minimum stimulation intensity that was able to
distinguish I or II wave patterns was determined as a threshold,
and was repeated twice.
Isolation and in vitro culture of
SGNs
The protocol for SGN isolation and culture was based
on a previous study (24). As the
previously described protocol (24) was performed on rats, and the
experiment in the present study was performed on mice, necessary
modifications were therefore made according to the similar anatomic
structures of the Corti organ, but different sizes of the
corresponding organs. A total of 60 specific pathogen-free mice
(male, 2 months old, weighing 25–35 g) were purchased from the
Department of Experimental Animal Science of Central South
University. Half of the mice were used directly for SGN isolation,
while the remaining mice were used for the establishment of the
deafness models by kanamycin sulfate-induced hearing impairment as
described above, followed by SGN isolation. SGNs were dissected
from the cochleae of all the mice as previously described (25). In detail, SGNs were isolated from
the modiolus, followed by the removal of the sensory epithelium
from the organ of Corti. The SGNs were then treated with 0.25%
trypsin for 15 min at 37°C, followed by trypsin neutralization with
10% fetal bovine serum (FBS) in Dulbecco's modified Eagle's medium
(DMEM). Subsequently, the SGNs were collected by centrifugation at
4°C, 500 x g for 7 min, and triturated to form a single-cell
suspension in DMEM/F12 supplemented with N2 (primary rat embryonic
hippocampal neurons, tumor cell lines of neuronal origin) and B27
(primary rat embryonic hippocampal neurons, primary rat neurons
from the striatum, substantia nigra, septum) cells. Morphological
observation of the SGNs was performed using a DMI3000B microscope
(Leica, Ernst-Leitz). The in vivo experimental protocol was
approved by the Animal Care and Use Committee at Xiangya School of
Medicine from Central South University. Euthanasia was performed by
an intraperitoneal injection of sodium pentobarbital (100 mg/kg,
prepared with physiological saline) (26), and the means used to verify mouse
death were as follows: No spontaneous breathing for a continuous
duration of 2–3 min, and no blinking reflex.
miRNA target predictions
Predicted binding targets of miR-204-5p were
analyzed by searching online databases, such as mirTarget2
(http://mirdb.org/), microRNA.org
(www.microrna.org) and starBase v2.0 (starbase.sysu.edu.cn/star-base2).
Plasmid construction and cell
transfection
Human EBLN3P genes were amplified by reverse
transcription-PCR (RT-PCR). In detail, the total RNAs were
extracted from the SGNs cells using TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.). The RNA concentrations were determined by
a NanoDrop ND-1000 instrument (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.). cDNA was produced by reverse transcription using
the PrimeScript™ One Step RT-PCR kit Ver.2 according to the
manufacturer's instructions (Takara). PCR amplification was
performed using the following primers: lncRNA EBLN3P sense,
5′-GTGTTGTCCCGGAAGTGCCTTCTC-3′ and antisense,
5′-TTGAAGGTTTGCCTTCTCTGAATAG-3′. The parameters for PCR
amplification were as follows: 5 min at 95°C, followed by 35 cycles
of 60 sec at 95°C, 60 sec at 58°C and 90 sec at 72°C, 8 min at
72°C. The PCR amplification were performed on an Eppendorf
Mastercycler X50h instrument (Eppendorf). The EBLN3P gene was then
subcloned into the expression vector, pcDNA3.1 (Invitrogen; Thermo
Fisher Scientific), named pcDNA3.1-EBLN3P. The 1,200-nt gene
fragment at the 5′ end of either EBLN3P containing the two
predicted binding sites (regions '314-322' and '973-981' in the
1,200-nt EBLN3P fragment) for miR-204-5p or EBLN3P-mutant (mut)
(with point mutations in the miR-204-5p response elements) was
amplified using a common PCR method, and then subcloned into the
pmirGLO vector (Promega Corp.) for subsequent luciferase reporter
analyses. The plasmids pcDNA3.1-MS2, pcDNA3.1-MS2-EBLN3P and
pcDNA3.1-MS2-EBLN3P-mut (miR-204-5p) were constructed for the
following RNA immunoprecipitation (RIP) assay.
The double-stranded miR-204-5p mimics, negative
control miRNAs, miR-204-5p inhibitors and recombinant adenoviruses
containing the overexpression vector of EBLN3P or the control
vector were produced by GeneChem, Inc. The recombinant plasmids
pLV4-small hairpin RNA (shRNA)/EBLN3P and pLV4-shRNA/argonaute 2
(AGO2), with their respective non-targeting control
pLV4-shRNA-negative control (NC), were constructed. All transient
transfections were performed using Lipofectamine 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's instructions. To improve the transfection efficiency
in neuronal cells, the positively transfected cells were
subsequently screened by flow cytometry following transfection with
a GFP-labeled for 48 h, and the follow-up experiment then
commences. The transfection efficiency was evaluated by
fluorescence microscopy observation.
Reverse transcription-quantitative PCR
(RT-qPCR)
For TMPRSS3 gene quantification, and the
determination of lncRNA EBLN3P and miR-204-5p expression, total
RNA, which contained all miRNAs and lncRNAs, was extracted using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). RNA
concentrations were measured with a NanoDrop ND-1000 instrument
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.), and the
final samples were stored at −80°C. cDNA was produced by reverse
transcription using the SuperScript® III RT-PCR kit
according to the manufacturer's instructions (Thermo Fisher
Scientific, Inc.). RT-qPCR was performed using the following
primers: TMPRSS3 sense, 5′-AGTGGGGTAGACGGAGACCT-3′ and antisense,
5′-CACTGAACCCTTCCTGGTTT-3′; lncRNA EBLN3P sense,
5′-TACGCGTTTTGGTCCCTGTT-3′ and antisense,
5′-GCCACTTGGCTCAAAAGACTG-3′; miR-204-5p sense,
5′-ACACTCCAGCTGGGTTCCCTTTGTCAT-3′ and anti-sense,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGCATGG-3′; β-actin sense,
5′-AGCAGCATCGCCCCAAAGTT-3′ and antisense,
5′-GGGCACGAAGGCTCATCATT-3′; and U6 sense, 5′-CTCGCTTCGGCAGCACA-3′
and antisense, 5′-AACGCTTCACGAATTTGCGT-3′. The parameters for PCR
quantification were as follows: 2 min at 95°C, followed by 40
cycles of 15 sec at 95°C and 30 sec at 60°C. The results of qPCR
were defined using the quantification cycle (Cq), and
2−ΔΔCq was used to calculate the relative expression
levels (27). All the PCR assays
were performed on an ABI 7500 instrument (Applied Biosystems;
Thermo Fisher Scientific, Inc.).
RIP assay
SGNs were co-transfected with pMS2-GFP and
pcDNA3.1-MS2, or pMS2-GFP and pcDNA3.1-MS2-EBLN3P, or pMS2-GFP and
pcDNA3.1-MS2-EBLN3P-mut (miR-204-5p). After 48 h, the
above-mentioned SGNs were subjected to RIP assay using an anti-GFP
antibody (dilution: 1:100, cat. no. 11814460001; Roche Diagnostics)
and the Magna RIP™ RNA-Binding Protein Immunoprecipitation kit (EMD
Millipore) in accordance with the manufacturer's instructions.
RNA pull - d own assay
lncRNA EBLN3P or lncRNA-EBLN3P-mut (miR-204-5p) were
in vitro transcribed from the vector pSPT19-EBLN3P or
pSPT19-EBLN3P-mut (miR-204-5p), respectively, and biotin-labeled
with Biotin RNA Labeling mix and T7 RNA polymerase (Roche
Diagnostics), followed by treatment with RNase-free DNase I and
purificatiob with RNeasy Mini kit (Qiagen GmbH). Cell lysates from
SGNs (1 µg) were co-incubated with 3 µg biotinylated
transcripts for 1 h at 25°C, and then the co-incubated complexes
were separated with streptavidin agarose beads (Invitrogen; Thermo
Fisher Scientific, Inc.), followed by RT-qPCR analysis.
Luciferase reporter assay
293T cells (ATCC, ~8,000 cells per well) or primary
isolated SGNs (12,000 cells per well) were seeded into 96-well
plates, and co-transfected with 50 nmol/l miR-204-5p mimic (or NC),
50 ng luciferase reporter plasmid and 5 ng pRLCMV Renilla
luciferase reporter plasmid using Lipofectamine 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Following 48 h of
incubation, the Firefly and Renilla luciferase activities
were measured using a Dual-Luciferase® Reporter Assay
kit (Promega Corp.). Data are presented as the relative ratio of
Firefly luciferase activities to Renilla luciferase
activities.
Western blot analysis
SGNs subjected to the different treatments were
firstly washed twice with PBS solution, and then lysed in RIPA
lysis buffer (Beyotime Institute of Biotechnology). The
concentration of the protein samples was determined by the BCA
method. A total of 40 µg protein for each sample was then
loaded and separated by 10% SDS/PAGE, followed by transfer onto
PVDF membranes (EMD Millipore). The membranes with targeted
proteins were blocked with 5% BSA at 37°C for 2 h, and then
incubated with primary antibodies overnight at 4°C. The membranes
were then washed 3 times, and incubated at 37°C for 1 h with an
HRP-conjugated anti-rabbit secondary antibody (1:5,000, ab97051,
Abcam), followed by the detection of the immunoreactive protein
bands with an Odyssey Scanning system. The anti-AGO2 rabbit
monoclonal antibody (mAb) (#2897, 1:1,000) and anti-cleaved
caspase-3 rabbit mAb (#9664, 1:1,000) were purchased from Cell
Signaling Technology, Inc., while the anti-TMPRSS3 rabbit
polyclonal antibody (#PA5-35325, 1:1,000) was purchased from
Invitrogen (Thermo Fisher Scientific, Inc.). Anti-β-actin rabbit
monoclonal antibody (1:5,000, ab179467, Abcam) was used as the
internal control. Densitometric analysis was performed using the
Scion Imaging application (4.0, Scion Corp.).
Cell viability assay
The SGN suspension (5x104 cells/ml) was
seeded into a 96-well plate and incubated overnight at 37°C. After
being subjected to the different treatments for 48 h, the SGNs were
incubated with MTT solution for 1 h. Subsequently, 100% DMSO were
added to each well, and the absorbance of each well was measured at
570 nm (28,29) using a microplate enzyme-linked
immunosorbent assay reader (Labsystems Dragon, Wellscan). Cell
viability was also measured with Trypan blue staining with the
Trypan blue staining kit (Sangon Biotech) at 37°C for 3 min.
Lactate dehydrogenase (LDH) release was quantified to determine
cell death using the CytoTox 96® Non-Radioactive
Cytotoxicity assay (Promega Corp.) according to manufacturer's
instructions and as previously described (30). Values were normalized to protein
concentrations to account for differences in cell numbers per well.
All the experiments were performed ≥3 times.
Flow cytometry
SGNs were seeded into 6-well plates at a density of
2x105 cells/well with DMEM supplemented with 10% FBS.
Subsequently, the cells were harvested by trypsinization and washed
twice with cold PBS solution. To measure cell apoptosis, the
pre-treated SGNs were fixed in ice-cold 70% ethanol at 4°C for 1 h.
After washing once with cold PBS, the SGNs were subsequently
stained with propidium iodide (P1304MP, Invitrogen; Thermo Fisher
Scientific) and anti-Annexin-V antibody (563973, BD Biosciences) at
4°C for 1 h. Finally, the SGNs were analyzed with a
fluorescence-activated cell sorting instrument (BD
Biosciences).
Adenovirus injection
A capillary with a tip of ~100 µm was made
with a P-97 puller. After the mice were anesthetized by an
intraperitoneal injection of 10% of chloral hydrate (300 mg/kg of
mouse body weight, and no mice exhibited signs of peritonitis, pain
or discomfort) (31), the round
window membrane was pierced with a capillary and fixed in
situ. Adenovirus for EBLN3P overexpression (2 µl) was
injected into the inner ear for 14 days by capillary using a
Hamilton microsyringe at an injection rate of 0.1 µl/min to
avoid damage to the inner ear tissue (32). Herein, another batch of specific
pathogen-free mice (male, 2 months old, weighing 25–35 g) was
purchased (total number, 12) from the Department of Experimental
Animal Science of Central South University. They were used for the
establishment of the deafness models by kanamycin sulfate-induced
hearing impairment as described above, followed by adenovirus
injection treatment. The 12 mice were divided into 2 groups as
follows: 6 mice were treated with control adenovirus, and another 6
mice were treated with adenovirus for EBLN3P overexpression. In
each group, 6 mice were used for quantification. Euthanasia was
performed as described above.
Hematoxylin and eosin (H&E) staining
and immunohistochemistry (IHC)
Paraffin-embedded cochlear SGN tissue samples from
mouse cochleae were selected for H&E staining and IHC. The
H&E staining kit was purchased from Abcam (ab245880). In
detail, the sections were deparaffinized if needed and hydrated in
distilled water. Hematoxylin [Mayer's (Lillie's Modification)] was
then applied to completely cover the tissue sections followed by
incubation for 5 min. The slides were then rinsed in two changes of
distilled water to remove the excess stain. Bluing reagent was
applied to completely cover the tissue sections followed by
incubation for 10–15 sec. The sections were again rinsed in two
changes of distilled water. The slides were then dipped in absolute
alcohol and the excess was blotted off. Adequate Eosin Y Solution
(Modified Alcoholic) was then applied to completely cover the
tissue sections followed by incubation for 2–3 min. The slices were
subsequently rinsed using absolute alcohol, followed by dehydration
in 3 changes of absolute alcohol. The slides were then cleared and
mounted in synthetic resin. IHC for β-tubulin (1:250, ab179513,
Abcam), TMPRSS3 (1:200, ab167160, Abcam) and neurotrophin-3 (NT-3,
1:100, ab216491, Abcam) was carried out using a rabbit monoclonal
or polyclonal primary antibody at 4°C overnight, an HRP-conjugated
goat anti-rabbit IgG secondary antibody (1:1,000, ab150077, Abcam)
at 37°C for 1 h, and DAB staining solution (ab64238, Abcam) at room
temperature for 10 min. The staining results were assessed with a
DMI3000B microscope (Leica, Ernst-Leitz, Wetzlar, Germany).
Fluorescence in situ hybridization
(FISH)
Paraffin-embedded cochlear spiral ganglion neuron
tissue samples from mouse cochleae were selected for in situ
hybridization with FITC-labeled lncRNA EBLN3P DNA probe
(ShineGene). The paraffin-embedded tissues were cut into
5-µm-thick tissue sections for FISH using a Leica RM2235
Manual Rotary Microtome (Leica Microsystems GmbH). After dewaxing
the slides with xylem (2x10 min) and methanol (2x5 min). The slides
were exposed to microwaves (720 W) in 10 mM citric acid monohydrate
(pH 6.0) for 30 sec, followed by immersion in 1 M sodium
thiocyanate for 10 min at 80℃, and in protease solution for 10 min.
The tissue sections were then washed with pure H2O,
air-dried and dehydrated in ascending gradients of alcohol. The
lncRNA EBLN3P probe mixtures (10 ml) were added to the air-dried
tissue sections. The ThermoBrite System (Abbott Molecular) was used
for denaturation and hybridization. Subsequently, the tissue
sections were washed with 0.4X saline sodium citrate solution for 2
min at 70°C, and 2X saline sodium citrate solution for a further 5
min at room temperature. Finally, 10 ml DAPI (Beyotime Institute of
Biotechnology) was added to the tissue sections, followed by
fluorescence microscopy (Leica, Ernst-Leitz) evaluation. The final
data were evaluated by two investigators.
TUNEL assay
Using a DeadEnd™ Colorimetric TUNEL System kit
(Promega Corp.), TUNEL assay was performed to detect the apoptosis
of the SGNs in accordance with the manufacturer's instructions. The
observation and capture of digital images were conducted using a
Nikon E80i microscope (Nikon Corp.).
Statistical analysis
Data are presented as the means ± standard deviation
from ≥3 independent experiments. A normality test
(D'Agostino-Pearson) was used to analyze the distribution of all
datasets. A two-tailed Student's t-test and one-way ANOVA were
performed to analyze the data using SPSS 20.0 (IBM Corp.). The
paired-samples t-test was performed to compare the mean of two
matched groups of cases. ANOVA was used for multiple comparisons,
and Tukey's post hoc test was performed after ANOVA. P<0.05 was
considered to indicate a statistically significant difference.
Results
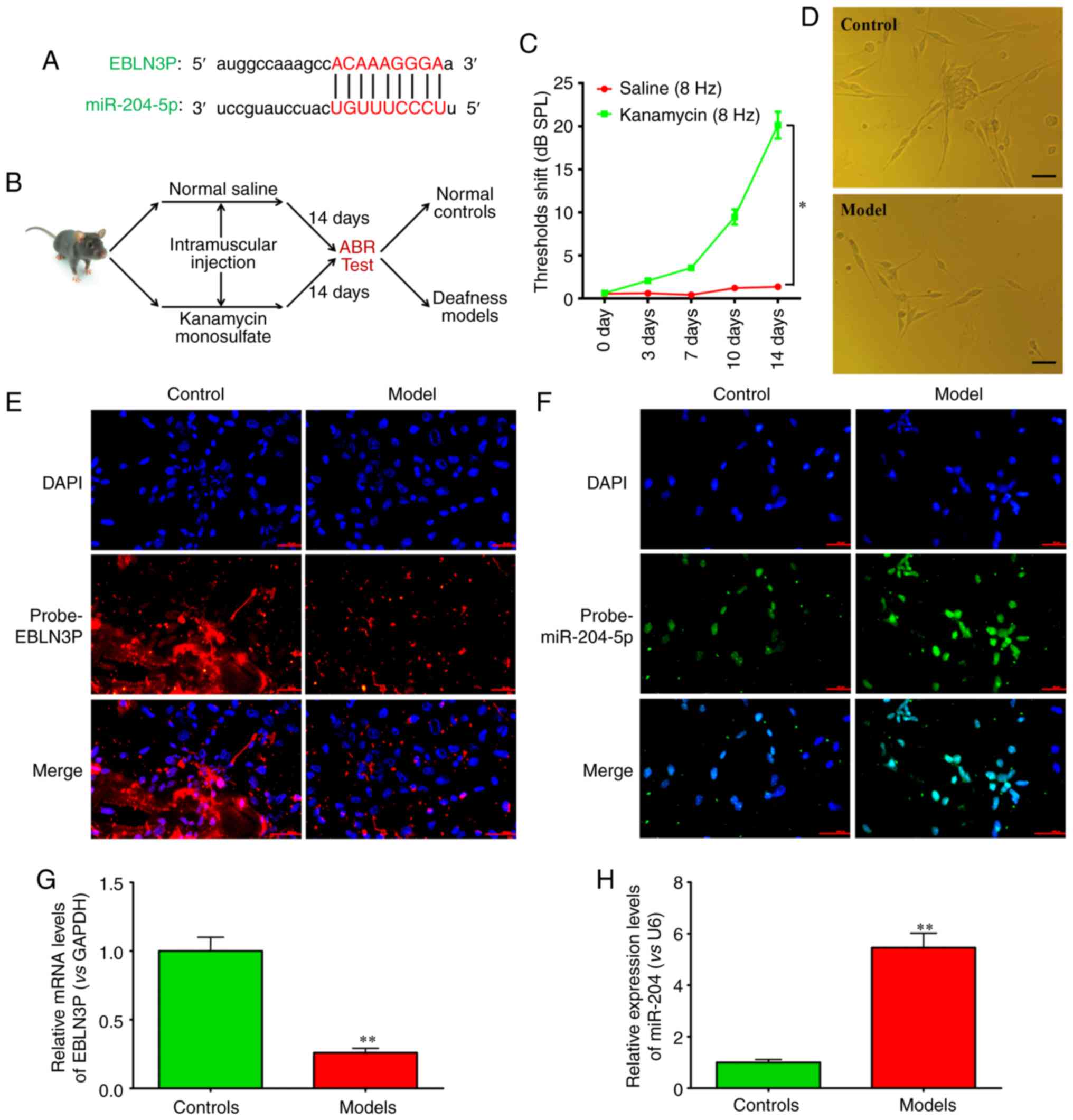
Expression of lncRNA EBLN3P and
miR-204-5p in models of deafness
By bioinformatics analysis using online databases,
such as mirTarget2, microRNA.org
and starBase v2.0, it was revealed that lncRNA EBLN3P could
effectively bind to miR-204-5p (Fig.
1A). Therefore, models of deafness were established (Fig. 1B). The ARB test indicated that
compared with the control saline-treated group, kanamycin
significantly increased the threshold (Fig. 1C). Subsequently, mouse primary
SGNs were separated (Fig. 1D). By
FISH, the expression of lncRNA EBLN3P and miR-204-5p in mouse
primary SGNs was detected. The results demonstrated that lncRNA
EBLN3P was overexpressed, while miR-204-5p was expressed at low
levels in primary SGNs in the control mice. However, in the primary
SGNs from the mice in the deafness models, lncRNA EBLN3P was
expressed at low levels, while miR-204-5p was overexpressed
(Fig. 1E and F). RT-qPCR further
confirmed the results of FISH (Fig.
1G and H).
lncRNA EBLN3P functions as a ceRNA,
regulating miR-204-5p in normal SGNs
Numerous mRNAs or lncRNAs have been reported to
function as ceRNAs by binding to targeted miRNAs and regulating
downstream gene expression. To confirm the direct binding between
lncRNA EBLN3P and miR-204-5p, RIP assay was performed to pull down
the miRNAs binding to endogenous lncRNA EBLN3P (Fig. 2A). RT-qPCR demonstrated that
lncRNA EBLN3P RIP in the SGNs was significantly enriched for
miR-204-5p, in contrast to the empty vector (MS2), IgG,
non-targeting miRNA (miR-67) and lncRNA EBLN3P with mutations in
miR-204-5p targeting sites (Fig.
2B). To further validate the strong interaction between lncRNA
EBLN3P and miR-204-5p, lncRNA EBLN3P was transcribed, labeled with
biotin and used to successfully pull down miR-204-5p (Fig. 2C). To validate the specific
association between lncRNA EBLN3P and miR-204-5p, dual-luciferase
reporters (pmirGLO) containing the 5′end 1,200-nt of lncRNA-EBLN3P,
either wild-type (WT) or mutated miR-204-5p binding sites were
generated. When miR-204-5p was co-transfected with pmirGLO-EBLN3P,
the luciferase activities of pmirGLO-EBLN3P decreased, while the
luciferase activities of empty vector pmirGLO and
pmirGLO-EBLN3P-mut (miR-204-5p) were not affected (Fig. 2D).
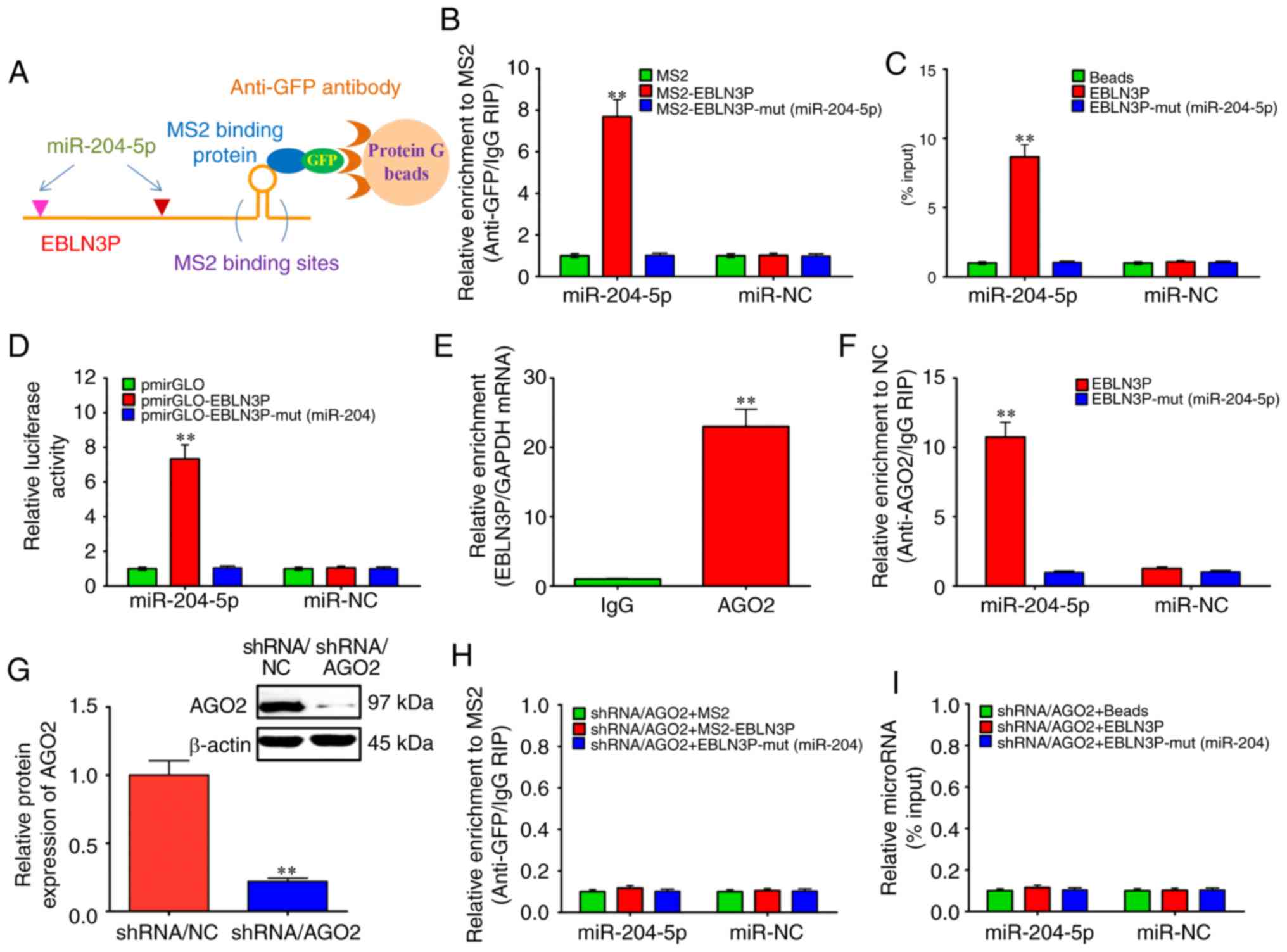 | Figure 2lncRNA EBLN3P functions as a
competing endogenous RNA by regulating miR-204-5p in normal SGNs.
(A and B) MS2-RIP assay followed by RT-qPCR to detect endogenous
miRNAs associated tightly with lncRNA EBLN3P. (C) Cell lysates of
normal SGNs were incubated with biotin-labeled EBLN3P. After
pull-down, the miRNAs were separated and assayed by RT-qPCR. (D)
Luciferase activities in normal SGNs cells co-transfected with
miR-204-5p and empty luciferase reporters, lncRNA EBLN3P wild
transcript or lncRNA EBLN3P mutant transcript. Data are presented
as the relative ratio of Firefly luciferase activity to
Renilla luciferase activity. (E) RIP assay of the binding
between lncRNA EBLN3P and AGO2 using control IgG and AGO2 special
antibody. lncRNA EBLN3P and GAPDH expression was quantified using
RT-qPCR, and analyzed as enrichment in RNA-binding protein RIP in
contrast to control IgG RIP. (F) Anti-AGO2 RIP was performed in
SGNs that were transiently transfected with miR-204-5p mimics,
followed by RT-qPCR to detect lncRNA EBLN3P or lncRNA EBLN3P-mut
(miR-204-5p) associated with AGO2. (G) After silencing AGO2 in
normal SGNs for 48 h, the protein expression level of AGO2 was
assessed by western blotting. (H) After silencing the AGO2 gene in
normal SGNs for 48 h, MS2-RIP analysis followed by RT-qPCR was
performed to detect miR-204-5p endogenously associated with lncRNA
EBLN3P. (I) After silencing AGO2 in normal SGNs for 48 h, the cells
were lysed, and the cell lysates were incubated with biotin-labeled
lncRNA EBLN3P. After pull-down, the whole miRNAs were extracted,
and miR-204-5p was measured by RT-qPCR. **P<0.01
(ANOVA with Tukey's post hot-test). lncRNA, long non-coding RNA;
miR, microRNA; miRNA, microRNA; SGN, spiral ganglion neuron; RIP,
RNA immunoprecipitation; RIP, RNA immunoprecipitation; AGO2,
argonaute 2. |
In general, miRNAs bind to the 3′-untranslated
region (UTR) of their targeted mRNA to repress the translation of
the target mRNA or promote mRNA degradation through an important
protein, AGO2 (33). Mature
miRNAs can be transported to the nucleus from the cytoplasm in the
presence of AGO2 protein, as well as TNRC6A, importin and other
proteins (34–36). In the present study, to
investigate whether miR-204-5p is regulated by lncRNA EBLN3P via
such a mechanism, the strong interaction between lncRNA EBLN3P and
AGO2 was further confirmed using an anti-AGO2-specific binding
antibody (Fig. 2E). An anti-AGO2
RIP experiment was then performed using the SGNs by transiently
overexpressing miR-204-5p. As shown in Fig. 2F, endogenous lncRNA EBLN3P
pull-down by AGO2 was specifically enriched in the
miR-204-5p-transfected SGNs, indicating that miR-204-5p is a bona
fide lncRNA EBLN3P targeting miRNA. This result demonstrated that
miR-204-5p bound to lncRNA EBLN3P without affecting the degradation
of lncRNA EBLN3P. In the presence of AGO2 protein, lncRNA EBLN3P
maintained a stable and strong interaction with miR-204-5p
(Fig. 2B and C). However, in the
absence of AGO2 protein, a negative association was observed
between lncRNA EBLN3P and miR-204-5p (Fig. 2G-I).
lncRNA EBLN3P regulates the expression of
TMPRSS3 through miR-204-5p in normal SGNs
Due to the complex associations between lncRNA
EBLN3P, miR-204-5p and TMPRSS3, the regulatory effects of lncRNA
EBLN3P on miR-204-5p and TMPRSS were further evaluated. The results
revealed that lncRNA EBLN3P knockdown successfully down-regulated
the expression of lncRNA EBLN3P (Fig.
3A), and decreased the activities of the TMPRSS3 3′-UTR
reporter (Fig. 3B). Furthermore,
lncRNA EBLN3P knockdown significantly downregulated the mRNA and
protein expression of TMPRSS3 (Fig.
3C and D). In addition, the results demonstrated that
overexpression of full-length lncRNA EBLN3P significantly
upregulated the expression of lncRNA EBLN3P (Fig. 3E), and markedly elevated the
activity of the TMPRSS3 3′-UTR reporter (Fig. 3F). However, when the SGNs were
preliminarily treated with the inhibitor of miR-204-5p
(anti-miR-204-5p), the overexpression of the full-length lncRNA
EBLN3P had no obvious effect on the activity of the TMPRSS3 3′-UTR
reporter (Fig. 3F). Similarly,
the overexpression of full-length lncRNA EBLN3P markedly elevated
the mRNA and protein expression levels of TMPRSS3. However, when
the SGNs were preliminarily treated with an inhibitor of
miR-204-5p, the overexpression of full-length lncRNA EBLN3P had no
obvious effects on the mRNA or protein expression of TMPRSS3
(Fig. 3G-I).
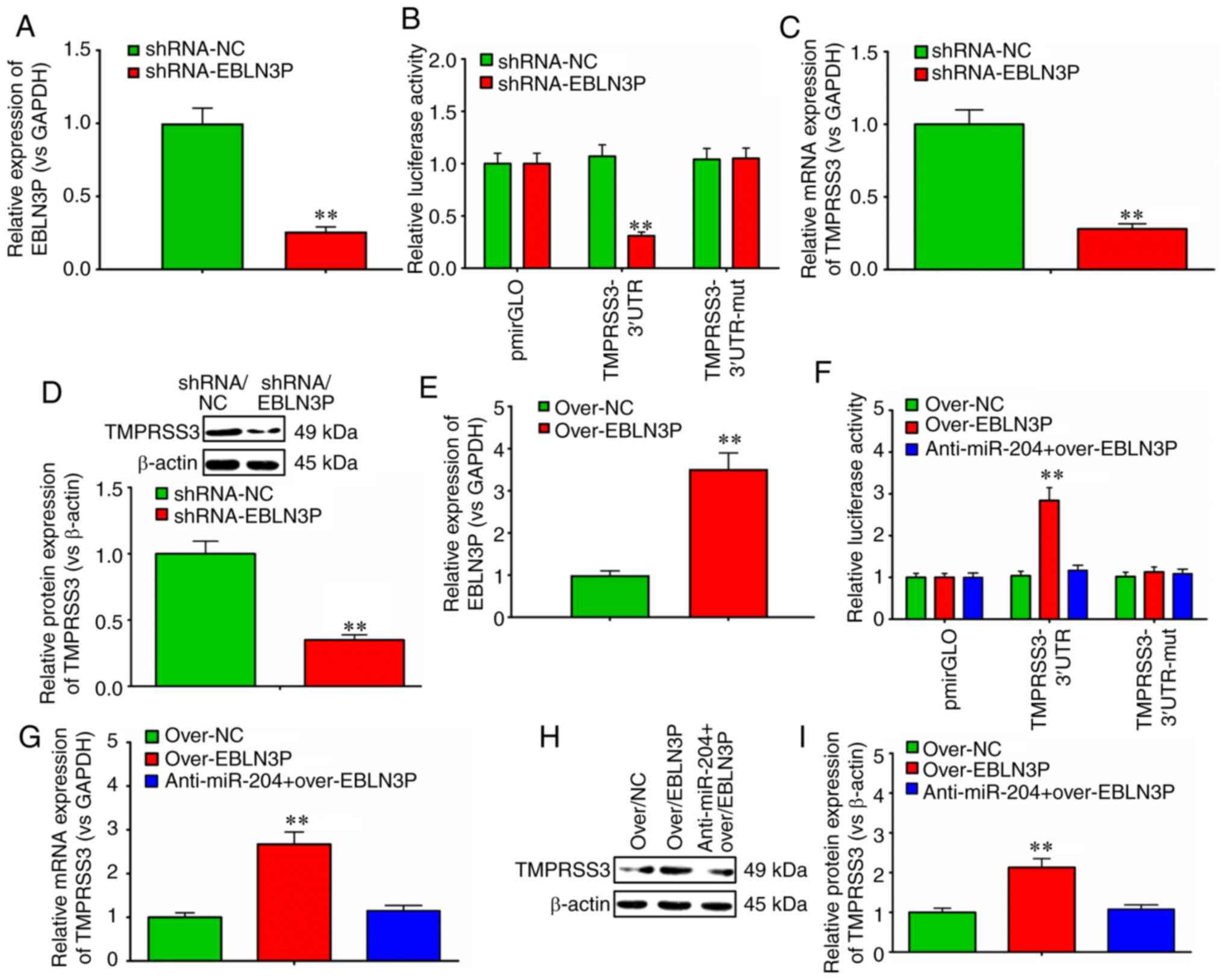 | Figure 3lncRNA EBLN3P regulates the
expression of TMPRSS3 through miR-204-5p in normal SGNs. (A)
RT-qPCR was performed to detect the effects of lncRNA EBLN3P
knockdown on the expression of EBLN3P in primary normal SGNs. (B)
Luciferase activity was evaluated to observe the effect of lncRNA
EBLN3P knockdown on the psiCHECK-2/TMPRSS3 wild-type 3′-UTR,
psiCHECK-2/TMPRSS3 mutated 3′-UTR and empty 3′-UTR vectors
(control). (C) RT-qPCR was performed to detect the effects of
lncRNA EBLN3P knockdown on the mRNA expression of TMPRSS3 in
primary normal SGNs. (D) Western blot analysis was performed to
detect the effects of lncRNA EBLN3P knockdown on the protein
expression of TMPRSS3 in primary normal SGNs. (E) RT-qPCR was
performed to detect the effects of lncRNA EBLN3P overexpression on
the expression of EBLN3P in primary normal SGNs. (F) Normal SGNs
were treated with over-NC, over-lncRNA EBLN3P or anti-miR-204-5p +
over-lncRNA EBLN3P, and the luciferase activity was evaluated.
(G-I) Normal SGNs were treated with control over-NC, over-lncRNA
EBLN3P or anti-miR-204-5p + over-lncRNA EBLN3P. RT-qPCR and western
blot analysis was performed to detect the mRNA and protein
expression of TMPRSS3 in primary normal SGNs.
**P<0.01 (Student's t-test) vs. NCs in panels A, B,
C, D and E. **P<0.01 (ANOVA with Tukey's post hoc
test) in panels F, G and I. lncRNA, long non-coding RNA; SGN,
spiral ganglion neuron; TMPRSS3, transmembrane protease, serine 3;
UTR, untranslated region; NC, negative control; miR, microRNA. |
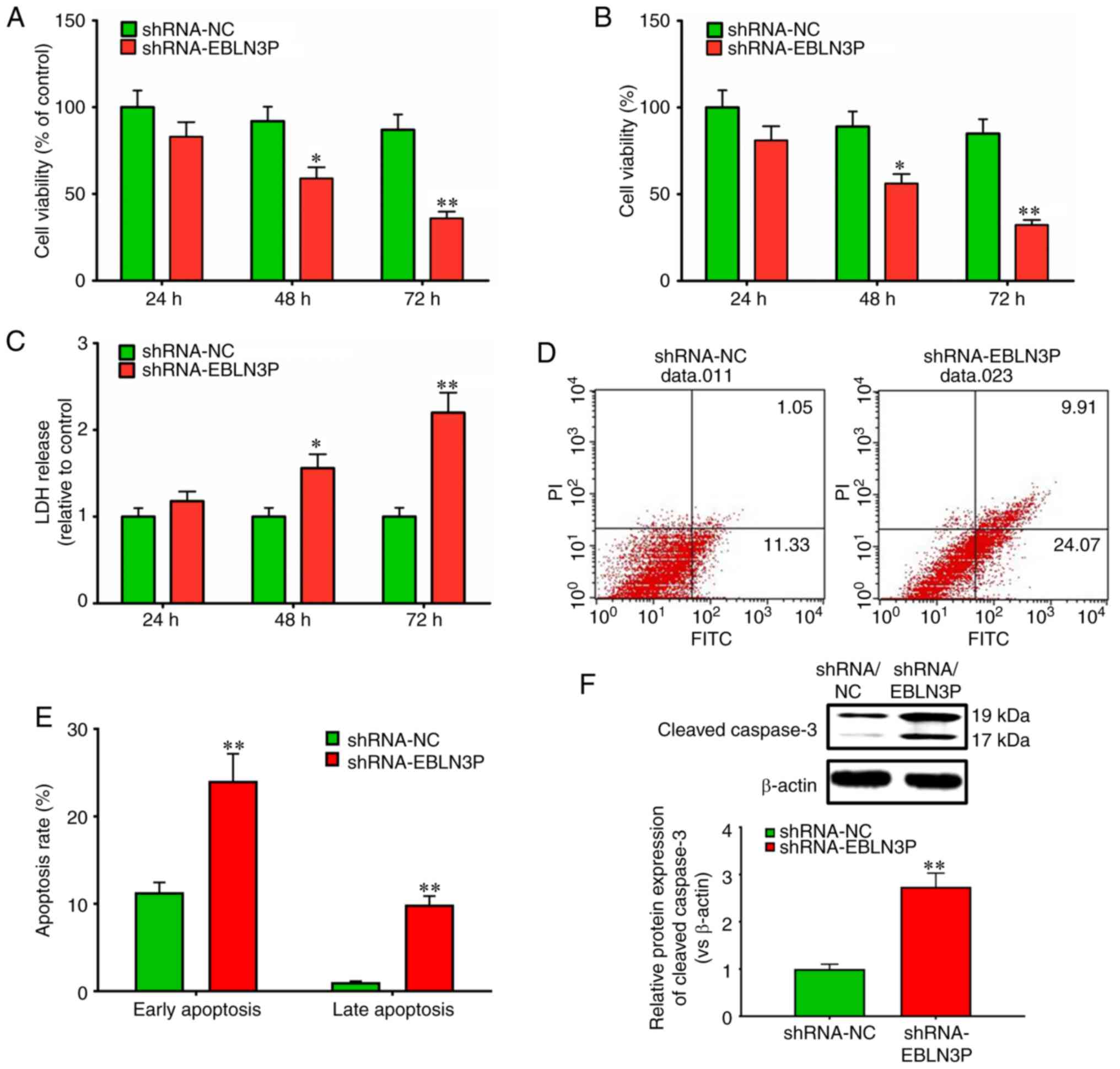
lncRNA EBLN3P knockdown decreases the
viability and promotes the apoptosis of normal SGNs
To determine the effects of lncRNA EBLN3P on the
viability of normal SGNs, the targeted knockdown of lncRNA EBLN3P
by shRNA was performed. The efficiency of plasmid transfection in
neuronal cells was 20-25%, as shown by fluorescence microscopy
observation (data not shown). The positively transfected cells were
further screened by flow cytometry following transfection with a
GFP-labeled plasmid for 48 h, prior to being subjected to
subsequent experiments. Transfection with lncRNA EBLN3P shRNA
markedly suppressed the viability of the SGNs at 48 and 72 h, in
contrast to transfection with negative control shRNA (P<0.05)
(Fig. 4A). Trypan blue staining
of the SGNs also revealed a significantly lower viability of the
SGNs transfected with lncRNA EBLN3P shRNA relative to that of the
control SGNs (Fig. 4B), which was
confirmed by a significantly higher LDH release from the cultures
of lncRNA EBLN3P shRNA-treated SGNs, indicating increased necrosis
(Fig. 4C). Subsequently, the
effect of lncRNA EBLN3P on the apoptosis of SGNs was further
investigated. Transfection of the SGNs with lncRNA EBLN3P shRNA
markedly promoted the early and late apoptosis of the SGNs compared
with those transfected with shRNA NC (P<0.05) (Fig. 4D and E). Furthermore, the
expression of caspase-3 was detected, which is closely associated
with cell apoptosis. Transfection of the normal SGNs with lncRNA
EBLN3P shRNA markedly promoted the expression of cleaved caspase-3
in SGNs, in comparison with the SGNs transfected with shRNA NC
(Fig. 4F). Thus, these results
demonstrated that the knockdown of lncRNA EBLN3P contributed to the
reduced survival of normal SGNs and to the increased apoptosis of
normal SGNs.
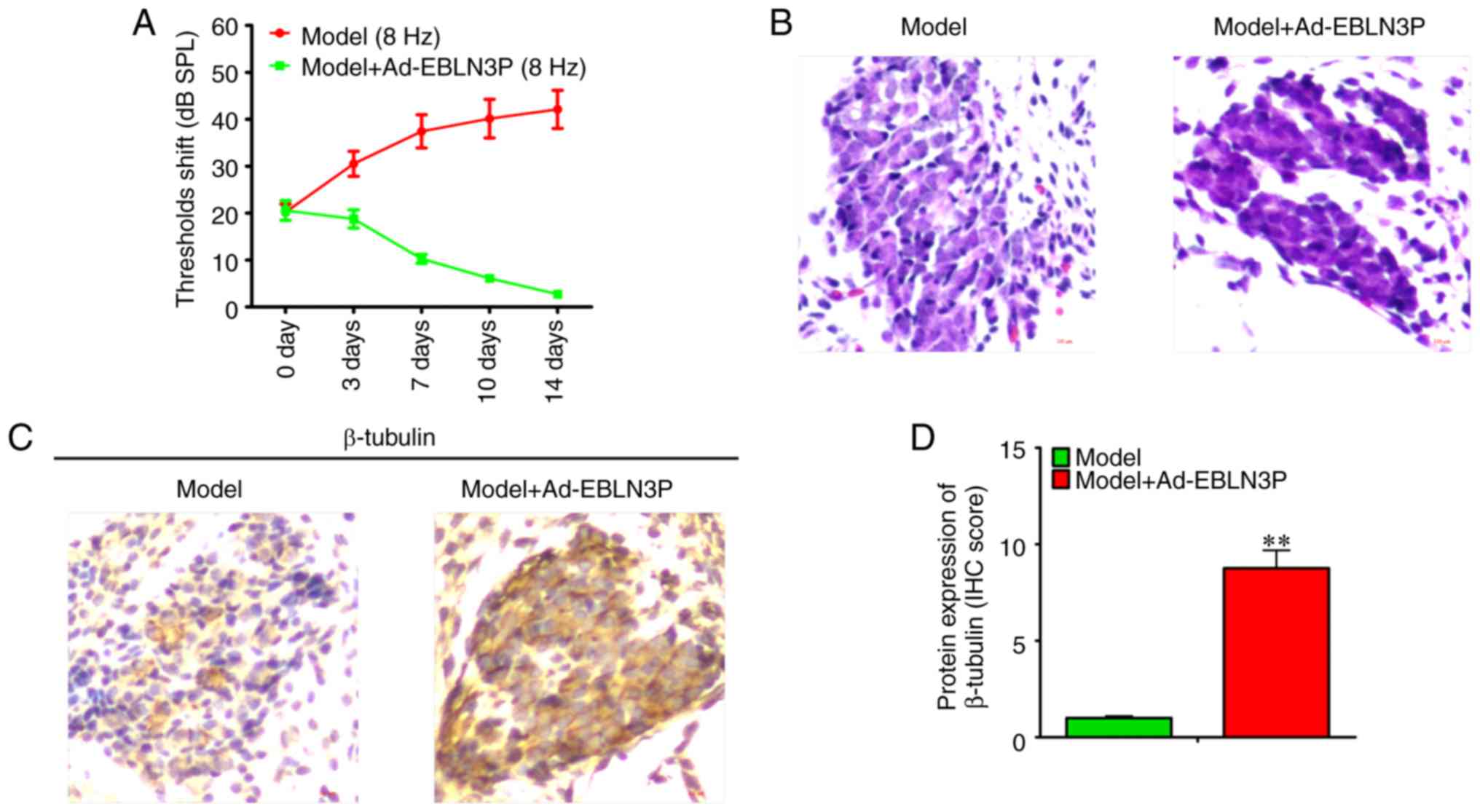
Recovery-promoting effect of lncRNA
EBLN3P on the structure and function of impaired organs of Corti
and SGNs in a model of deafness
Due to the positive regulatory effect of lncRNA
EBLN3P on the survival of SGNs, the healing effect of lncRNA EBLN3P
in models of deafness was then investigated. Adenovirus comprising
overexpression vectors of lncRNA EBLN3P was injected into the mice
in the models of deafness. The results of the ARB test revealed
that compared with the model group, Ad-EBLN3P significantly
decreased the threshold (Fig.
5A). H&E staining demonstrated that Ad-EBLN3P promoted the
recovery of the structure of the impaired organ of Corti and SGNs
(Fig. 5B). Subsequently, by
detecting the expression of β-tubulin in cochlear SGNs, it was
observed that Ad-EBLN3P increased the viability of neurons in
vivo (Fig. 5C and D).
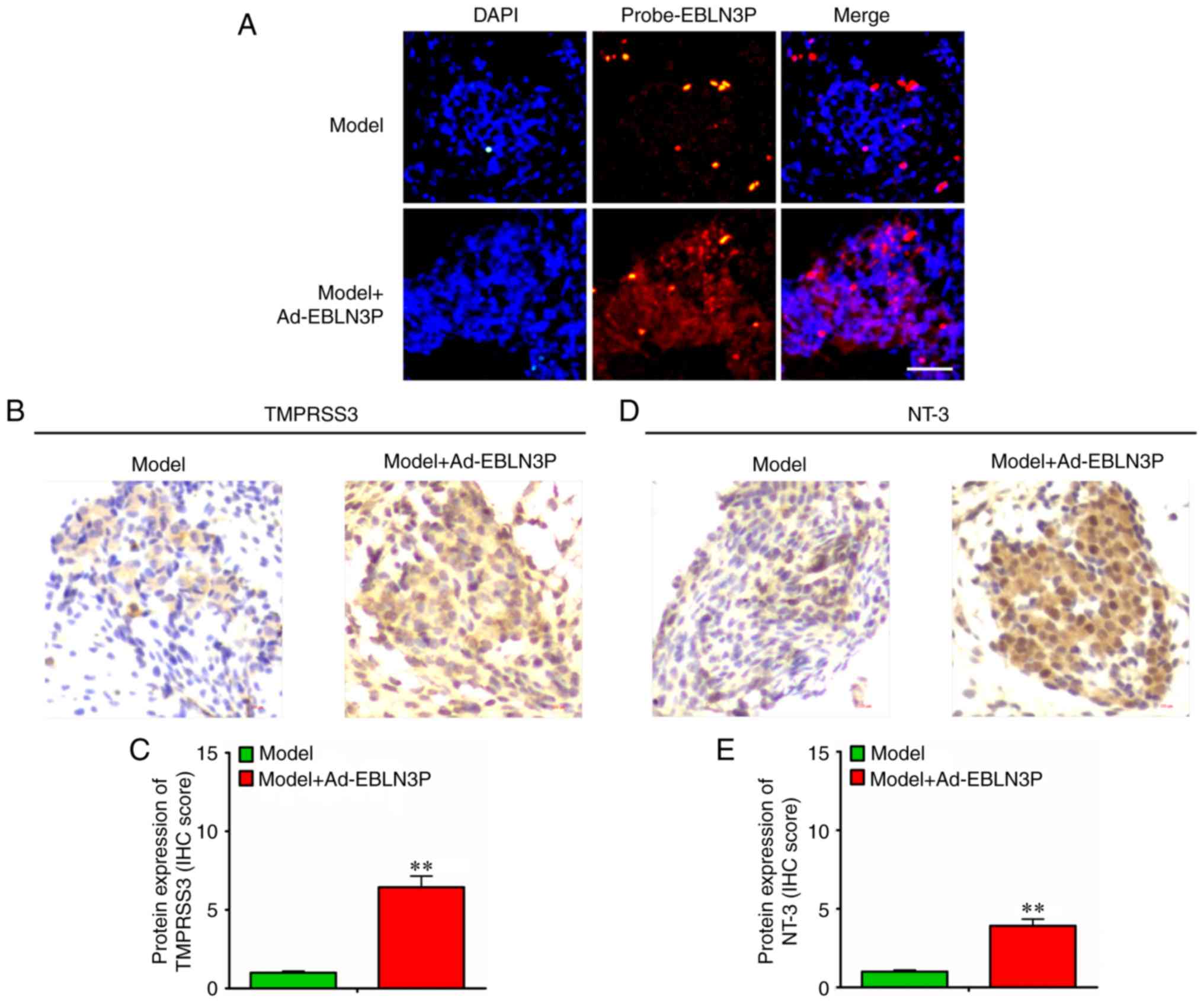
In subsequent experiments, the expression of
Ad-EBLN3P in cochlear SGNs was detected, and it was observed that
lncRNA EBLN3P and TMPRSS3 protein were overexpressed in the neurons
(Fig. 6A-C). Furthermore, NT-3
was also over-expressed in the neurons (Fig. 6D and E).
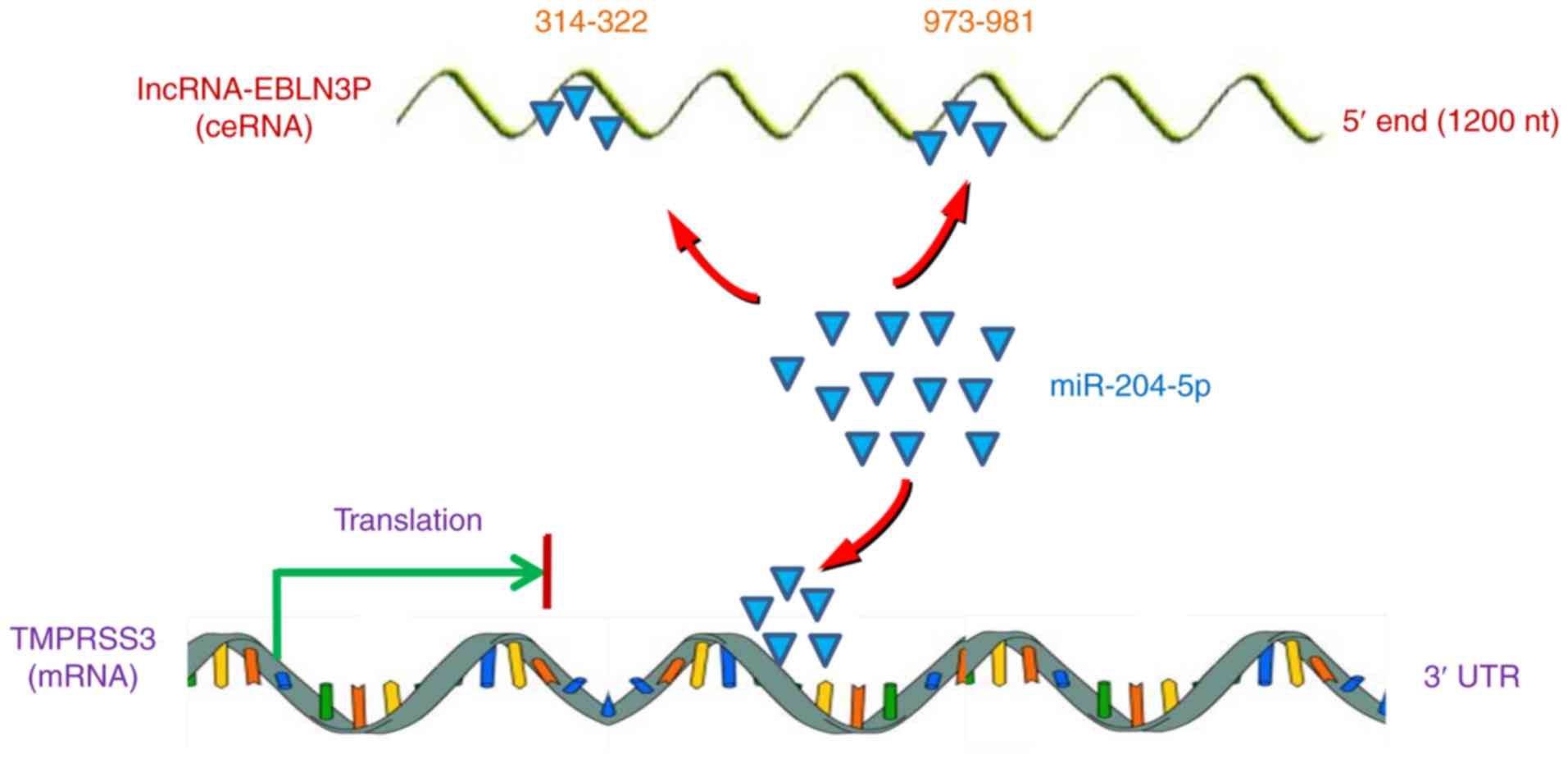
All the above-mentioned results indicated that
lncRNA EBLN3P promoted the recovery of the function of impaired
SGNs by competitively binding to miR-204-5p and regulating the
targeted gene TMPRSS3 expression (Fig. 7).
Discussion
Hearing loss is a frequent disorder and a highly
heterogeneous disease, with an incidence rate of 1/1,000
individuals worldwide. Multiple genetic and environmental causes
can lead to hearing loss, and SNHL is among the most common ones.
Several studies have reported the degeneration of SGNs and/or
sensory hair cells in SNHL (1,37,38). Moreover, SGNs are poorly
regenerated in the mammalian inner ear (39). Therefore, numerous studies have
focused on the prevention of SGN progressive degeneration.
Recently, numerous molecular genetic studies have been conducted on
the role of the TMPRSS3 gene in autosomal recessive non-syndromic
hearing loss (40-42). Currently, 16 different TMPRSS3
gene mutations have been identified to play key roles in the
development of the inner ear (40–45). Lee et al reported TMPRSS3
gene mutations in populations of East Asia (46). Previous studies have demonstrated
that kanamycin injection can lead to a reduction in TMPRSS3
expression in the cochlea, implying vital roles of the TMPRSS3 gene
in normal cochlea function (47).
A previous by the authors also suggested a high expression of the
TMPRSS3 gene in SGNs of mouse cochleae (13). It has also been demonstrated that
miR-204-5p suppresses the survival of cochlear SGNs in vitro
by targeting the TMPRSS3 gene (14). However, the upstream regulatory
mechanism of miR-204-5p has not been fully elucidated to date.
lncRNA EBLN3P was discovered and characterized
during the Human Genome Project in 2002 (48), and was confirmed in another genome
characterization in 2004 (49).
Previous studies on the genomic characterization of functional
lncRNA loci in human cells also found the existence of lncRNA
EBLN3P (50). However, no
functional or mechanistic reports have been published thus far, at
least to the best of our knowledge. Some studies have suggested the
potential role of lncRNAs as ceRNAs (51-53); however, the mechanism of the
discovered lncRNAs was not well defined.
The present study firstly demonstrated that lncRNA
EBLN3P functions as a ceRNA by regulating miR-204-5p in normal
SGNs. In vitro functional experiments demonstrated that
lncRNA EBLN3P promoted the recovery of the viability of normal SGNs
and inhibited the apoptosis of normal SGNs. Moreover, the
recovery-promoting effect of lncRNA EBLN3P on the structure and
function of impaired SGNs from the mice in the models of deafness
was demonstrated, and its molecular mechanisms may involve binding
to miR-204-5p and the regulation of the expression of its targeted
gene TMPRSS3. NT-3 is one of the important growth factors that
helps to stimulate and control neurogenesis. Its overexpression in
neurons also confirmed the recovery-promoting function of lncRNA
EBLN3P on the structure and function of impaired SGNs in deafness
models.
In conclusion, the findings of the present study
demonstrated that lncRNA EBLN3P promoted th e recovery of the
function of impaired SGNs by competitively binding to miR-204-5p
and regulating TMPRSS3 expression, suggesting that lncRNA EBLN3P
may be a potential therapeutic target in SNHL.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81570928).
Availability of data and materials
All the data generated or analyzed during this study
are included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
WJ, AP and ZZ designed the study. WJ, AP and ZZ also
conducted the reviewing and editing of the manuscript. WJ, AP, YC
and BP performed the experiments and data analysis. All authors had
full access to all the data in the study and take responsibility
for the integrity of the data and the accuracy of the data
analysis. WJ and AP wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were approved by Animal Care
and Use Committee at Xiangya School of Medicine from Central South
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Charizopoulou N, Lelli A, Schraders M, Ray
K, Hildebrand MS, Ramesh A, Srisailapathy CR, Oostrik J, Admiraal
RJ, Neely HR, et al: Gipc3 mutations associated with audiogenic
seizures and sensorineural hearing loss in mouse and human. Nat
Commun. 2:2012011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clark GM: Cochlear Implants: Fundamentals
and applications Graeme Clark. Springer; New York: 2003, View Article : Google Scholar
|
|
3
|
Feghali JG, Lefebvre PP, Staecker H, Kopke
R, Frenz DA, Malgrange B, Liu W, Moonen G, Ruben RJ and Van de
Water TR: Mammalian auditory hair cell regeneration/repair and
protection: A review and future directions. Ear Nose. Throat J.
77:276–280. 282–285. 1998. View Article : Google Scholar
|
|
4
|
Hardie NA and Shepherd RK: Sensorineural
hearing loss during development: Morphological and physiological
response of the cochlea and auditory brainstem. Hear Res.
128:147–165. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dodson HC and Mohuiddin A: Response of
spiral ganglion neurones to cochlear hair cell destruction in the
guinea pig. J Neurocytol. 29:525–537. 2000. View Article : Google Scholar
|
|
6
|
Dror AA and Avraham KB: Hearing loss:
Mechanisms revealed by genetics and cell biology. Annu Rev Genet.
43:411–437. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bindu LH and Reddy PP: Genetics of
aminoglycoside-induced and prelingual non-syndromic mitochondrial
hearing impairment: A review. Int J Audiol. 47:702–707. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wallrapp C, Hähnel S, Müller-Pillasch F,
Burghardt B, Iwamura T, Ruthenbürger M, Lerch MM, Adler G and Gress
TM: A novel transmembrane serine protease (TMPRSS3) overexpressed
in pancreatic cancer. Cancer Res. 60:2602–2606. 2000.PubMed/NCBI
|
|
9
|
Bugge TH, Antalis TM and Wu Q: Type II
transmembrane serine proteases. J Biol Chem. 284:23177–23181. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guipponi M, Toh MY, Tan J, Park D, Hanson
K, Ballana E, Kwong D, Cannon PZ, Wu Q, Gout A, et al: An
integrated genetic and functional analysis of the role of type II
transmembrane serine proteases (TMPRSSs) in hearing loss. Hum
Mutat. 29:130–141. 2008. View Article : Google Scholar
|
|
11
|
Guipponi M, Vuagniaux G, Wattenhofer M,
Shibuya K, Vazquez M, Dougherty L, Scamuffa N, Guida E, Okui M,
Rossier C, et al: The transmembrane serine protease (TMPRSS3)
mutated in deafness DFNB8/10 activates the epithelial sodium
channel (ENaC) in vitro. Hum Mol Genet. 11:2829–2836. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fasquelle L, Scott HS, Lenoir M, Wang J,
Rebillard G, Gaboyard S, Venteo S, François F, Mausset-Bonnefont
AL, Antonarakis SE, et al: Tmprss3, a transmembrane serine protease
deficient in human DFNB8/10 deafness, is critical for cochlear hair
cell survival at the onset of hearing. J Biol Chem.
286:17383–17397. 2001. View Article : Google Scholar
|
|
13
|
Ge S, Wang Q, Peng A, Wu W and Xie D:
Expression of proteinase TMPRSS3 in mouse cochlea. Zhong Nan Da Xue
Xue Bao Yi Xue Ban. 36:794–798. 2011.In Chinese. PubMed/NCBI
|
|
14
|
Li Y, Peng A, Ge S, Wang Q and Liu J:
MiR-204 suppresses cochlear spiral ganglion neuron survival in
vitro by targeting TMPRSS3. Hear Res. 314:60–64. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng J, Bi C, Clark BS, Mady R, Shah P and
Kohtz JD: The Evf-2 noncoding RNA is transcribed from the Dlx-5/6
ultraconserved region and functions as a Dlx-2 transcriptional
coactivator. Genes Dev. 20:1470–1484. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martianov I, Ramadass A, Serra Barros A,
Chow N and Akoulitchev A: Repression of the human dihydrofolate
reductase gene by a non-coding interfering transcript. Nature.
445:666–670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bächinger D, Horvath L, Eckhard A,
Goosmann MM, Honegger T, Gassmann M, Vogel J and Naldi AM: Neuronal
erythropoietin overexpression is protective against
kanamycin-induced hearing loss in mice. Toxicol Lett. 291:121–128.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ishikura F, Takano Y and Ueyama T: Acute
effects of beta-blocker with intrinsic sympathomimetic activity on
stress-induced cardiac dysfunction in rats. J Cardiol. 60:470–474.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lina IA and Lauer AM: Rapid measurement of
auditory filter shape in mice using the auditory brainstem response
and notched noise. Hear Res. 298:73–79. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gamma-aminobutyric acid A receptor and
N-methyl-D-aspartate receptor subunit expression in rat spiral
ganglion neurons. Neural Regen Res. 5:1–3. 2010.
|
|
25
|
Martinez-Monedero R, Corrales CE,
Cuajungco MP, Heller S and Edge AS: Reinnervation of hair cells by
auditory neurons after selective removal of spiral ganglion
neurons. J Neurobiol. 66:319–331. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Klahr AC, Dickson CT and Colbourne F:
Seizure activity occurs in the collagenase but not the blood
infusion model of striatal hemorrhagic stroke in rats. Transl
Stroke Res. 6:29–38. 2015. View Article : Google Scholar :
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Wang Y, Jiang SW, Liu X, Niu L, Ge XL,
Zhang JC, Wang HR, Fei AH, Gao CJ and Pan SM: Degradation of TRPML1
in neurons reduces neuron survival in transient global cerebral
ischemia. Oxid Med Cell Longev. 2018:46127272018. View Article : Google Scholar
|
|
29
|
Song L, Tang S, Dong L, Han X, Cong L,
Dong J, Han X, Zhang Q, Wang Y and Du Y: The neuroprotection of
KIBRA in promoting neuron survival and against amyloid β-induced
apoptosis. Front Cell Neurosci. 13:1372019. View Article : Google Scholar
|
|
30
|
Bogetofte H, Jensen P, Ryding M, Schmidt
SI, Okarmus J, Ritter L, Worm CS, Hohnholt MC, Azevedo C, Roybon L,
et al: PARK2 mutation causes metabolic disturbances and impaired
survival of human iPSC-derived neurons. Front Cell Neurosci.
13:2972019. View Article : Google Scholar :
|
|
31
|
Voss U, Sand E, Hellström PM and Ekblad E:
Glucagon-like peptides 1 and 2 and vasoactive intestinal peptide
are neuro-protective on cultured and mast cell co-cultured rat
myenteric neurons. BMC Gastroenterol. 12:302012. View Article : Google Scholar
|
|
32
|
Ishimoto S, Kawamoto K, Kanzaki S and
Raphael Y: Gene transfer into supporting cells of the organ of
Corti. Hear Res. 173:187–197. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chandradoss SD, Schirle NT, Szczepaniak M,
MacRae IJ and Joo C: A dynamic search process underlies MicroRNA
targeting. Cell. 162:96–107. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nishi K, Takahashi T, Suzawa M, Miyakawa
T, Nagasawa T, Ming Y, Tanokura M and Ui-Tei K: Control of the
localization and function of a miRNA silencing component TNRC6A by
argonaute protein. Nucleic Acids Res. 43:9856–9873. 2015.PubMed/NCBI
|
|
35
|
Schraivogel D, Schindler SG, Danner J,
Kremmer E, Pfaff J, Hannus S, Depping R and Meister G: Importin-β
facilitates nuclear import of human GW proteins and balances
cytoplasmic gene silencing protein levels. Nucleic Acids Res.
43:7447–7461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Weinmann L, Höck J, Ivacevic T, Ohrt T,
Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H and Meister G:
Importin 8 is a gene silencing factor that targets argonaute
proteins to distinct mRNAs. Cell. 136:496–507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim YR, Baek JI, Kim SH, Kim MA, Lee B,
Ryu N, Kim KH, Choi DG, Kim HM, Murphy MP, et al: Therapeutic
potential of the mitochondria-targeted antioxidant MitoQ in
mitochondrial-ROS induced sensorineural hearing loss caused by Idh2
deficiency. Redox Biol. 20:544–555. 2019. View Article : Google Scholar
|
|
38
|
Hu H, Ye B, Zhang L, Wang Q, Liu Z, Ji S,
Liu Q, Lv J, Ma Y, Xu Y, et al: Efr3a insufficiency attenuates the
degeneration of spiral ganglion neurons after hair cell loss. Front
Mol Neurosci. 10:862017. View Article : Google Scholar :
|
|
39
|
Zhang PZ, He Y, Jiang XW, Chen FQ, Chen Y,
Shi L, Chen J, Chen X, Li X, Xue T, et al: Stem cell
transplantation via the cochlear lateral wall for replacement of
degenerated spiral ganglion neurons. Hear Res. 298:1–9. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee K, Khan S, Islam A, Ansar M, Andrade
PB, Kim S, Santos-Cortez RL, Ahmad W and Leal SM: Novel TMPRSS3
variants in Pakistani families with autosomal recessive
non-syndromic hearing impairment. Clin Genet. 82:56–63. 2012.
View Article : Google Scholar
|
|
41
|
Walsh T, Abu Rayan A, Abu Sa'ed J, Shahin
H, Shepshelovich J, Lee MK, Hirschberg K, Tekin M, Salhab W,
Avraham KB, et al: Genomic analysis of a heterogeneous Mendelian
phenotype: Multiple novel alleles for inherited hearing loss in the
Palestinian population. Hum Genomics. 2:203–211. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wattenhofer M, Sahin-Calapoglu N,
Andreasen D, Kalay E, Caylan R, Braillard B, Fowler-Jaeger N,
Reymond A, Rossier BC, Karaguzel A and Antonarakis SE: A novel
TMPRSS3 missense mutation in a DFNB8/10 family prevents proteolytic
activation of the protein. Hum Genet. 117:528–535. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Charif M, Abidi O, Boulouiz R, Nahili H,
Rouba H, Kandil M, Delprat B, Lenaers G and Barakat A: Molecular
analysis of the TMPRSS3 gene in Moroccan families with
non-syndromic hearing loss. Biochem Biophys Res Commun.
419:643–647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hutchin T, Coy NN, Conlon H, Telford E,
Bromelow K, Blaydon D, Taylor G, Coghill E, Brown S, Trembath R, et
al: Assessment of the genetic causes of recessive childhood
non-syndromic deafness in the UK-implications for genetic testing.
Clin Genet. 68:506–512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Masmoudi S, Antonarakis SE, Schwede T,
Ghorbel AM, Gratri M, Pappasavas MP, Drira M, Elgaied-Boulila A,
Wattenhofer M, Rossier C, et al: Novel missense mutations of
TMPRSS3 in two consanguineous Tunisian families with non-syndromic
autosomal recessive deafness. Hum Mutat. 18:101–108. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee J, Baek JI, Choi JY, Kim UK, Lee SH
and Lee KY: Genetic analysis of TMPRSS3 gene in the Korean
population with autosomal recessive nonsyndromic hearing loss.
Gene. 532:276–280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Eppsteiner RW, Shearer AE, Hildebrand MS,
Deluca AP, Ji H, Dunn CC, Black-Ziegelbein EA, Casavant TL, Braun
TA, Scheetz TE, et al: Prediction of cochlear implant performance
by genetic mutation: The spiral ganglion hypothesis. Hear Res.
292:51–58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Strausberg RL, Feingold EA, Grouse LH,
Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler
GD, Altschul SF, et al: Generation and initial analysis of more
than 15,000 full-length human and mouse cDNA sequences. Proc Natl
Acad Sci USA. 99:16899–16903. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu SJ, Horlbeck MA, Cho SW, Birk HS,
Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et
al: CRISPRi-based genome-scale identification of functional long
noncoding RNA loci in human cells. Science. 355:pii: aah7111. 2017.
View Article : Google Scholar
|
|
51
|
Liang H, Pan Z, Zhao X, Liu L, Sun J, Su
X, Xu C, Zhou Y, Zhao D, Xu B, et al: LncRNA PFL contributes to
cardiac fibrosis by acting as a competing endogenous RNA of let-7d.
Theranostics. 8:1180–1194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Qiu YY, Wu Y, Lin MJ, Bian T, Xiao YL and
Qin C: LncRNA-MEG3 functions as a competing endogenous RNA to
regulate Treg/Th17 balance in patients with asthma by targeting
microRNA-17/RORγt. Biomed Pharmacother. 111:386–394. 2019.
View Article : Google Scholar
|
|
53
|
Ji Q, Cai G, Liu X, Zhang Y, Wang Y, Zhou
L, Sui H and Li Q: MALAT1 regulates the transcriptional and
translational levels of proto-oncogene RUNX2 in colorectal cancer
metastasis. Cell Death Dis. 10:3782019. View Article : Google Scholar : PubMed/NCBI
|