Introduction
Non-small cell lung carcinoma (NSCLC) is a growing
threat to humans and the dominant cause of cancer-related deaths
worldwide (1). The five-year
overall survival is not optimistic due to metastasis and
chemoresistance (2,3). Hence, it is pressing to find novel
molecular targets for NSCLC treatment.
Circular RNAs (circRNAs), a type of single-stranded
RNAs which form a covalently closed continuous loop, are produced
by backsplicing (4) and they are
resistant to exonuclease-mediated degradation (5). Growing evidence has proved that
circRNAs are associated with colon cancer (6), gastric cancer (7), bladder cancer (8) and NSCLC (9,10).
Microarray data showed that hsa_circ_0106705 (circ-ACACA) was
upregulated in human lung cancer according to Yang et al
(11). However, the regulatory
mechanism of circ-ACACA in NSCLC is hardly reported and needs to be
investigated further.
MicroRNAs (miRNAs/miRs) are short (~22 nucleotides)
and highly conserved noncoding RNAs, which mediate gene expression
by binding to the 3′-untranslated region of mRNA at the
post-transcriptional level (12).
Numerous studies have emphasized the core position of miRNAs in
governing cancer development (13,14). miR-1183 was reported to be
dysregulated in numerous human diseases (15-17) and Zhou et al (18) reported that miR-1183 functioned in
the tumorigenesis of NSCLC. Nevertheless, the precise mechanism of
miR-1183 in NSCLC progression remains to be studied.
The phosphoinositide 3-kinases/protein kinase B
(PI3K/PKB) pathway is essential for cell survival and apoptosis
(19) and their aberrant
activation is usually correlated with malignancy (20). A previous study indicated that the
PI3K/PKB pathway was associated with the initiation of endometrial
cancer (21) and
cisplatin-resistance in NSCLC (22). Therefore, in-depth studies of the
PI3K/PKB signaling pathway could contribute to the development of
new effective therapeutic methods for NSCLC.
In this study, the expression level of circ-ACACA in
NSCLC tissues and cells was first measured. Afterwards, the
function and potential regulatory mechanism of circ-ACACA in NSCLC
were further investigated by subsequent experiments.
Materials and methods
Specimens and cell culture
A total of 60 NSCLC tissues and paired nearby
healthy tissues were sourced from patients with NSCLC (36 males and
24 females; 20-65 years old) who had undergone surgical resection
between April 2015 to August 2017 at Liaocheng People's Hospital,
and the 60 NSCLC tissues included 37 early stages (I and II)
tissues and 23 advanced stages (III and IV) tissues or contained 21
lymphoid node metastasis tissues and 39 controls. Tumor, node and
metastasis (TNM) staging was classified according to the 7th
edition of the American Joint Committee on Cancer TNM
classification based on information obtained regarding the tumor
during tumor surgery and histological or imaging studies. Every
patient signed the informed consent and the official approval from
the Ethics Committee of Liaocheng People's Hospital was obtained in
this study. Human normal bronchus epithelium cell line (BEAS-2B)
and NSCLC cell lines (A549, H1975, H1395, H1793, H1299 and H1792)
were purchased from the American Type Culture Collection. McCoy's
5A medium (Sigma-Aldrich; Merck KGaA), containing 5% CO2
and 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA) was
used to culture cells.
Cell transfection
Small interfering (si) RNA against circ-ACACA
(si-circ-ACACA; 5′-GAA AGA CUU UGA AAU ACU CGU-3′), negative
control of siRNA (si-NC; 5′-AAG ACA UUG UGU GUC CGC CTT-3′),
miR-1183 mimic (named as miR-1183; 5′-CAC UGU AGG UGA UGG UGA GAG
UGG GCA-3′) and mimic negative control (miR-NC; 5′-ACG UGA CAC GUU
CGG AGA ATT-3′), and miR-1183 inhibitor (anti-miR-1183; 5′-UGC CCA
CUC UCA CCA UCA CCU ACA GUG-3′) and corresponding negative control
(anti-NC; 5′-UGA GCU GCA UAG AGU AGU GAU UA-3′) were obtained from
Shanghai GenePharma Co., Ltd. A549 and H1299 cells were transfected
with the above oligonucleotides (at a final concentration of 50 nM)
using Lipofectamine 2000 Transfection Reagent (Beijing Solarbio
Science & Technology Co., Ltd.) for 24 h referring to the
manufacturer's protocol.
RNA isolation, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
RNase R treatment
NSCLC tissues and cells were collected and total RNA
was isolated by the TriQuick Reagent (Beijing Solarbio Science
& Technology Co., Ltd.). Then, RNA was reverse transcribed to
cDNA (37°C for 15 min; 85°C for 5 sec; hold at 4°C) by PrimeScript™
RT Master Mix kit (Takara Biotechnology, Co., Ltd.). qPCR was
conducted (95°C for 5 min, then 40 cycles of 95°C for 15 sec, 60°C
for 30 sec and 72°C for 45 sec) with SYBR Green Realtime PCR Master
Mix (Beijing Solarbio Science & Technology Co., Ltd.) and data
were analyzed using 2-ΔΔCq method (23). β-actin and U6 were introduced as
the inner references. Primers in the present research: circ-ACACA
(forward 5′-GTG GCT TTG AAG GAG CTG TC-3′, reverse 5′-CAG ACA TGC
TGG ACC TTG AA-3′); miR-1183 (forward, 5′-ACT GAC CAC TGT AGG TGA
TGG T-3′, reverse 5′-GCG AGC ACA GAA TTA ATA CGA CTC ACT ATA
GG-3′); β-actin (forward 5′-GCA CCA CAC CTT CTA CAA TG-3′, reverse,
5′-TGC TTG CTG ATC CAC AT C TG-3′); U6 (forward, 5′-TCC GGG TGA TGC
TTT TCC TAG-3′ and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′).
Purified RNAs were treated with RNase R (Beijing Solarbio Science
& Technology Co., Ltd.) for subsequent experiments.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was tested with CCK-8 reagent
(Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. After transfection, A549 and H1299 cells
were seeded into 96-well plates and then incubated with 10
µl CCK-8 solution for 2 h. Optical density values were
examined at 450 nm wavelength using a microplate reader (Bio-Rad
Laboratories, Inc.).
Transwell assay
A Transwell chamber was employed to check the
capacity of cell migration. Transfected cells (5×104
cells) were seeded into the upper chamber and medium containing FBS
was placed in the lower chamber. After being treated with 0.1%
crystal violet for 20 min at room temperature (Beijing Solarbio
Science & Technology Co., Ltd.) following incubation for 24 h,
migrated cells were analyzed under an inverted light microscope
(MTX Lab Systems).
Glycolysis analysis
Glycolysis was evaluated by using Seahorse XF
glycolytic rate assay kit (Agilent Technologies, Inc.) on Seahorse
XFe96 analyzer (Agilent Technologies, Inc.). Cells
(2×104 cells/well) transfected with si-circ-ACACA,
anti-miR-1183, si-circ-ACACA + anti-miR-1183 or their matched
controls were seeded in the XF96 well plate. After the probes were
calibrated, 10 mmol glucose, 10 µmol oligomycin and 50 mmol
2-deoxyglucose were serially injected to measure the extracellular
acidification rate (ECAR). Data were analyzed with Seahorse XFe24
Wave software version Wave 2.2 (Agilent Technologies, Inc.).
Western blotting
Proteins from samples were isolated using RIPA
buffer (Vazyme) and protein concentration was checked by Detergent
Compatible Bradford Protein Quantification kit (Vazyme). Proteins
(20 µg/lane) were separated by 10% SDS-PAGE and then
transferred onto the polyvinylidene difluoride membranes (Vazyme).
The membranes were blocked with 5% skimmed milk (Vazyme) for 1 h at
room temperature and washed by phosphate-buffered saline.
Afterwards, the membranes were incubated at 4°C overnight with the
primary antibodies: Cellular-myelocytomatosis (c-myc; 1:1,000; cat.
no. ab32072; Abcam), matrix metallopeptidase 9 (MMP9; 1:1,000; cat.
no. ab38898; Abcam), glucose transporter 1 (GLUT-1; 1:1,000; cat.
no. ab652; Abcam), phosphatase and tensin homolog (PTEN; 1:3,000;
cat. no. ab32199; Abcam), phosphoinositide 3-kinases (PI3K;
1:2,000; cat. no. ab151549; Abcam), phosphorylated PI3K (p-PI3K;
1:1,000; cat. no. ab138364; Abcam), PKB (1:1,000; cat. no. ab8805;
Abcam), phosphorylated PKB (p-PKB; 1:1,000; cat. no. ab38449;
Abcam) or β-actin (1:3,000; cat. no. ab8227; Abcam) overnight.
After being rewashed, the membranes were incubated with the
horse-radish peroxidase-conjugated goat anti-rabbit secondary
antibody (1:3,000; cat. no. ab205718; Abcam) for 3 h at
37°C. The membranes were analyzed by the ChemiDoc™ MP
Imaging System with Image Lab™ Software version5.2 (Bio-Rad
Laboratories, Inc.) after being treated with an Enhanced ECL
Chemiluminescence Detection kit (Vazyme).
Dual-luciferase reporter assay
The potential complementary sequences of circ-ACACA
and miR -1183 were forecasted by circular RNA Interactome (24). The wild type (WT) sequence of
circ-ACACA harboring the binding sites of miR-1183 was inserted
into the pGL3 vector (Promega Corporation) to establish the
luciferase reporter vector WT-circ-ACACA. Similarly, the mutant
(MUT)-circ-ACACA reporter vector was established by mutating the
potential target sites of miR-1183. Then, the luciferase reporter
vectors (100 ng) were cotransfected with 50 ng miR-1183 or miR-NC
into A549 and H1299 cells for 24 h using Lipofectamine 2000
(Beijing Solarbio Science & Technology, Co., Ltd.). Firefly
luciferase activities were normalized by comparison with
Renilla luciferase. The Dual-Glo Luciferase Assay System kit
(Promega Corporation) was utilized to measure luciferase
activity.
RNA immunoprecipitation (RIP) assay
RIP was carried out using Magna RIP RNA-Binding
Protein Immunoprecipitation kit (EMD Millipore) following the
manufacturer's protocols. Briefly, harvested cells were lysed with
RIPA lysis buffer (Beyotime Institute of Biotechnology) and
incubated with magnetic beads conjugated with anti-Argonaute 2
(Anti-Ago2) antibody (1:5,000; cat. no. MABE253; EMD Millipore) for
8 h at 37°C, and immunoglobulin G (IgG; 1:5,000; cat. no. 12-370;
EMD Millipore) was used as a negative control. The protein was
removed by Proteinase K. The immune precipitated RNA was purified
and analyzed by RT-qPCR.
RNA pull-down assay
A biotin-labeled probe against miR-1183 (named as
Bio-miR-1183) and its negative control (named as Bio-NC) were
obtained from Sangon Biotech Co., Ltd. Transfected cells were lysed
with RIPA lysis buffer (Beyotime Institute of Biotechnology) and
incubated with streptavidin-coupled beads (Sangon Biotech Co.,
Ltd.). After being treated by proteinase K, circ-ACACA was isolated
and checked by RT-qPCR.
Xenograft mice model
BALB/c nude mice (male; 5 weeks old; ~18-23 g; n=10)
were acquired from Shanghai LingChang Biotech Co., Ltd. (Shanghai
SLAC Laboratory Animal Co., Ltd.) and kept in specific pathogen
free conditions (temperature 23±2°C, relative humidity 55±5%, 12
h/12 h light/dark cycle with ad libitum access to water and
food). The mice were randomly grouped into two groups (n=5 each).
Lentivirus harboring short hairpin RNA targeting circ-ACACA (named
as sh-circ-ACACA) and negative control (sh-NC) were constructed by
GeneCopoeia, Inc. A549 cells at a density of 1×106
cells/well in 6-well culture plates were infected with 4 µg
of filtered lentivirus plus 8 µg/ml polybrene
(Sigma-Aldrich; Merck KGaA) for 48 h and subsequently selected by
2.5 µg/ml puromycin (Invitrogen; Thermo Fisher Scientific,
Inc.) for at least 3 days to establish stable cell lines. A total
of ~2×106 A549 cells infected with sh-circ-ACACA or
sh-NC plasmid DNA in 200 µl of FBS-free culture medium were
injected subcutaneously into the flank of the nude mice. The tumor
volume was calculated every 7 days according to the formula: 0.5 ×
length × width2. The tumor weight was measured after the
mice were euthanized. The mRNA or protein levels of corresponding
genes in tumors were checked by RT-qPCR or western blotting,
respectively. The animal experiment was approved by the Animal Care
and Use Committee of Liaocheng People's Hospital and executed
referring to the instructions of the National Animal Protection and
Ethics Institute.
Statistical analysis
Experimental data were calculated by GraphPad Prism
8.0 (GraphPad Software, Inc.) and presented as the mean ± standard
deviation. Two independent groups were compared by using Student's
t-test. For more than two groups, the one-way analysis of variance
followed by Tukey post hoc test was utilized to assess the
difference. Receiver operating characteristic (ROC) curve analysis
was performed referring to a previous study (25). The Kaplan-Meier method was
utilized to assess overall survival and the log-rank test was used
to analyze the differences between survival curves. Pearson's
correlation coefficient was applied to analyze the correlation
between circ-ACACA and miR-1183 in NSCLC tissues. Every experiment
was repeated at least three times independently. P<0.05 was
considered to indicate a statistically significant difference.
Results
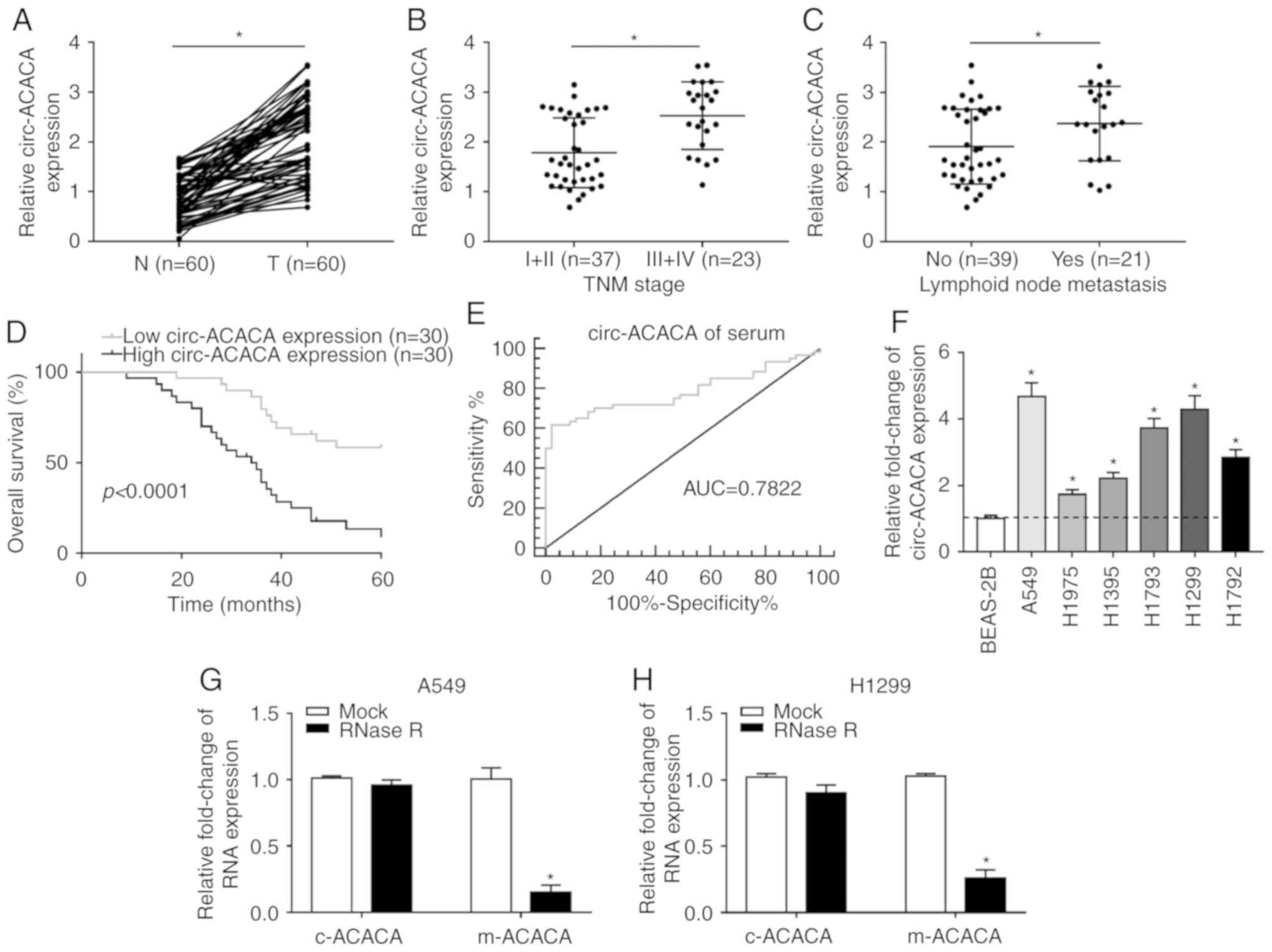
circ-ACACA is significantly upregulated
in NSCLC and correlates with the poor prognosis
To explore the role of circ-ACACA in NSCLC, the
expression patterns were first checked. The RT-qPCR data showed
that circ-ACACA was significantly upregulated in NSCLC tissues
compared with paired normal tissues (Fig. 1A). Next, the correlation between
circ-ACACA expression and NSCLC progression was evaluated and the
results showed that the level of circ-ACACA was increased in
advanced stages (III and IV) compared with early stages (I and II;
Fig. 1B). In addition, metastatic
samples displayed an upregulated level of circ-ACACA (Fig. 1C) and the higher level of
circ-ACACA led to the lower survival rate (Fig. 1D). Also, the diagnostic accuracy
of circ-ACACA was assessed using the ROC curve analysis and the
data showed that the area under the ROC curve was 0.7822 (Fig. 1E), which indicated that circ-ACACA
might be a hallmark of NSCLC. Moreover, circ-ACACA was also
significantly upregulated in NSCLC cells (Fig. 1F). Further analysis indicated that
the mRNA of circ-ACACA (c-ACACA) was conspicuously resistant to
RNase R compared with the mRNA of ACACA (m-ACACA; Fig. 1G and H). Collectively, these
results illuminated that circ-ACACA might act an oncogene in NSCLC
and have clinical diagnostic value.
Knockdown of circ-ACACA inhibits
proliferation and migration and retards glycolysis rate
The levels of c-ACACA and m-ACACA were checked and
the data showed that they were abundant in the cytoplasm of NSCLC
cells (Fig. 2A and B). To
investigate the function of circ-ACACA in NSCLC, A549 and H1299
cells were first infected with si-circ-ACACA or si-NC and then the
knockdown efficiency was confirmed (Fig. 2C and D). The CCK-8 assay showed
that downregulation of circ-ACACA inhibited proliferation of NSCLC
cells (Fig. 2E and F) and the
transwell assay indicated that knock-down of circ-ACACA weakened
the ability of migration of NSCLC cells (Fig. 2G and H). Glycolysis analysis
indicated that circ-ACACA silencing decreased ECAR in NSCLC cells
(Fig. 2I and J). Afterwards, the
protein levels of c-myc (related to proliferation), MMP9 (related
to migration) and GLUT-1 (related to glycolysis) were measured and
the results indicated that downregulation of circ-ACACA
significantly reduced the expression of these proteins in NSCLC
cells (Fig. 2K and L).
Altogether, these results demonstrated that circ-ACACA silencing
suppressed proliferation and migration and alleviated the Warburg
effect in NSCLC cells.
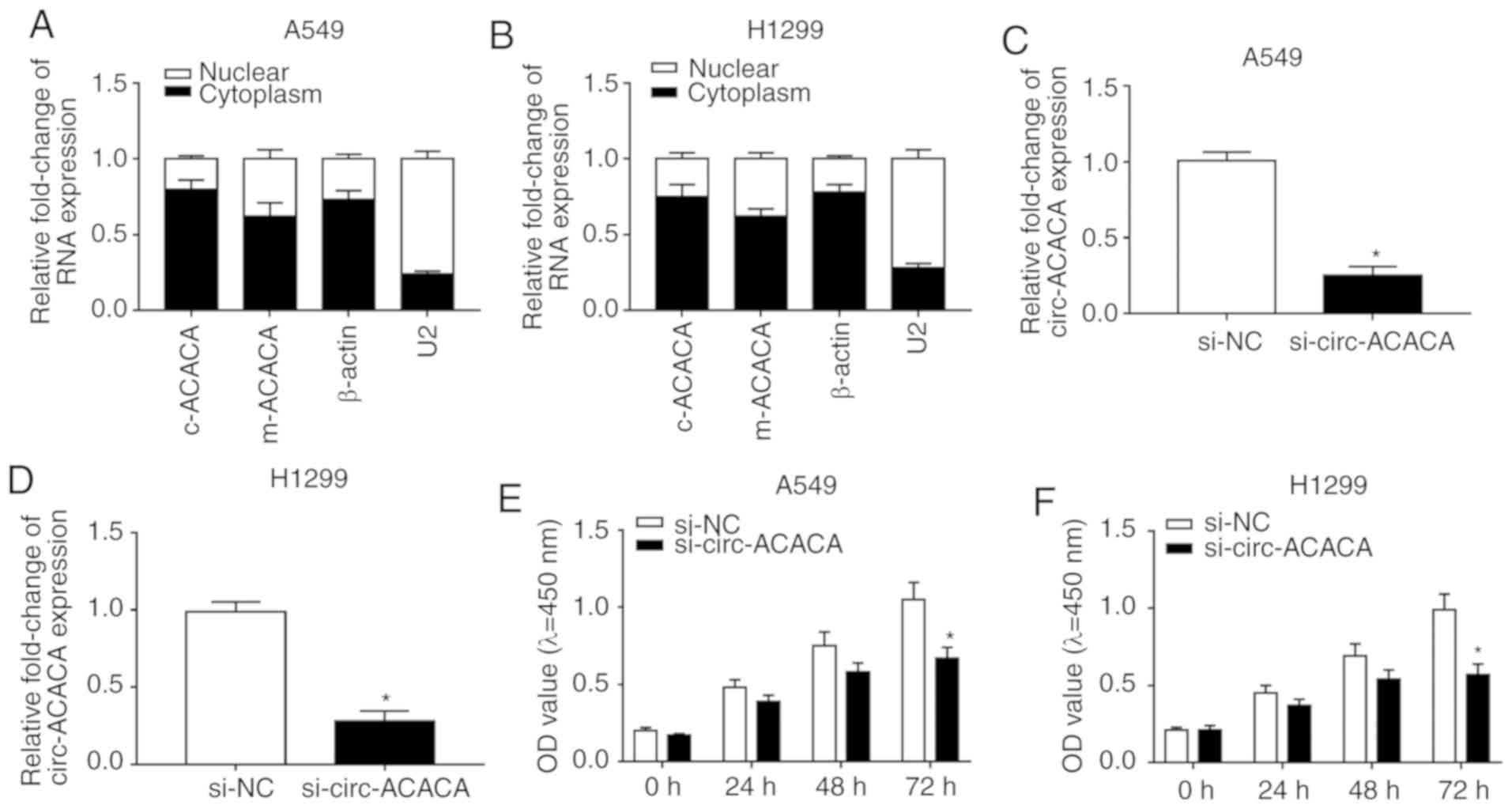 | Figure 2circ-ACACA silencing hampers the
progression of NSCLC. The levels of c-ACACA and m-ACACA in nucleus
and cytoplasm were detected by RT-qPCR of (A) A549 and (B) H1299
cells. β-actin and U2 were applied as positive controls in the
cytoplasm and nucleus, respectively. The level of circ-ACACA in
NSCLC cells infected with si-circ-ACACA or si-NC was checked by
RT-qPCR in (C) A549 and (D) H1299 cells. Cell Counting Kit-8 assay
was used to check proliferation of infected (E) A549 and (F) H1299
cells. A Transwell assay was performed to evaluate the ability of
migration of infected (G) A549 and (H) H1299 cells. The glycolysis
rate was detected by Seahorse XFe96 analyzer in (I) A549 and (J)
H1299 cells. The protein levels of c-myc, MMP9 and GLUT-1 in
infected (K) A549 and (L) H1299 cells were checked by western
blotting. *P<0.05 vs. si-NC. RT-q, reverse
transcription-quantitative; circ, circular; NSCLC, non-small cell
lung cancer; si, small interfering; NC, negative control; MMP,
matrix metalloproteinases; GLUT, glucose transporter; c-myc,
cellular-myelocytomatosis; OD, optical density; ECAR, extracellular
acidification rate; Glc, glucose; O, oligomycin; 2-DG,
2-deoxyglucose. |
Downregulation of miR-1183 promotes
proliferation and migration and elevates glycolysis rate
To probe the function of miR-1183 in NSCLC, its
expression was detected and the data showed that miR-1183 was
significantly downregulated in NSCLC tissues and cells compared
with normal tissues and cells (Fig.
3A and B). Thereafter, NSCLC cells were transfected with
anti-miR-1183 or anti-NC and the results indicated that miR-1183
was significantly decreased in the anti-miR-1183 group (Fig. 3C and D). Next, a CCK-8 assay and
transwell assay were performed and the data showed that
downregulation of miR-1183 promoted proliferation (Fig. 3E and F) and enhanced the migration
ability of NSCLC cells (Fig. 3G and
H). In addition, miR-1183 inhibitor increased ECAR in NSCLC
cells (Fig. 3I and J).
Simultaneously, the protein levels of c-myc, MMP9 and GLUT1 were
significantly elevated in the anti-miR-1183 group (Fig. 3K and L). To sum up, these results
demonstrated that miR-1183 might act as a tumor suppressor in NSCLC
progression in vitro.
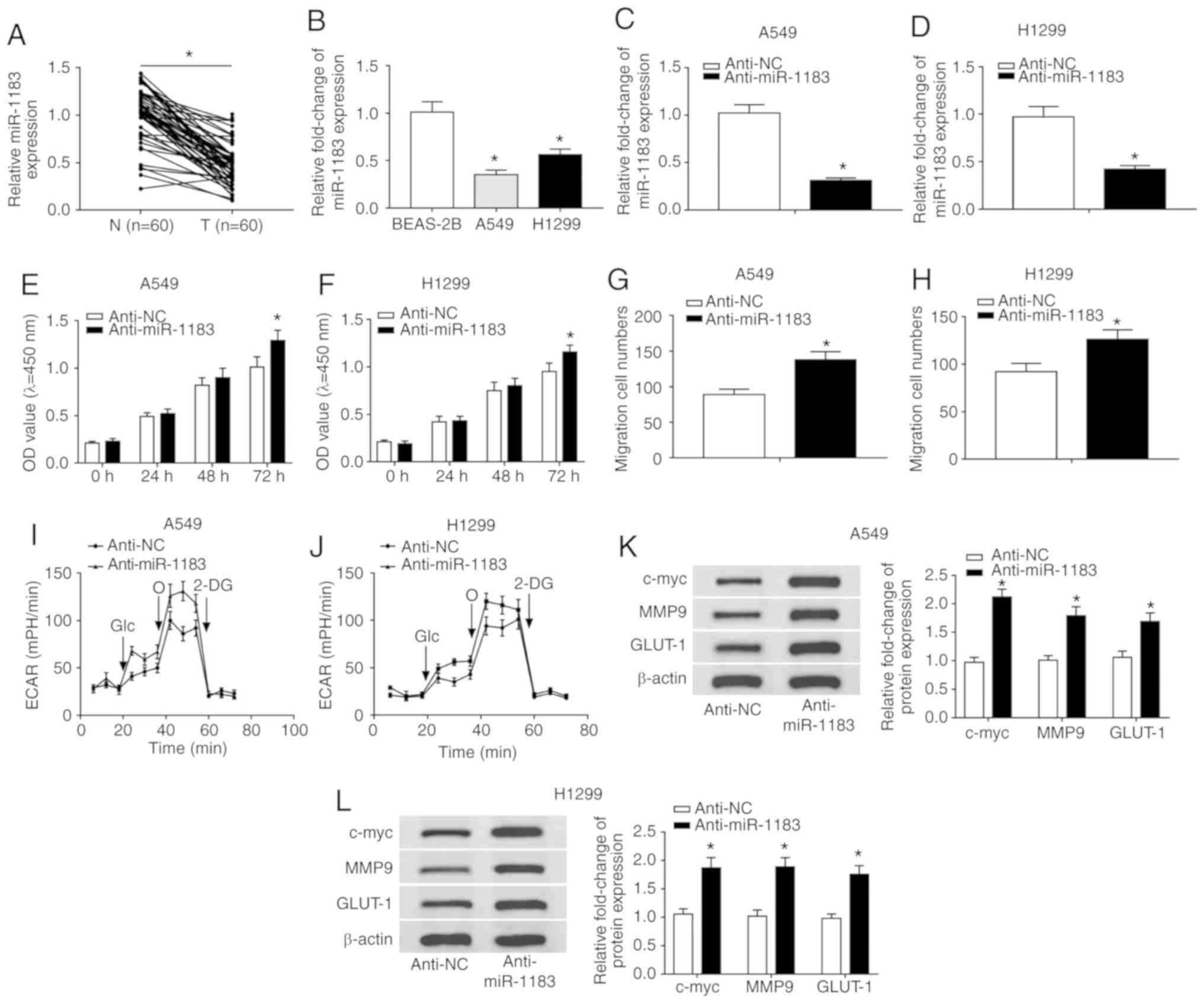 | Figure 3miR-1183 inhibitor promotes the
progression of NSCLC. (A) The expression level of miR-1183 in NSCLC
tissues (n=60) and normal tissues (n=60) was detected by RT-qPCR.
(B) The level of miR-1183 in normal cell line and NSCLC cell lines
was detected by RT-qPCR. The level of miR-1183 in (C) A549 and (D)
H1299 cells infected with anti-miR-1183 or anti-NC was checked by
RT-qPCR. The Cell Counting Kit-8 assay was used to check
proliferation of infected (E) A549 and (F) H1299 cells. Transwell
assay was utilized to check the ability of migration of infected in
(G) A549 and (H) H1299 cells. The glycolysis rate was detected by
Seahorse XFe96 analyzer in (I) A549 and (J) H1299 cells. The
protein levels of c-myc, MMP9 and GLUT-1 in infected NSCLC cells
were measured by western blotting in (K) A549 and (L) H1299 cells.
*P<0.05 vs. BEAS-2B or anti-NC. RT-q, reverse
transcription-quantitative; circ, circular; NSCLC, non-small cell
lung cancer; si, small interfering; NC, negative control; MMP,
matrix metalloproteinases; GLUT, glucose transporter; c-myc,
cellular-myelocytomatosis; OD, optical density; ECAR, extracellular
acidification rate; miR, microRNA; Glc, glucose; O, oligomycin;
2-DG, 2-deoxyglucose. |
circ-ACACA targets and negatively
regulates miR-1183 in NSCLC
The interaction between circRNAs and miRNAs in
cancer is documented in numerous reports (8,9).
To probe the relationship between the two, Pearson's correlation
coefficient was analyzed and the result indicated that the
expression of miR-1183 was negatively associated with circ-ACACA in
NSCLC tissues (Fig. 4A).
Afterwards, the miR-1183 mimic was introduced to NSCLC cells and
the overexpression efficiency was verified (Fig. 4B). By using circular RNA
Interactome, circ-ACACA was found to harbor the binding sites of
miR-1183 (Fig. 4C). The
Dual-luciferase reporter assay showed that miR-1183 significantly
diminished the luciferase activity of WT-circ-ACACA in NSCLC cells,
rather than MUT-circ-ACACA (Fig. 4D
and E). The RIP assay indicated that the relative enrichment of
circ-ACACA and miR-1183 was increased in the Anti-Ago2 group
compared with the control group (Fig.
4F and G). Simultaneously, RNA pull-down assay showed that
circ-ACACA was notably enriched in NSCLC cells transfected with
Bio-miR-1183 (Fig. 4H and I).
Further study indicated that knockdown of circ-ACACA significantly
increased the expression of miR-1183 in NSCLC cells (Fig. 4J and K). All in all, these results
demonstrated that miR-1183 was a target of circ-ACACA and
negatively modulated by circ-ACACA in NSCLC cells.
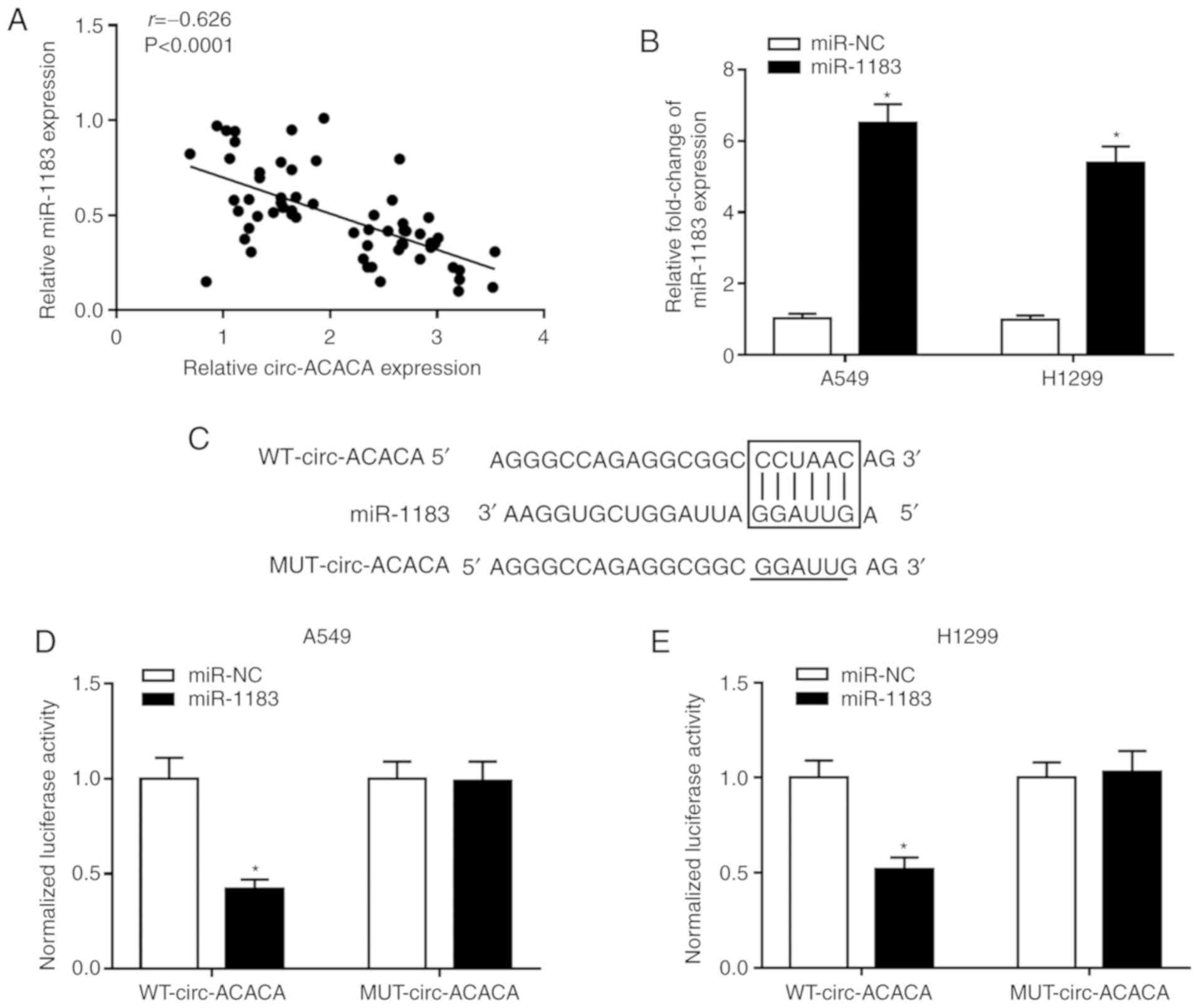 | Figure 4miR-1183 is a target of circ-ACACA
and is negatively regulated by circ-ACACA in NSCLC. (A) The
correlation between circ-ACACA and miR-1183 in NSCLC tissues was
analyzed using Pearson's correlation coefficient. (B) The level of
miR-1183 in NSCLC cells transfected with miR-1183 or miR-NC was
determined by RT-qPCR. (C) The putative binding sites between
circ-ACACA and miR-1183 were predicted by circular RNA Interactome.
The dual-luciferase reporter assay was used to check the luciferase
activity of (D) A549 and (E) H1299 cells cotransfected with the
miR-1183 and WT-circ-ACACA or MUT-circ-ACACA. The RIP assay was
conducted in (F) A549 and (G) H1299 cells using Anti-Ago2 to
investigate the relationship between circ-ACACA and miR-1183 and
Anti-IgG was used as the control. RNA pull-down was performed in
(H) A549 and (I) H1299 cells and the relative enrichment of
circ-ACACA in samples was detected by RT-qPCR. miR-1183 is a target
of circ-ACACA and is negatively regulated by circ-ACACA in NSCLC.
The level of miR-1183 in (J) A549 and (K) H1299 cells transfected
with si-circ-ACACA and si-NC was measured by RT-qPCR.
*P<0.05 vs. miR-NC, Anti-IgG, Bio-NC or si-NC. RT-q,
reverse transcription-quantitative; circ, circular; NSCLC,
non-small cell lung cancer; si, small interfering; NC, negative
control; WT, wild-type; MUT, mutant; miR, microRNA; RIP, RNA
immunoprecipitation; IgG, immunoglobulin. |
Downregulation of miR-1183 reverses
circ-ACACA silencing-mediated effects on proliferation, migration
and glycolysis rate
To further dissect the impact of the interaction
between circ-ACACA and miR-1183 on NSCLC progression, the level of
miR-1183 in NSCLC cells infected with si-circ-ACACA or
si-circ-ACACA + anti-miR-1183 was first measured, as well as the
corresponding controls. The result showed that miR-1183 was
significantly upregulated in the si-circ-ACACA group, while its
expression was significantly decreased following infection with
anti-miR-1183 (Fig. 5A and B).
The CCK-8 assay indicated that downregulation of miR-1183 inverted
the circ-ACACA silencing-mediated inhibitory effect on
proliferation of NSCLC cells (Fig. 5C
and D). The Transwell assay revealed that circ-ACACA
silencing-mediated the repressive impact on migration of NSCLC
cells and was reversed by the miR-1183 inhibitor (Fig. 5E and F). Similarly, the decreased
ECAR in the si-circ-ACACA group was reversed after infection with
anti-miR-1183 (Fig. 5G and H).
Meanwhile, the declined protein levels of c-myc, MMP9, GLUT-1 in
si-circ-ACACA group were reversed by the miR-1183 inhibitor
(Fig. 5I and J). From these
results, it could be concluded that circ-ACACA mediated the
progression of NSCLC by interacting with miR-1183 in
vitro.
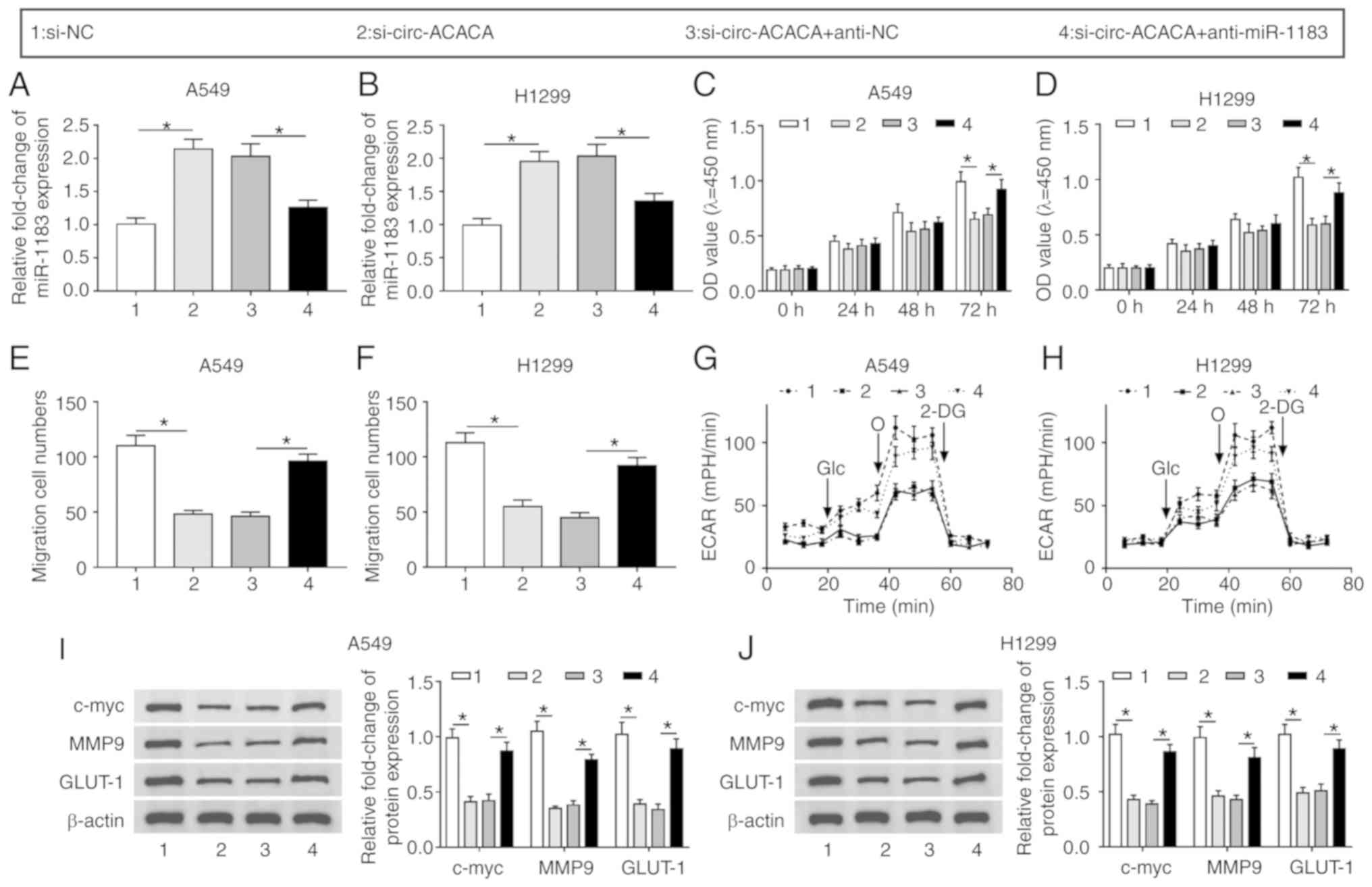 | Figure 5circ-ACACA/miR-1183 axis regulates
the progression of NSCLC. The level of miR-1183 in (A) A549 and (B)
H1299 cells infected with si-circ-ACACA or si-circ-ACACA +
anti-miR-1183, as well as matched controls was checked by reverse
transcription-quantitative PCR. Cell Counting Kit-8 assay was
employed to measure proliferation of infected in (C) A549 and (D)
H1299 cells. The ability of migration of infected (E) A549 and (F)
H1299 cells was evaluated by Transwell assay. The glycolysis rate
was detected by Seahorse XFe96 analyzer in (G) A549 and (H) H1299
cells. The protein levels of c-myc, MMP9 and GLUT-1 in infected (I)
A549 and (J) H1299 cells were checked by western blotting.
*P<0.05. circ, circular; NSCLC, non-small cell lung
cancer; si, small interfering; NC, negative control; MMP, matrix
metalloproteinases; GLUT, glucose transporter; c-myc,
cellular-myelocytomatosis; OD, optical density; ECAR, extracellular
acidification rate; miR, microRNA; Glc, glucose; O, oligomycin;
2-DG, 2-deoxyglucose. |
circ-ACACA regulates the PI3K/PKB
signaling pathway by interacting with miR-1183 in NSCLC cells
To investigate whether circ-ACACA could affect the
PI3K/PKB pathway, the protein levels of PTEN, total PI3K (t-PI3K),
p-PI3K, total PKB (t-PKB) and p-PKB in NSCLC cells infected with
si-circ-ACACA, si-circ-ACACA + anti-miR-1183 or matched controls
were detected. The results indicated that PTEN was significantly
upregulated and the levels of p-PI3K/t-PI3K and p-PKB/t-PKB were
notably downregulated in the si-circ-ACACA group, whereas the
situation was reversed after infection with anti-miR-1183 (Fig. 6A and B). In summary, these results
demonstrated that the circ-ACACA/miR-1183 axis regulated the
PI3K/PKB pathway in NSCLC.
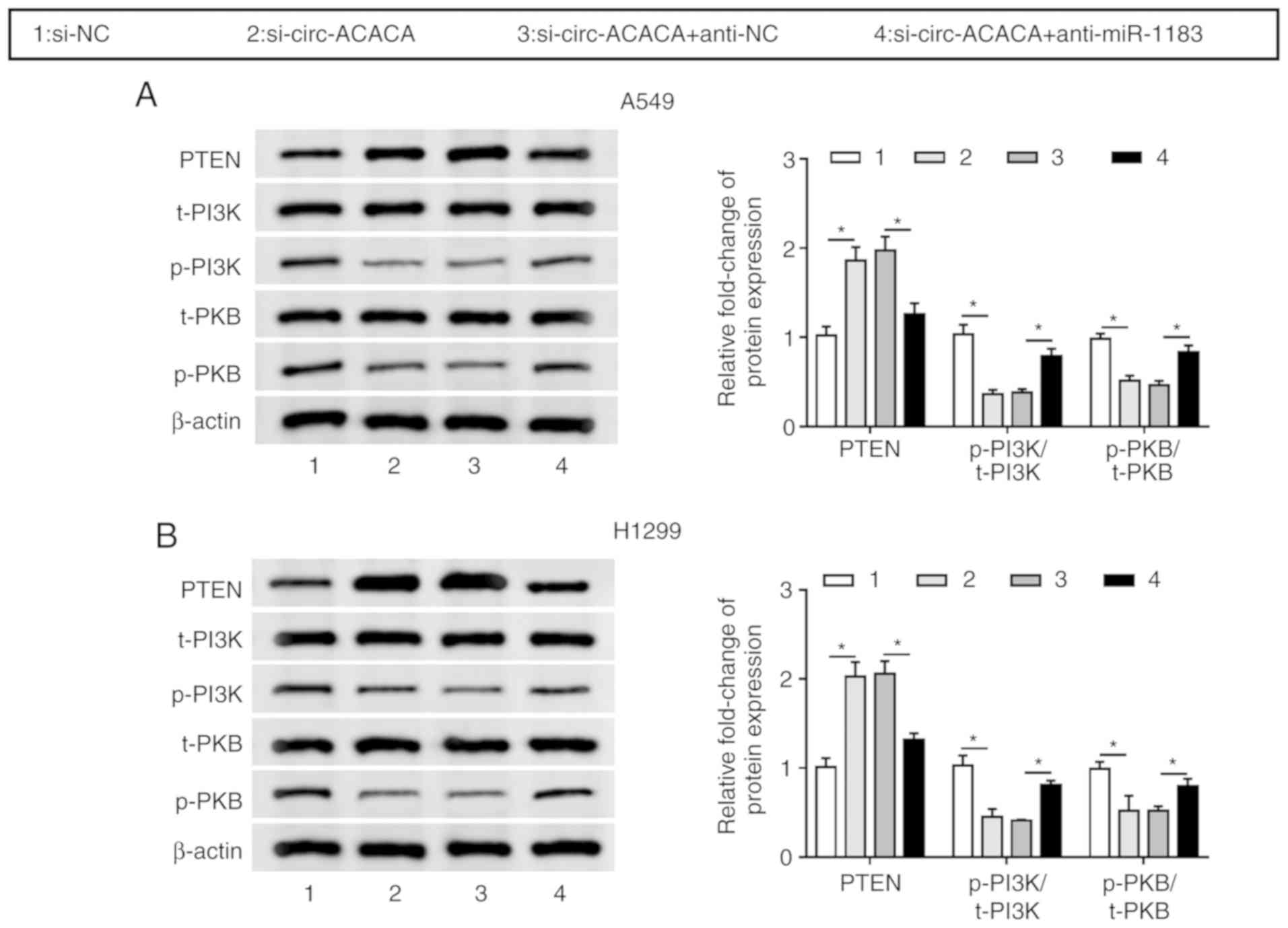 | Figure 6circ-ACACA/miR-1183 axis regulates
PI3K/PKB pathway. The protein levels of PTEN, t-PI3K, p-PI3K, t-PKB
and p-PKB in (A) A549 and (B) H1299 cells transfected with
si-circ-ACACA or si-circ-ACACA + anti-miR-1183, as well as matched
controls were checked by western blotting. *P<0.05.
circ, circular; si, small interfering; NC, negative control; miR,
microRNA; t-PI3K, total-phosphoinositide 3 kinase; p-PKB,
phosphorylated-protein kinase B. |
circ-ACACA silencing inhibits tumor
growth in vivo
To verify the function of circ-ACACA in NSCLC cells
in vivo, the xenograft mouse model was established using
A549 cells transfected with sh-circ-ACACA or sh-NC. The data showed
that knockdown of circ-ACACA led to an obvious shrink in tumor
volume (Fig. 7A) and decline in
tumor weight (Fig. 7B). Also, the
level of circ-ACACA was significantly decreased in the
sh-circ-ACACA group (Fig. 7C),
the opposite effect to the expression of miR-1183 (Fig. 7D). In addition, knockdown of
circ-ACACA reduced the protein levels of c-myc, MMP9 and GLUT-1 in
tumors (Fig. 7E). Similarly, the
protein level of PTEN was conspicuously elevated and the protein
levels of p-PI3K/t-PI3K and p-PKB/t-PKB were clearly declined in
sh-circ-ACACA group (Fig. 7F).
Taken together, these results suggested that downregulation of
circ-ACACA suppressed NSCLC progression in vivo.
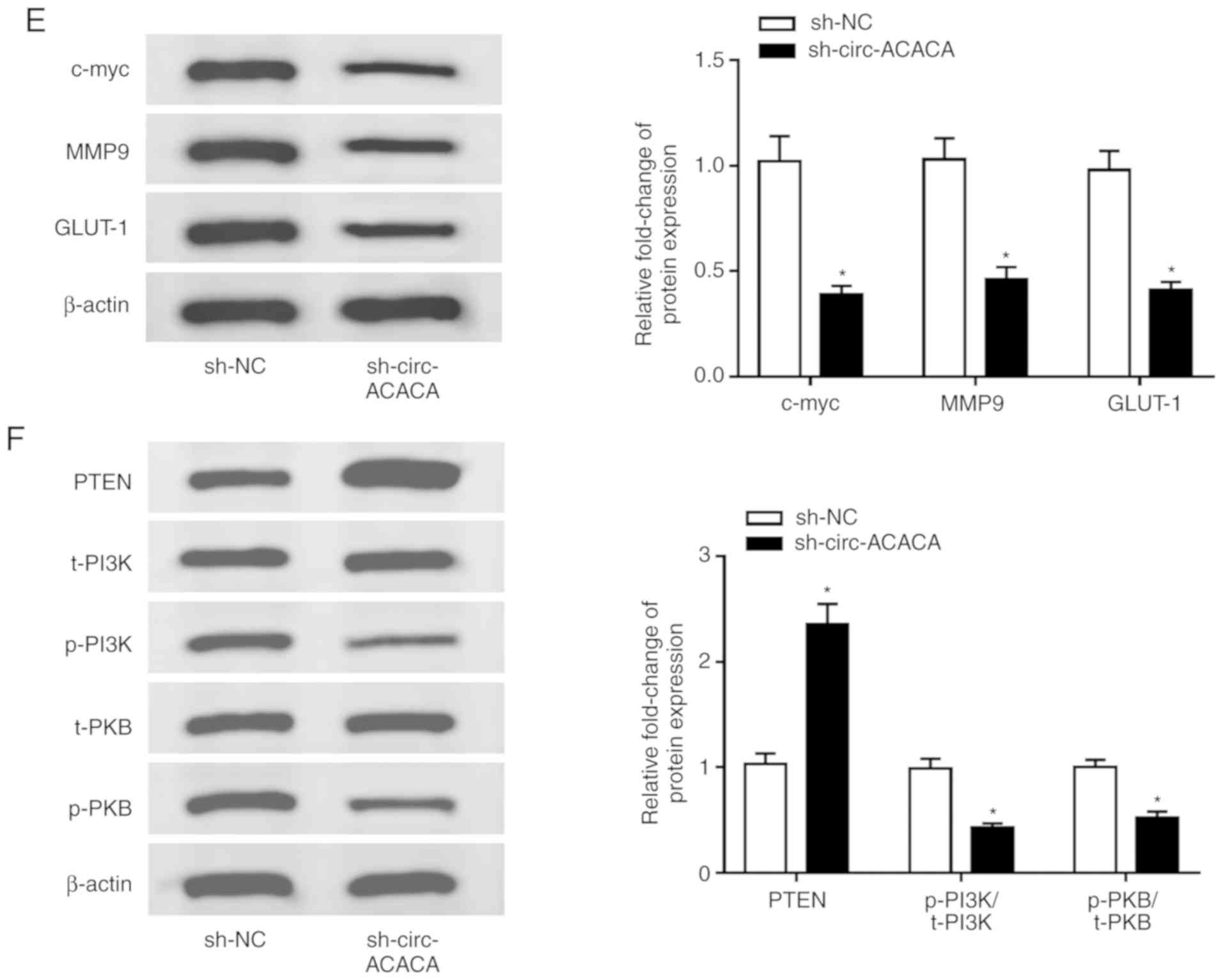 | Figure 7Downregulation of circ-ACACA
represses tumor growth. (A) The tumor volume in two groups (n=5)
was measured every 7 days. (B) Weight of the resected tumor was
examined after the mice were killed. (C) The level of circ-ACACA in
NSCLC cells infected with sh-circ-ACACA or sh-NC was measured by
RT-qPCR. (D) The level of miR-1183 in NSCLC cells infected with
sh-circ-ACACA or sh-NC was measured by RT-qPCR. (E) The protein
levels of c-myc, MMP9 and GLUT-1 in tumors were determined by
western blotting. (F) The protein levels of PTEN, t-PI3K, p-PI3K,
t-PKB and p-PKB in infected NSCLC cells were measured by western
blotting. *P<0.05 vs. sh-NC. circ, circular; si,
small interfering; NC, negative control; miR, microRNA; t-PI3K,
total-phosphoinositide 3 kinase; p-PKB, phosphorylated-protein
kinase B; RT-q, reverse transcription-quantitative; NSCLC,
non-small cell lung cancer; PTEN, phosphatase and tensin homolog;
MMP, matrix metalloproteinases; GLUT, glucose transporter; c-myc,
cellular-myelocytomatosis. |
Discussion
NSCLC accounts for ~85% of all lung cancers and the
five-year survival rate can be very low due to metastasis and
drug-resistance (2,3). Therefore, it is essential to find
new molecular targets and investigate potential mechanisms.
Recently, circRNAs have been verified to regulate the progression
of numerous cancers. Liu et al (7) found that circular RNA YAP1 inhibited
gastric cancer progression via modulating miR-367-5p. Lu et
al (8) reported that
circSLC8A1 suppressed bladder cancer progression via regulating
PTEN. Chen et al (9) found
that circRNA 100146 acted as an oncogene in NSCLC. To explore the
function of circ-ACACA in NSCLC, its expression level was checked
and it was found that circ-ACACA was upregulated in NSCLC tissues
and cells and contributed to poor prognosis. Also, ROC curve
analysis indicated that circ-ACACA could be a biomarker for NSCLC.
Further analysis showed that downregulation of circ-ACACA hindered
proliferation and migration of NSCLC cells. Alteration of energy
metabolism, especially abnormal activation of the glycolysis
pathway was observed in diverse human cancers (26,27). circRNAs are reported to be
involved in the regulation of the Warburg effect in human cancers
(28,29). Hence, the level of ECAR were
checked in NSCLC cells and it was found that circ-ACACA silencing
reduced the glycolysis rate. In addition, the protein levels of
c-myc, MMP9 and GLUT-1 also declined in NSCLC cells infected with
si-circ-ACACA. Furthermore, in vivo experiments showed that
knockdown of circ-ACACA repressed tumor growth. All in all, these
results suggested that circ-ACACA might function as an oncogene and
could be a potential therapeutic target in NSCLC.
Growing evidence has clarified the fact that
circRNAs could serve as the sponges of miRNAs to function in
numerous cancers (30,31) and the present research showed that
circ-ACACA was mainly expressed in the cytoplasm in NSCLC cells. In
this study, miR-1183 was forecasted to be a target of circ-ACACA
and this interaction was confirmed. A previous report showed that
miR-1183 was involved in the regulation of NSCLC progression
(18). In this study, the level
of miR-1183 was decreased in NSCLC tissues and cells. Moreover, the
miR-1183 inhibitor repressed proliferation and migration of NSCLC
cells and reduced the glycolysis rate. In-depth studies illustrated
that the repressive impact of circ-ACACA silencing-mediated NSCLC
progression was reversed by downregulating miR-1183. A previous
report indicated that the PI3K/PKB signaling pathway participated
in the regulation of NSCLC (32).
To investigate whether circ-ACACA affected this pathway, the
expression of related proteins in NSCLC cells infected with
si-circ-ACACA, si-circ-ACACA + anti-miR-ACACA or corresponding
controls was checked. The results indicated circ-ACACA silencing
elevated the expression level of PTEN, a major antagonist of PI3K
activity and also decreased the levels of p-PI3K/t-PI3K and
p-PKB/t-PKB, while downregulation of miR-1183 reversed the effect.
These data indicated that the silencing of circ-ACACA inactivating
the PI3K/PKB pathway, whereas this effect was abolished by the
miR-1183 inhibitor. Taken together, these results suggested that
circ-ACACA mediated the proliferation, migration and glycol-ysis
via sponging miR-1183 and the circ-ACACA/miR-1183 axis regulated
the PI3K/PKB pathway.
In conclusion, the current research demonstrated
that circ-ACACA was upregulated in NSCLC tissues and cells. Also,
downregulation of circ-ACACA restrained NSCLC progression via
interacting with miR-1183 and inactivating the PI3K/PKB pathway.
This novel mechanism may provide a theoretical basis for research
into circRNA-directed treatment in NSCLC.
Acknowledgement
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WW and XY designed the study and drafted the paper.
WX and HL performed experiments. MY analyzed the data. All authors
were involved in interpreting the results and reviewing the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Every patient signed the informed consent and the
official approval from the Ethics Committee of Liaocheng People's
Hospital was obtained in this study. The animal experiment was
approved by the Animal Care and Use Committee of Liaocheng People's
Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu G, Pei F, Yang F, Li L, Amin AD, Liu
S, Buchan JR and Cho WC: Role of autophagy and apoptosis in
non-small-cell lung cancer. Int J Mol Sci. 18:E3672017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cai J, Fang L, Huang Y, Li R, Xu X, Hu Z,
Zhang L, Yang Y, Zhu X, Zhang H, et al: Simultaneous overactivation
of Wnt/β-catenin and TGFβ signalling by miR-128-3p confers
chemoresistance-associated metastasis in NSCLC. Nat Commun.
8:158702017. View Article : Google Scholar
|
|
3
|
Cheng H and Perez-Soler R: Leptomeningeal
metastases in non-small-cell lung cancer. Lancet Oncol. 19:e43–e55.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
6
|
Ju HQ, Zhao Q, Wang F, Lan P, Wang Z, Zuo
ZX, Wu QN, Fan XJ, Mo HY, Chen L, et al: A circRNA signature
predicts postoperative recurrence in stage II/III colon cancer.
EMBO Mol Med. 11:e101682019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu H, Liu Y, Bian Z, Zhang J, Zhang R,
Chen X, Huang Y, Wang Y and Zhu J: Circular RNA YAP1 inhibits the
proliferation and invasion of gastric cancer cells by regulating
the miR-367-5p/p27 kip1 axis. Mol Cancer. 17:1512018. View Article : Google Scholar :
|
|
8
|
Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen
W, Jiang B, Qin H, Guo X, Liu M, et al: Circular RNA circSLC8A1
acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer
progression via regulating PTEN. Mol Cancer. 18:1112019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen L, Nan A, Zhang N, Jia Y, Li X, Ling
Y, Dai J, Zhang S, Yang Q, Yi Y and Jiang Y: Circular RNA 100146
functions as an oncogene through direct binding to miR-361-3p and
miR-615-5p in non-small cell lung cancer. Mol Cancer. 18:132019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu K, Liao X, Gong Y, He J, Zhou JK, Tan
S, Pu W, Huang C, Wei YQ and Peng Y: Circular RNA F-circSR derived
from SLC34A2-ROS1 fusion gene promotes cell migration in non-small
cell lung cancer. Mol Cancer. 18:982019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang L, Wang J, Fan Y, Yu K, Jiao B and Su
X: Hsa_circ_0046264 up-regulated BRCA2 to suppress lung cancer
through targeting hsa-miR-1245. Respir Res. 19:1152018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar :
|
|
13
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan W, Liu B, Qu S, Liang G, Luo W and
Gong C: MicroRNAs and cancer: Key paradigms in molecular therapy.
Oncol Lett. 15:2735–2742. 2018.PubMed/NCBI
|
|
15
|
Antônio LGL, Freitas-Lima P,
Pereira-da-Silva G, Assirati JA Jr, Matias CM, Cirino MLA,
Tirapelli LF, Velasco TR, Sakamoto AC, Carlotti CG Jr and Tirapelli
DPDC: Expression of MicroRNAs miR-145, miR-181c, miR-199a and
miR-1183 in the blood and hippocampus of patients with mesial
temporal lobe epilepsy. J Mol Neurosci. 69:580–587. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng X, Ander BP, Jickling GC, Zhan X,
Hull H, Sharp FR and Stamova B: MicroRNA and their target mRNAs
change expression in whole blood of patients after intracerebral
hemorrhage. J Cereb Blood Flow Metab. April 9–2019.Epub ahead of
print.
|
|
17
|
Prahm KP, Høgdall C, Karlsen MA,
Christensen IJ, Novotny GW and Høgdall E: Identification and
validation of potential prognostic and predictive miRNAs of
epithelial ovarian cancer. PLoS One. 13:e02073192018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou Y, Zheng X, Xu B, Chen L, Wang Q,
Deng H and Jiang J: Circular RNA hsa_circ_0004015 regulates the
proliferation, invasion, and TKI drug resistance of non-small cell
lung cancer by miR-1183/PDPK1 signaling pathway. Biochem Biophys
Res Commun. 508:527–535. 2019. View Article : Google Scholar
|
|
19
|
Nicholson KM and Anderson NG: The protein
kinase B/Akt signalling pathway in human malignancy. Cell Signal.
14:381–395. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mahajan K and Mahajan NP: PI3K-independent
AKT activation in cancers: A treasure trove for novel therapeutics.
J Cell Physiol. 227:3178–3184. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dong P, Konno Y, Watari H, Hosaka M,
Noguchi M and Sakuragi N: The impact of microRNA-mediated PI3K/AKT
signaling on epithelial-mesenchymal transition and cancer stemness
in endometrial cancer. J Transl Med. 12:2312014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen K, Abuduwufuer A, Zhang H, Luo L,
Suotesiyali M and Zou Y: SNHG7 mediates cisplatin-resistance in
non-small cell lung cancer by activating PI3K/AKT pathway. Eur Rev
Med Pharmacol Sci. 23:6935–6943. 2019.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar :
|
|
25
|
Zhang X, Zhuang J, Liu L, He Z, Liu C, Ma
X, Li J, Ding X and Sun C: Integrative transcriptome data mining
for identification of core lncRNAs in breast cancer. PeerJ.
7:e78212019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hua Q, Jin M, Mi B, Xu F, Li T, Zhao L,
Liu J and Huang G: LINC01123, a c-Myc-activated long non-coding
RNA, promotes proliferation and aerobic glycolysis of non-small
cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol.
12:912019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang J, Ren B, Yang G, Wang H, Chen G, You
L, Zhang T and Zhao Y: The enhancement of glycolysis regulates
pancreatic cancer metastasis. Cell Mol Life Sci. 77:305–321. 2020.
View Article : Google Scholar
|
|
28
|
Li Q, Pan X, Zhu D, Deng Z, Jiang R and
Wang X: Circular RNA MAT2B promotes glycolysis and malignancy of
hepatocellular carcinoma through the miR-338-3p/PKM2 axis under
hypoxic stress. Hepatology. 70:1298–1316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang J, Cong X, Ren M, Sun H, Liu T, Chen
G, Wang Q, Li Z, Yu S and Yang Q: Circular RNA hsa_circRNA_0007334
is predicted to promote MMP7 and COL1A1 expression by functioning
as a miRNA sponge in pancreatic ductal adenocarcinoma. J Oncol.
2019:76308942019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pérez-Ramírez C, Cañadas-Garre M, Molina
M, Faus-Dáder MJ and Calleja-Hernández M: PTEN and PI3K/AKT in
non-small-cell lung cancer. Pharmacogenomics. 16:1843–1862. 2015.
View Article : Google Scholar : PubMed/NCBI
|