Introduction
Patients who have abnormal pulmonary immune
responses, such as those with atopic asthma or cystic fibrosis
(CF), are prone to fungal colonization of the respiratory tract
(1). The ubiquitous environmental
mold Aspergillus fumigatus (Af) is the most common
cause (2). A clinical study in
America found that ~80% of children with CF also have IgG
antibodies against Asp f 1, an immunodominant Aspergillus
peptide antigen (3). Af
antigen exposure following persistent fungal colonization of the
lungs produces allergic bronchopulmonary aspergillosis (ABPA).
There is a high prevalence (28%) of Aspergillus
hypersensitivity and ABPA in patients with bronchial asthma,
worldwide from a meta-analysis of observational studies between
1965 and 2008 (4). The
pathogenesis of ABPA is not well understood; however, it is known
that patients with ABPA have immunoglobulin (Ig)E, IgA, and IgG
anti-Af serum antibodies (5).
The pulmonary immune response in patients with ABPA
includes a higher than normal T helper 2 (Th2) response, in
addition to elevated levels of IgE targeting the colonizing fungus
(6). In human bronchial
epithelium, Af exposure-triggered promotion of Th2 response
is associated with inhibition of interferon-β signaling through the
JAK-STAT1 signaling pathway, which shifts epithelial responses from
type Th1 to type Th2 (7,8), as well as, activation of
protease-activated receptor-2 and tyrosine-protein phosphate
nonreceptor type 11, which reduces CXCL10 expression, further
favoring induction of a Th2 response (9). In addition, Af has been
reported to promote Th2 responses through thymic stromal
lymphopoietin production by human corneal epithelial cells
(10). Sera from patients with
ABPA show increased IgE reactivity to Asp f 2 and Af crude
extract; and it has been hypothesized that the Af antigens,
Asp f 1 or Asp f 2, may underlie upregulation of Th2 (11). However, it has been reported that
an ABPA-associated Th2 response can be triggered in the absence of
specific Aspergillus antigens (12). Thus, the mechanisms by which
Af induces Th2 responses remain unknown. In particular, it
is unclear whether the immunomodulatory effects of Af
antigens are associated with the development of ABPA.
Th2 immune responses can be produced by
differentiation of macrophages toward an M2 type (13). Induction of pro-inflammatory
responses in human macrophages with Af has been shown to
result in upregulation of tumor necrosis factor-α and interleukin
(IL)-6 (14). In addition,
Af produces a metabolite, gliotoxin, which downregulates
vitamin D receptor expression on macrophages and airway epithelial
cells, which has been shown to lead to increased production of the
Th2 cytokines IL-5 and IL-13 (15). Notably, the T2 ribo-nuclease
(RNASET2) protein was found to be a major inducer of Th2
polarization. ω-1, a glycosylated RNASET2 protein, which is
secreted by Schistosoma mansoni (Sm) eggs and is
abundant in soluble egg antigen extract, has been shown to
condition dendritic cells to prime Th2 responses (16,17). The glycosylation and ribonuclease
activity of Sm ω-1 are both essential to the conditioning of
dendritic cells for Th2 polarization (17). In addition, Schistosoma
japonicum (Sj) CP1412, which is a ribonuclease T2 family
member, has been shown to promote M2-type macrophage
differentiation (18) and
recombinant Sj CP1412 has been reported to increase
expression of CD206, arginase 1 (ARG1), and IL-10 in mouse
macrophages (18).
The aim of the present study was to investigate the
hypothesis that Af-induced Th2 responses, which are observed
in ABPA, may involve RNASET2-mediated M2 polarization. The
recombinant Af RNASET2 (rAf RNASET2) was expressed
and purified in a bacterial pET system. Th2 cytokine expression was
evaluated in vivo in mice immunized with rAf RNASET2.
M2-type macrophage differentiation was examined in RAW264.7
macrophages incubated with rAf RNASET2 to further
investigate whether Af RNASET2 may be an important immune
regulatory factor in ABPA.
Materials and methods
Expression system components and
reagents
The following reagents were purchased for
recombinant protein expression: Escherichia coli (E.
coli) BL21 (DE3) plysS cells (Merck KGaA), pMD 19-T vectors
(Takara Biotechnology Co., Ltd.), pET-His vectors (Wuhan Miaoling
Bioscience & Technology Co., Ltd.), and primer STAR HS DNA
polymerase (Takara Biotechnology Co., Ltd.). Af RNASET2
cDNAs were synthesized by Nanjing GenScript Biotech Corp. Lysozyme
(Sangon Biotech Co., Ltd.) and blot membranes (nitrocellulose and
polyvinylidene fluoride; Merck KGaA) were used for pre-purification
cell lysis and electrophoresis analysis, respectively.
Mouse model
A total of 18 female BALB/c mice (6 weeks of age;
20-22 g) were purchased from Guangdong Medical Laboratory Animal
Center, and housed in a specific pathogen free facility with six
mice per cage under a stable temperature (24±1°C) and humidity
(55±10%). Mice were kept in open polypropylene cages with clean
chip bedding under a 12-h light/dark cycle with free access to a
standard rodent diet. The animals were acclimatized to the
laboratory for at least 1 week prior to the start of the
experiments. The health status of experimental mice was monitored
twice daily and humane endpoints were used to determine if mice met
the criteria for euthanasia prior to the study end point, namely
weight loss exceeding 10-15% of the body weight, lethargy,
inability to stand, or anorexia. All surgical procedures were
performed under 1.5-2% isoflurane anesthesia. At the end of the
experiments, animals were humanely sacrificed using CO2
asphyxiation. The flow rate of CO2 displaced ~50% of the
chamber volume per minute for euthanasia. The gas flow was
maintained for at least 1 min following clinical death, and death
was evaluated monitoring the lack of movement or visible breathing
for 5 min. Both the breeder of the animals and the assessor of the
results are blinded. All studies involving mice were performed
according to protocols approved by the Animal Ethical and Welfare
Committee of the School of Medicine of Shenzhen University. All
efforts were made to minimize discomfort and suffering of the
animals.
Cell culture
The RAW264.7 murine macrophages were purchased from
Guangzhou Cellcook Biotechnology Co., Ltd., and cultured in
complete DMEM with 10% FBS and antibiotics (100 U/ml penicillin and
100 µg/ml streptomycin), at 37°C in a humidified incubator
with 5% CO2.
Homology analysis of RNASET2 genes
The analysis of the amino acid sequence alignment
was performed to investigate the sequence conservation of Af
RNASET2 gene in the ribonuclease T2 family. GenBank (https://www.ncbi.nlm.nih.gov/genbank)
was used to download the amino acid sequences of Af RNASET2
[GenBank ID, XP754496.2; based on the Af AF293 genome
(Genome ID, 18)], Sj CP1412 (GenBank ID, AY570741.2),
Sm ω-1 (GenBank ID, DQ013207.1), and Clonorchis
sinensis (Cs) RNASET2 (GenBank ID, GAA50115.1) were
imported into DNAMAN software (version 8.0; Lynnon Biosoft) for
alignment. The sequences were saved in FASTA format for multiple
alignment analysis. The homologous alignment was
calculated according to the ratio of the number of conserved
amino acids to total number of amino acids.
Recombinant protein expression and
purification
The Af RNASET2 cDNA was synthesized following
codon optimization and submitted to the National Center for
Biotechnology Information (NCBI) database. The Af RNASET2
gene was amplified using PCR with Primer STAR HS DNA polymerase and
the aforementioned cDNA template. The following PCR primers were
used: Forward, 5′-AGA TGG ATC CAT GAA ATT CAA CAT AAC TAT CGC-3′
and reverse 5′-AGC GAT AAG CTT ATG TAC ATG TTA ATT CTT TTT CA-3′;
and the following thermocycling conditions: Initial denaturation at
94°C for 30 sec, and 30 cycles of 55°C for 30 sec, and 72°C for 50
sec. The PCR product, confirmed by Sanger DNA sequencing (GenScript
Biotech Corporation), was subcloned into a pET-His vector with
BamHI and HindIII restriction enzymes. The
recombinant expression vector pET-His-Af RNASET2 was
confirmed by double enzyme digestion with
BamHI/HindIII. Following double enzyme digestion with
BamHI/HindIII for 2 h at 37°C, the products were
analyzed using a 1% agarose gel.
The recombinant plasmid pET-His-Af RNASET2
was then transformed into E. coli BL21 (DE3) plysS cells.
The transformed E. coli were grown overnight in
Luria-Bertani medium containing 100 mg/l ampicillin at 37°C, and
induction was performed using isopropyl-β-D-thiogalactopyranoside
(IPTG) to a final concentration of 0.5 mM. Following cultivation
and an additional 3 h at 37°C, E. coli cells were harvested
by centrifugation at 9,600 x g for 5 min at 4°C. Following mixing
with protein extraction buffer (20 mM Tris HCl, 150 mM NaCl and 1
mg/ml lysozyme), harvested cells were sonicated (50 kHz; 3 sec
bursts, 5 sec inter-burst interval for a total of 20 min) in an ice
bath, and then centrifuged at 9,600 x g for 20 min at 4°C.
Subsequently, 10% SDS-PAGE was performed to analyze the status of
recombinant protein expression. The recombinant protein in the
soluble fraction was purified with a Ni-NTA column (cat. no.
17040303) and gel filter (Hi Load Superdex 16/600; cat. no.
28-9893-33) (both from GE Healthcare). Specifically, rAf
RNASET2 was expressed in the inclusion bodies, which were dissolved
in 6 M urea with 25 mM β-mercaptoethanol. Denatured proteins were
refolded and diluted 10-fold with 0.5 M L-arginine (pH 8.0)
(19). Arginine buffer was added
to the denatured protein solution slowly (flow rate of 0.5 ml/min)
and incubated overnight at 4°C. For renaturation, rAf
RNASET2 was dialyzed with PBS at 4°C for 24 h via a dialysis
membrane (Sangon Biotech Co., Ltd.) (19). Following renaturation, protein
concentrations were determined using the Bradford method (Sangon
Biotech Co., Ltd). The rAf RNASET2 protein endotoxin was
removed using an endotoxin removal kit following the manufacturer's
instructions and quantitation was performed using an endotoxin
assay kit, which revealed that the levels were reduced to <0.25
endotoxin units/ml (both Genscript Biotech Corporation).
Enzymatic activity analysis
Yeast RNA (Thermo Fisher Scientific, Inc.) was
dissolved in reaction buffer (50 mmol/l Tris HCl, 50 mmol/l NaCl,
pH 7.0) to prepare a stock solution (1 mg/ml). The rAf
RNASET2 (1 µg) aliquots were mixed thoroughly with 50
µg yeast RNA at 37°C for 20, 40, 60, 80, and 100 min. In
addition, a 30-min pretreatment with an RNase inhibitor (RNasin;
Thermo Fisher Scientific, Inc.) served as a positive control for
confirmation of digestion by RNASET2. Ribonuclease activity was
observed using the analysis of enzymolysis products in 1% agarose
electrophoresis gels, which was stained with Gelview dye (BioTeke
Corporation). Bands were visualized on a UV transilluminator
(Analytik Jena US LLC).
In vitro analysis of macrophage
stimulation
RAW264.7 macrophages (1×106) were
incubated with the indicated concentrations (20 or 40 µg/ml)
of rAf RNASET2 (pretreated with RNasin) in PBS at 37°C under 5%
CO2. After 24 h of co-culture, the cells and supernatant
were collected and centrifuged at 300-400 × g for 5 min at 4°C. The
cells were collected and incubated with the labeled antibodies
(FITC-CD16/32 (cat. no. 11-0161-81; 1:500; BioLegend, Inc.) or
FITC-CD206 (cat. no. MA5-16870; 1:500; Invitrogen; Thermo Fisher
Scientific, Inc.) for M2 or M1 macrophage detection, respectively,
and APC-F4/80 (cat. no. 17-4801-80; 1:500; BioLegend, Inc.) in PBS
containing 1% bovine serum albumin (Amresco LLC) for flow
cytometry. All incubations were performed at room temperature for
30 min in the dark and subsequently, the cells were washed with
PBS. Macrophage surface markers were detected using a Cytoflex flow
cytometer and analyzed using the CytExpert software (version 2.0)
(both Beckman Coulter, Inc.). IL-10 levels were measured in the
culture supernatant of RAW264.7 cells stimulated by incubation with
rAf RNASET2 (20 µg/ml) alone or in the presence of MAPK
signal inhibitors (U0126, 20 µM; SP600125, 20 µM and
SB203580, 1 µM) for 24 h. The cells were centrifuged at
300-400 x g for 5 min at 4°C and subsequently analyzed using ELISA
kits according to the manufacturer's instructions (cat. no.
HZ-IL-10-Mu; Shanghai and Shanghai Zhen Biotech Co., Ltd.).
RNA isolation and reverse transcription
quantitative PCR (RT-qPCR)
RAW264.7 macrophages were co-cultured with
rAf RNASET2 (20 µg/ml), inactive rAf RNASET2
(pretreated with RNasin (2 U/µl)), and PBS and LPS controls
at 37°C with 5% CO2 for 24 h, and with rAf
RNASET2 (20 µg/ml) alone or in the presence of MAPK signal
inhibitors (U0126, 20 µM; SP600125, 20 µM and
SB203580, 1 µM) for 24 h, under normal cell culture
conditions. RNA was extracted after the cells were centrifuged at
300-400 x g for 5 min at 4°C, using TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Total RNA was subsequently reverse transcribed into cDNA
using a TIANScript II RT kit (Tiangen Biotech, Co., Ltd.) according
to the manufacturer's protocol: 42°C for 50 min, followed by 95°C
for 5 min then cooled on the ice at 4°C for 10 min. RT-qPCR
analysis was performed with SYBRGreen (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with the primers listed in Table I.
 | Table ISequences of the primers used in for
reverse transcription-quantitative PCR. |
Table I
Sequences of the primers used in for
reverse transcription-quantitative PCR.
| Gene | Primer sequences
(5′→3′) | Accession
number |
|---|
| 18S
rRNA | Forward:
CGAGGGGTTCGGGATTTGTG | M35283.1 |
| Reverse:
AAAGCCAACCCGAGCGTC | |
| Arg-1 | Forward:
TGCGCCACATGAAAACCATC | NM_007482.3 |
| Reverse:
TTGGGAGGAGAAGGCGTTTG | |
| iNos | Forward:
GGTGAAGGGACTGAGCTGTTA | NM_007482.3 |
| Reverse:
TGAAGAGAAACTTCCAGGGGC | |
| IL-10 | Forward:
AAGGGTTACTTGGGTTGCCA | NM_010548.2 |
| Reverse:
CCTGGGGCATCACTTCTACC | |
| IL-13 | Forward:
CCTGGCTCTTGCTTGCCTT | NM_008355.3 |
| Reverse:
GGTCTTGTGTGATGTTGCTCA | |
| TGF-β | Forward:
GATCACCACAACCCACACCT | NM_009368.3 |
| Reverse:
AGGTTCGTGGACCCATTTCC | |
For the qPCR, 5 µl 2X SYBRGreen PCR buffer,
0.2 µl forward and reverse primers (both 10 µmol),
and 5 ng template were used to a total volume of 10 µl with
sterile water. The thermocycling conditions used were as follows:
Initial denaturation 95°C for 10 min, followed by 40 cycles of 95°C
for 15 sec and 60°C for 1 min. The real-time PCR was performed on a
qTOWER 2.2 system (Analytik Jena US LLC). All qPCRs were performed
in triplicate with 18S rRNA gene as a reference housekeeping
gene. Relative expression was calculated using the
2−ΔΔCq method (20).
Western blot analysis
RAW264.7 macrophages were co-cultured with
rAf RNASET2 (20 µg/ml), inactive rAf RNASET2
(pretreated with RNasin (2 U/µl), and PBS and LPS controls
at 37°C in a humidified incubator with 5% CO2 for 24 h.
The cells were centrifuged at 300-400 × g for 5 min at 4°C, and
subsequently lysed in RIPA buffer (Beyotime Insitute of
Biotechnology) with a protease-inhibitor cocktail (MedChemExpress
LLC). The protein concentration was determined using the Bradford
assay (Sangon Biotech Co., Ltd.) and the protein samples were
subsequently diluted to 1 mg/ml, then separated (20 µg/lane)
using 10 or 12% SDS-PAGE, following which the proteins were
transferred onto PVDF membranes. The membranes were blocked
overnight at 4°C with 5% skimmed milk in TBS +0.05% Tween-20. The
following primary antibodies were used: Phosphorylated (p)-ERK1/2
(cat. no. 4370; Cell Signaling Technology Inc.), ERK1/2 (cat. no.
46951; Cell Signaling Technology Inc.), p-p38 (cat. no. 4511; Cell
Signaling Technology Inc.), p-38 (cat. no. 8690; Cell Signaling
Technology Inc.) p-JNK (cat. no. 4668; Cell Signaling Technology
Inc.), JNK (cat. no. 9252; Cell Signaling Technology Inc.), IL-10
(cat. no. ab33471; Abcam), induced nitric oxide synthase (iNOS;
cat. no. ab213987; Abcam), ARG1 (cat. no. ab124917; Abcam), and
cyclooxygenase 2 (COX2; ab179800; Abcam) (all 1:1,000) were used.
The primary antibody for the loading control, GAPDH (cat. no.
sc-365062; Cell Signaling Technology, Inc.) was used at a dilution
of 1:2,000. After washing with PBS, the membranes were incubated
with horseradish peroxidase-conjugated secondary antibodies (cat.
nos. sc-2004 and sc-2005) (both 1:2,000 and Santa Cruz
Biotechnology, Inc.) for 1 h at room temperature. Proteins were
visualized using an enhanced chemiluminescence kit (Dalian Meilun
Biology Technology Co., Ltd.) according to the manufacturer's
protocol.
Effect of rAf RNASET2 on immune responses
in mice
Purified rAf RNASET2 was mixed with Imject
Aluminum adjuvant (Thermo Fisher Scientific, Inc.) to form a
water-in-oil adjuvant antigen. Mice were randomly divided into
immunized, adjuvant control, and PBS control groups (n=6 per
group), and injected intraperitoneally with 100 µg
rAf RNASET2 and 2 mg of aluminum adjuvant (Af RNASET2
+ adjuvant), 2 mg of aluminum adjuvant alone (adjuvant), or with
PBS only (control) once a week for 3 weeks. A total of 7 days
following the third immunization injection, about 50 µl
orbital venous blood samples were collected and the sera were
isolated following centrifugation at 3,000 x g for 20 min at 4°C.
The levels of IL-4, IL-13, IL-10, and interferon-γ (INF-γ) in mouse
sera were detected using cytokine ELISA kits (IL-4, cat. no.
HZ-IL-4-Mu; IL-10, cat. no. HZ-IL-10-Mu; IL-13, cat. no.
HZ-IL-13-Mu and INF-γ, cat. no. HZ-INF-γ-Mu) (all Shanghai and
Shanghai Zhen Biotech Co., Ltd.) according to the manufacturer's
instructions.
Statistical analysis
Quantitative data are presented as mean ± SDs.
Analyses were performed using Prism 7 software (GraphPad Software,
Inc.). Student's t-tests or one-way ANOVA followed by Dunnett's
post hoc test were performed, to detect statistical significance
between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Characteristics of Af RNASET2
The amino acid sequence of Af RNASET2
(GenBank XP_754496.2) was obtained based on the Af AF293
genome (Genome ID 18) in the NCBI database. The Af RNASET2
gene encodes 401 amino acids with a 15-amino-acid N-terminal
secretory signal; the mature RNASET2 protein is 382 amino acids
long. As shown in Fig. 1, a
homology comparison based on amino acid sequence alignment revealed
that Af RNASET2 was 11.84, 13.20, and 12.17% homologous with
Sj CP1412, Sm ω-1, and Cs RNASET2,
respectively. Notably, Af RNASET2 was predicted to contain
two conserved amino acid sequence (CAS) domains
(83WTIHGLWP89 and
140WEHEWNKHG148), namely CAS-1 and CAS-2, and
a conserved pair of cysteine residues (cysteine100 and
cysteine158) characteristic of T2 RNase-glycosylation
sites were also found.
 | Figure 1Amino acid sequence homology of
Af RNASET2, Sj CP1412, Sm ω-1, and Cs
RNASET2. Common amino acids are indicated by the grey shading.
Black solid lines indicate conserved functional domains, CAS-1 and
CAS-2. Solid black triangles indicate conserved cysteine residues.
Af, Aspergillus fumigatus; Sj, Schistosoma japonicum;
Sm, Schistosoma mansoni; Cs, Clonorchis
sinensis; CAS, conserved amino acid sequences; RNASET2, T2
ribonuclease. |
Expression and RNase activity of rAf
RNASET2
To obtain the rAf RNASET2 protein, cDNA
encoding mature Af RNASET2 (glutamicacid 26-arginine 407
region), without its signaling peptide (methionine 1-phenylalanine
25 domain) was synthesized, submitted to the NCBI (GenBank
MN593022), and subcloned to a pET-His prokaryotic expression vector
for expression and purification. cDNA from Af RNASET2 were
generated using PCR amplification and subcloned into pET-His
vectors using BamHI/HindIII restriction enzymes. The
band at 1,200 bp was consistent with the length of the Af
RNASET2 gene (Fig. 2A). The
pET-His-Af RNASET2 plasmid was confirmed using DNA
sequencing. Subsequently, the pET-His-Af RNASET2 plasmid was
transformed into E. coli BL21(DE3) plysS cells for
expression and purification. SDS-PAGE revealed that rAf
RNASET2 (~45 kDa) was expressed in the sediment following IPTG
induction (Fig. 2B; Lane 3). A
portion of the rAf RNASET2 protein sample was purified using
Ni-NTA-resin and renatured into soluble proteins by gradient
dialysis with urea (Fig. 2C).
Purified soluble rAf RNASET2 lysed yeast RNA in a
time-dependent manner and RNA digestion activity of rAf
RNASET2 was effective with ribonuclease A (Fig. 2D) and could be inhibited with the
ribonuclease inhibitor RNasin (Fig.
2E).
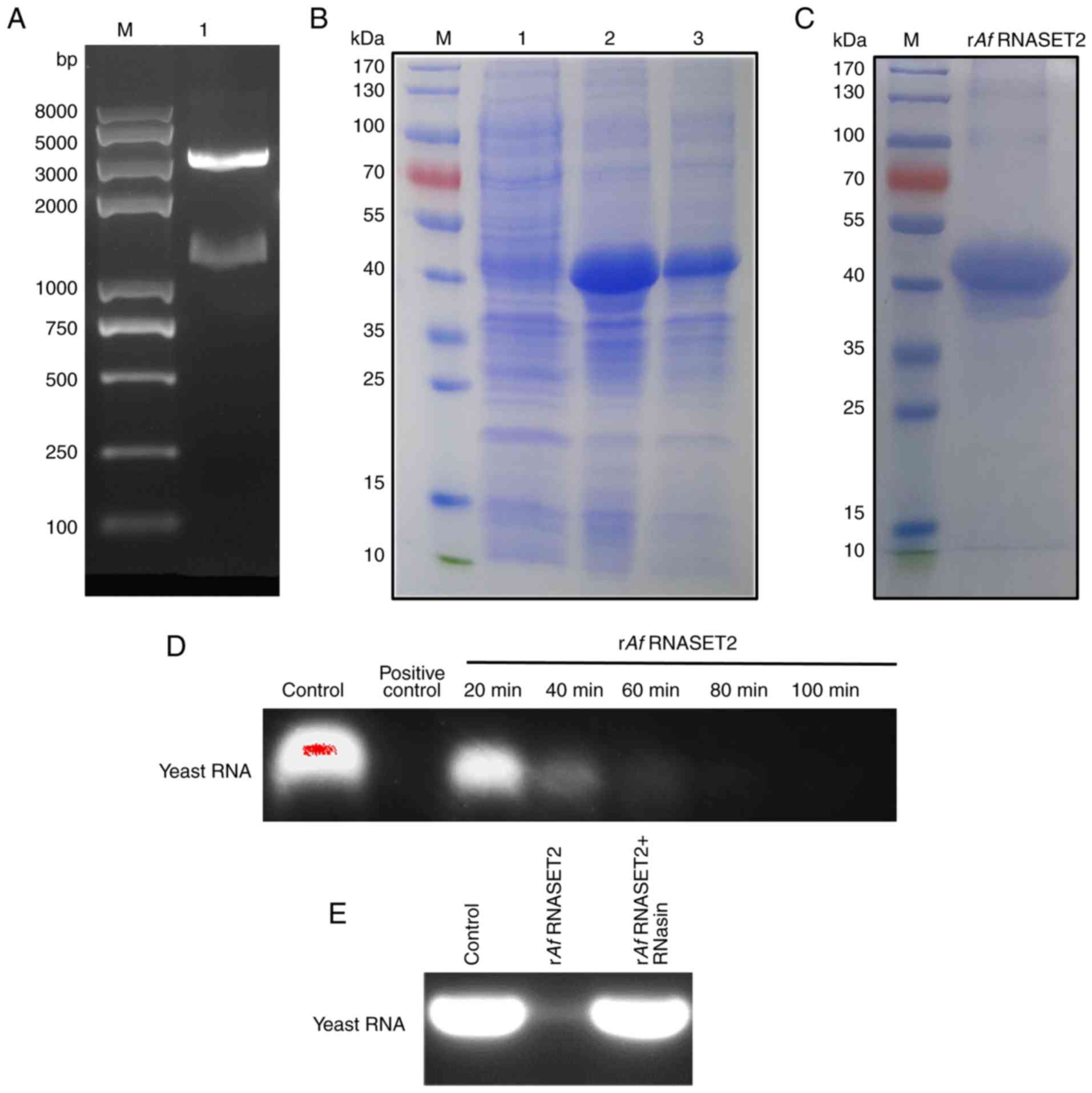 | Figure 2Expression, purification, and RNase
activity of rAf RNASET2. (A) Analysis of the prokaryotic
recombinant expression vector pET-His-Af RNASET2 (without
signal peptides, Glutamic acid 26-Arginine 407) by double enzyme
digestion. Lane M indicates the DNA marker. The 1,200 bp band
indicates the Af RNASET2 cDNA insert. (B) SDS-PAGE analysis
of pET-His-Af RNASET2 expression. Lane M, protein marker; lane 1,
E. coli transformed with pET-His-Af RNASET2 prior to IPTG
induction; lane 2, E. coli transformed with
pET-His-Af RNASET2 following IPTG induction; lane 3,
sediment from E. coli transformed with pET-His-Af
RNASET2 plasmid following IPTG induction, and ultrasonication. (C)
SDS-PAGE of renatured rAf RNASET2 protein following
purification using Ni-NTA affinity chromatography. Lane M indicates
the protein marker. (D) Analysis of RNase activity of rAf
RNASET2 (1 µg) with yeast RNA (50 µg) at 37°C at 20,
40, 60, 80 and 100 min). (E) Analysis of RNase activity of
rAf RNASET2 with or without RNasin (RNAse inhibitor)
pretreatment. RNase enzymolysis products were analyzed using 1%
agarose gel electrophoresis. E. coli, Escherichia coli; r,
recombinant; Af, Aspergillus fumigatus; RNASET2, T2
ribonuclease; IPTG, isopropyl-β-d-thiogalactopyranoside. |
rAf RNASET2 promotes the Th2 response in
vivo
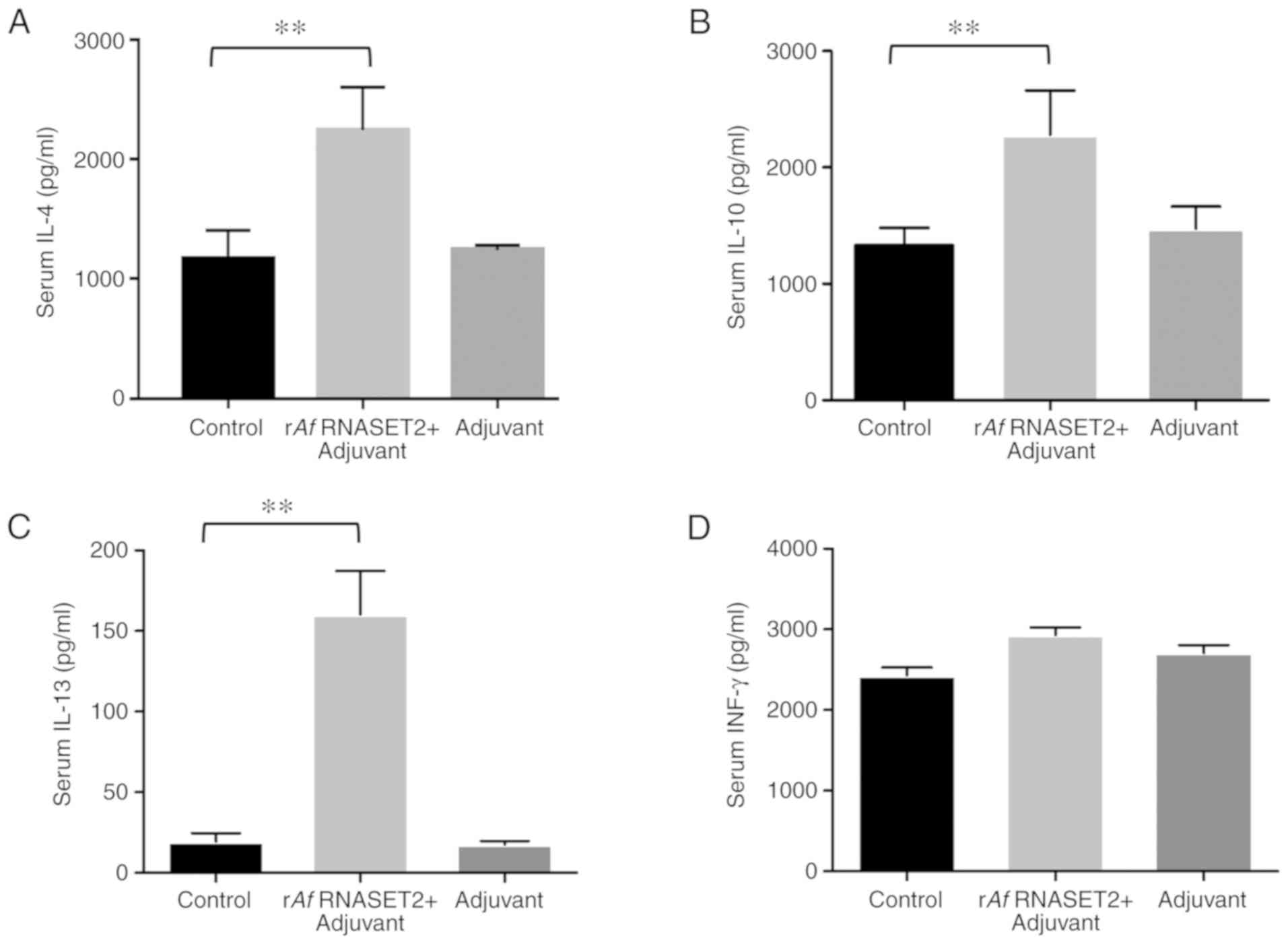
The levels of the Th2 response-related cytokines
IL-4 (P>0.01; Fig. 3A), IL-10
(P<0.01; Fig. 3B), and IL-13
(P<0.01; Fig. 3C) were
significantly increased in rAf RNASET2-immunized mouse sera,
collected 7 days following the third immunization compared with
that in PBS control groups. In addition, there was no change in the
expression of the Th1 response-related cytokine, INF-γ, between
Af RNASET2+ adjuvant and the control group (P>0.05;
Fig. 3D). This indicates that
rAf RNASET2 enhanced Th2 responses in vivo.
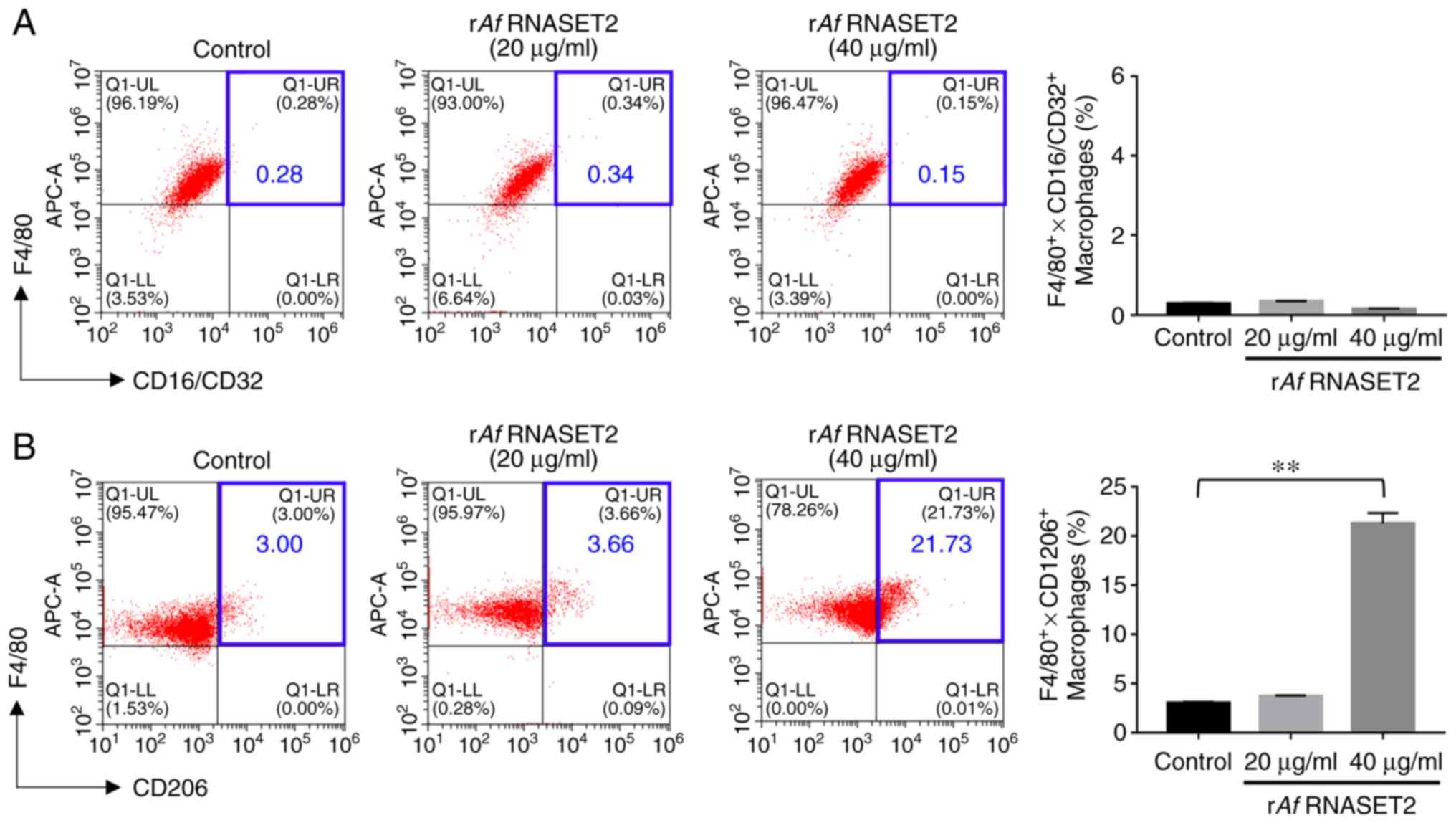
rAf RNASET2 promotes M2 macrophage
polarization
The flow cytometry results revealed that in
vitro stimulation of RAW264.7 macrophages with purified
rAf RNASET2 significantly increased the expression level of
the M2 surface marker CD206 (Fig.
4B), Notably, CD206 expression was increased from 3.00 to
21.73% following treatment with 40 µg/ml rAf RNASET2
(P<0.01). However, there was no significant change in the level
of the M1 surface marker CD16/32 (P<0.01; Fig. 4A).
The RT-qPCR results revealed that RAW264.7 cells
treated with rAf RNASET2 for 24 h had significantly
increased expression levels of M2-related genes, including
Arg1 (P<0.01; Fig. 5A),
Il-10 (P<0.01; Fig.
5C), and Il-13 (P<0.01; Fig. 5D), without a significant change in
the expression level of the M2 marker iNOS (P>0.05; Fig. 5B). Western blot analysis revealed
marked increases in ARG1 and IL-10 protein levels, with no changes
in iNOS and M1-related gene COX2 levels, in RAW264.7 cells treated
with rAf RNASET2 (Fig.
5E). LPS upregulated the protein expression level of COX2 in
RAW264.7 cells. An ELISA also revealed that the levels of IL-10 in
the culture supernatant of RAW264.7 cells stimulated by rAf
RNASET2 were also increased (Fig.
6B). These results indicate that rAf RNASET2 promoted M2
macrophage polarization in RAW264.7 cells.
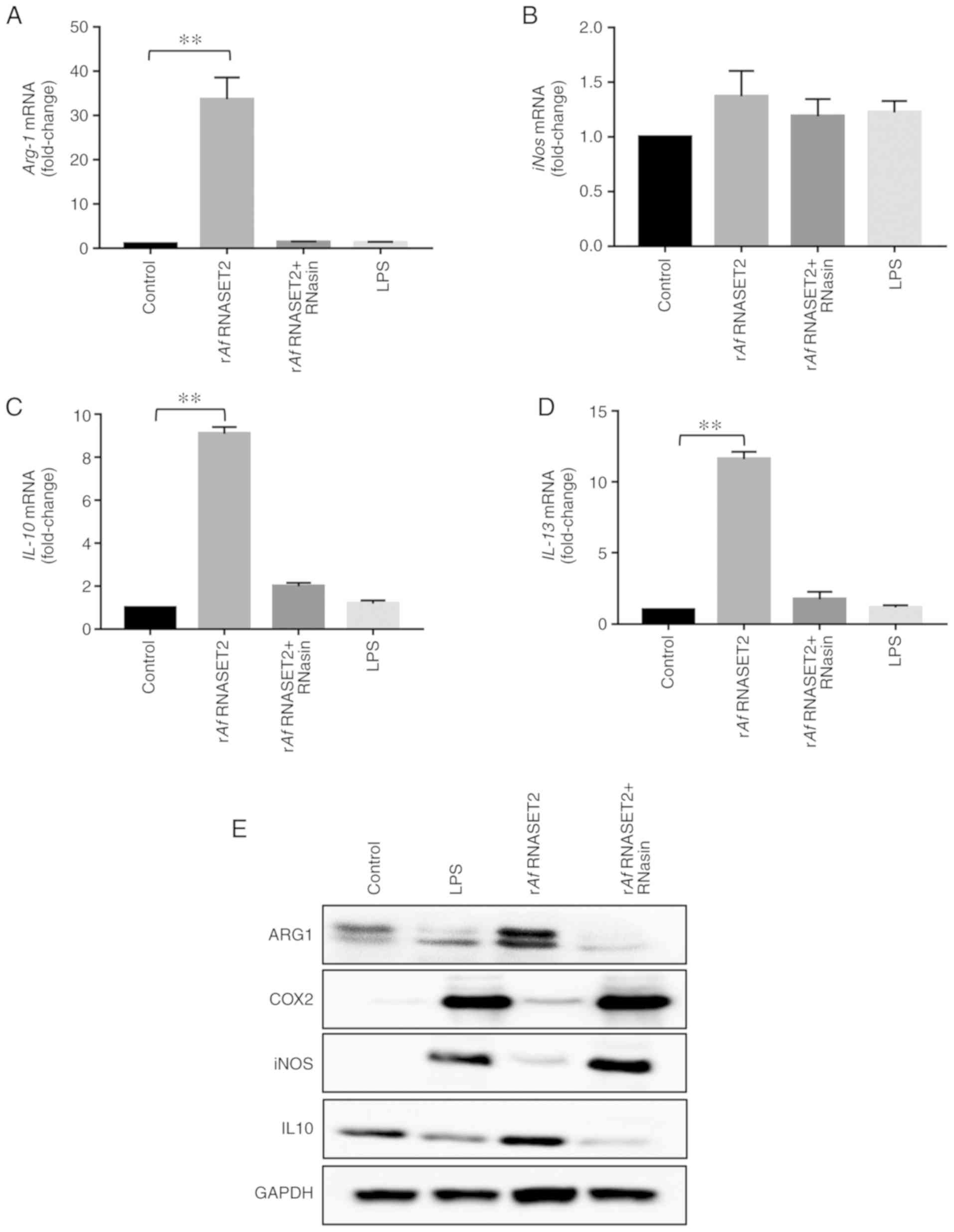 | Figure 5rAf RNASET2 increases
expression of M2 polarization markers. Reverse
transcription-quantitative PCR and western blot analysis were
conducted with RAW264.7 macrophages co-cultured with rAf
RNASET2 (20 µg/ml), inactive rAf RNASET2 (pretreated
with RNasin), and PBS and LPS controls at 37°C with 5%
CO2 for 24 h. mRNA expression levels of (A)
Arg-1, (B) iNos, (C) IL-10, and (D)
IL-13. Data are presented as mean ± SD from 3 independent
experiments. **P<0.01. (E) Western blot
analysis of ARG-1, COX2, iNOS, IL-10, and GAPDH in RAW264.7
macrophages following rAf RNASET2 (20 µg/ml)
treatment for 24 h. r, recombinant; Af, Aspergillus
fumigatus; RNASET2, T2 ribonuclease; LPS, lipopolysaccharide;
iNOS, induced nitric oxide synthase; IL, interleukin; Arg-1,
arginase-1. |
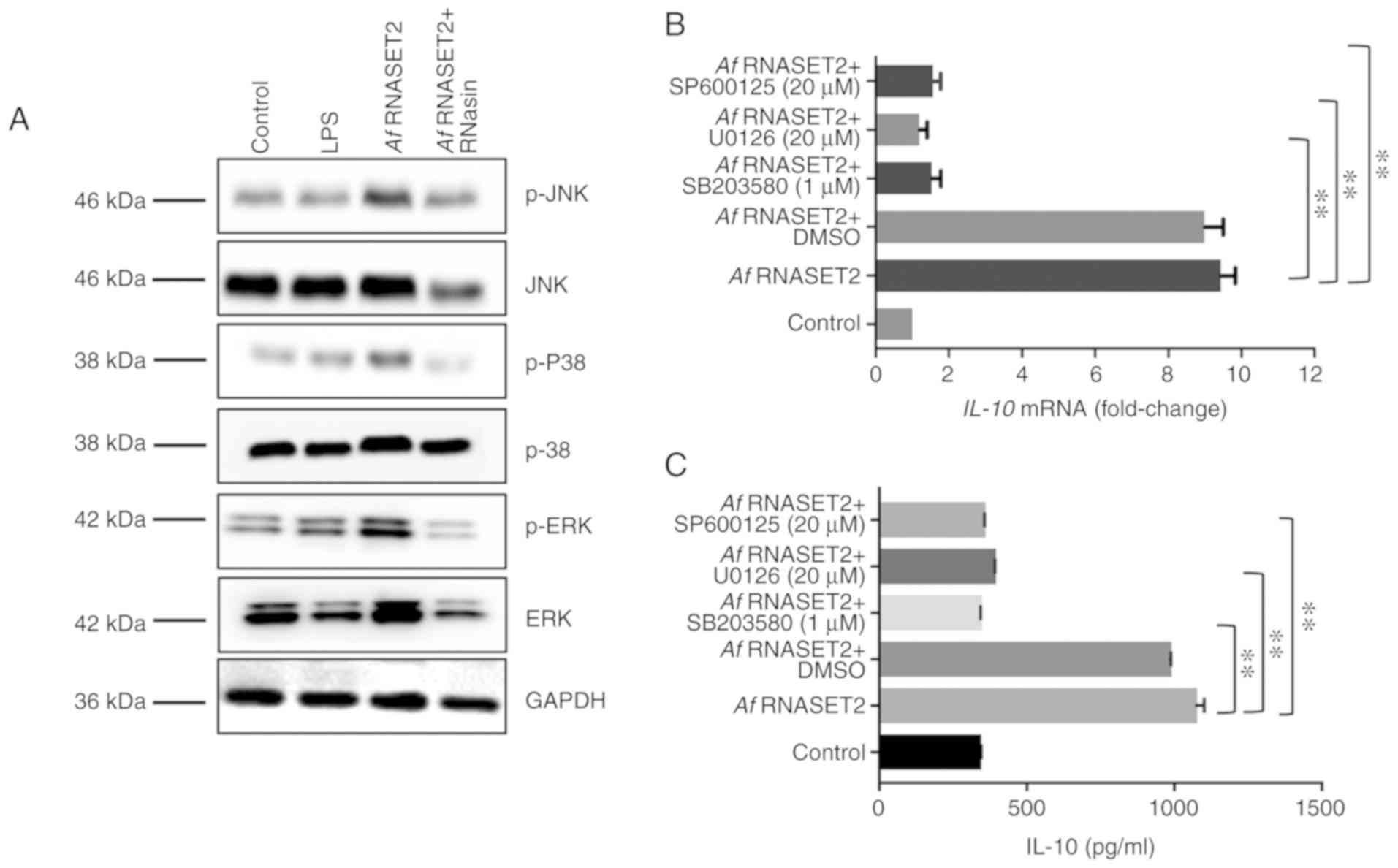 | Figure 6rAf RNASET2 upregulates proteins in
the MAPK signaling pathway in macrophages. (A) Western blot
analysis of JNK, p-JNK, p38, p-p38, ERK1/2, and p-ERK1/2 in
RAW264.7 macrophages following treatment with rAf RNASET2
(20 µg/ml) or (24-h) RNasin-inactivated rAf RNASET2.
Levels of IL-10 in RAW264.7 macrophages treated with rAf
RNASET2 (20 µg/ml) alone or in the presence of MAPK signal
inhibitors (U0126, SP600125 and SB203580 for ERK, JNK and p38,
respectively) were determined using (B) reverse
transcription-quantitative PCR and (C) ELISA. Data are represented
as mean ± SD from 3 independent experiments.
**P<0.01. r, recombinant; Af, Aspergillus
fumigatus; RNASET2, T2 ribonuclease; p, phosphorylated; LPS,
lipopolysaccharide. |
Conversely, in a parallel experiment conducted with
RNasin-inactivated rAf RNASET2, the aforementioned M2-like
changes were not observed, as evidenced by no significant changes
in the mRNA levels of ARG1, IL-10 and IL-13 (P>0.01 vs. control
group; Fig. 5). These results
indicated that high rAf RNASET2 enzyme activity was required for
the M2 polarization/Th2 response of macrophages to rAf
RNASET2.
rAf RNASET2 upregulated MAPK signaling in
macro- phages
Western blot analysis revealed that active, but not
inactive, rAf RNASET2 upregulated the expression levels of
p-ERK1/2, p-p38, and p-JNK in RAW264.7 cells (Fig. 6A). In addition, as IL-10 is
important for Th2 responses (21), the transcription and protein level
of IL-10 was subsequently detected to reflect the effect of MAPK
inhibitors on the rAf RNASET2-induced M2 macrophage polarization.
RT-qPCR revealed that MAPK inhibitors (U0126, SP600125 and SB203580
for ERK, JNK and p38, respectively) significantly reduced the
rAf RNASET2-induced increases in the protein expression
levels of IL-10 in RAW264.7 cells (P<0.01; Fig. 6B). In addition, ELISA revealed
that rAf RNASET2-induced IL-10 secretion in the cell
supernatant was similarly decreased by MAPK inhibitors (P<0.01;
Fig. 6C).
Discussion
In the present study, an E. coli expression
system was used to produce rAf RNASET2 with robust
ribonuclease activity. The amino acid sequence homology analysis
revealed conservation of the T2 ribonuclease functional domains,
CAS-1 and CAS-2. Glycosylation of rAf RNASET2 may have been
unaffected by ribonuclease activity due to the lack of modification
in a prokaryotic host. Similarly, recombinant Sj CP1412
protein (from pET bacterial transformation) mediated enzymolysis of
yeast RNA, affirming its ribonuclease activity (18). Glycosylation mutants of Sm
ω-1, in which N-linked glycosylation sites (N71/176Q) were altered,
did not have altered ribonuclease activity (16). These results suggest that
glycosylation may be independent of RNASET2 protein activity and
that prokaryotic expression methods are suitable for obtaining
recombinant ribonucleases. Hence, although previously published
data suggested that glycosylation of Sm ω-1 may be necessary
for conditioning dendritic cells for Th2 polarization (17), the findings in the present study
indicate that glycosylation of Af RNASET2 was not associated
with the observed immunoregulatory effect on M2 macrophage
polarization.
Af antigen exposure induces ABPA in the
majority of patients with atopic asthma or CF (22-24), as evidenced by a Th2 response and
elevated IgE levels (25).
Af proteases [i.e. Alp1 (26), Asp f 2 (27), Asp f 5 (28), and Asp f 13 (28)] have been shown to cause airway
inflammation and remodeling in a murine inhalation model. The
present study demonstrated that rAf RNASET2 immunization
upregulated the expression of the Th2 cytokines, IL-4, IL-10, and
IL-13, in sera, indicating a novel method of demonstrating whether
there are specific Af molecules required to promote Th2
responses in vivo. The T2 ribonuclease proteins Sj
CP1412 (18), Sm ω-1
(16), and Cs RNASET2
(29) can elicit Th2 immune
responses in vivo. Moreover, the rAf RNASET2 produced
in the present study elicited Th2 immune responses, supporting the
hypothesis that Af RNASET2 is a Th2 response regulator in
ABPA.
M2 macrophages can promote Th2 responses that could
lead to ABPA by secreting IL-4, IL-10, or IL-13 cytokines or the
chemokine CCL17 (30). Ke et
al. (18) revealed that
Sj CP1412 induced M2 polarization of macrophages as
evidenced by increased expression levels of CD206, ARG1, and IL-10.
Notably, in the present study, stimulating macrophages with
rAf RNASET2 also increased the protein expression levels of
CD206, ARG1, and IL-10, with no changed in the M1
macrophage-related surface marker CD16/32, suggesting that
rAf RNASET2 could promote M2 polarization in RAW264.7
macrophages. In addition, RNasin-inactivated rAf RNASET2 did
not produce this M2 response, which is consistent with previous
results with inactivated Sm ω-1 (16). These results suggest that a high
level of rAf RNASET2 enzyme activity is required for M2
polarization and an associated Th2 response. Thus, suppression of
Af RNASET2 activity may downregulate the immunoregulatory
effect on M2 polarization of macrophages and prevent Th2 responses
in vivo.
The effects of rAf RNASET2 on MAPK signaling
were investigated as the MAPK signaling pathway has been shown to
be involved in M2 polarization (31). The results from the present study
reveal that rAf RNASET2 upregulated p-ERK1/2, p-p38, and
p-JNK protein expression levels in RAW264.7 macrophages and that
MAPK inhibition attenuated rAf RNASET2-induced IL-10
secretion detected in the supernatant of RAW264.7 cell cultures.
These data indicate that MAPK signaling pathway is involved in the
production of M2 macrophages following rAf RNASET2
induction.
The results from the present study support the
hypothesis that Af RNASET2 may induce M2-type macrophage
polarization, thereby leading to a Th2 response in ABPA
development, and these effects were found to be dependent on
ribonuclease activity. As ribonuclease proteins are relatively
stable and resistant to degradation in the normal cell environment,
the mechanism of the Af RNASET2-induced Th2 response effect
provides important details regarding the predominant prevalence of
Af promotion of ABPA development. Moreover, the current
study supports the use of environmental intervention of
ribonuclease inhibitors to suppress the effects of Af
RNASET2-induced immune responses, as a prophylaxis against ABPA
development. However, the efficacy of this method requires further
investigation.
In conclusion, the results from the present study
provide important evidence regarding the molecular pathogenesis of
ABPA induced by Af exposure, which has not yet been
elucidated, and supports the hypothesis that Af RNASET2 is
an important immune regulatory factor in ABPA, primarily through
augmentation of M2 macrophages polarization and thus induction of a
Th2 immune response. These findings provide novel evidence
pertinent to understanding the immune regulatory role of RNase T2
family proteins during Af exposure.
Acknowledgments
The authors would like to thank the other members
of the Kunmei Ji laboratory for their critical comments regarding
the study design and the performing of the experiments.
Funding
The present study was supported in part by research
funding from the National Natural Science Foundation of China
(grant no. 81571570 and 81602595), Guangdong Province (grant nos.
2016A030313039, 2017A010105014 and 2018A050506083), and Shenzhen
City 2016 Biochemistry Discipline Construction, the Natural Science
Foundation of Shenzhen City (grant no. JCYJ20170818142053544).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
KJ and ZFZ conceived and designed the study. JJC,
YSH, ZLC and YSL analyzed the data. JJC, YSH, XJZ and ZLC performed
the studies. JJC and KJ contributed reagents, materials and
analysis tools. JJC, ZFZ and KJ wrote the manuscript. All authors
approved the final version of the manuscript.
Ethics approval and consent to
participate
All studies involving mice were performed according
to protocols approved by the Animal Ethical and Welfare Committee
of the School of Medicine of Shenzhen University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
ABPA
|
allergic bronchopulmonary
aspergillosis
|
|
Af
|
Aspergillus fumigatus
|
|
ANOVA
|
one-way analysis of variance
|
|
ARG1
|
arginase-1
|
|
CAS
|
conserved amino acid sequences
|
|
COX2
|
cyclooxygenase 2
|
|
Cs
|
Clonorchis sinensis
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
Ig
|
immunoglobulin
|
|
IL
|
interleukin
|
|
iNOS
|
induced nitric oxide synthase
|
|
IPTG
|
isopropyl-β-D-thiogalactopyr
anoside
|
|
JNK
|
c-Jun N-terminal kinase
|
|
MAPK
|
mitogen-activated protein kinases
|
|
NCBI
|
National Center for Biotechnology
Information
|
|
RNASET2
|
T2 ribonuclease
|
|
RNasin
|
ribonuclease inhibitor
|
|
Sj
|
Schistosoma japonicum
|
|
Sm
|
Schistosoma mansoni
|
|
Th1
|
T helper 1
|
|
Th2
|
T helper 2
|
References
|
1
|
Donaldson SH and Boucher RC: Update on
pathogenesis of cystic fibrosis lung disease. Curr Opin Pulm Med.
9:486–491. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nayak AP, Croston TL, Lemons AR, Goldsmith
WT, Marshall NB, Kashon ML, Germolec DR, Beezhold DH and Green BJ:
Aspergillus fumigatus, viability drives allergic responses to
inhaled conidia. Ann Allergy Asthma Immunol. 121:200–210e2. 2018.
View Article : Google Scholar
|
|
3
|
el-Dahr JM, Fink R, Selden R, Arruda LK,
Platts-Mills TA and Heymann PW: Development of immune responses to
Aspergillus at an early age in children with cystic fibrosis. Am J
Respir Crit Care Med. 150:1513–1518. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Agarwal R, Aggarwal AN, Gupta D and Jindal
SK: Aspergillus hypersensitivity and allergic bronchopulmonary
aspergillosis in patients with bronchial asthma: Systematic review
and meta-analysis. Int J Tuberc Lung Dis. 13:936–944.
2009.PubMed/NCBI
|
|
5
|
Fricker-Hidalgo H, Coltey B, Llerena C,
Renversez JC, Grillot R, Pin I, Pelloux H and Pinel C: Recombinant
allergens combined with biological markers in the diagnosis of
allergic bronchopulmonary aspergillosis in cystic fibrosis
patients. Clin Vaccine Immunol. 17:1330–1336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nguyen NL, Pilewski JM, Celedón JC,
Mandalapu S, Blanchard ML, DeRicco A, Hartigan E, Alcorn JF and
Kolls JK: Vitamin D supplementation decreases Aspergillus fumigatus
specific Th2 responses in CF patients with aspergillus
sensitization: A phase one open-label study. Asthma Res Pract.
1:32015. View Article : Google Scholar
|
|
7
|
Homma T, Bhushan B, Kato A, Norton J, Suh
L, Sha Q, Gupta DS and Schleimer RP: Aspergillus fumigatus may
promote Th2 activation by suppression of interferon signaling. J
Allergy Clin Immunol. 133:AB1362014. View Article : Google Scholar
|
|
8
|
Bhushan B, Homma T, Norton JE, Sha Q,
Siebert J, Gupta DS, Schroeder JW Jr and Schleimer RP: Suppression
of epithelial signal transducer and activator of transcription 1
activation by extracts of Aspergillus fumigatus. Am J Respir Cell
Mol Biol. 53:87–95. 2015. View Article : Google Scholar :
|
|
9
|
Homma T, Kato A, Bhushan B, Norton JE, Suh
LA, Carter RG, Gupta DS and Schleimer RP: Role of Aspergillus
fumigatus in triggering protease-activated receptor-2 in airway
epithelial cells and skewing the cells toward a T-helper 2 bias. Am
J Respir Cell Mol Biol. 54:60–70. 2016. View Article : Google Scholar :
|
|
10
|
Wang L, Wang L and Wu X: Aspergillus
fumigatus promotes T helper type 2 responses through thymic stromal
lymphopoietin production by human corneal epithelial cells. Clin
Exp Ophthalmol. 44:492–501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Knutsen AP, Hutcheson PS, Slavin RG and
Kurup VP: IgE antibody to Aspergillus fumigatus recombinant
allergens in cystic fibrosis patients with allergic
bronchopulmonary aspergillosis. Allergy. 59:198–203. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jolink H, De Boer R, Willems LN, van
Dissel JT, Falkenburg JH and Heemskerk MH: T helper 2 response in
allergic broncho-pulmonary aspergillosis is not driven by specific
Aspergillus antigens. Allergy. 70:1336–1339. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Furukawa S, Moriyama M, Miyake K,
Nakashima H, Tanaka A, Maehara T, Iizuka-Koga M, Tsuboi H,
Hayashida JN, Ishiguro N, et al: Interleukin-33 produced by M2
macrophages and other immune cells contributes to Th2 immune
reaction of IgG4-related disease. Sci Rep. 7:424132017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Øya E, Becher R, Ekeren L, Afanou AKJ,
Øvrevik J and Holme JA: Pro-inflammatory responses in human
bronchial epithelial cells induced by spores and hyphal fragments
of common damp indoor molds. Int J Environ Res Public Health.
16:10852019. View Article : Google Scholar :
|
|
15
|
Coughlan CA, Chotirmall SH, Renwick J,
Hassan T, Low TB, Bergsson G, Eshwika A, Bennett K, Dunne K, Greene
CM, et al: The effect of Aspergillus fumigatus infection on vitamin
D receptor expression in cystic fibrosis. Am J Respir Crit Care
Med. 186:999–1007. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Steinfelder S, Andersen JF, Cannons JL,
Feng CG, Joshi M, Dwyer D, Caspar P, Schwartzberg PL, Sher A and
Jankovic D: The major component in schistosome eggs responsible for
conditioning dendritic cells for Th2 polarization is a T2
ribonuclease (omega-1). J Exp Med. 206:1681–1690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Everts B, Hussaarts L, Driessen NN,
Meevissen MH, Schramm G, van der Ham AJ, van der Hoeven B, Scholzen
T, Burgdorf S, Mohrs M, et al: Schistosome-derived omega-1 drives
Th2 polarization by suppressing protein synthesis following
internalization by the mannose receptor. J Exp Med. 209:1753–1767.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen Ke XD, Song S, Yu LJ, Kikuchi CX,
Hirayama M, Gao K, Wang H, Yin J, Yao X Y, et al: Characterization
of Schistosoma japonicum CP1412 protein as a novel member of the
ribonuclease T2 molecule family with immune regulatory function.
Parasit Vectors. 10:892017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Z, Cai Z, Hou Y, Hu J, He Y, Chen J
and Ji K: Enhanced sensitivity of capture IgE-ELISA based on a
recombinant Der f 1/2 fusion protein for the detection of IgE
antibodies targeting house dust mite allergens. Mol Med Rep.
19:3497–3504. 2019.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 5:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Laouini D, Alenius H, Bryce P, Oettgen H,
Tsitsikov E and Geha RS: IL-10 is critical for Th2 responses in a
murine model of allergic dermatitis. J Clin Invest. 112:1058–1066.
2013. View Article : Google Scholar
|
|
22
|
Denning DW, O'Driscoll BR, Powell G, Chew
F, Atherton GT, Vyas A, Miles J, Morris J and Niven RM: Randomized
controlled trial of oral antifungal treatment for severe asthma
with fungal sensitization: The fungal asthma sensitization trial
(FAST) study. Am J Respir Crit Care Med. 179:11–18. 2009.
View Article : Google Scholar
|
|
23
|
Chishimba L, Niven RM, Cooley J and
Denning DW: Voriconazole and posaconazole improve asthma severity
in allergic bronchopulmonary aspergillosis and severe asthma with
fungal sensitization. J Asthma. 49:423–433. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pasqualotto AC, Powell G, Niven R and
Denning DW: The effects of antifungal therapy on severe asthma with
fungal sensitization and allergic bronchopulmonary aspergillosis.
Respirology. 14:1121–1127. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Murrison LB, Brandt EB, Myers JB and
Hershey GKK: Environmental exposures and mechanisms in allergy and
asthma development. J Clin Invest. 129:1504–1515. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Redes JL, Basu T, Ram-Mohan S, Ghosh CC,
Chan EC, Sek AC, Zhao M, Krishnan R, Rosenberg HF and Druey KM:
Aspergillus fumigatus-secreted alkaline protease 1 mediates airway
hyperresponsiveness in severe asthma. Immunohorizons. 3:368–377.
2019.PubMed/NCBI
|
|
27
|
Kurup VP, Xia JQ, Shen HD, Rickaby DA,
Henderson JD Jr, Fink JN, Chou H, Kelly KJ and Dawson CA: Alkaline
serine proteinase from Aspergillus fumigatus has synergistic
effects on Asp-f-2-induced immune response in mice. Int Arch
Allergy Immunol. 129:129–137. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Namvar S, Warn P, Farnell E, Bromley M,
Fraczek M, Bowyer P and Herrick S: Aspergillus fumigatus proteases,
Asp f 5 and Asp f 13, are essential for airway inflammation and
remodelling in a murine inhalation model. Clin Exp Allergy.
45:982–993. 2015. View Article : Google Scholar
|
|
29
|
Xu Y, Lin J, Bian M, Chen W, Liang P, Wang
X, Shang M, Qu H, Wu Z, Huang Y and Yu X: CsRNASET2 is an important
component of Clonorchis sinensis responsible for eliciting Th2
immune response. Parasitol Res. 114:2371–2379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang N, Liang H and Zen K: Molecular
mechanisms that influence the macrophage m1-m2 polarization
balance. Front Immunol. 5:6142014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Neamatallah T: Mitogen-activated protein
kinase pathway: A critical regulator in tumor-associated macrophage
polarization. J Microsc Ultrastruct. 7:53–56. 2019. View Article : Google Scholar : PubMed/NCBI
|