Introduction
As one of the most prevalent types of human cancer,
lung cancer is responsible for one-quarter of cancer-associated
mortalities worldwide (1). Lung
cancer rates and trends differ considerably according to sex, age,
race, socioeconomic status and geography, due to alterations in
historical smoking patterns (2).
Non-small cell lung cancer is divided into three different
subtypes: Lung adenocarcinoma; lung squamous cell carcinoma (LUSC);
and large cell lung carcinoma. LUSC contributes to ~30% of the
total lung cancer cases and results in ~400,000 mortalities each
year worldwide (3). There have
been many advances in the development of molecular lung
adenocarcinoma-targeted therapies, while limited progress has been
made in the development of treatments for LUSC, with the exception
of immunotherapy (4).
Cancer-associated fibroblasts (CAFs) have been
documented to stimulate cancer cell proliferation and invasion,
thereby supporting tumorigenesis (5). CAFs are defined by the acquisition
of an α-smooth muscle actin (α-SMA)-positive, 'activated'
myofibroblast phenotype (6). CAFs
and cancer cells have been revealed to have the ability to release
exosomal microRNAs (miRNAs) to exert effects over each other, and
the extracellular vesicles (EVs) can be categorized into three
types, on the basis of biogenesis and size: Exosomes (30-100 nm);
microvesicles (100-1000 nm); and oncosomes (1-10 μm)
(7). Exosomes could modulate
local and systemic cell communication through the transmission of
information via microRNAs, proteins, mRNAs and other substances
(8). For example, exosomal
miR-382-5p released by CAFs promoted the cell migration and
invasion in oral squamous cell carcinoma (9). Notably, overexpression of miR-369-3p
was identified in cisplatin-resistant non-small cell lung cancer
(NSCLC) tissues and cells and associated with the malignancy of
these cells (10). Using StarBase
(11) and TargetScan (12), miR-369 was predicted to bind to
neurofibromin 1 (NF1) in LUSC cells in the current study. NF1,
located on chromosome 17q11.2, is an important tumor suppressor and
encodes a rat sarcoma (Ras)-GTPase activating protein identified as
neurofibromin (13). Glomus cells
with inactivated NF1 exhibited elevated Ras-mitogen-activated
protein kinase (MAPK) signals (14). Neurofibromin accelerates the
transfer of the Ras family to their inactive state and acts as a
suppressor of the MAPK signaling pathway (15). In addition, NF1 deficiency
facilities murine Kirsten rat sarcoma viral oncogene homolog
gene-driven tumorigenesis of lung adenocarcinomas (16), which may validate the functional
activity of NF1 mutations in NSCLC and the efficacy of targeted
suppression of downstream Ras signaling pathway (17). In the present study, miR-369
secreted by CAF-EVs is reported to activate the MAPK signaling
pathway by downregulating NF1, initiating development and
progression of LUSC.
Materials and methods
Tissue collection
A total of 52 paired LUSC and matching paratumor
tissues (>5 cm from the LUSC tissues) were harvested by
resection from patients diagnosed and treated in the Affiliated
Hospital of Yan'an University from November 2012 to August 2013.
The cohort consisted of 39 males and 13 females, ranging from 49-79
years, with an average age of 64.9±7.3 years. Follow-up was
conducted every 3 months for 5 years. All the patients were
diagnosed with LUSC on the basis of histopathological evaluation
and had complete clinical data. No patients had been subjected to
radiotherapy or chemotherapy prior to surgical treatment. Patients
combined with chronic system disorders, such as acquired immune
deficiency syndrome, autoimmune diseases and diabetes, or other
malignancies were excluded. All human tissue donors provided
written informed consent prior to tissue donation. The study
protocol was approved by the Institutional Review Board of
Affiliated Hospital of Yan'an University.
Bioinformatics analysis
To screen differentially expressed miRNAs in cells
co-cultured with EVs, miRCURY LNA™ Universal RT microRNA PCR Human
panel (Exiqon; Qiagen GmbH) was used. miRNAs differentially
expressed in H520 cells co-cultured with PBS and EVs were analyzed,
according to the manufacturer's protocol. One-way analysis of
variance was applied for comparisons with threshold settings as:
P<0.05 and fold-change >2.5 or <-2.5. StarBase (http://starbase.sysu.edu.cn/) platform was
subsequently used to detect miR-369 expression in lung cancer
patients in The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) (18). Subsequently, StarBase and
TargetScan (version 7.2; http://www.targetscan.org/vert_72/) were used to
predict the targeting mRNAs of miR-369. The targeting mRNAs of
miR-369 were subjected to Kyoto Encyclopedia of Genes and Genomes
enrichment analysis (https://www.kegg.jp/) (19) using a bioinformatics software
DAVID (version 6.7; https://david.ncifcrf.gov/).
Culture and identification of CAFs
The CAFs were isolated and extracted from surgically
removed tissues from 6 patients with LUSC treated in the Affiliated
Hospital of Yan'an University from July to December 2017.
Subsequent to detachment through collagenase/trypsin, the cells
were cultured until they reached >85% confluence, and then
detached with trypsin again for passage. Isolated CAFs were
maintained in Dulbecco's modified Eagle's medium (DMEM)-F12
(Sigma-Aldrich; Merck KGaA) containing 10% FBS depleted of exosomes
(exo-FBS; System Biosciences, LLC), blocked with 1% bovine serum
albumin (Beijing Solarbio Life Sciences Co., Ltd.) for 30 min at
37°C and treated with 1X antibiotic-antimycotics (Thermo Fisher
Scientific, Inc.). The CAFs (1×106 cells/ml) were
stained for 30 min with phycoerythrin-conjugated monoclonal
antibodies against prolyl endopeptidase FAP (FAP; cat. no.
ab207178; 1:50; Abcam), vimentin (cat. no. ab92547; 1:50; Abcam)
and α-SMA (cat. no. ab32575; 1:50; Abcam) at 37°C in the dark.
Following phosphate buffered saline (PBS) washes, the cells were
examined using a FACSCaliburTM flow cytometer (BD Biosciences) and
analyzed using FlowJo V11.0 (BD Biosciences).
Extraction and identification of EVs
Following CAF identifi-cation, the cells between
passages 3 and 12 were used for EV extraction. EVs were extracted
from CAFs culture medium and centrifuged for 10 min at 3,000 × g at
4°C. The medium was then subjected to two rounds of centrifugation
to remove cell debris, firstly at 3,000 × g for 10 min and then at
10,000 × g for 30 min at 4°C. Subsequently, 1 ml culture medium was
centrifuged at 4°C at 100,000 × g for 2 h using Class H, R, and S
Preparative Ultracentrifuges with a Type 50.4 Ti Rotor (Beckman
Coulter, Inc.) to obtain the precipitate. This precipitate was
washed with PBS, then filtered through a 0.22 μm filter, and
then ultra-centrifuged at 4°C for 2 h at 100,000 × g to precipitate
the EVs. The precipitate resuspended in 1 ml PBS or lysis buffer
(Beyotime Institute of Biotechnology) was used for protein
measurement. The extracted EVs were then maintained at −80°C or
treated with 10% sodium dodecyl sulfate (SDS) to damage EV membrane
structure.
The isolated EV suspension was resuspended and
dropped onto the sealing membrane. The resuspended pellets were
deposited on electron microscopy grids. The grids were adsorbed for
20 min, transferred to glutaraldehyde droplet for about 5 min, and
then transferred to the appropriate washing buffer. Repeat
transfers to fresh drops of washing buffer was performed seven
times. Then, the grids were dried and transferred to a 4% hydrogen
peroxide droplet for 10 min, and then transferred to a 1% methyl
cellulose droplet for 5 min (all steps were carried out on ice).
After being air-dried for 30 min, images were captured using a
transmission electron microscope (80 kV) and analyzed by
DigitalMicrograph 3.9 (magnification, ×40,000; Gatan, Inc.).
CAFs were transfected with 10 nM miR-369 inhibitor
(5′-AGAUCGACCGUGUUAUAUUCGC-3′) or scramble inhibitor
(5′-GGUUGAUCUUUUCUCAGUA-3′; Tebu-bio) using Oligofectamine
(Invitrogen; Thermo Fisher Scientific, Inc.). A total of 6 h after
transfection, the cells were cultured in fresh DMEM-F12
supplemented with 1% A/A and 10% exo-FBS for 1 day. The media were
harvested for exosome preparation. The H520 and SK-MES-1 cells were
then co-cultured with exosomes at a concentration of
1×109 particles/ml. The inhibitors or scramble
inhibitors utilized were synthesized by Shanghai GenePharma Co.,
Ltd.
Western blot analysis
Total protein was extracted from CAFs-derived
exosomes or supernatant using radioimmunoprecipitation assay lysis
buffer (Beyotime Institute of Biotechnology). Briefly, 40 mg
protein were loaded on each lane, separated using 10% SDS-PAGE and
transferred onto polyvinylidene fluoride membranes. Following
blocking with 5% skim milk at room temperature for 30 min, the
membranes were subsequently incubated with the primary antibodies
(all 1:1,000) against CD9 antigen (cat. no. ab92726; Abcam), CD63
antigen (cat. no. 55051; Cell Signaling Technologies, Inc.), tumor
susceptibility gene 101 protein (cat. no. ab125011; Abcam), heat
shock protein 70 (HSP70; cat. no. 4873; Cell Signaling
Technologies, Inc.), CD81 antigen (cat. no. 10037; Cell Signaling
Technologies, Inc.) overnight at 4°C, and with horseradish
peroxidase (HRP)-labeled goat anti-rabbit IgG (cat. no. G-21234;
Invitrogen; Thermo Fisher Scientific, Inc.; 1:5,000) at room
temperature for 2 h. Blots were visu-alized using chemiluminescence
detection (GE Healthcare) and analyzed by ImageJ 1.8.0 software
(National Institutes of Health). Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH; cat. no. D190090; Shanghai Sangon Biological
Engineering Technology & Services Co., Ltd.) was applied as an
internal reference.
Cell culture and grouping
Human LUSC H520 and SK-MES-1 cell lines were
obtained from the Institute of Biochemistry and Cell Biology,
Shanghai Institutes for Biological Sciences, Chinese Academy of
Sciences. The aforementioned cells were seeded into cell culture
dishes at 1×105 cells/cm2 and maintained in
Roswell Park Memorial Institute (RPMI)-1640 (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) for 48 h at 37°C with 5% CO2. When the
cells reached 80-90% confluence, they were detached with 0.25%
trypsin (Gibco; Thermo Fisher Scientific, Inc.), and then
passaged.
Subsequently, H520 cells and SK-MES-1 cells at a
confluence >75% were co-cultured with 20 μl PBS and 20
μM exosomes from un-transfected cells or cells transfected
with miR-369 inhibitor or inhibitor control.
Reverse transcription-quantitative PCR
(RT-qPCR)
The total RNA was extracted from tissues and cells
using RNAiso Plus (Takara Bio, Inc.) or TRIzol® LS
Reagent (Takara Bio, Inc.). Subsequently, 10 mg RNA was subjected
to a 30-min at 10 V/cm voltage using 2% agarose gel at room
temperature to verify the reliability of RNA. Then, a PrimeScript
TM RT kit (Takara Bio, Inc.) was used to perform the RT strictly
according to the protocol of the manufacturer. qPCR analyses were
performed by 30 cycles of denaturation at 95°C and annealing at
45°C, followed by extension at 72°C using SYBR Premix Ex Taq
(Takara Bio, Inc.). The obtained quantification data were
normalized to the U6 expression. The sequences of the primers used
were as follows: miR-369 forward, 5′-GGGACCCAGTTCAAGTAATTCAGG-3′;
miR-369 reverse, 5′-TTTGGCACTAGCACATT-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′; and U6 reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Data were determined using the 2−ΔΔCq method (20).
Cell counting kit-8 (CCK-8)
Cells in a logarithmic growth phase were detached
with trypsin, diluted into a single cell suspension at
1×104 cells/ml using RPMI-1640 containing 10% FBS, and
then added to a 96-well plate, with 2×103 cells each
well in 200 μl solution. A blank well free of cells
containing the culture medium was used as a blank control. A total
of 5 identical wells were prepared for each group. Following
inoculation, the cells were grown under 5% CO2 at 37°C
for 3 days. At 0, 24, 48 and 72 h, cells in each well were
supplemented with 20 μl CCK-8 solution (5 mg/ml) and
cultured for an additional 4 h. Subsequently, 200 μl
dimethyl sulfoxide was added to the cells in each well and
oscillated for 10 min to thoroughly dissolve the crystals, followed
by the measurement of optical density (OD) value at the wavelength
of 490 nm.
5-Ethynyl-2′-deoxyuridine (EdU)
staining
The DNA replication capacity of H520 and SK-MES-1
cells at passage 3 was detected by a Cell-light EdU luminous
detection kit (Guangzhou RiboBio Co., Ltd.) according to the
instructions of the manufacturer. A total of 5 visual fields were
randomly selected for analysis using a FSX100 fluorescence
microscope (magnification, ×200; Olympus Corporation). Blue
fluorescence represented total cells, while the red fluorescence
denoted the replicating cells that had incorporated the EdU.
Finally, the percentage of EdU-positive cells was calculated.
Immunofluorescence
Cells were fixed for 15 min in 4% paraformaldehyde
at 4°C and treated for 20 min with 0.5% Triton X-100. Antibodies
against epithelial-cadherin (1:200; cat. no. ab92552; Abcam) and
neural-cadherin (1:100; cat. no. ab133504; Abcam) were utilized as
primary antibodies, while Alexa Fluor 594-labeled goat anti-rabbit
(1:5,000; cat. no. ab150088; Abcam) used as a secondary antibody.
The cells were subsequently counter-stained with
4′,6-Diamidino-2-Phenylindole and detected using a Leica DM3000
fluorescence microscope (magnification, ×400; Leica Microsystems,
Inc.).
Transwell assays for cell migration and
invasion
For the invasion assays, 60 μl Matrigel (BD
Biosciences) diluted in 300 μl serum-free medium at 4°C was
added under aseptic condition and cultured in the apical chamber
for 30 min. RPMI-1640 medium (30 μl) was then added in the
apical chamber and cultured in the 5% CO2 incubator at
37°C for 30 min. Following a 10-min centrifugation (160 × g at
4°C), the cell suspension was diluted at a density of
5×105 cells/ml. Next, 500 μl RPMI-1640 medium
containing 10% FBS was placed in the Transwell basolateral chamber,
with 200 μl cell suspension in the apical chamber. The plate
was placed in a 37°C incubator under 5% CO2 for 48 h and
then subjected to 1% crystal violet staining for 10 min at room
temperature. The cells that had not invaded were removed with
cotton balls, and images were captured under a light microscope
(magnification, ×200) to calculate the number of cells. The
migration capacity was assessed using a Transwell assay without the
addition of Matrigel in the apical chamber. During this protocol,
the chambers were removed for staining after 24 h.
ELISAs
NF1 (cat. no. HEN020), MAPK/ERK kinase 1 (MEK1; cat.
no. HEM016), pMEK1 (cat. no. HEM017), ERK1/2 (cat. no. HEE013) and
pERK1/2 (cat. no. HEE014) ELISA kits (Shanghai Bogoo Biological
Technology Co., Ltd.) were used in strict accordance with the
protocols of the manufacturer. Briefly, the cells were lysed with
radioimmunoprecipitation assay lysis buffer containing proteinase
inhibitors. Following centrifugation, the supernatants were added
into the 96-well plate, followed by blocking for 2 h with a
blocking buffer at room temperature. Antibodies against NF1, MEK1,
pMEK1, ERK1/2 and pERK1/2 were added to the medium and incubated
for 1 h. Following sequential introduction of substrate solution
and termination solution, the OD value at 450 nm of each well was
measured using an automated microplate reader (BioTek Instruments,
Inc.) within 30 min.
Dual-luciferase assay
StarBase (http://starbase.sysu.edu.cn/), a bioinformatic
software (11), was chosen to
predict the binding sequence of miR-369 and NF1-3′ untranslated
region (UTR). NF1 wild type (WT) and 3′UTR binding sequence mutant
(MT) were synthesized by Shanghai Sangon Biological Engineering
Technology & Services Co., Ltd. and inserted into pMIR-REPORTTM
(Thermo Fisher Scientific, Inc.). WT plasmids, MT plasmids and
miR-369 mimic and miRNA negative control (NC) were co-transfected
into 293T cells using a Lipofectamine® 3000 transfection
kit (Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. The cells were lysed 24 h after
transfection, and the luciferase activity was detected by a
Dual-Luciferase Reporter Assay System (Promega Corporation), as
previously described (21), and
normalized to Renilla luciferase activity.
Tumor xenograft model
A total of 40 specific-pathogen- free-grade BALB/c
nude mice (age 4-6 weeks, weight 20±2 g) were purchased from
Beijing Vital River Laboratory Animal Technology Co., Ltd. [permit
number: SCXK (Beijing) 2015-0001], and then randomized into 4
groups, with 10 animals in each group. A total of 4×106
H520 and SK-MES-1 cells in 100 μl saline were injected into
nude mice subcutaneously. All mice were housed in a temperature-
(22±1°C) and humidity-controlled (45-55%) room with a 12:12 h
light: Dark cycle and ad libitum access to chow and drinking water.
No mice were died during the treatment process. The tumor volume
(V) was measured every week after 7 days of injection and
calculated using calipers with the formula:
m12xm2x0.5236, in which
m1 represents the shortest diameter and m2
the longest diameter (22). A
total of 28 days following implantation, the mice were euthanized,
and tumors were excised for histological analysis. For mice
receiving EVs treatment, 100 μl EVs (20 μM) were
injected every 5 days to the site of subcutaneous tumors as
previously described (23). At
the end of the experiment (28 days after the injection of cells),
mice were euthanized by intraperitoneal injection of 1%
pentobarbital sodium at 100 mg/kg. Following euthanasia, animal
death was confirmed by observing the lack of heartbeat, respiratory
arrest, pupil dilation, and lack of nerve reflex.
Immunohistochemical staining
The tumor tissues from each group were
paraffin-embedded, dewaxed (in xylene I and xylene II for 20 min,
respectively) and hydrated (twice in anhydrous ethanol, once in 95%
ethanol and once in 80% ethanol; 5 min each time). A total of 5
sections were collected from each tumor. The sections were then
incubated with H2O2 for 15 min at room
temperature to eliminate the activity of endogenous peroxide.
Following sealing with normal goat serum at room temperature for 15
min, the sections were probed at 4°C overnight with 50 μl
rabbit anti-human antibody against proliferation marker protein
Ki-67 (Ki-67; 1:500; cat. no. ab15580; Abcam) and the
HRP-conjugated secondary goat anti-rabbit antibody against IgG
(1:500; cat. no. GTX213110-01; GeneTex, Inc.) at 37°C for 15 min.
Following treatment with 40 μl HRP-conjugated streptomyces
ovalbumin working solution at 37°C for 15 min, the sections were
stained with 0.01 M 3,3′-diaminobenzidine at room temperature for
30 min. Finally, the sections were counterstained with 2%
hematoxylin in ethanol at room temperature for 30 sec, dehydrated
and mounted. The cells with brownish yellow or brown nucleus were
regarded as Ki-67-positive. A total of 5 fields were chosen
randomly and observed under a light microscope (magnification,
×200), and the positive rate was calculated as the number of
positive cells/the number of total cells ×100%.
In vivo metastasis test
A total of 4×106 HT29 and SW116 cells
were injected into 40 mice through the tail veins (n=10/group). A
total of 100 μl EVs (20 μM) were injected every 5
days with a total of 8 injections through the tail veins as
previously described (24). No
mice were died during the process. In addition, the health and
behavior of mice were observed every day in the experiment, and the
mice exhibited good appetite, normal activity and good mental
condition prior to the end of the experiment. At the end of the
experiment, 45 days after the injection of cells, mice were
euthanized by intraperitoneal injection of 1% pentobarbital sodium
at 100 mg/kg. Following euthanasia, animal death was confirmed by
observing the absence of a heartbeat, respiratory arrest, pupil
dilation, and lack of nerve reflex. The lung and liver tissues were
removed for hematoxylin & eosin staining as previously
described (25).
Statistical analysis
Statistical analyses were performed with SPSS 21.0
(SPSS, Inc.). Kolmogorov-Smirnov testing was used to assess
normality. The results were expressed as mean ± standard deviation
(SD). Statistical analysis of differences between experimental
groups was performed using paired two-tailed t-tests, whereas
differences among multiple groups were analyzed by one-way or
two-way analysis of variance with Tukey's post hoc test. Fisher's
exact test was used to compare the enumeration data. P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of the CAF-EVs
Flow cytometry was first used to identify the
presence of CAFs surface markers α-SMA and FAP, and the fibroblast
marker vimentin, in the cells (Fig.
1A). Then, the CAF-derived EVs were identified by transmission
electron microscopy, nanoparticle tracking analysis and western
blot analysis assays. The results showed that the extracted EVs
corresponded to the definition of the EVs of the CAFs from the
Minimal information for studies of extracellular vesicles 2018
(MISEV2018) guidelines (26)
(Fig. 1B-D).
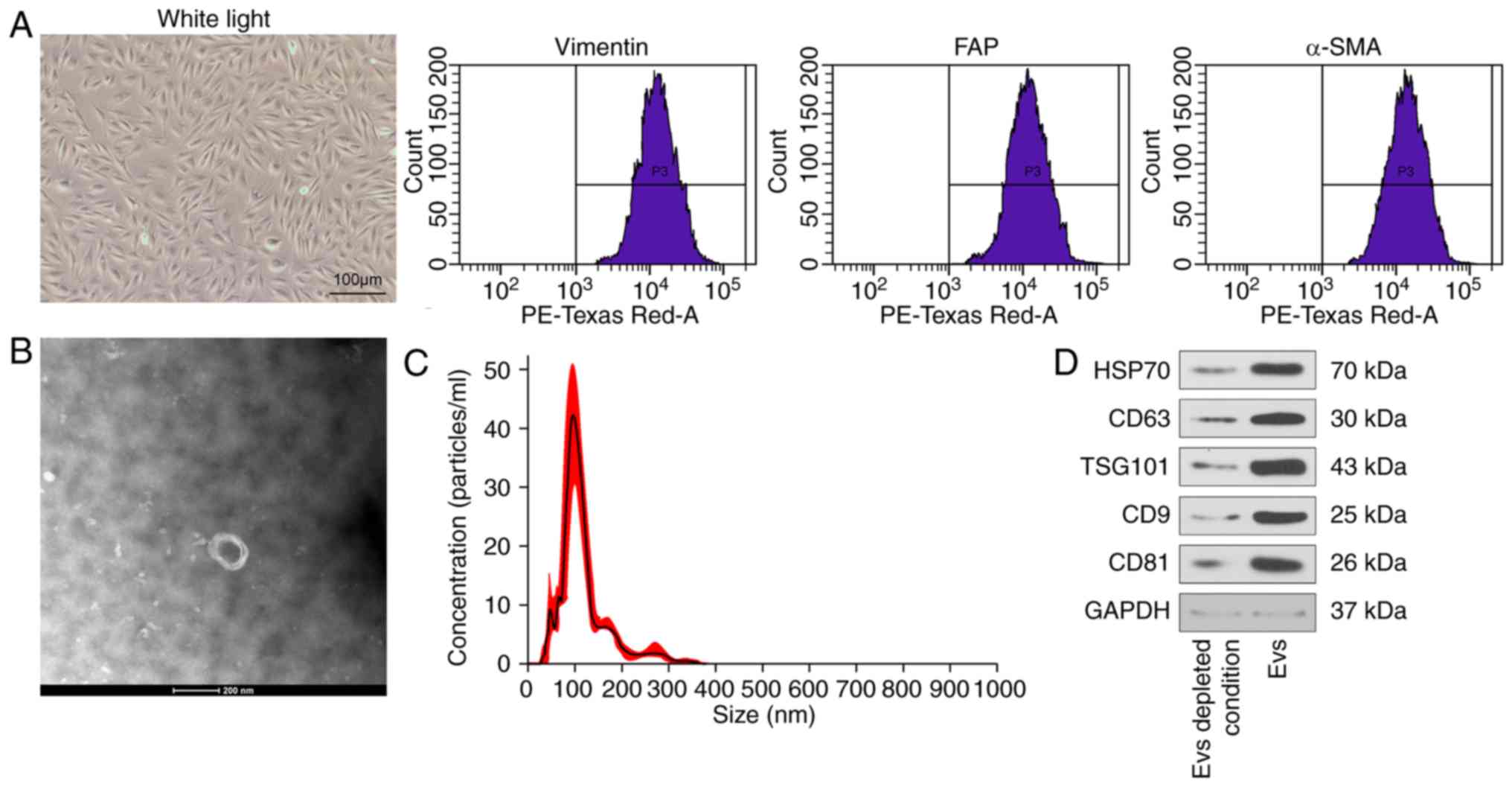 | Figure 1CAFs-derived EVs are successfully
isolated and identified. (A) Representative images of CAFs (scale
bar=100 μm) and identification of CAFs using antibodies
against vimentin, α-SMA, and FAP determined by flow cytometry. (B)
Transmission electron microscopy indicating EVs isolated from
CAFs-conditioned medium (scale bar=200 nm). (C) Detection of the
diameter of CAFs-EVs by nanosight tracking analysis. (D) The bands
of EVs markers CD63, HSP70, CD9, CD81 and TSG101 in EV-enriched
conditioned medium evaluated by western blot analysis, normalized
to GAPDH. CAFs, cancer-associated fibroblasts; EVs, extracellular
vesicles; α-SMA, α-smooth muscle actin; FAP, prolyl endopeptidase
FAP; CD63, CD63 antigen; HSP70, heat shock protein 70; CD9, CD9
antigen; CD81, CD81 antigen; TSG101, tumor susceptibility gene 101
protein. |
Malignancies of LUSC cells are promoted
by CAF-derived EVs treatment
Then, a CCK-8 kit was used to evaluate the cells
treated with different concentrations of EVs, and it was identified
that the optimum concentration was 20 μmol/l (Fig. 2A). The LUSC H520 and SK-MES-1 cell
lines were then treated with PBS, 20 μmol/l EVs and
SDS-treated EVs, respectively. It was identified that, following
EVs treatment, the viability of H520 and SK-MES-1 cells increased
significantly (Fig. 2B-C),
accompanied by significantly induced epithelial-mesenchymal
transition (EMT) (Fig. 2D and E),
and the number of invasive and migratory H520 and SK-MES-1 cells
elevated significantly (Fig. 2F and
G). However, following destruction of the membrane structure of
EVs using SDS treatment, the promotive roles of EVs were not
observed, which indicated that CAFs-derived EVs could promote the
malignant biological behavior of LUSC cells.
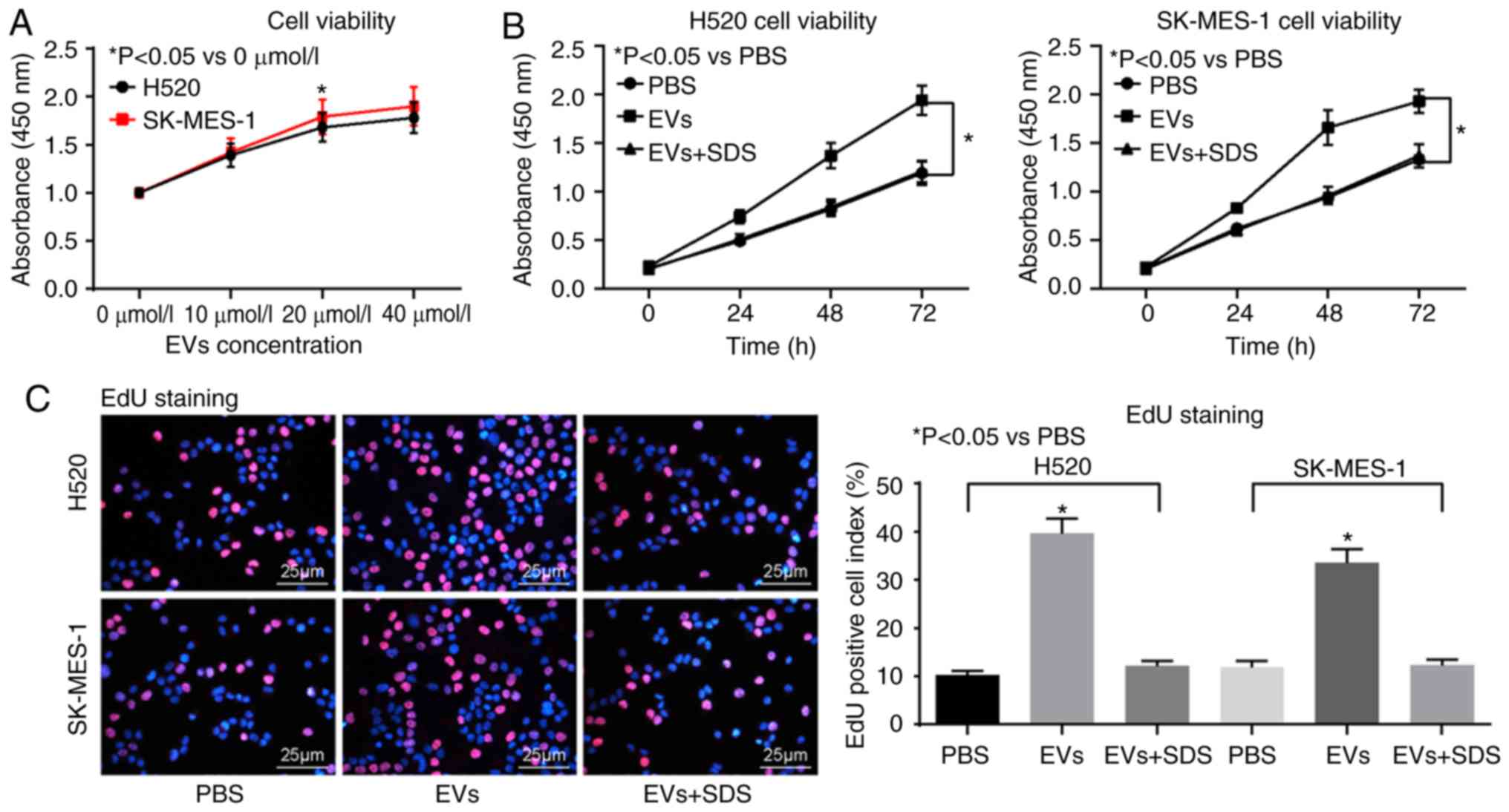 | Figure 2CAFs-derived EVs promote lung
squamous cell carcinoma cells malignancy. (A) The concentration of
EVs determined by Cell Counting Kit-8 assay. (B) The viability of
H520 cells assessed by EdU assay. (C) The viability of SK-MES-1
cells assessed by EdU assay. (D) Immunofluorescence staining of EMT
marker E-cadherin. (E) Immunofluorescence staining of EMT marker
N-cadherin. (F) Cell migration ability measured by Transwell assay.
(G) Cell invasion ability measured by Transwell assay. Data are
expressed as mean ± SD. In panel A and B, a two-way ANOVA and
Tukey's multiple comparisons test was used to determine statistical
significance, while in panel C to G, a one-way ANOVA was used.
*P<0.05 vs. PBS treatment. CAFs, cancer-associated
fibroblasts; EVs, extracellular vesicles; EdU,
5-ethynyl-2′-deoxyuridine; EMT, epithelial-mesenchymal transition;
E-cadherin, epithelial cadherin; N-cadherin, neural cadherin;
ANOVA, analysis of variance. |
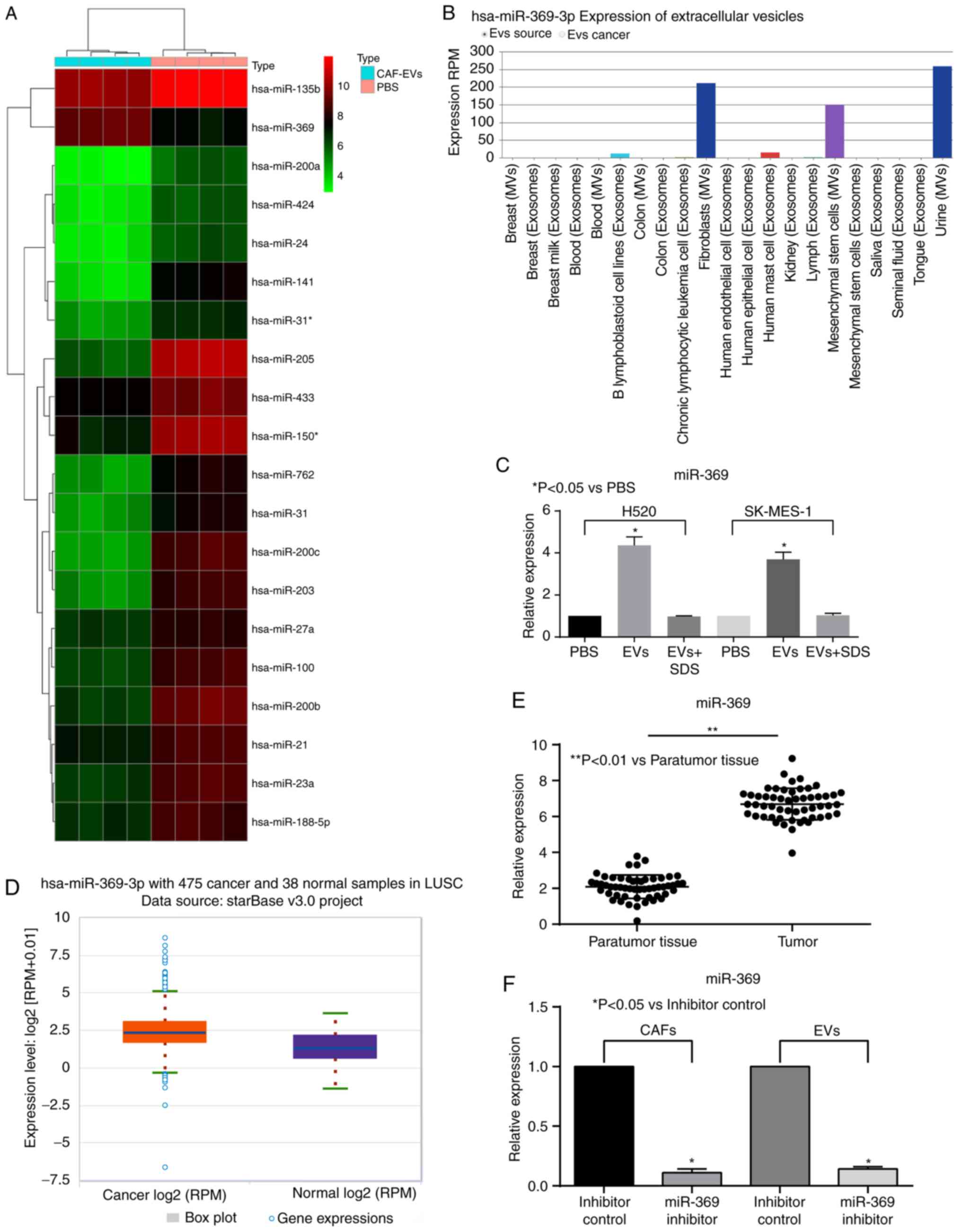
Expression of miR-369 in LUSC cells is
enhanced following treatment with CAFs-EVs
Then, a miRNA microarray analysis was used to
examine the miRNA expression of H520 cells treated with EVs. It was
identified that miR-369 increased significantly following EVs
treatment. It was revealed that miR-369 was highly expressed in the
EVs extracted from CAFs in the bioinformatics database EvimiR
(Fig. 3A and B). The expression
levels of miR-369 in H520 and SK-MES-1 cells were then detected by
RT-qPCR, which was enhanced by treatment with EVs (Fig. 3C). In addition, according to TCGA
database, miR-369 was significantly overexpressed in patients with
LUSC (Fig. 3D). Therefore, the
miR-369 expression levels were detected in 52 cases of LUSC tissues
and the corresponding paratumor tissues. miR-369 was observed to be
highly expressed in the LUSC tissues (Fig. 3E). The expression of miR-369 was
then downregulated in CAFs and the EVs were extracted for
subsequent experiments. RT-qPCR was used to verify the transfection
efficiency (Fig. 3F).
Downregulated miR-369 carried by CAFs-EVs
partially inhibits the malignancy of LUSC cells
Following the treatment of miR-369 inhibitor in the
CAFs-EVs, the malignant biological behaviors of H520 and SK-MES-1
cells promoted by EVs were significantly decreased (Fig. 4A-F).
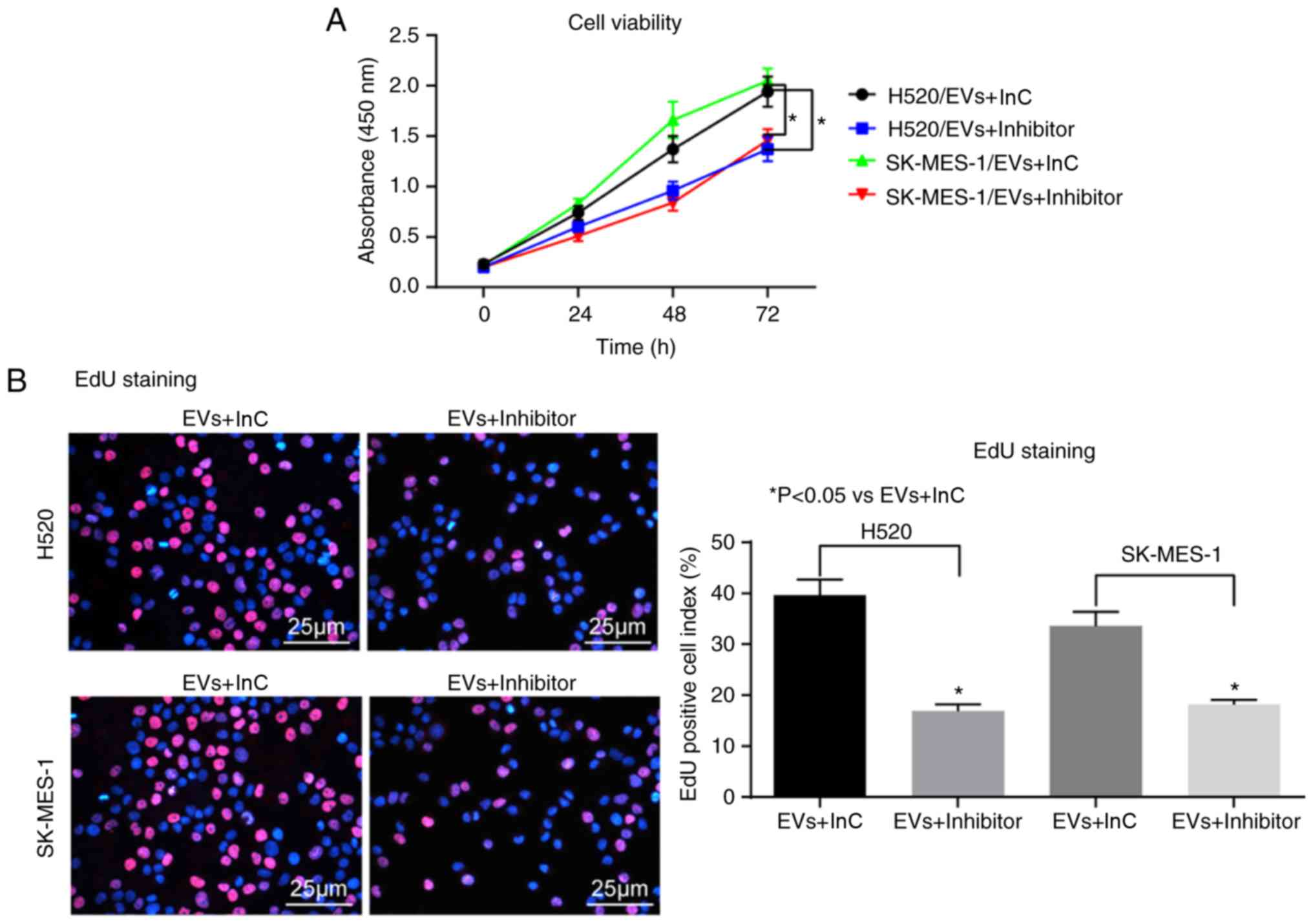 | Figure 4miR-369 inhibition in CAFs-derived
EVs partially reverses the effects of EVs on LUSC cells malignancy.
(A) The concentration of EVs determined by Cell Counting Kit-8
assay. (B) The viability of H520 and SK-MES-1 cells assessed by EdU
assay. (C) Immunofluorescence staining of EMT marker E-cadherin in
H520 and SK-MES-1 cells. (D) Immunofluorescence staining of EMT
marker N-cadherin in H520 and SK-MES-1 cells. (E) H520 and SK-MES-1
cell migration ability measured by Transwell assay. (F) H520 and
SK-MES-1 cell invasion ability measured by Transwell assay. Data
are expressed as mean ± SD. In panel A, two-way ANOVA and Tukey's
multiple comparisons test was used to determine statistical
significance. In panel B to F, one -way ANOVA was used.
*P<0.05 vs. EVs + InC treatment. miR, microRNA; CAFs,
cancer-associated fibroblasts; EVs, extracellular vesicles; EdU,
5-ethynyl-2′-deoxyuridine; EMT, epithelial-mesenchymal transition;
E-cadherin, epithelial cadherin; N-cadherin, neural cadherin; InC,
Inhibitor control; ANOVA, analysis of variance. |
miR-369 targets NF1 to activate the MAPK
signaling pathway
A large number of mRNAs that were targeted by
miR-369 were screened using the StarBase and TargetScan databases.
A Kyoto Encyclopedia of Genes and Genomes enrichment analysis was
performed for the 471 mRNAs identified, and a total of 61 enriched
signaling pathways were identified. The MAPK signaling pathway was
screened out, and NF1 was located upstream of the pathway (Fig. 5A-C). It was reported previously
that the NF1 mutation could promote or lead to the occurrence of
lung adenocarcinomas (16).
Therefore, a dual luciferase assay was used to validate the binding
connection between miR-369 and NF1 (Fig. 5D). Subsequently, the expression of
NF1 in the LUSC tissues and the corresponding paratumor tissues was
examined, and the NF1 mRNA expression in the LUSC tissues was
significantly decreased (Fig.
5E). Then, the expression of NF1, MEK1 and ERK1/2, and the
extent of MEK1 and ERK1/2 phosphorylation in cells, were detected
by ELISAs. It was identified that CAFs-EVs treatment could inhibit
NF1 protein expression and increase the extent of MEK1 and ERK1/2
phosphorylation. Then, miR-369 expression in EVs was inhibited
using transfection, and it was observed that that the expression of
NF1 was significantly increased, while the MEK1 and ERK1/2
phosphorylation levels significantly decreased (Fig. 5F).
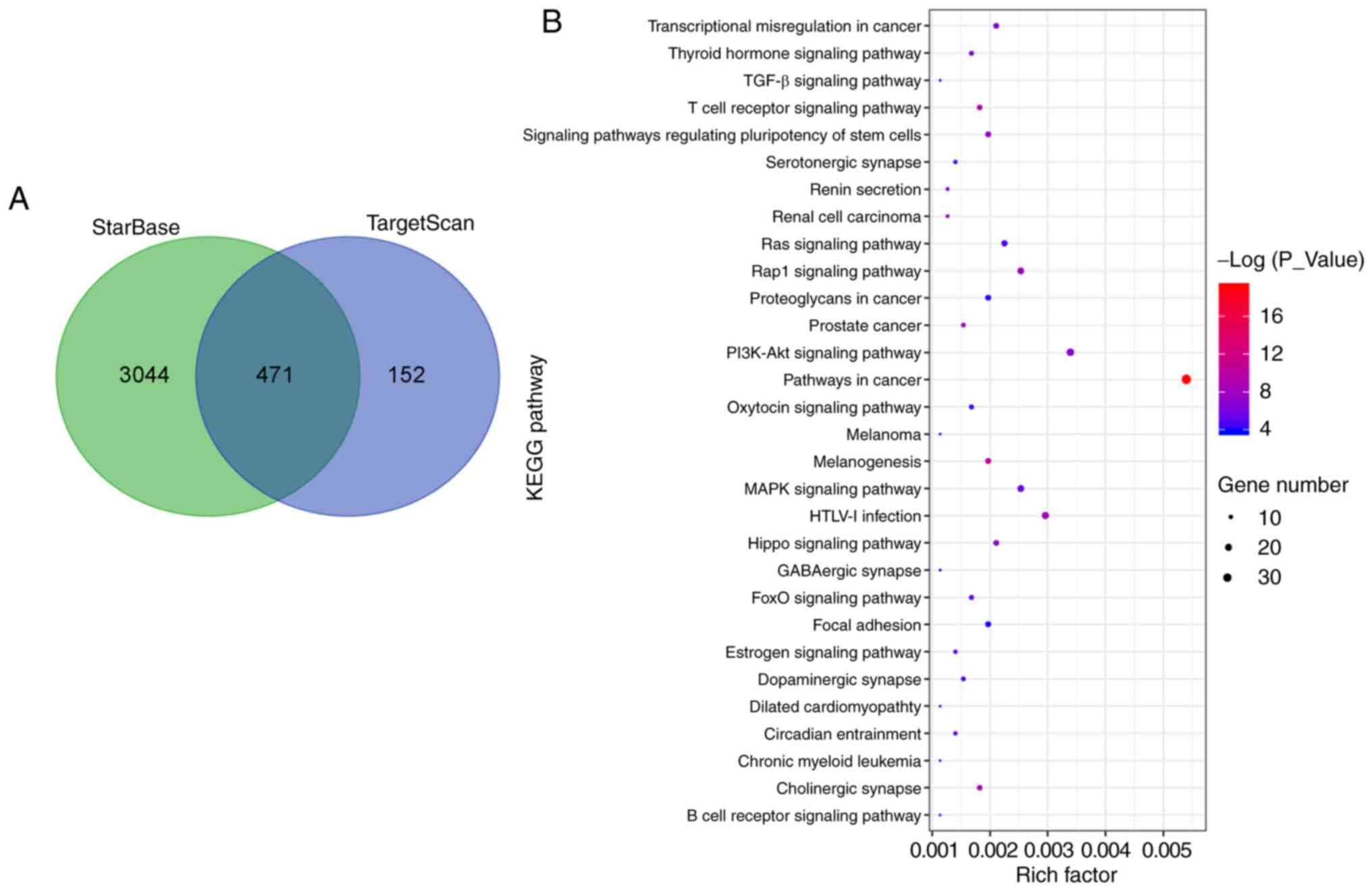 | Figure 5miR-369 activates the MAPK signaling
pathway by targeting the tumor suppressor NF1. (A) The mRNA targets
of miR-369 were predicted by TargetScan and StarBase, and 471 mRNA
was screened out. (B) Signaling pathways enriched by targeting
mRNAs of miR-369 analyzed by KEGG. (C) The MAPK signaling pathway
enriched by targeting mRNAs of miR-369 analyzed by KEGG. (D)
Predicted miRNA binding sites within the 3'-UTR of NF1 mRNA by
StarBase. The pairing between miR-369 and the putative binding
sites in the 3'-UTR of APC mRNA are shown. Data were analyzed using
a one-way ANOVA followed by Tukey's post hoc test. (E) The NF1
expression levels in 52 LUSC tumor tissues and paratumor tissues
were measured by reverse transcription-quantitative PCR. Data were
analyzed using a paired t-test. (F) The protein expression levels
of NF1, MEK1, pMEK1, ERK1/2 and pERK1/2 were determined by ELISA.
Data are expressed as mean ± SD. Data were analyzed using a one-way
ANOVA followed by Tukey's post hoc test. *P<0.05 vs.
PBS treatment. #P<0.05 vs. EVs + InC treatment.
**P<0.01 compared with paratumor tissues. miR,
microRNA; NF1, neurofibromin 1; UTR, untranslated region; MEK1,
mitogen-activated protein kinase/ERK kinase 1; ERK1/2,
extracellular signal-regulated kinase 1/2; p, phosphorylated; InC,
inhibitor control; KEGG, Kyoto Encyclopedia of Genes and
Genomes. |
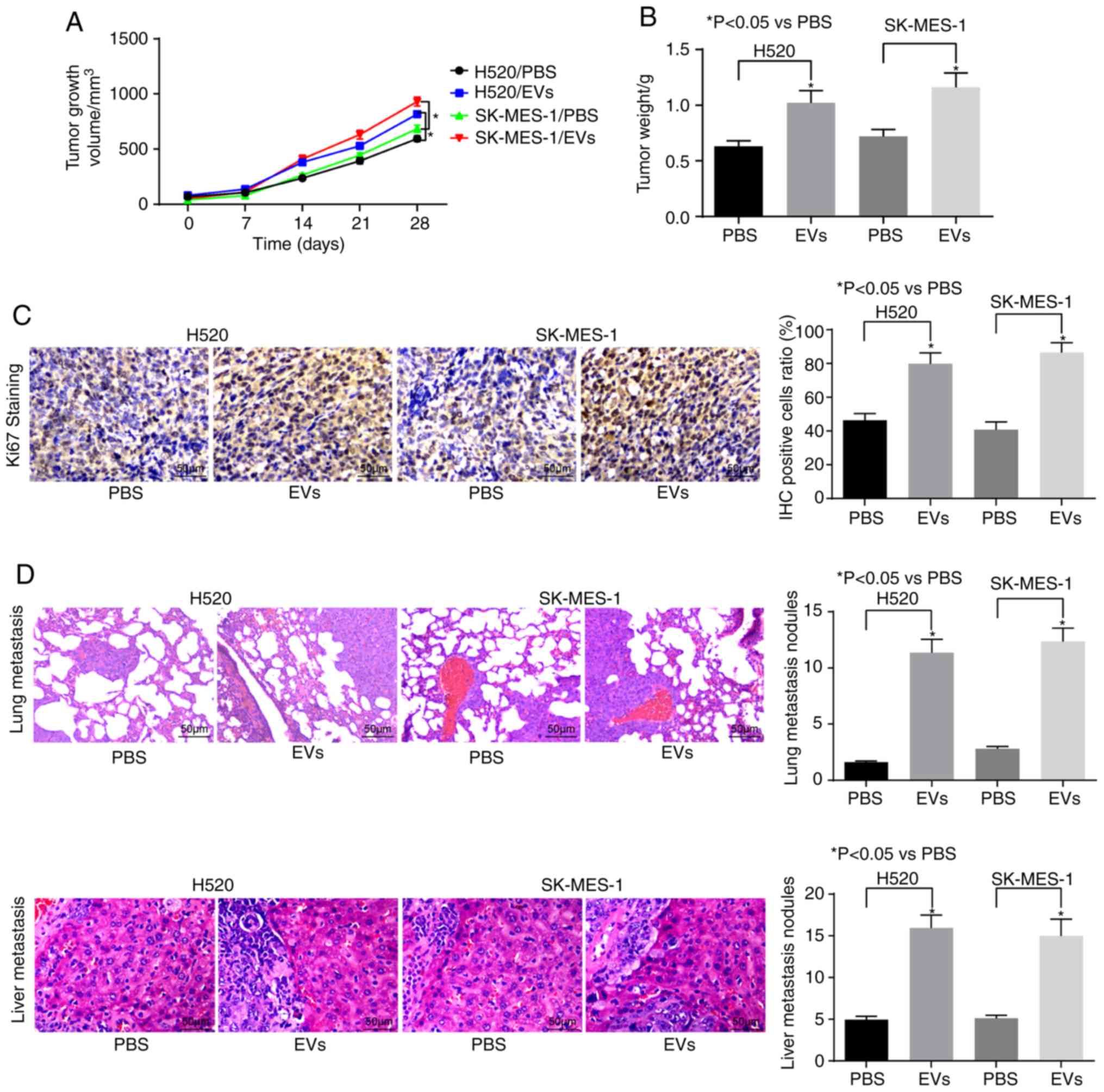
CAFs-EVs treatment promotes the growth
and metastasis of LUSC in vivo
With the aim of verifying the role of CAFs-derived
EVs on the development and metastasis of LUSC in vivo, in
vivo tumorigenesis and in vivo metastasis models were
established in nude mice. In the in vivo tumorigenesis
experiment, it was identified that EVs treatment could promote the
tumor volume and weight in vivo and increase the
Ki-67-positive cell rate of transplanted tumors (Fig. 6A-C). Furthermore, in the in
vivo metastasis model, it was identified that the number of
liver metastases and lung metastases increased significantly
following EVs treatment (Fig.
6D).
Discussion
Cancer is closely associated with fibroblast
activity at all stages of disease progression, including
metastasis, and CAFs may responsible for the generation of various
tumor components (27). The
primary result of the present study is the marked upregulation of
miR-369 in LUSC tissues compared with the paratumor tissues. In
addition, miR-369 inhibition in the CAF-derived EVs decreased LUSC
cell proliferation, migration and invasion in vitro, and
lung and liver metastases in vivo. Additionally, it was
demonstrated that miR-369 in CAF-derived EVs activated the MAPK
signaling pathway by directly binding to NF1. These results suggest
that the dysregulation of miR-369 may serve a paramount role in
accelerating tumorigenesis and metastases in LUSC.
According to EvimiR, an important miRNA database,
miR-369 was significantly upregulated in CAF-derived EVs. EVs have
been demonstrated to carry nucleic acids including miRNA, mRNA and
non-coding RNAs (28). Baffa
et al (29) observed that
miR-369-3p was overexpressed in metastatic NSCLC tissues. The
results of the present study demonstrated an association between
the upregulation of miR-369 expression in CAF-EVs and LUSC cell
migration, invasion and tumor progression, and suggested that the
NF1-mediated MAPK signaling pathway may be involved. By using the
publicly available databases StarBase and TargetScan, a conserved
binding site of miR-369 on the 3′UTR of NF1 gene was identified,
which was further confirmed by luciferase reporter assays.
Therefore, miR-369 may act as a positive regulator of LUSC cell
migration and invasion via specific down-regulation of NF1. NF1 was
reported to be highly expressed in NSCLC tissues and A549 and
HCC823 cells compared with the controls (30). Aberrations in NF1 contribute to
the dysregulation of the RAS/MAPK signaling pathway, culminating in
disfunction of cell growth and proliferation (31).
Furthermore, CAFs-EVs promoted the migration,
invasion and EMT of LUSC cells in vitro in the present
study. Exosomes extracted from the human sera of samples from
late-stage lung cancer enhanced vimentin expression and stimulated
the migration, invasion and EMT of human bronchial epithelial cells
(32). In addition, CAFs induced
EMT in lung cancer cells via exosomes in a zinc finger protein
SNAI1-dependent manner (33).
Exosomes containing miR-499a released from a highly metastatic cell
line increased cell proliferation, migration and EMT in lung
adenocarcinoma samples, and the trends were reversed by the
suppression of miR-499a-5p (34).
Vimentin has been confirmed to participate in cancer tumorigenesis,
EMT and cellular metastatic properties (32). Notably, CAFs-EVs exhibited
stimulatory effects on the growth of the H520 and SK-MES-1 cell
lines in vivo, suggesting that the inhibition of CAFs-EVs
may be a potential therapeutic strategy in LUSC. Consistent with
the data from the present study, Verset et al (35) provided quantitative data
demonstrating the increased expression of CAFs in the
cancer-associated stroma in rectal cancer. Exosomes antagonized the
protective effect of mesothelial cells and facilitated the
metastasis of tumor cells, particularly in fluid ascites, implying
that exosomes may induce the transformation of mesothelial cells
into CAFs to promote metastasis (36). Similar to the results of the
present study, melanoma cells were demonstrated to control the
creation of the dermal tumor niche by inducing EVs overexpressing
miR-211, which directly interacted with the insulin-like growth
factor 2 receptor, contributing to the potentiation of the MAPK
signaling pathway that encourages melanoma cell growth (37). In addition, gastric cancer
cell-induced exosomes promoted the proliferation and migration of
pericytes and enhanced the CAFs marker expression in pericytes,
during which the MEK/ERK pathway was activated by tumor-derived
exosomes (38). The present study
demonstrated that NF1 was expressed at decreased levels in the LUSC
tissues compared with the matched paratumor tissues, while the
treatment of EVs with the miR-369 inhibitor significantly enriched
the expression of NF1 and decreased the extent of MEK1 and ERK1/2
phosphorylation.
In summary, the present study identified miR-369 as
a novel potential oncogene in LUSC that acts by inducing EMT and
the metastasis process. It was observed that the silencing of
miR-369 in CAF-EVs significantly inhibited tumor growth and
metastasis in vivo, suggesting that miR-369 inhibition may
be a capable therapeutic target for LUSC through CAF-EVs. Further
studies should examine the association between EVs and LUSC cells
further, which may optimize the current therapeutic strategies for
LUSC.
Funding
This study was supported by the University Level
Scientific Research Project of Yan'an University (grant nos.
YDQ2019-35, D2017197 and YDJGY19-02).
Availability of data and materials
All the data generated or analyzed during this study
are included in this published article.
Authors' contributions
LG contributed to the conception and design of the
study. BL contributed to the experiments and clinical studies. JY
contributed to the data and statistical analyses. JS was involved
in manuscript preparation and acquisition of experimental data. JJ
and MM contributed to analysis and interpretation of data, and
reviewed the manuscript. All authors are responsible for the
integrity of the data and have read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were completed following the
protocol that was approved by the Animal Care and Use Committee of
Affiliated Hospital of Yan'an University. All efforts were made to
minimize the number of animals applied in the experiments and their
suffering. All human tissue donors provided written informed
consent prior to tissue donation. The study protocol was approved
by the Institutional Review Board of Affiliated Hospital of Yan'an
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The results published here are in whole or part
based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Siegel RL and Jemal A: Lung
Cancer Statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar
|
|
3
|
Xu F, Zhang H, Chen J, Lin L and Chen Y:
Immune signature of T follicular helper cells predicts clinical
prognostic and therapeutic impact in lung squamous cell carcinoma.
Int Immunopharmacol. 81:1059322020. View Article : Google Scholar
|
|
4
|
Shi Y, Li Y, Yan C, Su H and Ying K:
Identification of key genes and evaluation of clinical outcomes in
lung squamous cell carcinoma using integrated bioinformatics
analysis. Oncol Lett. 18:5859–5870. 2019.PubMed/NCBI
|
|
5
|
Erez N, Truitt M, Olson P, Arron ST and
Hanahan D: Cancer-associated fibroblasts are activated in incipient
neoplasia to orchestrate tumor-promoting inflammation in an
NF-kappaB-dependent manner. Cancer Cell. 17:135–147. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hanley CJ, Mellone M, Ford K, Thirdborough
SM, Mellows T, Frampton SJ, Smith DM, Harden E, Szyndralewiez C,
Bullock M, et al: Targeting the Myofibroblastic Cancer-Associated
Fibroblast Phenotype Through Inhibition of NOX4. J Natl Cancer
Inst. 110:2018. View Article : Google Scholar :
|
|
7
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang X, Li Y, Zou L and Zhu Z: Role of
exosomes in crosstalk between cancer-associated fibroblasts and
cancer cells. Front Oncol. 9:3562019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun LP, Xu K, Cui J, Yuan DY, Zou B, Li J,
Liu JL, Li KY, Meng Z and Zhang B: Cancer-associated
fibroblast-derived exosomal miR3825p promotes the migration and
invasion of oral squamous cell carcinoma. Oncol Rep. 42:1319–1328.
2019.PubMed/NCBI
|
|
10
|
Hao GJ, Ding YH, Wen H, Li XF, Zhang W, Su
HY, Liu DM and Xie NL: Attenuation of deregulated miR-369-3p
expression sensitizes non-small cell lung cancer cells to cisplatin
via modulation of the nucleotide sugar transporter SLC35F5. Biochem
Biophys Res Commun. 488:501–508. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42(Database issue): D92–D97. 2014. View Article : Google Scholar
|
|
12
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
13
|
Tlemsani C, Pecuchet N, Gruber A,
Laurendeau I, Danel C, Riquet M, Le Pimpec-Barthes F, Fabre E,
Mansuet-Lupo A, Damotte D, et al: NF1 mutations identify molecular
and clinical subtypes of lung adenocarcinomas. Cancer Med.
8:4330–4337. 2019.PubMed/NCBI
|
|
14
|
Brems H, Park C, Maertens O, Pemov A,
Messiaen L, Upadhyaya M, Claes K, Beert E, Peeters K, Mautner V, et
al: Glomus tumors in neurofibromatosis type 1: Genetic, functional,
and clinical evidence of a novel association. Cancer Res.
69:7393–7401. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stites EC, Trampont PC, Haney LB, Walk SF
and Ravichandran KS: Cooperation between Noncanonical Ras Network
Mutations. Cell Rep. 10:307–316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Min S, Liu H, Wu N, Liu X, Wang T,
Li W, Shen Y, Wang H, Qian Z, et al: Nf1 loss promotes Kras-driven
lung adenocarcinoma and results in Psat1-mediated glutamate
dependence. EMBO Mol Med. 11:e98562019. View Article : Google Scholar
|
|
17
|
Redig AJ, Capelletti M, Dahlberg SE, Sholl
LM, Mach S, Fontes C, Shi Y, Chalasani P and Jänne PA: Clinical and
molecular characteristics of NF1-mutant lung cancer. Clin Cancer
Res. 22:3148–3156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Modak JM, Roy-O'Reilly M, Zhu L, Staff I
and McCullough LD: Differential MicroRibonucleic acid expression in
cardioembolic stroke. J Stroke Cerebrovasc Dis. 28:121–124. 2019.
View Article : Google Scholar
|
|
19
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar :
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Zhang Y, Guo L, Li Y, Feng GH, Teng F, Li
W and Zhou Q: MicroRNA-494 promotes cancer progression and targets
adenomatous polyposis coli in colorectal cancer. Mol Cancer.
17:12018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ji Y, Han Z, Shao L and Zhao Y: Evaluation
of in vivo antitumor effects of low-frequency ultrasound-mediated
miRNA-133a microbubble delivery in breast cancer. Cancer Med.
5:2534–2543. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei S, Peng L, Yang J, Sang H, Jin D, Li
X, Chen M, Zhang W, Dang Y and Zhang G: Exosomal transfer of
miR-15b-3p enhances tumorigenesis and malignant transformation
through the DYNLT1/Caspase-3/Caspase-9 signaling pathway in gastric
cancer. J Exp Clin Cancer Res. 39:322020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang XY, Huang ZL, Huang J, Xu B, Huang
XY, Xu YH, Zhou J and Tang ZY: Exosomal circRNA-100338 promotes
hepatocellular carcinoma metastasis via enhancing invasiveness and
angiogenesis. J Exp Clin Cancer Res. 39:202020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu Y, Liu H, Shi X, Yao Y, Yang W and Song
Y: The long non-coding RNA HNF1A-AS1 regulates proliferation and
metastasis in lung adenocarcinoma. Oncotarget. 6:9160–9172. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thery C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extra-cellular vesicles 2018 (MISEV2018): A position statement of
the International Society for Extracellular Vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar
|
|
27
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ab Razak NS, Ab Mutalib NS, Mohtar MA and
Abu N: Impact of chemotherapy on extracellular vesicles:
Understanding the chemo-EVs. Front Oncol. 9:11132019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baffa R, Fassan M, Volinia S, O'Hara B,
Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM and
Rosenberg A: MicroRNA expression profiling of human metastatic
cancers identifies cancer gene targets. J Pathol. 219:214–221.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qian B, Wang DM, Gu XS, Zhou K, Wu J,
Zhang CY and He XY: LncRNA H19 serves as a ceRNA and participates
in non-small cell lung cancer development by regulating
microRNA-107. Eur Rev Med Pharmacol Sci. 22:5946–5953.
2018.PubMed/NCBI
|
|
31
|
Philpott C, Tovell H, Frayling IM, Cooper
DN and Upadhyaya M: The NF1 somatic mutational landscape in
sporadic human cancers. Hum Genomics. 11:132017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rahman MA, Barger JF, Lovat F, Gao M,
Otterson GA and Nana-Sinkam P: Lung cancer exosomes as drivers of
epithelial mesenchymal transition. Oncotarget. 7:54852–54866. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
You J, Li M, Cao LM, Gu QH, Deng PB, Tan Y
and Hu CP: Snail1-dependent cancer-associated fibroblasts induce
epithelial-mesenchymal transition in lung cancer cells via
exosomes. QJM. 112:581–590. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
He S, Li Z, Yu Y, Zeng Q, Cheng Y, Ji W,
Xia W and Lu S: Exosomal miR-499a-5p promotes cell proliferation,
migration and EMT via mTOR signaling pathway in lung
adenocarcinoma. Exp Cell Res. 379:203–213. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Verset L, Tommelein J, Moles Lopez X,
Decaestecker C, Boterberg T, De Vlieghere E, Salmon I, Mareel M,
Bracke M, De Wever O and Demetter P: Impact of neoadjuvant therapy
on cancer-associated fibroblasts in rectal cancer. Radiother Oncol.
116:449–454. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wei M, Yang T, Chen X, Wu Y, Deng X, He W,
Yang J and Wang Z: Malignant ascites-derived exosomes promote
proliferation and induce carcinoma-associated fibroblasts
transition in peritoneal mesothelial cells. Oncotarget.
8:42262–42271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng X, Bahr M and Doeppner TR: From
tumor metastasis towards cerebral ischemia-extracellular vesicles
as a general concept of intercellular communication processes. Int
J Mol Sci. 20:E59952019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ning X, Zhang H, Wang C and Song X:
Exosomes released by gastric cancer cells induce transition of
pericytes into cancer-associated fibroblasts. Med Sci Monit.
24:2350–2359. 2018. View Article : Google Scholar : PubMed/NCBI
|