Introduction
Juvenile idiopathic arthritis (JIA) is the most
common childhood rheumatic disease and is highly debilitating. The
disease often persists into adulthood, causing obvious functional
disabilities, including joint deformation, abnormal growth and
development, osteoporosis, pain, psychological abnormalities and
difficulty with self-care (1-3).
According to the International League of Associations for
Rheumatology criteria, there are seven types of JIA. One of these
specific types is rheumatoid factor-positive poly-arthritis
(RF-positive pJIA), which is defined by the presence of arthritis
in >4 joints and a positive rheumatoid factor (4-6).
RF-positive pJIA and adult rheumatoid arthritis (RA) have similar
clinical manifestations, as well as serological and immunogenetic
profiles. Patients with RA exhibit an activation of cells secreting
pro-inflammatory interleukin (IL)-17 cytokine (Th17), the
activation of which is normally suppressed by regulatory T
lymphocytes (Tregs) (7,8). An imbalance in the Th17/Treg cell
ratio along with disruptions in the cytokine environment have been
reported to be involved in synovial hyperplasia and joint
destruction in patients with RA (9,10).
A number of in vivo studies have demonstrated that primary
CD4+ T cells can differentiate into different subtypes
of helper T (Th) cells under the regulation of various antigens,
cytokines and other factors. Alterations in the Th cell subgroup
ratio plays a key role in the immunopathology of JIA (11-15). However, the process surrounding
the shift in the ratio of Th17/Treg cells is complex and dynamic,
and the specific mechanisms involved remain unclear.
Long non-coding RNAS (lncRNAs) play an important
role in biological processes and disease development by regulating
chromosome recombination, gene modification, gene transcription,
post-transcriptional modification and other mechanisms (16-18). The importance of lncRNAs has been
studied in the immune system. Specifically, lncRNA insulin receptor
precursor (INSR) has been shown to function through an
INSR-independent mechanism to enhance Treg differentiation and
promote immune suppression in the immune microenvironment of
pediatric acute lymphoblastic leukemia (ALL) (19). The involvement of lncRNAs in the
development and differentiation of CD4+ T cells has also
been reported (20). For example,
lncRNA Tmevpg1 has been reported to be specifically expressed by
the Th1 phenotype via T-bet, a T-box transcription factor (21). linc-MAF-4 has also been confirmed
to be a chromatin-associated lncRNA that is specific to the Th1
subtypes (22). However, the
specific mechanisms through which lncRNAs mediate immune
abnormalities and promote the development of JIA remain
unclear.
In the present study, sequence-based screening was
conducted in patients with JIA and healthy volunteers to explore
potential interactions between lncRNAs and mRNAs. A specific
lncRNA, RP11-340F14.6, was identified. It was reported that this
lncRNA induced the expression of P2X7R and may promote the immune
microenvironment that is associated with JIA.
Materials and methods
Clinical samples
Blood samples were obtained through the Department
of Children's Healthcare from 30 healthy volunteers with no
personal or family history of chronic autoimmune, cancer,
metabolic, or infectious diseases. The volunteers included 9 males
and 21 females, with an average age of 8.82±3.77 years. Blood
samples from 30 RF-positive patients with JIA were obtained between
May, 2017 and May, 2019 including 11 males and 19 females, with an
average age of 8.64±3.58 years at the Children's Hospital of
Nanjing Medical University. Peripheral lymphocytes were isolated
from blood samples. In brief, peripheral blood was collected from
all patients before receiving any therapeutic drugs, and blood was
collected from the healthy controls during a physical examination.
Peripheral lymphocytes were isolated by adopting the Ficoll-Hypaque
density gradient centrifugation method. Children who had previously
received disease-modifying anti-rheumatic drug (DMARD) therapy or
steroid therapy were excluded. Clinical characteristics were
classified according to the detailed diagnostic information
obtained from the medical records and physical examinations. All
experiments were performed in compliance with government policies
and the Helsinki Declaration. All patients or healthy controls had
the consent of their legal guardians or parents who signed an
informed consent form before collecting blood samples. The present
study was approved by the Ethics Committee of the Children's
Hospital of Nanjing Medical University.
Cell culture
Human T cells were filtered through a 75 µm
strainer and separated by Ficoll centrifugation (800 × g for 20 min
at 4°C). The mononuclear cells were resuspended in RPMI-1640
supplemented with 10% FBS. The anti-CD4 magnetic Dynabeads
(Invitrogen; Thermo Fisher Scientific, Inc.) were applied to sort
the T cells.
Microarray detection
Total RNA was isolated from 1×106 T cells and used for the lncRNA/mRNA
integrated microarray analysis (CapitalBio). Each group included 3
samples. Sample preparation and microarray hybridization were
performed according to the manufacturer's instructions with minor
modifications. Briefly, mRNA was purified from total RNA following
the removal of rRNA (using the mRNA-ONLY™ Eukaryotic mRNA Isolation
kit, EPICENTRE Biotechnologies), amplified and transcribed into
fluorescent cRNA along the entire length of the transcripts without
3' bias utilizing a random priming method. The arrays were scanned
using an Agilent Scanner (Agilent Technologies, Inc.). Agilent
Feature Extraction software (version 11.0.1.1) was used to analyze
the acquired array images. Quantile normalization and subsequent
data processing were performed using the GeneSpring GX v12.0
software package (Agilent Technologies, Inc.). Following quartile
normalization of the raw data, lncRNAs which had flags in present
or marginal ('All Targets Value') were selected for further
analysis. lncRNA expression patterns were revealed via Hierarchical
analysis using Cluster 3.0 software (Stanford University).
Mutagenesis of lncRNA and P2X7R, and
lentiviral packaging
The full-length construct of P2X7R, as well as the
full-length and mutant constructs of RP11-340F14.6 were synthesized
and cloned into pGC-LV plasmid purchased from GenScript Co. Ltd
(Nanjing, China). shRNA technology was employed to knockdown the
target genes or lncRNA. shRNAs targeting RP11-340F14.6 or P2X7R
were designed constructed by GenScript Co. Ltd. and were cloned
into the PLL.3.7 vector purchased from GenScript Co. Ltd. and
further packaged to produce lentiviral particles, as previously
described (23).
Transfection
The target vectors (20 µg) were mixed with
lentiviral packaging 15 µg Δ8.91 (GenScript Co. Ltd.) and
envelope expressing 10 µg VSV-G (GenScript Co. Ltd.)
plasmids to generate lentiviral particles in 1.2×107/20 ml 293T cells (ATCC) using 100
µl Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). Viral particles were concentrated by ultracentrifugation and
expression vector titers were determined. The plasmids were
constructed and transfected using lentivirus. Cells were cultured
with 100 µg/ml of human-derived IL-2. The 24-well plates
were placed in a cell incubator at 37°C 5% CO2 and
cultured for 48 h. Cells increased in volume after 24 h of
stimulation, indicating activation, and were transfected with the
lentivirus after 48 h of stimulation. Transfection was performed at
a multiplicity of infection (MOI) of 100, using polyberene at a
concentration of 5 µg/ml. When the cells reached the optimal
transfection state 72 h later, to transfected cells were screened
using puromycin. The transfection efficiency was determined by
fluorescence intensity and RT-PCR assay.
Flow cytometry
Following CD4+ T cell enrichment, the
cells were incubated with human anti-CD3-FITC (cat. no. 557832) and
anti-CD4-PerCP monoclonal antibodies (mAbs, cat. no. 564419) in 4°C
for 30 min (BD Pharmingen). Cells were fixed and permeabilized with
Cytofix/Cytoperm (cat. no. 56422 Human Fc Block from BD Pharmingen)
and then intracellularly stained with IL-17A-Phycoerythrin
(IL-17A-PE) or IgG-PE as an isotype control. To detect the Treg
cell frequency, cells were labeled with anti-CD4-FITC and
anti-CD25-APC antibodies (cat. no. 555434, BD Pharmingen).
Following fixation and permeabilization, the cells were stained
using anti-forkhead box protein 3 (Foxp3)-PE mAb (cat. no. 560046,
BD Pharmingen) or IgG-PE control at 4°C for 30 min and were then
analyzed using a BD FACSCanto II flow cytometer (BD Pharmingen).
The data were analyzed using Cell Quest analysis software, version
5.1 (BD Pharmingen).
RNA immunoprecipitation (RIP) assay
RIP assay was carried out using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore), as
previously described (24).
Anti-HA antibodies (1:50, ab9110; Abcam) were used for RIP. T cells
were either transduced with fixed or varying doses of lentivirus
containing RP11-340F14.6 along with lentivirus containing P2X7R.
The coprecipitated RNAs were detected by reverse transcription PCR
and quantitative (real-time) PCR. Total RNA (input control) and IgG
were assayed simultaneously.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from the cells using TRIzol
reagent (Invitrogen Life Technologies; Thermo Fisher Scientific,
Inc.) and purified using the RNeasy MinElute Clean up kit (Qiagen)
according to the manufacturer's instructions. cDNA was synthesized
from total RNA using the random priming method using the One step
PrimeScript kit (RR064A, Takara Bio, Inc.). Transcript levels were
measured in duplicate by qPCR (ABI 7900; Life Technologies; Thermo
Fisher Scientific, Inc.). The primer for RP11-340F14.6 was as
follows: Forward, 5'-GCC AAG CTT CTT GAA AGG CC-3' and reverse,
5'-TTC CAC GGA GTA GAG CGA GTC-3'. Primer sequences synthesized by
GenScript Co. Ltd. The amplification procedure was 95°C for
pre-denaturation for 30 sec; 95°C for 5 sec, 60°C for 31 sec (45
cycles); dissolution curves 95°C for 15 sec, 60°C for 60 sec, 95°C
15 sec. The relative expression of lncRNA and mRNA was normalized
to GAPDH and was calculated using the 2−ΔΔCq method as
previously described (25). The
primer sequences for GAPDH were as follows: Forward, 5'-AAG GTG AAG
GTC GGA GTC AAC-3' and reverse, 5'-GGG GTC ATT GAT GGC AAC AAT
A-3'.
Western blot analysis
Whole cell lysates were prepared as previously
described (24). Protein was
extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology). Protein concentration was determined by the BCA
method (Beyotime Institute of Biotechnology). Equal amounts of
proteins (20 µg) were boiled, separated on 10% SDS-PAGE and
transferred onto PVDF membranes. After blocking with 5% (w/v)
non-fat dry milk, the membranes were probed with the primary
antibody overnight at 4°C. The secondary antibodies were both
horseradish peroxidase (HRP)-conjugated IgG including anti-mouse
IgG (ab97040), anti-rabbit IgG (ab7090) and anti-sheep IgG (ab6747)
(all from Abcam) and used at a dilution of 1:1,000 and incubation
at room temperature for 1 h. Signals were detected by the
chemiluminescence procedure (Pierce; Thermo Fisher Scientific,
Inc.) with BioMax films (Kodak) and visualized using an ECL kit
(EMD Millipore). GAPDH was applied as the reference protein.
Antibodies, including P2X7R (ab48871), GAPDH (ab181602), HA-tag
(ab18181) were purchased from Abcam and used at a dilution of
1:1,000.
Statistical analysis
Data are presented as the means ± SEM. Differences
between 2 groups were analyzed using the Student's t-test. ANOVA
was performed to evaluate differences between multiple groups
followed by Tukey's post hoc test. Expression experiments were
repeated at least 3 times with samples in triplicates. Pearson's
correlation analysis was used to analyze the correlation between
the expression of RP11-340F14.6 and that of associated factors
[such as retinoic acid-related orphan receptor gamma t (RORγt),
Foxp3 and P2X7R]. Statistical analysis was performed using STATA
10.0 software and presented using GraphPad Prism software (GraphPad
Software). In all cases, P<0.05 was considered to indicate a
statistically significant difference.
Results
Abnormal shift in Th17/Treg in ratio in
immune microenvironment in JIA
It has been previously demonstrated that an
imbalance in the immune microenvironment is highly associated with
the risk of developing JIA, particularly as regards T cell
reprogramming. In the present study, peripheral blood T cells were
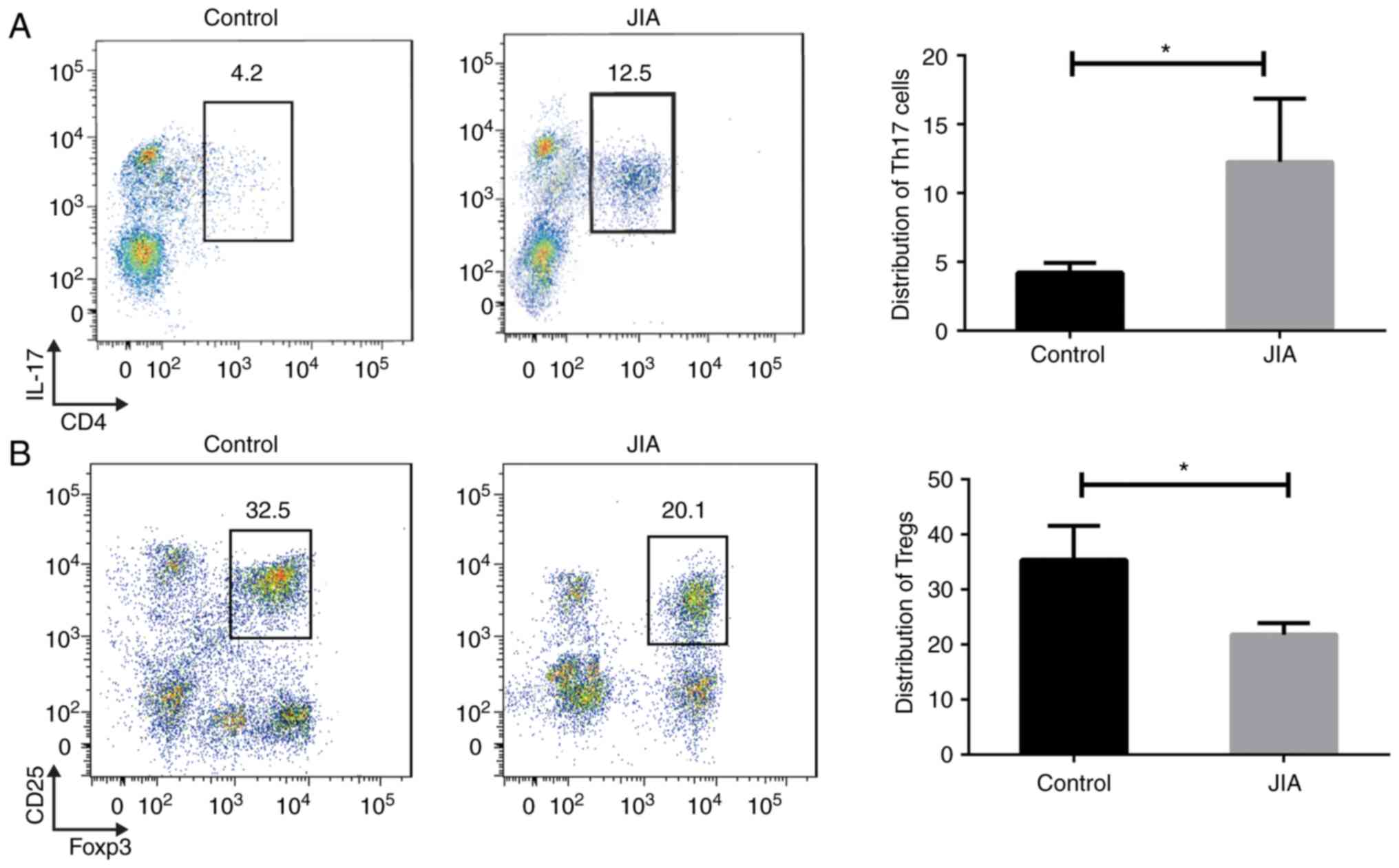
first extracted from patients with JIA and healthy controls. The
distribution of human Th17 and Treg immune cells sorted from JIA
blood samples was first analyzed. Using CD4 and IL-17 as markers, a
marked increase was identified in the percentage of Th17 cells
among the total number of T cells in the JIA samples compared to
those from healthy children (Fig.
1A). On the contrary, there was a reduced percentage of Tregs
labeled with CD25 and Foxp3 presented in the children with JIA
compared to the healthy controls (Fig. 1B). These findings suggest an
increase in the Th17/Treg ratio in the immune microenvironment of
children with JIA.
Transcriptome landscape of lncRNA in the
JIA immune microenvironment
Peripheral blood mononuclear cells (PBMCs) were
isolated from the JIA samples and matched with the healthy
controls. Using anti-CD4 magnetic beads, T cells were sorted in
PBMCs that were extracted from 6 pools of paired samples. A
high-throughput microarray of lncRNAs was applied to screen for
differential expression profiles between the JIA and control
samples. The aberrant expression of lncRNA was presented by
hierarchical clustering using a heatmap. The profile of the
differential expression of lncRNAs in T cells of children with JIA
was obtained (Fig. 2A). Among
these, lncRNAs were further filtered using the following criteria:
i) A fold-change cut-off of 4/0.25; ii) Cq value >25 for PCR
detection; iii) detection of at least 75% in all samples. There
were 138 lncRNAs that met these criteria. Finally, 20 of these 138
lncRNAs with the most significant P-values and q values were
labeled as candidates. A larger sample size including 20 JIA and 20
paired controls was used for further validation. Among the 20
candidate lncRNAs, LINC01225 presented no expression and therefore
the data column for this candidate was removed (Fig. 2B). A total of 4 lncRNAs
(LINC00471-001, ZPAS1-002, NEAT1 and RP11-340F14.6) exhibited a
significantly altered expression in the JIA samples compared to the
normal controls, with RP11-340F14.6 expression exhibiting the most
significant difference.
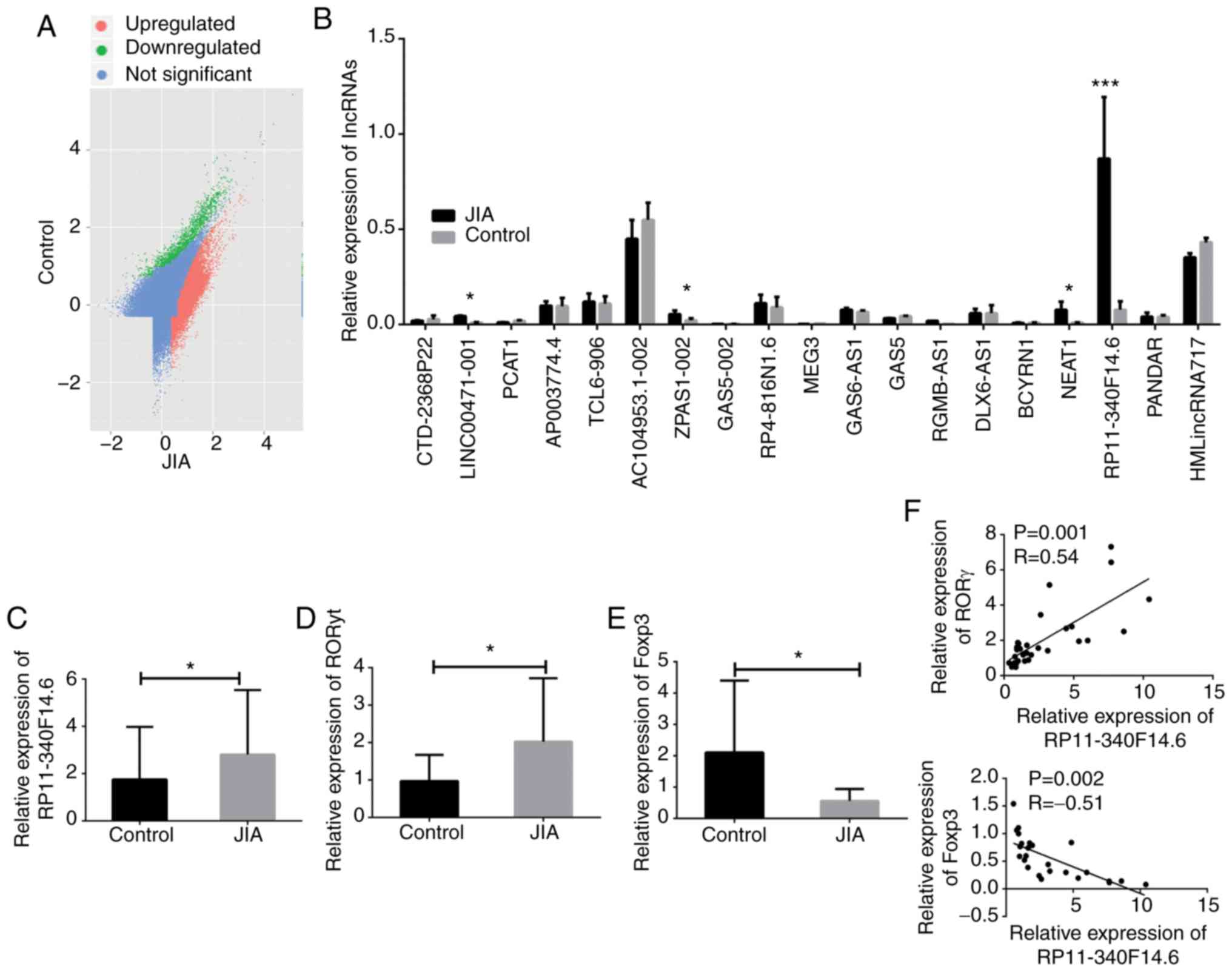 | Figure 2Aberrant expression of RP11-340F14.6
and its association with RORγt and Foxp3. (A) Scatter plot of the
differentially expressed lncRNAs in CD4+ T cells of
patients with JIA and healthy controls. Red does indicate
upregulated lncRNAs, while green dots indicate downregulated
lncRNAs. (B) Relative expression of candidate lncRNAs in human
CD4+ T cell samples, n=20. (C) Relative expression of
RP11-340F14.6 in human CD4+ T cell samples of patients
with JIA and paired controls, n=30. (D) Relative expression of
RORγt in human CD4+ T cell samples of patients with JIA
and paired controls, n=30. (E) Relative expression of Foxp3 in
human CD4+ T cell samples of patients with JIA and
paired controls, n=30. (F) Pearson's correlation analysis of the
correlation between RP11-340F14.6/RORγt and RP11-340F14.6/Foxp3
expression. Data are presented as the means ± SEM,
*P<0.05, ***P<0.001. JIA, juvenile
idiopathic arthritis; RORγt, retinoic acid-related orphan receptor
gamma t; Foxp3, forkhead box protein 3. |
To further investigate the aberrant expression of
RP11-340F14.6 in JIA, a case-control experiment as performed with
30 RNA samples extracted from children with JIA and 30 paired
control samples. An RT-qPCR assay was conducted and the increased
expression of RP11-340F14.6 in patients with JIA was validated
(Fig. 2C). Furthermore, a higher
presentation of RORγt combined with a decreased Foxp3 expression
were observed in the JIA group (Fig.
2D and E). Pearson's correction analysis also revealed that
RP11-340F14.6 expression positively correlated with RORγt
expression and negatively correlated with Foxp3 expression
(Fig. 2F). Based on the
preliminary data, it was thus hypothesized that RP11-340F14.6 may
be associated with the increase in the Th17/Treg ratio in the
immune microenvironment of JIA.
RP11-340F14.6 expression increases the
Th17/Treg ratio in the JIA immune microenvironment
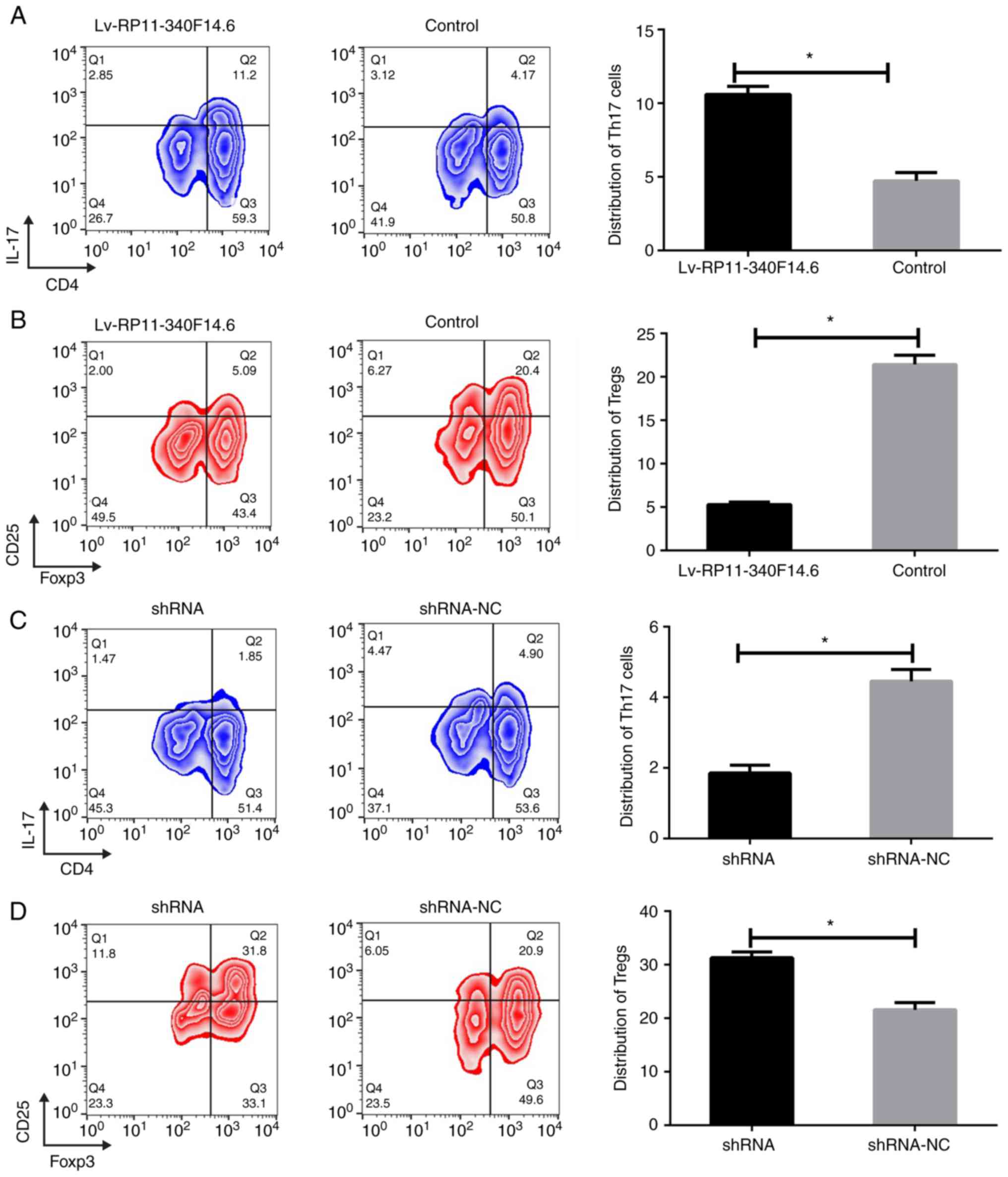
Subsequently, a series of in vitro
experiments were conducted to investigate the function of
RP11-340F14.6 in CD4+ T cells. CD4+ T cells
were sorted by flow cytometry from PBMCs of patients with JIA and
RP11-340F14.6 was overexpressed using a lentivirus in these cells.
It was found that the overexpression of RP11-340F14.6 induced the
expression of IL-17 and increased the percentage of Th17 cells,
which is defined by a CD4-positive cell subgroup (Fig. 3A). The ectopic expression of
RP11-340F14.6 also resulted in a decreased expression of Foxp3 and
a decrease in the number of Tregs which were labeled with CD25
(Fig. 3B). In the
RP11-340F14.6high CD4+ T cells from PBMCs,
endogenous RP11-340F14.6 expression was silenced using an shRNA
lentivirus. The cells in which RP11-340F14.6 was silenced exhibited
a lower percentage of Th17 cells and a greater distribution of
Tregs (Fig. 3C and D).
RP11-340F14.6 increases the Th17/Treg
ratio by specifically binding with P2X7R
According to the results described above, a
functional role of RP11-340F14.6 in JIA was identified. However,
since RP11-340F14.6 has been poorly investigated in human diseases,
the detailed mechanisms of action of this lncRNA remain unclear.
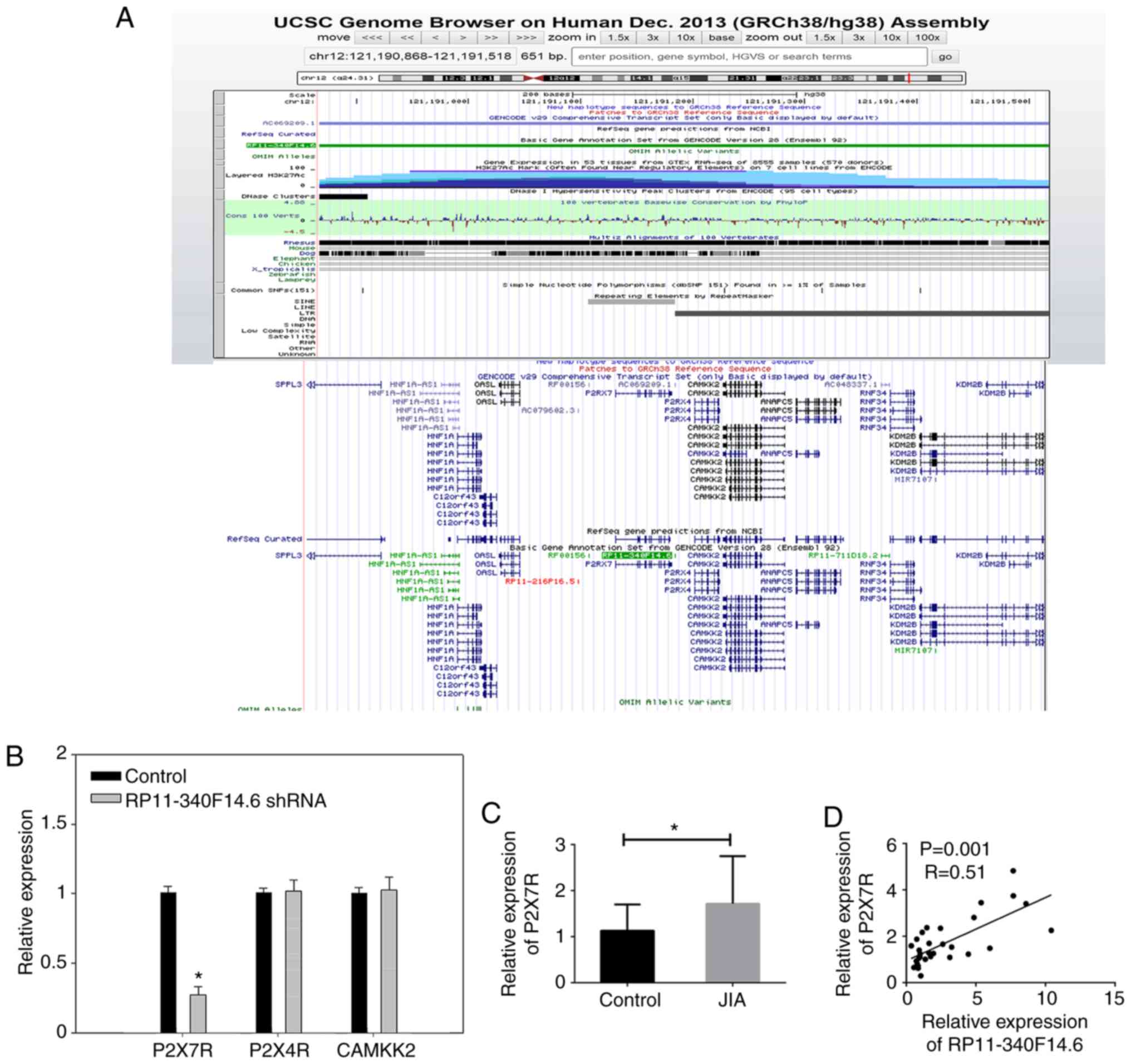
Thus, in the present study, detailed information on RP11-340F14.6
was obtained using the UCSC genome browser (http://genome.ucsc.edu/). Based on the FLANK10K theory
of lncRNA-mRNA interaction (26),
neighbors of RP11-340F14.6 labeled with the 10K region were
identified, including P2X7R, P2X4R and CAMKK2 (Fig. 4A). These neighbors are likely to
interact with RP11-340F14.6 through a cis-regulation
approach. Subsequently, the expression of these neighbors was
detected in RP11-340F14.6high CD4+ T cells
derived from PBMCs following transfection with RP11-340F14.6 shRNA.
Of note, it was found that P2X7R expression was decreased with the
loss of RP11-340F14.6 (Fig. 4B).
In human clinical samples, the mRNA expression of P2X7R was
increased in the JIA immune microenvironment (Fig. 4C) and its expression positively
correlated with RP11-340F14.6 expression (Fig. 4D).
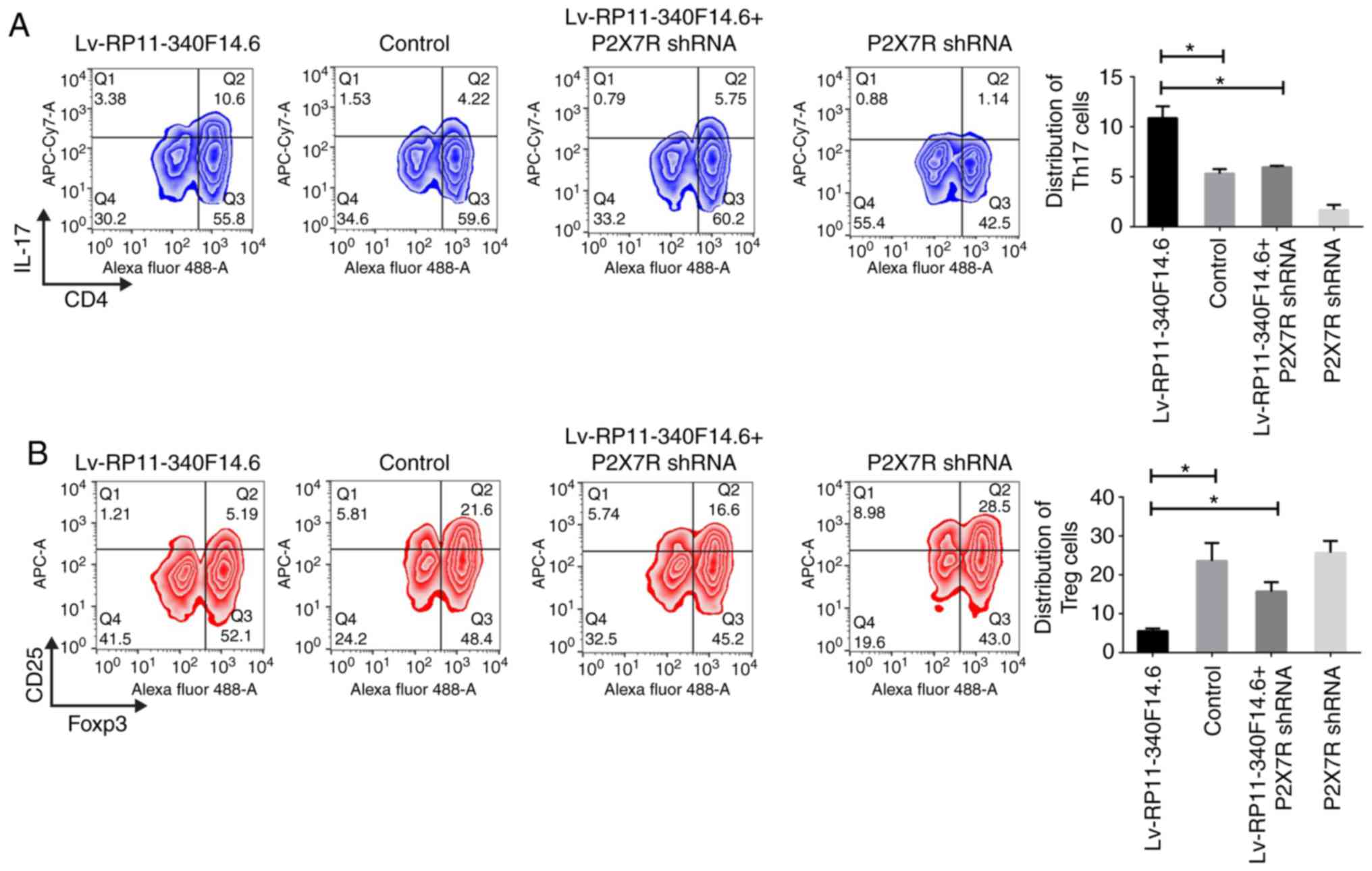
The phenotype for RP11-340F14.6 expression in T cell
differentiation was also measured to examine whether the function
of RP11-340F14.6 was mediated through interaction with P2X7R. The
expression of P2X7R was knocked down using shRNA technology and the
percentage of Th17 and Treg cells was determined. The
overexpression of RP11-340F14.6 increased the amount of Th17 cells;
however, this increase was attenuated by the loss of P2X7R
expression (Fig. 5A). The
percentage of Tregs was decreased by the overexpression of
RP11-340F14.6, but was restored with the loss of P2X7R expression
(Fig. 5B). These results
indicated that the overexpression of RP11-340F14.6 may have
increased the Th17/Treg ratio via a P2X7R-dependent mechanism.
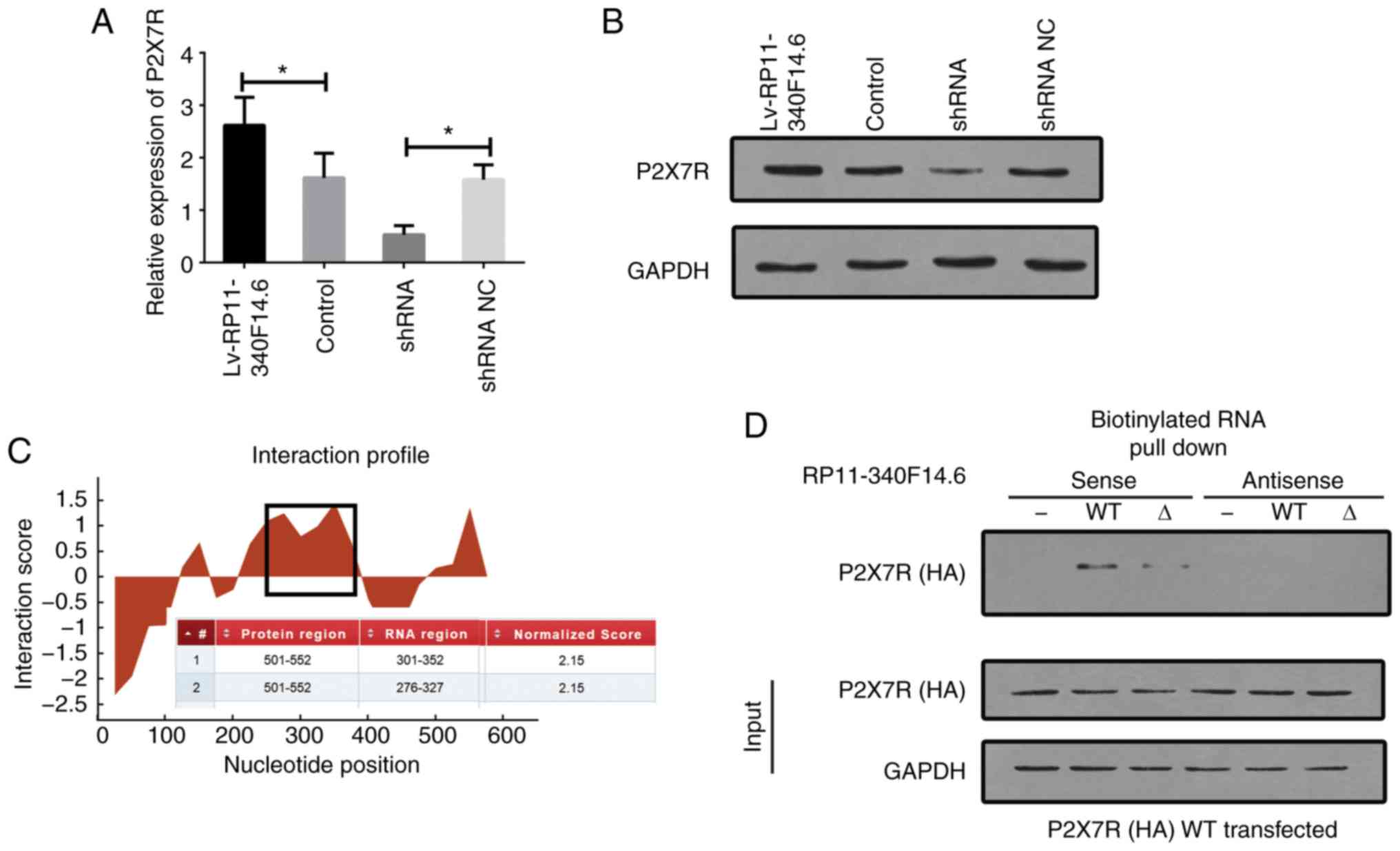
The expression of P2X7R mRNA and protein was
detected in cells following overexpression and/or silencing of
RP11-340F14.6. The increased expression of P2X7R was observed in
cells which overexpressed RP11-340F14.6, and this increase in P2X7R
mRNA expression was suppressed by the knockdown of RP11-340F14.6
expression (Fig. 6A and B).
CatRAPID, a bioinformatics software (http://service.tarta-glialab.com/page/catrapid_group),
was used to predict the potential binding fragment of P2X7R in
RP11-340F14.6 (Fig. 6C). The full
length of lncRNA nucleotide sequence and the full length of P2X7R
amino acid peptide were used as input. The RP11-340F14.6 region
between amino acids 301 to 352 was predicted to bind to P2X7R. For
P2X7R, the intracellular (IC) domain and amino acids between 501
and 522 were suggested to be the most probable binding domain for
RP11-340F14.6. RIP assays revealed that anti-HA (wild-type P2X7R)
antibodies specifically precipitated RP11-340F14.6; however,
deletions in the amino acids 501-522 of P2X7R abrogated its ability
to bind to RP11-340F14.6 (Fig.
6D). These results suggested that RP11-340F14.6 interacted with
P2X7R in a highly specific manner and that the predicted region
(301-352) of RP11-340F14.6 is crucial for its ability to interact
with P2X7R.
Discussion
JIA is a systemic autoimmune disease characterized
by persistent synovial inflammation accompanied bythe destruction
of bone and articular cartilage. The phenotypic variability
reflects the underlying fundamental biological diversity, as well
as differences in PBMC gene expression patterns and serum cytokine
profiles (27). lncRNAs play a
critical role in regulating the differentiation and function of
CD4+ T cells. This is evident as T cell subsets exhibit
specific lncRNA expression that defines their transcriptional
procedures and pedigree (28). It
has been reported that lncRNAs regulate the differentiation of T
helper cells by epigenetic and transcriptional reprogramming
mechanisms (29). The lncRNA NeST
(formally known as Tmevpg1) has been shown to promote Th1 cell
differentiation by increasing WDR5 expression (30). lnc-EFGR has also been shown to
enhance EGFR expression and result in the shift of Tregs and
CD8+ T cells (23). In
the present study, sample screening revealed the potential
importance of RP11-340F14.6. Loss-of-function and gain-of-function
assays confirmed that the expression of the closest neighbor of
this lncRNA, P2X7R, was modulated by RP11-340F14.6. Furthermore,
the expression of RP11-340F14.6 inhibited the differentiation of
Tregs and stimulated the differentiation of Th17 cells.
RF-positive pJIA has similar clinical manifestations
and pathogenesis with adult RA. The development of RA is highly
associated with Th17/Treg redistribution, specifically for an
inflammatory-associated, cytokine-induced immune micro-environment.
Tregs maintain immune tolerance and prevent autoimmunity by
inhibiting activation and proliferation of immune effector cells
(31). However, a number of
questions remain unanswered as to the mechanisms through which Th17
and Tregs actively regulate JIA (32). The present study also demonstrated
that the percentage of Th17 cells was markedly increased and was
accompanied by a decrease in the Treg population in patients with
JIA compared to the healthy controls. The increased expression of
lncRNA NEAT1 was previously found to be associated with the
development of tissue inflammation in RA, and the abundance of Th17
cells in PBMCs was increased in patients with RA; consistently,
NEAT1 knockdown inhibited the differentiation of Th17 cells, thus,
preventing the development of RA (33). In the present study, it was found
that specific expression of RP11-340F14.6 in JIA was positively
associated with the ratio of Th17/Tregs. The silencing of
RP11-340F14.6 significantly inhibited the differentiation of
CD4+ T cells into Th17 cells. The silencing of
RP11-340F14.6 also promoted the differentiation of JIA
CD4+ T cells into Tregs, which proved to be helpful for
the understanding of the mechanisms of JIA CD4+ T cell
differentiation.
In the present study, RIP assays revealed a specific
interaction between RP11-340F14.6 and P2X7R. This indicated that
RP11-340F14.6 may induce an increase in the Th17/Treg ratio via a
P2X7R-dependent mechanism. P2X7R is a distinct ligand-gated ion
channel and is a member of the purinergic type 2 receptor family
(34). P2X7R has been confirmed
to promote Treg differentiation, and thus its expression is closely
associated with Treg abundance (35). In previous studies, the authors
demonstrated a novel function of P2X7R signaling in regulating
CII-induced differentiation of Th17 cells (36). P2X7R can activate NLRP3, resulting
in caspase-1 meditated maturation and the release of
pro-inflammatory cytokines, such as IL-1β and IL-18 (37). In patients with RA, PBMCs can
release large amounts of IL-1β and IL-18. With ATP stimulation,
high levels of P2X7R and NLRP3 inflammatory bodies can also be
released (38). The P2X7R/NLRP3
pathway may play an important role in the regulation of
CD4+ T cell differentiation. It remains to be determined
whether RP11-340F14.6 promotes a shift in the Th17/Treg ratio by
altering the signaling of P2X7R/NLRP3 inflammatory corpuscles.
In conclusion, the present study identified the 651
bp lncRNA RP11-340F14.6, a neighbor of P2X7R (within 10 kb). The
present study aimed to investigate whether RP11-340F14.6 plays a
role in stimulating Th17 differentiation and suppressing Treg
distribution through a P2X7R-independent approach. A limitation to
this study was the small sample size that was restricted due to
time constraints. Nonetheless, the effect of the P2X7R signaling
pathway on cell phenotype requires further investigation. At
present, these findings suggest that lncRNA RP11-340F14.6 may serve
as a novel prospective intervention target for the treatment of
JIA.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (no. 81771762), the
National Natural Science Foundation of China (no. 81800438) and the
Nanjing Science and Technology Development Program (no.
YKK16179).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
NH contributed to the conception and design of the
research and drafted the manuscript. ZF and LM contributed to the
acquisition and analysis of the data. HM and HH contributed to the
interpretation of the data. HY and XZ equally contributed to the
design of the research. All authors critically revised the
manuscript, agree to be fully accountable for ensuring the
integrity and accuracy of the work, and read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were performed in compliance with
government policies and the Helsinki Declaration. All patients or
healthy controls had the consent of their legal guardians or
parents who signed an informed consent form before collecting blood
samples. The present study was approved by the Ethics Committee of
the Children's Hospital of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Glerup M, Herlin T and Twilt M: Clinical
outcome and long-term remission in JIA. Curr Rheumatol Rep.
19:752017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Selvaag AM, Aulie HA, Lilleby V and Flato
B: Disease progression into adulthood and predictors of long-term
active disease in juvenile idiopathic arthritis. Ann Rheum Dis.
75:190–195. 2016. View Article : Google Scholar
|
|
3
|
Ruperto N and Martini A: Current and
future perspectives in the management of juvenile idiopathic
arthritis. Lancet Child Adolesc Health. 2:360–370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Berntson L, Fasth A, Andersson-Gäre B,
Kristinsson J, Lahdenne P, Marhaug G, Nielsen S and Pelkonen P;
Svensson E; Nordic Study Group: Construct validity of ILAR and
EULAR criteria in juvenile idiopathic arthritis: A population based
incidence study from the Nordic countries. International league of
associations for rheumatology. European league against rheumatism J
Rheumatol. 28:2737–2743. 2001.
|
|
5
|
Horneff G, Klein A, Ganser G, Sailer-Höck
M, Günther A, Foeldvari I and Weller-Heinemann F: Protocols on
classification, monitoring and therapy in children's rheumatology
(PRO-KIND): Results of the working group polyarticular juvenile
idiopathic arthritis. Pediatr Rheumatol Online J. 15:782017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hinks A, Marion MC, Cobb J, Comeau ME,
Sudman M, Ainsworth HC, Bowes J; Juvenile Idiopathic Arthritis
Consortium for Immunochip; Becker ML, Bohnsack JF, et al: Brief
report: The genetic profile of rheumatoid factor-positive
polyarticular juvenile idiopathic arthritis resembles that of adult
rheumatoid arthritis. Arthritis Rheumatol. 70:957–962. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Komatsu N, Okamoto K, Sawa S, Nakashima T,
Oh-hora M, Kodama T, Tanaka S, Bluestone JA and Takayanagi H:
Pathogenic conversion of Foxp3+ T cells into TH17 cells
in autoimmune arthritis. Nat Med. 20:62–68. 2014. View Article : Google Scholar
|
|
8
|
Niu Q, Cai B, Huang ZC, Shi YY and Wang
LL: Disturbed Th17/Treg balance in patients with rheumatoid
arthritis. Rheumatol Int. 32:2731–2736. 2012. View Article : Google Scholar
|
|
9
|
Alunno A, Manetti M, Caterbi S,
Ibba-Manneschi L, Bistoni O, Bartoloni E, Valentini V, Terenzi R
and Gerli R: Altered immunoregulation in rheumatoid arthritis: The
role of regulatory T cells and proinflammatory Th17 cells and
therapeutic implications. Mediators Inflamm. 2015:7517932015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boissier MC, Assier E, Falgarone G and
Bessis N: Shifting the imbalance from Th1/Th2 to Th17/treg: The
changing rheumatoid arthritis paradigm. Joint Bone Spine.
75:373–375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stelmaszczyk-Emmel A, Jackowska T,
Rutkowska-Sak L, Marusak-Banacka M and Wasik M: Identification,
frequency, activation and function of CD4+ CD25(high)FoxP3+
regulatory T cells in children with juvenile idiopathic arthritis.
Rheumatol Int. 32:1147–1154. 2012. View Article : Google Scholar
|
|
12
|
Szymańska-Kałuża J, Cebula-Obrzut B,
Smolewski P, Stanczyk J and Smolewska E: Imbalance of Th17 and
T-regulatory cells in peripheral blood and synovial fluid in
treatment naïve children with juvenile idiopathic arthritis. Cent
Eur J Immunol. 39:71–76. 2014. View Article : Google Scholar
|
|
13
|
Nistala K, Moncrieffe H, Newton KR,
Varsani H, Hunter P and Wedderburn LR: Interleukin-17-producing T
cells are enriched in the joints of children with arthritis, but
have a reciprocal relationship to regulatory T cell numbers.
Arthritis Rheum. 58:875–887. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu SA, Yeh KW, Lee WI, Yao TC and Huang
JL: Persistent improper upregulation of Th17 and TReg cells in
patients with juvenile idiopathic arthritis. J Microbiol Immunol
Infect. 49:402–408. 2016. View Article : Google Scholar
|
|
15
|
Bending D, Pesenacker AM, Ursu S, Wu Q,
Lom H, Thirugnanabalan B and Wedderburn LR: Hypomethylation at the
regulatory T cell-specific demethylated region in CD25hi T cells is
decoupled from FOXP3 expression at the inflamed site in childhood
arthritis. J Immunol. 193:2699–2708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen YG, Satpathy AT and Chang HY: Gene
regulation in the immune system by long noncoding RNAs. Nat
Immunol. 18:962–972. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Yang X, Sun X, Rong L, Kang M, Wu
P, Ji X, Lin R, Huang J, Xue Y and Fang Y: Bone marrow infiltrated
Lnc-INSR induced suppressive immune microenvironment in pediatric
acute lymphoblastic leukemia. Cell Death Dis. 9:10432018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu G, Tang Q, Sharma S, Yu F, Escobar TM,
Muljo SA, Zhu J and Zhao K: Expression and regulation of intergenic
long noncoding RNAs during T cell development and differentiation.
Nat Immunol. 14:1190–1198. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Collier SP, Henderson MA, Tossberg JT and
Aune TM: Regulation of the Th1 genomic locus from Ifng through
Tmevpg1 by T-bet. J Immunol. 193:3959–3965. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ranzani V, Rossetti G, Panzeri I, Arrigoni
A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M,
et al: The long intergenic noncoding RNA landscape of human
lymphocytes highlights the regulation of T cell differentiation by
linc-MAF-4. Nat Immunol. 16:318–325. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie
Y, Wang K, Jia W, Chu WM and Sun B: The long noncoding RNA lnc-EGFR
stimulates T-regulatory cells differentiation thus promoting
hepatocellular carcinoma immune evasion. Nat Commun. 8:151292017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan SX, Wang J, Yang F, Tao QF, Zhang J,
Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, et al: Long noncoding RNA
DANCR increases stemness features of hepatocellular carcinoma by
derepression of CTNNB1. Hepatology. 63:499–511. 2016. View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Jia H, Osak M, Bogu GK, Stanton LW,
Johnson R and Lipovich L: Genome-wide computational identification
and manual annotation of human long noncoding RNA genes. RNA.
16:1478–1487. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hinze C, Gohar F and Foell D: Management
of juvenile idiopathic arthritis: Hitting the target. Nat Rev
Rheumatol. 11:290–300. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Roy S and Awasthi A: Emerging roles of
noncoding RNAs in T cell differentiation and functions in
autoimmune diseases. Int Rev Immunol. 38:232–245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Collier SP, Collins PL, Williams CL,
Boothby MR and Aune TM: Cutting edge: Influence of Tmevpg1, a long
intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J
Immunol. 189:2084–2088. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pesenacker AM and Wedderburn LR: T
regulatory cells in childhood arthritis-novel insights. Expert Rev
Mol Med. 15:e132013. View Article : Google Scholar
|
|
32
|
Lawson CA, Brown AK, Bejarano V, Douglas
SH, Burgoyne CH, Greenstein AS, Boylston AW, Emery P, Ponchel F and
Isaacs JD: Early rheumatoid arthritis is associated with a deficit
in the CD4+CD25high regulatory T cell population in peripheral
blood. Rheumatology (Oxford). 45:1210–1217. 2006. View Article : Google Scholar
|
|
33
|
Shui X, Chen S, Lin J, Kong J, Zhou C and
Wu J: Knockdown of lncRNA NEAT1 inhibits Th17/CD4+ T
cell differentiation through reducing the STAT3 protein level. J
Cell Physiol. 234:22477–22484. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Burnstock G: Pathophysiology and
therapeutic potential of purinergic signaling. Pharmacol Rev.
58:58–86. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schenk U, Frascoli M, Proietti M, Geffers
R, Traggiai E, Buer J, Ricordi C, Westendorf AM and Grassi F: ATP
inhibits the generation and function of regulatory T cells through
the activation of purinergic P2X receptors. Sci Signal. 4:ra122011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fan ZD, Zhang YY, Guo YH, Huang N, Ma HH,
Huang H and Yu HG: Involvement of P2X7 receptor signaling on
regulating the differentiation of Th17 cells and type II
collagen-induced arthritis in mice. Sci Rep. 6:358042016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shen HH, Yang YX, Meng X, Luo XY, Li XM,
Shuai ZW, Ye DQ and Pan HF: NLRP3: A promising therapeutic target
for autoimmune diseases. Autoimmun Rev. 17:694–702. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Choulaki C, Papadaki G, Repa A, Kampouraki
E, Kambas K, Ritis K, Bertsias G, Boumpas DT and Sidiropoulos P:
Enhanced activity of NLRP3 inflammasome in peripheral blood cells
of patients with active rheumatoid arthritis. Arthritis Res Ther.
17:2572015. View Article : Google Scholar : PubMed/NCBI
|