Introduction
Intervertebral disc degeneration (IDD) is a common
degenerative disease, as a total of 266 million individuals
worldwide have degenerative spine disease annually (1). IDD features include accelerated
extracellular matrix (ECM) degradation and abnormal ECM
biosynthesis, decreased hydration, reduced intervertebral disc
height and decreased ability to absorb external mechanical
compression (2,3). IDD is the predominant cause of
chronic lower back pain and spine-related ailments, imposing
significant economic and social burden worldwide (4). According to previous reports, 84% of
the world population experiences lower back pain during their
lifetime, with 10% being chronically disabled (5). However, the application of current
strategies for IDD treatment is hampered by an incomplete
understanding of its pathogenesis. IDD treatment is limited to
symptomatic interventions, which do not adequately improve outcomes
since no disease-modifying drugs are available (6). Consequently, the clinical management
of diseases related to IDD remains severely limited. Therefore,
determining the pathophysiological events and molecular mechanisms
underlying IDD is urgently needed for the development of new
treatments.
Intervertebral discs are composed of a central
nucleus pulposus (NP), a peripheral annulus fibrosus (AF) and
cartilaginous end plates, which connect overlying capillary beds
cranially and caudally (7). NP
cells are the predominant cell type in the NP tissue, which forms
the main component of the ECM by synthesizing type II collagen
(collagen II) and aggrecan, the major functional components of
intervertebral discs, to maintain normal disc height and to absorb
various mechanical loads (7).
Multiple adverse factors such as Substance P, chemokine ligand 5
and chronic overload of the disc (8–10)
enhance the levels of inflammatory cytokines (ICs) in the NP,
including interleukin-1β (IL-1β) and tumor necrosis factor-α
(TNF-α) (11,12). IL-1β contributes to IDD
development by accelerating the degradation of ECM components,
e.g., via increased production of catabolic factors such as matrix
metalloproteinases (MMPs) (11,12). TNF-α influences catabolic pathways
in a manner similar to IL-1β; TNF-α and IL-1β have been
demonstrated to induce degenerative changes to the ECM, which is a
major characteristic of disc degeneration (13). These ICs have been demonstrated to
induce an imbalance between anabolic and catabolic activities in NP
cells and to inhibit the expression of anabolic factors such as
collagen II and aggrecan, which initiate or accelerate the
development of IDD (8,11,13,14). Thus, it is necessary to develop an
effective tool to attenuate the inflammatory response and to
reverse the imbalance between anabolism and catabolism within the
NP microenvironment.
Multiple molecular inducers of IDD have been
reported in previous studies, with non-coding RNAs emerging as key
factors affecting IDD pathogenesis (15,16). Circular RNAs (circRNAs) are a
class of RNAs with circular structures without 5'-3' polarity and
polyadenylation tails, and are mostly produced by one or more exons
through reverse splicing (17).
Most circRNAs are endogenous non-coding RNAs conserved across
species and exhibit higher stability compared with linear mRNAs
(18). Competing endogenous RNAs
(ceRNAs) induce mRNA silencing by binding to the 3'-untranslated
region response element, also termed the microRNA (miRNA) response
element (17,19). circRNAs can also bind miRNAs to
participate in the regulatory network of ceRNAs (19). Thus, circRNAs act as
post-transcriptional regulators and interact with miRNAs as miRNA
sponges and ceRNAs in the cytoplasm (21-23). miRNA sponges are circRNAs with
miRNA binding capacity that absorb miRNAs and inhibit their
repressive effects on respective targets (24). A previous study has revealed that
miR-185-5p is downregulated in degenerative NP tissues (15). In addition, bioinformatics
analysis has demonstrated the ability of miR-185-5p to interact
with several circRNAs (15).
However, the mechanisms by which miR-185-5p affects IDD development
and progression remain unclear.
Our previous study demonstrated that circGRB10
promoted the survival of NP cells during nutrient deprivation by
upregulating Erb-B2 receptor tyrosine kinase through the
inactivation of miR-328-5p, which suppressed IDD development
(25). Therefore, it was
hypothesized in the present study that circ-RNAs may be involved in
the pathogenesis of IDD through by regulating miR-185-5p. The
present study aimed to examine the role of circ-RNAs in
degenerative NP tissues and to validate their functions in cultured
human NP cells.
Materials and methods
Ethics statement
This study was approved by the ethics committees of
Tianjin Medical University General Hospital (Tianjin, China) and
Hebei Province Cangzhou Hospital of Integrated Traditional and
Western Medicine (Cangzhou, China). Human NP tissue samples were
obtained from patients undergoing surgery between January 2018 and
May 2019 at Tianjin Medical University General Hospital and Hebei
Province Cangzhou Hospital of Integrated Traditional and Western
Medicine. Written informed consent was obtained from all patients
for the use of their tissue specimens for research purposes.
Clinical specimens
Human lumbar degenerative NP specimens were obtained
from 10 patients with IDD undergoing discectomy. Control samples
were obtained from 10 age- and sex-matched patients with fresh
traumatic lumbar fracture undergoing anterior decompressive surgery
due to neurological deficits. The characteristics of the patients
are presented in Table I. The
degree of IDD was determined by magnetic resonance imaging and
graded using the following grading system for the assessment of
lumbar disc degeneration: i) Grade I, homogeneous disc structure
with a bright hyper-intense white signal and a normal disc height;
ii) grade II, inhomogeneous disc structure with a hyperintense
white signal, clear distinction between the nucleus and annulus,
and normal disc height, with or without horizontal gray bands; iii)
grade III, inhomogeneous disc structure with an intermediate gray
signal intensity, unclear distinction between the nucleus and
annulus, and normal or slightly decreased disc height; iv) grade
IV, inhomogeneous disc structure with a hypointense dark gray
signal intensity, no distinction between the nucleus and annulus,
and normal or moderately decreased disc height; v) grade V,
inhomogeneous disc structure with a hypointense black signal
intensity, no distinction between the nucleus and annulus, and
collapsed disc space. Grading was performed using T2-weighted
midsagittal fast spin echo images (repetition time, 5,000 ms; echo
time, 130 ms) (26-28). The lesions in the control group
were of grade I or II, whereas those in patients with IDD were of
grade III, IV or V.
 | Table IClinicopathological features of the
study population. |
Table I
Clinicopathological features of the
study population.
| Variable | Normal (n=10) | IDD (n=10) | P-value |
|---|
| Age, years | 36.2±10.4 | 34.6±10.3 | 0.733a |
| Body mass index,
kg/m2 | 23.9±2.2 | 24.0±2.2 | 0.982a |
| Sex, n (%) | | | |
| Male | 7 (70) | 5 (50) | 0.650b |
| Female | 3 (30) | 5 (50) | |
Isolation and culture of human NP
cells
The NP was separated from AF samples under a
stereotactic microscope and sliced at 2-3 mm3 as
previously described (20). NP
cells were obtained after digestion with 0.25 mg/ml type II
collagenase (Invitrogen; Thermo Fisher Scientific, Inc.) for 12 h
at 37°C in Dulbecco's modified Eagle's medium/nutrient mixture F-12
(DMEM/F12; Gibco; Thermo Fisher Scientific, Inc.) and resuspended
at 2x105 cells/ml in DMEM/F12 supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100
mg/ml streptomycin, 100 U/ml penicillin and 1% L‑glutamine at 37°C
in a humidified atmosphere containing 5% CO2. At
confluency, the cells were trypsinized and passaged, with the
medium changed every other day. Cells at the second passage were
assessed in the experiments. In the IC treatment experiments, cells
were stimulated by 5 ng/ml of TNF-α and IL-1β (Sigma-Aldrich; Merck
KGaA) in the culture medium for 12 h. Unstimulated cells were used
as controls.
Bioinformatics analysis
Using the starBase platform (http://starbase.sysu.edu.cn), miR-185-5p was predicted
to have binding sites for a circRNA derived from tissue inhibitor
of metalloproteinases 2 (circ-TIMP2).
Small interfering RNA (siRNA) and
circ‑TIMP2 overexpression plasmid construction
According to the circRNA sequences of circ-TIMP2
(hsa_circ_0045942) in circBase (http://www.circbase.org/), circ-TIMP2 siRNA (each
siRNA had three pairs of sequences) and negative control (NC) siRNA
were designed and synthesized by Guangzhou Geenseed Biotech Co.,
Ltd. To construct a circ-TIMP2 overexpression vector, front and
back circular frames of TIMP2 were generated and added to a
pLCDH-ciR vector (Geneseed Biotech Co., Ltd.) for transcript
circularization. The front and back circular frames comprised
endogenous flanking genomic sequences with EcoRI and
BamHI restriction sites, respectively (24). The 3,122 bp target sequence
comprised the EcoRI site, the splice acceptor AG, the
circ-TIMP2 sequence, the splice donor GT, and the BamHI
site. The PCR product was cloned between the two frames. The
specific divergent primers for the back‑splice junction of
circ-TIMP2 were used to amplify the circRNA. A mock vector solely
containing a nonsense sequence between both circular frames without
circ-TIMP2 cDNA was generated. Vector construction was performed by
Guangzhou Geenseed Biotech Co. miR-185-5p and MMP2 siRNAs, as well
as the corresponding negative controls, were synthesized from
Guangzhou RiboBio Co., Ltd.
Cell transfection
NP cells of the second generation underwent
transfection with respective plasmids or siRNAs using
Lipofectamine® 3000 (Invitrogen) according to the
manufacturer's instructions. NP cells at 80% confluence were
transfected with the siRNAs using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 6 h, and
subsequently the media was replaced. The siRNA concentration for
each transfection was 135 ng/µl, and the plasmid
concentration was 100 ng/ml. The cells were harvested at 48 h
post‑transfection. The transfection efficiency was determined by
Reverse transcription-quantitative (RT-q) PCR.
RT‑qPCR
Total RNAs were extracted using the miRNeasy Mini
kit (Qiagen GmbH) on isolated NP cells. For circRNA, total RNAs
were incubated with or without 3 U/µg of RNase R (Epicentre;
Illumina, Inc.) at 37°C for 20 min, and the resulting RNA was
subsequently purified using the RNeasy MinElute Cleanup kit (Qiagen
GmbH). circ-TIMP2 levels were assessed by SYBR®
Green-based qPCR (Sigma-Aldrich; Merck KGaA). miRNA levels were
determined using stem-loop miRNA RT-PCR Quantitation kit (Shanghai
GenePharma Co., Ltd.). RNA was reverse-transcribed using a
PrimeScipt RT kit (Takara Bio, Inc.) at 37°C for 15 min followed by
inactivation of reverse transcriptase with heat treatment at 85°C
for 5 sec. cDNA was used to perform qRT-PCR on the 7500 Sequence
Detection System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using SYBR® Premix Ex Taq (Takara Bio, Inc.). All
primers used in the present study are listed in Table II. Adaptor primers were designed
for the reverse splice site of circTIMP2. PCR amplification was
performed in 10‑µl reaction mixtures comprising 2 µl
cDNA, 5 µl 2X master mix, 0.5 µl forward and reverse
primers (10 µM) and 2 µl water at 95°C for 10 min,
followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. GAPDH
was used for normalization, with the exception of miRNAs, for which
U6 was used. The relative expression levels of each circRNA, mRNA
or miRNA were determined by the 2-ΔΔCq method (29).
 | Table IIPrimers and sequences used in this
study. |
Table II
Primers and sequences used in this
study.
A, PCR primers
|
|---|
| Name | Sequence
(5'→3') |
|---|
| circ-TIMP2 | F:
TGCGCATGTCTCTGATGCTT(br1/)R: GGCCCTTTGAACATTTCTCTTTGA |
| miR-185-5p | F:
ACACTCCAGCTGGGTGGAGAGAAAGGCAGT
R: TGGTGTCGTGGAGTCG |
| MMP2 | F:
TACAGGATCATTGGCTACACACC
R: GGTCACATCGCTCCAGACT |
| Collagen-II | F:
TGGACGATCAGGCGAAACC
R: GCTGCGGATGCTCTCAATCT |
| Aggrecan | F:
CCCCTGCTATTTCATCGACCC
R: GACACACGGCTCCACTTGAT |
| GAPDH | F:
GCACCGTCAAGGCTGAGAAC
R: GGATCTCGCTCCTGGAAGATG |
| U6 | F:
CTCGCTTCGGCAGCACA
R: AACGCTTCACGAATTTGCGT |
| B, miRNA mimics and
siRNAs |
| Name | Sequence
(5'→3') |
| miR-185-5p
mimic |
UGGAGAGAAAGGCAGUUCCUGA |
| miR-185-5p si |
UCAGGAACUGCCUUUCUCUCCA |
| circ-TIMP2 si |
AGAGAAATGTTCAAAGGGCC |
| MMP2 si |
AGTTGGCAGTGCAATACCTGA |
| C, Probes for
pull-down assay |
| Name | Sequence
(5'→3') |
| circ-TIMP2
pull-down probe |
GGCCCTTTGAACATTTCTCTTTGATAAT |
| Random pull-down
probe |
TACGGATGTCTAGCGCTCTTGGGCTTTG |
| D, Probes for
fluorescence in situ hybridization |
| Name | Sequence
(5'→3') |
| circ-TIMP2 |
GATTCTCCTTATCATTACCGAGAAAGTTCTTC |
| miR-185-5p |
TCAGGAACTGCCTTTCTCTC CA |
| E, Probes for
northern blotting |
| Name | Sequence
(5'→3') |
| miR-185-5p |
TCAGGAACTGCCTTTCTCTCCA |
| U6 |
TGGAACGCTTCACGAATTTG |
Fluorescence in situ hybridization
(FISH)
FISH was performed to detect the subcellular
localization of circ-TIMP2 and miR-185-5p in NP cells according to
a method described by Vautrot et al (30). A FISH probe labeled with Alexa
Fluor® 488 for circ-TIMP2 (Thermo Fisher Scientific,
Inc.) was designed to detect the splice junction of two exons.
After prehy-bridization using 1X PBS and 0.5% Triton X-100, NPCs
were hybridized in hybridization buffer (50% formamide, 5X SSC, 500
µg/ml yeast tRNA, 10% Dextran sulfate, 1X Denhardt's
solution, 10 mM DDT, 1 mg/ml sheared salmon sperm DNA) with
specific probes at 55°C for 15‑17 h in a humidified chamber. The
probe sequence of circ-TIMP2 was 5'-GAT TCT CCT TAT CAT TAC CGA GAA
AGT TCT TC-3'. The probe for miR-185-5p was labeled with Cy3, and
the sequence was 5'-TCA GGA ACT GCC TTT CTC TCC A-3' (Wuhan
Servicebio Technology Co., Ltd.). Images were acquired using an
Nikon Eclipse TI‑SR fluorescence microscope (Nikon
Corporation).
Dual‑luciferase reporter assay
The binding site of circ-TIMP2 (31) (wild-type or mutated) was inserted
into the KpnI and SacI sites of the pGL3 promoter
vector (Shanghai Realgene Biotech, Inc.). First, cells were seeded
into 24-well plates (2x105 cells/well), followed by
transfection with 80 ng plasmid, 5 ng Renilla luciferase
vector pRL-SV40 and 50 nM miR-185-5p mimics or respective NCs using
1.5 µl/well Lipofectamine® 3000 by incubation for
10-15 min at room temperature. Cell collection was performed after
48 h, and the Dual-Luciferase Reporter Assay system (Promega
Corporation) was used to measure luciferase activity according to
the manufacturer's instructions. Firefly luciferase activity was
normalized to that of Renilla luciferase.
Western blotting
Western blotting was performed as previously
described (25). Cell lysis was
performed in a buffer containing 0.25 M Tris-HCl, 20% glycerol, 4%
sodium dodecyl sulfate (SDS) and 10% mercaptoethanol (pH 6.8)
supplemented with protease and phosphatase inhibitors. The protein
contents were measured using the Micro BCA Protein Assay kit
(Thermo Fisher Scientific, Inc.). Equal amounts of total protein
(10 µg) were separated on 10-12% SDS-polyacrylamide gels and
electro-transferred onto polyvinylidene fluoride membranes. After
blocking with 5% non-fat milk in Tris-buffered saline with 0.1%
Tween-20 (TBST) at room temperature for 1 h, the membranes were
incubated with primary antibodies (1:3,000; collagen-II, cat. no.
ab34712, Abcam; aggrecan, cat. no. ab194594, Abcam; MMP-2, cat. no.
ab97779, Abcam; GAPDH, cat. no. ab9485, Abcam) in TBST containing
5% non-fat milk overnight at 4°C. Secondary horseradish
peroxidase‑conjugated antibodies (1:6,000; Goat Anti-Rabbit IgG
H&L, cat. no. ab6721; Goat Anti-Mouse IgG H&L, cat. no.
ab205719, Abcam) were added at room temperature for 1 h, and
immunoblots were developed using an enhanced chemiluminescence
system (Cytiva). ImageJ 1.47 (National Institutes of Health) was
used for densitometry analysis.
Pull‑down assay with a biotinylated DNA
probe
The pull-down assay was performed as previously
described (32). The biotinylated
DNA probe complementary to circ-TIMP2 (Table II) was added to 500 µl
lysis buffer (0.5 M NaCl, 20 mM Tris-HCl and 1 mM EDTA, pH 7.5),
followed by incubation with streptavidin‑coated magnetic beads at
25°C for 3 h. Subsequently, the cell pellets were re-suspended with
the lysis buffer (1 ml/100 mg of cell pellet), and the cell lysates
were incubated with the probe-coated beads. Finally, the RNA
mixture was eluted and extracted for northern blot analysis by
using a magnetic support to separate the beads from cell lysate,
discarding the supernatant and washing the beads with 900 µl
wash buffer (0.5% SDS, 2X SSC) five times interspersed with 5 min
agitation on a rotator at room temperature.
Northern blotting
Digoxin-labeled probes were prepared with DIG
Northern Starter kit (Roche Diagnostics GmbH) as previously
described (33). Total RNA
samples were resolved on 2% agarose or 15% polyacrylamide-urea gels
and transferred to Hybond-N+ membranes (Amersham; Cytiva). The
membranes were dried, crosslinked by ultraviolet irradiation (265
nm; 0.15 J/cm2), and subjected to hybridization with
digoxin‑labeled miR‑185‑5p probes at 65°C or 42°C overnight;
digoxin-labeled U6 probes were used as controls (Table II). The blot was visualized on a
ChemiDoc XRS system (Bio-Rad Laboratories, Inc.).
RNA immunoprecipitation (RIP)
RIP was performed using a Magna RIP kit (EMD
Millipore) according to the manufacturer's instructions. Briefly,
2x107 NP cells were submitted to UV-crosslinking (600
mJ/cm2) and lysed with 100 µl RIP lysis buffer
containing a proteinase inhibitor cocktail (Roche Diagnostics GmbH)
and RNase inhibitor (Promega Corporation). Following incubation
with DNase I (Roche Diagnostics GmbH) at 37°C for 10 min, the
lysates were centrifuged at 12,000 x g at 4°C for 30 min. The
resulting supernatants were added to 900 µl RIP
immunoprecipitation buffer and incubated with 5 µg
anti-argonaute 2 (AGO2) antibodies pre-bound to magnetic beads for
3 h. Subsequently, 20% of the immunoprecipitate was evaluated by
western blotting, and the remaining 80% was treated with proteinase
K at 37°C for 30 min.
Statistical analysis
Each experiment was repeated at least three times,
and cells in every experiment were harvested from a single
isolation process. Continuous data are presented as the mean ±
standard deviation. GraphPad Prism version 7.0 (GraphPad Software,
Inc.) or SPSS version 22.0 (IBM Corp.) was used for statistical
analysis. Comparisons between two groups were performed by unpaired
Student's t-test. Categorical data were analyzed by the
χ2 test. Multi-group comparisons were performed using
one-way ANOVA, and Tukey's test was used for the post hoc analysis.
Pearson's correlation coefficients were determined to assess
correlations of continuous data, and Spearman's rank correlation
was used for ordinal data. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR‑185‑5p is downregulated in
degenerative NP tissues and regulates ECM synthesis
RT-qPCR results demonstrated that miR-185-5p was
significantly downregulated in degenerative NP tissues compared
with that in the controls (Fig.
1A). Spearman's correlation analysis revealed that miR-185-5p
was significantly negatively correlated with IDD grade (ρ=-0.926;
P<0.001; Fig. 1B). Next, the
functions of miR-185-5p in NP cells were assessed. The results
demonstrated that miR-185-5p mimics significantly increased the
expression level of miR-185-5p in NP cells, whereas siRNA markedly
decreased the expression level of miR-185-5p (Fig. 1C). NP cells transfected with the
miR-185-5p mimic displayed increased expression levels of collagen
II and aggrecan (Fig. 1D). These
results indicated a pro-anabolic role of miR-185-5p in NP cells.
Following treatment with TNF-α and IL-1β, cultured NP cells
exhibited significantly reduced expression levels of miR-185-5p
compared with those in the untreated control group (Fig. 1E). The miR-185-5p mimic
counteracted the TNF-α- and IL-1β-induced inhibition of ECM
production in NP cells (Fig. 1F and
G). These results indicated that miR-185-5p positively
regulated NP cell metabolism.
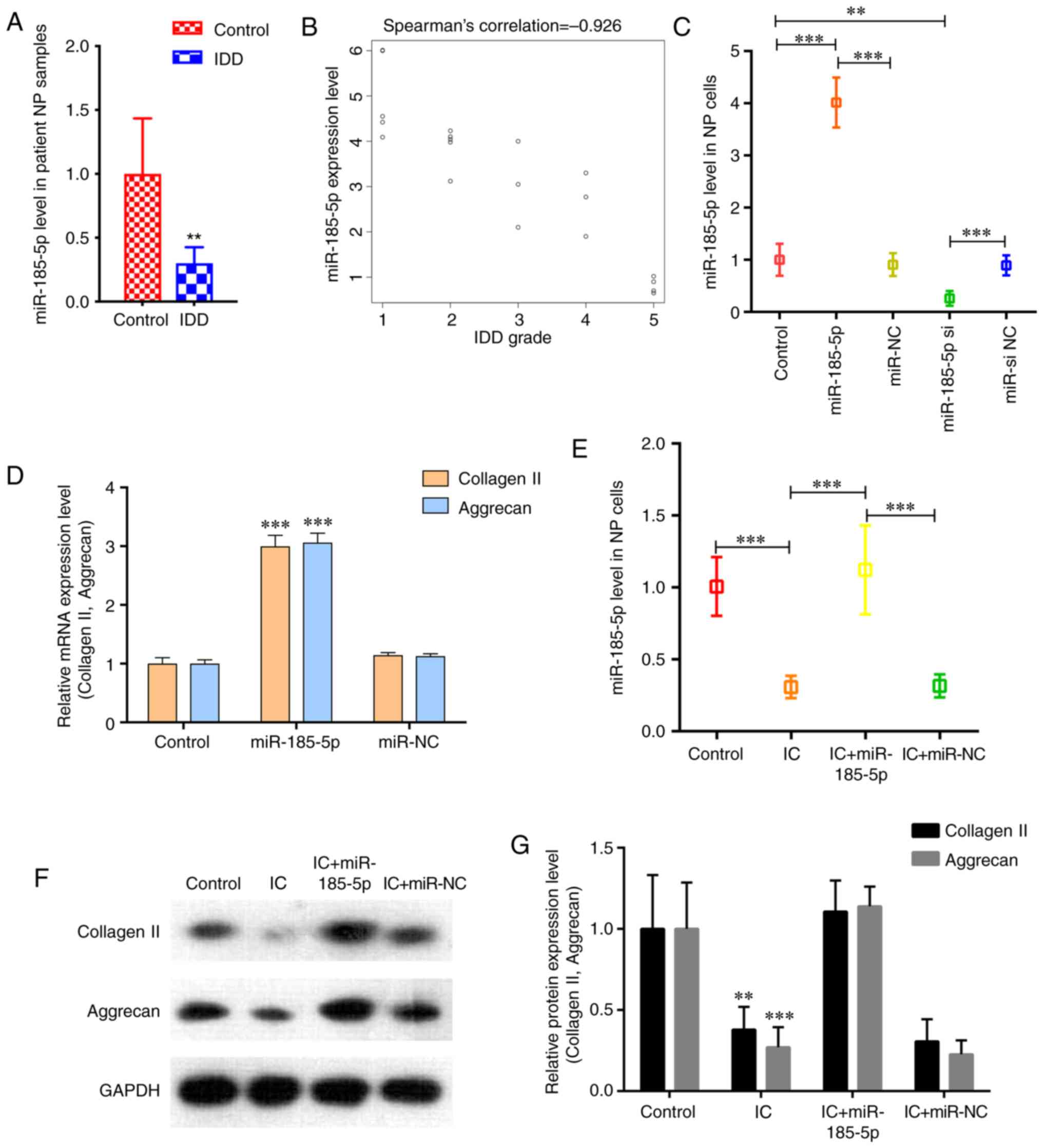 | Figure 1miR‑185‑5p is downregulated in
degenerative NP tissues and involved in the regulation of NP cell
function. (A) RT‑qPCR analysis confirmed the downregulation of
miR-185-5p in degenerative NP samples from patients with IDD
compared with the control group. n=10. **P<0.01 vs.
control. (B) miR-185-5p expression was significantly negatively
correlated with IDD grade (P<0.001). (C) NP cells were
transfected with the miR‑185‑5p mimic, miR‑NC, miR‑185‑5p si or
miR-si NC, and miR-185-5p levels in NP cells were analyzed by
RT-qPCR. **P<0.01, ***P<0.001. (D)
RT-qPCR was performed to analyze the mRNA expression levels of
collagen II and aggrecan in NP cells after transfection with
miR-185-5p. (E) NP cells were transfected with the miR-185-5p mimic
or miR-NC and treated with ICs (interleukin 1β and tumor necrosis
factor-α). RT-qPCR demonstrated decreased miR-185-5p levels in NP
cells treated with ICs, which was reversed by transfection with
miR-185-5p mimic. ***P<0.001. (F and G) Western blot
analysis demonstrated that miR-185-5p attenuated the catabolic
response and reversed the decreased expression of ECM components
induced by IC treatment in NP cells compared with the control
group. **P<0.01, ***P<0.001 vs.
control. miR, microRNA; NP, nucleus pulposus; IDD, intervertebral
disc degeneration; RT-qPCR, reverse transcription-quantitative PCR;
si, small interfering RNA; NC, negative control; IC, inflammatory
cytokine. |
miR‑185‑5p exerts its function in NP
cells via MMP2 inhibition
A previous study has demonstrated that miR-185
overexpression in vitro reduces MMP-2 levels (31). An increase in MMP2 expression was
observed in degenerative NP tissues compared with that in the
control samples (Fig. 2A). In
addition, Pearson correlation analysis revealed that MMP‑2
expression was significantly negatively correlated with miR-185-5p
(r=-0.870; P<0.001; Fig. 2B).
As predicted by bioinformatics, MMP2, which is a catabolic factor,
was a potential target of miR-185-5p (Fig. 2C). Transfection with the
miR‑185‑5p mimic significantly decreased the luciferase activity of
the wild-type MMP2 reporter compared with that in the control
groups, whereas introducing mutations in the target site abolished
this inhibitory effect (Fig. 2D).
In addition, western blotting demonstrated a significant increase
in MMP2 protein expression levels following treatment of NP cells
with TNF-α and IL-1β compared with that in untreated cells, and
this effect was attenuated by transfection with the miR-185-5p
mimic (Fig. 2E and F). Next, the
present study examined whether miR-185-5p and MMP2 were
functionally related in NP cells. Fig. S1 demonstrates the transfection
efficiency of the MMP‑2 overexpression vector and MMP‑2 siRNA
analyzed by RT-qPCR in NP cells. As demonstrated in Fig. 2G and H, overexpression of MMP2
significantly attenuated the protective effects of miR-185-5p in NP
cells treated with TNF-α and IL-1β, suggesting that miR-185-5p
regulated NP cell function by targeting MMP2.
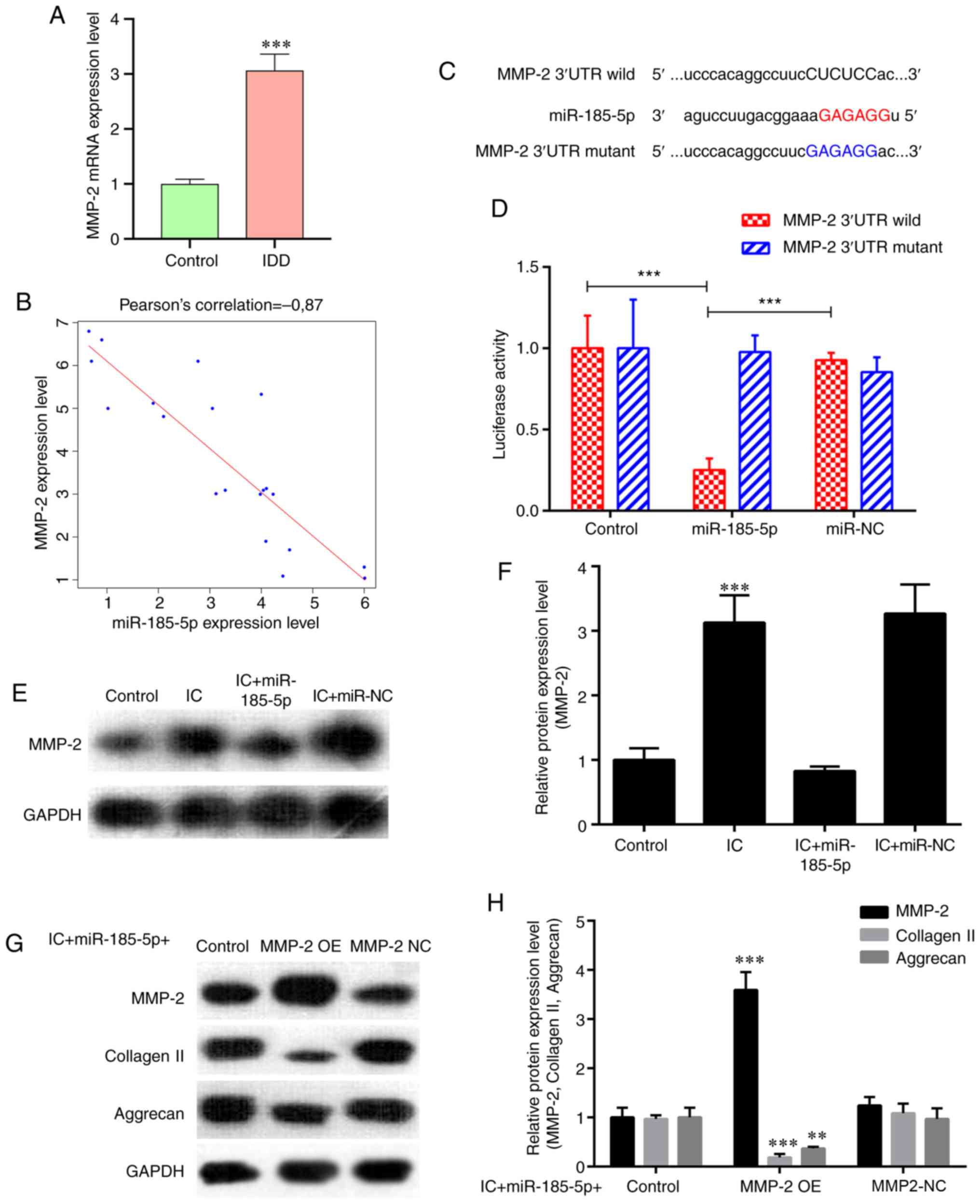 | Figure 2miR-185-5p regulates NP cell function
by inhibiting its target MMP2. (A) RT-qPCR analysis revealed higher
expression of MMP2 in degenerative NP tissues from patients with
IDD compared with that in the controls. ***P<0.001
vs. control. (B) MMP‑2 expression was significantly negatively
correlated with miR-185-5p expression (P<0.001). (C) Sequence
alignment of human miR-185-5p and the 3'-UTR region of MMP2 mRNA.
Mutations were introduced in the 3'-UTR of the MMP2 sequence to
create mutant luciferase reporter constructs. (D) Luciferase
reporter assay in NP cells after transfection with miR-NC or the
miR-185-5p mimic, Renilla luciferase vector pRL‑SV40 and the
reporter constructs. Firefly and Renilla luciferase
activities were measured in the same sample, and firefly luciferase
signals were normalized to Renilla luciferase signals.
***P<0.001. (E and F) NP cells were transfected with
the miR-185-5p mimic or miR-NC and treated with ICs (interleukin 1β
and tumor necrosis factor α). Western blot analysis demonstrated an
increase in MMP2 expression in NP cells treated with ICs compared
with that in the control group, which was alleviated by
transfection with the miR-185-5p mimic. ***P<0.001.
(G and H) Western blotting was used to analyze the protein
expression of extracellular matrix components (collagen II and
aggrecan) in NP cells. Overexpression of MMP2 interfered with the
protective effects of miR-185-5p mimic on the expression of these
functional proteins in IC-treated NP cells. **P<0.01,
***P<0.001. MMP2, matrix metallopeptidase 2; miR,
microRNA; NP, nucleus pulposus; IDD, intervertebral disc
degeneration; RT-qPCR, reverse transcription-quantitative PCR; UTR,
untranslated region; OE, overexpression vector; NC, negative
control; IC, inflammatory cytokine. |
circ‑TIMP2 acts as an miR‑185‑5p
sponge
miR-185-5p was predicted to have binding sites for
circ-TIMP2. RT-qPCR results demonstrated that circ-TIMP2 expression
levels were significantly higher in NP tissues from patients with
IDD compared with those in the control samples (Fig. 3A and B). Pearson correlation
analysis revealed that circ-TIMP2 expression was significantly
negatively correlated with miR‑185‑5p (r=-0.717; Fig. 3C), and Spearman's correlation
analysis demonstrated that circ-TIMP2 was positively correlated
with IDD grade (ρ=0.818; Fig.
3D). RNA-FISH analysis demonstrated that the fluorescent value
of circ‑TIMP2 was evaluated in NP samples from patients with IDD,
which indicated the circ-TIMP2 expression level was increased in
IDD NP samples compared with that in the normal controls (Fig. 3E). To further assess the
interaction between circ-TIMP2 and miR-185-5p, the predicted
binding sites of circ-TIMP2 and a mutated sequence were constructed
downstream of the firefly lucif-erase gene to yield wild-type and
mutant pGL3-circ-TIMP2, respectively. These plasmids were
co-transfected with the miR-185-5p mimic or miR-NC into NP cells,
and the results demonstrated that transfection with the miR-185-5p
mimic reduced luciferase activity of wild-type pGL3-circ-TIMP2
compared with that in NP cells transfected with miR-NC. However,
luciferase activities of the mutant pGL3-circ-TIMP2 were comparable
between the miR-185-5p mimic and NC groups (Fig. 3F and G). RNA-FISH results
demonstrated that circ-TIMP2 and miR-185-5p were co-localized in
the NP cell cytoplasm (Fig. 3H).
RIP confirmed that anti-AGO2 antibodies immunoprecipitated
circ-TIMP2 (Fig. 3I). In
addition, Northern blot analysis revealed that circ-TIMP2 pulled
down miR-185-5p (Fig. 3J). Taken
together, these results suggested that circ-TIMP2 could directly
bind miR-185-5p in NP cells.
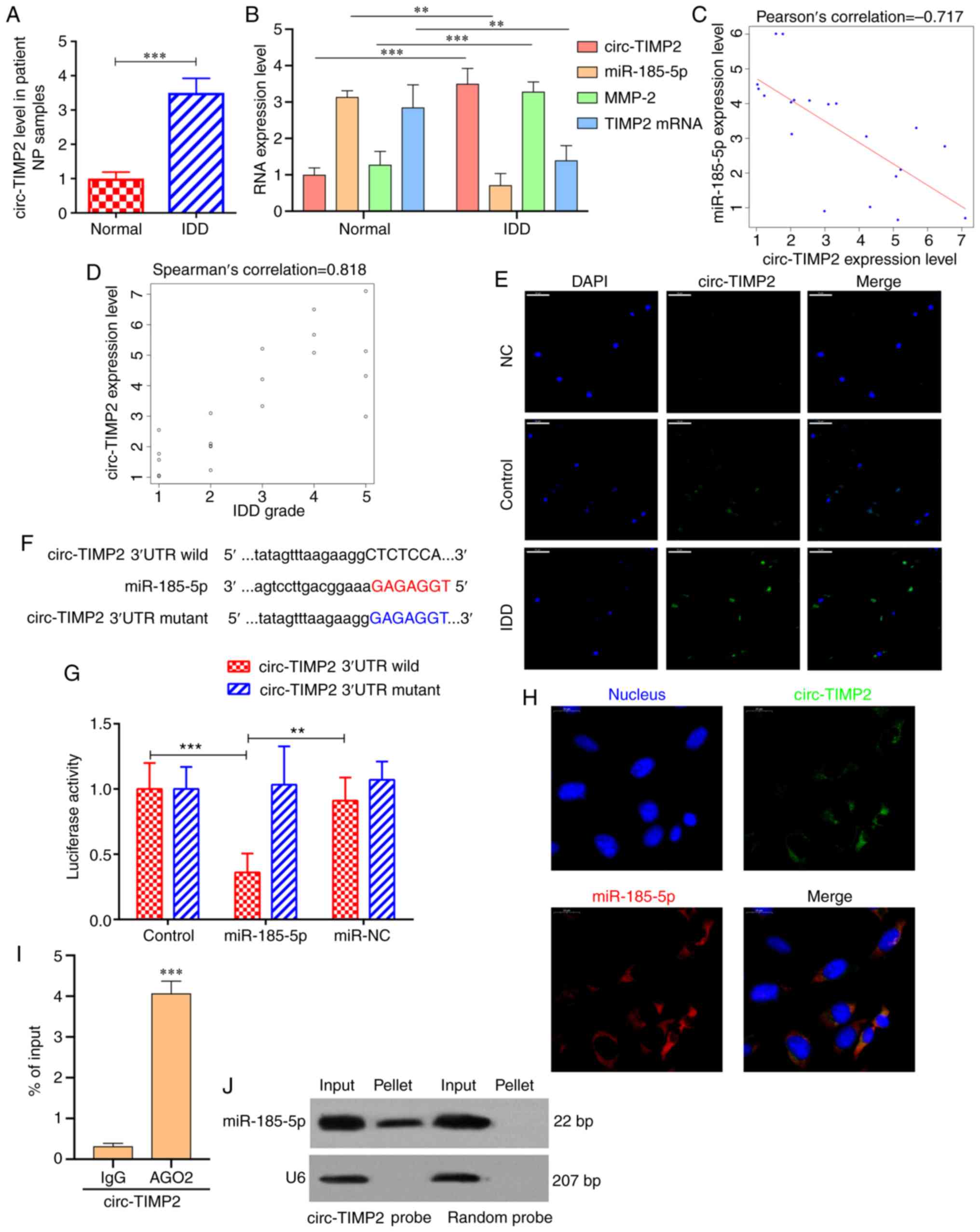 | Figure 3circ-TIMP2 acts as a miR-185-5p
sponge. (A) circ-TIMP2 levels in NP samples from patients with IDD
or controls were determined by RT-qPCR. n=10.
***P<0.001. (B) mRNA expression levels of circ-TIMP2,
miR-185-5p, MMP2 and TIMP2 in patients with IDD and controls were
determined by RT-qPCR. **P<0.01,
***P<0.001. (C) circ‑TIMP2 expression was
significantly negatively correlated with miR‑185‑5p expression
level (P<0.001). (D) circ‑TIMP2 expression was significantly
negatively correlated with IDD grade (P<0.001). (E) The
expression of circ‑TIMP2 was evaluated in NP samples from patients
with IDD and controls by RNA-FISH. The circ-TIMP2 probe was labeled
with Alexa Fluor® 488. Nuclei were stained with DAPI.
Scale bar, 50 µm. (F) Sequence alignment of human miR-185-5p
with circ-TIMP2. Mutations were introduced in the circ-TIMP2
sequence to create the mutant luciferase reporter constructs. (G)
Luciferase reporter assay in NP cells after transfection with
miR-NC or the miR-185-5p mimic, Renilla luciferase vector
pRL-SV40 and the reporter constructs. Firefly and Renilla
luciferase activities were measured in the same sample, and firefly
luciferase signals were normalized to Renilla luciferase
signals. **P<0.01, ***P<0.001. (H)
RNA-FISH analysis of co-localization of circ-TIMP2 and miR-185-5p
in the cytoplasm of NP cells. circ-TIMP2 and miR-185-5p probes were
labeled with Alexa Fluor® 488 and Cy3, respectively.
Nuclei were stained with DAPI. Scale bar, 20 µm. (I) RNA
immunoprecipitation confirmed that anti-AGO2 antibodies
immunoprecipitated circ-TIMP2. ***P<0.001 vs. IgG.
(J) miR-185-5p was pulled down by the circular probe for circ-TIMP2
but not by a random probe. The levels of miR-185-5p were evaluated
by northern blotting. Input, 20% samples were loaded; pellet, all
samples were loaded. MMP2, matrix metallopeptidase 2; miR,
microRNA; NP, nucleus pulposus; IDD, intervertebral disc
degeneration; RT-qPCR, reverse transcription-quantitative PCR;
circ, circular RNA; TIMP2, tissue inhibitor of metalloproteinases
2; FISH, fluorescence in situ hybridization; UTR,
untranslated region; NC, negative control; IgG, immunoglobulin G;
AGO2, argonaute 2. |
circ‑TIMP2 modulates NP cell function by
targeting miR‑185‑5p and MMP2
As presented in Fig.
4A, RT-qPCR results demonstrated that compared with the levels
in NP cells transfected with circ-NC, transfection with a
circ-TIMP2 overexpression plasmid resulted in increased circ-TIMP2
expression levels, which were significantly reduced by siRNA
silencing. Next, western blotting was used to assess the effect of
circ-TIMP2 on MMP2 expression. Overexpression of circ-TIMP2 in NP
cells increased MMP2 expression, whereas circ-TIMP2 knockdown had
the opposite effect (Fig. 4B). In
addition, circ-TIMP2 overexpression reversed the inhibitory effect
of miR-185-5p on MMP2 expression in NP cells (Fig. 4C). Taken together, these results
indicated that circ-TIMP2 functioned as an miR-185-5p sponge to
regulate MMP2 expression in NP cells. The function of circ-TIMP2 in
NP cells after treatment with TNF-α and IL-1β was further examined;
following administration of these ICs, RT-qPCR demonstrated
upregulation of circ-TIMP2 expression and downregulation of
miR-185-5p expression in NP cells; however, both effects were
reversed by circ-TIMP2 knockdown (Fig. 5A and B). Knockdown of circ-TIMP2
decreased the protein expression levels of MMP2 in IC-treated NPs
compared with those in NPs treated with IC and si NC (Fig. 5C and D). To assess whether MMP2
was a downstream mediator of circ-TIMP2 in IC-treated NP cells, NP
cells were co-transfected with circ-TIMP2 and MMP2 siRNA; the
results demonstrated that MMP2 silencing significantly attenuated
the effects of circ-TIMP2 on inhibiting collagen-II and aggrecan
expression in NP cells treated with TNF-α and IL-1β (Fig. 5E and F). Collectively, these
results suggested that circ-TIMP2 functioned in NP cells via
modulation of miR-185-5p and MMP2 expression.
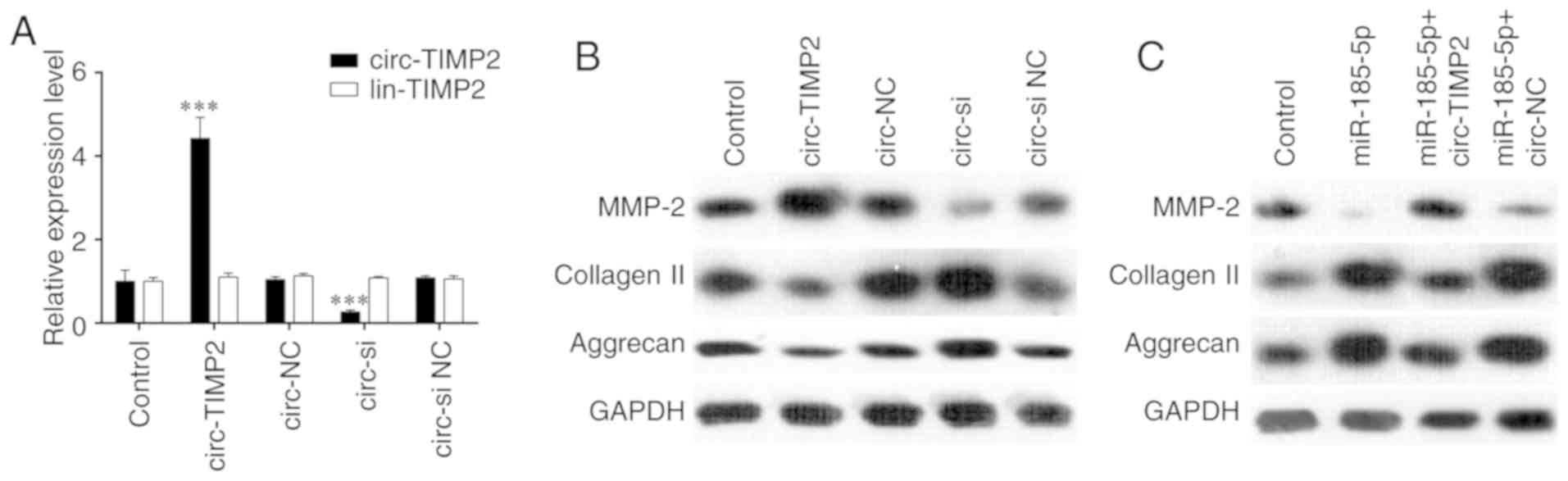 | Figure 4circ-TIMP2 functions in NP cells by
targeting miR-185-5p and MMP2. (A) NP cells were transfected with
circ-TIMP2, circ-NC, circ-si or circ-si NC, and circ-TIMP2 levels
were analyzed by reverse transcription-quantitative PCR.
***P<0.001 circ-TIMP2 vs. circ-NC or circ-si vs.
circ-si NC. (B) NP cells were transfected with circ-TIMP2, circ-NC,
circ-si or circ-si NC, and MMP2 expression was analyzed by western
blotting. The protein expression of MMP2 was enhanced after
circ-TIMP2 overexpression and reduced following circ-TIMP2
knockdown. (C) NP cells were co-transfected with miR-185-5p and
circ-TIMP2 or circ-NC. Western blot analysis demonstrated that
circ-TIMP2 blocked the inhibitory effect of miR-185-5p on MMP2
expression. MMP2, matrix metallopeptidase 2; miR, microRNA; NP,
nucleus pulposus; circ, circular RNA; lin, linear RNA; TIMP2,
tissue inhibitor of metalloproteinases 2; si, small interfering
RNA; circ-si, circ-TIMP2 siRNA; NC, negative control. |
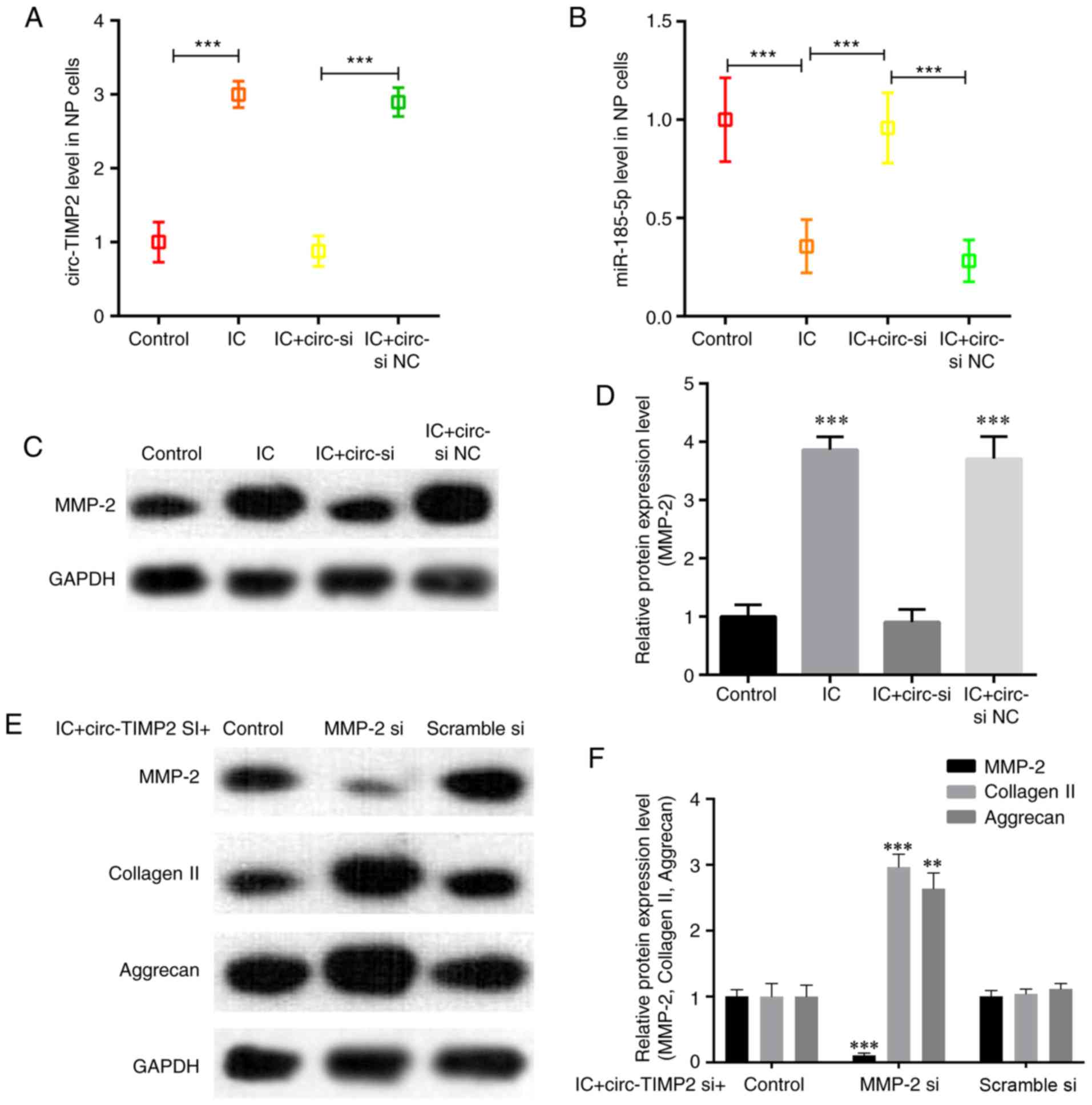 | Figure 5Function of circ-TIMP2 in NP cells
after treatment with tumor necrosis factor α and interleukin-1β.
(A) RT-qPCR analysis demonstrated an increase in circ-TIMP2
expression in NP cells treated with IC, which was reversed by
transfection with circ-si. ***P<0.001. (B) RT-qPCR
analysis demonstrated a decrease in miR-185-5p levels in NP cells
treated with IC, which was reversed by transfection with circ-si.
***P<0.001. (C and D) Western blotting demonstrated
that transfection with circ-si reduced the IC-induced increase of
MMP2 expression compared with that in si NC-transfected cells.
***P<0.001 vs. IC + circ-si NC. (E and F) Western
blotting was used to analyze the protein expression of MMP2 and
extracellular matrix components (collagen II and aggrecan) in NP
cells. Knockdown of MMP2 impaired the effect of circ-si on the
expression of these functional proteins in IC-treated NP cells.
**P<0.01, ***P<0.001 vs. scramble si.
circ-TIMP2, circular RNA derived from tissue inhibitor of
metalloproteinases 2; MMP2, matrix metallopeptidase 2; miR,
microRNA; NP, nucleus pulposus; si, small interfering RNA; circ‑si,
circ‑TIMP2 siRNA; NC, negative control; IC, inflammatory
cytokine. |
Discussion
Previous studies have indicated that certain miRNAs
are dysregulated in the development and progression of IDD and
serve vital roles by targeting distinct genes that regulate NP cell
function (16,34,35). The present study firstly
identified miR-185-5p as a key miRNA involved in IDD that regulates
the balance between anabolic and catabolic factors in NP cells. Of
note, the results of the present study revealed that MMP2 was a
direct target gene of miR-185-5p in NP cells. Next, whether
circRNAs affected the regulatory functions of miR-185-5p in NP
cells was examined by RT-qPCR, and the results demonstrated that
circ-TIMP2 levels were decreased in NP tissues from patients with
IDD compared with controls. In addition, the results demonstrated
that circ‑TIMP2 signifi-cantly inhibited the effects of miR-185-5p
and suppressed its function by direct binding. These findings
suggested that circ-TIMP2 may act as a miR-185-5p sponge to promote
ECM catabolism and suppress ECM anabolism in NP cells, which may
consequently accelerate IDD progression.
Dysregulation of miR-185-5p has been observed in
various types of tumors, such as colon and breast cancer (36,37). To date, the role of miR-185-5p in
the development and progression of IDD remains unclear. In the
present study, miR-185-5p expression levels were significantly
lower in degenerative NP tissues compared with those in the control
tissues, as confirmed by RT-qPCR analysis. Accumulating evidence
suggests that a variety of cellular events are dysregulated in IDD
progression, including NP cell apoptosis, ECM degradation and
proinflam-matory cytokine expression (38-41). A previous study has demonstrated
that excessive apoptosis of intervertebral disc cells serves a
crucial role in IDD pathogenesis (38). In addition, loss of collagen II is
an early sign of IDD (42,43).
Aggrecan, as the main proteoglycan in NP tissues, is also crucial
to normal disc function (44,45).
The hallmark of IDD is a progressive loss of the ECM
macromolecules aggrecan and collagen II (42). To further examine the function of
miR-185-5p in IDD pathogenesis, a series of experiments were
performed to assess the association between miR-185-5p and NP cell
function. Transfection with the miR-185-5p mimic markedly enhanced
collagen II and aggrecan levels in NP cells, which suggested that
miR-185-5p may serve a role in the development of IDD by affecting
ECM composition. The present study also aimed to identify the
possible target genes of miR-185-5p that are implicated in IDD
pathogenesis. Of note, high expression levels of MMP2 were observed
in degenerative NP tissues, and bioinformatics analysis
demonstrated that MMP2 was a potential target gene of miR-185-5p.
In addition, previous studies have reported that MMP2 degrades ECM
components such as collagen II and aggrecan (34,46). In the present study, MMP2 was
confirmed as a direct target of miR-185-5p by luciferase assay and
western blotting. MMPs are crucial players in tissue remodeling and
repair, and elevated MMP levels in the intervertebral disc
catabolize ECM constituents, causing IDD (47,48). MMPs (such as MMP2), which are
zinc- and calcium-dependent endopeptidases, participate in ECM
degradation and remod-eling (49). Similar to TNF-α, overexpression of
MMP2 has been demonstrated to enhance the degradation of ECM
components in NP cells (12,50). The present study demonstrated that
reduced miR-185-5p levels resulted in enhanced MMP2 expression,
with subsequent degradation of ECM constituents, including collagen
II and aggrecan.
As predicted by starBase, circ-TIMP2 was
demonstrated to possess binding sites for miR‑185‑5p, which was
confirmed in the present study by luciferase reporter, RNA
pull-down, RIP and RNA-FISH assays. In addition, the mRNA
expression of the miR-185-5p target MMP2 was positively regulated
by circ-TIMP2. In the present study, TNF-α and IL-1β were selected
as agents to induce a range of pathogenic responses in NP cells, as
they serve central roles in the pathological process of IDD
(11,14,51). Stimulation of NP cells with these
cytokines causes a pattern of changes similar to that observed in
patients with IDD (12). TNF-α
and IL-1β accumulation is considered to facilitate ECM degradation
via extrinsic and intrinsic pathways (14,51). In the present study, stimulation
of NP cells with TNF-α and IL-1β led to similar effects to those
previously observed in patients with IDD, including increased
expression levels of the ECM-degrading enzyme MMP2 and decreased
levels of the ECM components collagen II and aggrecan (12,52). MMP2 silencing remarkably impaired
the proinflammatory ability of circ‑TIMP2, confirming MMP2 as a
direct target gene of circ-TIMP2 and miR-185-5p. In addition, MMP2
silencing inhibited circ-TIMP2, which enhanced the TNF-α- and
IL-1β-induced effects in NP cells.
The role of mir-185-5p in IDD is relatively unclear.
A recent study by Zhang et al (53) demonstrated that miR-185-5p and
other miRNAs may affect IDD development by co-regulating the
expression of glycogen synthase kinase 3β. In the present study,
mir‑185‑5p was identified as a key miRNA in the IDD process,
providing a theoretical basis for developing novel optimized
treatment regimens for IDD.
The present study had certain limitations. First,
the newest classification method of disc degeneration described by
Riesenburger et al (54)
was not applied. The novel method is more elaborate and complex
compared with the one applied in the present study, and its
implementation is difficult and error-prone. In addition, the
present study did not assess fragments per kilobase of transcript
per million mapped reads values, since no RNA-seq was performed.
Furthermore, this was a single-center study, with potential
selection bias. Additionally, the mechanism underlying circ-TIMP2
upregulation in IDD remains largely unclear. Therefore,
well-designed multicenter studies are warranted to confirm the
present findings and to comprehensively assess the role of
circ-TIMP2 in IDD.
In summary, the results of the present study
demonstrated that circ-TIMP2 promotes TNF-α- and IL-1β-induced NP
cell imbalance between ECM anabolism and catabolism via
miR-185-5p-MMP2 signaling. These results provide a potential
therapeutic option for the treatment of IDD.
Supplementary Data
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 881802197) and the Natural
Science Foundation of Hebei (grant no. H2019110028).
Availability of data and materials
The data generated or analyzed during the present
study are included in this published article with the exception of
the siRNA sequences, which are patented.
Authors' contributions
WG conceived the study. BZ, KM, CS, HQD and WXL
conducted the experiments. WG, LZ and HRL analyzed the data. WG,
ZYD and QC interpreted the data. WG and BZ obtained funding. WG
wrote the manuscript with the help of the other authors. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committees of
Tianjin Medical University General Hospital and Hebei Province
Cangzhou Hospital of Integrated Traditional and Western Medicine.
Written informed consent was obtained from all patients for the use
of their tissue specimens for research purpose.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
References
|
1
|
Ravindra VM, Senglaub SS, Rattani A, Dewan
MC, Härtl R, Bisson E, Park KB and Shrime MG: Degenerative lumbar
spine disease: Estimating global incidence and worldwide volume.
Global Spine J. 8:784–794. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buckwalter JA: Aging and degeneration of
the human interverte-bral disc. Spine (Phila Pa 1976).
20:1307–1314. 1995. View Article : Google Scholar
|
|
3
|
Costi JJ, Stokes IA, Gardner-Morse MG and
Iatridis JC: Frequency-dependent behavior of the intervertebral
disc in response to each of six degree of freedom dynamic loading:
Solid phase and fluid phase contributions. Spine (Phila Pa 1976).
33:1731–1738. 2008. View Article : Google Scholar
|
|
4
|
Kalichman L and Hunter DJ: The genetics of
intervertebral disc degeneration. Familial predisposition and
heritability estimation. Joint Bone Spine. 75:383–387. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Samartzis D, Karppinen J, Mok F, Fong DY,
Luk KD and Cheung KM: A population-based study of juvenile disc
degeneration and its association with overweight and obesity, low
back pain, and diminished functional status. J Bone Joint Surg Am.
93:662–670. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Friedman BW, O'Mahony S, Mulvey L, Mulvey
L, Davitt M, Choi H, Xia S, Esses D, Bijur PE and Gallaghe EJ:
One-week and 3-month outcomes after an emergency department visit
for undifferentiated musculoskeletal low back pain. Ann Emerg Med.
59:128–133e3. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berg EJ and Ashurst JV: Anatomy, back,
cauda equina. StatPearls; Treasure Island (FL): 2020
|
|
8
|
Kepler CK, Markova DZ, Hilibrand AS,
Vaccaro AR, Risbud MV, Albert TJ and Anderson DG: Substance P
stimulates production of inflammatory cytokines in human disc
cells. Spine (Phila Pa 1976). 38:E1291–E1299. 2013. View Article : Google Scholar
|
|
9
|
Kepler CK, Markova DZ, Dibra F, Yadla S,
Vaccaro AR, Risbud MV, Albert TJ and Anderson DG: Expression and
relationship of proinflammatory chemokine RANTES/CCL5 and cytokine
IL-1β in painful human intervertebral discs. Spine (Phila Pa 1976).
38:873–880. 2013. View Article : Google Scholar
|
|
10
|
Adams MA, Freeman BJ, Morrison HP, Nelson
IW and Dolan P: Mechanical initiation of intervertebral disc
degeneration. Spine (Phila Pa 1976). 25:1625–1636. 2000. View Article : Google Scholar
|
|
11
|
Dudek M, Yang N, Ruckshanthi JP, Williams
J, Borysiewicz E, Wang P, Adamson A, Li J, Bateman JF, White MR, et
al: The intervertebral disc contains intrinsic circadian clocks
that are regulated by age and cytokines and linked to degeneration.
Ann Rheum Dis. 76:576–584. 2017. View Article : Google Scholar :
|
|
12
|
Wang J, Markova D, Anderson DG, Zheng Z,
Shapiro IM and Risbud MV: TNF-α and IL-1β promote a
disintegrin-like and metalloprotease with thrombospondin type I
motif-5-mediated aggrecan degradation through syndecan-4 in
intervertebral disc. J Biol Chem. 286:39738–39749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johnson ZI, Schoepflin ZR, Choi H, Shapiro
IM and Risbud MV: Disc in flames: Roles of TNF‑α and IL-1β in
intervertebral disc degeneration. Eur Cell Mater. 30:104–116. 2015.
View Article : Google Scholar
|
|
14
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar
|
|
15
|
Lan PH, Liu ZH, Pei YJ, Wu ZG, Yu Y, Yang
YF, Liu X, Che L, Ma CJ, Xie YK, et al: Landscape of RNAs in human
lumbar disc degeneration. Oncotarget. 7:63166–63176. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu YQ, Zhang ZH, Zheng YF and Feng SQ:
Dysregulated miR-133a mediates loss of type II collagen by directly
targeting matrix metalloproteinase 9 (MMP9) in human intervertebral
disc degeneration. Spine (Phila Pa 1976). 41:E717–E724. 2016.
View Article : Google Scholar
|
|
17
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang B, Hao Z, Zhu Y, Zhang H and Li G:
Genome-wide identification and functional analysis of circRNAs in
Zea mays. PLoS One. 13:e02023752018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vicens Q and Westhof E: Biogenesis of
circular RNAs. Cell. 159:13–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo W, Zhang B, Mu K, Feng SQ, Dong ZY,
Ning GZ, Li HR, Liu S, Zhao L, Li Y, et al: Circular RNA GRB10 as a
competitive endogenous RNA regulating nucleus pulposus cells death
in degenerative intervertebral disk. Cell Death Dis. 9:3192018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila Pa 1976).
26:1873–1878. 2001. View Article : Google Scholar
|
|
27
|
Middendorp M, Vogl TJ, Kollias K,
Kafchitsas K, Khan MF and Maataoui A: Association between
intervertebral disc degeneration and the oswestry disability index.
J Back Musculoskelet Rehabil. 30:819–823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Urrutia J, Besa P, Campos M, Cikutovic P,
Cabezon M, Molina M and Cruz JP: The Pfirrmann classification of
lumbar interverte-bral disc degeneration: An independent inter- and
intra-observer agreement assessment. Eur Spine J. 25:2728–2733.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Vautrot V, Aigueperse C, Branlant C and
Behm-Ansmant I: Fluorescence in situ hybridization of small
non-coding RNAs. Methods Mol Biol. 1296:73–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42(Database Issue): D92–D97. 2014. View Article : Google Scholar
|
|
32
|
Barnes C and Kanhere A: Identification of
RNA‑protein interactions through in vitro RNA pull-down assays.
Methods Mol Biol. 1480:99–113. 2016. View Article : Google Scholar
|
|
33
|
Wang M and Pestov DG: Quantitative
northern blot analysis of mammalian rRNA processing. Methods Mol
Biol. 1455:147–157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ji ML, Lu J, Shi PL, Zhang XJ, Wang SZ,
Chang Q, Chen H and Wang C: Dysregulated miR-98 contributes to
extracellular matrix degradation by targeting IL-6/STAT3 signaling
pathway in human intervertebral disc degeneration. J Bone Miner
Res. 31:900–909. 2016. View Article : Google Scholar
|
|
35
|
Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis
D, Jia LT, Wu SX, Huang J, Chen J and Luo ZJ: Deregulated miR-155
promotes Fas-mediated apoptosis in human intervertebral disc
degeneration by targeting FADD and caspase-3. J Pathol.
225:232–242. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu ZJ, Lu LG, Tao KZ, Chen DF, Xia Q, Weng
JJ, Zhu F, Wang XP and Zheng P: MicroRNA-185 suppresses growth and
invasion of colon cancer cells through inhibition of the
hypoxi-ainducible factor-2α pathway in vitro and in in vivo. Mol
Med Rep. 10:2401–2408. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yin C, Zhang G, Sun R, Pan X, Wang X, Li H
and Sun Y: MiR1855p inhibits Factin polymerization and reverses
epithelial mesenchymal transition of human breast cancer cells by
modulating RAGE. Mol Med Rep. 18:2621–2630. 2018.PubMed/NCBI
|
|
38
|
Zhao CQ, Jiang LS and Dai LY: Programmed
cell death in inter-vertebral disc degeneration. Apoptosis.
11:2079–2088. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kozaci LD, Guner A, Oktay G and Guner G:
Alterations in biochemical components of extracellular matrix in
intervertebral disc herniation: Role of MMP-2 and TIMP-2 in type II
collagen loss. Cell Biochem Funct. 24:431–436. 2006. View Article : Google Scholar
|
|
40
|
Matsui Y, Maeda M, Nakagami W and Iwata H:
The involvement of matrix metalloproteinases and inflammation in
lumbar disc herniation. Spine (Phila Pa 1976). 23:863–868. 1998.
View Article : Google Scholar
|
|
41
|
Crean JK, Roberts S, Jaffray DC,
Eisenstein SM and Duance VC: Matrix metalloproteinases in the human
intervertebral disc: Role in disc degeneration and scoliosis. Spine
(Phila Pa 1976). 22:2877–2884. 1997. View Article : Google Scholar
|
|
42
|
Kepler CK, Ponnappan RK, Tannoury CA,
Risbud MV and Anderson DG: The molecular basis of intervertebral
disc degeneration. Spine J. 13:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tran CM, Smith HE, Symes A, Rittié L,
Perbal B, Shapiro IM and Risbud MV: Transforming growth factor beta
controls CCN3 expression in nucleus pulposus cells of the
intervertebral disc. Arthritis Rheum. 63:3022–3031. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sivan SS, Wachtel E and Roughley P:
Structure, function, aging and turnover of aggrecan in the
intervertebral disc. Biochim Biophys Acta. 1840:3181–3189. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Perera RS, Dissanayake PH, Senarath U,
Wijayaratne LS, Karunanayake AL and Dissanayake VH: Single
nucleotide variants of candidate genes in aggrecan metabolic
pathway are associated with lumbar disc degeneration and modic
changes. PLoS One. 12:e01698352017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rutges JP, Kummer JA, Oner FC, Verbout AJ,
Castelein RJ, Roestenburg HJ, Dhert WJ and Creemers LB: Increased
MMP-2 activity during intervertebral disc degeneration is
correlated to MMP-14 levels. J Pathol. 214:523–530. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vo NV, Hartman RA, Yurube T, Jacobs LJ,
Sowa GA and Kang JD: Expression and regulation of
metalloproteinases and their inhibitors in intervertebral disc
aging and degeneration. Spine J. 13:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rutges JP, Nikkels PG, Oner FC, Ottink KD,
Verbout AJ, Castelein RJ, Creemers LB and Dhert WJ: The presence of
extracellular matrix degrading metalloproteinases during fetal
development of the intervertebral disc. Eur Spine J. 19:1340–1346.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Boxler S, Djonov V, Kessler TM, Hlushchuk
R, Bachmann LM, Held U, Markwalder R and Thalmann GN: Matrix
metallopro-teinases and angiogenic factors: Predictors of survival
after radical prostatectomy for clinically organ-confined prostate
cancer? Am J Pathol. 177:2216–2224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Song J, Wu C, Korpos E, Zhang X, Agrawal
SM, Wang Y, Faber C, Schäfers M, Körner H, Opdenakker G, et al:
Focal MMP-2 and MMP-9 activity at the blood-brain barrier promotes
chemokine-induced leukocyte migration. Cell Rep. 10:1040–1054.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Phillips KL, Cullen K, Chiverton N,
Michael AL, Cole AA, Breakwell LM, Haddock G, Bunning RA, Cross AK
and Le Maitre CL: Potential roles of cytokines and chemokines in
human intervertebral disc degeneration: Interleukin-1 is a master
regulator of catabolic processes. Osteoarthritis Cartilage.
23:1165–1177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang J, Tian Y, Phillips KL, Chiverton N,
Haddock G, Bunning RA, Cross AK, Shapiro IM, Le Maitre CL and
Risbud MV: Tumor necrosis factor α- and interleukin-1β-dependent
induction of CCL3 expression by nucleus pulposus cells promotes
macrophage migration through CCR1. Arthritis Rheum. 65:832–842.
2013. View Article : Google Scholar :
|
|
53
|
Zhang H, Zhang M, Meng L, Guo M, Piao M,
Huang Z and Yu H: Investigation of key miRNAs and their target
genes involved in cell apoptosis during intervertebral disc
degeneration development using bioinformatics methods. J Neurosurg
Sci. Feb 4, 2020 (Epub ahead of print).
|
|
54
|
Riesenburger RI, Safain MG, Ogbuji R,
Hayes J and Hwang SW: A novel classification system of lumbar disc
degeneration. J Clin Neurosci. 22:346–351. 2015. View Article : Google Scholar
|



















